Abstract
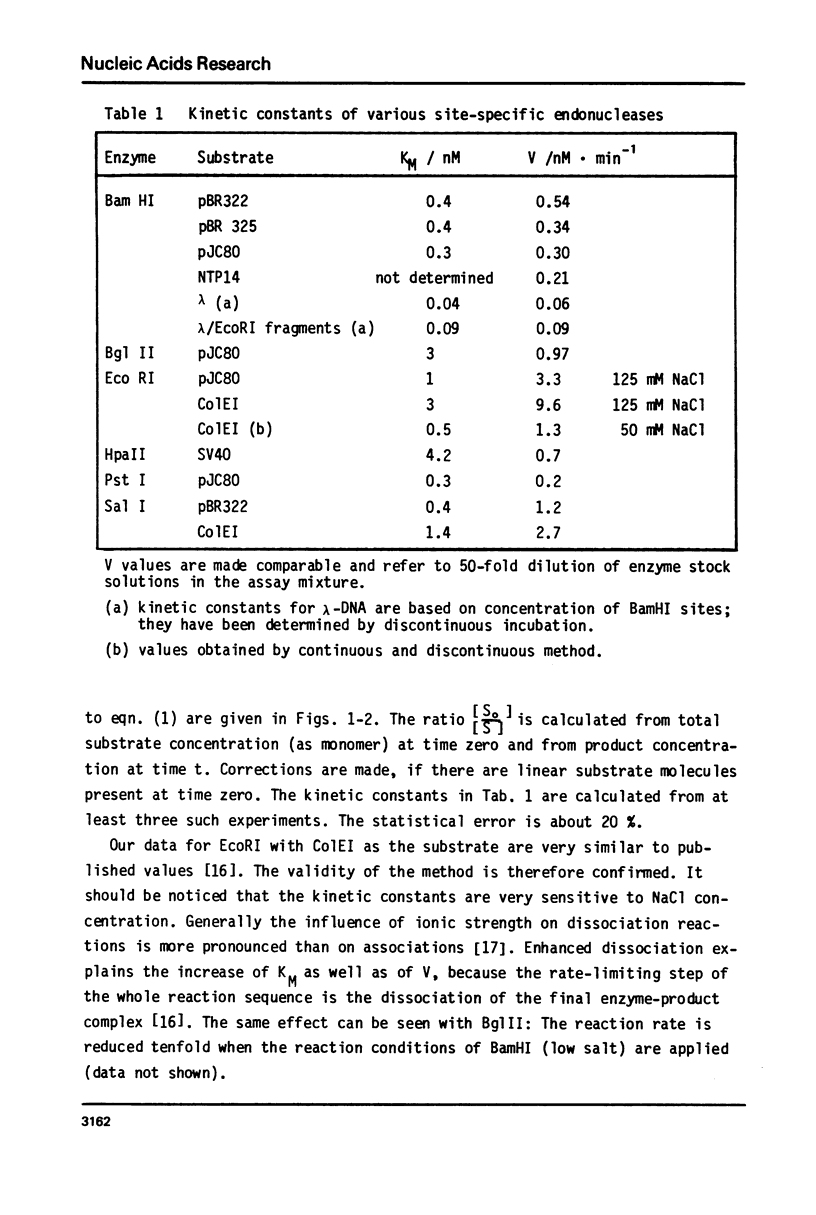
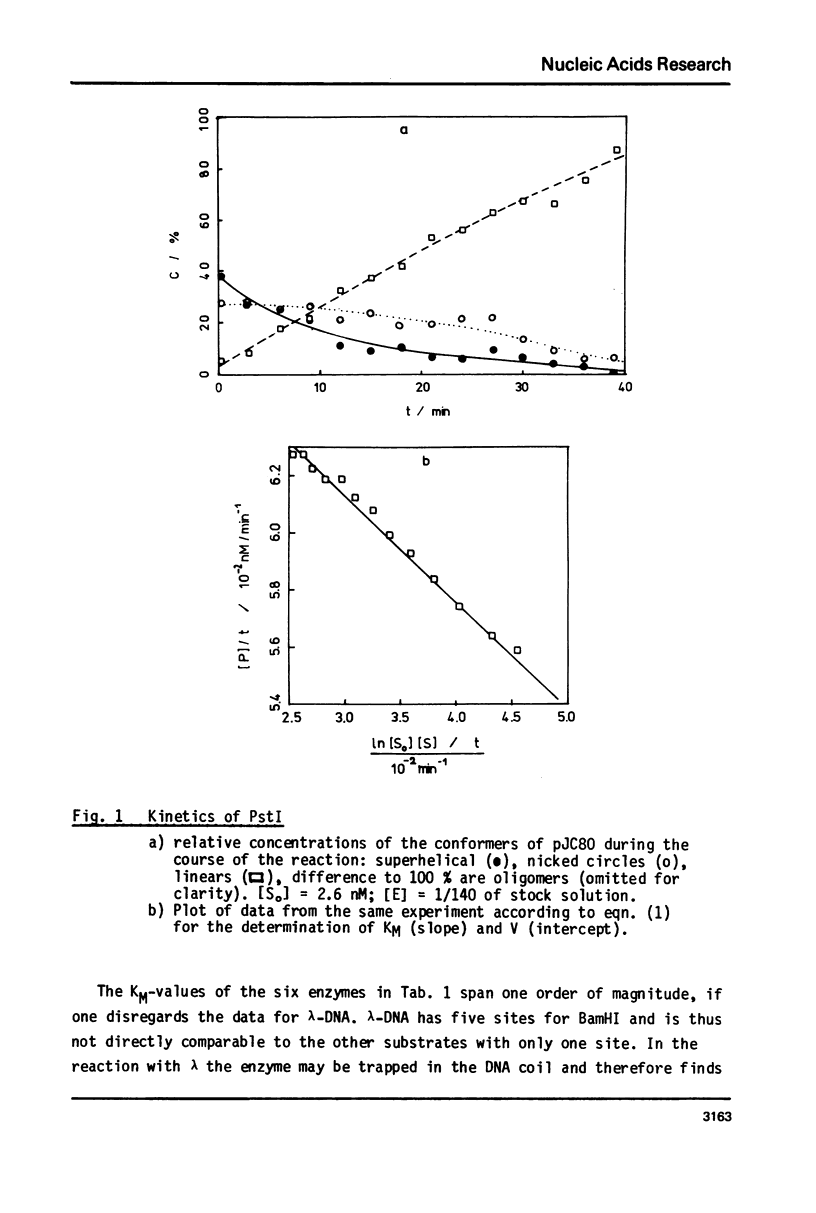
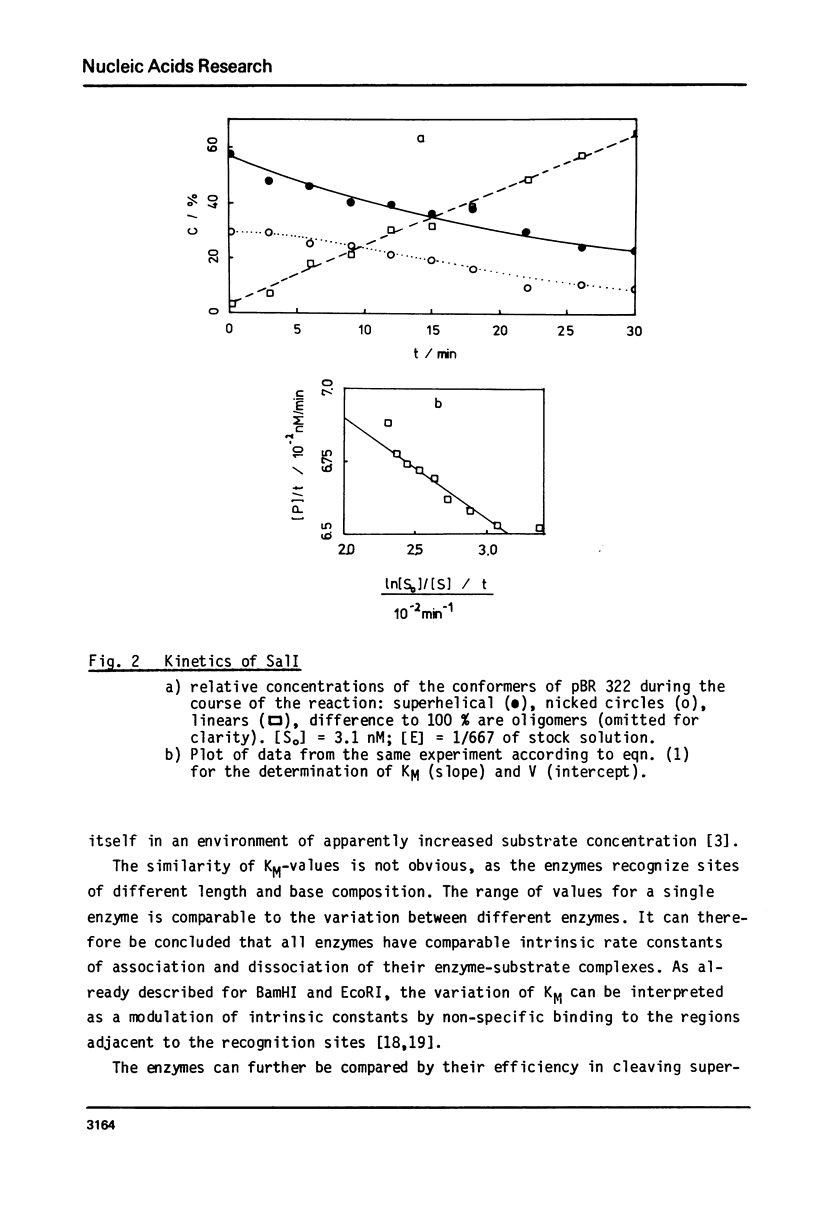
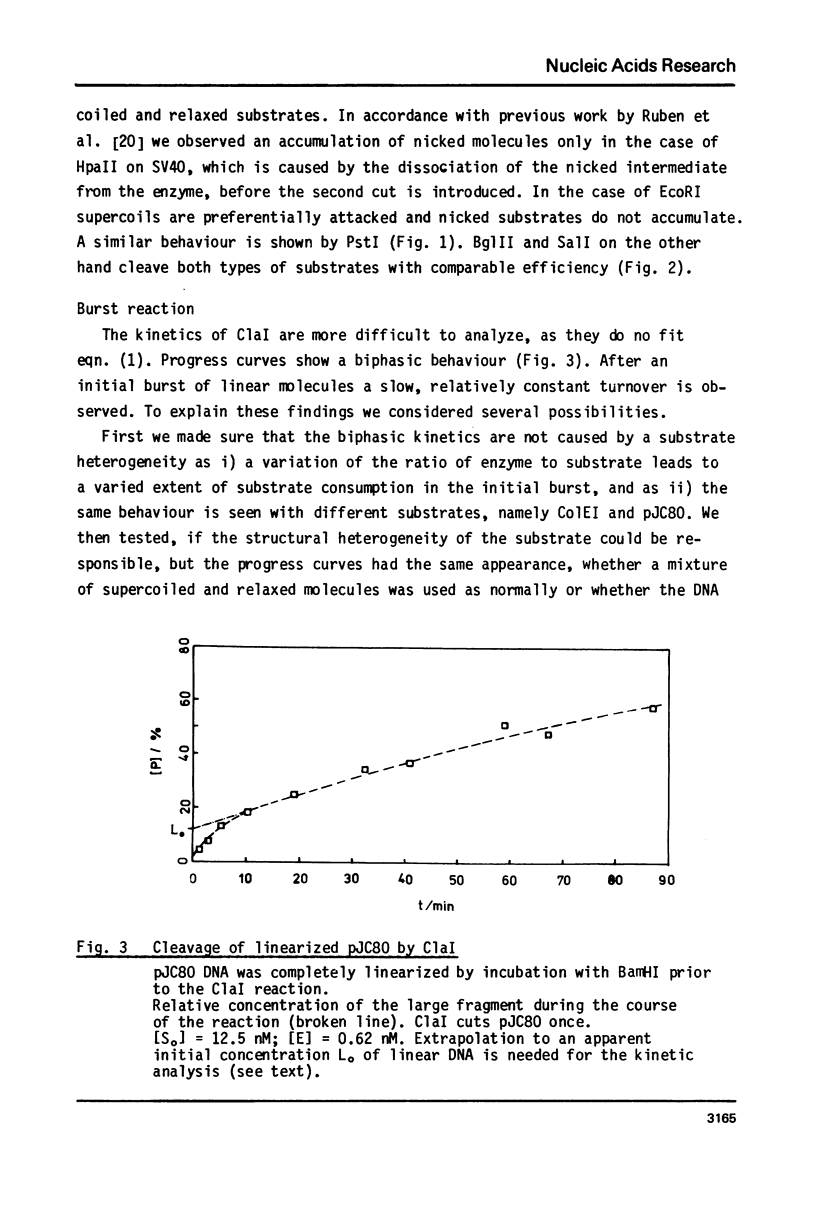
Reaction kinetics of the site-specific endonucleases BamHI, BgIII, C1aI, EcoRI, HpaII, PstI, SaII, SmaI, and XorII were investigated employing some frequently used substrates. Six of these enzymes could be analyzed under steady-state conditions. Kinetic data were obtained from progress curves applying an integrated Michaelis-Menten equation. KM ranged from 4 x 10(-9) M to 4 x 10(-11) M. Activities also spanned two orders of magnitude. In the case of C1aI the analysis of the pre-steady-state kinetics ("burst reaction") allowed the assessment of several rate constants. The rate-limiting step is the very slow dissociation of the enzyme-product complex (0.22 min(-1)). This complex is formed from the enzyme-bound nicked intermediate at a rate of 1.7 min(-1). The introduction of the first cut is again faster by a factor of about 6. SmaI and XorII resembled C1aI in their kinetics. The burst reaction can be used for the easy and unambiguous determination of molar concentrations of site-specific endonucleases in any preparation, which is free of non-specific DNases.
