Abstract
A remarkable amount of theoretical research has been carried out to elucidate the physical origins of the recently observed long-lived quantum coherence in the electronic energy transfer process in biological photosynthetic systems. Although successful in many respects, several widely used descriptions only include an effective treatment of the protein-chromophore interactions. In this work, by combining an all-atom molecular dynamics simulation, time-dependent density functional theory, and open quantum system approaches, we successfully simulate the dynamics of the electronic energy transfer of the Fenna-Matthews-Olson pigment-protein complex. The resulting characteristic beating of populations and quantum coherences is in good agreement with the experimental results and the hierarchy equation of motion approach. The experimental absorption, linear, and circular dichroism spectra and dephasing rates are recovered at two different temperatures. In addition, we provide an extension of our method to include zero-point fluctuations of the vibrational environment. This work thus presents, to our knowledge, one of the first steps to explain the role of excitonic quantum coherence in photosynthetic light-harvesting complexes based on their atomistic and molecular description.
Introduction
Recent experiments suggest the existence of long-lived quantum coherence during the electronic energy transfer process in photosynthetic light-harvesting complexes under physiological conditions (1–3). This has stimulated many researchers to seek for the physical origin of such a phenomenon. The role and implication of quantum coherence during the energy transfer have been explored in terms of the theory of open quantum systems (4–16), and in the context of quantum information and entanglement (17–20). However, the characteristics of the protein environment, and especially its thermal vibrations or phonons, have not been fully investigated from the molecular viewpoint. A more detailed description of the bath in atomic detail is desirable to investigate the structure-function relationship of the protein complex and to go beyond the assumptions used in popular models of photosynthetic systems.
Protein complexes constitute one of the most essential components in every biological organism. They remain one of the major targets of biophysical research due to their tremendously diverse and, in some cases, still unidentified structure-function relationship. Many biological units have been optimized through evolution and the presence of certain amino acids rather than others is fundamental for functionality (21–23). In photosynthesis, one of the most well-characterized pigment-protein complexes is the Fenna-Matthews-Olson (FMO) complex, which is a light-harvesting complex found in green sulfur bacteria. It functions as an intermediate conductor for exciton transport located between the antenna complex where light is initially absorbed and the reaction center. Since the resolution of its crystal structure more than 30 years ago (24), the FMO trimer, composed of three units each comprising eight bacteriochlorophylls, has been extensively studied both experimentally (25–28) and theoretically (29,30). For instance, regarding the structure-function relationship, it has been shown (31) that amino-acid residues cause considerable shifts in the site energies of bacteriochlorophyll a (BChl) molecules of the FMO complex and in turn causes changes to the energy transfer properties.
Have photosynthetic systems adopted interesting quantum effects to improve their efficiency in the course of evolution, as suggested by the experiments? In this article, we provide a first step to answer this question by characterizing the protein environment of the FMO photosynthetic system to identify the microscopic origin of the long-lived quantum coherence. We investigate the quantum energy transfer of a molecular excitation (exciton) by incorporating an all-atom molecular dynamics (MD) simulation. The molecular energies are computed with time-dependent density functional theory (TDDFT) along the MD trajectory. The evolution of the excitonic density matrix is obtained as a statistical ensemble of unitary evolutions by a time-dependent Schrödinger equation. Thus, this work is in contrast to many studies based on quantum master equations in that it includes atomistic detail of the protein environment into the dynamical description of the exciton. We also introduce a novel, to our knowledge, approach to add quantum corrections to the dynamics. Furthermore, a quantitative comparison to the hierarchical equation of motion and the Haken-Strobl-Reineker (HSR) method is presented. As the main result, the time evolution of coherences and populations shows characteristic beatings on the timescale of the experiments. Surprisingly, we observe that the cross correlation of site energies does not play a significant role in the energy transfer dynamics.
The article is structured as follows. The partitioning of the system and bath Hamiltonian in classical and quantum degrees of freedom and details of the MD simulations and calculation of site energies are discussed in Molecular Dynamics Simulations. The exciton dynamics of the system under the bath fluctuations is then presented in Exciton Dynamics. In Quantum Jump Correction to MD Method, we introduce a quantum correction to the previous exciton dynamics. Using the discussed methods, we evaluated site energies and their distribution at 77 and 300 K in Site Energy Distributions. We also computed the linear absorption spectrum of the FMO complex in Simulated Spectra. The site basis dephasing rates are discussed in their own subsection. From the exciton dynamics of the system, we obtained populations and coherences and compared these to the quantum jump correction to molecular dynamics (QJC-MD) approach in Population Dynamics and Long-Lived Quantum Coherence. We then compare the MD and quantum-corrected MD methods to the hierarchical equation of motion (HEOM) and Haken-Strobl-Reineker (HSR) methods in Comparison Among MD, QJC-MD, HEOM, and HSR Methods. In Correlation Functions and Spectral Density, we determined the spectral density for each site from the energy time bath-correlator and studied the effect of auto and cross correlations on the exciton dynamics by introducing a comparison to first-order autoregressive processes. We then summarize our results and present our conclusions.
Theoretical and Computational Approach
Molecular dynamics simulations
A computer simulation of the quantum evolution of the entire FMO complex is certainly unfeasible with the currently available computational resources. However, we are only interested in the electronic energy transfer dynamics among BChl molecules embedded in the protein support. This suggests a decomposition of the total system Hamiltonian operator into three parts: the relevant system, the bath of vibrational modes, and the system-bath interaction Hamiltonians. The system Hamiltonian operates on the excitonic system alone, which is defined by a set of two-level systems. Each two-level system represents the ground and first excited electronic state of a BChl molecule. In addition, the quantum mechanical state of the exciton is assumed to be restricted to the single-exciton manifold because the exciton density is low. On the other hand, factors affecting the system site energies have intractably large degrees of freedom, so it is reasonable to treat all those degrees of freedom as the bath of an open quantum system.
