Abstract
Nanopores could potentially be used to perform single molecule DNA sequencing at low cost and with high throughput1–4. Although single-base resolution and differentiation have been demonstrated with nanopores using ionic current measurements5–7, direct sequencing has not been achieved due to difficulties in recording very small (~pA) ionic current at a bandwidth consistent with fast translocation speeds1–3. Here we show that solid-state nanopores can be combined with silicon nanowire field-effect transistors (FETs) to create sensors in which detection is localised and self-aligned at the nanopore. Well-defined FET signals associated with DNA translocation are recorded when an ionic strength gradient is imposed across the nanopores. Measurements and modelling show that FET signals are generated by highly-localized changes in the electrical potential during DNA translocation and that the nanowire-nanopore sensors could enable large-scale integration with a high intrinsic bandwidth.
Current nanopore technology is based on detecting a modulation in the ionic current due to the partial blockade of a nanopore during DNA translocation1–4. Significant progress has been made towards realizing the goal of direct DNA sequencing during translocation through protein nanopore engineering5,6 and novel membrane materials7–9, although challenges remain1,3. For example, the DNA translocation speed, ca. 1 µsec/base, is faster than the bandwidth electronics available to amplify the small ionic current, and it is difficult to record ionic current from individual nanopores in a highly-parallel multiplexed format. To overcome these issues, methods have been developed for better control of translocation4,10–13, thus enabling potential reductions in translocation speed that could facilitate ionic current detection. Simultaneously, new detection designs have been proposed that could potentially enable recording larger and local signals from sensors integrated with the nanopore1–3. These integrated sensors include devices based on measurement of capacitive coupling14 and tunneling currents15–17, although none have yet improved upon traditional ionic current detection in experiments. FETs, including nanowire and carbon nanotube FETs, have shown high intrinsic speed18,19 and high sensitivity as chemical and biological sensors20–22 and thus might also function as integrated detectors for nanopores. However, the lack of a clear mechanism for FET-based detection of DNA during nanopore translocation, where the relatively high solution ionic strength is expected to screen the detection of molecular charge previously used in sensing experiments20,21, has left these detectors largely unexplored.
We investigated the possibility of integrating a FET with a nanopore using synthesized silicon nanowires as the nanoscale FETs20–22, where the integrated nanowire-nanopore FET sensor uses a short-channel silicon nanowire FET on a silicon nitride (SiNx) membrane based chip with the nanopore through both the nanowire edge and the SiNx membrane support (Fig. 1a and Fig. S1). Fabrication of nanowire-nanopore FET sensors involved several key steps (see SI Methods). First, p-type silicon nanowires FETs were fabricated on SiNx membranes with nickel-metal source/drain contacts with a typical spacing of 1–2 µm. To minimize the signal attenuation due to FET channel series resistance, the active length of the silicon nanowire was further reduced to less than 200 nm by solid-state diffusion to form metallic nickel silicide (NiSi) contacts19 (inset, Fig. 1b). Second, a focused electron beam in a transmission electron microscopy (TEM) was used to form the nanopore23 through the edge of the silicon nanowire and the underlying membrane (Fig. 1b).
Figure 1. nanowire-nanopore transistor.
a, Schematic of the nanowire-nanopore measurement setup. Inset: Zoom-in view around the nanopore. b, High-resolution TEM image of a silicon nanowire with the nanopore off-axis at the nanowire edge; scale bar = 10 nm. Inset: A larger scale TEM image of a nanowire-nanopore FET device showing the central silicon nanowire connected to darker NiSi contacts, which are indicated by the white dashed line. The region where the high-resolution TEM image was recorded is indicated by the yellow dashed square. Scale bar = 50 nm. c, SGM image of a Si nanowire-nanopore device recorded with the tip voltage at −10 V. Scale bar = 1 µm. The nanopore position is indicated by the open black circle, the Ni contacts are indicated by white dashed lines and the nanowire between the two contacts is indicated by the black dashed line. Inset: AFM topographic image of this device, where the SGM image area is indicated by the white dotted square. The colour scale (−100 – 200 nS) corresponds to the conductance change. d, Scanning gate sensitivity profile of the same device before and after nanopore formation, where the profile was taken along the black dashed line in panel C, and averaged over ~100 nm width perpendicular to the dashed line.
