Abstract
The synaptic ribbon is an electron-dense structure found in hair cells and photoreceptors. The ribbon is surrounded by neurotransmitter-filled vesicles and considered to play a role in vesicle release. We generated an objective, quantitative analysis of the protein composition of the ribbon complex using a mass spectrometry-based proteomics analysis. Our use of affinity-purified ribbons and control IgG immunoprecipitations ensure that the identified proteins are indeed associated with the ribbon complex. The use of mouse tissue, where the proteome is complete, generated a comprehensive analysis of the candidates. We identified 30 proteins (comprising 56 isoforms and subunits) associated with the ribbon complex. The ribbon complex primarily comprises proteins found in conventional synapses, which we categorized into 6 functional groups: vesicle handling (38.5%), scaffold (7.3%), cytoskeletal molecules (20.6%), phosphorylation enzymes (10.6%), molecular chaperones (8.2%), and transmembrane proteins from the presynaptic membrane firmly attached to the ribbon (11.3%). The 3 CtBP isoforms represent the major protein in the ribbon whether calculated by molar amount (30%) or by mass (20%). The relatively high quantity of phosphorylation enzymes suggests a very active and regulated structure. The ribbon appears to comprise a concentrated cluster of proteins dealing with vesicle creation, retention and distribution, and consequent exocytosis.
Keywords: synapse, synaptic ribbon, proteomics, vesicle release, active zone, retina, cochlea
Introduction
The synaptic ribbon is one of the great enigmas of sensory biology. It sits at the core of some of the most remarkable synapses in the body, yet little is known of its molecular structure or function. Surrounded by neurotransmitter-filled vesicles, it is thought to generate a readily releasable pool of vesicles at the synapse 1–3. The ribbon is found in photoreceptors and in hair cells – cell types specialized for graded synaptic transmission; they are adapted to release measured amounts of neurotransmitter in response to small changes in membrane potential. Most other synapses in the nervous system respond to transient presynaptic voltages in the form of action potentials. This has led to speculation that the synaptic ribbon may exist to provide a transient response capability to a synapse otherwise optimized for graded responses 4. This idea is supported by one of the few functional analyses of the ribbon’s role in stimulus coding: Bassoon knockout mice have fewer synaptic ribbons, and most are detached from the presynaptic membrane region. The primary deficit in the auditory responses of these mice is a loss of rapid onset responses to stimuli. A secondary deficit is a reduction in both spontaneous and evoked discharge rate, suggesting an additional role enabling sustained high rates of exocytosis 5,6.
Despite an abundance of immunohistochemical and other candidate analyses, there is no quantitative or objective data on the protein composition of the ribbon complex. Uthaiah and Hudspeth recently presented an extensive biochemical analysis of isolated synaptic ribbon complexes that combined immunoblotting, immunohistochemistry, and LC-MS (liquid chromatography-mass spectrometry) proteomics to generate a broad array of strong candidates for function at the ribbon complex 7. We have taken a complementary approach to identifying the proteins in the ribbon complex, which is to affinity-purify very large quantities of ribbon complex from adult mouse retinas to generate an objective assessment of the major proteins in the synaptic ribbon complex by LC-MS/MS. The affinity purification likely yielded both the ribbon and portions of the presynaptic density, but few of the associated vesicles. The project was by necessity completed on retina ribbons because the number of ribbons required for a quantitative mass spectrometry analysis is not achievable from hair cells, but it is reasonable to assume that the ribbon composition will apply to the auditory system as well.
Experimental Procedures
Animals
Mice, of various strains, 3 – 52 weeks of age were used. Animal procedures were approved by the Animal Care Committee of the Massachusetts Eye and Ear Infirmary.
Purification of synaptic ribbon complexes
Mice were anesthetized with pentobarbital (300 mg/Kg). Eyeballs were removed; the retinas scraped off and kept at −80 °C. Frozen bovine retinas were obtained from PelFreeze (Arkansas). A total of 30 bovine retinas or 1000 murine retinas were utilized for a set of immunoprecipitations (IPs). We modified Schmitz et al’s 8,9 ribbon purification protocol, avoiding the last steps, namely the 70–20% sucrose step gradient, and the consequent treatment with 2 M NaCl and high pH. For the mouse retinas we also skipped the preceding 30%–50% sucrose gradient step and reduced the Triton buffer incubation to 5 min in order to increase our protein yield. The final pellets of the murine and bovine ribbon isolation were recovered in IP buffer (50mM Tris pH 7.5, 0.5% Triton X100,150 mM NaCl, 10% Glycerol, 1 mM EGTA, 2 mM MgCl2 and 1 mM of PMSF (phenylmethanesulfonylflouride), and sonicated. The protein yield, amounting to 30 mg of murine material and 22.5 mg of bovine material, was adjusted to 3 μg/ul. Most importantly, we introduced an affinity-purification step with anti-CtBP2 monoclonal antibody (Ab), which recognizes both Ribeye and CtBP2. As a control we used mouse immunoglobulin G (IgG).
Immunoprecipitation and gel separation of ribbon-associated proteins for sequencing
Dynabeads® M-450 Tosylactivated (Invitrogen) were coupled to antibodies according to manufacturer’s protocol and cross-linked with dimethyl pimelimidate dihydrochloride (Sigma) for 20 min. Partially purified ribbon fractions (described above) were incubated with 500 μl beads coupled to Ribeye/CtBP2 Ab or normal IgG at 4°C for 4.5h followed by washing in IP buffer. Beads were heated at 80°C in Novex® Tris-Glycin SDS sample buffer (Invitrogen) and the released proteins loaded on NuPAGE 10% Bis-Tris Gel 1.0 mm (Invitrogen) and separated electrophoretically. Samples were stained with the Novex® colloidal blue kit (Invitrogen). The gels were sliced across the horizontal axis of the lanes into 6 pieces each, the sizes of which varied slightly in accordance of the observed amount of protein bands in them.
Protein Identification
After destaining, gel sections were reduced, carboxyamidomethylated, and digested with trypsin in gel. To identify proteins specifically present in the ribbon complex, peptides from each sample section were subjected to microcapillary reverse-phase HPLC directly coupled to the nano-electrospray ionization source of a LTQ-Orbitrap Velos hybrid mass spectrometer (LC/MS/MS). The Orbitrap repetitively surveyed an m/z range from 395 to 1600, while data-dependent MS/MS spectra on the ten most abundant ions in each survey scan were acquired in the linear ion trap. MS/MS spectra were acquired with relative collision energy of 30%, 2.5-Da isolation width, and recurring ions dynamically excluded for 60 s. Preliminary evaluation of peptide-spectrum matches (PSMs) was facilitated using SEQUEST with a 30 ppm mass tolerance against the mouse or bovine subset of the Uniprot Knowledgebase. Using a custom version of the Harvard Proteomics Browser Suite (Thermo Scientific, San Jose CA), PSMs were accepted with mass error <3.0 ppm and score thresholds to attain an estimated false discovery rate of ~1% using a reverse decoy database strategy.
Stringent criteria were applied to all proteins in order for them to be considered ribbon constituents: we required that the protein be present in the CtBP2 pull-down material in quantities al least 4 times higher than in the IgG control and be identified by a minimum of four unique peptides in the CtBP2 immunoprecipitate alone. Protein assignments were designed to be most parsimonious: 1) no peptide spectrum was assigned to more than one protein and, 2) when a peptide sequence could be assigned to more than one protein, the assignment was made to the protein that had accumulated the most peptide spectra. For that reason we have grouped together proteins with highly related sequences such as CtBP1 and CtBP2, ERC1 and ERC2 (also called ELKS and CAST), and Bassoon with Piccolo. When performing the calculations we considered the number of potentially shared peptides to estimate the average number of unique peptides per protein. Each peptide is given a value of 1 if it is not potentially shared with other sequence-related protein isoforms and that value is assigned to the only protein it could belong to. If a peptide is potentially shared by more than one protein in a group, the value of 1 is divided by the number of proteins in the group that could potentially give rise to that peptide upon trypsin digest (see Supplementary Table 1). For example if two isoforms in a group share a peptide each isoform is assigned 0.5 unique peptides, and if there are 3 isoforms sharing a common peptide each isoform is given a value of 0.33. Upon establishing the potential commonality and respective values for each peptide, we calculated the total number of unique peptides per protein isoform. As a simple example if two proteins A and B have been assigned 8 and 2 unique peptides respectively by the algorithm, and 4 of the peptides assigned to protein A are theoretically observable in protein B, we recalculate the proteins A and B to have 6 and 4 unique peptides respectively. Highly related proteins like actins and proteins forming functional protein oligomers such as the subunits voltage-dependent calcium channel were left in the final list of top likely specific co-precipitates (even if they had less than 4 unique peptides or low ratios).
Protein Abundance Determination
We have adapted a method described previously for estimation of relative protein abundance 10. Our sequencing data has provided the number of unique peptides per protein in both IgG and CtBP fractions. To calculate the number of observable unique peptides per protein, protein sequences were digested in silico using the Expasy Peptide Mass tool: (http://ca.expasy.org/tools/peptide-mass.html) taking into account the experimental conditions used: allowing for two missed cleavages, oxidized methionines, treatment with iodoacetamide, and selecting for molecular weight range of the peptides between 750–4000 Da (comparable with the scan range of the mass spectrometer 700–3500).
The PAI (protein abundance index) is defined as:
where Nobsd and Nobsbl are the number of unique observed peptides and the number of unique observable peptides per protein, respectively.
The emPAI (exponentially modified protein abundance index) is defined as:
The protein contents in molar fraction percentages are calculated as:
where and Σ (emPAI) is the sum of emPAI values for all the proteins selected by us to be included in the analysis.
Calculation for the number of unique observable peptides selecting for masses ranging 500–3000 or 500–4000 Da gained similar ratios for the proteins in the mix. Generally the same holds true if all theoretical peptides were included.
Western blots: performed as described previously 11.
