Abstract
According to recent experiments, the molecular-motor myosin behaves like a strain sensor, exhibiting different functional responses when loads in opposite directions are applied to its tail. Within an elastic-network model, we explore the sensitivity of the protein to the forces acting on the tail and find, in agreement with experiments, that such forces invoke conformational changes that should affect filament binding and ADP release. Furthermore, conformational responses of myosin to the application of forces to individual residues in its principal functional regions are systematically investigated and a detailed sensitivity map of myosin-V is thus obtained. The results suggest that the strain-sensor behavior is involved in the intrinsic operation of this molecular motor.
Introduction
Myosin motors play a fundamental role in biological cells and are responsible for a variety of functions, including force generation in muscles and intracellular transport (1). Because of their importance, these proteins have been a subject of extensive experimental and theoretical investigation (2). Nonetheless, complete understanding of their functional mechanisms has not yet been achieved. In recent experiments (3–5), such molecular motors have been probed by applying external forces. It has been shown that by varying the direction and magnitude of the forces acting on the myosin tail, binding to the actin filament (3) and ADP release (4,5) can be controlled. Thus, the myosin molecule operates as a strain sensor, with well-defined responses in its different parts induced by specific mechanical perturbations. The aim of this study is to explore the origins of such selective mechanical sensitivity and to explain the experimental results. Proceeding further, we perform a systematic analysis of the responses to arbitrary perturbations in the nucleotide-binding pocket and the actin-binding region. The results suggest that the strain-sensing mechanism is intrinsic for functional operation of this molecular motor.
Different theoretical approaches can be applied to study motor proteins. Molecular dynamics (MD) simulations intend to trace the motions of all atoms and, thus, provide a full account of myosin operation. However, these computational experiments do not permit resolution of the behavior on characteristic millisecond scales, and thus, further acceleration techniques, such as guided MD simulations, should be used (6,7). Although computational speed may be increased considerably through the use of specialized hardware (8,9), simplified descriptions would still be needed to provide orientation within the great amounts of generated numerical data. As general experience in dealing with complex systems reveals (see, e.g., Mikhailov and Calenbuhr (10)), the very existence of robust ordered collective motions often implies that some reduced dynamical descriptions should be possible. It is therefore remarkable that, to a large extent, conformational motions in protein machines can already be reproduced within a rough elastic-network (EN) approximation.
In the EN approach (11–15), a protein is described as a purely mechanical object formed by a network of identical point particles that are connected by elastic strings. All of these particles, which typically correspond to entire residues, are identical. Moreover, the strings have different natural lengths and, it is often assumed, the same stiffness. The pattern of network connections is constructed on the basis of experimentally known equilibrium conformations of the protein being used. Thus, the model is stripped of almost all chemical details and the dynamics of the network is only governed by the shape, i.e., the network structure, of a macromolecule. In this study, a variant of the EN approach, known as the anisotropic-network model (16,17), will be used.
Despite their apparent simplicity, EN approaches have turned out to be surprisingly successful. Special investigations have shown that such models can predict ligand-induced conformational changes and describe thermal fluctuations (B-factors) in a variety of proteins (11,14,15,18–21). Although the studies are often carried out within the linear normal-mode approximation, nonlinear equations of relaxational EN dynamics have also been explored (22–26). EN models have furthermore been extended to include the possibility of partial unfolding of proteins in ligand-induced conformational transitions (27–30). Moreover, in a recent dynamical EN simulation, it was possible to follow entire operation cycles of the molecular-motor hepatitis C virus helicase, including its interactions with DNA (26).
Myosins have been previously studied in terms of ENs. Zheng and Doniach (31) analyzed overlaps of nucleotide-dependent conformational changes of myosin and EN normal modes. They were able to explain the observed changes with only a small number of slow normal modes (see also Zheng and Brooks (32)). Takano et al. introduced electrostatic interactions with actin and demonstrated fluctuation-induced biased motion of myosin along the actin filament (33). Nonlinear conformational relaxation processes in myosin-II (23) and myosin-V (24) were considered, and the existence of special attractive relaxation paths in these proteins was shown. Furthermore, kinetic coordination in the myosin-V dimer has been investigated by following transition pathways between different structural states modeled as ENs (34).
In this study, an EN will be used as a model of the myosin-V molecule. It is known that a single macromolecule of myosin naturally exhibits coordinated movements of its parts that would require complicated machinery in a comparable macroscopic device. ATP binding and hydrolysis, followed by product release, are localized near the ATP-binding site in the region comprising the so-called front and back doors (35,36). These local events, however, affect the attachment of myosin to the actin filament, which takes place in the region of the actin cleft (37–40). In addition, binding of myosin to the filament has effects on the release of the hydrolysis products (Pi and ADP) (41).
In a recent theoretical investigation of mechanical coupling in myosin-V based on guided MD simulations, it was demonstrated that significant conformational changes in the converter and actin-binding cleft could be induced by imposing a local perturbation of the protein configuration in the region of the ATP-binding pocket (6). Allosteric communication between different functional parts of myosin-V has also been studied by block normal-mode (BNM) analysis (42). Moreover, correlation normal-mode analysis, based on the EN description and combined with MD simulations, has been performed (43).
In contrast to such publications, which consider responses to local perturbations generated in accordance with the known structural data, we shall examine how the protein responds to the application of an external force of arbitrary orientation to each single residue in important functional regions. Our aim is to identify those residues that are particularly sensitive to strain in such a way that their perturbation invokes strong conformational responses. Thus, it is possible to obtain a detailed pattern of intrinsic intramolecular communication.
We find here that applying static forces to the myosin tail induces opening and closing of the front door and in this way provides an effective control mechanism for nucleotide release. Moreover, backward strain in the tail region leads to conformational changes in the actin cleft that are suited to facilitate binding to the filament. These findings explain the experiments (3–5) in terms of conformational changes inside the molecule. We furthermore see that the nucleotide-binding region is divided into two functional parts: application of forces in the front-door region, where the adenylate ring of ADP binds, leads to movement of the tail, whereas forces in the back-door region, where phosphate is trapped after hydrolysis, induce conformational changes in the actin-binding cleft. We monitor the effects of perturbations in the actin-binding region and observe communication between the hypertrophic cardiomyopathy (HCM) loop and the tail, indicating a connection between binding to the filament and tail motion. Thus, a complex pattern of intramolecular communication, based on elastic deformations and strain-sensor behavior, is revealed in myosin-V.
