Abstract
Background
The association of rs16996148 single nucleotide polymorphism (SNP) in NCAN/CILP2/PBX4 and serum lipid levels is inconsistent. Furthermore, little is known about the association of rs16996148 SNP and serum lipid levels in the Chinese population. We therefore aimed to detect the association of rs16996148 SNP and several environmental factors with serum lipid levels in the Guangxi Mulao and Han populations.
Method
A total of 712 subjects of Mulao nationality and 736 participants of Han nationality were randomly selected from our stratified randomized cluster samples. Genotyping of the rs16996148 SNP was performed by polymerase chain reaction and restriction fragment length polymorphism combined with gel electrophoresis, and then confirmed by direct sequencing.
Results
The levels of apolipoprotein (Apo) B were higher in Mulao than in Han (P < 0.001). The frequencies of G and T alleles were 87.2% and 12.8% in Mulao, and 89.9% and 10.1% in Han (P <0.05); respectively. The frequencies of GG, GT and TT genotypes were 76.0%, 22.5% and 1.5% in Mulao, and 81.2%, 17.4% and 1.4% in Han (P <0.05); respectively. There were no significant differences in the genotypic and allelic frequencies between males and females in both ethnic groups. The levels of HDL-C, ApoAI, and the ratio of ApoAI to ApoB in Mulao were different between the GG and GT/TT genotypes in males but not in females (P < 0.01 for all), the subjects with GT/TT genotypes had higher serum levels of HDL-C, ApoAI, and the ratio of ApoAI to ApoB than the subjects with GG genotype. The levels of TC, TG, LDL-C, ApoAI, and ApoB in Han were different between the GG and GT/TT genotypes in males but not in females (P < 0.05-0.001), the T allele carriers had higher serum levels of TC, TG, LDL-C, ApoAI, and ApoB than the T allele noncarriers. The levels of HDL-C, ApoAI, and the ratio of ApoAI to ApoB in Mulao were correlated with the genotypes in males (P < 0.05-0.01) but not in females. The levels of TC, TG, HDL-C, LDL-C, ApoAI and ApoB in Han were associated with the genotypes in males (P < 0.05-0.001) but not in females. Serum lipid parameters were also correlated with several enviromental factors in both ethnic groups (P < 0.05-0.001).
Conclusions
The genotypic and allelic frequencies of rs16996148 SNP and the associations of the SNP and serum lipid levels are different in the Mulao and Han populations. Sex (male)-specific association of rs16996148 SNP in the NCAN/CILP2/PBX4 and serum lipid levels is also observed in the both ethnic groups.
Introduction
Coronary artery disease (CAD) is the leading cause of morbidity and mortality in industrialized countries, and the prevalence of this disease is increasing rapidly in developing countries [1]. Consistent and compelling evidence has demonstrated association between dyslipidemia and CAD incidence worldwide [2-4]. It is well-established that dyslipidemia is a complex trait caused by multiple environmental and genetic factors [5-7] and their interactions [8,9]. Family studies suggest that in many populations, about half of the variation in serum lipid profiles is genetically determined [10,11], and it is clear that serum lipid levels are strongly influenced by the genetic constitution of each individual.
Recent genome-wide association studies (GWAS) in different populations have identified more than 95 loci associated with serum lipid levels [5,12-46]. Common variants at these loci together explain < 10% of variation in each lipid trait [23,24,40]. Rare variants with large individual effects may also contribute to the heritability of lipid traits [40]. In addition, GWAS also discovered a number of novel loci that influence serum lipid phenotypes [15,17,21,39]. One of these newly identified single nucleotide polymorphisms (SNPs) is rs16996148 SNP in the NCAN/CILP2/PBX4. rs16996148 is located on chromosome 19p13 in an intergenic region between CILP2 and PBX4, which encodes a cartilage intermediate layer protein and a putative transcription factor expressed in testis, respectively [47,48]. Neurocan (NCAN) is a nervous system-specific proteoglycan involved in neuronal pattern formation, remodeling of neuronal networks and regulation of synaptic plasticity [49], with no obvious relation to LDL-C or TG concentrations. The roles of CILP2 and PBX4 in lipid metabolism are also unclear at this time [50,51]. In Europeans, however, rs16996148 in NCAN/CILP2/PBX4 has been showed significant associations with LDL-C and TG concentrations [15,17]. Tai et al. [51] also reported that rs16996148 SNP was significantly associated with LDL-C and HDL-C concentrations in the Malays under a recessive model of inheritance. Nevertheless, Nakayama et al. [50] did not observe significant associations between rs16996148 SNP and blood lipid profiles in the Japanese population. These results suggest that the ability of associations to generalize across other racial/ethnic populations varied greatly, some of these GWAS-identified variants may not be functional and are more likely to be in linkage disequilibrium with the functional variants.
China is a multiethnic country with 56 ethnic groups. Han nationality is the largest ethnic group, and Mulao nationality is one of the 55 minorities with population of 207,352 according to the fifth national census statistics of China in 2000. Ninety percent of them live in the Luocheng Mulao Autonomous County, Guangxi Zhuang Autonomous Region, People's Republic of China. The history of this minority can be traced back to the Jin Dynasty (AD 265-420). In a previous study, Xu et al. [52] showed that the genetic relationship between Mulao nationality and other minorities in Guangxi was much closer than that between Mulao and Han or Uighur nationality. To the best of our knowledge, however, the association of rs16996148 SNP and serum lipid levels has not been previously reported in the Chinese population. Therefore, the aim of the present study was to detect the association of rs16996148 SNP in the NCAN/CILP2/PBX4 and several environmental factors with serum lipid profiles in the Mulao and Han populations.
Materials and Methods
Participants
Participants in the present study included 712 individuals of Mulao nationality living in Luocheng Mulao Autonomous County, Guangxi Zhuang Autonomous Region, People's Republic of China. They were randomly selected from our previous stratified randomized cluster samples [53]. The ages of the participants ranged from 15 to 86 years, with an average age of 51.81 ± 14.76 years. There were 330 males (46.3%) and 382 females (53.7%). All participants were rural agricultural workers. During the same period, a total of 736 people of Han nationality who reside in the same villages were also randomly selected from our previous stratified randomized cluster samples. The average age of the subjects was 51.77 ± 14.96 years (range 15 to 86). There were 308 men (41.8%) and 428 women (58.2%). All of them were also rural agricultural workers. All study subjects were essentially healthy and had no evidence of any chronic illness, including hepatic, renal, or thyroid. The participants with a history of heart attack or myocardial infarction, stroke, congestive heart failure, diabetes or fasting blood glucose ≥ 7.0 mmol/L determined by glucose meter were excluded from the analyses. The participants were not taking medications known to affect serum lipid levels (lipid-lowering drugs such as statins or fibrates, beta-blockers, diuretics, or hormones). The experimental design was approved by the Ethics Committee of the First Affiliated Hospital, Guangxi Medical University. All participants in this study provided written informed consent.
Epidemiological survey
The survey was carried out using internationally standardized methods [54]. All participants underwent a complete history, physical examination, and laboratory assessment of cardiovascular risk factors, including cigarette smoking, family history of myocardial infarction, blood pressure, presence of diabetes mellitus. Information on demographics, socioeconomic status, and lifestyle factors was collected with standardized questionnaires. The alcohol information included questions about the number of liangs (about 50 g) of rice wine, corn wine, rum, beer, or liquor consumed during the preceding 12 months. Alcohol consumption was categorized into groups of grams of alcohol per day: ≤ 25 and > 25. Smoking status was categorized into groups of cigarettes per day: ≤ 20 and > 20. At the physical examination, several parameters were measured. Sitting blood pressure was measured three times with the use of a mercury sphygmomanometer after the subjects had a 5-minute rest, and the average of the three measurements was used for the level of blood pressure. Systolic blood pressure was determined by the first Korotkoff sound, and diastolic blood pressure by the fifth Korotkoff sound. Body weight, to the nearest 50 grams, was measured using a portable balance scale. Subjects were weighed without shoes and in a minimum of clothing. Height was measured, to the nearest 0.5 cm, using a portable steel measuring device. From these two measurements body mass index (BMI, kg/m2) was calculated.
Determination of serum lipid levels
Venous blood samples were collected after an overnight (at least 12 hours) fast. A part of the sample (2 mL) was collected into glass tubes and allowed to clot at room temperature, and used to determine serum lipid levels. Another part of the sample (3 mL) was transferred to tubes with anticoagulate solution (4.80 g/L citric acid, 14.70 g/L glucose, and 13.20 g/L tri-sodium citrate) and used to extract DNA. Serum TC, TG, HDL-C, and LDL-C levels in the samples were measured according to standard enzymatic methods. Serum ApoAI and ApoB levels were detected by the immunoturbidimetric immunoassay. All determinations were performed with an autoanalyzer (Type 7170A; Hitachi Ltd., Tokyo, Japan) in the Clinical Science Experiment Center of the First Affiliated Hospital, Guangxi Medical University [6,7].
DNA preparation and genotyping
Genomic DNA was isolated from peripheral blood leukocytes using the phenol-chloroform method [8,9]. The extracted DNA was stored at -80°C until analysis. Genotyping of the rs16996148 SNP was performed by polymerase chain reaction and restriction fragment length polymorphism (PCR-RFLP). PCR amplification was performed using 5'-CATCCAGCATTTAGAGGTGTGA-3' and 5'-CTAGGGCAAAGGAAGTGTTTC-3' (Sangon, Shanghai, People's Republic of China) as the forward and reverse primer pairs; respectively. Each amplification reaction was performed using 100 ng (2 μL) of genomic DNA in 25 μL of reaction mixture consisting of 1.0 μL of each primer (10 μmo1/L), 12.5 μL 2 × Taq PCRMasterMix (constituent: 0.1 U Taq polymerase/μL, 500 μM dNTP each, 20 mM Tris-HCl, pH 8.3, 100 mM KCl, 3 mM MgCl2, and stabilizers), and nuclease-free water 8.5 μL. After initial denaturizing at 95°C for 5 min, the reaction mixture was subjected to 33 cycles of denaturation at 95°C for 30 s, annealing at 60°C for 45 s and extension 1 min at 72°C, followed by a final 5 min extension at 72°C. After electrophoresis on a 2.0% agarose gel with 0.5 μg/mL ethidium bromide, the amplification products were visualized under ultraviolet light. Then 2.5 U of Hin1II restriction enzyme, 8 μL nuclease-free water and 1 μL of 10 × buffer solution were added directly to the PCR products (5 μL) and digested at 37°C overnight. After restriction enzyme digestion of the amplified DNA, genotypes were identified by electrophoresis on 2.5% agarose gel and visualized with ethidium-bromide staining ultraviolet illumination. Genotypes were scored by an experienced reader blinded to epidemiological data and serum lipid levels.
DNA sequencing
Six samples (GG, GT and TT genotypes in two; respectively) detected by the PCR-RFLP were also confirmed by direct sequencing. The PCR products were purified by low melting point gel electrophoresis and phenol extraction, and then the DNA sequences were analyzed in Shanghai Sangon Biological Engineering Technology & Services Co., Ltd., People's Republic of China.
Diagnostic criteria
The normal values of serum TC, TG, HDL-C, LDL-C, ApoAI, ApoB levels and the ratio of ApoAI to ApoB in our Clinical Science Experiment Center were 3.10-5.17, 0.56-1.70, 1.16-1.42, 2.70-3.10 mmol/L, 1.20-1.60, 0.80-1.05 g/L, and 1.00-2.50; respectively [53]. The individuals with TC > 5.17 mmol/L and/or TG > 1.70 mmol/L were defined as hyperlipidemic [6,7,53]. Hypertension was diagnosed according to the criteria of 1999 World Health Organization-International Society of Hypertension Guidelines for the management of hypertension [55,56]. The diagnostic criteria of overweight and obesity were according to the Cooperative Meta-analysis Group of China Obesity Task Force. Normal weight, overweight and obesity were defined as a BMI < 24, 24-28, and > 28 kg/m2; respectively [57].
