I. General Introduction/Background
Successful fertilization is fundamentally important to a sexually reproducing species and requires a series of well-coordinated events including gamete activation, recognition, signaling, adhesion and fusion (Wassarman 1999; Singson, Zannoni et al. 2001; Primakoff and Myles 2002). Although our current understanding of these processes comes largely from work in marine invertebrates and vertebrate model systems, C. elegans has emerged as another powerful system for fertilization studies (Singson 2001; Yamamoto, Kosinski et al. 2006; Singson, Hang et al. 2008; Nishimura and L’Hernault 2010).
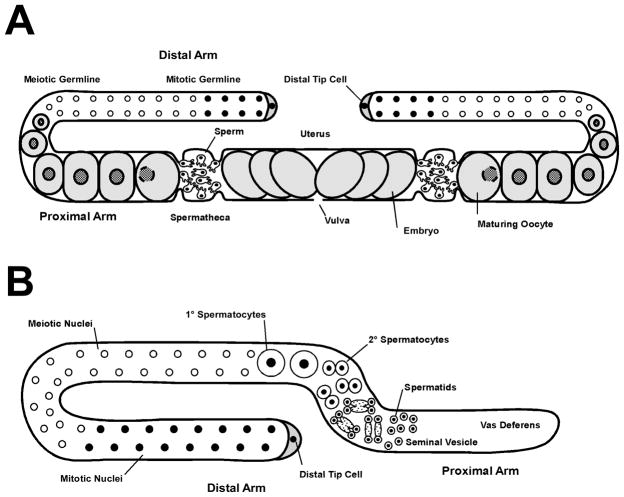
Fertilization in C. elegans takes place in the spermatheca, the site of sperm storage in the hermaphrodite. The hermaphrodite reproductive tract consists of a bi-lobed gonad (Fig. 1A) in which a separate spermatheca connects each lobe to the shared uterus (the male gonad is single-lobed [Fig. 1B]). Within the hermaphrodite gonad, both gamete types are produced in a sequential manner. Sperm are produced first during the last larval stage of development and stored in the spermatheca. The gonad switches to oocyte production in the adult hermaphrodite. Oocytes undergo meiotic maturation as they move towards the uterus and are ovulated into the spermatheca, where they immediately contact multiple spermatozoa. Blocks to polyspermy exist, as only a single sperm fertilizes each oocyte (Parry, Velarde et al. 2009). The coordination of events leading to sperm/oocyte contact ensures highly efficient sperm utilization as virtually all functional sperm fertilize oocytes (Ward and Carrel 1979; Kadandale and Singson 2004). The hermaphrodite’s own sperm can be supplemented by mating to males; male sperm are deposited in the uterus and immediately travel to the spermatheca to await oocyte passage. A sperm-sensing mechanism ensures that metabolically costly oocytes are not wasted; when hermaphrodites lack sperm, they ovulate at a very low basal level. Conversely, the presence of sperm within the hermaphrodite spermatheca causes a dramatic increase in ovulation rate (McCarter, Bartlett et al. 1999)(Miller, Nguyen et al. 2001). After fertilization, the zygote secretes a multi-layered egg shell and begins embryonic development. Eggs then pass through the uterus and are laid before hatching (Fig. 2).
Figure 1.
C. elegans adult gonads. (A) Hermaphrodite gonad showing general scheme of oocyte development. (B) Male gonad showing general scheme of spermatogenesis.
Figure 2.
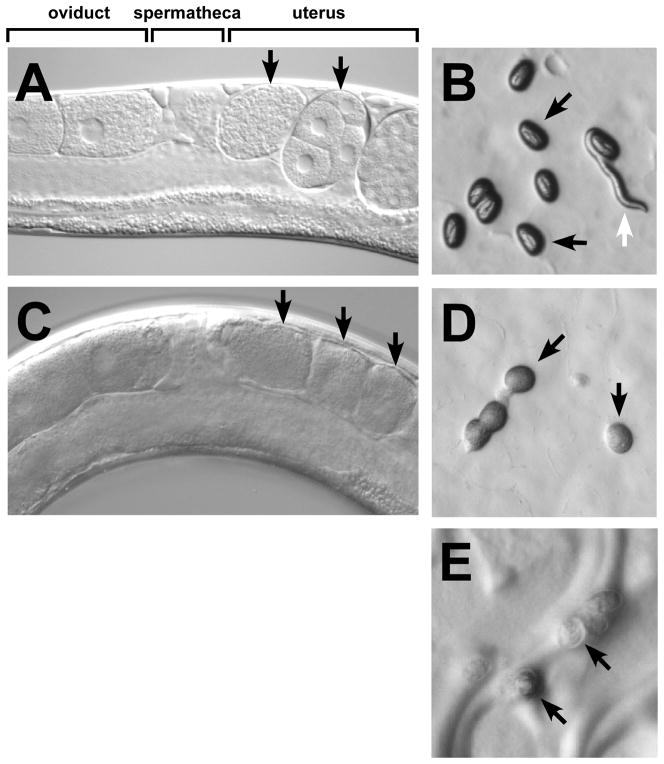
Examples of the sterile phenotypes observed when fertilization or proper egg activation does not occur. (A) In wild-type hermaphrodites, embryos (black arrows) can be observed developing in the uterus. (B) Oblong shelled embryos (black arrows) are laid at the 50+ cell stage and ultimately hatch into larvae (white arrow). (C) In sterile mutants (e.g. spe-9(hc52) shown here) unfertilized oocytes (black arrows) are seen in the uterus. (D) These soft, flattened and opaque unfertilized oocytes (black arrows) are also laid. (E) Egg activation mutants (e.g. spe-11(hc77) shown here) lay more rounded thinly-shelled eggs or “pebbles” (black arrows) that neither develop nor hatch.
C. elegans offers several advantages over other model systems for studying fertilization. Although its amoeboid sperm possess neither a flagellum nor an acrosome (Fig. 3), these sperm successfully perform the same tasks required of all spermatozoa (e.g. migration to the fertilization site, species-specific oocyte recognition, fusion). In addition, the events of fertilization can be directly observed in living animals through the worm’s transparent cuticle (McCarter, Bartlett et al. 1999). It is also possible to isolate large quantities of sperm and oocytes, though this is more challenging to do than in some other model systems (L’Hernault and Roberts 1995; Aroian, Field et al. 1997; Miller 2006). Fertilization studies in C. elegans routinely use molecular and genetic tools that are unavailable or difficult to use in other systems. The complete sequencing of the worm genome and the availability of microarrays greatly simplifies the identification and analysis of genes required for fertility (Singson 2001; Singson, Hang et al. 2008). Perhaps the greatest advantage of C. elegans is the ease with which one can perform forward genetics to screen for fertilization-defective mutants (discussed below). Such screens have identified many of these mutants, which may be classified broadly into those mutations affecting sperm (spe or fer mutants, for spermatogenesis or fertilization defective) and those affecting eggs/oocytes (egg mutants). Note that the fer designation has been discontinued and all new sperm development or function mutants are now given the spe designation.
Figure 3.
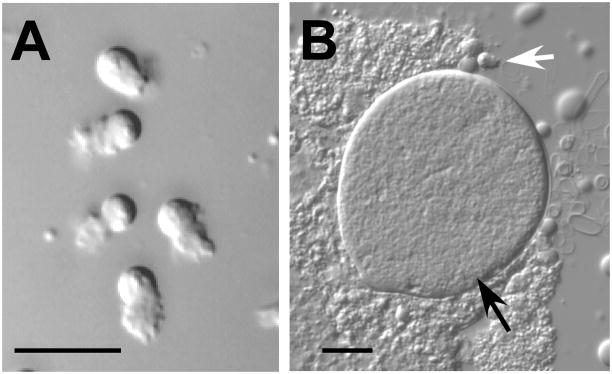
Gamete morphology. (A) in vitro pronase-activated male-derived sperm. (B) Hermaphrodite dissection showing relative size of in vivo activated sperm and oocyte. The oocyte (black arrow) is approximately 160 times the volume of the sperm (white arrow). Bars = 10μm
Although the majority of characterized spe/fer mutations affect sperm development, a subset of these mutations specifically affect sperm function (i.e. fertilization). spe-9 class mutants, for example, produce sperm that are unable to fertilize oocytes despite exhibiting normal morphology, motility and gamete contact (L’Hernault, Shakes et al. 1988; Singson, Mercer et al. 1998; Xu and Sternberg 2003; Chatterjee, Richmond et al. 2005; Kroft, Gleason et al. 2005). Recent studies have also uncovered egg mutations that specifically influence fertilization and/or egg activation (Kadandale, Stewart-Michaelis et al. 2005). A partial listing of characterized genes required for fertilization is given in Table 1. Many of the experimental tools and techniques discussed in this chapter were developed for the study of these genes.