Methods
This study is based on the EN modeling of conformational dynamics in the presence of external forces. The EN model of myosin-V heavy chain is constructed by using as a reference state its post-rigor equilibrium conformation with Mg·ADP·BeFx, an ATP analog, as yielded by x-ray diffraction analysis (Protein Data Bank ID 1W7J, chain A) (40). The model is coarse-grained, with each particle in the network corresponding to a residue in the protein. The equilibrium conformation is defined by a set of coordinates , representing equilibrium positions of the particles and determined by the coordinates of α-carbons in the PDB data. The nucleotide is not explicitly included in the model. The EN is constructed by examining all pair distances between the particles in the equilibrium conformation and connecting with elastic springs those pairs separated by distances shorter than the cutoff length l0. The natural length of each spring is equal to the equilibrium distance , so that the equilibrium conformation should represent the energy minimum. This approach corresponds to the anisotropic network model (16,17). The elastic energy of the network is
| (1) |
Here, is the distance between two particles i and j. The elements of the adjacency matrix A are Aij = 1 if , and Aij = 0 otherwise. The cutoff length l0 = 10 Å has been chosen. All elastic links are assumed to have the same stiffness constant; this parameter is removed from Eq. 1 by an appropriate rescaling of the elastic energy. Note that after such rescaling the forces become measured in units of length (Å). The force with the strength of 1 Å applied to a single elastic link induces its elongation by 1 Å.
Fig. 1, a and b, shows the equilibrium conformation (PDB ID 1W7J) and the respective EN. Note that the residues missing in the PDB data are not included in the constructed network. Most of them belong to a loop (loop 2) that is highly flexible and whose conformation is strongly affected by thermal fluctuations. In Fig. 1 a, we have schematically outlined the principal parts of myosin-V: the tail, the region of the nucleotide-binding pocket (NBP), and the region of the actin-binding cleft.
Figure 1.
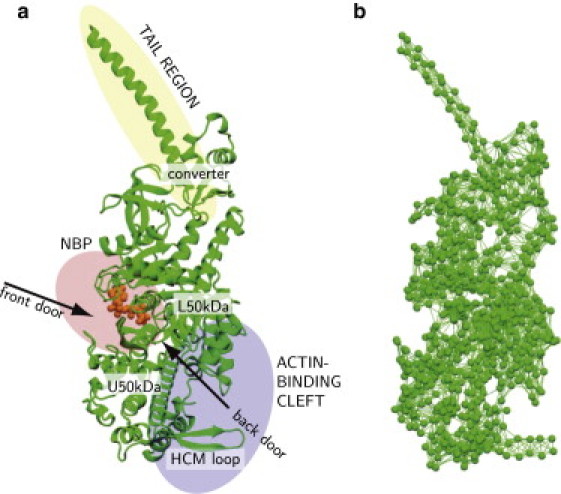
Myosin-V (a) and its EN (b). In panel a, three principal functional regions of the protein are schematically shown; the ATP-analog is also displayed (orange). Moreover, some important structural elements, such as the front and the back doors and the HCM loop, are also indicated here. In panel b, each particle corresponds to a residue, the links represent elastic interactions between them.
When external forces Fi are introduced, the network energy becomes . The equilibrium configuration of the network in the presence of static forces corresponds to the minimum of the energy, E. Therefore, it can be found by direct integration of the relaxation equations
| (2) |
where γ is the viscose friction coefficient, which can be eliminated (as we always do below) by appropriate rescaling of time. In this study, we are interested only in static responses to the application of constant forces. Therefore, only the final states of the relaxation processes, described by Eq. 2, are important here. Note, however, that Eq. 2 can also be used to consider slow conformational relaxation processes on the scale of milliseconds, where inertial effects are negligible (44).
Explicitly, we have
| (3) |
These relaxation equations are generally nonlinear in terms of the coordinates Ri. The nonlinearity is due to the nonlinear dependence of the distances dij on the coordinates of the particles, . Because the adjacency matrix is assumed constant, only elastic deformations of the network not accompanied by breaking of the links or by establishment of new connections are described by Eq. 3. In our investigations, external probing forces were always applied to only one residue.
If deviations of the particles from their equilibrium positions are small, Eq. 3 can be further linearized in terms of ri, leading to a description in terms of the normal modes. In this study, however, we do not perform the linearization, because the conditions under which nonharmonicity and interaction between normal modes become significant are not a priori known. In a previous EN study of conformational relaxation in myosin-V, it was shown that even motions of the tail may be nonharmonical (24).
Our intention is to systematically probe mechanical responses of the EN of myosin-V to the application of static forces, with various orientations, to single residues in three principal regions of this protein. To find the stationary conformation in the presence of a force, Eq. 3 was numerically integrated. The integration using the explicit Euler method was continued until a stationary state was found.
Generally, when a static force is applied to a residue, it would not only induce an internal deformation of the network, but also lead to rigid translational and rotational motions. To monitor only intramolecular conformational changes, immobilization is needed to exclude such motions. In our study, immobilization was implemented by applying to all particles in the network forces whose magnitudes and directions were recomputed at each integration step so that they compensated for possible rigid translations and rotations. The details of the immobilization procedure are given in the Supporting Material. Note that these additional compensating forces were introduced in such a way that they could not induce internal deformations.
The entire EN of myosin-V includes 752 particles. For probing of mechanical responses, a subset of 82 particles was selected (see Fig. 2 a). Residue 792 was located in the tail region and used to apply forces to the tail. The second group of residues was from the NBP region (Table S2 in the Supporting Material, first column). All residues in this group are ATP neighbors, i.e., they are adjacent to ATP. The third group (Table S3, first column) included the residues located on the surface of the actin-binding cleft.
Figure 2.