The sensitivity of nanowire-nanopore FET sensors fabricated in this way was characterized by scanning gate microscopy (SGM) (see SI Methods). A SGM map of the conductance change versus biased tip position for a silicon nanowire FET device after nanopore formation (Fig. 1c) shows a pronounced peak of conductance change localised around the nanopore position and no response from the NiSi region of the nanowire device. The sensitivity (i.e., conductance change / tip voltage) along the nanowire (Fig. 1d, red line) exhibits a maximum, 18 nS/V, sharply localised and aligned with the nanopore position at ca. the midpoint along the length of the semiconductor channel. We note that the sensitivity of this device before nanopore formation is relatively constant, 5–8 nS/V (Fig. 1d, black line), along the entire active Si channel. The >2 fold sensitivity enhancement can be explained qualitatively by the increase in channel resistance of the nanopore portion of the silicon nanowire FET where Si is removed. Although additional work will be needed to quantify factors contributing to the observed enhancement, this localised sensitivity makes the nanowire-nanopore FET attractive for monitoring translocation events through the nanopore.
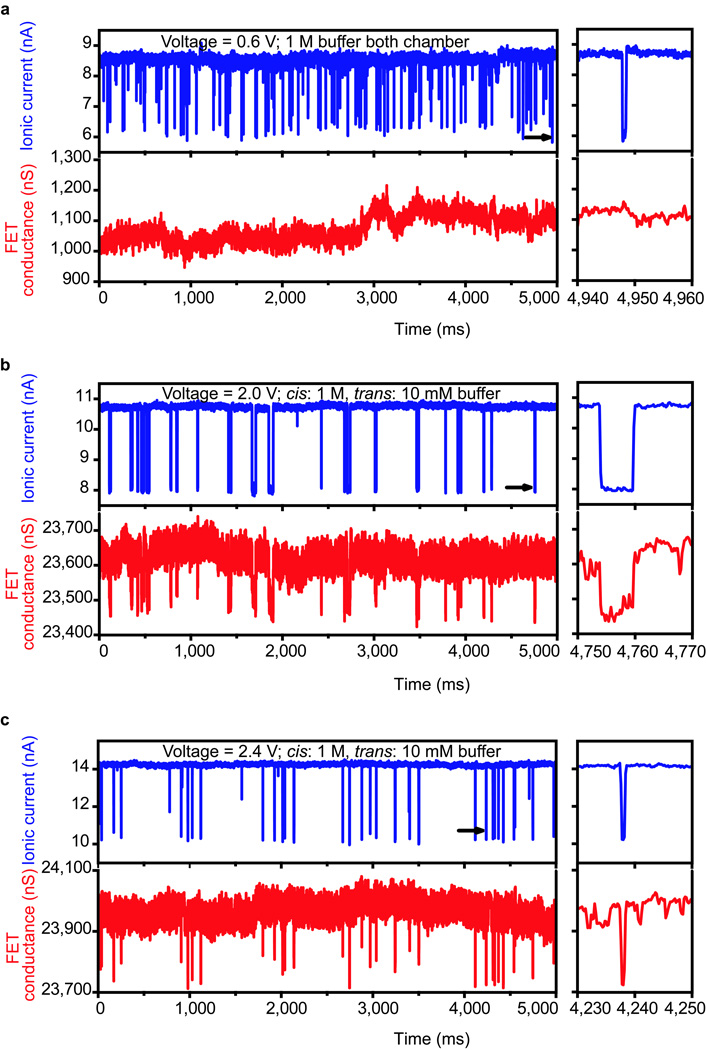
Single channel DNA translocation measurements were carried out with two polydimethylsiloxane (PDMS) solution chambers as trans and cis reservoirs above and below the SiNx membrane, respectively, where both chambers were filled with 1 M KCl buffer as typically used in nanopore experiments24,25. Following injection of 6 nM, ~2.6 kbps linear double-stranded DNA (dsDNA), pUC19 (see SI Methods), into the cis chamber, we observed translocation events in the ionic current channel when the trans-membrane voltage reached ~0.6 V (Fig. 2a, top-panel). Simultaneous recording of the nanowire FET conductance (Fig. 2a, lower-panel) showed no noticeable translocation signals and only small and slow conductance baseline shifts. However, if we fill trans (nanowire-nanopore FET side) and cis (back side) chambers with different ionic strength solutions, e.g. 10 mM in the trans chamber and 1 M in cis chamber, clear FET conductance signals with perfect time correlation to ionic current events can be observed (Fig. 2b) for a voltage of ~2 V. Qualitatively, the larger voltage is expected given the lower solution resistance thus lower electric field in the cis side, which determines the DNA entrance of the nanopore3. Further increases in the voltage to 2.4 V (Fig. 2c), the duration and frequency of translocation events in both ionic current and FET channels decreased and increased, respectively. These changes in duration and frequency are consistent with previous results reported for ionic current events recorded in other nanopore experiments10,24. Importantly, the change in the FET signal during translocation – a decrease in conductance – is opposite that expected for charge-based sensing of the DNA with a p-type semiconductor20, thus implying a new detection mechanism.