Antibodies
CtBP2 and KIF3A (BD Transduction Lab cat # 612044 and 611508, respectively), Bassoon (Assays Designs, cat # SAP7F407), Piccolo (Abcam, cat# ab76186), mouse IgG1κ (Sigma Aldrich, cat # M9269), ATP Synthase 5A1 (ProteinTech Group, cat # 14676-1-AP), secondary antibodies (#A11001, A10042 Invitrogen).
Immunolabeling of mouse retinas and purified/immunoprecipitated synaptic ribbons
Murine eyes or post- IP beads were fixed for 10–20 min in 3.7% paraformaldehyde. The eyes were treated with 30% sucrose/PBS overnight and cryosectioned at 8 μm. An overnight incubation with primary antibodies (diluted 1:200) in 10% normal goat serum was followed PBS wash and incubation with secondary antibodies (dilution1:500) for 1.5 h.
Results and Discussion
Affinity pull-down yielded large quantities of highly purified ribbons
We began by repeating the differential- and density-gradient centrifugation protocol developed by Schmitz et al. 8,9 using cow retinas. Because we observed major losses (99%) of the Ribeye protein (an alternative transcription product of Ctbp2 considered to be a signature protein of the ribbon 9) we developed an affinity pull-down procedure to reduce the loss of material and quantity of impurities (Figures 1 and 2). The efficiency of the IP procedure suggested it should be possible to obtain purified ribbons at earlier steps in the purification protocol- fractions 3 or even 2 which left us with about 30-fold more protein compared with the original procedure using the last (sixth) fraction (Figure 1, right panel).
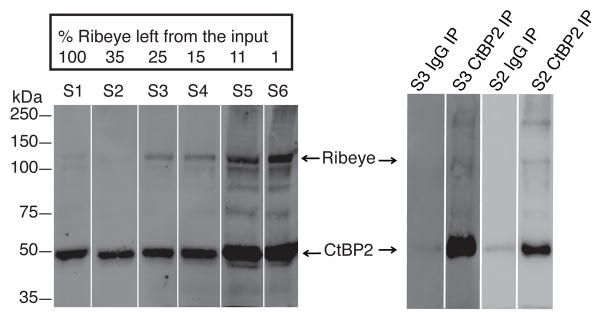
Figure 1.
Left panel: Loss of Ribeye during the purification scheme used for ribbon isolation based on the protocol developed originally by Schmitz. Western blots were used to separate Ribeye from CtBP2 (short) allowing calculation of the ramaining amount of Ribeye, based on sample volume, intensity of Ribeye’s Western blot signal, and percent loaded material for each sample. CtBP2 Ab detects both CtBP2 and Ribeye running at 50 and 120 kDa respectively. With each additional purification step, the Ribeye signal became more pronounced relative to the CtBP2 isoform.
Right panel: Immunoprecipitation with CtBP2/Ribeye or control IgG antibody on fractions 3 (S3) and 2 (S2) from the ribbon purification. The control IgG Ab has little background binding of CtBP2. The CtBP2 pull-down lanes have a very strong band around 50 kDa corresponding to the CtBP2 (short) isoform and a weakly stained, longer form running at 120 kDa and corresponding to the Ribeye isoform. The high molecular weight band above might represent multimers of the CtBP2 isoforms for it appears specifically in the CtBP2/Ribeye IP lanes only.
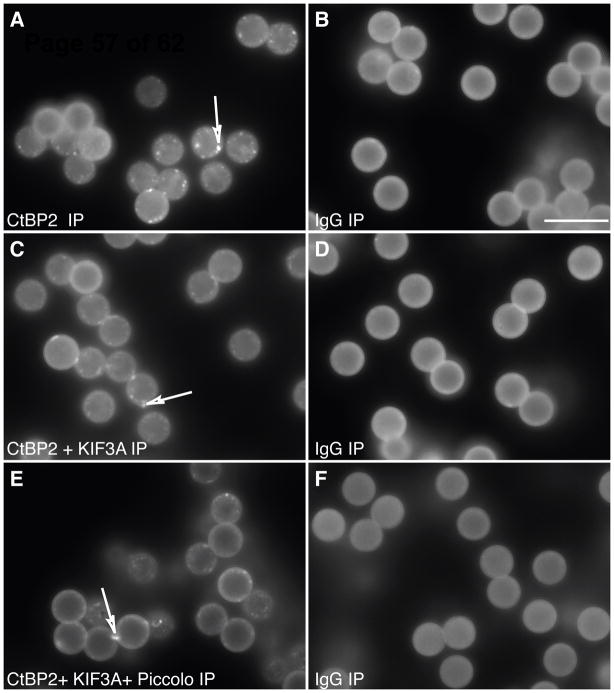
Figure 2. Affinity purified ribbons attached to magnetic beads.
Antibodies for proteins known to be in the ribbon were coupled to magnetic beads. The results were optimized by visualizing the precipitated ribbons with immunolabeling for CtBP2/Ribeye. Ribbons could be pulled down with antibodies for CtBP2 (corresponding to the B domain of Ribeye isoform), KIF3A, and Piccolo. The quantity of ribbons pulled down with CtBP2/Ribeye was not increased by adding antibodies to other ribbon proteins such as Piccolo and/or KIF3A (A, C, E). As a control for nonspecific binding, we used mouse IgG (B, D and F). The beads exhibit a degree of autofluorescence. The bright dots (indicated by arrows) in A, C, and E are partially broken ribbons. Due to the spherical structure of the beads, only the periphery is in focus at these pictures. In reality the entire bead is covered with ribbons. Scale bar (in B) 10 μm.
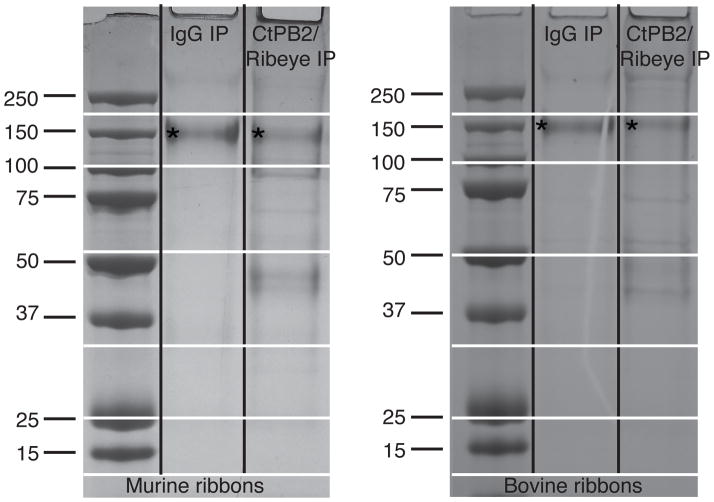
The purification and immunoprecipitation procedure was scaled up to purify a large quantity of ribbons for proteomics analysis (1000 mouse retinas or 30 cow retinas, see Experimental Procedures). Eluates from the control beads contained mostly antibodies; in the non-reducing conditions used in this experiment they appear as bands of 150 kDa. Eluates from the CtBP2/Ribeye immunoprecipitates had a number of bands that did not appear in the control (Figure 3).
Figure 3. Coomassie Blue staining of the affinity purified ribbons using CtBP2 antibody and control IgG.
The 150 kDa non-reduced Ab molecule is indicated with an asterisk. A small fraction is also broken down to heavy and light chains −50 and 25 kDa respectively. The more abundant additional bands are clearly visible in the CtPB2 IP lanes, although the scanned image does not depict less abundant proteins segregating into faintly stained bands. The horizontal lines indicate the cutting sites when slicing the gel into discrete portions for sequencing.
Mass spectrometry based proteomics analysis
LC tandem mass spectrometry proteomics analyses were performed on each slice shown in figure 3 for both the mouse and the bovine retina preparations. Because the bovine proteome is incomplete, and due to the unquantified time between the cow’s death and retina freezing, the cow data was used only for corroboration. The description that follows is for the mouse ribbon proteome.
We identified over 2,500 peptides attributed to over 206 different proteins. Quantification of the individual protein amounts was accomplished by calculating the exponentially modified protein abundance index 10. This method is based on a linear relationship between the logarithm of protein abundance and the protein coverage, the latter calculated as the number of observed vs. observable unique peptides. The algorithm is useful for estimating the relative ratios of proteins in a complex sample (see calculation details in Experimental Procedures section). The amount of material needed (1000 retinas) to generate the large numbers of unique peptide fragments for quantification made repeated measurements impractical, limiting our ability to assess the statistical significance of the stoichiometric findings. We have chosen to focus only on the most robustly identified proteins, so our false positive rate should be very low, but of course we may have missed many low stoichiometry proteins.
Of the 206 proteins, 30 were also present in similar quantities in the control IgG immunoprecipitates and thus eliminated from further consideration. Another 6 proteins were eliminated because they were less than four-fold higher in the CtBP2/Ribeye vs. control immunoprecipitates. Finally, we excluded from our quantitative analysis 102 proteins that were exclusively present in the CtBP2/Ribeye immunoprecipitates but were represented by less than 4 unique peptide sequences. A few exceptions were made to this selection process for quaternary proteins and highly homologous isoforms (see Experimental Procedures). Because we have used such a conservative selection process, we may have excluded from quantification some proteins present in small amounts in the ribbon (listed in Table 3), but chose to emphasize instead the major components for quantification.
Table 3. Minor co-purified proteins (including nuclear).