Table 1.
Genes Required for Fertilization and Egg Activation in C. elegans
| Fertilization
| |||
|---|---|---|---|
| Gene | Protein | Localization | Reference |
| spe-9 | single-pass transmembrane protein with 10 EGF repeats | spermatozoa pseudopod | Singson et al, 1998 |
| spe-38 | novel four-pass integral membrane protein | spermatozoa pseudopod | Chatterjee et al, 2005 |
| spe-41/trp-3 | TRPC channel subunit | spermatozoa plasma membrane | Xu and Sternberg, 2003 |
| spe-42 | novel seven-pass integral membrane protein | unknown | Kroft et al, 2005 |
| egg-1/2 | type-II transmembrane protein with LDL-receptor repeats | oocyte plasma membrane | Kadandale et al, 2005 |
| Egg Activation
| |||
|---|---|---|---|
| Gene | Protein | Localization | Reference |
| spe-11 | soluble protein | perinuclear in sperm | Browning and Strome, 1996 |
| egg-3 | protein tyrosine phosphatase-like (PTPL) | oocyte cortex | Maruyama et al, 2007 |
| egg-4/5 | protein tyrosine phosphatase-like (PTPL) | oocyte cortex | Parry et al, 2009 |
C. elegans also enables the evolutionary assessment of fertilization molecules and can help elucidate major molecular themes. For example, EGF-repeat-containing molecules have been implicated in fertilization across a wide evolutionary spectrum, from HrVC70 in ascidians (Sawada, Tanaka et al. 2004) to SPE-9 in worms (Singson, Mercer et al. 1998) and SED-1 in mammals (Ensslin and Shur 2003).
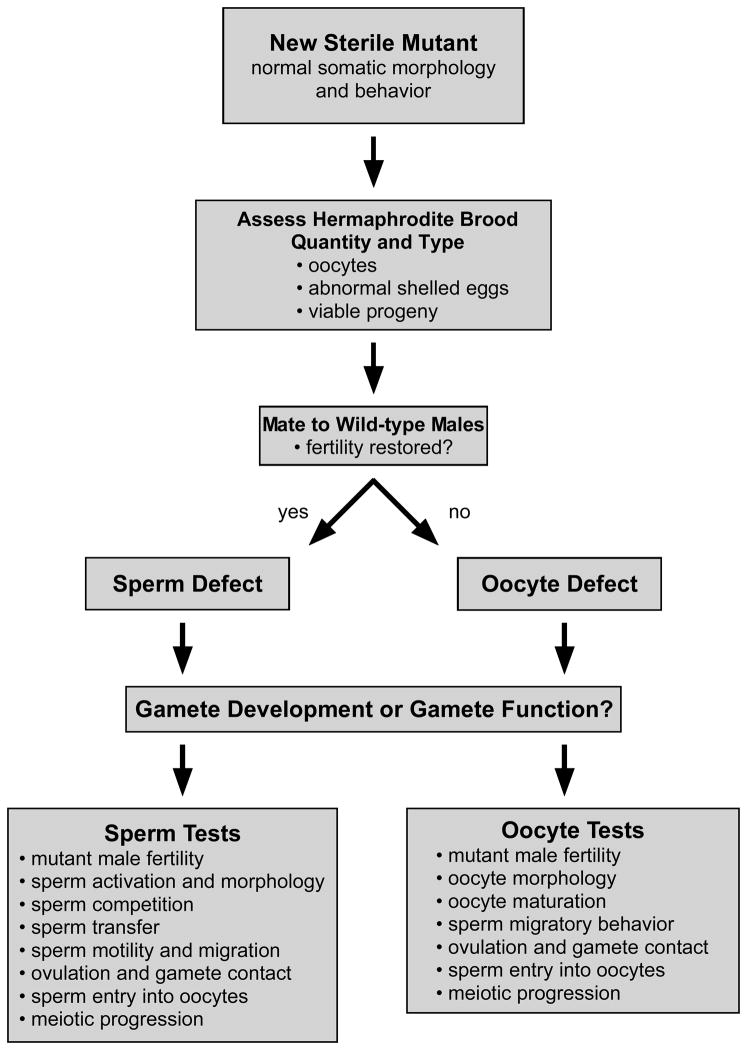
In this chapter we introduce the major experimental approaches/implications to consider when using C. elegans to study fertilization. A general scheme for the study of sterility mutants in C. elegans is presented in Fig. 4. Detailed protocols can be found in the original literature and in L’Hernault and Roberts (1995).
Figure 4.
A general scheme for working with C. elegans sterility mutants. Once the general class of mutant is determined (spe or egg) by crossing a new sterile hermaphrodite to wild-type males, one can begin to assess whether the gene is specifically for fertilization, or more generally for gamete development. See text for information regarding specific tests.
II. Finding Sterile Mutants
A major advantage of using C. elegans for fertilization study is the ability to use forward genetic screens to isolate fertility mutants. Mutant screening in C. elegans is typically carried out using ethyl methane sulphonate (EMS) to induce mutations in the germline of wild-type hermaphrodites. In the subsequent F2 generation, homozygous mutants are screened for the phenotype of interest (L’Hernault, Shakes et al. 1988). Using standard concentrations of EMS, investigators can expect to find one null mutation per 2,000 gene copies examined (Jorgensen and Mango 2002) and approximately one in every 30 F2 generation animals will display a Spe phenotype (S. L’Hernault, personal communication).
The power of these genetic screens lies in the ability to isolate mutations in sperm-specific genes by simply selecting hermaphrodites that are self-sterile but whose oocytes can be still be fertilized by wild-type male sperm (Fig. 5). More than 60 sperm-specific mutants have been identified this way, enabling the genetic delineation of sperm development and function pathways (L’Hernault, Shakes et al. 1988; L’Hernault 1997; L’Hernault 2006; Nishimura and L’Hernault 2010). C. elegans is particularly amenable to such sperm-specific screens because they can be carried out with hermaphrodites (rather than in males as in gonochoristic systems such as mice or flies) and thus remain independent of confounding issues such as male mating behavior or secondary sex characteristics (L’Hernault, Shakes et al. 1988; Lessard, Pendola et al. 2004; Wakimoto, Lindsley et al. 2004).
Figure 5.