More formally, to describe the system-bath interplay by including atomistic detail of the bath, we start from the total Hamiltonian operator and decompose it in a general way such that no assumptions on the functional form of the system-bath Hamiltonian are necessary (32):
| (1) |
Here, R corresponds to the nuclear coordinates of the FMO complex including BChl molecules, protein, and enclosing water molecules. The set of states |m〉⊗|R〉 denote the presences of the exciton at site m given that the FMO complex is in the configuration R, εm(R) represents the site energy of the mth site, and Jmn(R) is the coupling constant between the mth and nth sites. Note that the site energies and coupling terms can be modulated by R. |1〉〈1| is the identity operator in the excitonic subspace, is the kinetic operator for the nuclear coordinates of the FMO complex, and Vm(R) is the potential energy surface for the complex when the exciton is at site m under Born-Oppenheimer approximation. Given multiple Born-Oppenheimer surfaces, one would need to carry out a coupled nonadiabatic propagation. However, as a first approximation, we assume that the change of Born-Oppenheimer surfaces does not affect the bath dynamics significantly. This approximation becomes better at small reorganization energies. Indeed, BChl molecules have significantly smaller reorganization energies than other chromophores (33). With this assumption, we can ignore the dependence on the excitonic state in the V term, thus the system-bath Hamiltonian only contains the one-way influence from the bath to the system. We also adopted the Condon approximation so that the J terms do not depend on R:
| (2) |
Based on this decomposition of the total Hamiltonian, we set up a model of the FMO complex with the AMBER 99 force field (34,35) and approximate the dynamics of the protein complex bath by classical mechanics. The initial configuration of the MD simulation was taken from the x-ray crystal structure of the FMO complex of Prosthecochloris aestuarii (PDB ID: 3EOJ.). Shake constraints were used for all bonds containing hydrogen, and the cutoff distance for the long-range interaction was chosen to be 12 Å. After a 2-ns-long equilibration run, the production run was obtained for a total time of 40 ps with a 2-fs timestep. For the calculation of the optical gap, snapshots were taken every 4 fs. Two separate simulations at 77 K and 300 K were carried out with an isothermal-isobaric (NPT) ensemble to investigate the temperature dependence of the bath environment. Then, parameters for the system and the system-bath Hamiltonian were calculated using quantum chemistry methods along the trajectory obtained from the MD simulations.
We chose not to include the newly resolved eighth BChl molecule (31) in our simulations because, up to now, the large majority of the scientific community has focused on the seven-site system that is, therefore, a better benchmark to compare our calculations to previous work. It is important to note, however, that this eighth site may have an important role in the dynamics. In particular, as suggested in Ritschel et al. (36) and Moix et al. (37), this eighth site is considered to be the primary entering point for the exciton in the FMO complex and its position dictates a preferential exciton transport pathway rather than two independent ones. Also when starting with an exciton on this eighth site, the oscillations in the coherences are largely suppressed.
The time-dependent site energy ϵm was evaluated as the excitation energy of the Qy transition of the corresponding BChl molecule. We employed the time-dependent density functional theory (TDDFT) with BLYP functional within the Tamm-Dancoff approximation (TDA) using the Q-Chem quantum chemistry package (38). The basis set was chosen to be 3-21G after considering a trade-off between accuracy and computational cost. The Qy transition was identified as the excitation with the highest oscillator strength among the first 10 singlet excited states. Then, the transition dipole of the selected state was verified to be parallel to the y molecular axis. Every atom that did not belong to the TDDFT target molecule was incorporated as a classical point charge to generate the external electric field for the quantum mechanical/molecular mechanical (QM/MM) calculation. Given that the separation between BChl molecules and the protein matrix is quite clear, employing this simple QM/MM method with classical external charges to calculate the site energies is a good approximation. The external charges were taken from the partial charges of the AMBER force field (34,35). The coupling terms, Jmn, can also be obtained from quantum chemical approaches like transition density cube or fragment-excitation difference methods (39,40). However, in this case we employed the MEAD values of the couplings of the Hamiltonian presented in Adolphs and Renger (30) and considered them to be constant in time. The value was straightforwardly chosen as the time-averaged site energy for the mth site.
Exciton dynamics
In this section, we describe the method for the dynamics of the excitonic reduced density matrix within our molecular dynamic simulation framework. It is based on a simplified version of the quantum-classical hybrid method described in May and Kühn (32). The additional assumption on Hamiltonian (2) is that the bath coordinate R is a classical variable, denoted by a superscript cl. As discussed above, the time-dependence of these variables arises from the Newtonian MD simulations. The additional force on the nuclei due to the electron-phonon coupling (32) is neglected. Hence, the Schrödinger equation for the excitonic system is given by
| (3) |
The system-environment coupling leads to an effective time-dependent Hamiltonian Heff(t) = HS + HSB(Rcl(t)). This equation suggests a way to propagate the reduced density matrix as an average of unitary evolutions given by Eq. 3. First, short MD trajectories (in our case, 1 ps long) are uniformly sampled from the full MD trajectory (40 ps). Then, for each short MD trajectory, the excitonic system can be propagated under unitary evolution with a simple time-discretized exponential integrator. The density matrix is the classical average of these unitary evolutions,
| (4) |
where M is the number of sample short trajectories. Each trajectory is subject to different time-dependent fluctuations from the bath, which manifests itself as decoherence when averaged to the statistical ensemble. Compared to many methods based on the stochastic unraveling of the master equation, e.g., Dalibard et al. (41) and Piilo et al. (42), our formalism directly utilizes the fluctuations generated by the MD simulation. Therefore, the detailed interaction between system and bath is captured. The temperature of the bath is set by the thermostat of the MD simulation, thus no further explicit temperature dependence is required in the overall dynamics. The dynamics obtained by this numerical integration of the Schrödinger equation will also be compared to the HEOM approach. The HEOM is briefly described in the Supporting Material along with a discussion on the differences with respect to the MD method.
Quantum jump correction to MD method
The MD/TDDFT simulation above leads to crucial insights into the exciton dynamics. However, it does not capture quantum properties of the vibrational environment such as zero-point fluctuations. At zero temperature all the atoms in the MD simulation are completely frozen. Moreover, similarly to an infinite-temperature model, at long times of the quantum dynamical simulation the exciton is evenly distributed among all molecules, as we will see below. To obtain a more realistic description, we modify the stochastic simulation by introducing quantum jumps derived from the zero-point (zp) fluctuations of the modes in the vibrational environment. We refer to this corrected version of the MD propagation as “quantum jump correction to molecular dynamics” (QJC-MD).
Introducing harmonic bath modes, explicitly we reformulate the system-bath Hamiltonian as
| (5) |
Here, each gmξ represents the coupling strength of a site m to a particular mode ξ and bξ(b†ξ) is the annihilation (creation) operator for that mode and the usual commutation relations hold. We now formulate our correction by separating the bath operators into two parts, ; the first part is due to zero-point fluctuations and the second comes from our MD simulations (analogous to the creation operator). The commutator relations for these newly defined operators (and their conjugate counterparts) arise naturally, given that the zp-fluctuations are quantum mechanical and the MD fluctuations are classical, i.e., and . In addition, the zp-fluctuations are not populated .