Figure 2. Single channel nanowire-nanopore FET detection of DNA translocation.
Left panels of a–c: a, Simultaneously recorded ionic current and FET conductance signals with both chambers filled with 1 M KCl buffer, voltage 0.6 V and 6 nM pUC 19 dsDNA in the cis chamber. b, Simultaneously recorded ionic current and FET conductance signals at 2 V voltage. c, Simultaneously recorded ionic current and FET conductance signals at 2.4 V voltage. Measurements in panel b and c were carried out with a trans chamber KCl buffer concentration of 10 mM, cis chamber KCl buffer concentration of 1 M and 1.4 nM pUC19 DNA. Right panels of a–c are zoom-in views of single ionic current and FET conductance events at the time indicated by black arrows on the ionic current traces of the corresponding left panels.
The nanowire-nanopore FET signal amplitude at non-balanced buffer salt concentration can be compared to the ionic current signal by converting the FET conductance to a current. This conversion shows that the FET has a current change of ~30 nA compared to the ~3 nA ionic current changes during DNA translocation. Although the noise in this nanowire-nanopore FET (Fig. 2) is relatively high, other nanowire-nanopore FETs (e.g. see Fig. 4b below) and other silicon nanowire FET sensors26,27 demonstrate that much lower noise (and correspondingly higher signal to noise ratio) can be achieved in general for nanowire-nanopore FETs. In addition, the relatively large (30 nA) translocation signal from the FET suggests the potential for higher bandwidth recording than with smaller ionic current detection schemes.
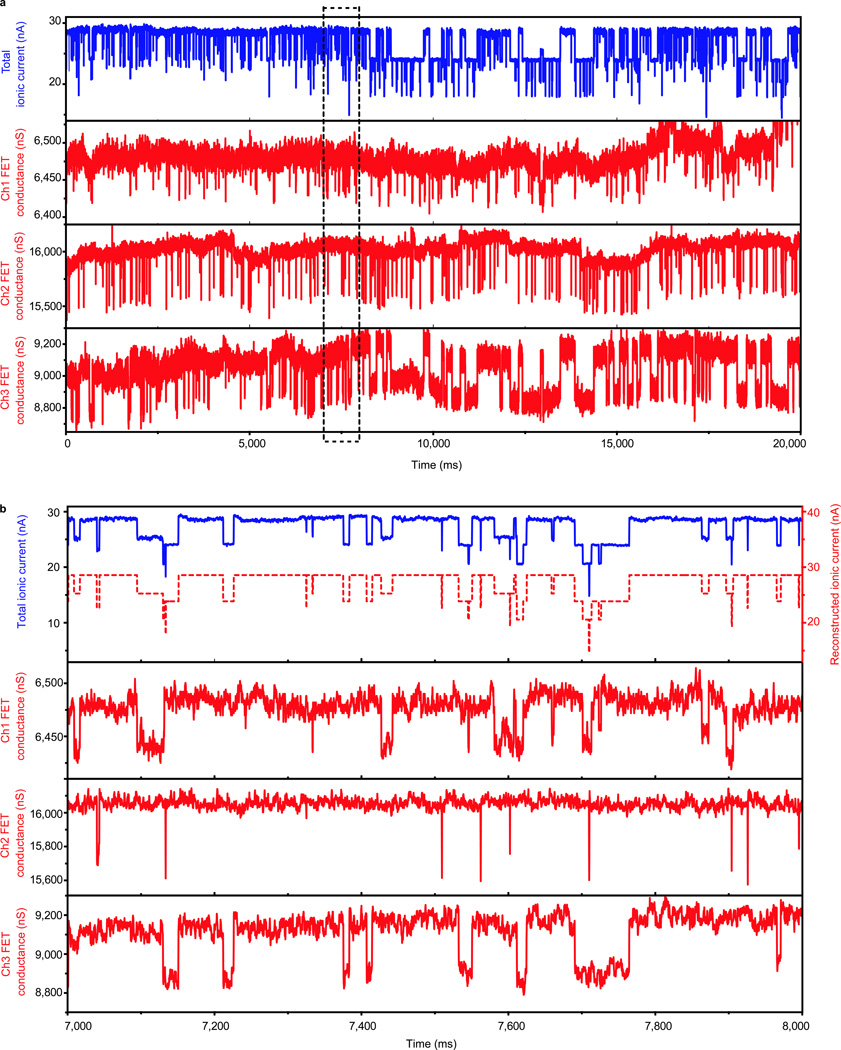
Figure 4. Multi-channel recording of DNA translocation with three nanowire-nanopore FET sensors.