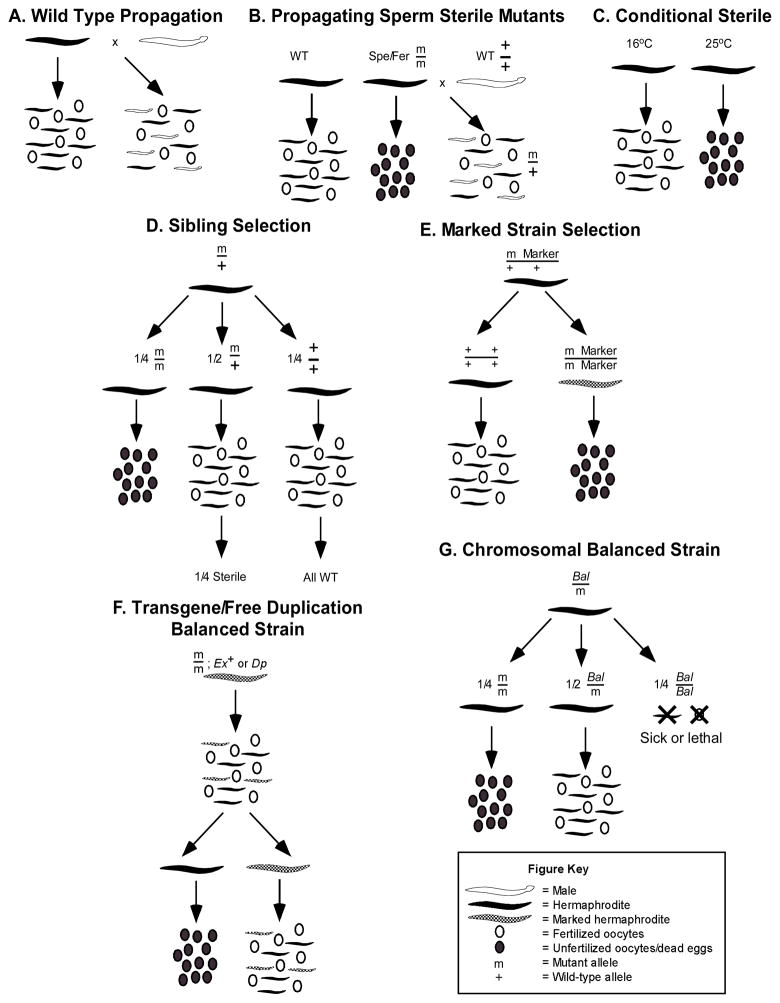
Common schemes for the maintenance/propagation of sterile mutations in C. elegans. (A) Wild type strain propagation. (B) Sterility mutant propagation. (C) Conditional sterility mutant propagation. (D) Sibling Selection for non-conditional sterility mutants. (E) Marked strain selection. (F) Chromosomally balanced strains. (G) Transgene/free duplication-balanced strains. See text for details.
Genetic screens for conditional egg-sterile mutants have likewise yielded many interesting candidate genes. (G. Singaravelu, D. Shakes and A. Singson, unpublished). These conditional screens are especially useful for the isolation of fertility mutants as they allow the propagation of homozygous mutations in fertility genes. Isolating egg sterile mutants in standard non-conditional F2 screens requires considerably more work since progeny cannot be recovered from homozygous egg-sterile mutants. As a result, the mutant chromosome must be recovered from heterozygous siblings.
Conditional fertility screens are similar to the conditional maternal effect lethal (mel) screens conducted in C. elegans (O’Connell, Leys et al. 1998; Jorgensen and Mango 2002). In a typical mel screen, worms with a pre-existing mutation that blocks normal egg-laying are mutagenized, and the uteri of the resulting F2 hermaphrodites are examined for the presence of dead F3 progeny. The screen is first carried out at 25 C, and any F2 animals that are filled with dead F3 embryos are subsequently shifted to 16 °C to recover homozygous temperature-sensitive (ts) mutants. This same screen can be modified to isolate fertility mutants simply by looking for worms containing unfertilized oocytes rather than dead embryos. Oocytes that have been fertilized and have successfully completed egg-activation encase themselves in a rigid eggshell (Fig. 2A,B). Oocytes that remain unfertilized or that have defects in egg-activation lack this shell and are visibly distinct (the phenotype known as “squashy,” “mushy,” or “ugly brown mels”) (Fig 2. C,D). Because such “squashy” oocytes were routinely discarded in most large-scale mel screens, it is likely that new screens that focus specifically on this phenotype can identify many new fertility genes (G. Singaravelu, D. Shakes and A. Singson, unpublished).
The ts alleles isolated in conditional screens can be used to determine the temporal requirements for a given protein’s function or synthesis, and their availability can greatly simplify genetic manipulations (L’Hernault, Shakes et al. 1988; Putiri, Zannoni et al. 2004). However, although their phenotypes may represent a wide range of severities, ts alleles are rarely amorphic (null) and investigators must therefore be cautious when relying on them to make inferences about gene functions/interactions. Null phenotypes are best defined using molecularly verified amorphic mutants.
Reverse genetic methods may also be used to study the function of fertility genes (Geldziler, Kadandale et al. 2004). Microarray analysis in C. elegans has helped identify numerous candidates for reverse genetic approaches; 1343 spermatogenesis-enriched and 1652 oogenesis-enriched genes were initially identified (Reinke, Gil et al. 2004) and subsequent studies have built upon to this original list (Reinke and Cutter 2009). Many of these genes may function in the events of fertilization.
RNAi is often used as a quick, loss-of-function, reverse genetic approach (Maeda, Kohara et al. 2001; Kamath and Ahringer 2003) (Also see chapter by Cipriani and Piano). Injecting, feeding or soaking worms to introduce dsRNA can systemically trigger gene-specific silencing in both the treated animal and its F1 progeny (Fire, Xu et al. 1998; Timmons and Fire 1998; Timmons, Court et al. 2001; Kamath and Ahringer 2003). RNAi has been successfully used to study C. elegans genes with early sperm or oocyte development functions (Sumiyoshi, Sugimoto et al. 2002), but most C. elegans sperm genes have proven refractory to this method for unknown reasons (Geldziler, Kadandale et al. 2004). Consequently, RNAi has not been a productive method for studying gene candidates with fertilization functions, and systematic RNAi screens have undoubtedly failed to identify many key molecules required for spermatogenesis and sperm function. Nevertheless, injection and soaking-based RNAi approaches may be useful in identifying at least some egg-sterile mutants (Maeda, Kohara et al. 2001; Kadandale, Stewart-Michaelis et al. 2005).
RNAi and similar knockdown methods are relatively easy ways to assess gene function, but RNAi-induced phenotypes are not genetic mutations and therefore require careful interpretation. Genes differ greatly in the extent to which they can be functionally reduced by RNAi; RNAi phenotypes range from no effect to total loss of function. RNAi phenotypes are also highly sensitive to experimental subtleties such as genetic background and exposure method, time, and temperature. For instance, the effective RNAi knockdown of the egg-1 and egg-2 genes requires injection or soaking treatment at 25°C to see a complete fertilization defect (Lee and Schedl 2001; Maeda, Kohara et al. 2001; Kadandale, Stewart-Michaelis et al. 2005). Feeding RNAi yields only incomplete knockdown phenotypes at best. Such variability can confound analyses and makes proper controls critically important.
Since RNAi methods do not provide actual germline mutations, other methods are required for further genetic studies/manipulations (Liu, Spoerke et al. 1999). Deletion library screening is one technique used extensively in C. elegans as a powerful large-scale method for deriving true knockout mutations in target genes known only via their sequence (Jansen, Hazendonk et al. 1997). Pools of mutagenized worms are split for long-term viable freezing and DNA extraction. The DNA pools are then screened for deletions in the gene of interest via PCR and populations containing such deletions are sequentially sub-divided to isolate a pure strain homozygous for the deletion. Such a strategy was used to demonstrate an essential role for various genes in fertilization (Xu and Sternberg 2003; Kadandale, Stewart-Michaelis et al. 2005; Parry, Velarde et al. 2009). New methods for gene knockout based on DNA transposition will also be important for generating fertility gene mutations (see chapter by Robert and Bessereau).
III. Maintaining Sterile Mutants
Once sterility mutations have been generated and identified it becomes necessary to maintain them. Non-conditional recessive spe hermaphrodites cannot be maintained as self-fertile homozygotes, but the mutant chromosome can be propagated by crossing spe hermaphrodites with wild-type males. These crosses are referred to in the literature variously as “rescue,” propagation, complementation or maintenance crosses (Fig. 5). Since these maintenance crosses are nothing more than standard genetic backcrosses (Fig. 5B), many spe mutant strains have been cleared of extraneous mutations. Non-conditional egg-sterile mutants cannot be complemented by mating, however, and must be maintained using labor intensive “sib selection” (Fig. 5D) prior to genetic balancing (see below); the mutation of interest is recovered from a heterozygous sibling of the homozygous sterile mutant. Since sibling heterozygotes are often phenotypically wild-type, worms must be individually plated and followed for the segregation of sterile progeny to assure the correct genotype and maintain the strain. This must be done with each generation.