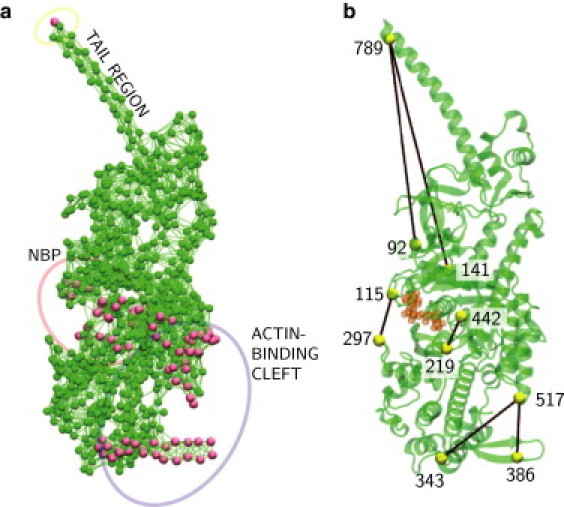
(a) Set of residues probed by application of static forces. (b) Labels, and the distances between them, used to monitor conformational changes.
Although integration of Eq. 3 gave positions of all particles in the state with a force applied, this information was too detailed, and to quantify induced deformations some measures (or “order parameters”, cf. Ovchinnikov et al. (6)). were needed. In our analysis, we chose a set of 10 particles as the labels and monitored pair distances between them. The labels and selected distances are shown in Fig. 2 b.
The first three labels, corresponding to residues 789, 141, and 92, were selected to specify motion of the tail. Here, the first label is in the tail and the other two are attached to stiff parts of the protein.
Four labels were used to measure conformational changes in the NBP region. The labels, corresponding to residue 115 in the N-terminal subdomain and residue 297 in the upper 50-kDa subdomain, were chosen to characterize the opening of the front door. They are in contact with the adenylate ring, gating the front door through which ATP enters and ADP leaves the reaction site (35). The back-door opening and closing were characterized by the distance between residues 219 and 442 in switches 1 and 2, respectively, in the upper 50-kDa subdomain. These two amino acids form a salt bridge that hinders the phosphate from leaving the NBP after hydrolysis (7,36).
The last three residues belonged to the actin-binding region. Distances between residue 517 in the lower 50-kDa and residues 343 and 386 in the upper 50-kDa subdomain were monitored. These labels lie in the actin-binding site and characterize opening and closing of the cleft, as well as the motion of the HCM loop.
To examine responses of the network, static forces were applied to one of the chosen particles, and after integration of Eq. 3, changes of the distances between the labels with respect to their equilibrium values were determined.
Results
In this section, we report the results of systematic investigations of mechanical responses of myosin-V to the application of forces at individual residues. The effects are separately presented in three sections, depending on the region where the forces were applied.
Forces acting on the tail
In this section, we intend to reproduce computationally the experiments (3–5) in which external forces were acting on the myosin tail. We apply forces to one residue in the tail and look at the conformational responses in the NBP and in the actin-binding cleft, depending on the force orientation and amplitude.
Experimentally, forward and backward force directions are defined with respect to the direction of the processive motion of myosin along the actin filament. To determine filament orientation with respect to myosin in the considered model, a special analysis had to be performed. First, we built a filament by using the structural data for F-actin (PDB ID 2ZWH) (45). Then, we needed to determine how the filament was positioned with respect to myosin in the actin-myosin complex. To do this, we used the results of recent guided MD simulations (39) in which several sites for binding of myosin to actin were identified. By keeping the filament stiff, we anchored it to the myosin network by elastic links at these sites. After that, relaxation in the actomyosin system was followed by integration of dynamical equations, and the final equilibrium configuration of the complex was determined. Thus, the relative filament direction, used in subsequent numerical investigations, could be identified. The details are given in the Supporting Material.
In the simulations, external forces were always applied to residue 792. Since the tail is a relatively stiff structure, the exact position of the force application point was not important. The forces were parallel to the filament direction, which had been previously identified. Their amplitudes were varied from 0 to 6.5 Å, and both forward and backward directions were considered. For each force, Eq. 3 was numerically integrated until a stationary protein configuration was achieved.
The responses of the EN are displayed in Fig. 3. As we see, changes in the NBP are induced. The distance between residues 115 and 297, defining the opening of the front door, grows under forward strain and decreases if backward forces are applied. However, the change of the pair distance between residues 219 and 442, characterizing the salt bridge, remains <0.1 Å. Hence, we conclude that although the front-door configuration is controlled by the force acting on the tail, the responses in the back-door region of the NBP are negligible.
Figure 3.
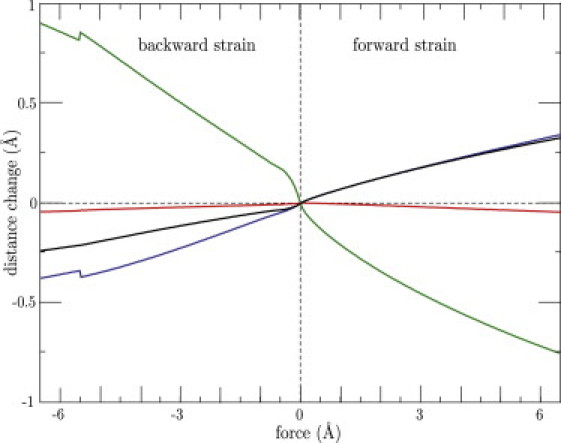
Responses of pair distances between labels 115 and 297 (black), 219 and 442 (red), 343 and 517 (blue), and 386 and 517 (green) as functions of the amplitude of the force applied to the tail. Small abrupt changes observed at large negative forces are due to local buckling effects.
As evidenced by Fig. 3, external forces furthermore lead to conformational changes in the actin-binding region. When backward strain is applied, the actin cleft, characterized by the distance between residues 343 and 517, closes. At the same time, the distance between residues 386 and 517 increases, and thus, the HCM loop moves away from the lower 50-kDa domain. The opposite conformational changes are observed when the forward strain is applied.