Cytoplasmic and nuclear proteins, that did not pass our threshold criteria, are listed in the 1st (Uniprot abbreviation) and 2nd columns. In the 3rd and 4th columns show the number of unique peptides in the CtBP2 and IgG precipitates respectively. Column 5 shows overlap between mouse and cow data. The exact matches for mouse proteins present in our bovine list of co-precipitates are indicated with two dots, while those represented by close protein homologues are indicated with one dot. Only a few proteins were absent in the bovine data set (no dots).
| Uniprot name | Protein | # uniq. pept. in CtPB2 IP | # uniq. pept. in IgG IP | Bovine Overlap |
|---|---|---|---|---|
| ACAD9 | Acyl-CoA Dehydrogenase Family Member 9 | 2 | 0 | • |
| CLTC | Clathrin Heavy Chain 1 | 2 | 0 | •• |
| DPYSL2 | Dihydropyrimidinase-Related Protein 2 | 3 | 0 | •• |
| GNB1 | Guanine Nucleotide-Binding Protein Subunit β-1 | 2 | 0 | •• |
| HTRA1 | Serine Protease HTRA1 | 3 | 0 | |
| IMMT | Mitochondrial Inner Membrane Protein | 2 | 0 | •• |
| INA | Alpha-Internexin | 2 | 0 | •• |
| MYO6 | Myosin-VI | 2 | 0 | •• |
| NLGN2 | Neuroligin-2 | 2 | 0 | •• |
| NSF | Vesicle-Fusing ATPase | 2 | 0 | •• |
| RPH3A | Rabphilin-3A | 3 | 0 | •• |
| RPLP0 | 60S Acidic Ribosomal Protein P0 | 2 | 0 | •• |
| RPSA | 40S Ribosomal Protein SA | 2 | 0 | • |
| SYN1 | Synapsin-1 | 3 | 0 | •• |
| SYN2 | Synapsin-2 | 2 | 0 | • |
| SYNGR3 | Synaptogyrin-3 | 2 | 0 | •• |
| SYT1 | Synaptotagmin-1 | 2 | 0 | •• |
| TRAPPC9 | Trafficking Protein Particle Complex subunit 9 | 2 | 0 | |
| — NUCLEAR PROTEINS — | ||||
| DDX17 | Probable ATP-dependent RNA Helicase DDX17 | 2 | 0 | • |
| EIF4A3 | Eukaryotic Initiation Factor 4A-III | 3 | 0 | |
| FMR1 | Fragile X Mental Retardation Protein 1 homolog | 2 | 0 | • |
| HDAC2 | Histone Deacetylase 2 | 2 | 0 | |
| PURB | Transcriptional Activator Protein Pur- β | 3 | 0 | • |
| ZNF516 | Zinc Finger Protein 516 | 2 | 0 | |
We quantified, with emPAI, the remaining 68 proteins, including isoforms, that were represented by more than 4 unique peptides in CtBP2/Ribeye immunoprecipitates and were either absent or represented by at least 4-fold fewer unique peptides in the control Of these 68 proteins, 12 are typically found in the nucleus. This is not surprising, for CtBP2 is a transcriptional co-repressor and despite our efforts to eliminate the nuclear fraction early in the purification, some contamination remained. We eliminated these 12 as likely candidates to be in the ribbon, and have treated them separately (Table 2). Many of the remaining 56 proteins were either subunits of a quaternary structure or represented by closely related isoforms, allowing the dataset to be compressed to describe 30 proteins that were present in high enough amounts to warrant quantification (Table 1).
Table 2. Nuclear proteins co-purified with CtBP2 Ab.
The names of the nuclear proteins, which passed our threshold criteria, are listed in the 1st (Uniprot abbreviation) and 2nd columns. In the 3rd and 4th columns are shown the number of unique peptides in the CtBP2 and IgG precipitates respectively. Column 5 shows overlap between mouse and cow data. The exact matches for mouse proteins present in our bovine list of co-precipitates are indicated with two dots, while those represented by close protein homologues are indicated with one dot. Only a few proteins were absent in the bovine data set (no dots).
| Uniprot name | Protein | # uniq. pept. in CtPB2 IP | # uniq. pept. in IgG IP | Bovine Overlap |
|---|---|---|---|---|
| AOF2 | Lysine-Specific Histone Demethylase | 9 | 0 | |
| DDX5 | RNA Helicase DDX5 | 8 | 0 | •• |
| HIST1H1C | Histone H1.2 | 4 | 0 | •• |
| HIST1H2BF | Histone H2B type | 5 | 1 | • |
| HNRNPM | Heterogeneous Nuclear Ribonucleoprotein M | 5 | 0 | •• |
| LMNB1 | Lamin-B1 | 16 | 1 | •• |
| MYEF2 | Myelin Expression Factor 2 | 4 | 0 | |
| PABPC1 | Polyadenylate-Binding Protein 1 | 5 | 1 | • |
| RBM14 | RNA- Binding Protein 14 | 16 | 1 | •• |
| RBM9 | RNA- Binding Protein 9 | 4 | 0 | • |
| SCAI | Suppressor of Cancer cell Invasion Protein | 10 | 0 | •• |
| C20ORF112 HOMOLOG | Similar to Nucleolar Protein 4 Isoform 1 | 9 | 0 | •• |
Table 1. Quantified proteins in the ribbon complex.
The names the proteins, that passed our criteria, are listed in the 1st (Uniprot abbreviation) and 2nd columns. Columns 3 and 4 describe the number of unique peptides observed and observable for each protein. Those numbers were used to calculate the emPAI value given in the 5th column and derive the molar % ratio for individual proteins and protein groups of closely related isoforms and/or quaternary structures (columns 6 and 7 respectively). The 8th column shows the number of unique peptides in the IgG precipitates. Column 9 shows overlap between mouse and cow data. Most mouse proteins were also present in our bovine list of ribbon candidates as an exact match (indicated with two dots), or were represented by protein homologues (indicated with one dot), and only a few were absent in the bovine data set, which can be attributed at least partially to poor data annotation (for example Bassoon protein).
| Uniprot name | Protein | Calc. uniq. pept. | Observable | emPAI | % by individ. prot. | % group proteins | # pept. in Contr. | Bovine Overlap |
|---|---|---|---|---|---|---|---|---|
| ACTA2 | α-Actin-2 | 3.75 | 73 | 0.13 | 1.09 | 5.88 | 0 | • |
| ACTB | Cytoplasmic Actin 1 | 4.75 | 75 | 0.16 | 1.36 | 2 | • | |
| ACTB | Putative Actin | 5.75 | 74 | 0.2 | 1.7 | 0 | • | |
| ACTG1 | γ Actin-Like Protein | 5.75 | 73 | 0.2 | 1.73 | 0 | •• | |
| ATP1A1 | Na/K- ATPase α1 | 2 | 185 | 0.03 | 0.22 | 1.92 | 0 | •• |
| ATP1A3 | Na/K- ATPase α3 | 3 | 180 | 0.04 | 0.34 | 1 | •• | |
| ATP1B1 | Na/K- ATPase α1 | 5 | 79 | 0.16 | 1.36 | 0 | •• | |
| ATP5A1 | ATP Synthase, subunit α | 16 | 143 | 0.29 | 2.55 | 5.78 | 3 | •• |
| ATP5B | ATP Synthase, subunit β | 14 | 102 | 0.37 | 3.23 | 2 | •• | |
| BSN | Bassoon | 8 | 675 | 0.03 | 0.24 | 1.45 | 0 | |
| PCLO | Piccolo | 53 | 933 | 0.14 | 1.21 | 0 | •• | |
| CACNA2D4 | VCa Channel α-2/δ-4 | 3 | 162 | 0.04 | 0.38 | 1.12 | 0 | • |
| CACNB3 | VCa Channel β-3 | 4 | 113 | 0.08 | 0.74 | 0 | • | |
| CAMK2A | CaM Kinase II α | 3.5 | 99 | 0.08 | 0.74 | 2.97 | 0 | •• |
| CAMK2B | CaM Kinase II β | 3.5 | 100 | 0.08 | 0.73 | 0 | •• | |
| CAMK2D | CaM Kinase II δ | 7 | 101 | 0.17 | 1.5 | 0 | •• | |
| CASK | Cask | 8 | 211 | 0.09 | 0.79 | 0.79 | 0 | •• |
| CtBP1 | C-terminal Binding Protein | 23.5 | 69 | 1.19 | 10.34 | 30 | 0 | •• |
| CtBP2 | C-terminal Binding Protein | 37.5 | 73 | 2.26 | 19.66 | 0 | •• | |
| CTNNA2 | Catenin α-2 | 12 | 234 | 0.13 | 1.09 | 2.07 | 0 | •• |
| CTNNB1 | Catenin β-1 | 6 | 129 | 0.11 | 0.98 | 0 | •• | |
| EML5 | EML5 | 13 | 435 | 0.07 | 0.62 | 0.62 | 0 | •• |
| EPB41 | Band 4.1-Protein | 5.5 | 222 | 0.06 | 0.51 | 1.36 | 0 | •• |
| EPB41L2 | Band 4.1-Like Protein 2 | 7.5 | 272 | 0.07 | 0.57 | 0 | • | |