Because fertile males are required to maintain sperm-sterile mutants as described above, a ready supply is essential. In C. elegans, males naturally arise at too low a frequency (1 in 1000, due to non-disjunction of the X chromosome) to be useful for this purpose (although heat shocking hermaphrodites at 25–30 °C for >6 hours will increase this frequency) (Hodgkin 1999). Consequently, many researchers keep daily matings of wild-type worms (which generate 50% male progeny) to ensure a sufficient quantity (Fig. 5A). Alternatively, specific him (high incidence of males) strains may be used (Hodgkin 1997). These mutants show increased X chromosome loss and therefore generate many males. him-3, him-5, and him-8 do not affect spermatogenesis and are routinely used for this purpose (Nelson and Ward 1980; Zannoni, L’Hernault et al. 2003). They do affect chromosome number and ploidy, however, so their use must be monitored to ensure they are not affecting the original function/process-of-interest. Although these are the most widely used methods for obtaining males, other methods such as heat-shock, ethanol treatment and RNAi against him genes can generate males for crossing (Hodgkin 1999; Fay 2006).
Maintaining sterile strains as heterozygotes is tedious and requires manual selection, making it impractical for large-scale operations. Fortunately, a variety of genetic balancers allows mutants of interest to be maintained and propagated without manual manipulation (Fig. 5E–G). We will only briefly mention genetic balancers here, referring the reader to Chapter 7 in the previous edition of this text and the chapter by Jones et al. in this edition for a more thorough discussion (Edgley, Baillie et al. 1995).
Many types of balancers can be used to maintain sterile mutations, including chromosomal rearrangements (translocations, duplications and inversions) and transgenes specifically constructed for this purpose. Regardless of type, all good balancers used for existing strain maintenance share common characteristics; heterozygotes (or transgene-containing animals) must be viable and fertile as well as have a unique, easy-to-score phenotype or mutations that effectively eliminate homozygote balancers from the population (see chapter by Jones et al.) (Edgley, Baillie et al. 1995).
When working with fundamental processes like fertility, one must take care to ensure that balanced strains are not lost. Fertility is highly selected, and any reversions or mutations conferring a fertility advantage will quickly take over a stock. Further, we have observed weakly fertile stocks become more fertile over many generations (I. Chatterjee and A. Singson, unpublished). Conversely, spontaneous mutations that further negatively affect fertility may arise in the balancer strains, confounding analyses. In addition, balancers themselves can occasionally stop functioning to DNA rearrangements, resulting in the loss of balancing. Therefore it is very important to carefully monitor balanced strains and to keep frozen stocks soon after their construction.
DNA injected into the worm gonad can form heritable extrachromosomal arrays (Mello and Fire 1995) that can be used to balance mutations. Because these balancing transgenes can often be lost, mosaic animals can be studied to determine whether a given sterile gene is required in the germline for its action. For example, spe-19 is maintained via an extrachromosomal balancing array that also contains a GFP marker (myo3::gfp, which expresses in body wall muscle) (Geldziler, Chatterjee et al. 2005). Studies of mosaic (glowing sterile and non-glowing fertile) animals revealed that glowing Spe animals failed to carry the transgene in the germline while rare non-glowing fertile animals sired glowing progeny (and therefore contained the transgene in the germline). These results strongly suggest that spe-19 is required in the germline for function, but do not rule out an additional requirement in somatic tissue. Note that transgene expression is often repressed in the germline (so-called “germline silencing”), somewhat limiting the usefulness of this type of analysis for fertility study (Putiri, Zannoni et al. 2004).
Maintenance of spe-8 mutations provides another balancer example. Using the free duplication sDp2 which contains wild-type copies of both dpy-5 and spe-8, the strain is maintained as a homozygous spe-8 dpy-5 double mutant and affords an easy visual assay for the spe-8 mutation. Animals complemented by the duplication are phenotypically wild-type; animals that do not contain the duplication are Spe and Dpy (Singson, Hill et al. 1999; Edgley, Baillie et al. 2006).
Of course, many sterile mutations are conditional and do not require balancers. Temperature-sensitive (ts) alleles of many sterility genes (both sperm and egg) exist or can be generated by appropriate screening strategies (L’Hernault, Shakes et al. 1988; Putiri, Zannoni et al. 2004; Kadandale, Stewart-Michaelis et al. 2005). Although, by definition, ts sterility alleles produce progeny at the permissive temperature, the fertility of these strains may not be as robust as in wild-type animals; they are sometimes leaky, giving low brood sizes at the permissive temperature.
In some instances, sterile mutants can be stably maintained as homozygotes through mating because their mutant phenotype is sex-specific. All members of the spe-8 class of genes, for example, are specifically required for hermaphrodite spermiogenesis (Shakes and Ward 1989; L’Hernault 1997; Geldziler, Chatterjee et al. 2005). However because the mutant males are fertile, homozygous populations can be maintained simply by crossing mutant males to mutant hermaphrodites. Mutants with male-specific spermatogenesis- defects also exist (Stanfield and Villeneuve 2006). Although ideal for maintaining mutant populations, these types of sex-specific sterile mutations are rare.
IV. Fecundity Analysis
Once mutant strains of interest have been isolated, out-crossed and genetically stabilized, they can be characterized in detail. Quantifying fertility using brood analysis, for example, is of fundamental importance for determining a mutation’s functional severity. In hermaphrodites this is done by allowing individually-plated wild-type and mutant L4 animals to self fertilize and then counting, averaging, and comparing the numbers of eggs/oocytes laid. In practice, eggs/oocytes are usually counted daily while the hermaphrodite is moved to new plate to facilitate counting the next day’s totals. Animals are typically transferred to new plates until no new eggs/oocytes are laid within a 24h period. Alternatively, the eggs/oocytes may be removed each day to avoid damaging delicate mutants via unnecessary manipulations. Brood sizes may also be examined over shorter time periods (an approach that is less traumatic for the worm and less labor intensive for the investigator). The total number of lifetime progeny can be assessed using the “mate to death” assay (Kimble and Ward 1988); the daily introduction of new males constantly replenishes the experimental hermaphrodite’s sperm supply. Brood results are most usefully expressed as the percentage of wild-type numbers; absolute numbers are less informative due to the wide variation in actual brood sizes among animals resulting from unknown or difficult-to-control factors. In studies of maternal-effect lethal mutants, quantitative assessments of fertility should include the percentage hatched as well as distinct categories for non/weak-shelled “oocytes” (oocyte or egg steriles), hard-shelled dead embryos and viable larvae (Fig. 2).
Male fertility is typically assessed by crossing mutant males to marked strains and counting the number of outcrossed (wild-type looking) progeny. dpy-5 is often used as a marker as it is easily scorable, but any easily identified recessive morphological marker will suffice. Another method obviates marked strains; one simply counts the number of F1 males and multiplies by two, since males sire 50% male progeny. Markers/mutations may affect male mating behavior/efficiency; some him mutants, for example, have smaller broods than wild-type animals due to aneuploidy, and almost all morphologically marked males have at least partially compromised mating efficiencies (Hodgkin 1997). Mating behavior/efficiency issues must therefore be distinguished from fertility issues when assessing overall fecundity.
Minimizing brood size variability due to environmental and genetic factors is important. Because fertility is age dependent, age-matched wild-type controls are essential. Nutritional effects can be minimized by using growth plates made at the same time and seeded from the same bacterial culture. Any plate contamination will complicate oocyte counts. For consistency of measurements, a single investigator should collect all data and proper statistical analysis must be performed to determine the significance of observed differences between groups. When assessing transgenic lines, it is important to remember that transgenic worms frequently produce smaller broods due to the unrelated effects of germline silencing (Putiri, Zannoni et al. 2004). The brood sizes of several different lines should be assessed as individual transgenic arrays frequently exhibit distinct levels of expression.
Directionality to sperm migration has been observed; sperm sometimes move to a single spermatheca rather than both, resulting in a mated Spe hermaphrodite laying a mix of eggs and unfertilized oocytes. If this is seen, the hermaphrodite should be examined to determine whether sperm are differentially localized in this manner.