All measurements are made with 1 M KCl buffer in cis chamber and 10 mM KCl buffer in trans chamber, voltage of 3 V and 1.4 nM pUC19 DNA. a, Simultaneous recording of the total ionic current and three nanowire-nanopore FET conductance channels. b, Higher-resolution view of the multiplexed recording from dashed rectangular area in panel a. The dashed red trace in the upper panel corresponds to the reconstructed ionic current trace (offset 10 nA for clarity) calculated from the three FET traces.
To understand the nanowire-nanopore detection mechanism we first consider basic experimental facts. First and as discussed above, it is possible to exclude direct charge sensing by the nanowire-nanopore FET20,21 because the negative charge on the DNA backbone should produce an increase in conductance for the p-type device during translocation instead of the observed decrease. Second, the importance of the differential buffer salt concentration suggests that solution resistance plays an important role in the signal generation. Specifically, under balanced buffer conditions (1 M / 1 M), the nanopore dominates the solution resistance and the voltage drops primarily across the nanopore. The potential around the nanowire-nanopore sensor is very close to ground regardless of the change in the solution resistance during DNA translocation. However, when the buffer concentration in the trans chamber containing the nanowire-nanopore sensor is lower than the cis chamber, the nanopore and the trans chamber solution resistances are comparable while the resistance of cis chamber will be negligible. Hence, changes in the solution resistance of nanopore and trans chamber during DNA translocation can result in a change of the potential around the nanowire-nanopore sensor that is detected.
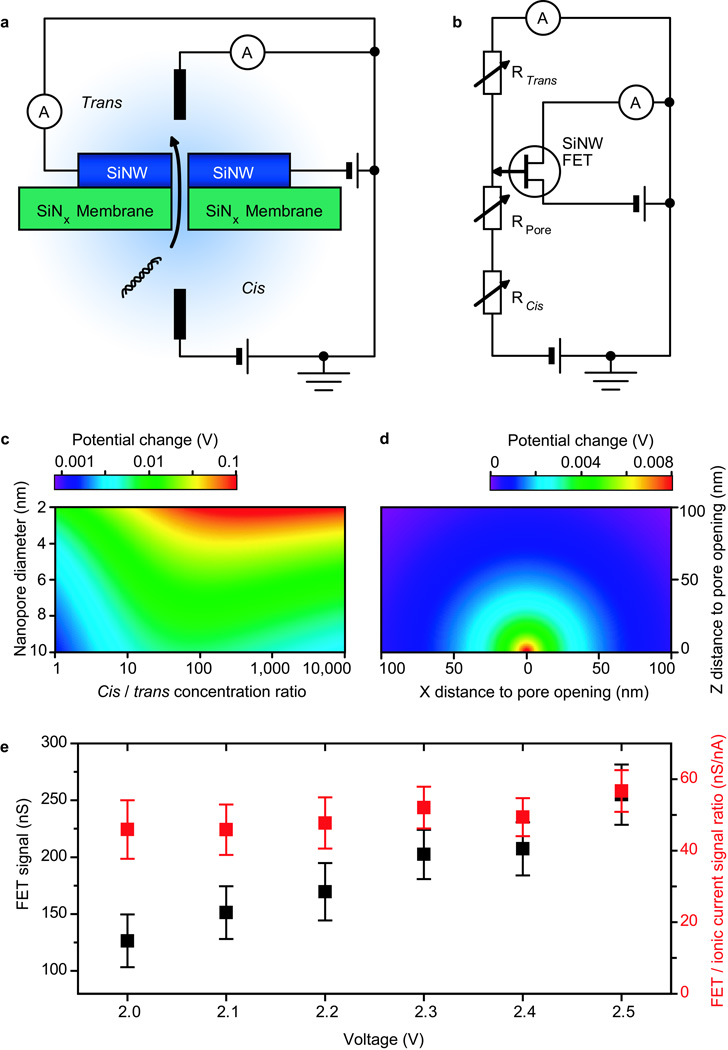
To understand quantitatively this proposed and unexpected detection mechanism for the nanowire-nanopore FET, we have modelled the buffer concentration, electric potential and electric field distributions inside the solution of the nanopore and solution chamber system (Fig. 3a). The equivalent circuit (Fig. 3b) separates the total solution resistance into nanopore resistance, cis and trans chamber access resistances. The nanowire-nanopore FET sensor is simplified as a point-like potential detector at the nanopore opening on the trans side. Translocation of DNA molecules through the nanopore will partially block the nanopore thus leading to a transient change in nanopore resistance and both chamber access resistances. Detailed calculations (see SI Methods and Fig. S2) provide the solution electrical potential change around the trans chamber nanopore opening (potential change signal) during DNA translocation as:
| (1) |
Here V, A, l, d, CCis, CTrans and r are the voltage, cross-sectional area of the DNA, membrane thickness, nanopore diameter, cis and trans chamber buffer salt concentrations and distance to the nanopore opening respectively.
Figure 3. The nanowire-nanopore sensing mechanism.
a, Schematic of the sensing circuit. b, Equivalent circuit diagram of a. SiNW stands for silicon nanowire in a and b. c, Calculated potential change at the nanopore opening in the trans chamber, by supplementary equation (9), due to one dsDNA molecule translocation at 1 V voltage as a function of nanopore diameter and cis / trans chamber buffer concentration ratio. d, Calculated potential change distribution in trans chamber for a 10 nm diameter nanopore at 1 V voltage by equation (1). e, Experimental values of the FET signal (black data points) and FET signal / ionic current signal ratio (red data points) under different voltages.