| EPB41L3 | Band4.1-Like Protein 3 | 3 | 220 | 0.03 | 0.28 | 0 | •• | |
| ERC1 | ELKS1/CASK2 | 31.5 | 365 | 0.22 | 1.91 | 6.17 | 0 | •• |
| ERC2 | CAST1/ELKS2 | 56.5 | 326 | 0.49 | 4.26 | 0 | •• | |
| GABRA1 | GABA Rec. α 1 | 6 | 110 | 0.13 | 1.16 | 2.5 | 0 | •• |
| GABRB2 | GABA Rec. 2β | 2.5 | 99 | 0.06 | 0.52 | 0 | •• | |
| GABRB3 | GABA Receptor β3 | 2.5 | 88 | 0.07 | 0.59 | 0 | •• | |
| GABRG2 | GABA Receptor γ2 | 1 | 87 | 0.03 | 0.23 | 0 | • | |
| GLUL | Glutamine Synthetase | 4 | 76 | 0.13 | 1.12 | 1.12 | 0 | •• |
| GPHN | Gephyrin | 9 | 158 | 0.14 | 1.22 | 1.22 | 0 | •• |
| GRIP1 | Glutamate Rec. Inter. Prot. 1 | 8 | 207 | 0.09 | 0.81 | 0.81 | 0 | •• |
| HSPA1B | Heat Shock 70 kDa Prot. 1 | 8.83 | 156 | 0.14 | 1.21 | 8.18 | 0 | •• |
| HSPA5 | Heat Shock 70 kDa Prot. 5 | 13.33 | 157 | 0.22 | 1.88 | 0 | •• | |
| HSPA8 | Heat Shock 70 kDa Prot. 8 | 17.83 | 163 | 0.29 | 2.49 | 2 | •• | |
| HSPA9 | Heat Shock 70 kDa Prot. 9 | 7.5 | 188 | 0.1 | 0.84 | 0 | •• | |
| HSPD1 | Heat Shock Protein 60 | 12.5 | 155 | 0.2 | 1.77 | 0 | •• | |
| MYH10 | Myosin10 | 7 | 648 | 0.03 | 0.22 | 0.22 | 0 | •• |
| NEFL | Neurofilament Light | 6 | 159 | 0.09 | 0.79 | 0.79 | 0 | •• |
| PGAM5 | Ser/Thr-Phosphatase | 10 | 82 | 0.32 | 2.82 | 2.82 | 0 | |
| PPFIA2 | Liprin-α2 | 7 | 356 | 0.05 | 0.4 | 0.4 | 0 | •• |
| PPP2R1A | Ser/Thr Phosphatase Regulatory subunit α | 9 | 121 | 0.19 | 1.62 | 1.62 | 0 | •• |
| RAPGEF4 | Rap Guanine Nucleotide Exchange Factor 4 | 33 | 245 | 0.36 | 3.16 | 3.16 | 0 | •• |
| RIMBP2 | RIM-Binding Protein 2 | 4 | 180 | 0.05 | 0.46 | 0.46 | 0 | |
| SAG | S-arrestin | 5 | 88 | 0.14 | 1.21 | 1.21 | 1 | •• |
| SPNA2 | Spectrin α | 22 | 763 | 0.07 | 0.6 | 1.08 | 0 | • |
| SPTBN1 | Spectrin β | 16 | 679 | 0.06 | 0.48 | 0 | •• | |
| SRCIN1 | SRC Kinase Inhibitor 1 | 5 | 340 | 0.03 | 0.3 | 0.3 | 0 | •• |
| TRIM21 | Trim21 | 5 | 118 | 0.1 | 0.89 | 0.89 | 1 | •• |
| TUBA1B | Tubulin α-1B | 7.5 | 63 | 0.32 | 2.74 | 10.94 | 2 | •• |
| TUBA4A | Tubulin α-4A | 4.5 | 76 | 0.15 | 1.27 | 0 | •• | |
| TUBB2A | Tubulin β-2A | 7.17 | 68 | 0.27 | 2.39 | 2 | •• | |
| TUBB2C | Tubulin β-2C | 6.67 | 68 | 0.25 | 2.2 | 1 | •• | |
| TUBB5 | Tubulin β-5 | 7.17 | 69 | 0.27 | 2.35 | 2 | •• |
Proteins in the ribbon complex
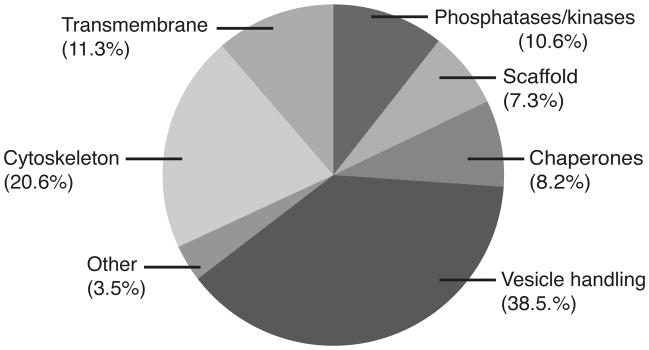
Of the 30 proteins quantified within the ribbon complex, 10 comprised nearly 4/5 (78.5%) of the total molar quantity, and thus are considered major constituents. Each major protein constitutes more than 2% of the total molar protein content. These proteins are: CtBP (30%), Actin (5.9%), Tubulin (10.9%), heat shock proteins (HSPA1b, 5, 8 and 9) (8.2%), ERC (ELKS/CAST) (6.2%), ATP Synthase (5.8%), CaM Kinase II (3%), RAPGEF4 (3.2%), PGAM5 (2.8%), and the GABA-A Receptor (2.5%). The ribbon complex (the ribbon core, a portion of the presynaptic membrane, and any other firmly attached proteins) is for the most part comprised of protein groups found in conventional synapses (Figure 4).
Figure 4.
Schematic representation of protein groups’ molar quantities.
Many of the ribbon proteins have well-established roles in vesicle handling: CtBP, ERC, Piccolo, Bassoon, Liprin and RIMBP. They also have well-documented interactions with one other 12–16. CtBP isoforms dominate this category. Because the sequence homology of the B domain of the CtBP isoforms is very high, and because the A domain of Ribeye was not present in the Uniprot library with which we generated the proteomics analysis, we had to infer the relative proportions of the CtBP isoforms. This was feasible because Ribeye has a much higher molecular weight than the other CtBP isoforms, and thus ran in a different position in the gel, which was consequently analyzed in a separate mass spec run. We estimate that the Ribeye comprised between one third and one half of the total molar amount of the CtBP isoforms. Bassoon is known to play a role in tethering the ribbon to the synaptic complex 6. A number of other known synaptic proteins were identified exclusively in ribbon immunoprecipitates, but with fewer than 4 unique peptides, so they did not meet the criteria for quantification. These include Synapsin (1 and 2), Rabphilin 3a, Synaptogyrin3, Synaptotagmin1, Clathrin Heavy Chain1, SNAP25, β-2-Syntropihin, Syntaxin-Binding Protein1, and Synaptic Vesicle Glycoprotein 2a (see also Table 3). Our purification procedure (specifically a tritonization step and subsequent sonication) probably stripped attached vesicles from the ribbon complex so that vesicle-associated proteins were absent or under-represented in the mix.
We identified several transmembrane proteins that are most likely associated with the synaptic membrane contacting the ribbon core. They include Na/K ATPase, GABA A Receptor, the L-TYPE Calcium Channel, and ATP Synthase. The L-Type Calcium Channel can bind Piccolo and RIMBP17,18. GABA-A Receptor α and β subunits were present in equimolar quantities with lesser quantities of the γ subunit. The receptor is a pentamer, and it likely comprises 2 α1 subunits, 2 β (β2 and β3) subunits and a γ subunit 19. ATP Synthase, once thought of as a mitochondrial enzyme is now known to be present in a variety of other locations in cells 20. It is unlikely that the ATP Synthase in our ribbon immunoprecipitates is from mitochondria. First, we only identified the alpha and beta (soluble) subunits while the mitochondrial ATP Synthase contains as many as 14 subunits both membrane bound and soluble, located at the inner mitochondrial membrane 21,22. Second, we co-precipitated very few other mitochondrial proteins, further decreasing the possibility of mitochondrial origin for the ATP synthase. We examined the distribution of ATP Synthase, and CaM Kinase II and observed them located proximal to, though not within, the ribbon, while CtBP1 is located within the ribbon core (Figure 5). GABAA receptors have been localized in the bipolar cells in the retina along with Gephryn, its binding partner 23–27. The bipolar cells contain part of the CtBP2/Ribeye protein content in the retina 28, thus probably accounting for this co-purification. The localization of PGAM5 protein has been found both in the nucleus and mitochondria 29–31, raising the possibility that it may have been pulled down as a nuclear contaminate.
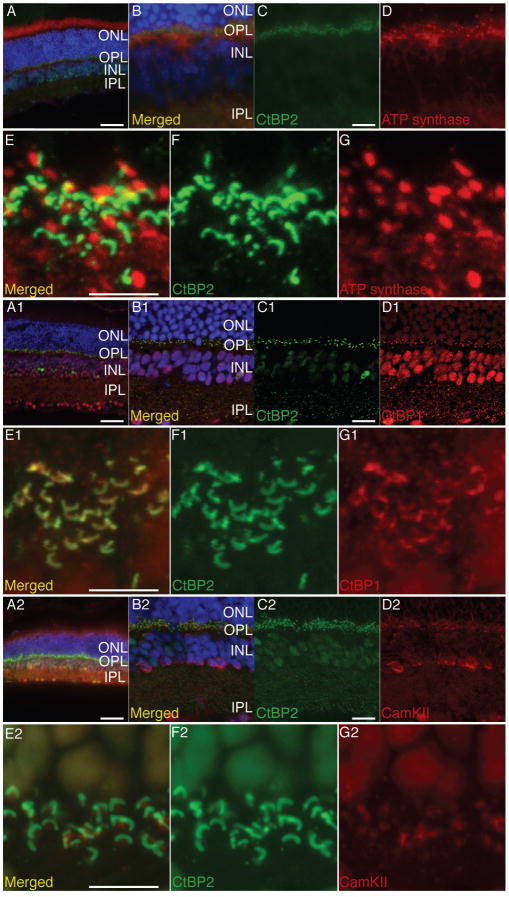
Figure 5. IHC staining for ATP Synthase, CtBP1 and CaM Kinase II in relation to CtBP2 on adult mouse retina.
Panels A, A1 and A2 show an overview of the retinal staining, while panels (B, B1, B2, C, C1 and C2) show medium magnification of the inner and outer plexiform layers. Panels EG, E1-G1 and E2-G2 display high magnification the spatial relation between CtBP2 in green and ATP Synthase CtBP1 and CaM Kinase respectively in red at the outer plexiform layer. Note that even when the two proteins are not colocalized they are juxtaposed. Scale bar in A, A1 and A2 is 50 μm, scale in C, C1 and C2 in 10 μm and in E, E1 and E2 is 5 μm.
