Abstract
Tumor recurrence is the major cause of death in lung cancer treatment. To date, there is no clinically applied gene expression-based model to predict the risk for tumor recurrence in non-small cell lung cancer (NSCLC). We sought to embed crosstalk with major signaling pathways into biomarker identification. Three approaches were used to identify prognostic gene signatures from 442 lung adenocarcinoma samples. Candidate genes co-expressed with 6 or 7 major NSCLC signaling hallmarks were identified from genome-wide coexpression networks specifically associated with different prognostic groups. From these candidate genes, the first approach selected genes significantly associated with disease-specific survival using univariate Cox model. The second approach used random forests to refine the gene signatures; and the third approach used Relief algorithm to form the final gene sets. A total of 21 gene signatures were identified using these three approaches. These gene signatures generated significant prognostic stratifications (log-rank P < 0.05 in Kaplan-Meier analyses; Hazard Ratio >1, P< 0.05) in all tumors, stage I only, and in stage I patients not receiving chemotherapy in all training and test sets. In multivariate analyses with age, gender, race, smoking history, cancer stage, and tumor differentiation, a 10-gene signature had a hazard ratio of 3.23 (95% CI: [1.48, 7.06]), which was a more significant prognostic factor than other clinical factors, except cancer stage (III vs. I; with no significant difference). All identified 21 gene signatures outperformed other lung cancer signatures evaluated in the Director's Challenge Study. This study is an important step toward personalized prognosis of tumor recurrence and patient selection for adjuvant chemotherapy, with significant impact on down-stream clinical applications.
Keywords: lung adenocarcinoma, gene co-expression networks, biomarker identification, signaling pathways, prognostic stratification, tumor recurrence, metastasis, non-small cell lung cancer
Introduction
Lung cancer is a complex disease and remains the leading cause of cancer-related mortality world-wide. Non-small cell lung cancer (NSCLC) accounts for about 80% of lung cancer cases. In the current practice, surgical resection is the major treatment option for stage IA NSCLC patients. However, about 35-50% of stage I NSCLC patients will develop and die from tumor recurrence within five years following the surgery (1;2), and adjuvant chemotherapy of stage II and stage III disease has resulted in very modest survival benefits (3). It is not currently feasible to identify specific patients at high risk for recurrence who might benefit from adjuvant chemotherapy, especially for stage I patients. The emerging use of biomarkers may enable physicians to make treatment decisions based on the specific characteristics of individual patients and their tumors, instead of population statistics (4). To date, there is no fully-validated and clinically applied model for predicting lung cancer recurrence (5).
There have been many studies on lung cancer prognosis by transcriptional profiling (6-12). In these studies, genes are ranked according to their association with the clinical outcome, and the top-ranked genes are included in the classifier. However, rank-based feature selection algorithms cannot model complex molecular interactions in disease. It has been noted that individual biomarkers showing strong association with disease outcome are not necessarily good classifiers (13-15). Because genes and proteins do not function in isolation, but rather interact with one another to form modular networks (16); thus, understanding these interactions is critical to unraveling the molecular basis of disease. Molecular network analyses have been shown to be useful in disease classification (17) and identification of novel therapeutic targets (18). Nevertheless, major challenges include the modeling of genome-scale coexpression networks and the identification of a particular set of markers, from among the enormous number of potential markers, that has the highest prognostic ability of disease outcome (19).
Genes implicated in cancer initiation and progression show dysregulated interactions with their molecular partners (20), and cancer genes are more likely to actively interact with signal proteins (21). In this study, we present a network-based methodology for the combined analysis of disease-mediated genome-wide coexpression networks, crosstalk with major NSCLC signaling pathways, and clinical approaches. This network-based methodology identified extensive gene signatures coexpressed with major NSCLC signaling proteins, which generated significant prognostic stratification in all cancer stages (I, II, and IIIA), stage I only, and in stage I patients not receiving chemotherapy in the Director's Challenge Study of lung adenocarcinoma (n = 442).
Materials and Methods
Microarray profiles and patient samples
Gene expression profiles were quantified with Affymetrix HG-U133A on 442 lung adenocarcinoma samples in the Director's Challenge Study (12). This study cohort is composed of 4 data sets (University of Michigan, H. Lee Moffitt Cancer Center, Memorial Sloan-Kettering Cancer Center, and Dana-Farber Cancer Institute) contributed by 6 institutions. None of the patients received preoperative chemotherapy or radiation and at least two years of follow-up information was available for each patient. The raw microarray data are available from the caArray website [https://array.nci.nih.gov/caarray/project/details.action?project.id=182]. The data used in this analysis was quantile-normalized and log2-transformed with dChip (22). A brief summary of the clinical characteristics of all patient samples is provided in Supplementary Table 1.
Implication induction algorithm
An implication induction algorithm based on prediction logic (23) was used to derive gene coexpression networks with the Software package Genet [available at: http://www.hsc.wvu.edu/mbrcc/fs/GuoLab/products.asp]. Prediction logic reveals the implication relationships among variables in a dataset and evaluates propositions in formal logic. A modified U-Optimality method (24) was used to derive the implication relation between each pair of genes (25). The six foremost implication rules could be used to represent the gene regulation relations in the biological context (Supplementary Fig. 3). In this study, the minimum scope and the minimum precision of a derived implication relation were significantly greater than zero (P < 0.05, one-sided z-tests).