Full text
PDF







Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Atkins G. L., Nimmo I. A. The reliability of Michaelis constants and maximum velocities estimated by using the integrated Michaelis-Menten equation. Biochem J. 1973 Dec;135(4):779–784. doi: 10.1042/bj1350779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkner K. L., Folk W. R. The effects of substituted pyrimidines in DNAs on cleavage by sequence-specific endonucleases. J Biol Chem. 1979 Apr 10;254(7):2551–2560. [PubMed] [Google Scholar]
- Goebel W., Bonewald R. Class of small multicopy plasmids originating from the mutant antibiotic resistance factor R1 drd-19B2. J Bacteriol. 1975 Aug;123(2):658–665. doi: 10.1128/jb.123.2.658-665.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershfield V., Boyer H. W., Yanofsky C., Lovett M. A., Helinski D. R. Plasmid ColEl as a molecular vehicle for cloning and amplification of DNA. Proc Natl Acad Sci U S A. 1974 Sep;71(9):3455–3459. doi: 10.1073/pnas.71.9.3455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinsch B., Kula M. R. Physical and kinetic properties of the site specific endonuclease Bam HI from Bacillus amylolique-faciens. Nucleic Acids Res. 1980 Feb 11;8(3):623–633. doi: 10.1093/nar/8.3.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinsch B., Mayer H., Kula M. R. Binding of non-substrate nucleotides to a restriction endonuclease: a model for the interaction of bam HI with its recognition sequence. Nucleic Acids Res. 1980 Jun 11;8(11):2547–2559. doi: 10.1093/nar/8.11.2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livshitz M. A., Gursky G. V., Zasedatelev A. S., Volkenstein M. V. Equilibrium and kinetic aspects of protein-DNA recognition. Nucleic Acids Res. 1979;6(6):2217–2236. doi: 10.1093/nar/6.6.2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modrich P., Zabel D. EcoRI endonuclease. Physical and catalytic properties of the homogenous enzyme. J Biol Chem. 1976 Oct 10;251(19):5866–5874. [PubMed] [Google Scholar]
- Roberts R. J. Restriction and modification enzymes and their recognition sequences. Nucleic Acids Res. 1981 Jan 10;9(1):r75–r96. doi: 10.1093/nar/9.1.213-c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruben G., Spielman P., Tu C. D., Jay E., Siegel B., Wu R. Relaxed circular SV40 DNA as cleavage intermediate of two restriction endonucleases. Nucleic Acids Res. 1977 Jun;4(6):1803–1813. doi: 10.1093/nar/4.6.1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith H. O. Nucleotide sequence specificity of restriction endonucleases. Science. 1979 Aug 3;205(4405):455–462. doi: 10.1126/science.377492. [DOI] [PubMed] [Google Scholar]
- Smith H. R., Humphreys G. O., Willshaw G. A., Anderson E. S. Characterisation of plasmids coding for the restriction endonuclease EcoRI. Mol Gen Genet. 1976 Feb 2;143(3):319–325. doi: 10.1007/BF00269410. [DOI] [PubMed] [Google Scholar]
- Sutcliffe J. G. pBR322 restriction map derived from the DNA sequence: accurate DNA size markers up to 4361 nucleotide pairs long. Nucleic Acids Res. 1978 Aug;5(8):2721–2728. doi: 10.1093/nar/5.8.2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas M., Davis R. W. Studies on the cleavage of bacteriophage lambda DNA with EcoRI Restriction endonuclease. J Mol Biol. 1975 Jan 25;91(3):315–328. doi: 10.1016/0022-2836(75)90383-6. [DOI] [PubMed] [Google Scholar]
- Wang R. Y., Shedlarski J. G., Farber M. B., Kuebbing D., Ehrlich M. Two sequence-specific endonucleases from Xanthomonas oryzae. Characterization and unusual properties. Biochim Biophys Acta. 1980 Feb 29;606(2):371–385. doi: 10.1016/0005-2787(80)90047-7. [DOI] [PubMed] [Google Scholar]