Statistical analysis
The statistical analyses were done with the statistical software package SPSS 13.0 (SPSS Inc., Chicago, Illinois). Quantitative variables were expressed as mean ± standard deviation (serum TG levels were presented as medians and interquartile ranges). Qualitative variables were expressed as percentages. Allele frequency was determined via direct counting, and the standard goodness-of-fit test was used to test the Hardy-Weinberg equilibrium. Difference in genotype distribution between the groups was obtained using the chi-square test. The difference in general characteristics between two ethnic groups was tested by the Student's unpaired t-test. The association of genotypes and serum lipid parameters was tested by analysis of covariance (ANCOVA). Age, sex, BMI, blood pressure, alcohol consumption, and cigarette smoking were included in the statistical models as covariates. Multiple linear regression analyses adjusted for age, sex, BMI, blood pressure, alcohol consumption, and cigarette smoking were also performed to assess the association of serum lipid levels with genotypes (GG = 1, GT = 2, TT = 3; or GG = 1, GT/TT = 2) and several environment factors. A P value of less than 0.05 was considered statistically significant.
Results
General characteristics and serum lipid levels
Table 1 shows the general characteristics and serum lipid levels of the study population. The levels of ApoB and the percentages of subjects who consumed alcohol were higher but the levels of BMI and diastolic blood pressure were lower in Mulao than in Han (P < 0.05-0.001). There were no significant differences in the levels of age, body height, weight, systolic blood pressure, pulse pressure, blood glucose, TC, TG, HDL-C, LDL-C, ApoAI; the ratio of ApoAI to ApoB; the percentages of subjects who smoked cigarettes; and the ratio of male to female between the two ethnic groups (P > 0.05 for all).
Table 1.
The general characteristics and serum lipid levels in the Mulao and Han populations
| Parameter | Mulao | Han Chinese | t (χ2) | P |
|---|---|---|---|---|
| Number | 712 | 736 | - | - |
| Male/female | 330/382 | 308/428 | 2.974 | 0.090 |
| Age (years) | 51.81 ± 14.76 | 51.77 ± 14.96 | 0.058 | 0.954 |
| Height (cm) | 155.44 ± 7.72 | 154.71 ± 7.93 | 1.774 | 0.076 |
| Weight (kg) | 52.88 ± 9.03 | 53.72 ± 8.85 | -1.803 | 0.072 |
| Body mass index (kg/m2) | 21.83 ± 3.00 | 22.43 ± 3.32 | -3.600 | 0.000 |
| Systolic blood pressure (mmHg) | 129.16 ± 21.47 | 129.58 ± 18.67 | -0.397 | 0.692 |
| Diastolic blood pressure (mmHg) | 80.90 ± 11.60 | 82.27 ± 11.05 | -2.298 | 0.022 |
| Pulse pressure (mmHg) | 48.25 ± 16.07 | 47.30 ± 14.19 | 1.187 | 0.235 |
| Blood glucose | 5.96 ± 1.54 | 5.96 ± 1.61 | -0.096 | 0.923 |
| Cigarette smoking [n (%)] | ||||
| Nonsmoker | 519 (72.9) | 532 (72.3) | ||
| ≤ 20 cigarettes/day | 159 (22.3) | 178 (24.2) | ||
| > 20 cigarettes/day | 34 (5.8) | 26 (3.5) | 1.901 | 0.386 |
| Alcohol consumption [n (%)] | ||||
| Nondrinker | 516 (72.5) | 565 (76.8) | ||
| ≤ 25 g/day | 67 (9.4) | 79 (10.7) | ||
| > 25 g/day | 129 (18.1) | 92 (12.5) | 9.007 | 0.011 |
| Total cholesterol (mmol/L) | 5.02 ± 1.29 | 5.02 ± 1.03 | 0.040 | 0.968 |
| Triglyceride (mmol/L) | 1.05 (0.81) | 1.07 (0.91) | -0.562 | 0.574 |
| HDL-C (mmol/L) | 1.75 ± 0.46 | 1.75 ± 0.53 | -0.171 | 0.864 |
| LDL-C (mmol/L) | 2.92 ± 0.88 | 2.91 ± 0.85 | 0.337 | 0.736 |
| Apolipoprotein (Apo) AI (g/L) | 1.33 ± 0.40 | 1.35 ± 0.26 | -1.456 | 0.146 |
| ApoB (g/L) | 0.97 ± 0.54 | 0.86 ± 0.20 | 5.202 | 0.000 |
| ApoAI/ApoB | 1.61 ± 0.95 | 1.66 ± 0.51 | -1.409 | 0.159 |
HDL-C, high-density lipoprotein cholesterol; LDL-C, low-density lipoprotein cholesterol. The value of Triglyceride was presented as median (interquartile range), the difference between the two ethnic groups was determined by the Wilcoxon-Mann-Whitney test.
Electrophoresis and genotypes
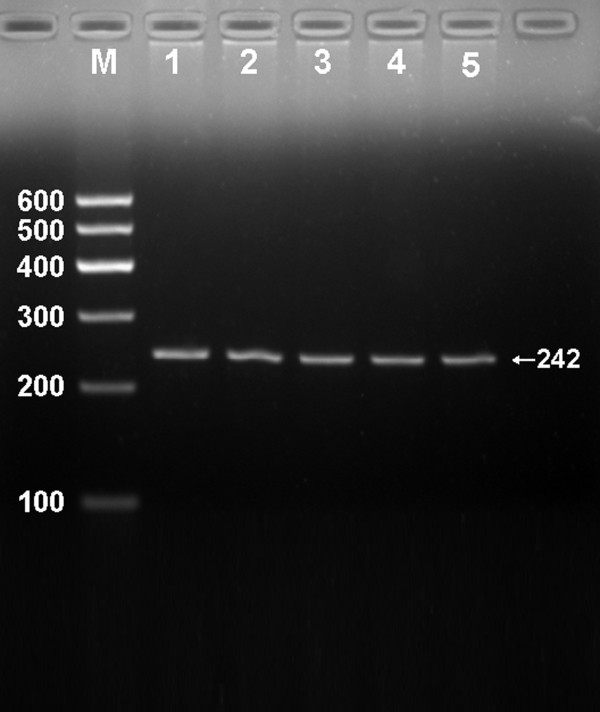
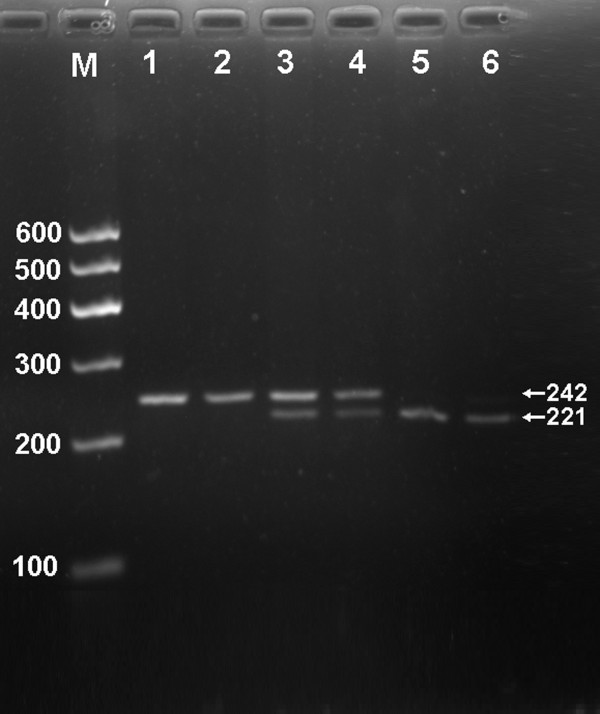
After the genomic DNA of the samples was amplified by PCR and imaged by 2.0% agarose gel electrophoresis, the PCR products of 242 bp nucleotide sequences could be seen in the samples (Figure 1). The genotypes identified were named according to the presence (T allele) or absence (G allele) of the enzyme restriction sites. Thus, GG genotype is heterozygote for the absence of the site (bands at 242 bp), GT genotype is heterozygote for the absence and presence of the site (bands at 242-, 221- and 21-bp), and TT genotype is homozygote for the presence of the site (bands at 221- and 21- bp; Figure 2). The 21 bp fragments were invisible in the gel owing to its fast migration speed. The genotype distribution of rs16996148 SNP followed the Hardy-Weinberg equilibrium.
Figure 1.
Electrophoresis of PCR products of the samples. Lane M, 100 bp marker ladder; lanes 1-5, samples. The 242 bp bands are the target genes.
Figure 2.
Genotyping of rs16996148 polymorphism in the NCAN gene. Lane M, 100 bp marker ladder; lanes 1 and 2, GG genotype (242 bp); lanes 3 and 4, GT genotype (242-, 221- and 21-bp); and lanes 5 and 6, TT genotype (221- and 21-bp). The 21 bp fragments were invisible in the gel owing to its fast migration speed.
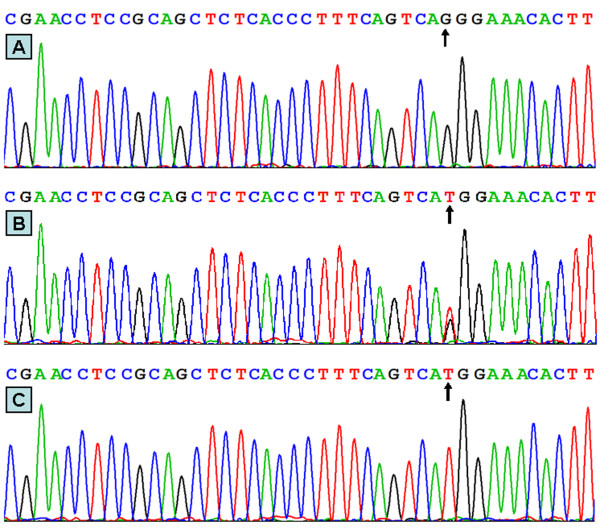
Nucleotide sequences
The results were shown as GG, GT and TT genotypes of the rs16996148 SNP by PCR-RFLP, the genotypes were also confirmed by sequencing (Figure 3); respectively.
Figure 3.
A part of the nucleotide sequence of rs16996148 SNP. (A) GG genotype; (B) GT genotype; (C) TT genotype.
Genotypic and allelic frequencies
The genotypic and allelic frequencies of rs16996148 SNP in the both ethnic groups are shown in Table 2. The G and T allele frequencies of rs16996148 SNP were 87.2% and 12.8% in Mulao, and 89.9% and 10.1% in Han (P <0.05); respectively. The frequencies of GG, GT and TT genotypes were 76.0%, 22.5% and 1.5% in Mulao, and 81.2%, 17.4% and 1.4% in Han (P <0.05); respectively. There were no significant differences in the genotypic and allelic frequencies between males and females in both groups.
Table 2.