Random forests
The random forests algorithm is an ensemble of tree classifiers (26). The basic step of random forests is to form diverse tree classifiers from a single training set. Each tree is built using a different bootstrap sample from the original data. About one-third of the cases are not used in the construction of a tree. These cases are called out-of-bag (OOB) cases. The feature selection experiments were performed using the varSelRF package (27) of R [http://www.r-project.org/]. The feature subset with the smallest OOB error was chosen as the optimal feature subset.
Relief feature selection algorithm
Relief evaluates the importance of a variable by repeatedly sampling an instance and checking the value of the given variable for the nearest instance from the same and different classes. The values of the attributes of the nearest neighbors are compared to the sampled instance and used to update the relevance scores for each attribute. As approximated in the following equation, Relief computes the weight of attribute A as:
Relief assigns more weight to those attributes that have the same value for instances from the same class and differentiate from instances in different classes (28;29).
Results
Network-based Methodology for signature identification
Patient samples from UM and HLM formed the training set (n = 256), whereas samples from MSK (n = 104) and DFCI (n = 82) constituted two independent test sets. Genes with missing measurements in at least half of the samples were removed from analysis. Furthermore, for genes measured using multiple probes, the average expression of the duplicates was used to represent the expression profile of the unique gene. This gave 12,566 unique genes for the implication network analysis.
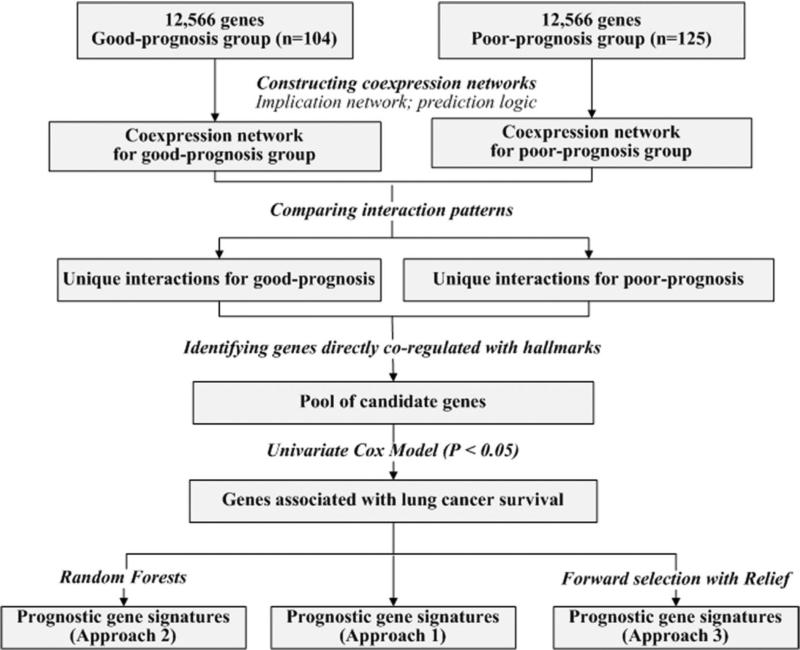
To construct implication networks, the mean expression of each gene in a patient cohort was used as a cut-off to partition the expression profiles. If the expression of a gene in a patient sample was greater than the mean in the cohort, this gene was denoted as up-regulated in this tumor sample; otherwise, it was denoted as down-regulated in the tumor sample. In the training set, patients who died within 5 years were labeled as poor-prognosis (n = 125), and those who survived 5 years after surgery were labeled as good-prognosis (n = 104). Censored cases (those with follow-up of less than 5 years) were removed from the analysis (n = 27). For each patient group in the training set, a genome-scale coexpression network was constructed using the implication induction algorithm. Between each pair of genes, possible significant (P < 0.05; one-sided z-tests) coexpression relations were derived in each patient group, constituting disease-mediated gene coexpression networks. By comparing the implication rules connecting each pair of nodes between the two networks, disease-specific differential network components were identified. These differential components contain the coexpression relations that were either present in the poor-prognosis group but missing in the good-prognosis group, or conversely, those present in the good-prognosis group but missing in the poor-prognosis group (Fig. 1).
Figure 1.
Overview of network-based methodology for identifying prognostic gene signatures.
Next, candidate genes were obtained by retrieving genes displaying a direct significant (P < 0.05, z-tests) co-regulation relation with major NSCLC signal proteins from the differential components associated with each prognosis group. From the human NSCLC signaling pathways delineated by the KEGG pathway database (available at: http://www.genome.jp/kegg/pathway/hsa/hsa05223.html), 11 signaling proteins (TP53, MET, RB1, EGF, EGFR, KRAS, E2F1, E2F2, E2F3, E2F4, and E2F5) were included in this study based on their reported clinical relevance in NSCLC progression. Candidate genes with significant coexpression relations with any combination of 6 or 7 signaling proteins were included for further analysis (Fig. 1).