Therefore, these fluctuations can only induce excitonic transitions from higher to lower exciton states in the instantaneous eigenbasis of the Hamiltonian, thus leading to relaxation of the excitonic system. The evolution of the populations PM of the instantaneous eigenstates |M(t)〉 due to the zero-point correction is expressed by a Pauli master equation as
| (6) |
and for the coherences as
| (7) |
The associated rate can be derived from a secular Markovian Redfield theory (43) to be
where the spectral density J(ω) is only nonzero for positive transition frequencies ωMN = EM – EN and taken to be as in Eq. 8, below. The coefficients cm(M) translate from site to energy basis. The time evolution given by Eqs. 6 and 7 is included in the dynamics simulation by introducing quantum jumps as in the Monte Carlo wavefunction method (41). We thus arrive at a hybrid classically averaged H(t) simulation with additional quantum transitions induced by the vacuum fluctuations of the vibrational modes.
Results and Discussion
Site energy distributions
Using the coupled QM/MD simulations, site energies were obtained for each BChl molecule. These energies and their fluctuations are reported in Fig. 1. We note that the magnitude of the fluctuations are of the order of hundreds of cm−1. Although the order of the site energies does not perfectly match previously reported results (30,44), the overall trend does not deviate much, especially considering that our result is purely based on ab initio calculations without fitting to the experimental result. The Qy transition energies calculated by TDDFT are known to be systematically blue-shifted with respect to the experiment (45). However, the scale of the fluctuations remains reasonable. Therefore, the comparison in Fig. 1 was made after shifting the overall mean energy to zero for each method.
Figure 1.
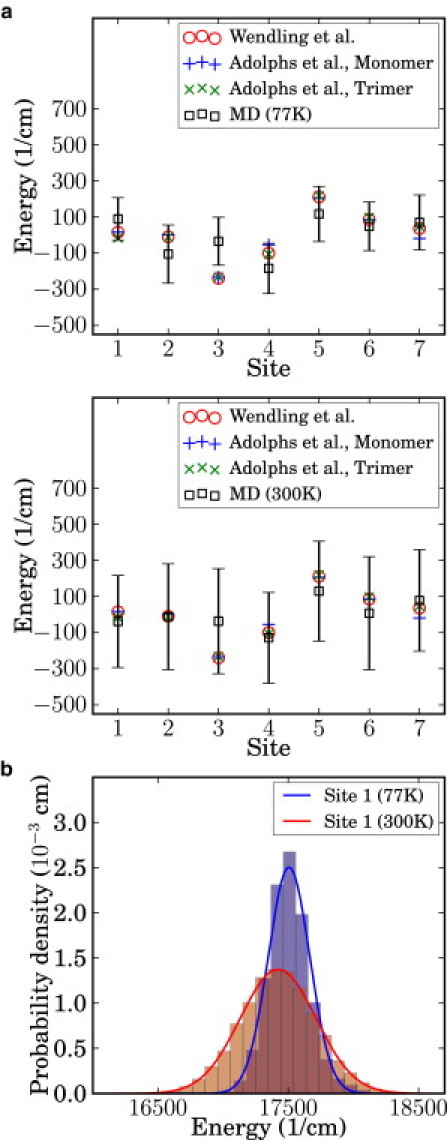
(a) Comparison of the calculated site energies for each BChl molecule to the previous works by Adolphs and Renger (30) and Wendling et al. (44). Our calculation, labeled as MD, was obtained using QM/MM calculations with the TDDFT/TDA method at 77 K and 300 K. (Vertical bars) Standard deviation for each site. (b) Marginal distribution of site 1 energy at 77 K and 300 K. (Histograms) Original data; (solid lines) estimated Gaussian distribution.
The excitation energy using TDDFT does not always converge when the configuration of the molecule deviates significantly from its ground-state structure. The number of points that failed to converge was, on average, <4% for configurations at 300 K, and <2% at 77 K. We interpolated the original time series to obtain smaller time steps and recover the missing points. Interpolation could lead to severe distortion of the marginal distribution when the number of available points is too small. However, in our case, the distributions virtually remained the same with and without interpolation.
Dephasing rates
In the Markovian approximation and assuming an exponentially decaying autocorrelation function, the dephasing rate γϕ is proportional to the variance of the site energy σ2ϵ (43),
| (8) |
where τ is a time decay parameter that we estimated through a comparison to first-order autoregressive processes, as described in Correlation Functions and Spectral Density. The dependence on the variance is clearly justified: states associated with large site energy fluctuations tend to undergo faster dephasing. Fig. 2 a presents the approximate site basis dephasing rates for each site with τ ≈ 5 fs. The averaged value of the slopes is 0.485 cm−1 K−1, which is in good agreement with the experimentally measured value of 0.52 cm−1 K−1 obtained from a closely related species Chlorobium tepidum in the exciton basis (3). From this plot, we note the presence of a positive correlation between temperature and dephasing rate. This correlation is plausible: as temperature increases so does the energy disorder, hence the coherences should decay faster. In fact, in the Markovian approximation, dephasing rates increase linearly with temperature (43,46). Calculations at other temperatures are underway to verify this and to obtain more information on the precise temperature dependence of the dephasing rates.
Figure 2.
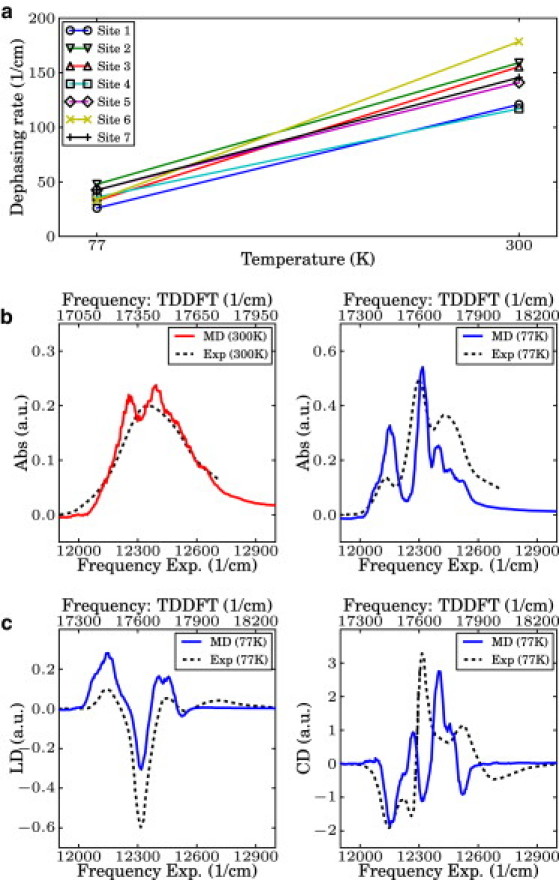
(a) Calculated dephasing rate for each site at 77 K and 300 K. (b) Simulated linear absorption spectra of the FMO complex at 77 K and 300 K. They were shifted to be compared to the experimental spectra as obtained by G. Engel (University of Chicago, personal communication, 2011). (c) Simulated linear dichroism (LD) and circular dichroism (CD) spectra at 77 K. Experimental spectra were obtained from Wendling et al. (44). Although TDDFT-calculated spectra shows systematically overestimated site energies, the width and overall shape of the spectra is in good agreement.