V. Ovulation/Ovulation Rates
Ovulation levels/rates are closely related to overall brood sizes and can be used to differentiate Spe mutant classes. Meiotic maturation/ovulation is stimulated by a sperm-derived signal: the major sperm protein (MSP) (Miller, Nguyen et al. 2001; Kosinski, McDonald et al. 2005). On average, sperm-containing wild-type hermaphrodites ovulate approximately 2.5 times per gonad arm per hour, while fog-2 females (which lack sperm) ovulate only 0.09 times per gonad arm per hour (McCarter, Bartlett et al. 1999; Miller, Ruest et al. 2003). Consequently, ovulation rates can be used to indirectly assess the presence of spermatids/spermatozoa. The ovulation rates of spermatocyte-arrest Spe mutants are significantly lower than those of Spe mutants whose genes affect spermiogenesis/sperm function (Singson, Mercer et al. 1998; Kadandale and Singson 2004). Presumably only the spermatid/spermatozoa-producing Spe mutants have the ability to produce and respond to the MSP signal and ovulation rates have been used to distinguish spe mutant classes (Singson, Mercer et al. 1998; Chatterjee, Richmond et al. 2005; Singson, Hang et al. 2008).
Measurement of ovulation rates can be made in several ways. Paralyzed or anesthetized animals can be directly observed under a compound microscope using time-lapse video, for example (Ward and Carrel 1979; McCarter, Bartlett et al. 1999), or one can use a dissecting scope to count oocytes on single worm plates at set time intervals (Miller, Ruest et al. 2003; Kadandale and Singson 2004). Reproducible plate counts require fresh uncontaminated bacterial lawns but are less traumatic for the worms and yield results comparable to the more labor-intensive direct observations. In either method, age-matched controls are absolutely essential because the rate-limiting factor for ovulation rate changes with age; for young adults, it is oocyte growth (Miller, Ruest et al. 2003) while for older animals it is sperm. As with brood assessments, relative differences are more important than absolute numbers, and appropriate statistical analysis is mandatory.
Ovulation rates can be measured in either self-crossed or male-mated hermaphrodites. Self-crossed hermaphrodites are easier to use but if they become sperm-depleted during the course of the assay, their ovulation rates will decrease over time and result in lower overall averages unrelated to the mutant-of-interest. To avoid sperm depletion effects, mutant hermaphrodites may be plated with wild-type males to maintain non-basal ovulation rates for the duration of the assay. Conversely, the ability of mutant sperm to induce ovulation can be assessed by measuring the ovulation rates of fem or fog “females” crossed with mutant males.
Many ovulation-defective mutants exhibit a secondary phenotype whereby oocytes undergo multiple rounds of DNA synthesis to become polyploid (Iwasaki, McCarter et al. 1996). The location of these endomitotic (EMO) oocytes offers clues to the nature of the mutation; EMO oocytes in the gonad arm suggest spermathecal and/or ovulation defects while EMO oocytes in the uterus suggest defects in fertilization itself or in egg activation following sperm entry. Note that in the absence of sperm, wild-type hermaphrodites also produce EMO oocytes in their uterus (Ward and Carrel 1979).
VI. Spermatogenesis
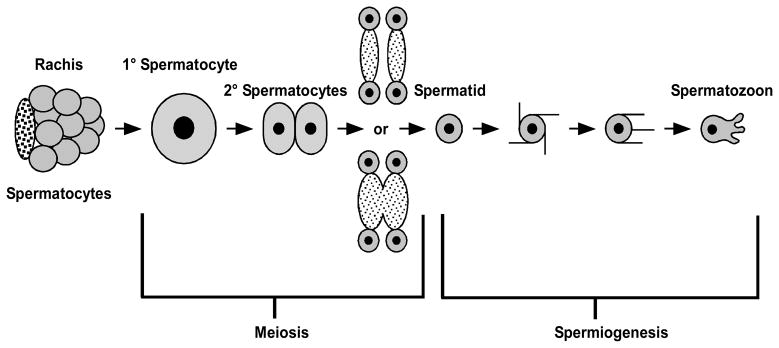
In C. elegans, gametogenesis and early spermatogenesis occur in a linear progression along the length of the tube-like gonad (Fig. 6, but also see Fig. 1). After leaving the distal mitotic zone, germ cells simultaneously enter meiosis and become committed to spermatogenesis (Jaramillo-Lambert, Ellefson et al. 2007). After homolog pairing, they then enter an extended period of pachytene. By mid to late pachytene, sperm-specific proteins begin to accumulate within the developing spermatocytes. As the most proximal spermatocytes exit pachytene, synaptonemal complexes between the paired homologs disassemble. Then global transcription ceases as the chromatin fully condenses and the spermatocytes enter a distinctive karyosome stage during which the chromosomes come together in single mass within the still-intact nuclear envelope (Shakes, Wu et al. 2009). Throughout this initial phase of spermatogenesis, the germ-cell nuclei are only partially encased within cellular membranes as the individual spermatocytes are syncytially connected to a common central cytoplasmic core called the rachis. Spermatocytes transition to the meiotic division stage of spermatogenesis as individual spermatocytes then detach from this rachis and their microtubule organization switches from a network to a centrosome-based pattern (Shakes, Wu et al. 2009).
Figure 6.
Spermatogenesis in C. elegans. Major developmental stages are indicated.
During the first meiotic division, 1° spermatocytes undergo a symmetrical, actin-based (and sometimes incomplete) division to form two 2° spermatocytes. The second round of meiotic chromosome segregation rapidly follows. Anaphase II is followed by the transient formation of a shallow cleavage furrow, but this furrow rapidly regresses. Cellular components unnecessary for subsequent sperm formation accumulate in a central residual body whereas individual spermatids bud and detach from this residual body using poorly understood, non-actin-based mechanisms of division (Ward, Argon et al. 1981; Shakes, Wu et al. 2009). Once spermatids detach from the residual body, they undergo a rapid maturation process that includes final compaction of the sperm chromatin, release of MSP from a paracrystalline assembly known as the fibrous body into the cytosol, and docking of the sperm-specific “membranous organelles (MOs) with the plasma membrane (Ward, Argon et al. 1981; Shakes, Wu et al. 2009).
Although some investigators are beginning to include both residual body formation and initial events of spermatid maturation as part of “spermiogenesis” (Wu, Nera et al. 2010), within the C. elegans literature, spermiogenesis is most frequently defined as the subsequent dramatic morphogenesis of spherical, sessile spermatids into amoeboid, motile, fertilization-competent spermatozoa (Shakes and Ward 1989). Cellular changes include plasma membrane flow to the site of the newly developing pseudopod, fusion of caveolae-like membranous organelles (MO) to the cell body plasma membrane, and the formation of a dynamic MSP pseudopod cytoskeleton. The entire transition takes less than ten minutes, and the formation of filopodia-like spikes precedes the formation of fully motile pseudopods (Shakes and Ward 1989; Singaravelu, Chatterjee et al. 2011). For male-derived spermatids, spermiogenesis occurs after ejaculation. For hermaphrodite-derived spermatids, the process begins as they are pushed into the spermatheca with the passage of the first oocyte (Singson 2006). In males, premature spermiogenesis is actively repressed by a protease inhibitor known as SWM-1 (Stanfield and Villeneuve 2006). Unlike many other examples of morphogenetic change, C. elegans spermiogenesis occurs without actin/tubulin cytoskeleton regulation or new protein synthesis; ribosomes, actin and tubulin are all discarded during the spermatid budding division.
It is important to distinguish between sperm function mutants (which affect fertilization directly via defects in sperm-egg interaction) and sperm development mutants (which affect the process only indirectly via defects in sperm morphogenesis or motility). Sperm development itself can be quickly assessed in microscope preparations of dissected worms. To study early spermatogenesis, “sperm squashes” are made: males are placed in sperm buffer and dissected at the pharynx-intestine junction. A coverslip is used to press the gonad into a monolayer of spermatocytes and spermatids that are then examined using DIC microscopy for spermatid number, shape and size. Lipid-soluble dyes such as Hoechst 33342 may be included to detect DNA via epifluorescence, although UV exposure causes the cells to deteriorate rapidly. For further analysis, one can quick freeze these sperm squash preparations on dry ice, remove the coverslip, and prepare the sample for immunofluorescence (for more details see the Immunofluorescence chapter in this volume by Shakes et al. and Shakes et al., 2009). Useful markers for the general staging of spermatogenesis include combinations of DAPI (DNA) with antibodies against α-tubulin (Sigma FITC-labeled DM1A), phosphorylated histone H3 (Ser10) (Upstate Biotechnology), and/or MSP (Developmental Studies Hybridoma Bank).