Three approaches were taken to identify gene signatures from the pool of candidate genes. In the first approach, candidate genes with significant association with disease-specific survival (P < 0.05, univariate Cox model) were identified as signature genes. In the second approach, random forests were used to obtain a refined set of signature genes from the significant probes (P < 0.05; univariate Cox model). In the third approach, significant probes (P < 0.05; univariate Cox model) were further ranked by the Relief algorithm, and the top ranked genes formed the final gene signatures in a step-wise forward selection. Specifically, starting from the top ranked gene, one gene was added at each step to the gene set, until the prognostic accuracy could not be improved by the addition of more genes. The final gene set was identified as the gene signature. Fig. 1 gives an overview of the methodology.
Evaluation of identified prognostic gene signatures
To evaluate if the identified signatures could provide accurate prognostic prediction for lung adenocarcinoma, a multivariate Cox proportional hazard model was used in prognostic stratification. The models and cutoff values defined using the training set were applied to the independent test sets without re-estimating the parameters. The prognostic performance of each identified gene signature was evaluated according to following criteria: log-rank tests in Kaplan-Meier analyses and hazard ratio of death from lung cancer for all cancer stages, for stage I only and for stage I without receiving chemotherapy in training and test cohorts.
In the first approach, among candidate genes that co-regulated with 6 signaling proteins, 9 gene signatures generated significant stratification (log-rank P <0.05) with significant hazard ratios (P < 0.05) in all three patient cohorts (Supplementary Table 2). Among these 9 gene signatures, 5 of them also had significant hazard ratios (P <0.05) on stage I patients in all three cohorts. Among the 5 gene signatures that could give accurate prognostic categorization in all stages and stage I tumors, 4 gene signatures (S1-S4; Supplementary Table 3) generated significant stratifications (log-rank P <0.05 in Kaplan-Meier analysis, with hazard ratio significantly greater than 1) for stage I patients without receiving chemotherapy (Supplementary Table 2). Similarly, among candidate genes co-regulated with 7 signaling proteins in the first approach, 4 gene signatures generated accurate prognostic stratification (log-rank P <0.05 in Kaplan-Meier analysis, with hazard ratio significantly greater than 1) in all three patient cohorts, and one of them also generated accurate prognostic prediction in stage I patients in all three datasets (Supplementary Table 2).
The second approach identified 1 gene signature (S5; Supplementary Table 4) that provided significant stratifications in patients with all resectable cancer stages, stage I only, and stage I without receiving chemotherapy (Supplementary Table 2). The third approach identified 16 such gene signatures (S6-S21; Supplementary Table 5). In summary, a total of 21 gene signatures were identified using the three approaches in this study, which, in turn, generated significant prognostic categorizations in lung adenocarcinomas with all cancer stages, stage I only, and stage I without chemotherapy (Supplementary Table 2).
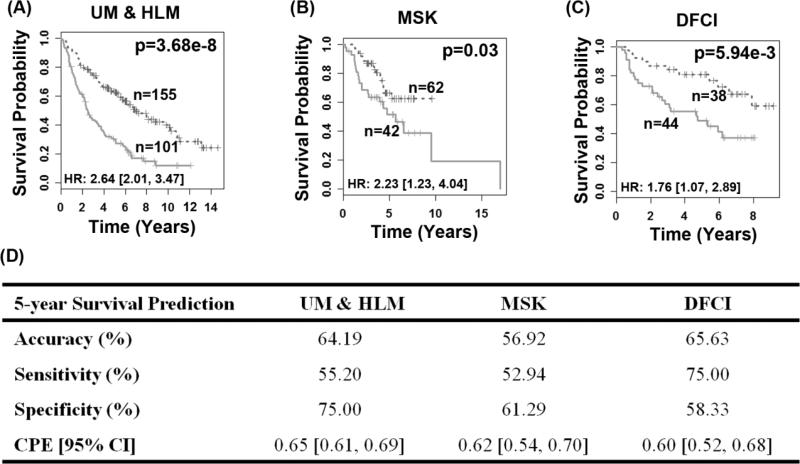
Survival prediction using the identified 10-gene prognostic signature
The identified 21 gene signatures had similar prognostic performance. As an example, a prognostic evaluation of a 10-gene signature identified using the third approach (S13; Supplementary Table 5) was provided.