Simulated spectra
The absorption, linear dichroism (LD), and circular dichroism (CD) spectra can be obtained from the Fourier transform of the corresponding response functions. The spectra can be evaluated for the seven BChl molecules using the following expressions (47,48),
| (9) |
where m and n are indices for the BChl molecules in the complex, is the transition dipole moment of the mth site, Umn(t,0) is the (m,n) element of the propagator in the site basis, is the unit vector in the direction of the rotational symmetry axis, is the coordinate vector of the site m, and 〈…〉 indicates an ensemble average. The ensemble average was evaluated by sampling and averaging over 4000 trajectories. We applied a low-pass filter to smooth out the noise originated from truncating the integration and due to the finite number of trajectories. Fig. 2, b and c, shows direct comparison of the calculated and experimental spectra at 77 K and 300 K. As discussed in Site Energy Distributions, TDDFT tends to systematically overestimate the excitation energy of the Qy transition (49), yet the fluctuation widths of the site energies are reasonable. In fact, the width and overall shape of the calculated spectrum is in good agreement with the experimental spectrum at each temperature. Calculated LD and CD spectra also reproduce well the experimental measurements, considering that no calibration to experiments was carried out. Because both LD and CD spectra are sensitive to the molecular structure, it appears that our microscopic model correctly captures these details.
Population dynamics and long-lived quantum coherence
The MD method is based on minimal assumptions and directly evaluates the dynamics of the reduced density matrix from the total density matrix as described in the Methods. The reduced density matrix was obtained after averaging over 4000 trajectories. Fig. 3 shows the population and coherence dynamics of each of the seven sites according to the dephasing induced by the nuclear motion of the FMO complex. In particular, the populations and the absolute value of the pairwise coherences, as defined in Sarovar et al. (19) (2·|ρ12(t)| and 2·|ρ56(t)|), are plotted at both 77 and 300 K starting with an initial state in site 1 (first row) and then in site 6 (second row). Until very recently (36,37), sites 1 and 6 have been thought as the entry point of an exciton in the FMO complex, therefore most of the previous literature chose the initial reduced density matrix to be either pure states |1〉〈1| or |6〉〈6| (10,15,50). However, our method could be applied to any mixed initial state without modification. We note that coherent beatings last for ∼400 fs at 77 K and 200 fs at 300 K. These timescales are in agreement with those reported for FMO (3,10) and with what was found in Dephasing Rates. Although quite accurate in the short time limit, the MD method populations do not reach thermal equilibrium at long times. This was verified by propagating the dynamics to twice the time shown in Fig. 3. This final classical equal distribution is similar to the HSR model result. The three central panels of Fig. 3 show the same populations and coherences obtained from the QJC-MD method. As discussed above, this method includes a zero point correction through relaxation transitions and predicts a more realistic thermal distribution at 77 K. At 300 K, the quantum correction is less important in the dynamics because the Hamiltonian fluctuations dominate over the zero temperature quantum fluctuations.
Figure 3.
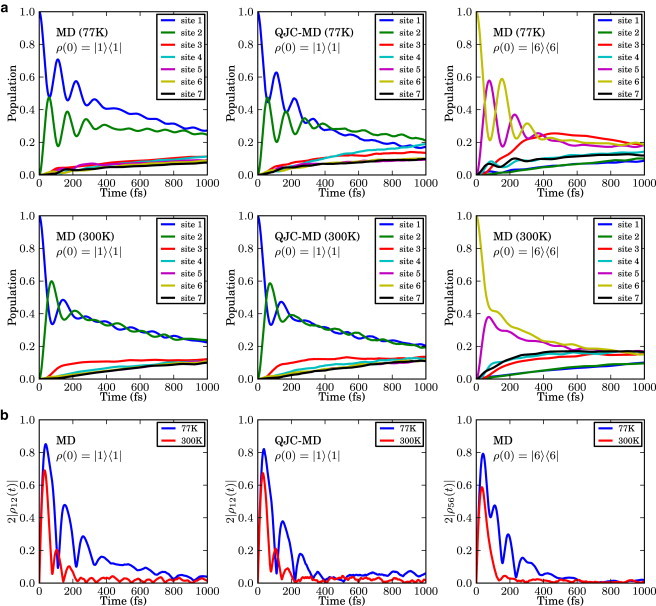
(a) Time evolution of the exciton population of each chromophore in the FMO complex at 77 K and 300 K. (b) Change of the pairwise coherence, or concurrence in time. Initial pure states, ρS(0) = |1〉〈1| for the top and center panels were propagated using the two formulations developed in this article, MD and QJC-MD, to utilize the atomistic model of the protein complex bath from the MD/TDDFT calculation. The initial state was set to |6〉〈6| and propagated using the MD method.
Comparison among MD, QJC-MD, HEOM, and HSR methods
Fig. 4 shows a direct comparison of the population dynamics of site 1 calculated using the HEOM method discussed by Ishizaki and Fleming (10,51), our MD and quantum-corrected methods at 77 K and 300 K, and the HSR model (52,53) with dephasing rates obtained from Eq. 8. We observe that the short-time dynamics and dephasing characteristics are surprisingly similar, considering that the methods originate from very different assumptions. Atomistic detail can allow for differentiation of the system-environment coupling for different chromophores. For example, at both temperatures (right panels), the MD populations of site 6 undergo faster decoherence than the corresponding HEOM results. We attribute this to the difference in energy gap fluctuations of site energy between sites 1 and 6 obtained from the MD simulation as can be seen in site 1. On one hand, in the HEOM method, site energy fluctuations are considered to be identical across all sites; on the other, in our method the fluctuations of each site are obtained from the MD simulation in which each site is associated with a different chromophore-protein coupling. Nevertheless, the fact that we obtain qualitatively similar results to the HEOM approach (at least when starting in ρ(0) = |1〉〈1|) without considering nonequilibrium reorganization processes suggests that such processes might not be dominant in the FMO. The quantum correction results (QJC-MD), for every temperature and initial state, are in between the HEOM and MD results. This is due to the induced relaxation from zero-point fluctuations of the bath environment, which are not included in the MD method but included in the QJC-MD and HEOM methods.
Figure 4.