Synapse-specific molecules are held in place and prevented from diffusion by additional protein interactions with scaffolding molecules, which eventually link them to the cytoskeleton. Scaffolding molecules we observed include Gephyrin, Catenin, GRIP1, CASK, Spectrin, Band 4.1 Protein, and Band 4.1 Like Proteins. Among the functionally related group of cytoskeletal proteins were Tubulin, Actin, Vimentin, EML5, NEFL, and Myosin10. CASK is known to anchor transmembrane synaptic proteins like Neurexin and Rabphilin. Gephyrin binds GABA receptors and links them to Tubulins 23,24. Catenins can bind GRIP1 and also link to the cytoskeleton via their interaction with Actin 32–34. Actin, Band 4.1 Proteins, Spectrins and Vimentin are known to associate directly 35–39 and Vimentin anchors the position of a number of organelles in the cytosol 40. Perhaps along with the Tubulins it plays a similar role for the ribbon. Vimentin has also been shown to bind to a SNARE protein SNAP23 41.
The complex interactions, localizations and activities of the proteins are regulated in part by kinases and phosphatases. Most synaptic constituents have putative phosphorylation sites, and for many proteins these sites are characterized 42,43. We found CaM Kinase II, RAPGEF4 (EPAC2), PPP2R1A and PGAM5 representing 10.6% of the protein, suggesting tight regulation of the ribbon function. CaM Kinase II (Ca-dependent phosphorylation enzyme) regulates Liprin 44 and appears to be located within the structure of the synaptic ribbon 7. We observed 3 isoforms (α, β, and δ) of CaM Kinase II, with equimolar amounts of α and β and twice as much of the δ isoform. EPAC2 is expressed in photoreceptors45; it is known to be involved in cAMP mediated vesicle release 46 and is a binding partner of Piccolo 17 and Rim 47. Piccolo also binds L-TYPE Ca Channels17 and an Actin-Binding Protein 48 providing a possible link between members in the presynaptic complex. Phosphoglyceraldehyde mutase (PGAM5) has no mutase activity, but instead serves as a serine-threonine phosphorylation enzyme 49. The proper folding and chaperoning of these proteins require some of the many chaperones in the ribbon precipitates: a number of heat shock proteins are listed in Table 1. In the category “other” we have put the S-Arrestin, SCRIN1, Glutamine Synthase and TRIM21 proteins. S-Arrestin participates in agonist-mediated desensitization of G-Protein-Coupled Receptors 50. Glutamine Synthase converts glutamate to glutamine. It was identified in a previous ribbon proteomics analysis 7. TRIM21 is a cytosolic IgG receptor and part of the intracellular antibody-mediated proteolysis pathway 51. We suspect it is a contamination/component in the anti-CtBP2 Ab reagent.
Of the 102 proteins that were excluded from the quantitative analysis above, 24 were present exclusively in the CtBP2/Ribeye immunoprecipitates and had at least 2 unique peptide fragments. These are listed in Table 3, and include a number of proteins known to be associated with synaptic vesicles, including Clathrin, NSF, Rabphilin, Synapsin, Synaptogyrin, and Synaptotagmin.
The synaptic ribbon complex, which consists of the ribbon core and portions of the presynaptic density is primarily composed of proteins associated with vesicle release at the cytomatrix active zone of conventional synapses, suggesting that the molecular mechanisms of vesicle handling and release here may be fundamentally conventional. We have quantified the relative amounts of many proteins that were previously identified in the ribbon through smaller scale proteomics analysis or through candidate analyses (i.e. immunohistochemistry or immunoblotting) 6–8,14, including CtBP isoforms, ERC 1 and 2 (i.e. CAST and ELKS), Bassoon, and Piccolo, CaM Kinase II, Spectrin, Vimentin, Tubulin, NA/K ATPase. Uthaiah and Hudspeth recently investigated the composition of presynaptic proteins isolated by differential and density-gradient centrifugation and affinity-purified CtBP2-containing complexes 7. The set of proteins isolated with anti-CtBP2 antibodies is partially overlapping with ours. In their work involving CtBP2/Ribeye pull-down they used chicken cochlea material to precipitate with mouse CtBP2 (or Ribeye B-domain) antibody and bovine retina to which they applied rabbit polyclonal Ab raised against the chicken Ribeye’s A domain. We view the study as a complementary to ours in which we used murine or bovine retinal material to precipitate with mouse anti CtBP2 (or Ribeye B-domain) monoclonal Ab. In addition our use of IgG as a control/nonreactive antibody allowed us to eliminate some proteins as not being necessarily associated with the ribbon. Our protocols also varied from those of Uthaiah and Hudspeth in our use of a relatively harsh sonication step, which we suspect may have broken filamentous structures linking the vesicles to the ribbon. As a result of that or simply low interaction strength with the ribbon many exocytosis proteins outlined in their paper were not identified in our analysis, suggesting they may be present in minor quantities or are primarily associated with vesicles.
Our analysis provides a quantitative assessment of the molecular composition of the synaptic ribbon complex based on an exponentially modified protein abundance index 10. More precise measurements of the absolute amount of particular proteins in the mix are possible, but require knowing the sample’s components of interest and the use of high-cost corresponding isotope-labeled peptides as internal standards in methods such as AQUA or QconCAT. We have specified amounts of proteins as molar ratios. Mass ratios for these proteins will differ depending on the molecular weight, and this becomes very different for proteins at the extremes of the spectrum. For this particular mix of proteins, a 71 kDa molecule will have equal molar and mass ratios. Bassoon and Piccolo, at 500 kDa were present collectively in a relatively small molar quantity (1.45%), but high mass ratio (10.5%) while PGAM5, with a molecular weight of 32 kDa, had a high molar ratio (2.82%) but a low mass ratio (1.2%).
The CtBP isoforms, CtBP2 (short), CtBP2 (Ribeye) and CtBP1 together constituted 30% of the total protein content on a molar basis, though our quantities may be exaggerated by the fact that these proteins are primary targets for the pull-down. Given that CtBP is primarily in the ribbon itself and not concentrated in the presynaptic density 6–8,52 it is likely that CtBP isoforms comprise an even greater percentage of the protein in the ribbon itself, corroborating Zenisek’s deduction that it could be as high as 2/3 of the protein in the ribbon proper 53. Quantification of the amounts of the CtBP isoforms in the ribbon is made difficult by their presence in the nucleus. While we know that Ribeye and CtBP1 are localized in the ribbon (Figure 5 and 7) it hard to confirm/exclude the presence of CtBP2 with immunohistochemistry, for CtBP2 is essentially the B-domain of Ribeye. However judged from our sequencing results where CtBP2 and Ribeye are separated by size in two different gel slices but give rise to shared B-domain peptides, we conclude that the amount of CtBP2 is as much as 1/3 of CtBP isoforms and about equal to Ribeye. Our purification procedure begins with cell lysis followed by nuclear fraction separation by differential centrifugation. Judged from the number of nuclear proteins co-purified with the ribbon, we think some of CtBP2 is of nuclear origin, but the relatively large quantities of CtBP2 we collected implies that CtBP2 in the ribbon as well.
The quantitative analysis of relative molar amounts of protein in the ribbon complex allows us to begin to contemplate the contributions of the major proteins to function. The high molar ratios of CtBP and ERC isoforms indicate prominent roles in ribbon function. CtBP protein family members play a critical role in membrane fission at several intracellular transport steps, including transfer from the Golgi to the epithelial basolateral membrane, fluid-phase endocytosis, macropinocytosis, and retrograde transport of receptors to the endoplasmic reticulum 54–56. Polymorphic tubular membrane enclosures operate between Golgi complex and the basolateral plasma membrane in epithelial cells. They protrude out of the Golgi complex along microtubules and then detach and migrate to the plasma membrane 57. When CtBP1 is inactivated (by dominant-negative mutants, blocking Abs or siRNA), these protruding tubules are unable to pinch off the Golgi complex 54. When Golgi transport is blocked by dominant-negative CtBP1 mutant, the necks of the buds accumulate the mutant protein 58. Injection of active recombinant CtBP1 leads to rapid formation of Golgi vesicles almost bypassing the formation of tubular intermediates. The intimate involvement of CtBP1 in vesicle fission in the Golgi suggests that CtBP1 may be acting similarly in the ribbon (see below).
ERC proteins are key to vesicle fusion in other vesicle release systems such as the pancreas 12. ERC1 binds Bassoon, Piccolo and Rim2 12 ERC1 is involved in intracellular membrane trafficking 59,60 and is important for the insulin secretion in the pancreatic B cells. It is located in the docking site of the insulin granules and the fusion events of the granules occurs mostly on ERC1 enriched membrane clusters. Overexpression of ERC1 increases exocytosis while knockdown of ERC1 decreases it 61. Some of our knowledge of ERC proteins also comes from analogous structures and proteins in invertebrates. The neuromuscular junction in Drosophila possesses a structure called T-bar, which appears to be an analog of the ribbon 62. It is known to contain an ERC2 homolog called Bruchpilot, large deletions in which lead to the disintegration of the T-bar 62–65. With smaller deletions the T-bar has normal size but is devoid of filament-tethered vesicles 66. These analogies become all the more significant considering that ERC proteins constitute over 6% of the ribbon. A functional link between vesicular proteins and the cytoskeleton can be found in the recently described filaments linking vesicles into discrete clusters at the presynaptic zone of brain neurons. The molecular composition of these filaments is still being investigated. ERC2 immunoreactivity, as well as that of Bassoon is associated with the filaments adjacent to the membrane 67,68. Bassoon, Piccolo, ERC and Bruchpilot proteins contain contiguous coiled-coil sequences 63,69–71 reminiscent of tethering factors in Golgi and endoplasmic reticulum 72 The coils typically form rod-like structures, believed to establish contacts between membranes at a distance, thereby increasing the specificity or efficiency of the initial attachment/tethering of vesicles 72. The presence of high proportion of Tubulins and Actins (together contributing close to 17%) suggests a role in vesicle movement, though we do not know how closely they are associated with the ribbon itself.