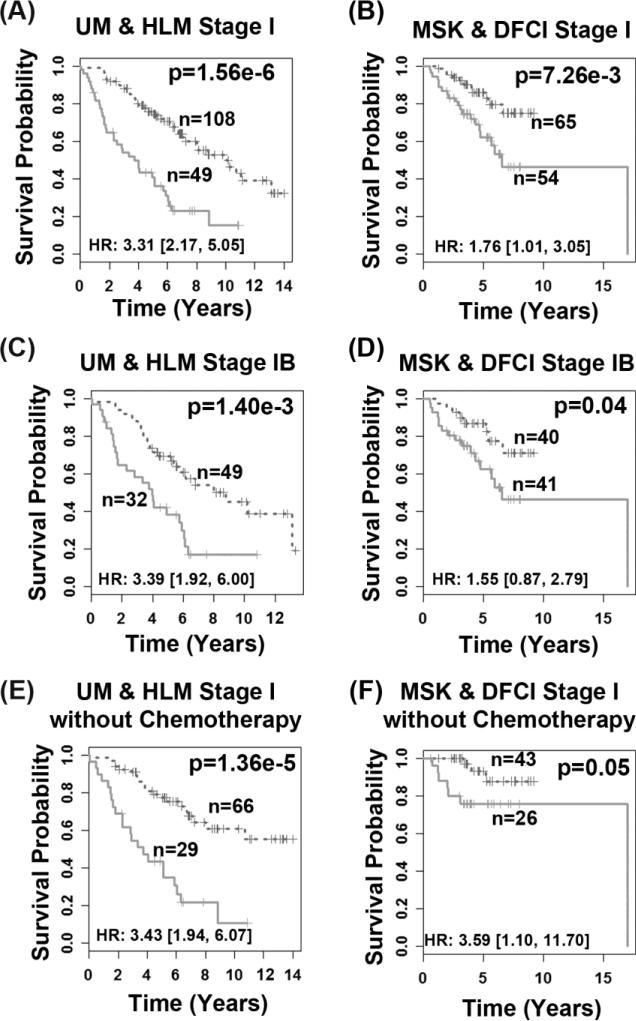
A multivariate Cox proportional hazard model was fitted with the 10 genes as covariates on bootstrapped training samples for 1,000 times. For each gene covariate, the average of the 1,000 bootstrapped coefficients was used in the training model. Using the training model, a survival risk score was generated for each patient. A risk score of -12.04 was identified as a cutoff value for patient stratification in the training set (Fig. 2A). This training model and cut-off value was then applied to the two validation sets (Fig. 2B-2C). In all three patient cohorts, this scheme stratified patients into prognostic groups with distinct post-operative overall survival (log-rank P < 0.03, Kaplan-Meier analyses). When the high-risk group is defined as a group of patients who died within 5 years, and the low-risk group is designated as a group of patients who survived 5 years or longer, this model achieved sensitivity (correctly predicted high-risk patients) of 55.20% on the training set, 52.94% on MSK, and 75% on DFCI. The specificity (correctly predicted low-risk patients) was 75% on the training set, 61.29 % on MSK, and 58.33% on DFCI (Fig. 2D). Furthermore, the 10-gene model could identify high-risk patients with stage I (log-rank P ≤ 0.007; Fig. 3A-3B) or stage IB (log-rank P ≤ 0.04; Fig. 3C-3D) cancers on both the training set and combined test sets. In stage I patients who did not receive chemotherapy, the prognostic model stratified high- and low-risk groups with distinct survival outcome in both training and test sets (log-rank P ≤ 0.05; Fig. 3E-3F). These results demonstrate that the 10-gene signature provides a more refined prognosis than the current AJCC staging system. Using this model, patients with stage I NSCLC could be advised to either receive or be spared from chemotherapy according to the expression profiles of the 10 prognostic genes.
Figure 2. Prognostication of disease-specific survival using the 10-gene signature in all stage lung adenocarcinoma patients.
The model stratified patients into two prognostic groups with significantly different (P < 0.03) survival outcome in the training set UM&HLM (A) and both test sets MSK (B) and DFCI (C) in Kaplan-Meier analyses. Log-rank tests were used to assess the difference in survival probability between the two prognostic groups. Performance of 5-year survival prediction on training and two test sets (D).
Figure 3. Prognostic performance of the 10-gene signature in stage I lung adenocarcinoma.
The model generated significant prognostic categorization for stage I patients in both training set UM&HLM (A) and combined test sets MSK&DFCI (B), for stage IB patients in training (C) and combined test sets (D), as well as for stage I patients without receiving chemotherapy in both training (E) and combined test sets (F). Statistical significance of the difference in survival probability between the two prognostic groups was assessed with log-rank tests in Kaplan-Meier analyses.
The constructed 10-gene risk score algorithm was evaluated using clinical factors, including lung cancer prognostic factors, and by using multivariate Cox analysis on the combined testing cohorts (Table 1). Without the 10-gene risk score, tumor stage was the only significant predictor of death from lung cancer (age was borderline significant). After the 10-gene risk score was included, the gene risk score became a highly significant prognostic factor with a hazard ratio of 3.23 (95% CI: [1.48, 7.06]). The hazard ratio of the gene risk score was higher than other clinical covariates, except cancer stage (III vs. I; with no significant difference). Similar results were obtained from a multivariate analysis of age, gender, cancer stage, and the gene risk score (Supplementary Table 6). These results demonstrate that the 10-gene signature is a more accurate prognostic factor than most commonly used clinical factors.
Table 1.
Multivariate Cox proportional analysis of all available clinical covariates and 10-gene risk score in the combined test cohorts (MSK and DFCI).