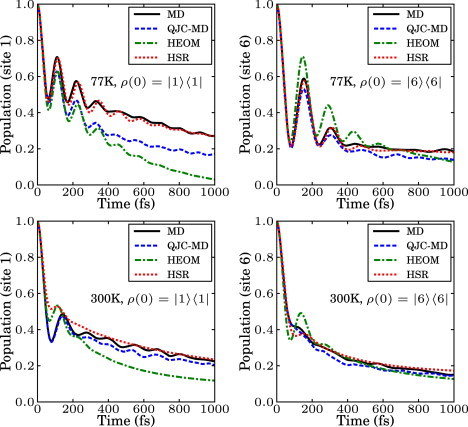
Comparison of the population dynamics obtained by using the MD method, the corrected MD, the hierarchy equation of motion approach, and the Haken-Strobl-Reineker model at 77 K and 300 K. (Right panels) Initial state in site 1; (left panels) initial state in site 6. All methods show similar short-time dynamics and dephasing, whereas the long time dynamics is different and the different increases as relaxation is incorporated in the various methods.
The HSR results take into account the site-dependence of the dephasing rates based on Eq. 8. The method is briefly described in the Supporting Material. Due to the Markovian assumption, this model shows slightly less coherence than the HEOM method and similarly to the MD method it converges to an equal classical mixture of all sites in the long time limit.
Correlation functions and spectral density
The bath autocorrelation function and its spectral density contain information on interactions between the excitonic system and the bath. The bath correlation function is defined as C(t) = 〈δϵ(t)δϵ(0)〉 with . For the MD simulation, C(t) is shown in Fig. 5 a for the two temperatures.
Figure 5.
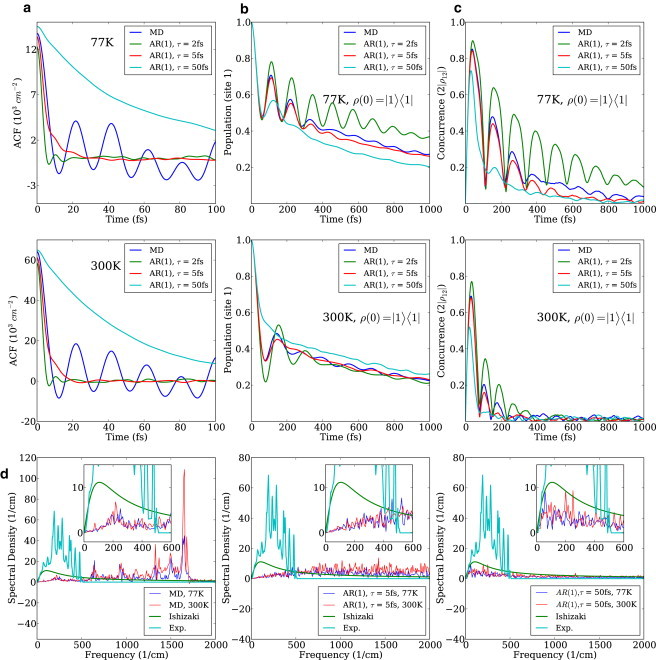
(a) Site 1 autocorrelation functions using MD and AR(1) processes generated with time constant equal to 2 fs, 5 fs, and 50 fs at 77 K and 300 K. (b) Site 1 population dynamics of MD and AR(1) processes with the different time constants at 77 K and 300 K. (c) The change of pairwise coherence between site 1 and 2 of MD and AR(1) processes with the different time constants at 77 K and 300 K. (d) Spectral density of site 1 of the FMO complex from the MD simulation at 77 K and 300 K. They clearly show the characteristic vibrational modes of the FMO complex. High-frequency modes are overpopulated due to the ultraviolet catastrophe observed in classical mechanics. The Ohmic spectral density used by Ishizaki and Fleming (10) was presented for comparison. The spectral densities of site 1 from AR(1) processes are also presented.
To study the effect of the decay rate of the autocorrelation function on the population dynamics, we modeled site energies using first-order autoregressive (AR(1)) processes (54). The marginal distribution of each process was tuned to have the same mean and variance as for the MD simulation. The autocorrelation function of the AR(1) process is an exponentially decaying function:
| (10) |
We generated three AR(1) processes with different time constants τ and propagated the reduced density matrix using the Hamiltonian corresponding to each process. As can be seen in Fig. 5 a, the autocorrelation function of the AR(1) process with τ ≈ 5 fs has a similar initial decay rate to that of the MD simulation at both temperatures. Therefore, as shown in the last three horizontal panels, its spectral density is in good agreement with the MD simulation result in the low frequency region, i.e., up to 600 cm−1. Modes in this region are known to be the most important in the dynamics and in determining the decoherence rate. And as Fig. 5, b and c, shows, that same AR(1) process with τ ≈ 5 fs exhibits similar population beatings and concurrences to those of the MD simulation. The relation of this 5-fs timescale to others reported in Ishizaki and Fleming (10) and Cho et al. (55) is presently unclear. We suspect that the discrepancy between the two results should decrease when one propagates the MD in the excited state. Work in this direction is in progress in our group.
The spectral density can be evaluated as the reweighted cosine transform of the corresponding bath autocorrelation function C(t) (48,49),
| (11) |
With our data, the spectral density exhibits characteristic phonon modes from the dynamics of the FMO complex (see Fig. 5 d, first panel). However, high-frequency modes tend to be overpopulated due to the limitation of using classical mechanics. Most of these modes are the local modes of the pigments, which can be seen from the pigment-only calculation in Ceccarelli et al. (35). There are efforts to incorporate quantum effects into the classical MD simulation in the context of vibrational coherence (56–58). We are investigating the possibilities of incorporating corrections based on a similar approach. Moreover, we also obtain a discrepancy of the spectral density in the low frequency region. On one hand, the origin could lie in the harmonic approximation of the bath modes leading to the tanh prefactor in Eq. 11 or in the force field used in this work. On the other, the form of the standard spectral density is from Wendling et al. (27) that measures fluorescence line-narrowing on a much longer timescale, approximately nanoseconds, than considered in our simulations (approximately picoseconds). Assuming correctness of our result, this implies that, for the simulation of fast exciton dynamics in photosynthetic light-harvesting complexes, a different spectral density than the widely used one has to be employed.
Site energy cross correlations between chromophores due to the protein environment have been postulated to contribute to the long-lived coherence in photosynthetic systems (2). Many studies have explored this issue, some quite recently (11,15,30,59–61). We tested this argument by decorrelating the site energies. For each unitary evolution, the site energies of different molecules at the same time were taken from different parts of the MD trajectory. In this way, we could significantly reduce potential cross correlation between sites while maintaining the autocorrelation function of each site. As can be seen in Fig. 6, no noticeable difference between the original and shuffled dynamics is observed.
Figure 6.