Spermatocytes/spermatids can be easily isolated from males, but naturally activated spermatozoa must usually be isolated from the gonads of mated hermaphrodites (Chatterjee, Richmond et al. 2005). The isolation of in vivo activated spermatozoa directly from self-fertile hermaphrodites is substantially more challenging as hermaphrodite spermatozoa are 50% smaller (LaMunyon and Ward 1998; Geldziler, Chatterjee et al. 2006) and significantly less numerous.
Spermiogenesis may also be assessed in vitro via the addition of known activators to sperm media before worm dissection. Pronase (a non-specific collection of proteases) can be used to examine activation using light microscopy, but other choices such as monesin (a cationic ionophore) or triethanolamine (a compound affecting intracellular pH) are more appropriate for studies using immunofluorescence (Roberts, Pavalko et al. 1986; Zannoni, L’Hernault et al. 2003; Chatterjee, Richmond et al. 2005; Singaravelu, Chatterjee et al. 2011).
VII. Sperm Migratory Behavior/Tracking
For successful C. elegans fertilization, male-derived spermatozoa must first migrate from the vulva (the site of insemination) to the spermatheca. Sperm (regardless of derivation) must then remain there, either by firmly attaching themselves to the spermatheca wall or by crawling back if dislodged by passing oocytes. Defects in hermaphrodite sperm retention can be assessed by comparing the number of sperm in whole-mount, DAPI-stained young adult hermaphrodites immediately before egg-laying with that of slightly older siblings that have already begun laying eggs (L’Hernault, Shakes et al. 1988). The morphology of DAPI-stained sperm nuclei is quite distinctive and can be easily scored within the spermatheca and uterus. Alternatively, the hermaphrodite gonad can be directly observed using DIC microscopy and time-lapse photography. Animals can be dissected on glass microscope slides or anesthetized on agar pads to study sperm number and morphology. It can be challenging, however, to follow the four-dimensional movements of an individual spermatozoon using DIC optics alone.
Fluorescent methods for sperm tracking experiments include the use of Nile Blue (Ward and Carrel 1979), SYTO17 (Singson, Mercer et al. 1998; Hill and L’Hernault 2001), or MITO tracker (Kubagawa, Watts et al. 2006; Stanfield and Villeneuve 2006). Males are labeled by soaking them in a dye-containing solution and are then mated to unlabeled hermaphrodites. These hermaphrodites are subsequently anesthetized and examined via fluorescent microscopy for the presence and location of sperm.
Care must be taken to ensure that the fluorescent dye used does not adversely affect the process/function under study. The use of mutant hermaphrodites with reduced gut autofluorescence (such as daf-4 or glo-1) facilitates the identification of male-derived sperm within the female reproductive tract (Hill and L’Hernault 2001; Artal-Sanz, Tsang et al. 2003; Kroft, Gleason et al. 2005).
VIII. Sperm Competition
Although hermaphrodite self-fertilization is the primary mode of C. elegans reproduction, mated hermaphrodites produce predominantly outcrossed progeny because of the competitive superiority of male-derived sperm (termed “sperm competition”). This phenomenon is a primary reason that C. elegans is such a powerful genetic system. The transition towards outcross progeny typically occurs within one day after male-introduction; by day two, progeny are almost exclusively outcrossed (Ward and Carrel 1979; LaMunyon and Ward 1995; LaMunyon and Ward 1997; LaMunyon and Ward 1998; LaMunyon and Ward 1999; Singson, Hill et al. 1999). Sperm competition is independent of the ability to fertilize; the sperm of fertilization-defective C. elegans mutants can effectively out-compete hermaphrodite sperm, reducing the self-fertility of these mated hermaphrodites (Singson, Hill et al. 1999). The dominance of male-derived sperm may be partially due to size; C. elegans male-derived sperm are approximately 50% larger than hermaphrodite-derived sperm, move faster, and are thought to displace them from the distal end of the spermatheca (LaMunyon and Ward 1998). However, size per se cannot be the complete explanation, as C. remanei male-derived sperm do not reduce self-fertility in mated C. elegans hermaphrodites, despite being two times larger in diameter (Hill and L’Hernault 2001). C. elegans hermaphrodites may also have an independent mechanism for actively selecting functional sperm (Kadandale and Singson 2004).
Sequential mating experiments with multiple males suggest sperm competition in C. elegans is limited to males vs. hermaphrodites. This is perhaps not surprising given that in wild populations male sperm are more likely to encounter hermaphrodite sperm within the spermatheca than sperm from another male (LaMunyon and Ward 1999).
The competitive ability of male-derived sperm has useful implications for the study of sterile mutants in C. elegans. To be competitive, the males themselves must successfully mate and transfer sperm to the hermaphrodite while their sperm must have successfully completed in vivo spermiogenesis, become motile, and migrated to the spermatheca. Sperm competition assays are therefore valuable tools for the characterization of sterile mutations in C. elegans, but these assumptions should be confirmed via other direct means (such as the sperm tracking methods described above). In a typical experiment, morphologically marked L4 hermaphrodites (e.g. dpy-5) are crossed with unmarked mutant males. The numbers of outcrossed (Non-Dpy) and selfed (Dpy) progeny are counted, and the percentage self-progeny is recorded (Singson, Hill et al. 1999; Xu and Sternberg 2003; Chatterjee, Richmond et al. 2005; Geldziler, Chatterjee et al. 2005; Kroft, Gleason et al. 2005).
In addition to unmated hermaphrodite and wild-type male controls, only healthy and age-matched worms should be used as male mating behavior and efficiency may be affected by age and overall health. To minimize variations in the microenvironment, all crosses should be done concurrently using the same food batch and incubator.
IX. Oogenesis and Oocyte Maturation
The earliest stages of oocyte and sperm development are indistinguishable in C. elegans hermaphrodites; a common pool of germ cells gives rise to both gamete types (Fig. 1). During oogenesis, the most proximal pachytene nuclei are induced to exit their arrest by a localized MAPK-mediated signal (Church, Guan et al. 1995). They then undergo programmed cell death or begin the late stage of oocyte growth and differentiation (Gumienny, Lambie et al. 1999). This developmental switch is partly regulated by GLD-1, which localizes to the pachytene region of the germline where it actively represses the expression of late stage oocyte differentiation markers (Lee and Schedl 2001). As non-apoptotic presumptive oocytes then pass through the loop region, they progress from diplotene to diakinesis. During this phase they rapidly enlarge while remaining connected to a progressively narrowing rachis. In wild-type hermaphrodites only the most proximal oocytes are fully cellularized, and only the most proximal oocyte undergoes oocyte maturation.
A variety of cell cycle and differentiation markers are available to check the developmental progression of proximal oocytes for those egg-sterile mutants that may have defects in late-stage oocyte differentiation. In the early stages of this post-loop differentiation process, wild-type oocytes begin to express two proteins whose levels continuously increase during oocyte development - OMA-1 (Detwiler, Reuben et al. 2001; Lin 2003) and the yolk protein receptor RME-2 (Grant and Hirsh 1999). The uptake of intestinally-synthesized yolk proteins can also be visualized using the vitellegenin GFP strain YP170::GFP (Grant and Hirsh 1999).
As oocytes progress closer to the spermatheca, they can be distinguished by their temporal response to the MSP maturation signal provided by sperm in the spermatheca (McCarter, Bartlett et al. 1999; Miller, Ruest et al. 2003). Approximately every 23 minutes, the most proximal oocyte in the gonad undergoes meiotic maturation in response to MSP via VAB-1, an Eph receptor protein tyrosine kinase (McCarter, Bartlett et al. 1999; Miller, Ruest et al. 2003). This progressive response can be monitored using antibodies against activated MAPK (MAPK-YT) and Ser10 phosphohistone H3 (Hsu, Sun et al. 2000; Miller, Nguyen et al. 2001).