Our finding of a significant quantity of enzymes (both phosphorylation and metabolic) and chaperone (heat shock) proteins indicates dynamic control of the ribbon. CaM Kinase II performs calcium-dependent phosphorylation to alter protein function while PGAM5 is a serine/threonine phosphatase that dephosphorylates proteins. Phosphorylation enzymes might regulate the availability of vesicles based on changes in intracellular calcium. Because the ribbon is intimately associated with calcium channels that control vesicle release, it is reasonable to suggest that CaM Kinase is activated under conditions that evoke vesicle release or depletion, and then perhaps plays a role in refilling the vesicle docking sites on the ribbon.
Conclusions
Based on our findings of the ribbon composition and known functions of ribbon complex proteins in other systems, we suggest that the synaptic ribbon may function to create, organize, release, and perhaps standardize the size of vesicles at the synaptic ribbon. CtBP may initiate vesicle fission to create vesicles near the ribbon from tubulo-vesicular structures endocytosed around the ribbon. In Bassoon knockout mice 6, the ribbon detaches from the synapse, leaving endocytic membrane retrieval normal, but an accumulation of tubular and cisternal membrane structures occurs at the presynaptic densities. Many membranous structures are also accumulated around the floating ribbons and they have vesicle-like appearance when located very close to the ribbon, and cisternal appearance when located away from the ribbon. These membrane structures are similar to those observed at synapses following strong stimulation 73,74. So perhaps CtBP proteins, via fission, parse these endocytic structures into new synaptic vesicles, which then remain associated with the ribbon and repopulate the readily-releasable pool. Additional support for this hypothesis comes from electron tomography on frog hair cells to recreate the fine structure the ribbon synapses 74. Prolonged maximal excitation of the synapse depleted 70% of the vesicles in its close vicinity, and led to the appearance of intracellular cisterns and membrane invaginations accounting for the most part of vesicular membrane loss. The authors conclude that formation of new synaptic vesicles from the observed cisterns and membrane invaginations is the rate-limiting step in the vesicle recycling, suggesting that even when functioning normally, the ribbon may not always be able to keep up with abnormally high rates of transmitter release and endocytosis.
Once vesicles are created, vesicle-handling proteins such as the ERC1 and ERC2, Bassoon, and Piccolo may organize them around the ribbon. Cytoskeletal proteins like Actin and Tubulin may organize the structure or participate in moving the vesicles about the surface of the ribbon. The presence of a large quantity of phosphorylation proteins may indicate a relatively active process, changing it performance characteristics with varying demand. In summary, proteomics analysis of the ribbon complex suggests this structure is a highly organized and concentrated cluster of proteins dealing with vesicle creation, retention and distribution and consequent exocytosis (this idea is illustrated in Figure 6). The relatively high quantities of CTBP and ERC isoforms suggest a significant role for those proteins in the ribbon function. Given observations in other systems that these proteins play roles in vesicle fission and handling, we hypothesize a similar function for these proteins in the synaptic ribbon.
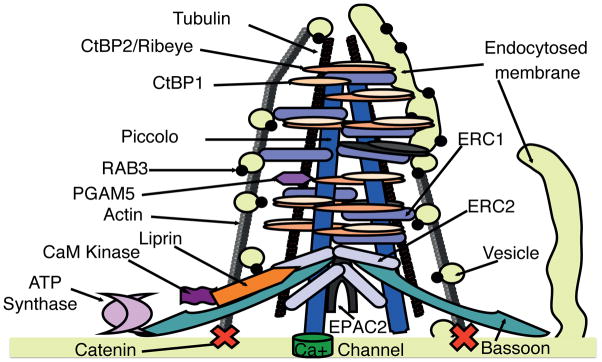
Figure 6. Synaptic ribbon composition and function. We have depicted the ribbon based in large part on known interactions and localizations of its protein constituents.<.
br>Binding of Liprin and ERC1 and Erc216, Liprin and Cam KInase II44, Ribeye and Bassoon13, CtBP1 and Bassoon13, ERC1 and Piccolo15, ERC1 and Bassoon15 have been described. Piccolo binds Actin-Binding Protein1, which in turn binds Actin, while actin also binds Catenin33,48. We have placed ATP Synthase and PGAM5 speculatively in our proposed model. The right hand portion of the figure illustrates the hypothesis that CtBP isoforms create vesicles by fission of endocytosed membranous cisterns.
Supplementary Material
Column A describes the protein group. Column B list all unique peptide identified in the mass spectrometry run for that protein group. Columns C to G contain the names of all individual proteins in a given group and provides information if unique peptides from column B could be potentially observed in them. A value of 0 indicates that the peptide can not be generated from the tryptic digest of the protein, while any value greater than 0 indicates that the particular peptide could be observed in this protein. A value of 1 means that the peptide could be theoretically observed in only one protein in the group. A value of 0.5 indicates that the peptide could be theoretically observed in two proteins within the group, while value of 0.33 indicates the peptide could be theoretically observed in three proteins within the group, while a value of 0.25 means the peptide is theoretically shared by four proteins within the group. At the bottom of columns C to G for each section separated by black line are summed the values for all unique peptides potentially assignable to a given protein.
Acknowledgments
The work was supported by the NIDCD (DC767 and DC9651). We appreciate the contributions of J. Evans of the U. Mass proteomics facility. We thank R. A. Eatock, M. C. Brown, and M. C. Liberman for comments on earlier versions of the manuscript.
Footnotes
Supporting Information available: This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Glowatzki E, Grant L, Fuchs P. Hair cell afferent synapses. Current opinion in neurobiology. 2008;18:389–95. doi: 10.1016/j.conb.2008.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moser T, Brandt A, Lysakowski A. Hair cell ribbon synapses. Cell and tissue research. 2006;326:347–59. doi: 10.1007/s00441-006-0276-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sterling P, Matthews G. Structure and function of ribbon synapses. Trends in neurosciences. 2005;28:20–9. doi: 10.1016/j.tins.2004.11.009. [DOI] [PubMed] [Google Scholar]
- 4.LoGiudice L, Matthews G. The role of ribbons at sensory synapses. Neuroscientist. 2009;15:380–91. doi: 10.1177/1073858408331373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buran BN, Strenzke N, Neef A, Gundelfinger ED, Moser T, Liberman MC. Onset coding is degraded in auditory nerve fibers from mutant mice lacking synaptic ribbons. J Neurosci. 2010;30:7587–97. doi: 10.1523/JNEUROSCI.0389-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Khimich D, Nouvian R, Pujol R, Tom Dieck S, Egner A, Gundelfinger ED, Moser T. Hair cell synaptic ribbons are essential for synchronous auditory signalling. Nature. 2005;434:889–94. doi: 10.1038/nature03418. [DOI] [PubMed] [Google Scholar]
- 7.Uthaiah RC, Hudspeth AJ. Molecular anatomy of the hair cell’s ribbon synapse. J Neurosci. 2010;30:12387–99. doi: 10.1523/JNEUROSCI.1014-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schmitz F, Bechmann M, Drenckhahn D. Purification of synaptic ribbons, structural components of the photoreceptor active zone complex. J Neurosci. 1996;16:7109–16. doi: 10.1523/JNEUROSCI.16-22-07109.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schmitz F, Konigstorfer A, Sudhof TC. RIBEYE, a component of synaptic ribbons: a protein’s journey through evolution provides insight into synaptic ribbon function. Neuron. 2000;28:857–72. doi: 10.1016/s0896-6273(00)00159-8. [DOI] [PubMed] [Google Scholar]
- 10.Ishihama Y, Oda Y, Tabata T, Sato T, Nagasu T, Rappsilber J, Mann M. Exponentially modified protein abundance index (emPAI) for estimation of absolute protein amount in proteomics by the number of sequenced peptides per protein. Mol Cell Proteomics. 2005;4:1265–72. doi: 10.1074/mcp.M500061-MCP200. [DOI] [PubMed] [Google Scholar]
- 11.Kantardzhieva A, Alexeeva S, Versteeg I, Wijnholds J. MPP3 is recruited to the MPP5 protein scaffold at the retinal outer limiting membrane. FEBS J. 2006;273:1152–65. doi: 10.1111/j.1742-4658.2006.05140.x. [DOI] [PubMed] [Google Scholar]
- 12.Ohara-Imaizumi M, Ohtsuka T, Matsushima S, Akimoto Y, Nishiwaki C, Nakamichi Y, Kikuta T, Nagai S, Kawakami H, Watanabe T, Nagamatsu S. ELKS, a protein structurally related to the active zone-associated protein CAST, is expressed in pancreatic beta cells and functions in insulin exocytosis: interaction of ELKS with exocytotic machinery analyzed by total internal reflection fluorescence microscopy. Mol Biol Cell. 2005;16:3289–300. doi: 10.1091/mbc.E04-09-0816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.tom Dieck S, Altrock WD, Kessels MM, Qualmann B, Regus H, Brauner D, Fejtova A, Bracko O, Gundelfinger ED, Brandstatter JH. Molecular dissection of the photoreceptor ribbon synapse: physical interaction of Bassoon and RIBEYE is essential for the assembly of the ribbon complex. J Cell Biol. 2005;168:825–36. doi: 10.1083/jcb.200408157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deguchi-Tawarada M, Inoue E, Takao-Rikitsu E, Inoue M, Ohtsuka T, Takai Y. CAST2: identification and characterization of a protein structurally related to the presynaptic cytomatrix protein CAST. Genes Cells. 2004;9:15–23. doi: 10.1111/j.1356-9597.2004.00697.x. [DOI] [PubMed] [Google Scholar]