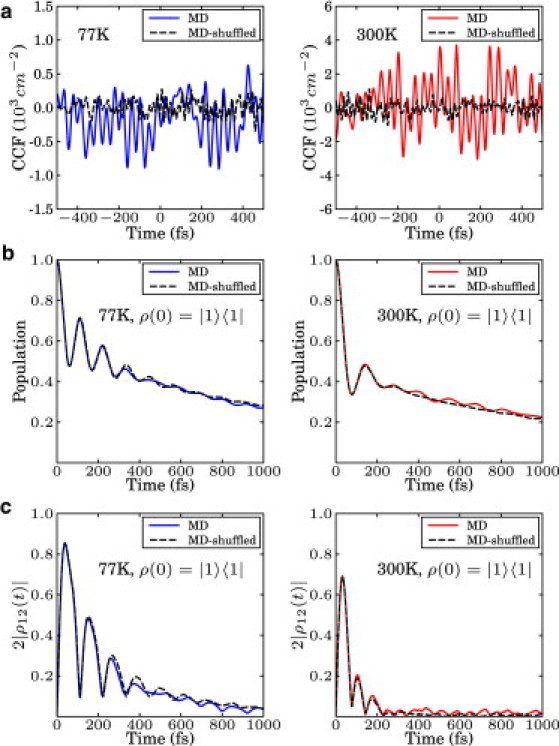
(a) Cross-correlation function of the original MD trajectory and a randomly shuffled trajectory between sites 1 and 2 at 77 K and 300 K. (b) Site 1 population dynamics of the original dynamics and the shuffled dynamics at 77 K and 300 K. (c) The pairwise coherence between sites 1 and 2. Original and shuffled dynamics are virtually identical at both temperatures.
Conclusion
The theoretical and computational studies presented in this article show that the long-lived quantum coherence in the energy transfer process of the FMO complex of P. aestuarii can be simulated with the atomistic model of the protein-chromophore complex. Unlike traditional master equation approaches, we propagate in a quantum/classical framework both the system and the environment state to establish the connection between the atomistic details of the protein complex and the exciton transfer dynamics. Our method combines MD simulations and QM/MM with TDDFT/TDA to produce the time evolution of the excitonic reduced density matrix as an ensemble average of unitary trajectories.
The conventional assumption of unstructured and uncorrelated site energy fluctuations is not necessary for our method. No ad hoc parameters were introduced in our formalism. The temperature and decoherence time were extracted from the site energy fluctuation by the MD simulation of the protein complex. The simulated dynamics clearly shows the characteristic quantum-wave-like population change and the long-lived quantum coherence during the energy transfer process in the biological environment. On this note it is worth mentioning that one has to be careful in the choice of force field and in the method used to calculated site energies. In fact, as presented in Olbrich et al. (62) a completely different energy transfer dynamics was obtained by using the semiempirical ZINDO-S/CIS to determine site energies.
Moreover, we determined the correlations of the site energy fluctuations for each site and between sites through the direct simulation of the protein complex. The spectral density shows the influence of the characteristic vibrational frequencies of the FMO complex. This spectral density can be used as an input for quantum master equations or other many-body approaches to study the effect of the structured bath. The calculated linear absorption spectrum we obtained is comparable to the experimental result, which supports the validity of our method. The characteristic beating of exciton population and pairwise quantum coherence exhibit excellent agreement with the results obtained by the HEOM method. It is also worth noting the remarkable agreement of the dephasing timescales of the MD simulations, the HEOM approach, and experiment.
Recently, characterization of the bath in the LH2 (48,49) and FMO (61) photosynthetic complexes were reported using MD simulation and quantum chemistry at room temperature. Those studies mostly focused on energy and spatial correlations across the sites, the linear absorption spectrum, and spectral density. The detailed study in Olbrich et al. (61) also suggests that spatial correlations are not relevant in the FMO dynamics.
This work opens the road to understanding whether biological systems employed quantum mechanics to enhance their functionality during evolution. We are planning to investigate the effects of various factors on the photosynthetic energy transfer process. These include: mutation of the protein residues, different chromophore molecules, and temperature dependence. Further research in this direction could elucidate the design principle of the biological photosynthesis process by nature, and could be beneficial for the discovery of more efficient photovoltaic materials and in biomimetics research.
Acknowledgments
S.S. thanks the Samsung Scholarship for financial support. P.R. thanks the Jacques-Emile Dubois Graduate Student Dissertation Fellowship for financial support. This work was supported by Defense Advanced Research Projects Agency Award No. N66001-10-4060 and by the Center of Excitonics, an Energy Frontier Research Center funded by the U.S. Department of Energy, Office of Science, and Office of Basic Energy Sciences under Award No. DE-SC0001088. A.A.-G. also acknowledges generous support from the Alfred P. Sloan and the Camille and Henry Dreyfus foundations.
Supporting Material
References
- 1.Engel G.S., Calhoun T.R., Fleming G.R. Evidence for wavelike energy transfer through quantum coherence in photosynthetic systems. Nature. 2007;446:782–786. doi: 10.1038/nature05678. [DOI] [PubMed] [Google Scholar]
- 2.Lee H., Cheng Y.-C., Fleming G.R. Coherence dynamics in photosynthesis: protein protection of excitonic coherence. Science. 2007;316:1462–1465. doi: 10.1126/science.1142188. [DOI] [PubMed] [Google Scholar]
- 3.Panitchayangkoon G., Hayes D., Engel G.S. Long-lived quantum coherence in photosynthetic complexes at physiological temperature. Proc. Natl. Acad. Sci. USA. 2010;107:12766–12770. doi: 10.1073/pnas.1005484107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rebentrost P., Mohseni M., Aspuru-Guzik A. Environment-assisted quantum transport. N. J. Phys. 2009;11:033003. doi: 10.1063/1.3002335. [DOI] [PubMed] [Google Scholar]
- 5.Plenio M.B., Huelga S.F. Dephasing-assisted transport: quantum networks and biomolecules. N. J. Phys. 2008;10:113019. [Google Scholar]
- 6.Jang S., Cheng Y.-C., Eaves J.D. Theory of coherent resonance energy transfer. J. Chem. Phys. 2008;129:101104. doi: 10.1063/1.2977974. [DOI] [PubMed] [Google Scholar]
- 7.Palmieri B., Abramavicius D., Mukamel S. Lindblad equations for strongly coupled populations and coherences in photosynthetic complexes. J. Chem. Phys. 2009;130:204512. doi: 10.1063/1.3142485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ishizaki A., Fleming G.R. Unified treatment of quantum coherent and incoherent hopping dynamics in electronic energy transfer: reduced hierarchy equation approach. J. Chem. Phys. 2009;130:234111. doi: 10.1063/1.3155372. [DOI] [PubMed] [Google Scholar]