To further analyse potential change signal, we first plot the signal at the nanopore opening as a function of nanopore diameter and cis / trans chamber buffer concentration ratio as shown in Fig. 3c. The potential change is predicted to increase with decreasing nanopore diameter, and can reach more than 10% of the applied voltage when the nanopore diameter is ca. 2 nm. The maximal potential change signal for a given diameter nanopore is also predicted to occur at intermediate buffer concentration ratios. Specifically, for the nanopore geometry used in our experiments (7 – 10 nm diameter, 50 nm thick membrane), the potential change signal can be larger than 1% of voltage at the optimal buffer concentration ratio (~100:1). We have tested the consistency of the model predictions with our experiments. Specifically, conversion of the FET conductance signal it into potential change by the measured solution-gated transconductance27, yields a potential change of 25 mV (Fig. 2c), which agrees with our model calculations, 31 – 16 mV, a 7 – 10 nm nanopore diameter with all other conditions the same as the experiment.
In addition, the predicted distribution of the potential change in the trans chamber at this optimal condition (Fig 3d) shows a highly localised signal within several tens of nanometers of the nanopore, thus suggesting the possibility of high density integration of nanowire-nanopore sensors without cross-talking. Moreover, analysis of the voltage drops across the nanopore and both chambers allows their solution resistances to be compared. To do so we plotted the potential distribution in both chambers at the optimal buffer concentration ratio (Fig. S3), and found the access resistance of the trans chamber and the resistance of the nanopore are indeed within the same order of magnitude while the access resistance of cis chamber is negligible. From the signal distribution, we can also roughly estimate the intrinsic bandwidth of the potential change signal (see SI Methods and Fig. S4). A simple RC model suggests a potential signal bandwidth of ~1 GHz under our experimental condition. Finally, the different voltages required to initiate translocation at 1:1 and 100:1 buffer concentration ratios (Fig. 2) can be explained quantitatively by our model: The onset of translocation is determined by the electric field at the nanopore opening in the cis chamber, which pulls the negatively charged DNA into the nanopore. We plotted the electric potential and field distribution inside the nanopore for both 1:1 and 100:1 buffer concentration ratio (Fig. S5). For 1:1 case, the field (~1.8×107 V/m) is uniformly distributed within the nanopore, but for 100:1 case, the field (~0.45×107 V/m) is smaller in cis side because the lower resistance (due to higher buffer concentration) on cis side. To produce the same electric field, the voltage applied for the 100:1 case should be ~4 times higher than the voltage for 1:1 case, which is quantitatively consistent with the observed onset of translocation events at ~2 V and 0.5–0.6 V, respectively (see Fig. 2).