The significance of the observed responses becomes clear if the operation mode of myosin is taken into account. ADP and Pi, the hydrolysis products, are exiting the nucleotide pocket in different directions, through the front and the back door, respectively. Therefore, closing of the front door, which, as we have found, is induced by backward strains, would hinder the ADP release and thus enhance the ADP affinity. This agrees with experiments (4,5) where dependence of the ADP affinity on force direction was observed. Movie S1 displays the dynamical response of the network to an external force applied in the forward direction. The induced tail motion and the pronounced changes in the actin-binding cleft region are clearly seen there; changes in the front door are weaker and less apparent.
The effects of strains on binding to the actin filament have been investigated in experiments (3) in which backward strain was shown to increase the probability of strong binding. The original interpretation (3) of these results was that such forces induce opening of the back door, which enhances the Pi release that should precede strong binding. Our simulations, however, do not reveal any considerable sensitivity of the back door to the forces acting on the tail, so this original interpretation is not supported by our data. Nonetheless, the experimentally observed behavior can still be understood by analyzing the responses induced in the actin-binding cleft.
Conformational changes accompanying a transition to strong binding of myosin to the actin filament can de identified by comparing the structures in the post-rigor state with the filament detached and in the nucleotide-free (or rigor) state of the same myosin (PDB ID 1OE9) (46), which corresponds to the conformation of myosin bound to the filament. Focusing on the changes in the actin-binding cleft, one can notice that the transition from the post-rigor to the rigor state is characterized by shortening of the distance between residues 343 and 517 and, at the same time, by an increase of the distance between the HCM loop and the lower 50-kDa domain (characterized by residues 386 and 517). However, these are exactly the changes we found to be induced by the application of backward strain. Hence, the experimental data (3) are explained by conformational changes that are invoked directly in the actin-binding cleft rather than being mediated through hypothetical responses in the back-door region of the nucleotide pocket.
Forces in the NBP
The NBP is an important functional region of myosin. It is known that ATP enters the pocket through the front door and that after the hydrolysis, the ADP product leaves through the same opening. In contrast, the phosphate, representing the second product, leaves the NBP region through the back door. The processes taking place in the NBP in turn affect filament binding and tail motions.
Responses to perturbations localized in the NBP have been investigated previously in theoretical studies using restrained targeted MD simulations (6), and by means of normal mode analysis (32). In these previous studies, it was demonstrated that global conformational transitions from the nucleotide-free to the ATP-bound states could be reproduced by applying special forces to a group of residues inside the NBP, with the directions chosen to correspond to the experimentally known local changes between the two conformations.
The approach we pursued is different. Our aim was not to reproduce a particular known response, but instead to systematically test the global sensitivity of the protein to arbitrary forces applied to various single residues in the nucleotide-binding region. In this manner, we aimed to construct a map of the NBP in which residues responsible for particular functional behavior could be identified.
To probe mechanical responses, 27 residues adjacent to the ATP in the considered equilibrium conformation were selected (Table S2, left column, and Fig. 2 a). For every chosen residue, a series of 200 simulations was performed. In all simulations, the magnitude of the applied force was the same (f0 = 1 Å), but its orientations were randomly varied. The simulations were always continued until a stationary state was found. After that, changes in the monitored pair distances between the labels in different regions were determined. The induced distance changes were analyzed, and for each pair of labels, the maximum absolute distance change over 200 force orientations was evaluated. This maximum distance change was then taken as the measure of the protein's sensitivity to forces applied to a particular residue. An example of the relaxation process leading to a new stationary state is shown in Movie S2. Here, forces were applied to residue 115 in the front door, and conformational changes, especially in the tail, are clearly visible.
The sensitivities of all 27 residues with respect to different pair distances are summarized in Table S2, and the directions of communication are schematically shown in Fig. 4.
Figure 4.
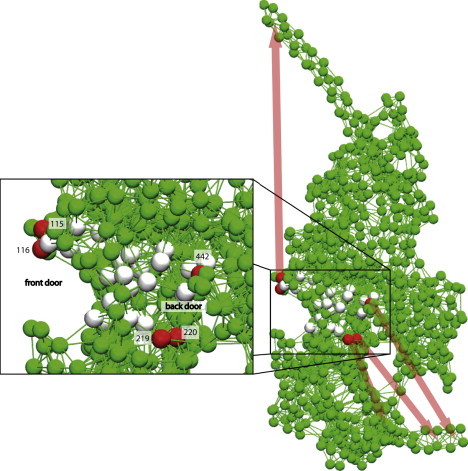
Sensitivity of residues in the NBP region. Directions of communication are indicated by colored arrows. (Inset) Enlargement of the region of the NBP outlined by the box. Residues most sensitive with respect to tail motion and conformational changes in the actin cleft are colored red. Responses induced by the forces applied at the gray colored residues are weaker.
As we have found, applying forces to the residues in the front-door region produces a strong effect on the tail (Fig. 4). However, the application of forces to residues near the back door or the P-loop only weakly affects the tail region. Thus, the tail responds mainly to the perturbations applied at the entrance of the front door.
Proceeding further, the sensitivity with respect to responses in the actin-binding cleft was investigated. As we observed (see Supporting Material), the actin-binding region is mainly affected by the application of forces in the back-door region. When force is applied to residue 442 in the back door, a strong response is produced in the distance between residues 343 and 517, which characterize the cleft opening. Moreover, application of forces in the back door has a pronounced effect on the HCM loop to which residue 386 belongs. The HCM loop is known to come into contact with actin and may play an important role in myosin-actin interactions (46–48). It should be noted, however, that some sensitivity of the actin-binding cleft to perturbations in the front door was also seen.
Thus, we have found that myosin behaves as a strain sensor not only with respect to the forces applied to its tail. The forces acting on some residues within the NBP are also producing well-defined conformational responses, which are functionally important.
We have furthermore discovered that in terms of the sensitivity of its residues, the NBP region is clearly divided into a front-door and a back-door domain (Fig. 4, inset). On the one hand, a pronounced effect on the actin cleft was observed when forces were applied to residues 219, 220, and 442 in the back-door region. Remarkably, it is exactly the salt bridge between the two sensitive residues 219 and 442 that hinders phosphate release after hydrolysis. On the other hand, we saw that the forces applied at the front door primarily affected the tail and could induce its motions. We found that the forces applied to the residues of the P-loop region, lying in the middle between the front and the back doors, do not lead to significant conformational changes either in the tail or in the actin cleft.