Maturation promoting factor (MPF) is necessary for allowing the oocyte to progress from mitotic division to metaphase I of the first meiosis (Schmitt and Nebreda 2002; Burrows, Sceurman et al. 2006). MPF is a complex of a cyclin-dependent kinase (Cdk1) and cyclin B and the activity of this complex is inhibited by WEE-1.3 (Doree and Hunt 2002; Burrows, Sceurman et al. 2006). MPF promotes a progression of events beginning as early as in the -3 oocyte (the third oocyte proximal to the spermatheca). One of the earliest events is the phosphorylation of serine 10 on histone H3 (Burrows, Sceurman et al. 2006). The subsequent sequence of events occur primarily in the -1 oocytes and can be visualized using DIC microscopy. These include the disappearance of the nucleolus, migration of the nucleus to the distal side of the oocyte, nuclear envelope breakdown, and rounding of the oocyte (McCarter, Bartlett et al. 1999). Other events including entry into metaphase I of meiosis and formation of the meiosis I spindle can be assessed in strains that include expression constructs with GFP:His2B and GFP:tubulin to visualize the chromatin and microtubule structures (Tenenhaus, Subramaniam et al. 2001; McNally, Audhya et al. 2006). The anaphase promoting complex subsequently promotes the rotation of the meiotic spindle and the metaphase to anaphase transition (McNally and McNally 2005).
X. Assessing Fertilization and Egg Activation in Egg-Sterile Mutants
The oocyte enters the spermatheca in metaphase I of the first meiotic division and meiotic resumption and fertilization occur concurrently (Ward and Carrel 1979; McNally and McNally 2005). As the oocyte enters the spermatheca, the meiosis I spindle assembles into in a pentagonal array of chromosomes. Next the DNA is translocated to the cortex, the spindle then rotates perpendicular to the cortex, and the chromosomes begin to separate (Albertson and Thomson 1993; McNally and McNally 2005). If fertilization occurs, the cell will proceed through anaphase I and half of the chromosomes will be deposited into the first polar body (McNally and McNally 2005). The oocyte chromosomes subsequently undergo the second meiotic division and a second polar body is extruded. Finally, pronuclei form around both the maternal and paternal chromatin (Sadler and Shakes 2000; McNally and McNally 2005).
In addition to triggering meiotic resumption, the newly fertilized egg undergoes a number of changes that are necessary for proper egg activation and embryo development. These include the activation and/or degradation of selected maternal mRNAs and proteins, release of cortical granules (CGs), secretion of a chitinous eggshell, and the mounting a membrane block to polyspermy to prevent fertilization by a second sperm (Stitzel, Cheng et al. 2007; Horner and Wolfner 2008; Singson, Hang et al. 2008; Marcello and Singson 2010). If all of these processes are coordinated and completed properly, the embryo will develop as it passes through the uterus and is eventually laid at approximately the 30-cell stage (Ringstad and Horvitz 2008; Singson, Hang et al. 2008).
In mutagenesis or RNAi screens, egg-sterile animals (egg) with defects in either sperm-egg fusion or egg activation are identified in the same way as spe mutants. Animals are screened for mutants that lay “eggs” that either possess weak, osmotically-sensitive eggshells (Fig. 2E) or lack eggshells altogether (Fig. 2D). To distinguish egg activation defect mutants from fertilization defective mutants, DAPI staining can be used to score young meiotic stage “eggs” within the spermatheca or uterus for the presence of sperm chromatin (sperm entry) (Fig. 7).
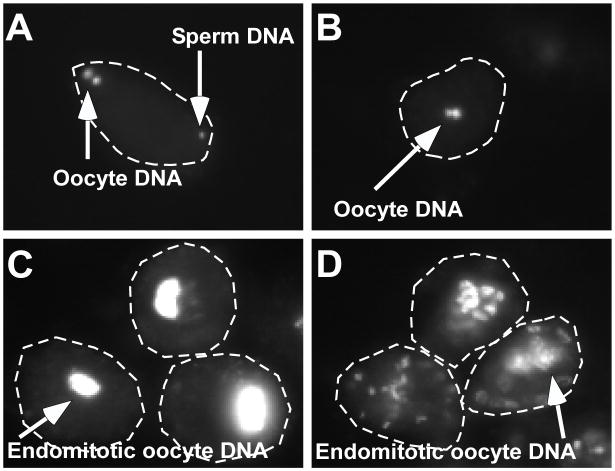
Figure 7.
DAPI-stained dissected oocytes. (A) Newly-fertilized oocyte with both oocyte DNA and a visible sperm chromatin mass. (B) Unfertilized oocytes lacking a sperm chromatin mass. (C) Older endomitotic unfertilized oocyte. (D) Older endomitotic egg activation mutant oocyte.
If mutant oocytes remain unfertilized, they will lack both a sperm chromatin mass and meiotic polar bodies (McNally and McNally 2005) (Fig. 7B). In unfertilized oocytes, the maternal chromosomes initiate the meiotic divisions and reach anaphase I but the resulting anaphase chromosome masses subsequently decondense to form two distinct pronuclei. These unfertilized oocytes fail to form polar bodies or attempt the second meiotic division (McNally and McNally 2005). During the subsequent rounds of endomitotic cell cycling they form a single, large, polyploid DNA mass (Doniach and Hodgkin 1984; Miller, Ruest et al. 2003; Chatterjee, Richmond et al. 2005) (Fig. 7C).
In contrast, newly fertilized oocytes from maternal or paternal egg activation mutants are expected to contain a visible sperm chromatin mass (Kadandale, Geldziler et al. 2005; Maruyama, Velarde et al. 2007; Parry, Velarde et al. 2009) (Fig. 7A). Further, in egg activation mutants, a distinct endomitotic phenotype is seen. Rather than having a single large polyploidy DNA mass, many smaller DNA masses are formed (Fig. 7D). This is likely due to the action of sperm-contributed centrosomes separating newly replicated DNA with each endomitotic cyclie. This difference in DNA morphology can be used to help determine whether sperm entry has occurred.
Post-fertilization defects in meiosis or subsequent cell divisions can be observed in fixed embryos by staining with combinations of DAPI, the M-phase marker anti-phosphohistone H3 (serine 10), and anti-β-tubulin DM1A antibodies (Golden 2000). These defects can also be seen in live embryos by crossing to strains containing either pie-1:Histone 2B:GFP (Praitis, Casey et al. 2001) or pie-1:GFP:β-tubulin (Strome, Powers et al. 2001).
EGG-1 and EGG-2 are two proteins in the plasma membrane of the oocyte that are necessary for fertilization (Kadandale, Stewart-Michaelis et al. 2005). These genes have proven to be extremely useful diagnostic tools for analyzing egg-sterile mutants with defects in fertilization (Kadandale, Stewart-Michaelis et al. 2005). Both genes encode type II transmembrane molecules and their extracellular domains include eight low-density lipoprotein (LDL)-receptor-repeats, which are known to function as receptors for a variety of ligands and mediate multiple cellular responses (Nykjaer and Willnow 2002; Kadandale, Stewart-Michaelis et al. 2005). Sperm are able to make contact with egg-1/2 deficient oocytes but are unable to fuse with them (Kadandale, Stewart-Michaelis et al. 2005). The localization of EGG-1 and EGG-2 can be visualized using GFP-tagged versions of the proteins. GFP:EGG-1 (Fig. 8G) and EGG-2:GFP are localized to the surface of developing oocytes but become undetectable after fertilization, presumably as a result of endocytosis (Kadandale, Stewart-Michaelis et al. 2005).