The genotypic and allelic frequencies of rs16996148 SNP the Mulao and Han populations [n (%)]
| Group | n | Genotype | Allele | |||
|---|---|---|---|---|---|---|
| GG | GT | TT | G | T | ||
| Mulao | 712 | 541 (76.0) | 160 (22.5) | 11 (1.5) | 1242 (87.2) | 182 (12.8) |
| Han | 736 | 598 (81.2) | 128 (17.4) | 10 (1.4) | 1324 (89.9) | 148 (10.1) |
| χ2 | - | 6.060 | 5.329 | |||
| P | - | 0.048 | 0.022 | |||
| Mulao | ||||||
| Male | 330 | 243 (73.6) | 79 (24.0) | 8 (2.4) | 565 (85.6) | 95 (14.4) |
| Female | 382 | 298 (78.0) | 81 (21.2) | 3 (0.8) | 677 (88.6) | 87 (11.4) |
| χ2 | - | 4.113 | 2.871 | |||
| P | - | 0.128 | 0.095 | |||
| Han | ||||||
| Male | 308 | 246 (79.9) | 60 (19.5) | 2 (0.6) | 552 (89.6) | 64 (10.4) |
| Female | 428 | 352 (82.2) | 68 (15.9) | 8 (1.9) | 772 (90.2) | 84 (9.8) |
| χ2 | - | 3.415 | 0.132 | |||
| P | - | 0.181 | 0.726 | |||
Genotypes and serum lipid levels
As shown in Table 3, the levels of HDL-C in Mulao were different among the three genotypes (P < 0.05), the T allele carriers had higher serum HDL-C levels than the T allele noncarriers. When serum lipid parameters in Mulao were analyzed according to sex, we found that the levels of HDL-C, ApoAI, and the ratio of ApoAI to ApoB in males but not in females were different between the GG and GT/TT genotypes (P < 0.01 for all), the subjects with GT/TT genotypes had higher serum levels of HDL-C, ApoAI, and the ratio of ApoAI to ApoB than the subjects with GG genotype.
Table 3.
The genotypes of rs16996148 SNP and serum lipid levels in the Mulao and Han populations
| Genotype | n | TC (mmol/L) |
TG (mmol/L) |
HDL-C (mmol/L) |
LDL-C (mmol/L) |
ApoAI (g/L) |
ApoB (g/L) |
ApoAI/ ApoB |
|---|---|---|---|---|---|---|---|---|
| Mulao | ||||||||
| GG | 541 | 5.03 ± 1.35 | 1.07(0.84) | 1.72 ± 0.44 | 2.94 ± 0.88 | 1.31 ± 0.40 | 0.98 ± 0.56 | 1.59 ± 1.01 |
| GT | 160 | 4.99 ± 1.11 | 1.02(0.73) | 1.83 ± 0.50 | 2.88 ± 0.91 | 1.37 ± 0.41 | 0.94 ± 0.49 | 1.68 ± 0.73 |
| TT | 11 | 4.91 ± 0.85 | 1.18(0.89) | 1.89 ± 0.20 | 2.78 ± 0.74 | 1.32 ± 0.47 | 0.93 ± 0.46 | 1.54 ± 0.62 |
| F | - | 0.108 | 1.338 | 3.910 | 0.284 | 1.593 | 0.564 | 0.685 |
| P | - | 0.897 | 0.512 | 0.020 | 0.752 | 0.204 | 0.569 | 0.505 |
| GG | 541 | 5.03 ± 1.35 | 1.07(0.84) | 1.72 ± 0.44 | 2.94 ± 0.88 | 1.31 ± 0.40 | 0.98 ± 0.56 | 1.59 ± 1.01 |
| GT/TT | 171 | 4.99 ± 1.09 | 1.03(0.74) | 1.83 ± 0.49 | 2.87 ± 0.90 | 1.37 ± 0.41 | 0.94 ± 0.49 | 1.67 ± 0.73 |
| F | - | 0.045 | -1.154 | 7.483 | 0.398 | 2.981 | 1.028 | 1.304 |
| P | - | 0.832 | 0.248 | 0.006 | 0.528 | 0.085 | 0.311 | 0.254 |
| Han | ||||||||
| GG | 598 | 4.95 ± 0,98 | 1.07(0.86) | 1.74 ± 0.55 | 2.86 ± 0.81 | 1.35 ± 0.26 | 0.85 ± 0.20 | 1.67 ± 0.52 |
| GT | 128 | 5.28 ± 1.16 | 1.15(1.23) | 1.76 ± 0.44 | 3.13 ± 0.94 | 1.37 ± 0.23 | 0.90 ± 0.21 | 1.59 ± 0.43 |
| TT | 10 | 5.40 ± 1.38 | 0.84(0.49) | 2.17 ± 0.41 | 3.05 ± 1.26 | 1.62 ± 0.33 | 0.83 ± 0.26 | 2.15 ± 0.79 |
| F | 7.467 | 6.323 | 2.473 | 5.504 | 5.845 | 2.909 | 1.940 | |
| P | 0.001 | 0.042 | 0.085 | 0.004 | 0.003 | 0.055 | 0.145 | |
| GG | 598 | 4.95 ± 0.98 | 1.07(0.86) | 1.74 ± 0.55 | 2.86 ± 0.81 | 1.35 ± 0.26 | 0.85 ± 0.20 | 1.67 ± 0.52 |
| GT/TT | 138 | 5.29 ± 1.17 | 1.15(1.18) | 1.79 ± 0.45 | 3.12 ± 0.96 | 1.38 ± 0.25 | 0.89 ± 0.21 | 1.63 ± 0.49 |
| F | - | 12.684 | -1.555 | 1.389 | 10.222 | 4.955 | 5.469 | 0.091 |
| P | - | 0.000 | 0.120 | 0.239 | 0.001 | 0.026 | 0.020 | 0.763 |
| Mulao/male | ||||||||
| GG | 243 | 5.18 ± 1.50 | 1.15(1.07) | 1.72 ± 0.45 | 2.96 ± 0.86 | 1.32 ± 0.44 | 1.04 ± 0.62 | 1.48 ± 0.65 |
| GT/TT | 87 | 5.08 ± 1.64 | 1.08(0.86) | 1.86 ± 0.58 | 2.84 ± 0.86 | 1.44 ± 0.40 | 0.98 ± 0.52 | 1.70 ± 0.77 |
| F | - | 0.276 | -0.341 | 7.151 | 1.375 | 7.126 | 0.574 | 9.195 |
| P | - | 0.600 | 0.733 | 0.008 | 0.245 | 0.008 | 0.449 | 0.003 |
| Mulao/female | ||||||||
| GG | 298 | 4.91 ± 1.21 | 1.03(0.65) | 1.72 ± 0.44 | 2.92 ± 0.89 | 1.31 ± 0.37 | 0.93 ± 0.50 | 1.68 ± 1.22 |
| GT/TT | 84 | 4.89 ± 1.01 | 0.95(0.60) | 1.80 ± 0.38 | 2.90 ± 0.95 | 1.29 ± 0.41 | 0.89 ± 0.46 | 1.63 ± 0.68 |
| F | - | 0.110 | -1.648 | 1.134 | 0.181 | 0.104 | 0.106 | 0.163 |
| P | - | 0.740 | 0.099 | 0.288 | 0.671 | 0.747 | 0.745 | 0.686 |
| Han/male | ||||||||
| GG | 246 | 5.08 ± 0.90 | 1.14(0.92) | 1.69 ± 0.42 | 2.90 ± 0.77 | 1.36 ± 0.28 | 0.90 ± 0.19 | 1.58 ± 0.47 |
| GT/TT | 62 | 5.59 ± 1.12 | 1.71(1.61) | 1.75 ± 0.45 | 3.26 ± 0.78 | 1.44 ± 0.28 | 0.97 ± 0.23 | 1.56 ± 0.46 |
| F | - | 13.935 | -2.649 | 2.569 | 8.390 | 7.843 | 4.675 | 0.248 |
| P | - | 0.000 | 0.008 | 0.110 | 0.004 | 0.005 | 0.031 | 0.619 |
| Han/female | ||||||||
| GG | 352 | 4.86 ± 1.02 | 1.00(0.81) | 1.78 ± 0.62 | 2.83 ± 0.84 | 1.33 ± 0.24 | 0.81 ± 0.20 | 1.74 ± 0.54 |
| GT/TT | 76 | 5.05 ± 1.16 | 0.93(0.80) | 1.81 ± 0.46 | 3.01 ± 1.08 | 1.34 ± 0.21 | 0.84 ± 0.18 | 1.69 ± 0.50 |
| F | - | 3.262 | -0.389 | 0.133 | 3.680 | 0.141 | 1.239 | 0.548 |
| P | - | 0.072 | 0.698 | 0.715 | 0.056 | 0.707 | 0.266 | 0.459 |
TC, total cholesterol; TG, triglyceride; HDL-C, high-density lipoprotein cholesterol; LDL-C, low-density lipoprotein cholesterol; ApoAI, apolipoprotein AI; ApoB, apolipoprotein B
In the Han population, the levels of TC, TG, LDL-C and ApoAI among the three genotypes, or the levels of TC, LDL-C, ApoAI, and ApoB between the GG and GT/TT genotypes were different (P < 0.05-0.001), the T allele carriers had higher serum TC, TG, LDL-C, ApoAI and ApoB levels than the T allele noncarriers. When serum lipid parameters in Han were stratified according to sex, we showed that the levels of TC, TG, LDL-C, ApoAI, and ApoB in males but not in females were different between the GG and GT/TT genotypes (P < 0.05-0.001), the subjects with GT/TT genotypes had higher serum levels of TC, TG, LDL-C, ApoAI, and ApoB than the subjects with GG genotype.
Risk factors for serum lipid parameters
The correlation between the genotypes of rs16996148 SNP and serum lipid parameters in Mulao and Han is shown in Table 4. The levels of HDL-C, ApoAI, and the ratio of ApoAI to ApoB in Mulao were correlated with the genotypes in males (P < 0.05-0.01) but not in females. The levels of TC, TG, HDL-C, LDL-C, ApoAI and ApoB in Han were associated with the genotypes in males (P < 0.05-0.001) but not in females.
Table 4.
Correlation between the genotypes of rs16996148 SNP and serum lipid parameters in the Mulao and Han populations
| Lipid parameter | Relative factor | Unstandardized coefficient | Std. error | Standardized coefficient | t | P |
|---|---|---|---|---|---|---|
| Mulao plus Han | ||||||
| TC | Genetype | 0.137 | 0.065 | 0.053 | 2.106 | 0.035 |
| HDL-C | Genetype | 0.082 | 0.028 | 0.075 | 2.924 | 0.004 |
| ApoAI | Genetype | 0.049 | 0.019 | 0.066 | 2.577 | 0.010 |
| Mulao | ||||||
| HDL-C | Genetype | 0.091 | 0.035 | 0.094 | 2.609 | 0.009 |
| Han | ||||||
| TC | Genetype | 0.322 | 0.082 | 0.136 | 3.934 | 0.000 |
| TG | Genetype | 0.335 | 0.161 | 0.074 | 2.082 | 0.038 |
| LDL-C | Genetype | 0.234 | 0.068 | 0.119 | 3.424 | 0.001 |
| ApoAI | Genetype | 0.057 | 0.020 | 0.096 | 2.802 | 0.005 |
| ApoB | Genetype | 0.038 | 0.015 | 0.081 | 2.482 | 0.013 |
| Mulao/male | ||||||
| HDL-C | Genetype | 0.133 | 0.050 | 0.137 | 2.647 | 0.009 |
| ApoAI | Genetype | 0.109 | 0.045 | 0.128 | 2.419 | 0.016 |
| ApoAI/ApoB | Genetype | 0.192 | 0.072 | 0.141 | 2.676 | 0.008 |
| Han/male | ||||||
| TC | Genetype | 0.513 | 0.122 | 0.223 | 4.203 | 0.000 |
| TG | Genetype | 0.860 | 0.345 | 0.139 | 2.493 | 0.013 |
| HDL-C | Genetype | 0.114 | 0.053 | 0.113 | 2.159 | 0.032 |
| LDL-C | Genetype | 0.327 | 0.101 | 0.176 | 3.241 | 0.001 |
| ApoAI | Genetype | 0.107 | 0.033 | 0.159 | 3.268 | 0.001 |
| ApoB | Genetype | 0.058 | 0.024 | 0.121 | 2.388 | 0.018 |
Serum lipid parameters were also correlated with several environment factors such as age, gender, alcohol consumption, cigarette smoking, blood pressure, blood glucose, and BMI in both ethnic groups (P < 0.05-0.001; Table 5).
Table 5.