It is important to note that the response of the actin cleft to perturbations in the NBP can be controlled by the forces applied to the tail. Fig. 5 shows how the change in the distance between residues 343 and 517, characterizing the width of the actin cleft, is dependent on the force applied to residue 442 in the back door. We chose to apply the force in the direction that maximizes the absolute distance change, and its amplitude was varied. In addition, analogous dependences on the presence of forward and backward strain in the tail are shown in the figure. One can see that backward strain enhances the closing of the actin cleft, whereas forward strain tends to prevent it. Thus, although the forces applied at the tail do not directly affect phosphate release through the back door, they can nonetheless modulate the effects of phosphate release on the opening or closing of the actin cleft, which controls interactions of myosin with the filament.
Figure 5.
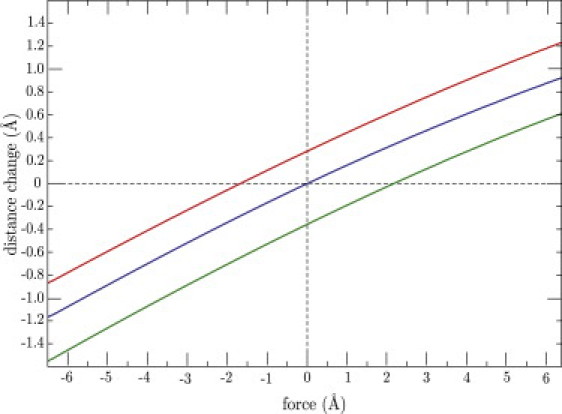
Changes in the distance between residues 343 and 517, which characterize opening of the actin-binding cleft, as functions of the force applied to residue 442 in the back-door region. The red curve corresponds to the forward force of 5 Å applied to the tail. The green curve is for the force with the same strength applied in the backward direction. The blue curve corresponds to the absence of force.
Forces in the actin-cleft region
Binding of myosin to an actin filament should lead to perturbations localized in the actin-binding cleft region. Such perturbations should have a specific form, determined by the organization of the actin-myosin interface and details of the myosin-actin interactions. As in the previous section, our aim is not to reproduce the responses to a particular local perturbation. Instead, we systematically investigate in this section the sensitivity of the protein to the application of forces with arbitrary orientations to individual residues in the actin-binding cleft region and observe how strains in the cleft can affect the tail or the front and back doors.
High-resolution experimental data of myosin bound to the actin filament are currently not available. Based on cryo-EM experiments, models for the actomyosin interface were proposed (35,38,39). Lorenz and Holmes (39) suggested possible electrostatic interactions and hydrogen bonds between myosin II and actin. Using these results, we have identified a set of 54 residues that may come in contact with the filament (Table S3, left column, and Fig. 2 a). To probe sensitivities in the actin cleft, 200 independent simulations were performed for each single residue in the set. Forces with strength f0 = 1 Å and randomly chosen orientation were applied to a given residue and integration was continued until the system relaxed to a new equilibrium. After that, changes in the characteristic pair distances between the labels were determined. For each residue and pair distance, the maximum response over the series of 200 simulations was recorded. The determined sensitivities are shown in Table S3 and systematically analyzed in the Supporting Material. The results are also schematically represented in Fig. 6, where arrows indicate the principal communication pathways.
Figure 6.
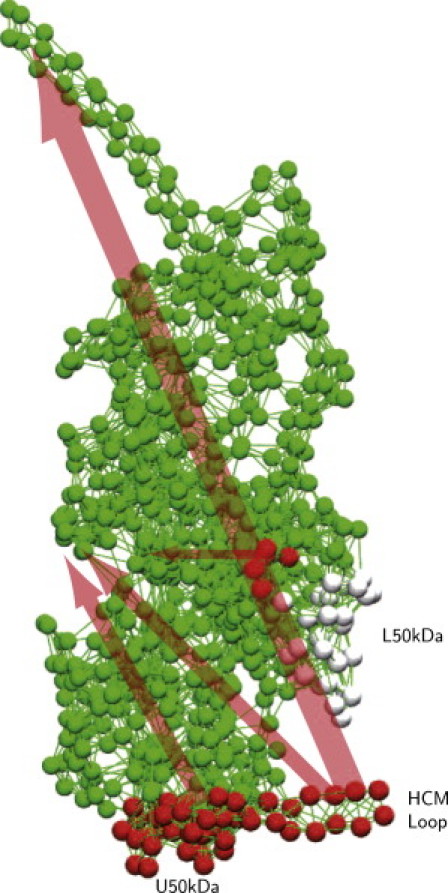
Responses induced by the application of forces to residues in the actin-binding cleft region. Red spheres indicate residues 377–390 in the HCM loop, 340–350 in the upper 50-kDa subdomain, and 540–544 in the lower 50-kDa subdomain. Gray spheres represent less sensitive residues in the lower 50-kDa subdomain. Arrows schematically indicate the directions and strength of intramolecular communication.
Strong responses of the tail were induced when forces were applied to the residues in the HCM loop (see, for example, Movie S3). There is also some sensitivity of the tail to perturbations applied in the upper 50-kDa domain (residues 340 to 350). In contrast, the tail is not significantly sensitive to the forces acting in the lower 50-kDa domain. Examining responses in the NBP region, we observed that the front door is strongly sensitive to the forces applied in the upper 50-kDa domain and the HCM loop. The back door is sensitive to perturbations acting in the lower 50-kDa domain.
Discussion
Functional responses of myosin motors to the application of external forces have been investigated experimentally in previous studies. Iwaki et al. (3) attached a bead to the tail of a myosin-VI monomer and dragged it along the actin filament in two opposite directions. It was found that the transition from weak to strong binding of myosin to the filament is affected by application of a dragging force and strongly depends on the direction of the force. The probability of strong binding increases when forces are applied in the direction opposite to that of the intrinsic processive motion (i.e., the backward direction for this molecular motor). In other experiments, effects of external loads on the ADP release from myosin-V attached to the actin filament were studied (4,5,49,50). It has been found that the release is hindered when forces are applied in the backward direction, opposite to the direction of processive motion for this myosin (49,50). Oguchi et al. (4,5) measured the magnitude of force that would need to be applied to the tail to unbind a myosin-V monomer from an actin filament for different force directions and ADP concentrations. These experiments indicated that ADP release is slowed down when a force is applied in the backward direction (see also Sakamoto et al. (51) and Komori et al. (52)).