A crucial difference between our FET-based local potential sensing and other proposed DNA direct sensing mechanisms8–17 is the dependence of the FET signal on the ionic current signal and the voltage. For DNA sensing mechanisms that rely on the direct interaction between DNA and the sensor through electrical coupling8 or quantum mechanical tunnelling15,16, the signal typically is not related directly to the ionic current or voltage and should not change significantly when the voltage changes. For the local potential sensing mechanism, however, the sensor signal is predicted to be proportional to the voltage and can be regarded as the linear amplification of the ionic current signal. Hence, the ratio between FET signal and ionic current signal should be a constant for a given nanopore geometry and buffer concentration (see SI Methods). This unique feature was experimentally tested by plotting the FET signal amplitude (black) and FET / ionic current signal ratio (red) at different voltage values (Fig. 3e). Notably, the data shows that the FET signal amplitude increases, but the signal ratio is approximately a constant with increasing voltage, and thus provides strong support for sensing by a local potential mechanism.
We have also carried out several additional experiments to test the validity of our new model. First, DNA translocation experiments were carried out at different buffer salt concentrations in the cis / trans chambers (see SI Methods and Figs. S6, S7). Measurements made at 3.3 M / 33mM, which is the same 100:1 ratio as results presented below in Fig. 4 for 1 M / 10 mM, show the signal amplitude is ca. constant when the buffer concentration changes proportionally in both chambers, in agreement with the model predictions and in contrast to conventional charged-based FET sensing mechanism. Furthermore, when the cis / trans concentration ratio is reduced to 10:1, the recorded translocation signal (Fig. S7) is reduced as predicted by the model in Fig. 3c. Last, translocation experiments carried using the formally-neutral polymer polyethylene glycol (see SI Methods and Fig. S8) show correlated ionic current and FET conductance signals similar to but smaller in amplitude that recorded for DNA translocation. The smaller FET signal is consistent with expectations for our model (given the smaller ionic current signal change), and more importantly, the fact that we observe the same signal polarity in the FET channel is inconsistent with a charge-based FET sensing mechanism but in complete agreement with our potential sensing mechanism.
A key advantage of nanowire-nanopore FET sensor is the potential for integration and multiplexing within a single analysis chamber without complex microfluidic systems28. Notably, simultaneous recording from three nanowire-nanopore devices (Fig. 4a) demonstrates that continuous translocation events are observed in all three FET channels as well as for the total ionic current channel. Closer examination of the three FET and total ionic current signals (Fig. 4b.) shows clearly that the three FET channels operate independently and every falling or rising edge apparent in the total ionic current channel can be uniquely correlated to a corresponding edge in one of the three FET channels. Significantly, the total ionic current signal reconstructed from the data for the three FET channels (dashed-red trace, top panel, Fig. 4b) exhibits nearly perfect agreement with the measured total ionic current (see SI Methods). In addition, the histogram of channel specific ionic currents (see SI Methods and Fig. S9) demonstrates that the ionic current signal amplitudes in different channels are also independent. Since previous studies have shown that it is possible to fabricate large numbers of nanowire-FET devices29 with reproducible properties, and the local potential signal decay length is as short as tens of nanometers, we expect that it will be possible to multiplex the nanowire-nanopore FETs at much higher number and density as well.
Direct sequencing of long single-stranded DNA molecules using FET-based nanopore sensors and the new potential change detection mechanism will require optimizing the signal-to-noise ratio associated with individual bases and improving signal spatial resolution. Recognizing that direct base differentiation by FET potential measurement is coupled to variations in the ionic current, suggests that concepts proposed and demonstrated for base-resolved ionic current measurements5–7 could be combined with our work for success. For example, it should be possible to extend our nanowire-nanopore FET to atomically-thin graphene membranes7–9 to achieve single base spatial resolution, although the graphene nanopore would require precise structure engineering to enable differentiation of the distinct bases. Alternatively, coupling an engineered protein nanopore to the nanowire-nanopore FET could provide both spatial and base-resolution necessary for direct sequencing due to the localised change of the potential at the nanopore opening (Fig. 3d). Our nanowire-nanopore FET sensor results and modelling strongly motivate such effort given advantages over direct ionic current and other sensor based detection schemes, including larger measurement signals, high signal bandwidth with attractive nanopore size scaling, and straightforward integration and multiplexing. We believe that this work provides a strong starting point for a new class of nanopore sequencing devices with capability for fast direct sequencing at large-scale integration.