- 9.Ishizaki A., Fleming G.R. On the adequacy of the Redfield equation and related approaches to the study of quantum dynamics in electronic energy transfer. J. Chem. Phys. 2009;130:234110. doi: 10.1063/1.3155214. [DOI] [PubMed] [Google Scholar]
- 10.Ishizaki A., Fleming G.R. Theoretical examination of quantum coherence in a photosynthetic system at physiological temperature. Proc. Natl. Acad. Sci. USA. 2009;106:17255–17260. doi: 10.1073/pnas.0908989106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rebentrost P., Mohseni M., Aspuru-Guzik A. Role of quantum coherence and environmental fluctuations in chromophoric energy transport. J. Phys. Chem. B. 2009;113:9942–9947. doi: 10.1021/jp901724d. [DOI] [PubMed] [Google Scholar]
- 12.Strümpfer J., Schulten K. Light harvesting complex II B850 excitation dynamics. J. Chem. Phys. 2009;131:225101. doi: 10.1063/1.3271348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Virshup A.M., Punwong C., Martínez T.J. Photodynamics in complex environments: ab initio multiple spawning quantum mechanical/molecular mechanical dynamics. J. Phys. Chem. B. 2009;113:3280–3291. doi: 10.1021/jp8073464. [DOI] [PubMed] [Google Scholar]
- 14.Caruso F., Chin A.W., Plenio M.B. Highly efficient energy excitation transfer in light-harvesting complexes: the fundamental role of noise-assisted transport. J. Chem. Phys. 2009;131:105106. [Google Scholar]
- 15.Wu J., Liu F., Silbey R.J. Efficient energy transfer in light-harvesting systems, I: Optimal temperature, reorganization energy and spatial-temporal correlations. N. J. Phys. 2010;12:105012. [Google Scholar]
- 16.Dijkstra A.G., Tanimura Y. Correlated fluctuations in the exciton dynamics and spectroscopy of DNA. N. J. Phys. 2010;12:055005. [Google Scholar]
- 17.Caruso F., Chin A., Plenio M.B. Entanglement and entangling power of the dynamics in light-harvesting complexes. Phys. Rev. A. 2010;81:1–8. [Google Scholar]
- 18.Fassioli F., Olaya-Castro A. Distribution of entanglement in light-harvesting complexes and their quantum efficiency. N. J. Phys. 2010;12:085006. [Google Scholar]
- 19.Sarovar M., Ishizaki A., Whaley K.B. Quantum entanglement in photosynthetic light-harvesting complexes. Nat. Phys. 2010;6:462–467. [Google Scholar]
- 20.Rebentrost P., Aspuru-Guzik A. Communication: Exciton-phonon information flow in the energy transfer process of photosynthetic complexes. J. Chem. Phys. 2011;134:101103. doi: 10.1063/1.3563617. [DOI] [PubMed] [Google Scholar]
- 21.Tamoi M., Tabuchi T., Shigeoka S. Point mutation of a plastidic invertase inhibits development of the photosynthetic apparatus and enhances nitrate assimilation in sugar-treated Arabidopsis seedlings. J. Biol. Chem. 2010;285:15399–15407. doi: 10.1074/jbc.M109.055111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jahns P., Graf M., Shikanai T. Single point mutation in the Rieske iron-sulfur subunit of cytochrome b6/F leads to an altered pH dependence of plastoquinol oxidation in Arabidopsis. FEBS Lett. 2002;519:99–102. doi: 10.1016/s0014-5793(02)02719-9. [DOI] [PubMed] [Google Scholar]
- 23.Pesaresi P., Sandonà D., Bassi R. A single point mutation (E166Q) prevents dicyclohexylcarbodiimide binding to the photosystem II subunit CP29. FEBS Lett. 1997;402:151–156. doi: 10.1016/s0014-5793(96)01518-9. [DOI] [PubMed] [Google Scholar]
- 24.Olson J.M. The FMO protein. Photosynth. Res. 2004;80:181–187. doi: 10.1023/B:PRES.0000030428.36950.43. [DOI] [PubMed] [Google Scholar]
- 25.Freiberg A., Lin S., Blankenship R.E. Exciton dynamics in FMO bacteriochlorophyll protein at low temperatures. J. Phys. Chem. B. 1997;101:7211–7220. doi: 10.1021/jp9633761. [DOI] [PubMed] [Google Scholar]
- 26.Savikhin S., Buck D.R., Struve W.S. Toward level-to-level energy transfers in photosynthesis: the Fenna-Matthews-Olson protein. J. Phys. Chem. B. 1998;102:5556–5565. [Google Scholar]
- 27.Wendling M., Pullerits T., van Amerongen H. Electron-vibrational coupling in the Fenna-Matthews-Olson complex of Prosthecochloris aestuarii determined by temperature-dependent absorption and fluorescence line-narrowing measurements. J. Phys. Chem. B. 2000;104:5825–5831. [Google Scholar]
- 28.Tronrud D.E., Wen J., Blankenship R.E. The structural basis for the difference in absorbance spectra for the FMO antenna protein from various green sulfur bacteria. Photosynth. Res. 2009;100:79–87. doi: 10.1007/s11120-009-9430-6. [DOI] [PubMed] [Google Scholar]
- 29.Vulto S.I.E., Neerken S., Aartsma T.J. Excited-state structure and dynamics in FMO antenna complexes from photosynthetic green sulfur bacteria. J. Phys. Chem. B. 1998;102:10630–10635. [Google Scholar]
- 30.Adolphs J., Renger T. How proteins trigger excitation energy transfer in the FMO complex of green sulfur bacteria. Biophys. J. 2006;91:2778–2797. doi: 10.1529/biophysj.105.079483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schmidt am Busch M., Müh F., Renger T. The eighth bacteriochlorophyll completes the excitation energy funnel in the FMO protein. J. Phys. Chem. Lett. 2011;2:93–98. doi: 10.1021/jz101541b. [DOI] [PubMed] [Google Scholar]
- 32.May V., Kühn O. Wiley-VCH Verlag; Weinheim, Germany: 2004. Charge and Energy Transfer Dynamics in Molecular Systems. [Google Scholar]
- 33.Gilmore J., McKenzie R.H. Quantum dynamics of electronic excitations in biomolecular chromophores: role of the protein environment and solvent. J. Phys. Chem. A. 2008;112:2162–2176. doi: 10.1021/jp710243t. [DOI] [PubMed] [Google Scholar]
- 34.Cornell W.D., Cieplak P., Kollman P.A. A second generation force field for the simulation of proteins, nucleic acids, and organic molecules. J. Am. Chem. Soc. 1995;117:5179–5197. [Google Scholar]