Our study provides a theoretical explanation for these results. We observed a direct effect of external forces on the front door, through which ADP leaves the protein. When backward strain is applied to the tail, this tends to close the front door and, accordingly, should make the ADP release less probable. Thus, external forces can regulate the ADP affinity through conformational changes in the front-door region. We did not find a direct effect of the strains on the back door, through which the phosphate leaves the protein, leading to filament attachment. However, we observed that strain directly influences the protein conformation in the actin-binding cleft region. Backward strains tend to favor closing of the actin cleft and to generate a movement of the HCM loop away from the lower 50-kDa domain, thus enhancing the effects of phosphate release through the back door (see Fig. 3) and making strong coupling more probable. In this way, an explanation of the experiments (3) is also provided.
Force-dependent release of ADP is important for the operation of the dimeric myosin-V, i.e., for its processive motion along the actin filament (53,54). Recent theoretical investigations using a reduced kinetic description of the cycles of the myosin-V dimer suggest that the transition rates of ATP and ADP binding to the leading head need to be load-dependent (55). Thus, the strain-sensor behavior of myosin-V, revealed in experiments in which external forces were applied to its tail and explained in this study, should play a profound role in natural functioning of this molecular motor.
Moreover, we systematically analyzed conformational responses of myosin-V to the application of forces to individual residues in the NBP and on the surface of the actin-binding cleft. Thus, a detailed intrinsic sensitivity map of myosin was constructed. As we have seen, by perturbing specific residues in the front- or back-door regions of the NBP, motions of the tail or opening and closure of the actin cleft can be induced. On the other hand, application of forces to residues in the actin-binding cleft can also invoke tail motions and have effects in the NBP region. In our numerical study, the responses were induced by external forces. Under natural conditions, however, mechanical perturbations in the NBP and the actin-binding cleft can easily arise as a consequence of ligand binding and reaction events.
Remarkably, both explanation of the experiments (3–5,49,50) with external loads and identification of intrinsic communication pathways inside the protein could already be obtained in our study by using a greatly simplified EN model. The model is stripped of chemical details, so that a protein is treated as a purely mechanical object, i.e., a network of particles connected by elastic strings. It turns out that generic elastic responses of such a network are sufficient to understand intramolecular communication responsible for the coordination of processes in different functional parts of the molecular motor. Our investigations were performed in the framework of the full nonlinear elastic model, without the linearization and transition to a normal-mode description. We checked (see Supporting Material) that nonlinear effects are indeed essential for the considered phenomena.
In the recent publication by Zheng (34), full nonlinear EN equations were used, and coordination between conformational changes in two coupled units in the myosin dimer, resulting from mechanical interactions through the common tail, were considered. However, the analysis was only performed along a particular conformational pathway connecting two experimentally known myosin states, and the responses were considered at the level of entire protein domains rather than that of individual residues. In contrast, we have systematically analyzed the responses of the myosin molecule to the application of arbitrary forces in various directions to a large number of individual residues located in three principal functional regions. We have monitored how and to what extent such forces affected individual residues in other functional residues.
Our results suggest that conformational changes induced in myosin-V by ATP binding, the hydrolysis reaction, and product release should not be very sensitive to chemical details of the respective microscopic processes. Such changes can be invoked, as we have shown, by the application of forces to the network nodes corresponding to specific residues in the NBP region. We also found that the tail and the NBP are sensitive to mechanical perturbations of single residues in the actin-binding cleft region, which may naturally arise when binding of myosin-V to the actin filament takes place. It should be noted that based on elastic responses of the macromolecule to mechanical perturbations in the ATP binding pocket, similar behavior has recently been reported (26) for a different molecular motor, the hepatitis C virus helicase; within the EN model, the entire operation cycle of this motor could be reproduced by applying forces to only a few residues.
Based on such observations, we conjecture that the strain-sensor behavior and the intramolecular communication relying on elastic deformations should be a general property of protein motors, essentially involved in the organization of their functional activity. It would be interesting to test this suggestion in additional specially designed experiments. In such experiments, forces can be applied to single selected residues (or small groups of them), and conformational responses can be monitored by using AFM or FRET techniques. Such experiments have been performed to study, for example, partial unfolding of single proteins (56,57). Moreover, once specific residues responsible for important conformational responses have been identified, one can try to modify them by using protein engineering methods and thus verify their functional roles.
Acknowledgments
We thank M. Iwaki and T. Komori for stimulating discussions. VMD (58) was used for the visualization of data.
Financial support from the Global Center of Excellence program System Dynamics of Biological Function (Ministry of Education, Culture, Sports, Science and Technology of Japan) and from the research training group (GRK 1558) Nonequilibrium Collective Dynamics in Condensed Matter and Biological Systems (Deutsche Forschungsgemeinschaft of Germany) is gratefully acknowledged.