Methods summary
The silicon nanowires were synthesized by chemical vapour deposition (CVD) methods described previously30. Electron-beam lithography (EBL) and solid-state diffusion of nickel contact were used to fabricate short channel devices on commercially available SiNx TEM membrane grid chips. SiNx conformal thin film deposited by plasma enhanced CVD was used to passivate all metal electrodes before final lift-off. nanopores were drilled by focusing the 200 keV electron beam in a JEOL 2010F field emission TEM. The nanowire-nanopore FET sensor chip was glued onto a home-made PCB chip carrier and electrically connected to the chip carrier by wire-bonding. The chip carrier was sandwiched by mechanically clamped-on PDMS chambers with a tight seal, which were filled with sterilized and filtered buffer solutions. DNA translocation measurements were made using linearized pUC19. FET and ionic current signals were amplified and digitized using standard electronics with the sensor set-up mounted in a Faraday box.
Full Methods and any associated references are available in the online version of the paper at www.nature.com/naturenanotechnology.
Supplementary Material
Acknowledgement
We thank A. Meller, M. Wanunu, D. Casanova, J. Huang, J. Cahoon and T.J. Kempa for helpful discussions. CML acknowledges support of this work by a NIH Director’s Pioneer Award 5DP1OD003900.
Footnotes
Author Contributions P.X., Q.X., Y.F. and C.M.L. designed the experiments. P.X., Q.X. and Y.F. performed the experiments. P.X. performed the modelling and calculation. P.X. and Q.Q. wrote the program for data processing. P.X., Q.X., Y.F., Q.Q. and C.M.L. analyzed the data and wrote the paper.
Additional information The authors declare no competing financial interests. Supplementary Information accompanies the paper on www.nature.com/naturenanotechnology. Reprints and permission information is available online at http://npg.nature.com/reprintsandpermissions.
References
- 1.Branton D, et al. The potential and challenges of nanopore sequencing. Nature Biotech. 2008;26:1146–1153. doi: 10.1038/nbt.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Venkatesan BM, Bashir R. Nanopore sensors for nucleic acid analysis. Nature Nanotech. 2011;6:615–624. doi: 10.1038/nnano.2011.129. [DOI] [PubMed] [Google Scholar]
- 3.Zwolak M, Ventra MD. Colloquium: Physical approaches to DNA sequencing and detection. Rev. Mod. Phys. 2008;80:141–163. [Google Scholar]
- 4.Dekker C. Solid-state nanopores. Nature Nanotech. 2007;2:209–215. doi: 10.1038/nnano.2007.27. [DOI] [PubMed] [Google Scholar]
- 5.Clarke J, et al. Continuous base identification for single-molecule nanopore DNA sequencing. Nature Nanotech. 2009;4:265–270. doi: 10.1038/nnano.2009.12. [DOI] [PubMed] [Google Scholar]
- 6.Derrington IM, et al. Nanopore DNA sequencing with MspA. Proc. Natl. Acad. Sci. U.S.A. 2010;107:16060–16065. doi: 10.1073/pnas.1001831107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garaj S, et al. Graphene as a subnanometre trans-electrode membrane. Nature. 2010;467:190–193. doi: 10.1038/nature09379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Merchant CA, et al. DNA translocation through graphene nanopores. Nano Lett. 2010;10:2915–2921. doi: 10.1021/nl101046t. [DOI] [PubMed] [Google Scholar]
- 9.Schneider GF, et al. DNA translocation through graphene nanopores. Nano Lett. 2010;10:3163–3167. doi: 10.1021/nl102069z. [DOI] [PubMed] [Google Scholar]