- 15.Takao-Rikitsu E, Mochida S, Inoue E, Deguchi-Tawarada M, Inoue M, Ohtsuka T, Takai Y. Physical and functional interaction of the active zone proteins, CAST, RIM1, and Bassoon, in neurotransmitter release. J Cell Biol. 2004;164:301–11. doi: 10.1083/jcb.200307101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ko J, Na M, Kim S, Lee JR, Kim E. Interaction of the ERC family of RIM-binding proteins with the liprin-alpha family of multidomain proteins. J Biol Chem. 2003;278:42377–85. doi: 10.1074/jbc.M307561200. [DOI] [PubMed] [Google Scholar]
- 17.Shibasaki T, Sunaga Y, Fujimoto K, Kashima Y, Seino S. Interaction of ATP sensor, cAMP sensor, Ca2+ sensor, and voltage-dependent Ca2+ channel in insulin granule exocytosis. J Biol Chem. 2004;279:7956–61. doi: 10.1074/jbc.M309068200. [DOI] [PubMed] [Google Scholar]
- 18.Hibino H, Pironkova R, Onwumere O, Vologodskaia M, Hudspeth AJ, Lesage F. RIM binding proteins (RBPs) couple Rab3-interacting molecules (RIMs) to voltage-gated Ca(2+) channels. Neuron. 2002;34:411–23. doi: 10.1016/s0896-6273(02)00667-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sigel E, Baur R, Boulineau N, Minier F. Impact of subunit positioning on GABAA receptor function. Biochem Soc Trans. 2006;34:868–71. doi: 10.1042/BST0340868. [DOI] [PubMed] [Google Scholar]
- 20.Hong S, Pedersen PL. ATP synthase and the actions of inhibitors utilized to study its roles in human health, disease, and other scientific areas. Microbiol Mol Biol Rev. 2008;72:590–641. doi: 10.1128/MMBR.00016-08. Table of Contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abrahams JP, Leslie AG, Lutter R, Walker JE. Structure at 2. 8 A resolution of F1-ATPase from bovine heart mitochondria. Nature. 1994;370:621–8. doi: 10.1038/370621a0. [DOI] [PubMed] [Google Scholar]
- 22.Collinson IR, Runswick MJ, Buchanan SK, Fearnley IM, Skehel JM, van Raaij MJ, Griffiths DE, Walker JE. Fo membrane domain of ATP synthase from bovine heart mitochondria: purification, subunit composition, and reconstitution with F1-ATPase. Biochemistry. 1994;33:7971–8. doi: 10.1021/bi00191a026. [DOI] [PubMed] [Google Scholar]
- 23.Saiepour L, Fuchs C, Patrizi A, Sassoe-Pognetto M, Harvey RJ, Harvey K. Complex role of collybistin and gephyrin in GABAA receptor clustering. J Biol Chem. 2010;285:29623–31. doi: 10.1074/jbc.M110.121368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Prior P, Schmitt B, Grenningloh G, Pribilla I, Multhaup G, Beyreuther K, Maulet Y, Werner P, Langosch D, Kirsch J, et al. Primary structure and alternative splice variants of gephyrin, a putative glycine receptor-tubulin linker protein. Neuron. 1992;8:1161–70. doi: 10.1016/0896-6273(92)90136-2. [DOI] [PubMed] [Google Scholar]
- 25.Sassoe-Pognetto M, Kirsch J, Grunert U, Greferath U, Fritschy JM, Mohler H, Betz H, Wassle H. Colocalization of gephyrin and GABAA-receptor subunits in the rat retina. J Comp Neurol. 1995;357:1–14. doi: 10.1002/cne.903570102. [DOI] [PubMed] [Google Scholar]
- 26.Greferath U, Grunert U, Fritschy JM, Stephenson A, Mohler H, Wassle H. GABAA receptor subunits have differential distributions in the rat retina: in situ hybridization and immunohistochemistry. J Comp Neurol. 1995;353:553–71. doi: 10.1002/cne.903530407. [DOI] [PubMed] [Google Scholar]
- 27.Yue L, Xie A, Bruzik KS, Frolund B, Qian H, Pepperberg DR. Potentiating action of propofol at GABAA receptors of retinal bipolar cells. Invest Ophthalmol Vis Sci. 2011;52:2497–509. doi: 10.1167/iovs.10-5991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ostergaard J, Hannibal J, Fahrenkrug J. Synaptic contact between melanopsin-containing retinal ganglion cells and rod bipolar cells. Invest Ophthalmol Vis Sci. 2007;48:3812–20. doi: 10.1167/iovs.06-1322. [DOI] [PubMed] [Google Scholar]
- 29.Lo SC, Hannink M. PGAM5, a Bcl-XL-interacting protein, is a novel substrate for the redox-regulated Keap1-dependent ubiquitin ligase complex. J Biol Chem. 2006;281:37893–903. doi: 10.1074/jbc.M606539200. [DOI] [PubMed] [Google Scholar]
- 30.Lo SC, Hannink M. PGAM5 tethers a ternary complex containing Keap1 and Nrf2 to mitochondria. Exp Cell Res. 2008;314:1789–803. doi: 10.1016/j.yexcr.2008.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Imai Y, Kanao T, Sawada T, Kobayashi Y, Moriwaki Y, Ishida Y, Takeda K, Ichijo H, Lu B, Takahashi R. The loss of PGAM5 suppresses the mitochondrial degeneration caused by inactivation of PINK1 in Drosophila. PLoS Genet. 2010;6:e1001229. doi: 10.1371/journal.pgen.1001229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li H, Kim JH, Koh SS, Stallcup MR. Synergistic effects of coactivators GRIP1 and beta-catenin on gene activation: cross-talk between androgen receptor and Wnt signaling pathways. J Biol Chem. 2004;279:4212–20. doi: 10.1074/jbc.M311374200. [DOI] [PubMed] [Google Scholar]
- 33.Rimm DL, Koslov ER, Kebriaei P, Cianci CD, Morrow JS. Alpha 1(E)-catenin is an actin-binding and -bundling protein mediating the attachment of F-actin to the membrane adhesion complex. Proc Natl Acad Sci U S A. 1995;92:8813–7. doi: 10.1073/pnas.92.19.8813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ozawa M, Ringwald M, Kemler R. Uvomorulin-catenin complex formation is regulated by a specific domain in the cytoplasmic region of the cell adhesion molecule. Proc Natl Acad Sci U S A. 1990;87:4246–50. doi: 10.1073/pnas.87.11.4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gerke V, Weber K. Identity of p36K phosphorylated upon Rous sarcoma virus transformation with a protein purified from brush borders; calcium-dependent binding to non-erythroid spectrin and F-actin. EMBO J. 1984;3:227–33. doi: 10.1002/j.1460-2075.1984.tb01789.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Burridge K, Kelly T, Mangeat P. Nonerythrocyte spectrins: actin-membrane attachment proteins occurring in many cell types. J Cell Biol. 1982;95:478–86. doi: 10.1083/jcb.95.2.478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Takeshita K, MacDonald RI, MacDonald RC. Band 4. 1 enhances spectrin binding to phosphatidylserine vesicles. Biochem Biophys Res Commun. 1993;191:165–71. doi: 10.1006/bbrc.1993.1198. [DOI] [PubMed] [Google Scholar]
- 38.Tyler JM, Hargreaves WR, Branton D. Purification of two spectrin-binding proteins: biochemical and electron microscopic evidence for site-specific reassociation between spectrin and bands 2.1 and 4. 1. Proc Natl Acad Sci U S A. 1979;76:5192–6. doi: 10.1073/pnas.76.10.5192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Correia I, Chu D, Chou YH, Goldman RD, Matsudaira P. Integrating the actin and vimentin cytoskeletons. adhesion-dependent formation of fimbrin-vimentin complexes in macrophages. J Cell Biol. 1999;146:831–42. doi: 10.1083/jcb.146.4.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Katsumoto T, Mitsushima A, Kurimura T. The role of the vimentin intermediate filaments in rat 3Y1 cells elucidated by immunoelectron microscopy and computer-graphic reconstruction. Biol Cell. 1990;68:139–46. doi: 10.1016/0248-4900(90)90299-i. [DOI] [PubMed] [Google Scholar]
- 41.Faigle W, Colucci-Guyon E, Louvard D, Amigorena S, Galli T. Vimentin filaments in fibroblasts are a reservoir for SNAP23: a component of the membrane fusion machinery. Mol Biol Cell. 2000;11:3485–94. doi: 10.1091/mbc.11.10.3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lonart G, Schoch S, Kaeser PS, Larkin CJ, Sudhof TC, Linden DJ. Phosphorylation of RIM1alpha by PKA triggers presynaptic long-term potentiation at cerebellar parallel fiber synapses. Cell. 2003;115:49–60. doi: 10.1016/s0092-8674(03)00727-x. [DOI] [PubMed] [Google Scholar]
- 43.Greengard P, Valtorta F, Czernik AJ, Benfenati F. Synaptic vesicle phosphoproteins and regulation of synaptic function. Science. 1993;259:780–5. doi: 10.1126/science.8430330. [DOI] [PubMed] [Google Scholar]
- 44.Hoogenraad CC, Feliu-Mojer MI, Spangler SA, Milstein AD, Dunah AW, Hung AY, Sheng M. Liprinalpha1 degradation by calcium/calmodulin-dependent protein kinase II regulates LAR receptor tyrosine phosphatase distribution and dendrite development. Dev Cell. 2007;12:587–602. doi: 10.1016/j.devcel.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 45.Whitaker CM, Cooper NG. Differential distribution of exchange proteins directly activated by cyclic AMP within the adult rat retina. Neuroscience. 2010;165:955–67. doi: 10.1016/j.neuroscience.2009.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sedej S, Rose T, Rupnik M. cAMP increases Ca2+-dependent exocytosis through both PKA and Epac2 in mouse melanotrophs from pituitary tissue slices. J Physiol. 2005;567:799–813. doi: 10.1113/jphysiol.2005.090381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ozaki N, Shibasaki T, Kashima Y, Miki T, Takahashi K, Ueno H, Sunaga Y, Yano H, Matsuura Y, Iwanaga T, Takai Y, Seino S. cAMP-GEFII is a direct target of cAMP in regulated exocytosis. Nat Cell Biol. 2000;2:805–11. doi: 10.1038/35041046. [DOI] [PubMed] [Google Scholar]