Supporting Material
References
- 1.Sellers J.R. 2nd ed. Oxford University Press; New York: 1999. Myosins. [Google Scholar]
- 2.Sweeney H.L., Houdusse A. Structural and functional insights into the myosin motor mechanism. Annu. Rev. Biophys. 2010;39:539–557. doi: 10.1146/annurev.biophys.050708.133751. [DOI] [PubMed] [Google Scholar]
- 3.Iwaki M., Iwane A.H., Yanagida T. Brownian search-and-catch mechanism for myosin-VI steps. Nat. Chem. Biol. 2009;5:403–405. doi: 10.1038/nchembio.171. [DOI] [PubMed] [Google Scholar]
- 4.Oguchi Y., Mikhailenko S.V., Ishiwata S. Robust processivity of myosin V under off-axis loads. Nat. Chem. Biol. 2010;6:300–305. doi: 10.1038/nchembio.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oguchi Y., Mikhailenko S.V., Ishiwata S. Load-dependent ADP binding to myosins V and VI: implications for subunit coordination and function. Proc. Natl. Acad. Sci. USA. 2008;105:7714–7719. doi: 10.1073/pnas.0800564105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ovchinnikov V., Trout B.L., Karplus M. Mechanical coupling in myosin V: a simulation study. J. Mol. Biol. 2010;395:815–833. doi: 10.1016/j.jmb.2009.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cecchini M., Alexeev Y., Karplus M. Pi release from myosin: a simulation analysis of possible pathways. Structure. 2010;18:458–470. doi: 10.1016/j.str.2010.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shaw D.E., Maragakis P., Wriggers W. Atomic-level characterization of the structural dynamics of proteins. Science. 2010;330:341–346. doi: 10.1126/science.1187409. [DOI] [PubMed] [Google Scholar]
- 9.Stone J.E., Hardy D.J., Schulten K. GPU-accelerated molecular modeling coming of age. J. Mol. Graph. Model. 2010;29:116–125. doi: 10.1016/j.jmgm.2010.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mikhailov A.S., Calenbuhr V. Springer; Berlin: 2007. From Cells to Societies. [Google Scholar]
- 11.Cui Q., Bahar I., editors. Normal Mode Analysis: Theory and Applications to Biological and Chemical Systems. Chapman & Hall/CRC; Boca Raton, FL: 2006. [Google Scholar]
- 12.Tirion M.M. Large amplitude elastic motions in proteins from a single-parameter, atomic analysis. Phys. Rev. Lett. 1996;77:1905–1908. doi: 10.1103/PhysRevLett.77.1905. [DOI] [PubMed] [Google Scholar]
- 13.Hinsen K. Analysis of domain motions by approximate normal mode calculations. Proteins. 1998;33:417–429. doi: 10.1002/(sici)1097-0134(19981115)33:3<417::aid-prot10>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 14.Bahar I., Atilgan A.R., Erman B. Direct evaluation of thermal fluctuations in proteins using a single-parameter harmonic potential. Fold. Des. 1997;2:173–181. doi: 10.1016/S1359-0278(97)00024-2. [DOI] [PubMed] [Google Scholar]
- 15.Haliloglu T., Bahar I., Erman B. Gaussian dynamics of folded proteins. Phys. Rev. Lett. 1997;79:3090–3093. [Google Scholar]
- 16.Doruker P., Atilgan A.R., Bahar I. Dynamics of proteins predicted by molecular dynamics simulations and analytical approaches: application to α-amylase inhibitor. Proteins. 2000;40:512–524. [PubMed] [Google Scholar]
- 17.Atilgan A.R., Durell S.R., Bahar I. Anisotropy of fluctuation dynamics of proteins with an elastic network model. Biophys. J. 2001;80:505–515. doi: 10.1016/S0006-3495(01)76033-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tama F., Sanejouand Y.-H. Conformational change of proteins arising from normal mode calculations. Protein Eng. 2001;14:1–6. doi: 10.1093/protein/14.1.1. [DOI] [PubMed] [Google Scholar]
- 19.Liao J.-L., Beratan D.N. How does protein architecture facilitate the transduction of ATP chemical-bond energy into mechanical work? The cases of nitrogenase and ATP binding-cassette proteins. Biophys. J. 2004;87:1369–1377. doi: 10.1529/biophysj.103.038653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chennubhotla C., Rader A.J., Bahar I. Elastic network models for understanding biomolecular machinery: from enzymes to supramolecular assemblies. Phys. Biol. 2005;2:S173–S180. doi: 10.1088/1478-3975/2/4/S12. [DOI] [PubMed] [Google Scholar]
- 21.Yang L., Song G., Jernigan R.L. How well can we understand large-scale protein motions using normal modes of elastic network models? Biophys. J. 2007;93:920–929. doi: 10.1529/biophysj.106.095927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Piazza F., De Los Rios P., Sanejouand Y.-H. Slow energy relaxation of macromolecules and nanoclusters in solution. Phys. Rev. Lett. 2005;94:145502. doi: 10.1103/PhysRevLett.94.145502. [DOI] [PubMed] [Google Scholar]
- 23.Togashi Y., Mikhailov A.S. Nonlinear relaxation dynamics in elastic networks and design principles of molecular machines. Proc. Natl. Acad. Sci. USA. 2007;104:8697–8702. doi: 10.1073/pnas.0702950104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Togashi Y., Yanagida T., Mikhailov A.S. Nonlinearity of mechanochemical motions in motor proteins. PLOS Comput. Biol. 2010;6:e1000814. doi: 10.1371/journal.pcbi.1000814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hayashi K., Takano M. Violation of the fluctuation-dissipation theorem in a protein system. Biophys. J. 2007;93:895–901. doi: 10.1529/biophysj.106.100487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Flechsig H., Mikhailov A.S. Tracing entire operation cycles of molecular motor hepatitis C virus helicase in structurally resolved dynamical simulations. Proc. Natl. Acad. Sci. USA. 2010;107:20875–20880. doi: 10.1073/pnas.1014631107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Higo J., Umeyama H. Protein dynamics determined by backbone conformation and atom packing. Protein Eng. 1997;10:373–380. doi: 10.1093/protein/10.4.373. [DOI] [PubMed] [Google Scholar]
- 28.Miyashita O., Onuchic J.N., Wolynes P.G. Nonlinear elasticity, proteinquakes, and the energy landscapes of functional transitions in proteins. Proc. Natl. Acad. Sci. USA. 2003;100:12570–12575. doi: 10.1073/pnas.2135471100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim M.K., Chirikjian G.S., Jernigan R.L. Elastic models of conformational transitions in macromolecules. J. Mol. Graph. Model. 2002;21:151–160. doi: 10.1016/s1093-3263(02)00143-2. [DOI] [PubMed] [Google Scholar]