Correlation between environmental factors and serum lipid parameters in the Mulao and Han populations
| Lipid parameter | Relative factor | Unstandardized coefficient | Std. error | Standardized coefficient | t | P |
|---|---|---|---|---|---|---|
| Mulao plus Han | ||||||
| TC | Body mass index | 0.053 | 0.010 | 0.145 | 5.530 | 0.000 |
| Age | 0.012 | 0.002 | 0.155 | 5.960 | 0.000 | |
| Alcohol consumption | 0.177 | 0.040 | 0.113 | 4.379 | 0.000 | |
| Diastolic blood pressure | 0.006 | 0.003 | 0.054 | 2.004 | 0.045 | |
| TG | Body mass index | 0.107 | 0.016 | 0.170 | 6.582 | 0.000 |
| Gender | -0.309 | 0.127 | -0.077 | -2.445 | 0.015 | |
| Alcohol consumption | 0.214 | 0.085 | 0.079 | 2.509 | 0.012 | |
| Blood glucose | 0.080 | 0.032 | 0.063 | 2.464 | 0.014 | |
| HDL-C | Body mass index | -0.030 | 0.004 | -0.191 | -7.412 | 0.000 |
| Alcohol consumption | 0.105 | 0.021 | 0.157 | 4.981 | 0.000 | |
| Gender | 0.105 | 0.031 | 0.105 | 3.348 | 0.001 | |
| Age | 0.002 | 0.001 | 0.058 | 2.261 | 0.024 | |
| LDL-C | Body mass index | 0.051 | 0.007 | 0.186 | 7.334 | 0.000 |
| Age | 0.010 | 0.002 | 0.174 | 6.673 | 0.000 | |
| Blood glucose | 0.030 | 0.014 | 0.054 | 2.082 | 0.038 | |
| ApoAI | Alcohol consumption | 0.131 | 0.014 | 0.287 | 9.143 | 0.000 |
| Body mass index | -0.010 | 0.003 | -0.090 | -3.483 | 0.001 | |
| Age | 0.002 | 0.001 | 0.073 | 2.863 | 0.004 | |
| Gender | 0.061 | 0.021 | 0.089 | 2.850 | 0.004 | |
| Nation | -0.045 | 0.017 | -0.066 | -2.586 | 0.010 | |
| ApoB | Body mass index | 0.021 | 0.003 | 0.165 | 6.425 | 0.000 |
| Nation | 0.121 | 0.021 | 0.147 | 5.768 | 0.000 | |
| Blood glucose | 0.024 | 0.007 | 0.092 | 3.611 | 0.000 | |
| Gender | -0.075 | 0.021 | -0.091 | -3.537 | 0.000 | |
| Pulse pressure | 0.001 | 0.001 | 0.054 | 2.109 | 0.035 | |
| ApoAI/ApoB | Body mass index | -0.047 | 0.006 | -0.197 | -7.634 | 0.000 |
| Blood glucose | -0.036 | 0.013 | -0.076 | -2.904 | 0.004 | |
| Gender | 0.207 | 0.048 | 0.135 | 4.320 | 0.000 | |
| Alcohol consumption | 0.118 | 0.032 | 0.115 | 3.663 | 0.000 | |
| Age | -0.003 | 0.001 | -0.066 | -2.526 | 0.012 | |
| Nation | -0.087 | 0.039 | -0.058 | -2.247 | 0.025 | |
| Mulao | ||||||
| TC | Body mass index | 0.066 | 0.016 | 0.153 | 4.163 | 0.000 |
| Age | 0.011 | 0.003 | 0.126 | 3.455 | 0.001 | |
| Alcohol consumption | 0.179 | 0.061 | 0.108 | 2.953 | 0.003 | |
| TG | Body mass index | 0.126 | 0.025 | 0.186 | 5.088 | 0.000 |
| Alcohol consumption | 0.407 | 0.095 | 0.156 | 4.275 | 0.000 | |
| HDL-C | Body mass index | -0.037 | 0.006 | -0.244 | -6.727 | 0.000 |
| Alcohol consumption | 0.066 | 0.021 | 0.113 | 3.115 | 0.002 | |
| LDL-C | Body mass index | 0.054 | 0.011 | 0.182 | 4.997 | 0.000 |
| Age | 0.009 | 0.002 | 0.146 | 3.998 | 0.000 | |
| ApoAI | Alcohol consumption | 0.095 | 0.019 | 0.184 | 5.009 | 0.000 |
| Age | 0.002 | 0.001 | 0.088 | 2.393 | 0.017 | |
| ApoB | Body mass index | 0.026 | 0.007 | 0.142 | 3.830 | 0.000 |
| Gender | -0.087 | 0.040 | -0.080 | -2.16 | 0.031 | |
| ApoAI/ApoB | Body mass index | -0.041 | 0.012 | -0.129 | -3.468 | 0.001 |
| Blood glucose | -0.052 | 0.023 | -0.084 | -2.260 | 0.024 | |
| Han Chinese | ||||||
| TC | Diastolic blood pressure | 0.014 | 0.003 | 0.147 | 3.981 | 0.000 |
| Age | 0.012 | 0.002 | 0.180 | 5.005 | 0.000 | |
| Body mass index | 0.041 | 0.011 | 0.133 | 3.688 | 0.000 | |
| Alcohol consumption | 0.188 | 0.052 | 0.127 | 3.610 | 0.000 | |
| TG | Body mass index | 0.082 | 0.022 | 0.138 | 3.744 | 0.000 |
| Blood glucose | 0.151 | 0.043 | 0.125 | 3.500 | 0.000 | |
| Gender | -0.474 | 0.144 | -0.119 | -3.301 | 0.001 | |
| Diastolic blood pressure | 0.017 | 0.006 | 0.094 | 2.560 | 0.011 | |
| HDL-C | Body mass index | -0.022 | 0.006 | -0.141 | -3.811 | 0.000 |
| Alcohol consumption | 0.110 | 0.034 | 0.144 | 3.250 | 0.001 | |
| Gender | 0.228 | 0.054 | 0.213 | 4.196 | 0.000 | |
| Cigarette smoking | 0.122 | 0.046 | 0.123 | 2.640 | 0.008 | |
| LDL-C | Age | 0.010 | 0.002 | 0.185 | 4.946 | 0.000 |
| Body mass index | 0.047 | 0.009 | 0.182 | 5.052 | 0.000 | |
| Blood glucose | 0.050 | 0.019 | 0.094 | 2.624 | 0.009 | |
| Diastolic blood pressure | 0.006 | 0.003 | 0.074 | 1.992 | 0.047 | |
| ApoAI | Alcohol consumption | 0.129 | 0.016 | 0.345 | 8.253 | 0.000 |
| Body mass index | -0.014 | 0.003 | -0.175 | -4.997 | 0.000 | |
| Cigarette smoking | 0.088 | 0.021 | 0.182 | 4.119 | 0.000 | |
| Gender | 0.099 | 0.025 | 0.189 | 3.942 | 0.000 | |
| ApoB | Body mass index | 0.018 | 0.002 | 0.300 | 8.877 | 0.000 |
| Blood glucose | 0.023 | 0.004 | 0.182 | 5.433 | 0.000 | |
| Gender | -0.046 | 0.016 | -0.112 | -2.819 | 0.005 | |
| Systolic blood pressure | 0.001 | 0.000 | 0.093 | 2.446 | 0.015 | |
| Age | 0.001 | 0.001 | 0.104 | 2.802 | 0.005 | |
| Alcohol consumption | 0.026 | 0.012 | 0.089 | 2.230 | 0.026 | |
| ApoAI/ApoB | Body mass index | -0.050 | 0.005 | -0.323 | -9.364 | 0.000 |
| Age | -0.004 | 0.001 | -0.112 | -3.234 | 0.001 | |
| Blood glucose | -0.026 | 0.011 | -0.082 | -2.347 | 0.019 | |
| Gender | 0.273 | 0.049 | 0.264 | 5.575 | 0.000 | |
| Cigarette smoking | 0.154 | 0.042 | 0.161 | 3.683 | 0.000 | |
| Alcohol consumption | 0.098 | 0.031 | 0.133 | 3.225 | 0.001 |
TC, total cholesterol; TG, triglyceride; HDL-C, high-density lipoprotein cholesterol; LDL-C, low-density lipoprotein cholesterol; ApoAI, apolipoprotein AI; ApoB, apolipoprotein B
Discussion
The present study shows that serum ApoB levels were higher in Mulao than in Han nationalities. There were no significant differences in the remaining serum lipid parameters between the two ethnic groups. It is well known that dyslipidemia is the result of a combination of genetic and environmental factors [6-9]. Both family and twin studies suggest that in many populations, about 40-60% of the variation in serum lipid profiles is genetically determined [10,11], and it is clear that LDL-C, HDL-C and TG concentrations are strongly influenced by the genetic constitution of each individual. The engagements of Mulao nationality were family-arranged in childhood, usually with the girl being four or five years older than the boy. There was a preference for marriage to mother's brother's daughter. Engagement and marriage were marked by bride-wealth payments. Marriage ceremonies were held when the girl reached puberty. She remained with her natal family until her first child was born. Till then she was free to join the young men and women who came together for responsive singing, flirtations, and courtships at festival times. Divorce and remarriage were permitted, with little restriction. The two-generation household is the most common unit of residence. Households are under the control of the father, and divide when the sons marry, with only the youngest son remaining with the parents. Therefore, we believe that the genetic background and some lipid-associated genetic variants in this population may be different from those in Han nationality.
The genotypic and allelic frequencies of rs16996148 SNP in the NCAN/CILP2/PBX4 in diverse racial/ethnic groups are inconsistent. The frequency of T allele was 8% in European Americans, 15% in African Americans, 4% in American Indians, 6% in Mexican Americans and Hispanics [40], and 12% in Japanese [50]. The minor allele frequency in the Malay population was 17% [51]. In the present study, we showed that the T allele frequency of rs16996148 SNP was higher in Mulao than in Han (12.8% vs. 10.1%, P <0.05). The frequencies of GG, GT and TT genotypes were also different between the two ethnic groups (P <0.05). There were no significant differences in the genotypic and allelic frequencies between males and females in both ethnic groups. These results indicate that the prevalence of the T allele variant of the rs16996148 SNP may have a racial/ethnic specificity.
The potential relationship between the rs16996148 SNP and plasma or serum lipid levels in humans has been evaluated in several previous studies (GWAS). However, previous findings on the association of this SNP with the changes in plasma lipid levels are inconsistent. The rs16996148 SNP in NCAN/CILP2/PBX4 has been shown significant associations with LDL-C and TG concentrations in Europeans [15,17]. The minor allele (T allele) of rs16996148 SNP was associated with lower concentrations of both LDL-C (by ~16 mg/dl) and TG [15]. Tai et al. [51] also reported that rs16996148 SNP was significantly associated with lower LDL-C and elevated HDL-C concentrations in the Malays under a recessive model of inheritance [51]. Nevertheless, Nakayama et al. [50] did not observe significant associations between rs16996148 and blood lipid profiles in the Japanese population. In the current study, we found that the levels of HDL-C, ApoAI, and the ratio of ApoAI to ApoB in Mulao were different between the GG and GT/TT genotypes in males but not in females, the subjects with GT/TT genotypes had higher serum levels of HDL-C, ApoAI, and the ratio of ApoAI to ApoB than the subjects with GG genotype. The levels of TC, TG, LDL-C, ApoAI, and ApoB in Han were different between the GG and GT/TT genotypes in males but not in females, the T allele carriers had higher serum levels of TC, TG, LDL-C, ApoAI, and ApoB than the T allele noncarriers. The levels of HDL-C, ApoAI, and the ratio of ApoAI to ApoB in Mulao were correlated with the genotypes in males but not in females. The levels of TC, TG, HDL-C, LDL-C, ApoAI and ApoB in Han were correlated with the genotypes in males but not in females. These findings suggest that there is a sex-specific association of rs16996148 SNP in the NCAN/CILP2/PBX4 and serum lipid levels in our study populations.