| Variable* | P-value | Hazard Ratio (95% CI)ψ |
|---|---|---|
| Analysis without 10-gene risk score | ||
| Gender (Male) | 0.43 | 1.22 (0.74,1.99) |
| Age at diagnosis (>60) | 0.05 | 1.70 (0.99,2.92) |
| Race | ||
| Others/Unknown | 0.28 | 0.43 (0.09,1.97) |
| White | 0.10 | 0.28 (0.06,1.28) |
| Smoking history | (0.00,0.00) | |
| Smokers | 0.62 | 0.84 (0.43,1.66) |
| Unknown | 0.91 | 0.89 (0.11,7.10) |
| Tumor differentiation | ||
| Moderately differentiated | 0.14 | 0.53 (0.23,1.24) |
| Poorly differentiated | 0.70 | 1.17 (0.53,2.61) |
| Cancer Stage | ||
| Stage II | 3.31E-04 | 2.72 (1.57,4.69) |
| Stage III | 2.38E-05 | 4.93 (2.35,10.33) |
| Analysis with 10-gene risk score | ||
| Gender (Male) | 0.37 | 1.25 (0.76, 2.04) |
| Age at diagnosis (>60) | 0.05 | 1.69 (0.99, 2.89) |
| Race | ||
| Others/ Unknown | 0.20 | 0.37 (0.08, 1.67) |
| White | 0.10 | 0.28 (0.06, 1.25) |
| Smoking history | ||
| Smokers | 0.81 | 0.92 (0.47, 1.80) |
| Unknown | 0.87 | 1.18 (0.15, 9.64) |
| Tumor differentiation | ||
| Moderately differentiated | 0.13 | 0.52 (0.23, 1.21) |
| Poorly differentiated | 0.81 | 1.10 (0.50, 2.41) |
| Cancer Stage | ||
| Stage II | 4.19E-04 | 2.66 (1.54, 4.58) |
| Stage III | 3.47E-05 | 4.79 (2.28, 10.05) |
| 10-gene risk score | 3.31E-03 | 3.23 (1.48, 7.06) |
Gender was a binary variable (0 for female and 1 for male); age at diagnosis was a binary variable (0 for < 60 years old and 1 otherwise); race was a categorical variable of 3 categories (African American [as the reference group], White, and Others [composed of Asian (5), Hawaiian or Pacific Islander (1), and unknown]); tumor grade was categorical variable of 3 categories (Well [as the reference group], Moderately, and Poorly differentiated); Smoking history was a categorical variable of 3 categories (Non-smokers, Smokers, and Unknown); cancer stage was a categorical variable with 3 categories (Stage I [as the reference group], Stage II, and Stage III).
denotes confidence interval.
Discussion
Lung cancer is a dynamic and diverse disease and associated with numerous somatic mutations, deletion and amplification events. The heterogeneous nature of lung cancer makes it a very difficult disease in the clinical managements. Tumor recurrence and metastasis is the major treatment failure and death of lung cancer. It remains a critical issue to reliably identify specific patients at high risk for recurrence and metastasis of lung cancer. Molecular prediction is a necessary step in the future direction of personalized cancer care.
This study presents a novel network-based methodology for modeling coexpression with major NSCLC signaling hallmarks for biomarker identification. Using this network-based approach, we previously identified a 14-gene (30) and a 13-gene signatures (25) with significant prognostic performance in patients with all cancer stages. Nevertheless, these two signatures did not generate significant stratification in stage I patients in all evaluated patient cohorts. Because tumors utilize different signaling pathways, we hypothesize that including a diverse set of pathways would perform more uniformly across heterogeneous tumor sets, particular, in stage I tumors. In this study, we used different combinations of the 11 NSCLC signaling hallmarks for the identification of co-expressed gene signatures. Based on the evaluation results of the prognostic performance, using a combination of 6 or 7 hallmarks could identify gene signatures with significant stratification in all patients, stage I patients, including those not receiving chemotherapy.