- 35.Ceccarelli M., Procacci P., Marchi M. An ab initio force field for the cofactors of bacterial photosynthesis. J. Comput. Chem. 2003;24:129–142. doi: 10.1002/jcc.10198. [DOI] [PubMed] [Google Scholar]
- 36.Ritschel G., Roden J., Eisfeld A. Absence of quantum oscillations and dependence on site energies in electronic excitation transfer in the Fenna-Matthews-Olson trimer. J. Phys. Chem. Lett. 2011;2:2912–2917. [Google Scholar]
- 37.Moix J., Wu J., Cao J. Efficient energy transfer in light-harvesting systems, III: The influence of the eighth bacteriochlorophyll on the dynamics and efficiency in FMO. J. Phys. Chem. Lett. 2011;2:3045–3052. [Google Scholar]
- 38.Shao Y., Molnar L.F., Head-Gordon M. Advances in methods and algorithms in a modern quantum chemistry program package. Phys. Chem. Chem. Phys. 2006;8:3172–3191. doi: 10.1039/b517914a. [DOI] [PubMed] [Google Scholar]
- 39.Krueger B.P., Scholes G.D., Fleming G.R. Calculation of couplings and energy-transfer pathways between the pigments of LH2 by the ab initio transition density cube method. J. Phys. Chem. B. 1998;102:9603–9604. [Google Scholar]
- 40.Hsu C.-P., You Z.-Q., Chen H.-C. Characterization of the short-range couplings in excitation energy transfer. J. Phys. Chem. C. 2008;112:1204–1212. [Google Scholar]
- 41.Dalibard J., Castin Y., Mølmer K. Wave-function approach to dissipative processes in quantum optics. Phys. Rev. Lett. 1992;68:580–583. doi: 10.1103/PhysRevLett.68.580. [DOI] [PubMed] [Google Scholar]
- 42.Piilo J., Maniscalco S., Suominen K.A. Non-Markovian quantum jumps. Phys. Rev. Lett. 2008;100:180402. doi: 10.1103/PhysRevLett.100.180402. [DOI] [PubMed] [Google Scholar]
- 43.Breuer H.-P., Petruccione F. Oxford University Press; New York: 2002. The Theory of Open Quantum Systems. [Google Scholar]
- 44.Wendling M., Przyjalgowski M.A., van Amerongen H. The quantitative relationship between structure and polarized spectroscopy in the FMO complex of Prosthecochloris aestuarii: refining experiments and simulations. Photosynth. Res. 2002;71:99–123. doi: 10.1023/A:1014947732165. [DOI] [PubMed] [Google Scholar]
- 45.Vokácová Z., Burda J.V. Computational study on spectral properties of the selected pigments from various photosystems: structure-transition energy relationship. J. Phys. Chem. A. 2007;111:5864–5878. doi: 10.1021/jp071639t. [DOI] [PubMed] [Google Scholar]
- 46.Leggett A.J., Chakravarty S., Zwerger W. Dynamics of the dissipative two-state system. Rev. Mod. Phys. 1987;59:1–85. [Google Scholar]
- 47.Pearlstein R.M. Theoretical interpretation of antenna spectra. In: Scheer H., editor. Chlorophylls. CRC Press; New York: 1991. pp. 1047–1078. [Google Scholar]
- 48.Damjanović A., Kosztin I., Schulten K. Excitons in a photosynthetic light-harvesting system: a combined molecular dynamics, quantum chemistry, and polaron model study. Phys. Rev. E. 2002;65:031919. doi: 10.1103/PhysRevE.65.031919. [DOI] [PubMed] [Google Scholar]
- 49.Olbrich C., Kleinekathöfer U. Time-dependent atomistic view on the electronic relaxation in light-harvesting system II. J. Phys. Chem. B. 2010;114:12427–12437. doi: 10.1021/jp106542v. [DOI] [PubMed] [Google Scholar]
- 50.Huo P., Coker D.F. Iterative linearized density matrix propagation for modeling coherent excitation energy transfer in photosynthetic light harvesting. J. Chem. Phys. 2010;133:184108. doi: 10.1063/1.3498901. [DOI] [PubMed] [Google Scholar]
- 51.Zhu J., Kais S., Aspuru-Guzik A. Modified scaled hierarchical equation of motion approach for the study of quantum coherence in photosynthetic complexes. J. Phys. Chem. B. 2011;115:1531–1537. doi: 10.1021/jp109559p. [DOI] [PubMed] [Google Scholar]
- 52.Haken H., Reineker P. The coupled coherent and incoherent motion of excitons and its influence on the line shape of optical absorption. Z. Phys. 1972;249:253–268. [Google Scholar]
- 53.Haken H., Strobl G. An exactly solvable model for coherent and incoherent exciton motion. Z. Phys. 1973;262:135–148. [Google Scholar]
- 54.Percival D.B., Walden A.T. Cambridge University Press; Cambridge, UK: 1993. Spectral Analysis for Physical Application. [Google Scholar]
- 55.Cho M., Vaswani H.M., Fleming G.R. Exciton analysis in 2D electronic spectroscopy. J. Phys. Chem. B. 2005;109:10542–10556. doi: 10.1021/jp050788d. [DOI] [PubMed] [Google Scholar]
- 56.Egorov S.A., Everitt K.F., Skinner J.L. Quantum dynamics and vibrational relaxation. J. Phys. Chem. A. 1999;103:9494–9499. [Google Scholar]
- 57.Skinner J.L., Park K. Calculating vibrational energy relaxation rates from classical molecular dynamics simulations: quantum correction factors for processes involving vibration-vibration energy transfer. J. Phys. Chem. B. 2001;105:6716–6721. [Google Scholar]
- 58.Stock G. Classical simulation of quantum energy flow in biomolecules. Phys. Rev. Lett. 2009;102:118301. doi: 10.1103/PhysRevLett.102.118301. [DOI] [PubMed] [Google Scholar]
- 59.Nazir A. Correlation-dependent coherent to incoherent transitions in resonant energy transfer dynamics. Phys. Rev. Lett. 2009;103:146404. doi: 10.1103/PhysRevLett.103.146404. [DOI] [PubMed] [Google Scholar]
- 60.Abramavicius D., Mukamel S. Exciton dynamics in chromophore aggregates with correlated environment fluctuations. J. Chem. Phys. 2011;134:174504. doi: 10.1063/1.3579455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Olbrich C., Strümpfer J., Kleinekathöfer U. Quest for spatially correlated fluctuations in the FMO light-harvesting complex. J. Phys. Chem. B. 2011;115:758–764. doi: 10.1021/jp1099514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Olbrich C., Jansen T.L.C., Kleinekathöfer U. From atomistic modeling to excitation transfer and two-dimensional spectra of the FMO light-harvesting complex. J. Phys. Chem. B. 2011;115:8609–8621. doi: 10.1021/jp202619a. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