It is well known that environmental factors such as dietary patterns, lifestyle, obesity, physical activity, and hypertension are all strongly related with serum lipid levels [6,7]. Furthermore, exposure to different lifestyles and environments in our populations resident in Guangxi may further modify the effect of genetic variation on blood lipids. In the present study, we also showed that serum lipid parameters were correlated with age, sex, alcohol consumption, cigarette smoking, BMI, and blood pressure in both ethnic groups. These data suggest that the environmental factors also play an important role in determining serum lipid levels in our populations. Although rice and corn are the staple foods in both ethnic groups, the people of Mulao nationality like to eat cold foods along with acidic and spicy dishes, so bean soy sauce and pickled vegetables are among their most popular dishes. They also like to eat animal offals which contain abundant saturated fatty acid. The effects of dietary macronutrients on serum lipid levels and their effects on CAD have been extensively studied [58-62]. Almost 40 y ago, the Puerto Rican Heart Study found lower mean concentrations of TC and TG in Puerto Ricans than in subjects in the Framingham Heart Study [63]. Among urban Puerto Rican men, TC was positively associated with the percentage of energy from total fat, saturated fatty acids (SFAs), simple sugars, and protein and negatively associated with the percentage of energy from polyunsaturated fatty acids (PUFAs), total carbohydrate, and PUFA/SFA. Overall, diet and relative weight can account for at most 6% of the variability in serum cholesterol observed, with at most 2.5% of the variability due diet alone [63].
Conclusion
The present study shows that the T allele frequency of rs16996148 SNP in the NCAN/CILP2/PBX4 is significantly higher in Mulao than in Han. The frequencies of GG, GT and TT genotypes are also different between the two ethnic groups. The subjects with GT/TT genotypes in Mulao had higher serum levels of HDL-C, ApoAI, and the ratio of ApoAI to ApoB than the subjects with GG genotype in males but not in females. The T allele carriers in Han had higher serum levels of TC, TG, LDL-C, ApoAI, and ApoB than the T allele noncarriers in males but not in females. These lipid parameters were also correlated with the genotypes in males but not in females. These results suggest that there is a sex-specific association of rs16996148 SNP in the NCAN/CILP2/PBX4 and serum lipid levels in our study populations.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
TTY participated in the design, undertook genotyping, and helped to draft the manuscript. RXY conceived the study, participated in the design, carried out the epidemiological survey, collected the samples, and drafted the manuscript. QL, PH, XNZ, KKH, LHHA and DFW carried out the epidemiological survey and collaborated to the genotyping. CWL and SLP carried out the epidemiological survey and collected the samples. All authors read and approved the final manuscript.
Contributor Information
Ting-Ting Yan, Email: yan083199@163.com.
Rui-Xing Yin, Email: yinruixing@yahoo.com.cn.
Qing Li, Email: liqing000116dsl@163.com.
Ping Huang, Email: huangping914@126.com.
Xiao-Na Zeng, Email: zengxiaona241@163.com.
Ke-Ke Huang, Email: 38872286@qq.com.
Lynn Htet Htet Aung, Email: lifeinnivana@gmail.com.
Dong-Feng Wu, Email: wulove26@tom.com.
Cheng-Wu Liu, Email: gxmulchw@126.com.
Shang-Ling Pan, Email: bitterlypan@yahoo.com.cn.
Acknowledgements
This study was supported by the National Natural Science Foundation of China (No: 30960130).
References
- Ezzati M, Lopez AD, Rodgers A, Vander Hoorn S, Murray CJ. Selected major risk factors and global and regional burden of disease. Lancet. 2002;360:1347–60. doi: 10.1016/S0140-6736(02)11403-6. [DOI] [PubMed] [Google Scholar]
- Law MR, Wald NJ, Rudnicka AR. Quantifying effect of statins on low density lipoprotein cholesterol, ischaemic heart disease, and stroke: systematic review and meta-analysis. BMJ. 2003;326:1423. doi: 10.1136/bmj.326.7404.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuulasmaa K, Tunstall-Pedoe H, Dobson A, Fortmann S, Sans S, Tolonen H, Evans A, Ferrario M, Tuomilehto J. Estimation of contribution of changes in classic risk factors to trends in coronary-event rates across the WHO MONICA Project populations. Lancet. 2000;355:675–87. doi: 10.1016/s0140-6736(99)11180-2. [DOI] [PubMed] [Google Scholar]
- Clarke R, Emberson JR, Parish S, Palmer A, Shipley M, Linksted P, Sherliker P, Clark S, Armitage J, Fletcher A, Collins R. Cholesterol fractions and apolipoproteins as risk factors for heart disease mortality in older men. Arch Intern Med. 2007;167:1373–8. doi: 10.1001/archinte.167.13.1373. [DOI] [PubMed] [Google Scholar]
- Teslovich TM, Musunuru K, Smith AV, Edmondson AC, Stylianou IM, Koseki M, Pirruccello JP, Ripatti S, Chasman DI, Willer CJ, Johansen CT, Fouchier SW, Isaacs A, Peloso GM, Barbalic M, Ricketts SL, Bis JC, Aulchenko YS, Thorleifsson G, Feitosa MF, Chambers J, Orho-Melander M, Melander O, Johnson T, Li X, Guo X, Li M, Shin Cho Y, Jin Go M, Jin Kim Y, Lee JY, Park T, Kim K, Sim X, Twee-Hee Ong R, Croteau-Chonka DC, Lange LA, Smith JD, Song K, Hua Zhao J, Yuan X, Luan J, Lamina C, Ziegler A, Zhang W, Zee RY, Wright AF, Witteman JC, Wilson JF, Willemsen G, Wichmann HE, Whitfield JB, Waterworth DM, Wareham NJ, Waeber G, Vollenweider P, Voight BF, Vitart V, Uitterlinden AG, Uda M, Tuomilehto J, Thompson JR, Tanaka T, Surakka I, Stringham HM, Spector TD, Soranzo N, Smit JH, Sinisalo J, Silander K, Sijbrands EJ, Scuteri A, Scott J, Schlessinger D, Sanna S, Salomaa V, Saharinen J, Sabatti C, Ruokonen A, Rudan I, Rose LM, Roberts R, Rieder M, Psaty BM, Pramstaller PP, Pichler I, Perola M, Penninx BW, Pedersen NL, Pattaro C, Parker AN, Pare G, Oostra BA, O'Donnell CJ, Nieminen MS, Nickerson DA, Montgomery GW, Meitinger T, McPherson R, McCarthy MI, McArdle W, Masson D, Martin NG, Marroni F, Mangino M, Magnusson PK, Lucas G, Luben R, Loos RJ, Lokki ML, Lettre G, Langenberg C, Launer LJ, Lakatta EG, Laaksonen R, Kyvik KO, Kronenberg F, König IR, Khaw KT, Kaprio J, Kaplan LM, Johansson A, Jarvelin MR, Janssens AC, Ingelsson E, Igl W, Kees Hovingh G, Hottenga JJ, Hofman A, Hicks AA, Hengstenberg C, Heid IM, Hayward C, Havulinna AS, Hastie ND, Harris TB, Haritunians T, Hall AS, Gyllensten U, Guiducci C, Groop LC, Gonzalez E, Gieger C, Freimer NB, Ferrucci L, Erdmann J, Elliott P, Ejebe KG, Döring A, Dominiczak AF, Demissie S, Deloukas P, de Geus EJ, de Faire U, Crawford G, Collins FS, Chen YD, Caulfield MJ, Campbell H, Burtt NP, Bonnycastle LL, Boomsma DI, Boekholdt SM, Bergman RN, Barroso I, Bandinelli S, Ballantyne CM, Assimes TL, Quertermous T, Altshuler D, Seielstad M, Wong TY, Tai ES, Feranil AB, Kuzawa CW, Adair LS, Taylor HA Jr, Borecki IB, Gabriel SB, Wilson JG, Holm H, Thorsteinsdottir U, Gudnason V, Krauss RM, Mohlke KL, Ordovas JM, Munroe PB, Kooner JS, Tall AR, Hegele RA, Kastelein JJ, Schadt EE, Rotter JI, Boerwinkle E, Strachan DP, Mooser V, Stefansson K, Reilly MP, Samani NJ, Schunkert H, Cupples LA, Sandhu MS, Ridker PM, Rader DJ, van Duijn CM, Peltonen L, Abecasis GR, Boehnke M, Kathiresan S. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466:707–13. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruixing Y, Yuming C, Shangling P, Fengping H, Tangwei L, Dezhai Y, Jinzhen W, Limei Y, Weixiong L, Rongshan L, Jiandong H. Effects of demographic, dietary and other lifestyle factors on the prevalence of hyperlipidemia in Guangxi Hei Yi Zhuang and Han populations. Eur J Cardiovasc Prev Rehabil. 2006;13:977–84. doi: 10.1097/01.hjr.0000239476.79428.25. [DOI] [PubMed] [Google Scholar]
- Ruixing Y, Qiming F, Dezhai Y, Shuquan L, Weixiong L, Shangling P, Hai W, Yongzhong Y, Feng H, Shuming Q. Comparison of demography, diet, lifestyle, and serum lipid levels between the Guangxi Bai Ku Yao and Han populations. J Lipid Res. 2007;48:2673–81. doi: 10.1194/jlr.M700335-JLR200. [DOI] [PubMed] [Google Scholar]
- Ruixing Y, Yiyang L, Meng L, Kela L, Xingjiang L, Lin Z, Wanying L, Jinzhen W, Dezhai Y, Weixiong L. Interactions of the apolipoprotein C-III 3238C > G polymorphism and alcohol consumption on serum triglyceride levels. Lipids Health Dis. 2010;9:86. doi: 10.1186/1476-511X-9-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin RX, Li YY, Liu WY, Zhang L, Wu JZ. Interactions of the apolipoprotein A5 gene polymorphisms and alcohol consumption on serum lipid levels. PLoS One. 2011;6:e17954. doi: 10.1371/journal.pone.0017954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilia G, Chen WM, Scuteri A, Orrú M, Albai G, Dei M, Lai S, Usala G, Lai M, Loi P, Mameli C, Vacca L, Deiana M, Olla N, Masala M, Cao A, Najjar SS, Terracciano A, Nedorezov T, Sharov A, Zonderman AB, Abecasis GR, Costa P, Lakatta E, Schlessinger D. Heritability of cardiovascular and personality traits in 6,148 Sardinians. PLoS Genet. 2006;2:e132. doi: 10.1371/journal.pgen.0020132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollin TI, Hsueh WC, Steinle NI, Snitker S, Shuldiner AR, Mitchell BD. A genome-wide scan of serum lipid levels in the Old Order Amish. Atherosclerosis. 2004;173:89–96. doi: 10.1016/j.atherosclerosis.2003.11.012. [DOI] [PubMed] [Google Scholar]