- 10.Fologea D, Uplinger J, Thomas B, McNabb DS, Li J. Slowing DNA translocation in a solid-state nanopore. Nano Lett. 2005;5:1734–1737. doi: 10.1021/nl051063o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peng H, Ling XS. Reverse DNA translocation through a solid-state nanopore by magnetic tweezers. Nanotech. 2009;20:185101. doi: 10.1088/0957-4484/20/18/185101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Olasagasti F, et al. Replication of individual DNA molecules under electronic control using a protein nanopore. Nature Nanotech. 2010;5:798–806. doi: 10.1038/nnano.2010.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luan B, et al. Base-by-base ratcheting of single stranded DNA through a solid-state nanopore. Phy. Rev. Lett. 2010;104:238103. doi: 10.1103/PhysRevLett.104.238103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gracheva ME, et al. Simulation of the electric response of DNA translocation through a semiconductor nanopore-capacitor. Nanotech. 2006;17:622–633. [Google Scholar]
- 15.King GM, Golovchenko JA. Probing nanotube-nanopore interactions. Phy. Rev. Lett. 2005;95:216103. doi: 10.1103/PhysRevLett.95.216103. [DOI] [PubMed] [Google Scholar]
- 16.Lagerqvist J, Zwolak M, Ventra MD. Fast DNA sequencing via transverse electronic transport. Nano Lett. 2006;6:779–782. doi: 10.1021/nl0601076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ivanov AP, et al. DNA tunnelling detector embedded in a nanopore. Nano Lett. 2011;11:279–285. doi: 10.1021/nl103873a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chaste J, et al. Single carbon nanotube transistor at GHz frequency. Nano Lett. 2008;8:525–528. doi: 10.1021/nl0727361. [DOI] [PubMed] [Google Scholar]
- 19.Hu Y, Xiang J, Liang G, Yan H, Lieber CM. Sub-100 nanometer channel length Ge/Si nanowire transistors with potential for 2 THz switching speed. Nano Lett. 2008;8:925–930. doi: 10.1021/nl073407b. [DOI] [PubMed] [Google Scholar]
- 20.Cui Y, Wei Q, Park H, Lieber CM. Nanowire Nanosensors for Highly Sensitive and Selective Detection of Biological and Chemical Species. Science. 2001;293:1289–1292. doi: 10.1126/science.1062711. [DOI] [PubMed] [Google Scholar]
- 21.Patolsky F, et al. Electrical detection of single viruses. Proc. Natl. Acad. Sci. U.S.A. 2004;101:14017–14022. doi: 10.1073/pnas.0406159101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Patolsky F, et al. Detection, stimulation, and inhibition of neuronal signals with high-density nanowire transistor arrays. Science. 2006;313:1100–1104. doi: 10.1126/science.1128640. [DOI] [PubMed] [Google Scholar]
- 23.Kim MJ, Wanunu M, Bell DC, Meller A. Rapid fabrication of uniformly sized nanopores and nanopore arrays for parallel DNA analysis. Adv. Mat. 2006;18:3149–3153. [Google Scholar]
- 24.Kasianowicz JJ, Brandin E, Branton D, Deamer DW. Characterization of individual polynucleotide molecules using a membrane channel. Proc. Natl. Acad. Sci. U.S.A. 1996;93:13770–13773. doi: 10.1073/pnas.93.24.13770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li J, et al. Ion-beam sculpting at nanometre length scales. Nature. 2001;412:166–169. doi: 10.1038/35084037. [DOI] [PubMed] [Google Scholar]
- 26.Cohen-Karni T, Timko BP, Weiss LE, Lieber CM. Flexible electrical recording from cells using nanowire transistor arrays. Proc. Natl. Acad. Sci. USA. 2009;106:7309–7313. doi: 10.1073/pnas.0902752106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tian B, Cohen-Karni T, Qing Q, Duan X, Xie P, Lieber CM. Three-dimensional, flexible nanoscale field-effect transistors as localised bioprobes. Science. 2010;329:831–834. doi: 10.1126/science.1192033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Osaki T, Suzuki H, Le Pioutle B, Takeuchi S. Multichannel simultaneous measurements of single-molecule translocation in α-hemolysin nanopore array. Anal. Chem. 2009;81:9866–9870. doi: 10.1021/ac901732z. [DOI] [PubMed] [Google Scholar]
- 29.Yan H, et al. Programmable nanowire circuits for nanoprocessors. Nature. 2011;470:240–244. doi: 10.1038/nature09749. [DOI] [PubMed] [Google Scholar]
- 30.Cui Y, Zhong Z, Wang D, Wang W, Lieber CM. High performance silicon nanowire field effect transistors. Nano Lett. 2003;3:149–152. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.