- 30.Maragakis P., Karplus M. Large amplitude conformational change in proteins explored with a plastic network model: adenylate kinase. J. Mol. Biol. 2005;352:807–822. doi: 10.1016/j.jmb.2005.07.031. [DOI] [PubMed] [Google Scholar]
- 31.Zheng W., Doniach S. A comparative study of motor-protein motions by using a simple elastic-network model. Proc. Natl. Acad. Sci. USA. 2003;100:13253–13258. doi: 10.1073/pnas.2235686100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zheng W., Brooks B.R. Probing the local dynamics of nucleotide-binding pocket coupled to the global dynamics: myosin versus kinesin. Biophys. J. 2005;89:167–178. doi: 10.1529/biophysj.105.063305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Takano M., Terada T.P., Sasai M. Unidirectional Brownian motion observed in an in silico single molecule experiment of an actomyosin motor. Proc. Natl. Acad. Sci. USA. 2010;107:7769–7774. doi: 10.1073/pnas.0911830107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zheng W. Coarse-grained modeling of conformational transitions underlying the processive stepping of myosin V dimer along filamentous actin. Proteins. 2011;79:2291–2305. doi: 10.1002/prot.23055. [DOI] [PubMed] [Google Scholar]
- 35.Holmes K.C., Angert I., Schröder R.R. Electron cryo-microscopy shows how strong binding of myosin to actin releases nucleotide. Nature. 2003;425:423–427. doi: 10.1038/nature02005. [DOI] [PubMed] [Google Scholar]
- 36.Lawson J.D., Pate E., Yount R.G. Molecular dynamics analysis of structural factors influencing back door pi release in myosin. Biophys. J. 2004;86:3794–3803. doi: 10.1529/biophysj.103.037390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Murphy C.T., Spudich J.A. The sequence of the myosin 50-20K loop affects Myosin's affinity for actin throughout the actin-myosin ATPase cycle and its maximum ATPase activity. Biochemistry. 1999;38:3785–3792. doi: 10.1021/bi9826815. [DOI] [PubMed] [Google Scholar]
- 38.Holmes K.C., Schröder R.R., Houdusse A. The structure of the rigor complex and its implications for the power stroke. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2004;359:1819–1828. doi: 10.1098/rstb.2004.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lorenz M., Holmes K.C. The actin-myosin interface. Proc. Natl. Acad. Sci. USA. 2010;107:12529–12534. doi: 10.1073/pnas.1003604107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Coureux P.D., Sweeney H.L., Houdusse A. Three myosin V structures delineate essential features of chemo-mechanical transduction. EMBO J. 2004;23:4527–4537. doi: 10.1038/sj.emboj.7600458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Howard J. Sinauer Associates; Sunderland, MA: 2001. Mechanics of Motor Proteins and Cytoskeleton. [Google Scholar]
- 42.Cecchini M., Houdusse A., Karplus M. Allosteric communication in myosin V: from small conformational changes to large directed movements. PLOS Comput. Biol. 2008;4:e1000129. doi: 10.1371/journal.pcbi.1000129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zheng W. Multiscale modeling of structural dynamics underlying force generation and product release in actomyosin complex. Proteins. 2010;78:638–660. doi: 10.1002/prot.22594. [DOI] [PubMed] [Google Scholar]
- 44.Kitao A., Hirata F., Gō N. The effects of solvent on the conformation and the collective motions of protein: normal mode analysis and molecular dynamics simulations of melittin in water and in vacuum. Chem. Phys. 1991;158:447–472. [Google Scholar]
- 45.Oda T., Iwasa M., Narita A. The nature of the globular- to fibrous-actin transition. Nature. 2009;457:441–445. doi: 10.1038/nature07685. [DOI] [PubMed] [Google Scholar]
- 46.Coureux P.D., Wells A.L., Houdusse A. A structural state of the myosin V motor without bound nucleotide. Nature. 2003;425:419–423. doi: 10.1038/nature01927. [DOI] [PubMed] [Google Scholar]
- 47.Sweeney H.L., Straceski A.J., Faust L. Heterologous expression of a cardiomyopathic myosin that is defective in its actin interaction. J. Biol. Chem. 1994;269:1603–1605. [PubMed] [Google Scholar]
- 48.Volkmann N., Lui H., Hanein D. The R403Q myosin mutation implicated in familial hypertrophic cardiomyopathy causes disorder at the actomyosin interface. PLoS ONE. 2007;2:e1123. doi: 10.1371/journal.pone.0001123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Veigel C., Schmitz S., Sellers J.R. Load-dependent kinetics of myosin-V can explain its high processivity. Nat. Cell Biol. 2005;7:861–869. doi: 10.1038/ncb1287. [DOI] [PubMed] [Google Scholar]
- 50.Purcell T.J., Sweeney H.L., Spudich J.A. A force-dependent state controls the coordination of processive myosin V. Proc. Natl. Acad. Sci. USA. 2005;102:13873–13878. doi: 10.1073/pnas.0506441102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sakamoto T., Webb M.R., Sellers J.R. Direct observation of the mechanochemical coupling in myosin Va during processive movement. Nature. 2008;455:128–132. doi: 10.1038/nature07188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Komori T., Nishikawa S., Yanagida T. Simultaneous measurement of nucleotide occupancy and mechanical displacement in myosin-V, a processive molecular motor. Biophys. J. 2009;96:L04–L06. doi: 10.1016/j.bpj.2008.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mehta A.D., Rock R.S., Cheney R.E. Myosin-V is a processive actin-based motor. Nature. 1999;400:590–593. doi: 10.1038/23072. [DOI] [PubMed] [Google Scholar]
- 54.Yildiz A., Forkey J.N., Selvin P.R. Myosin V walks hand-over-hand: single fluorophore imaging with 1.5-nm localization. Science. 2003;300:2061–2065. doi: 10.1126/science.1084398. [DOI] [PubMed] [Google Scholar]
- 55.Bierbaum V., Lipowsky R. Chemomechanical coupling and motor cycles of myosin V. Biophys. J. 2011;100:1747–1755. doi: 10.1016/j.bpj.2011.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dietz H., Berkemeier F., Rief M. Anisotropic deformation response of single protein molecules. Proc. Natl. Acad. Sci. USA. 2006;103:12724–12728. doi: 10.1073/pnas.0602995103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Eyal E., Bahar I. Toward a molecular understanding of the anisotropic response of proteins to external forces: insights from elastic network models. Biophys. J. 2008;94:3424–3435. doi: 10.1529/biophysj.107.120733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. 27–28. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