- Yang Q, Lai CQ, Parnell L, Cupples LA, Adiconis X, Zhu Y, Wilson PW, Housman DE, Shearman AM, D'Agostino RB, Ordovas JM. Genome-wide linkage analyses and candidate gene fine mapping for HDL3 cholesterol: the Framingham Study. J Lipid Res. 2005;46:1416–25. doi: 10.1194/jlr.M400382-JLR200. [DOI] [PubMed] [Google Scholar]
- Harrap SB, Wong ZY, Scurrah KJ, Lamantia A. Genome-wide linkage analysis of population variation in high-density lipoprotein cholesterol. Hum Genet. 2006;119:541–6. doi: 10.1007/s00439-006-0167-4. [DOI] [PubMed] [Google Scholar]
- Diabetes Genetics Initiative of Broad Institute of Harvard and MIT, Lund University, Novartis Institutes of BioMedical Research. Saxena R, Voight BF, Lyssenko V, Burtt NP, de Bakker PI, Chen H, Roix JJ, Kathiresan S, Hirschhorn JN, Daly MJ, Hughes TE, Groop L, Altshuler D, Almgren P, Florez JC, Meyer J, Ardlie K, Bengtsson Boström K, Isomaa B, Lettre G, Lindblad U, Lyon HN, Melander O, Newton-Cheh C, Nilsson P, Orho-Melander M, Råstam L, Speliotes EK, Taskinen MR, Tuomi T, Guiducci C, Berglund A, Carlson J, Gianniny L, Hackett R, Hall L, Holmkvist J, Laurila E, Sjögren M, Sterner M, Surti A, Svensson M, Svensson M, Tewhey R, Blumenstiel B, Parkin M, Defelice M, Barry R, Brodeur W, Camarata J, Chia N, Fava M, Gibbons J, Handsaker B, Healy C, Nguyen K, Gates C, Sougnez C, Gage D, Nizzari M, Gabriel SB, Chirn GW, Ma Q, Parikh H, Richardson D, Ricke D, Purcell S. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316:1331–6. doi: 10.1126/science.1142358. [DOI] [PubMed] [Google Scholar]
- Kathiresan S, Melander O, Guiducci C, Surti A, Burtt NP, Rieder MJ, Cooper GM, Roos C, Voight BF, Havulinna AS, Wahlstrand B, Hedner T, Corella D, Tai ES, Ordovas JM, Berglund G, Vartiainen E, Jousilahti P, Hedblad B, Taskinen MR, Newton-Cheh C, Salomaa V, Peltonen L, Groop L, Altshuler DM, Orho-Melander M. Six new loci associated with blood low-density lipoprotein cholesterol, high-density lipoprotein cholesterol or triglycerides in humans. Nat Genet. 2008;40:189–97. doi: 10.1038/ng.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandhu MS, Waterworth DM, Debenham SL, Wheeler E, Papadakis K, Zhao JH, Song K, Yuan X, Johnson T, Ashford S, Inouye M, Luben R, Sims M, Hadley D, McArdle W, Barter P, Kesäniemi YA, Mahley RW, McPherson R, Grundy SM. Wellcome Trust Case Control Consortium. Bingham SA, Khaw KT, Loos RJ, Waeber G, Barroso I, Strachan DP, Deloukas P, Vollenweider P, Wareham NJ, Mooser V. LDL-cholesterol concentrations: a genome-wide association study. Lancet. 2008;371:483–91. doi: 10.1016/S0140-6736(08)60208-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willer CJ, Sanna S, Jackson AU, Scuteri A, Bonnycastle LL, Clarke R, Heath SC, Timpson NJ, Najjar SS, Stringham HM, Strait J, Duren WL, Maschio A, Busonero F, Mulas A, Albai G, Swift AJ, Morken MA, Narisu N, Bennett D, Parish S, Shen H, Galan P, Meneton P, Hercberg S, Zelenika D, Chen WM, Li Y, Scott LJ, Scheet PA, Sundvall J, Watanabe RM, Nagaraja R, Ebrahim S, Lawlor DA, Ben-Shlomo Y, Davey-Smith G, Shuldiner AR, Collins R, Bergman RN, Uda M, Tuomilehto J, Cao A, Collins FS, Lakatta E, Lathrop GM, Boehnke M, Schlessinger D, Mohlke KL, Abecasis GR. Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat Genet. 2008;40:161–9. doi: 10.1038/ng.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kooner JS, Chambers JC, Aguilar-Salinas CA, Hinds DA, Hyde CL, Warnes GR, Gómez Pérez FJ, Frazer KA, Elliott P, Scott J, Milos PM, Cox DR, Thompson JF. Genome-wide scan identifies variation in MLXIPL associated with plasma triglycerides. Nat Genet. 2008;40:149–51. doi: 10.1038/ng.2007.61. [DOI] [PubMed] [Google Scholar]
- Wallace C, Newhouse SJ, Braund P, Zhang F, Tobin M, Falchi M, Ahmadi K, Dobson RJ, Marçano AC, Hajat C, Burton P, Deloukas P, Brown M, Connell JM, Dominiczak A, Lathrop GM, Webster J, Farrall M, Spector T, Samani NJ, Caulfield MJ, Munroe PB. Genome-wide association study identifies genes for biomarkers of cardiovascular disease: serum urate and dyslipidemia. Am J Hum Genet. 2008;82:139–49. doi: 10.1016/j.ajhg.2007.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López S, Buil A, Ordoñez J, Souto JC, Almasy L, Lathrop M, Blangero J, Blanco-Vaca F, Fontcuberta J, Soria JM. Genome-wide linkage analysis for identifying quantitative trait loci involved in the regulation of lipoprotein a (Lpa) levels. Eur J Hum Genet. 2008;16:1372–9. doi: 10.1038/ejhg.2008.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heid IM, Boes E, Müller M, Kollerits B, Lamina C, Coassin S, Gieger C, Döring A, Klopp N, Frikke-Schmidt R, Tybjaerg-Hansen A, Brandstätter A, Luchner A, Meitinger T, Wichmann HE, Kronenberg F. Genome-wide association analysis of high-density lipoprotein cholesterol in the population-based KORA study sheds new light on intergenic regions. Circ Cardiovasc Genet. 2008;1:10–20. doi: 10.1161/CIRCGENETICS.108.776708. [DOI] [PubMed] [Google Scholar]
- Coletta DK, Schneider J, Hu SL, Dyer TD, Puppala S, Farook VS, Arya R, Lehman DM, Blangero J, DeFronzo RA, Duggirala R, Jenkinson CP. Genome-wide linkage scan for genes influencing plasma triglyceride levels in the Veterans Administration Genetic Epidemiology Study. Diabetes. 2009;58:279–84. doi: 10.2337/db08-0491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aulchenko YS, Ripatti S, Lindqvist I, Boomsma D, Heid IM, Pramstaller PP, Penninx BW, Janssens AC, Wilson JF, Spector T, Martin NG, Pedersen NL, Kyvik KO, Kaprio J, Hofman A, Freimer NB, Jarvelin MR, Gyllensten U, Campbell H, Rudan I, Johansson A, Marroni F, Hayward C, Vitart V, Jonasson I, Pattaro C, Wright A, Hastie N, Pichler I, Hicks AA, Falchi M, Willemsen G, Hottenga JJ, de Geus EJ, Montgomery GW, Whitfield J, Magnusson P, Saharinen J, Perola M, Silander K, Isaacs A, Sijbrands EJ, Uitterlinden AG, Witteman JC, Oostra BA, Elliott P, Ruokonen A, Sabatti C, Gieger C, Meitinger T, Kronenberg F, Döring A, Wichmann HE, Smit JH, McCarthy MI, van Duijn CM, Peltonen L. ENGAGE Consortium. Loci influencing lipid levels and coronary heart disease risk in 16 European population cohorts. Nat Genet. 2009;41:47–55. doi: 10.1038/ng.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kathiresan S, Willer CJ, Peloso GM, Demissie S, Musunuru K, Schadt EE, Kaplan L, Bennett D, Li Y, Tanaka T, Voight BF, Bonnycastle LL, Jackson AU, Crawford G, Surti A, Guiducci C, Burtt NP, Parish S, Clarke R, Zelenika D, Kubalanza KA, Morken MA, Scott LJ, Stringham HM, Galan P, Swift AJ, Kuusisto J, Bergman RN, Sundvall J, Laakso M, Ferrucci L, Scheet P, Sanna S, Uda M, Yang Q, Lunetta KL, Dupuis J, de Bakker PI, O'Donnell CJ, Chambers JC, Kooner JS, Hercberg S, Meneton P, Lakatta EG, Scuteri A, Schlessinger D, Tuomilehto J, Collins FS, Groop L, Altshuler D, Collins R, Lathrop GM, Melander O, Salomaa V, Peltonen L, Orho-Melander M, Ordovas JM, Boehnke M, Abecasis GR, Mohlke KL, Cupples LA. Common variants at 30 loci contribute to polygenic dyslipidemia. Nat Genet. 2009;41:56–65. doi: 10.1038/ng.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiura Y, Shen CS, Kokubo Y, Okamura T, Morisaki T, Tomoike H, Yoshida T, Sakamoto H, Goto Y, Nonogi H, Iwai N. Identification of genetic markers associated with high-density lipoprotein-cholesterol by genome-wide screening in a Japanese population: the Suita study. Circ J. 2009;73:1119–26. doi: 10.1253/circj.cj-08-1101. [DOI] [PubMed] [Google Scholar]
- Li X, Monda KL, Göring HH, Haack K, Cole SA, Diego VP, Almasy L, Laston S, Howard BV, Shara NM, Lee ET, Best LG, Fabsitz RR, MacCluer JW, North KE. Genome-wide linkage scan for plasma high density lipoprotein cholesterol, apolipoprotein A-1 and triglyceride variation among American Indian populations: the Strong Heart Family Study. J Med Genet. 2009;46:472–9. doi: 10.1136/jmg.2008.063891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Ren J, Li W, Huang X, Yan X, Yang B, Zhao Y, Guo Y, Mao H, Huang L. A genome-wide scan for quantitative trait loci affecting serum glucose and lipids in a White Duroc × Erhualian intercross F(2) population. Mamm Genome. 2009;20:386–92. doi: 10.1007/s00335-009-9190-9. [DOI] [PubMed] [Google Scholar]
- Chasman DI, Paré G, Zee RY, Parker AN, Cook NR, Buring JE, Kwiatkowski DJ, Rose LM, Smith JD, Williams PT, Rieder MJ, Rotter JI, Nickerson DA, Krauss RM, Miletich JP, Ridker PM. Genetic loci associated with plasma concentration of low-density lipoprotein cholesterol, high-density lipoprotein cholesterol, triglycerides, apolipoprotein A1, and Apolipoprotein B among 6382 white women in genome-wide analysis with replication. Circ Cardiovasc Genet. 2008;1:21–30. doi: 10.1161/CIRCGENETICS.108.773168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kangas-Kontio T, Kakko S, Tamminen M, von Rohr P, Hoeschele I, Juvonen T, Kere J, Savolainen MJ. Genome scan for loci regulating HDL cholesterol levels in Finnish extended pedigrees with early coronary heart disease. Eur J Hum Genet. 2010;18:604–13. doi: 10.1038/ejhg.2009.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccolo SR, Abo RP, Allen-Brady K, Camp NJ, Knight S, Anderson JL, Horne BD. Evaluation of genetic risk scores for lipid levels using genome-wide markers in the Framingham Heart Study. BMC Proc. 2009;3:S46. doi: 10.1186/1753-6561-3-s7-s46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keebler ME, Sanders CL, Surti A, Guiducci C, Burtt NP, Kathiresan S. Association of blood lipids with common DNA sequence variants at 19 genetic loci in the multiethnic United States National Health and Nutrition Examination Survey III. Circ Cardiovasc Genet. 