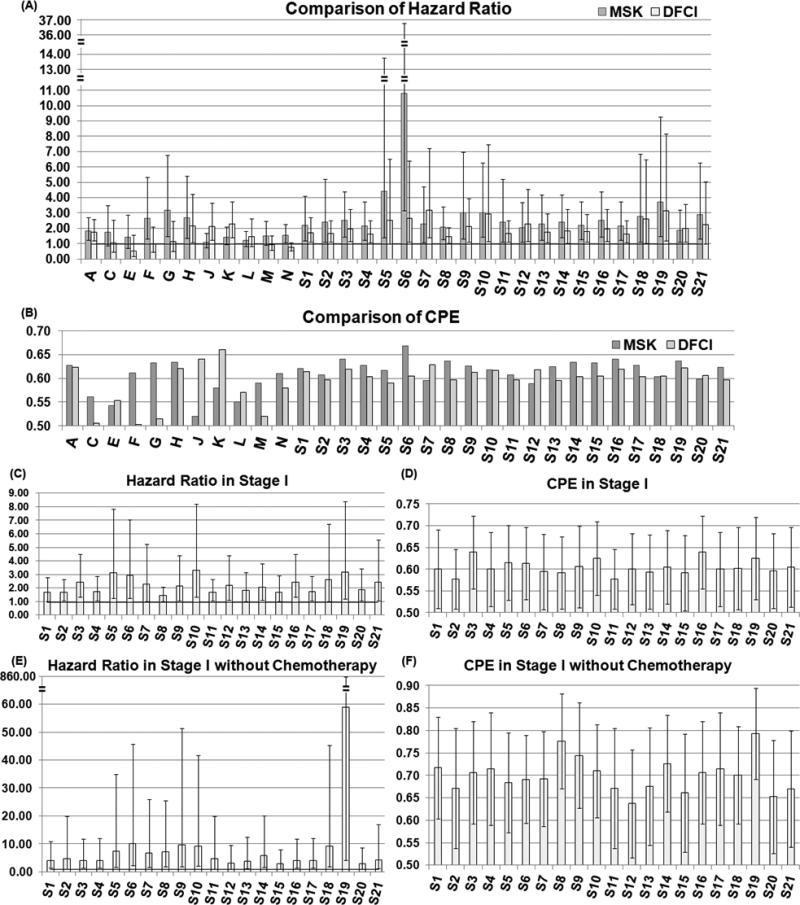
All 21 gene signatures identified in this study outperformed other lung cancer signatures reported in the literature on the same multi-center patient cohorts, not only in all tumor stages, but also in stage I patients and in stage I patients not receiving chemotherapy. Specifically, 11 lung cancer prognostic signatures were evaluated in the Director's Challenge Study (13) and the best signature was reported as “method A” (A in Fig. 4), which contains about 9,591 genes. These 11 signatures were identified using traditional statistical and machine learning methods. In comparison with the 11 gene signatures, our 21 gene signatures and “method A” are the only models with significant hazard ratio in all three patient cohorts (Fig. 4A). Furthermore, all 21 gene signatures had a significant hazard ratio and a concordance probability estimates (CPE) greater than 0.5 in stage I patients (Fig. 4C-4D), and stage I patients without receiving chemotherapy (Fig. 4E-4F), a prognostic capacity which has not been reported in previous studies (10;12). More importantly, the size of the 21 gene signatures ranges from 3 to 33 genes, which is feasible for clinical application. Previous gene signatures were identified using traditional rank-based gene selection algorithms, which did not account for complex molecular coexpressions and involvements of signaling pathways in lung cancer progression. Our study results indicate that identifying genes concurrently co-expressed with multiple NSCLC signaling pathways could enhance prognostic values. The identified biomarkers could reveal potential mechanisms underlying metastasis (see functional pathway analyses, Supplementary, Fig. 1-2). The novel implication networks could successfully model the disease-specific coexpression relations among signature genes (see disease-specific coexpression networks, Supplementary Fig. 3-23).
Figure 4. Comparison of 21 identified gene signatures with other lung cancer gene signatures.
The 21 prognostic gene signatures were compared with 11 gene signatures evaluated in the Director's Challenge Study (12) in two test sets in terms of hazard ratio (A) and concordance probability estimate [CPE] (B). The prognostic performance of the 21 gene signatures was evaluated for stage I patients by hazard ratio (C) and CPE (D), as well as for stage I patients without receiving chemotherapy in the combined test cohorts (E, F). The error bar in the charts represents 95% confidence interval of the measurement. Signatures A, C-N were from the Director's Challenge Study (12), and the details were summarized in Table S1 in our previous study (25).
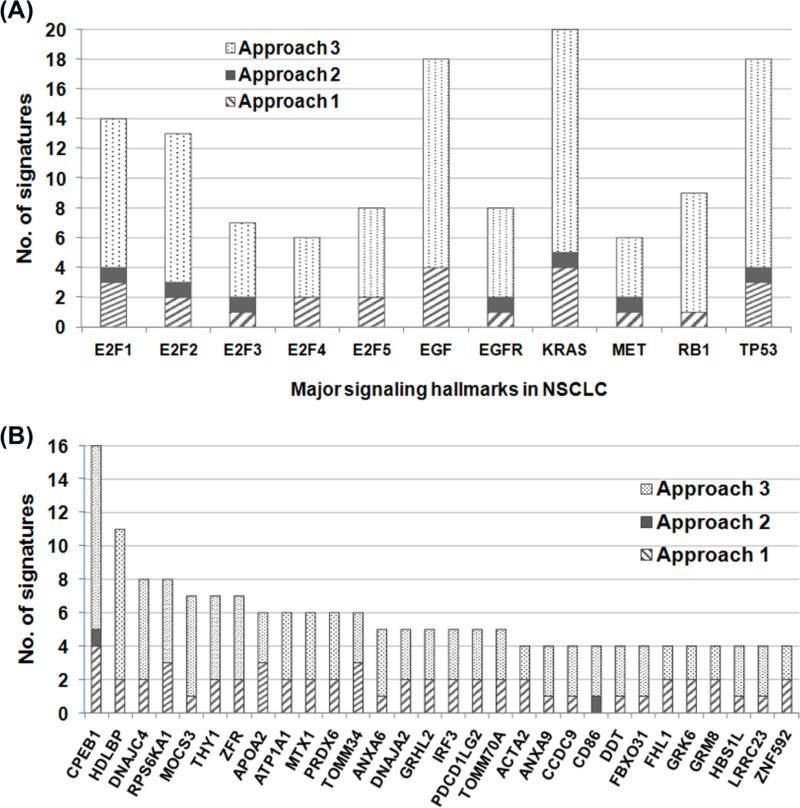
Summary of signaling hallmarks from the 21 signatures suggests that genes co-regulated with KRAS, EGF, or TP53 tend to have more prognostic capacity for lung cancer in the genomic space (Fig. 5A). Among the 132 marker genes of the 21 signatures, cytoplasmic polyadenylation element binding protein 1 (CPEB1) was present in 16 signatures (Fig 5B). CPEB1 regulates beta-catenin mRNA translation and cell migration (31), as well as human cellular senescence and bioenergetics by modulating p53 mRNA polyadenylation-induced translation (32). Our results imply that CPEB1 is an important prognostic biomarker for lung cancer and might be involved in cancer progression and metastasis.
Figure 5. Summary of major NSCLC signaling hallmarks involved in the signature identification and overlapping genes in the 21 prognostic gene signatures.