- 48.Fenster SD, Kessels MM, Qualmann B, Chung WJ, Nash J, Gundelfinger ED, Garner CC. Interactions between Piccolo and the actin/dynamin-binding protein Abp1 link vesicle endocytosis to presynaptic active zones. J Biol Chem. 2003;278:20268–77. doi: 10.1074/jbc.M210792200. [DOI] [PubMed] [Google Scholar]
- 49.Takeda K, Komuro Y, Hayakawa T, Oguchi H, Ishida Y, Murakami S, Noguchi T, Kinoshita H, Sekine Y, Iemura S, Natsume T, Ichijo H. Mitochondrial phosphoglycerate mutase 5 uses alternate catalytic activity as a protein serine/threonine phosphatase to activate ASK1. Proc Natl Acad Sci U S A. 2009;106:12301–5. doi: 10.1073/pnas.0901823106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Palczewski K, McDowell JH, Jakes S, Ingebritsen TS, Hargrave PA. Regulation of rhodopsin dephosphorylation by arrestin. J Biol Chem. 1989;264:15770–3. [PubMed] [Google Scholar]
- 51.Mallery DL, McEwan WA, Bidgood SR, Towers GJ, Johnson CM, James LC. Antibodies mediate intracellular immunity through tripartite motif-containing 21 (TRIM21) Proc Natl Acad Sci U S A. 2010;107:19985–90. doi: 10.1073/pnas.1014074107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dick O, tom Dieck S, Altrock WD, Ammermuller J, Weiler R, Garner CC, Gundelfinger ED, Brandstatter JH. The presynaptic active zone protein bassoon is essential for photoreceptor ribbon synapse formation in the retina. Neuron. 2003;37:775–86. doi: 10.1016/s0896-6273(03)00086-2. [DOI] [PubMed] [Google Scholar]
- 53.Zenisek D, Horst NK, Merrifield C, Sterling P, Matthews G. Visualizing synaptic ribbons in the living cell. J Neurosci. 2004;24:9752–9. doi: 10.1523/JNEUROSCI.2886-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bonazzi M, Spano S, Turacchio G, Cericola C, Valente C, Colanzi A, Kweon HS, Hsu VW, Polishchuck EV, Polishchuck RS, Sallese M, Pulvirenti T, Corda D, Luini A. CtBP3/BARS drives membrane fission in dynamin-independent transport pathways. Nat Cell Biol. 2005;7:570–80. doi: 10.1038/ncb1260. [DOI] [PubMed] [Google Scholar]
- 55.Corda D, Colanzi A, Luini A. The multiple activities of CtBP/BARS proteins: the Golgi view. Trends Cell Biol. 2006;16:167–73. doi: 10.1016/j.tcb.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 56.Haga Y, Miwa N, Jahangeer S, Okada T, Nakamura S. CtBP1/BARS is an activator of phospholipase D1 necessary for agonist-induced macropinocytosis. EMBO J. 2009;28:1197–207. doi: 10.1038/emboj.2009.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Polishchuk EV, Di Pentima A, Luini A, Polishchuk RS. Mechanism of constitutive export from the golgi: bulk flow via the formation, protrusion, and en bloc cleavage of large trans-golgi network tubular domains. Mol Biol Cell. 2003;14:4470–85. doi: 10.1091/mbc.E03-01-0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yang JS, Lee SY, Spano S, Gad H, Zhang L, Nie Z, Bonazzi M, Corda D, Luini A, Hsu VW. A role for BARS at the fission step of COPI vesicle formation from Golgi membrane. EMBO J. 2005;24:4133–43. doi: 10.1038/sj.emboj.7600873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Monier S, Jollivet F, Janoueix-Lerosey I, Johannes L, Goud B. Characterization of novel Rab6-interacting proteins involved in endosome-to-TGN transport. Traffic. 2002;3:289–97. doi: 10.1034/j.1600-0854.2002.030406.x. [DOI] [PubMed] [Google Scholar]
- 60.Wang Y, Liu X, Biederer T, Sudhof TC. A family of RIM-binding proteins regulated by alternative splicing: Implications for the genesis of synaptic active zones. Proc Natl Acad Sci U S A. 2002;99:14464–9. doi: 10.1073/pnas.182532999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nomura H, Ohtsuka T, Tadokoro S, Tanaka M, Hirashima N. Involvement of ELKS, an active zone protein, in exocytotic release from RBL-2H3 cells. Cell Immunol. 2009;258:204–11. doi: 10.1016/j.cellimm.2009.05.005. [DOI] [PubMed] [Google Scholar]
- 62.Fouquet W, Owald D, Wichmann C, Mertel S, Depner H, Dyba M, Hallermann S, Kittel RJ, Eimer S, Sigrist SJ. Maturation of active zone assembly by Drosophila Bruchpilot. J Cell Biol. 2009;186:129–45. doi: 10.1083/jcb.200812150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wagh DA, Rasse TM, Asan E, Hofbauer A, Schwenkert I, Durrbeck H, Buchner S, Dabauvalle MC, Schmidt M, Qin G, Wichmann C, Kittel R, Sigrist SJ, Buchner E. Bruchpilot, a protein with homology to ELKS/CAST, is required for structural integrity and function of synaptic active zones in Drosophila. Neuron. 2006;49:833–44. doi: 10.1016/j.neuron.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 64.Kittel RJ, Wichmann C, Rasse TM, Fouquet W, Schmidt M, Schmid A, Wagh DA, Pawlu C, Kellner RR, Willig KI, Hell SW, Buchner E, Heckmann M, Sigrist SJ. Bruchpilot promotes active zone assembly, Ca2+ channel clustering, and vesicle release. Science. 2006;312:1051–4. doi: 10.1126/science.1126308. [DOI] [PubMed] [Google Scholar]
- 65.Allport JR, Donnelly LE, Kefalas P, Lo G, Nunn A, Yadollahi-Farsani M, Rendell NB, Murray S, Taylor GW, MacDermot J. A possible role for mono (ADP-ribosyl) transferase in the signalling pathway mediating neutrophil chemotaxis. Br J Clin Pharmacol. 1996;42:99–106. doi: 10.1046/j.1365-2125.1996.37014.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hallermann S, Kittel RJ, Wichmann C, Weyhersmuller A, Fouquet W, Mertel S, Owald D, Eimer S, Depner H, Schwarzel M, Sigrist SJ, Heckmann M. Naked dense bodies provoke depression. J Neurosci. 2010;30:14340–5. doi: 10.1523/JNEUROSCI.2495-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Siksou L, Rostaing P, Lechaire JP, Boudier T, Ohtsuka T, Fejtova A, Kao HT, Greengard P, Gundelfinger ED, Triller A, Marty S. Three-dimensional architecture of presynaptic terminal cytomatrix. J Neurosci. 2007;27:6868–77. doi: 10.1523/JNEUROSCI.1773-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fernandez-Busnadiego R, Zuber B, Maurer UE, Cyrklaff M, Baumeister W, Lucic V. Quantitative analysis of the native presynaptic cytomatrix by cryoelectron tomography. J Cell Biol. 2010;188:145–56. doi: 10.1083/jcb.200908082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lupashin V, Sztul E. Golgi tethering factors. Biochim Biophys Acta. 2005;1744:325–39. doi: 10.1016/j.bbamcr.2005.03.013. [DOI] [PubMed] [Google Scholar]
- 70.Wang X, Hu B, Zieba A, Neumann NG, Kasper-Sonnenberg M, Honsbein A, Hultqvist G, Conze T, Witt W, Limbach C, Geitmann M, Danielson H, Kolarow R, Niemann G, Lessmann V, Kilimann MW. A protein interaction node at the neurotransmitter release site: domains of Aczonin/Piccolo, Bassoon, CAST, and rim converge on the N-terminal domain of Munc13-1. J Neurosci. 2009;29:12584–96. doi: 10.1523/JNEUROSCI.1255-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fenster SD, Chung WJ, Zhai R, Cases-Langhoff C, Voss B, Garner AM, Kaempf U, Kindler S, Gundelfinger ED, Garner CC. Piccolo, a presynaptic zinc finger protein structurally related to bassoon. Neuron. 2000;25:203–14. doi: 10.1016/s0896-6273(00)80883-1. [DOI] [PubMed] [Google Scholar]
- 72.Guo W, Sacher M, Barrowman J, Ferro-Novick S, Novick P. Protein complexes in transport vesicle targeting. Trends Cell Biol. 2000;10:251–5. doi: 10.1016/s0962-8924(00)01754-2. [DOI] [PubMed] [Google Scholar]
- 73.Holt M, Cooke A, Wu MM, Lagnado L. Bulk membrane retrieval in the synaptic terminal of retinal bipolar cells. J Neurosci. 2003;23:1329–39. doi: 10.1523/JNEUROSCI.23-04-01329.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lenzi D, Crum J, Ellisman MH, Roberts WM. Depolarization redistributes synaptic membrane and creates a gradient of vesicles on the synaptic body at a ribbon synapse. Neuron. 2002;36:649–59. doi: 10.1016/s0896-6273(02)01025-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Column A describes the protein group. Column B list all unique peptide identified in the mass spectrometry run for that protein group. Columns C to G contain the names of all individual proteins in a given group and provides information if unique peptides from column B could be potentially observed in them. A value of 0 indicates that the peptide can not be generated from the tryptic digest of the protein, while any value greater than 0 indicates that the particular peptide could be observed in this protein. A value of 1 means that the peptide could be theoretically observed in only one protein in the group. A value of 0.5 indicates that the peptide could be theoretically observed in two proteins within the group, while value of 0.33 indicates the peptide could be theoretically observed in three proteins within the group, while a value of 0.25 means the peptide is theoretically shared by four proteins within the group. At the bottom of columns C to G for each section separated by black line are summed the values for all unique peptides potentially assignable to a given protein.