2009;2:238–43. doi: 10.1161/CIRCGENETICS.108.829473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talmud PJ, Drenos F, Shah S, Shah T, Palmen J, Verzilli C, Gaunt TR, Pallas J, Lovering R, Li K, Casas JP, Sofat R, Kumari M, Rodriguez S, Johnson T, Newhouse SJ, Dominiczak A, Samani NJ, Caulfield M, Sever P, Stanton A, Shields DC, Padmanabhan S, Melander O, Hastie C, Delles C, Ebrahim S, Marmot MG, Smith GD, Lawlor DA, Munroe PB, Day IN, Kivimaki M, Whittaker J, Humphries SE, Hingorani AD. ASCOT investigators, NORDIL investigators, BRIGHT Consortium. Gene-centric association signals for lipids and apolipoproteins identified via the HumanCVD BeadChip. Am J Hum Genet. 2009;85:628–42. doi: 10.1016/j.ajhg.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Relf BL, Larkin EK, De Torres C, Baur LA, Christodoulou J, Waters KA. Genome-wide linkage of obstructive sleep apnoea and high-density lipoprotein cholesterol in a Filipino family: bivariate linkage analysis of obstructive sleep apnoea. J Sleep Res. 2010;19:349–57. doi: 10.1111/j.1365-2869.2009.00797.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissglas-Volkov D, Aguilar-Salinas CA, Sinsheimer JS, Riba L, Huertas-Vazquez A, Ordoñez-Sánchez ML, Rodriguez-Guillen R, Cantor RM, Tusie-Luna T, Pajukanta P. Investigation of variants identified in caucasian genome-wide association studies for plasma high-density lipoprotein cholesterol and triglycerides levels in Mexican dyslipidemic study samples. Circ Cardiovasc Genet. 2010;3:31–8. doi: 10.1161/CIRCGENETICS.109.908004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen CT, Wang J, Lanktree MB, Cao H, McIntyre AD, Ban MR, Martins RA, Kennedy BA, Hassell RG, Visser ME, Schwartz SM, Voight BF, Elosua R, Salomaa V, O'Donnell CJ, Dallinga-Thie GM, Anand SS, Yusuf S, Huff MW, Kathiresan S, Hegele RA. Excess of rare variants in genes identified by genome-wide association study of hypertriglyceridemia. Nat Genet. 2010;42:684–7. doi: 10.1038/ng.628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterworth DM, Ricketts SL, Song K, Chen L, Zhao JH, Ripatti S, Aulchenko YS, Zhang W, Yuan X, Lim N, Luan J, Ashford S, Wheeler E, Young EH, Hadley D, Thompson JR, Braund PS, Johnson T, Struchalin M, Surakka I, Luben R, Khaw KT, Rodwell SA, Loos RJ, Boekholdt SM, Inouye M, Deloukas P, Elliott P, Schlessinger D, Sanna S, Scuteri A, Jackson A, Mohlke KL, Tuomilehto J, Roberts R, Stewart A, Kesäniemi YA, Mahley RW, Grundy SM. Wellcome Trust Case Control Consortium. McArdle W, Cardon L, Waeber G, Vollenweider P, Chambers JC, Boehnke M, Abecasis GR, Salomaa V, Järvelin MR, Ruokonen A, Barroso I, Epstein SE, Hakonarson HH, Rader DJ, Reilly MP, Witteman JC, Hall AS, Samani NJ, Strachan DP, Barter P, van Duijn CM, Kooner JS, Peltonen L, Wareham NJ, McPherson R, Mooser V, Sandhu MS. Genetic variants influencing circulating lipid levels and risk of coronary artery disease. Arterioscler Thromb Vasc Biol. 2010;30:2264–76. doi: 10.1161/ATVBAHA.109.201020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acuña-Alonzo V, Flores-Dorantes T, Kruit JK, Villarreal-Molina T, Arellano-Campos O, Hünemeier T, Moreno-Estrada A, Ortiz-López MG, Villamil-Ramírez H, León-Mimila P, Villalobos-Comparan M, Jacobo-Albavera L, Ramírez-Jiménez S, Sikora M, Zhang LH, Pape TD, Granados-Silvestre Mde A, Montufar-Robles I, Tito-Alvarez AM, Zurita-Salinas C, Bustos-Arriaga J, Cedillo-Barrón L, Gómez-Trejo C, Barquera-Lozano R, Vieira-Filho JP, Granados J, Romero-Hidalgo S, Huertas-Vázquez A, González-Martín A, Gorostiza A, Bonatto SL, Rodríguez-Cruz M, Wang L, Tusié-Luna T, Aguilar-Salinas CA, Lisker R, Moises RS, Menjivar M, Salzano FM, Knowler WC, Bortolini MC, Hayden MR, Baier LJ, Canizales-Quinteros S. A functional ABCA1 gene variant is associated with low HDL-cholesterol levels and shows evidence of positive selection in Native Americans. Hum Mol Genet. 2010;19:2877–85. doi: 10.1093/hmg/ddq173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleheen D, Soranzo N, Rasheed A, Scharnagl H, Gwilliam R, Alexander M, Inouye M, Zaidi M, Potter S, Haycock P, Bumpstead S, Kaptoge S, Di Angelantonio E, Sarwar N, Hunt SE, Sheikh N, Shah N, Samuel M, Haider SR, Murtaza M, Thompson A, Gobin R, Butterworth A, Ahmad U, Hakeem A, Zaman KS, Kundi A, Yaqoob Z, Cheema LA, Qamar N, Faruqui A, Mallick NH, Azhar M, Samad A, Ishaq M, Rasheed SZ, Jooma R, Niazi JH, Gardezi AR, Memon NA, Ghaffar A, Rehman FU, Hoffmann MM, Renner W, Kleber ME, Grammer TB, Stephens J, Attwood A, Koch K, Hussain M, Kumar K, Saleem A, Kumar K, Daood MS, Gul AA, Abbas S, Zafar J, Shahid F, Bhatti SM, Ali SS, Muhammad F, Sagoo G, Bray S, McGinnis R, Dudbridge F, Winkelmann BR, Böehm B, Thompson S, Ouwehand W, März W, Frossard P, Danesh J, Deloukas P. Genetic determinants of major blood lipids in Pakistanis compared with Europeans. Circ Cardiovasc Genet. 2010;3:348–57. doi: 10.1161/CIRCGENETICS.109.906180. [DOI] [PubMed] [Google Scholar]
- Keebler ME, Deo RC, Surti A, Konieczkowski D, Guiducci C, Burtt N, Buxbaum SG, Sarpong DF, Steffes MW, Wilson JG, Taylor HA, Kathiresan S. Fine-mapping in African Americans of 8 recently discovered genetic loci for plasma lipids: the Jackson Heart Study. Circ Cardiovasc Genet. 2010;3:358–64. doi: 10.1161/CIRCGENETICS.109.914267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumitrescu L, Carty CL, Taylor K, Schumacher FR, Hindorff LA, Ambite JL, Anderson G, Best LG, Brown-Gentry K, Bůžková P, Carlson CS, Cochran B, Cole SA, Devereux RB, Duggan D, Eaton CB, Fornage M, Franceschini N, Haessler J, Howard BV, Johnson KC, Laston S, Kolonel LN, Lee ET, MacCluer JW, Manolio TA, Pendergrass SA, Quibrera M, Shohet RV, Wilkens LR, Haiman CA, Le Marchand L, Buyske S, Kooperberg C, North KE, Crawford DC. Genetic determinants of lipid traits in diverse populations from the population architecture using genomics and epidemiology (PAGE) study. PLoS Genet. 2011;7:e1002138. doi: 10.1371/journal.pgen.1002138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaess BM, Tomaszewski M, Braund PS, Stark K, Rafelt S, Fischer M, Hardwick R, Nelson CP, Debiec R, Huber F, Kremer W, Kalbitzer HR, Rose LM, Chasman DI, Hopewell J, Clarke R, Burton PR, Tobin MD, Hengstenberg C, Samani NJ. Large-scale candidate gene analysis of HDL particle features. PLoS One. 2011;6:e14529. doi: 10.1371/journal.pone.0014529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lettre G, Palmer CD, Young T, Ejebe KG, Allayee H, Benjamin EJ, Bennett F, Bowden DW, Chakravarti A, Dreisbach A, Farlow DN, Folsom AR, Fornage M, Forrester T, Fox E, Haiman CA, Hartiala J, Harris TB, Hazen SL, Heckbert SR, Henderson BE, Hirschhorn JN, Keating BJ, Kritchevsky SB, Larkin E, Li M, Rudock ME, McKenzie CA, Meigs JB, Meng YA, Mosley TH, Newman AB, Newton-Cheh CH, Paltoo DN, Papanicolaou GJ, Patterson N, Post WS, Psaty BM, Qasim AN, Qu L, Rader DJ, Redline S, Reilly MP, Reiner AP, Rich SS, Rotter JI, Liu Y, Shrader P, Siscovick DS, Tang WH, Taylor HA, Tracy RP, Vasan RS, Waters KM, Wilks R, Wilson JG, Fabsitz RR, Gabriel SB, Kathiresan S, Boerwinkle E. Genome-wide association study of coronary heart disease and its risk factors in 8,090 African Americans: the NHLBI CARe Project. PLoS Genet. 2011;7:e1001300. doi: 10.1371/journal.pgen.1001300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanghera DK, Been LF, Ralhan S, Wander GS, Mehra NK, Singh JR, Ferrell RE, Kamboh MI, Aston CE. Genome-wide linkage scan to identify loci associated with type 2 diabetes and blood lipid phenotypes in the Sikh Diabetes Study. PLoS One. 2011;6:e21188. doi: 10.1371/journal.pone.0021188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butterworth AS, Braund PS, Farrall M, Hardwick RJ, Saleheen D, Peden JF, Soranzo N, Chambers JC, Sivapalaratnam S, Kleber ME, Keating B, Qasim A, Klopp N, Erdmann J, Assimes TL, Ball SG, Balmforth AJ, Barnes TA, Basart H, Baumert J, Bezzina CR, Boerwinkle E, Boehm BO, Brocheton J, Bugert P, Cambien F, Clarke R, Codd V, Collins R, Couper D, Cupples LA, de Jong JS, Diemert P, Ejebe K, Elbers CC, Elliott P, Fornage M, Franzosi MG, Frossard P, Garner S, Goel A, Goodall AH, Hengstenberg C, Hunt SE, Kastelein JJ, Klungel OH, Klüter H, Koch K, König IR, Kooner AS, Laaksonen R, Lathrop M, Li M, Liu K, McPherson R, Musameh MD, Musani S, Nelson CP, O'Donnell CJ, Ongen H, Papanicolaou G, Peters A, Peters BJ, Potter S, Psaty BM, Qu L, Rader DJ, Rasheed A, Rice C, Scott J, Seedorf U, Sehmi JS, Sotoodehnia N, Stark K, Stephens J, van der Schoot CE, van der Schouw YT, Thorsteinsdottir U, Tomaszewski M, van der Harst P, Vasan RS, Wilde AA, Willenborg C, Winkelmann BR, Zaidi M, Zhang W, Ziegler A, de Bakker PI, Koenig W, Mätz W, Trip MD, Reilly MP, Kathiresan S, Schunkert H, Hamsten A, Hall AS, Kooner JS, Thompson SG, Thompson JR, Deloukas P, Ouwehand WH, Watkins H, Danesh J, Samani NJ, Barnes T, Rafelt S, Codd V, Tomaszewski M, Ouwehand WH, Bruinsma N, Dekker LR, Henriques JP, Koch KT, de Winter RJ, Alings M, Allaart CF, Gorgels AP, Verheugt FW, Braund PS, Thompson JR, Samani NJ, Mueller M, Meisinger C, DerOhannessian S, Mehta NN, Ferguson J, Hakonarson H, Matthai W, Wilensky R, Hopewell JC, Parish S, Linksted P, Notman J, Gonzalez H, Young A, Ostley T, Munday A, Goodwin N, Verdon V, Shah S, Cobb L, Edwards C, Mathews C, Gunter R, Benham J, Davies C, Cobb M, Cobb L, Crowther J, Richards A, Silver M, Tochlin S, Mozley S, Clark S, Radley M, Kourellias K, Silveira A, Söderholm B, Olsson P, Barlera S, Tognoni G, Rust S, Assmann