The number of gene signatures coexpressed with each signaling hallmark was summarized in (A). Genes appeared in at least 4 identified signatures were listed in (B), with the frequency that a gene was selected to form a prognostic signature using three approaches presented in this study.
Based on the current study results, the potential down-stream clinical applications could utilize a customized Affymetrix U133A array to contain the identified signature genes for prognostic categorization, similar to the development of a commercial prognostic gene test for breast cancer, MammaPrint© (33;34). In this case, the identified gene expression-based prognostic models could be used directly in prospective evaluation and future clinical applications. Alternatively, quantitative RT-PCR assays could be used to validate and refine the identified gene signatures for clinical applications, which is an approach taken in the development of another commercial prognostic gene product for breast cancer, Oncotype DX© (35).
Conclusion
This study demonstrates that modeling disease-mediated coexpression networks and crosstalk with major signaling hallmarks is key to identifying clinically important prognostic biomarkers from the genomic space. We believe that this approach is the most promising for effectively developing reliable and clinically useful marker panels. The identified 21 gene signatures could be used to predict the risk of tumor recurrence and advise patient selection for adjuvant chemotherapy, with significant impact on down-stream clinical applications.
Supplementary Material
Acknowledgements
We are grateful for the editing help from Rebecca Raese at West Virginia University.
Funding: This study was funded by NIH/NLM R01LM009500 (PI: Guo) and NIH/NCRR P20RR16440 and Supplement (PD: Guo).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest statement
The authors declare no conflict of interest including any financial, personal or other relationships with other people or organizations that could inappropriately influence (bias) this work.
Reference List
- 1.Hoffman PC, Mauer AM, Vokes EE. Lung cancer. Lancet. 2000 Feb 5;355(9202):479–85. doi: 10.1016/S0140-6736(00)82038-3. [DOI] [PubMed] [Google Scholar]
- 2.Naruke T, Goya T, Tsuchiya R, Suemasu K. Prognosis and survival in resected lung carcinoma based on the new international staging system. J.Thorac.Cardiovasc.Surg. 1988 Sep;96(3):440–7. [PubMed] [Google Scholar]
- 3.General Thoracic Surgery. Seventh ed. Lippincott Williams & Wilkins; Philadelphia: 2009. [Google Scholar]
- 4.Dalton WS, Friend SH. Cancer biomarkers--an invitation to the table. Science. 2006 May 26;312(5777):1165–8. doi: 10.1126/science.1125948. [DOI] [PubMed] [Google Scholar]
- 5.Subramanian J, Simon R. Gene expression-based prognostic signatures in lung cancer: ready for clinical use? J.Natl.Cancer Inst. 2010 Apr 7;102(7):464–74. doi: 10.1093/jnci/djq025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beer DG, Kardia SL, Huang CC, Giordano TJ, Levin AM, Misek DE, Lin L, Chen G, Gharib TG, Thomas DG, et al. Gene-expression profiles predict survival of patients with lung adenocarcinoma. Nat.Med. 2002 Aug;8(8):816–24. doi: 10.1038/nm733. [DOI] [PubMed] [Google Scholar]
- 7.Bhattacharjee A, Richards WG, Staunton J, Li C, Monti S, Vasa P, Ladd C, Beheshti J, Bueno R, Gillette M, et al. Classification of human lung carcinomas by mRNA expression profiling reveals distinct adenocarcinoma subclasses. Proc.Natl.Acad.Sci.U.S.A. 2001 Nov 20;98(24):13790–5. doi: 10.1073/pnas.191502998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bild AH, Yao G, Chang JT, Wang Q, Potti A, Chasse D, Joshi MB, Harpole D, Lancaster JM, Berchuck A, et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature. 2006 Jan 19;439(7074):353–7. doi: 10.1038/nature04296. [DOI] [PubMed] [Google Scholar]
- 9.Borczuk AC, Kim HK, Yegen HA, Friedman RA, Powell CA. Lung adenocarcinoma global profiling identifies type II transforming growth factor-beta receptor as a repressor of invasiveness. Am.J.Respir.Crit Care Med. 2005 Sep 15;172(6):729–37. doi: 10.1164/rccm.200504-615OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen HY, Yu SL, Chen CH, Chang GC, Chen CY, Yuan A, Cheng CL, Wang CH, Terng HJ, Kao SF, et al. A five-gene signature and clinical outcome in non-small-cell lung cancer. N.Engl.J.Med. 2007 Jan 4;356(1):11–20. doi: 10.1056/NEJMoa060096. [DOI] [PubMed] [Google Scholar]
- 11.Raponi M, Zhang Y, Yu J, Chen G, Lee G, Taylor JM, Macdonald J, Thomas D, Moskaluk C, Wang Y, et al. Gene expression signatures for predicting prognosis of squamous cell and adenocarcinomas of the lung. Cancer Res. 2006 Aug 1;66(15):7466–72. doi: 10.1158/0008-5472.CAN-06-1191. [DOI] [PubMed] [Google Scholar]
- 12.Shedden K, Taylor JM, Enkemann SA, Tsao MS, Yeatman TJ, Gerald WL, Eschrich S, Jurisica I, Giordano TJ, Misek DE, et al. Gene expression-based survival prediction in lung adenocarcinoma: a multi-site, blinded validation study. Nat.Med. 2008 Aug;14(8):822–7. doi: 10.1038/nm.1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baker SG, Kramer BS, Srivastava S. Markers for early detection of cancer: statistical guidelines for nested case-control studies. BMC.Med.Res.Methodol. 2002;2:4. doi: 10.1186/1471-2288-2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Emir B, Wieand S, Su JQ, Cha S. Analysis of repeated markers used to predict progression of cancer. Stat.Med. 1998 Nov 30;17(22):2563–78. doi: 10.1002/(sici)1097-0258(19981130)17:22<2563::aid-sim952>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 15.Pepe MS, Janes H, Longton G, Leisenring W, Newcomb P. Limitations of the odds ratio in gauging the performance of a diagnostic, prognostic, or screening marker. Am.J.Epidemiol. 2004 May 1;159(9):882–90. doi: 10.1093/aje/kwh101. [DOI] [PubMed] [Google Scholar]
- 16.Hartwell LH, Hopfield JJ, Leibler S, Murray AW. From molecular to modular cell biology. Nature. 1999 Dec 2;402(6761 Suppl):C47–C52. doi: 10.1038/35011540. [DOI] [PubMed] [Google Scholar]
- 17.Chuang HY, Lee E, Liu YT, Lee D, Ideker T. Network-based classification of breast cancer metastasis. Mol.Syst.Biol. 2007;3:140. doi: 10.1038/msb4100180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Csermely P, Agoston V, Pongor S. The efficiency of multi-target drugs: the network approach might help drug design. Trends Pharmacol.Sci. 2005 Apr;26(4):178–82. doi: 10.1016/j.tips.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 19.Sotiriou C, Piccart MJ. Taking gene-expression profiling to the clinic: when will molecular signatures become relevant to patient care? Nat.Rev.Cancer. 2007 Jul;7(7):545–53. doi: 10.1038/nrc2173. [DOI] [PubMed] [Google Scholar]
- 20.Mani KM, Lefebvre C, Wang K, Lim WK, Basso K, la-Favera R, Califano A. A systems biology approach to prediction of oncogenes and molecular perturbation targets in B-cell lymphomas. Mol.Syst.Biol. 2008;4:169. doi: 10.1038/msb.2008.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cui Q, Ma Y, Jaramillo M, Bari H, Awan A, Yang S, Zhang S, Liu L, Lu M, O'Connor-McCourt M, et al. A map of human cancer signaling. Mol.Syst.Biol. 2007;3:152. doi: 10.1038/msb4100200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li C. Automating dChip: toward reproducible sharing of microarray data analysis. BMC.Bioinformatics. 2008;9:231. doi: 10.1186/1471-2105-9-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guo L, Cukic B, Singh H. Predicting Fault Prone Modules by the Dempster-Shafer Belief Networks.. Proceedings of 18th IEEE International Conference on Automated Software Engineering (ASE'03).; 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hildebrand DK, Laing JD, Rosenthal H. Prediction Analysis of Cross Classifications. John Wiley & Sons; 1977. [Google Scholar]
- 25.Guo NL, Wan YW, Bose S, Denvir J, Kashon ML, Andrew ME. A novel network model identified a 13-gene lung cancer prognostic signature. Int.J.Comput.Biol.Drug Des. 2011;4(1):19–39. doi: 10.1504/IJCBDD.2011.038655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Breiman L. Random Forests. Machine Learning. 2001;45:5–32. [Google Scholar]
- 27.Diaz-Uriarte R, Alvarez dA. Gene selection and classification of microarray data using random forest. BMC.Bioinformatics. 2006;7:3. doi: 10.1186/1471-2105-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hall MA, Holmes G. Benchmarking Attribute Selection Techniques for Discrete Class Data Mining. IEEE Transactions on Knowledge and Data Engineering. 2003;15(3):1437–47. [Google Scholar]
- 29.Witten IH, Frank E. Data Mining: Practical Machine Learning Tools and Techniques. 2nd Edition Morgan Kaufmann; 2005. [Google Scholar]
- 30.Wan YW, Bose S, Denvir J, Guo NL. A Novel Network Model for Molecular Prognosis.. Proc.ACM International Conference on Bioinformatics and Computational Biology.; 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jones KJ, Korb E, Kundel MA, Kochanek AR, Kabraji S, McEvoy M, Shin CY, Wells DG. CPEB1 regulates beta-catenin mRNA translation and cell migration in astrocytes. Glia. 2008 Oct;56(13):1401–13. doi: 10.1002/glia.20707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Burns DM, Richter JD. CPEB regulation of human cellular senescence, energy metabolism, and p53 mRNA translation. Genes Dev. 2008 Dec 15;22(24):3449–60. doi: 10.1101/gad.1697808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van 't Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, Peterse HL, van der KK, Marton MJ, Witteveen AT, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002 Jan 31;415(6871):530–6. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 34.van de Vijver MJ, He YD, van 't Veer LJ, Dai H, Hart AA, Voskuil DW, Schreiber GJ, Peterse JL, Roberts C, Marton MJ, et al. A gene-expression signature as a predictor of survival in breast cancer. N.Engl.J.Med. 2002 Dec 19;347(25):1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 35.Paik S, Shak S, Tang G, Kim C, Baker J, Cronin M, Baehner FL, Walker MG, Watson D, Park T, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N.Engl.J.Med. 2004 Dec 30;351(27):2817–26. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.