G, Heath S, Zelenika D, Gut I, Green F, Farrall M, Peden J, Goel A, Ongen H, Franzosi MG, Lathrop M, Seedorf U, Clarke R, Collins R, Hamsten A, Watkins H, Aly A, Anner K, Björklund K, Blomgren G, Cederschiöld B, Danell-Toverud K, Eriksson P, Grundstedt U, Hamsten A, Heinonen M, Hellénius ML, van't Hooft F, Husman K, Lagercrantz J, Larsson A, Larsson M, Mossfeldt M, Mälarstig A, Olsson G, Sabater-Lleal M, Sennblad B, Silveira A, Strawbridge R, Söderholm B, Öhrvik J, Zaman KS, Mallick NH, Azhar M, Samad A, Ishaq M, Shah N, Samuel M, Schunkert H, König IR, Kathiresan S, Reilly M, Assimes TL, Holm H, Preuss M, Stewart AF, Barbalic M, Gieger C, Absher D, Aherrahrou Z, Allayee H, Altshuler D, Anand S, Andersen K, Anderson JL, Ardissino D, Ball SG, Balmforth AJ, Barnes TA, Becker LC, Becker DM, Berger K, Bis JC, Boekholdt SM, Boerwinkle E, Braund PS, Brown MJ, Burnett MS, Buysschaert I, Carlquist JF, Chen L, Codd V, Davies RW, Dedoussis G, Dehghan A, Demissie S, Devaney J, Do R, Doering A, El Mokhtari NE, Ellis SG, Elosua R, Engert JC, Epstein S, de Faire U, Fischer M, Folsom AR, Freyer J, Gigante B, Girelli D, Gretarsdottir S, Gudnason V, Gulcher JR, Tennstedt S, Halperin E, Hammond N, Hazen SL, Hofman A, Horne BD, Illig T, Iribarren C, Jones GT, Jukema JW, Kaiser MA, Kaplan LM, Kastelein JJ, Khaw KT, Knowles JW, Kolovou G, Kong A, Laaksonen R, Lambrechts D, Leander K, Li M, Lieb W, Diemert P, Lettre G, Loley C, Lotery AJ, Mannucci PM, Maouche S, Martinelli N, McKeown PP, Meisinger C, Meitinger T, Melander O, Merlini PA, Mooser V, Morgan T, Mühleisen TW, Muhlestein JB, Musunuru K, Nahrstaedt J, Nelson CP, Nöthen MM, Olivieri O, Peyvandi F, Patel RS, Patterson CC, Peters A, Qu L, Quyyumi AA, Rader DJ, Rallidis LS, Rice C, Roosendaal FR, Rubin D, Salomaa V, Sampietro ML, Sandhu MS, Schadt E, Schäfer A, Schillert A, Schreiber S, Schrezenmeir J, Schwartz SM, Siscovick DS, Sivananthan M, Sivapalaratnam S, Smith AV, Smith TB, Snoep JD, Soranzo N, Spertus JA, Stark K, Stefansson K, Stirrups K, Stoll M, Tang WH, Thorgeirsson G, Thorleifsson G, Tomaszewski M, Uitterlinden AG, van Rij AM, Voight BF, Wareham NJ, AWells G, Wichmann HE, Willenborg C, Witteman JC, Wright BJ, Ye S, Ziegler A, Cambien F, Goodall AH, Cupples LA, Quertermous T, März W, Hengstenberg C, Blankenberg S, Ouwehand WH, Hall AS, Deloukas P, Thorsteinsdottir U, Roberts R, Thompson JR, O'Donnell CJ, McPherson R, Erdmann J, Samani NJ, Onland-Moret NC, van Setten J, de Bakker PI, Verschuren WM, Boer JM, Wijmenga C, Hofker MH, Maitland-van der Zee AH, de Boer A, Grobbee DE, Attwood T, Belz S, Braund P, Cambien F, Cooper J, Crisp-Hihn A, Diemert P, Deloukas P, Foad N, Erdmann J, Goodall AH, Gracey J, Gray E, Gwilliams R, Heimerl S, Hengstenberg C, Jolley J, Krishnan U, Lloyd-Jones H, Lugauer I, Lundmark P, Maouche S, Moore JS, Muir D, Murray E, Nelson CP, Neudert J, Niblett D, O'Leary K, Ouwehand WH, Pollard H, Rankin A, Rice CM, Sager H, Samani NJ, Sambrook J, Schmitz G, Scholz M, Schroeder L, Schunkert H, Syvannen AC, Tennstedt S, Wallace C. Large-scale gene-centric analysis identifies novel variants for coronary artery disease. PLoS Genet. 2011;7:e1002260. doi: 10.1371/journal.pgen.1002260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surakka I, Isaacs A, Karssen LC, Laurila PP, Middelberg RP, Tikkanen E, Ried JS, Lamina C, Mangino M, Igl W, Hottenga JJ, Lagou V, van der Harst P, Mateo Leach I, Esko T, Kutalik Z, Wainwright NW, Struchalin MV, Sarin AP, Kangas AJ, Viikari JS, Perola M, Rantanen T, Petersen AK, Soininen P, Johansson A, Soranzo N, Heath AC, Papamarkou T, Prokopenko I, Tönjes A, Kronenberg F, Döring A, Rivadeneira F, Montgomery GW, Whitfield JB, Kähönen M, Lehtimäki T, Freimer NB, Willemsen G, de Geus EJ, Palotie A, Sandhu MS, Waterworth DM, Metspalu A, Stumvoll M, Uitterlinden AG, Jula A, Navis G, Wijmenga C, Wolffenbuttel BH, Taskinen MR, Ala-Korpela M, Kaprio J, Kyvik KO, Boomsma DI, Pedersen NL, Gyllensten U, Wilson JF, Rudan I, Campbell H, Pramstaller PP, Spector TD, Witteman JC, Eriksson JG, Salomaa V, Oostra BA, Raitakari OT, Wichmann HE, Gieger C, Järvelin MR, Martin NG, Hofman A, McCarthy MI, Peltonen L, van Duijn CM, Aulchenko YS, Ripatti S. for the ENGAGE Consortium. A Genome-Wide Screen for Interactions Reveals a New Locus on 4p15 Modifying the Effect of Waist-to-Hip Ratio on Total Cholesterol. PLoS Genet. 2011;7:e1002333. doi: 10.1371/journal.pgen.1002333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Tao L, Chen Z, Zhou D, Kan M, Zhang D, Li C, He L, Liu Y. Association of genetic Loci with blood lipids in the chinese population. PLoS One. 2011;6:e27305. doi: 10.1371/journal.pone.0027305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson K, Farley D, Hu SI, Terkeltaub R. One of two chondrocyte-expressed isoforms of cartilage intermediate-layer protein functions as an insulin-like growth factor 1 antagonist. Arthritis Rheum. 2003;48:1302–14. doi: 10.1002/art.10927. [DOI] [PubMed] [Google Scholar]
- Wagner K, Mincheva A, Korn B, Lichter P, Pöpperl H. Pbx4, a new Pbx family member on mouse chromosome 8, is expressed during spermatogenesis. Mech Dev. 2001;103:127–31. doi: 10.1016/s0925-4773(01)00349-5. [DOI] [PubMed] [Google Scholar]
- Rauch U, Feng K, Zhou XH. Neurocan: a brain chondroitin sulfate proteoglycan. Cell Mol Life Sci. 2011;58:1842–56. doi: 10.1007/PL00000822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama K, Bayasgalan T, Yamanaka K, Kumada M, Gotoh T, Utsumi N, Yanagisawa Y, Okayama M, Kajii E, Ishibashi S, Iwamoto S. Jichi Community Genetics Team (JCOG) Large scale replication analysis of loci associated with lipid concentrations in a Japanese population. J Med Genet. 2009;46:370–4. doi: 10.1136/jmg.2008.064063. [DOI] [PubMed] [Google Scholar]
- Tai ES, Sim XL, Ong TH, Wong TY, Saw SM, Aung T, Kathiresan S, Orho-Melander M, Ordovas JM, Tan JT, Seielstad M. Polymorphisms at newly identified lipid-associated loci are associated with blood lipids and cardiovascular disease in an Asian Malay population. J Lipid Res. 2009;50:514–20. doi: 10.1194/jlr.M800456-JLR200. [DOI] [PubMed] [Google Scholar]
- Xu L, Deng QY, Li SF, Zhou LN, Gong JC, Wei BY. Genetic analysis of Mulao nationality using 15 short tandem repeats. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2008;25:96–100. [PubMed] [Google Scholar]
- Li Q, Yin RX, Yan TT, Miao L, Cao XL, Hu XJ, Aung LH, Wu DF, Wu JZ, Lin WX. Association of the GALNT2 gene polymorphisms and several environmental factors with serum lipid levels in the Mulao and Han populations. Lipids Health Dis. 2011;10:160. doi: 10.1186/1476-511X-10-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- People's Republic of China-United States Cardiovascular and Cardiopulmonary Epidemiology Research Group. An epidemiological study of cardiovascular and cardiopulmonary disease risk factors in four populations in the People's Republic of China. Baseline report from the P.R.C.-USA Collaborative Study. Circulation. 1992;85:1083–96. doi: 10.1161/01.cir.85.3.1083. [DOI] [PubMed] [Google Scholar]
- Ruixing Y, Limei Y, Yuming C, Dezhai Y, Weixiong L, Muyan L, Fengping H, Jinzhen W, Guangqing Y, Zhenbiao N. Prevalence, awareness, treatment, control and risk factors of hypertension in the Guangxi Hei Yi Zhuang and Han populations. Hypertens Res. 2006;29:423–32. doi: 10.1291/hypres.29.423. [DOI] [PubMed] [Google Scholar]
- Ruixing Y, Weixiong L, Hanjun Y, Dezhai Y, Shuquan L, Shangling P, Qiming F, Jinzhen W, Jianting G, Yaju D. Diet, lifestyle, and blood pressure of the middle-aged and elderly in the Guangxi Bai Ku Yao and Han populations. Am J Hypertens. 2008;21:382–7. doi: 10.1038/ajh.2008.1. [DOI] [PubMed] [Google Scholar]
- Cooperative Meta-analysis Group of China Obesity Task Force. Predictive values of body mass index and waist circumference to risk factors of related diseases in Chinese adult population. Zhonghua Liu Xing Bing Xue Za Zhi. 2002;23:5–10. [PubMed] [Google Scholar]
- Grundy SM, Denke MA. Dietary influences on serum lipids and lipoproteins. J Lipid Res. 1990;31:1149–72. [PubMed] [Google Scholar]
- Watts GF, Jackson P, Burke V, Lewis B. Dietary fatty acids and progression of coronary artery disease in men. Am J Clin Nutr. 1996;64:202–9. doi: 10.1093/ajcn/64.2.202. [DOI] [PubMed] [Google Scholar]
- Mansfield E, McPherson R, Koski KG. Diet and waist-to-hip ratio: important predictors of lipoprotein levels in sedentary and active young men with no evidence of cardiovascular disease. J Am Diet Assoc. 1999;99:1373–9. doi: 10.1016/S0002-8223(99)00335-1. [DOI] [PubMed] [Google Scholar]
- Yu-Poth S, Zhao G, Etherton T, Naglak M, Jonnalagadda S, Kris-Etherton PM. Effects of National Cholesterol Education Program's Step I and Step II dietary intervention programs on cardiovascular disease risk factors: a meta-analysis. Am J Clin Nutr. 1999;69:632–46. doi: 10.1093/ajcn/69.4.632. [DOI] [PubMed] [Google Scholar]
- Bermudez OI, Velez-Carrasco W, Schaefer EJ, Tucker KL. Dietary and plasma lipid, lipoprotein, and apolipoprotein profiles among elderly Hispanics and non-Hispanics and their association with diabetes. Am J Clin Nutr. 2002;76:1214–21. doi: 10.1093/ajcn/76.6.1214. [DOI] [PubMed] [Google Scholar]
- Garcia-Palmieri MR, Tillotson J, Cordero E, Costas R Jr, Sorlie P, Gordon T, Kannel WB, Colon AA. Nutrient intake and serum lipids in urban and rural Puerto Rican men. Am J Clin Nutr. 1977;30:2092–100. doi: 10.1093/ajcn/30.12.2092. [DOI] [PubMed] [Google Scholar]