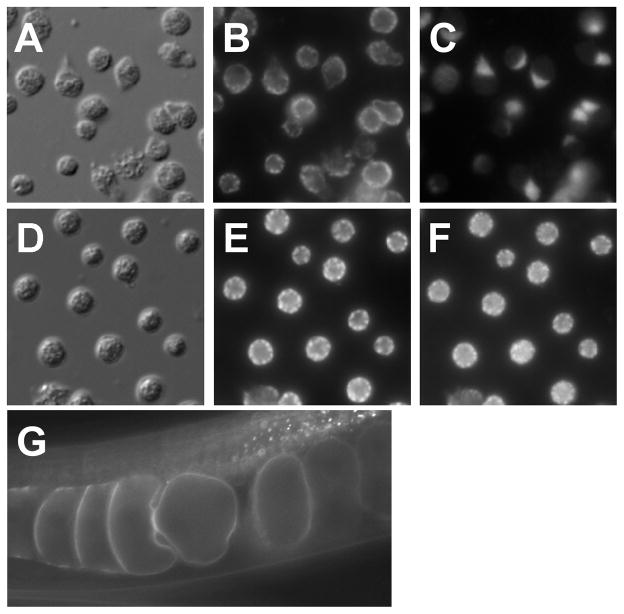
Figure 8.
Examples of sperm and oocyte protein localization. (A–C) Localization of 1CB4 and SPE-9. (A) Nomarski DIC micrograph of mature spermatozoa. (B) Localization of 1CB4 to MOs. (C) Localization of SPE-9 to pseudopods. (D–F) Co-localization of 1CB4 and SPE-38 in spermatids. (D) Nomarski DIC micrograph of spermatids. (E) Localization of 1CB4 to MOs. (F) Localization of SPE-38 to MOs. Note the identical distribution of staining in panels E and F. (G) GFP fluorescence of a GFP:EGG-1 fusion protein in oocytes.
Oocyte maturation, fertilization, and the completion of meiosis are overlapping and dependent processes that are coordinated by a number of important regulatory factors (Marcello and Singson 2010). Defects in any step can have deleterious effects on subsequent steps and ultimately result in improper embryogenesis. To understand how a specific gene or protein of interest functions in these processes, mutants with defects in specific stages of meiosis or related GFP marked strains can be used. For example, to analyze the relationship between cell cycle progression and egg activation, the activity of the anaphase-promoting complex (APC/C) was blocked using either temperature-sensitive mutants or RNAi. In oocytes depleted of the APC/C subunit mat-1, fertilization occurs, but the fertilized oocytes remain locked in a meiotic metaphase I state and subsequent events of the oocyte-to-embryo transition do not occur (Golden, Sadler et al. 2000). In other studies, cyclin B:GFP levels have been used to indicate meiotic progression. Degradation of maternally supplied cyclin B by the APC/C was shown to drive the transition from metaphase I to anaphase I (Furuta, Tuck et al. 2000; Golden, Sadler et al. 2000; Davis, Wille et al. 2002; Rahman and Kipreos 2010); however, in the absence of fertilization, oocytes progress to anaphase I but the drop in their cyclin B levels proved to be incomplete (McNally and McNally 2005). Destruction of the remaining cyclin B was found to require fertilization and passage through the second meiotic division and more specifically, a second E3 ubiquitin complex, Cul-2/Zyg-11, which allows progression to anaphase II (Liu, Vasudevan et al. 2004; Sonneville and Gonczy 2004). The use of mat-1-depleted animals and cyclin B:GFP will help place the functioning of many genes or proteins of interest in the context of meiotic progression. The coordination of the cell cycle, fertilization and egg activation are still poorly understood. The concurrent and dependent processes of meiotic resumption and fertilization and their subsequent events likely result in a multitude of branching cellular pathways (Singson, Hang et al. 2008; Marcello and Singson 2010).
Egg-activation mutants are likely to fall into different classes, and appropriate GFP strains and disruption of gene function by RNAi or mutation can be used to analyze genes or proteins of interest in egg activation. Sperm entry promotes the establishment of embryonic polarity and triggers egg activation (Goldstein and Hird 1996; Sadler and Shakes 2000). One example of a paternal egg activation mutant is spe-11; when spe-11 sperm fertilize wild-type oocytes, the oocytes fail to either complete meiosis or secrete an eggshell, but instead form multi-nucleate one-cell embryos (Hill, Shakes et al. 1989; Browning and Strome 1996). Other egg activation mutants may arise from mutations in maternally-required genes that regulate and coordinate egg activation events (Maruyama, Velarde et al. 2007; Stitzel, Cheng et al. 2007; Cheng, Klancer et al. 2009; Parry, Velarde et al. 2009). Such genes would include regulatory proteins at the oocyte cortex that couple fertilization with cell cycle progression through the second meiotic division and the events of egg activation (Govindan and Greenstein 2007; Maruyama, Velarde et al. 2007; Parry, Velarde et al. 2009; Parry and Singson 2011). For example, EGG-3, EGG-4, and EGG-5 are pseudo-phosphatases located in the oocyte cortex that coordinate fertilization with cell cycle resumption by regulating the activity of minibrain kinase-2 (MBK-2) (Maruyama, Velarde et al. 2007; Stitzel, Cheng et al. 2007; Stitzel and Seydoux 2007; Cheng, Klancer et al. 2009; Parry, Velarde et al. 2009). MBK-2 helps regulate early embryonic development after fertilization by phosphorylating maternal substrates for degradation. These MBK-2 targets include the katanin subunit MEI-1, the RNA/TAF-4 binding proteins OMA-1 and OMA-2, and the polarity factors MEX-5 and MEX-6 (Detwiler, Reuben et al. 2001; Pellettieri, Reinke et al. 2003; Quintin, Mains et al. 2003; Pang, Ishidate et al. 2004; Nishi and Lin 2005; Guven-Ozkan, Nishi et al. 2008). Appropriate GFP strains can be used to assess whether other egg activation mutants exhibit defects in the localization, sorting, or degradation of any of these known components.
XI. Eggshell Production
In response to fertilization and proper egg activation, a multi-layered eggshell is formed around the developing embryo to allow for completion of meiosis, polar body extrusion, embryo polarity and formation of an osmotic barrier. The developing oocyte is covered by a thin vitelline layer that can be observed by electron microscopy or staining with Malcura pomifera agglutinin (MPA) or Griffonia simplicifolia lectin I (GSL I). At the time of fertilization this vitelline layer begins to separate from the plasma membrane and becomes the outermost layer of the mature eggshell (Rappleye, Paredez et al. 1999; Bembenek, Richie et al. 2007). The second (middle) layer is formed when the oocyte membrane protein CHS-1 catalyses UDP-N-acteylglucosamine polymerization to produce chitin, the material that gives eggshells their mechanical strength. Chitin deposition can be assayed by staining with a rhodamine-conjugated chitin-binding probe. The proteolipid inner layer is formed immediately before the first zygotic cell division and provides a permeability and osmotic barrier. The fidelity of this osmotic/permeability barrier can be determined by staining with lipophilic fluorescent plasma membrane dye FM 4–64, which can only penetrate and stain embryos that have not yet formed an intact barrier or eggshell.
Unfertilized oocytes and activation-defective eggs are sensitive to osmotic strength due to the lack of a protective eggshell and must be handled with special care. Intact and dissected hermaphrodites should at a minimum be handled in osmotic egg buffers (Edgar 1995). Otherwise, unfertilized oocytes may burst under osmotic pressure and even intact embryos may swell within a weak eggshell and cause an embryo to be mistakenly scored as cytokinesis defective. In some cases, oocytes are so fragile that dissections from the uterus are impossible. These oocytes must be examined carefully in intact animals (Parry, Velarde et al. 2009).
Cortical granule (CG) exocytosis is necessary for proper eggshell formation (Bembenek, Richie et al. 2007). CGs transport chondroitin proteoglycans to the extracellular space surrounding the embryo, and can be detected by Wheat Germ Aggultinin (WGA) or the Golgi marker UGTP-1 (Bembenek, Richie et al. 2007). CAV-1 is prominent but non-essential component of CGs (Sato, Grant et al. 2008). CG exocytosis, which occurs during anaphase I, does not require fertilization. However, it does require a variety of cell cycle components including the APC/C and separase (sep-1) as well as the small GTPase RAB-11 and the target-SNARE SYN-4 (Bembenek, Richie et al. 2007). Additional exocyotic events must be required for proper eggshell formation; however, the exact nature of these events remains unclear (Bembenek, Richie et al. 2007).
Although cortical granule exocytosis is known to contribute to the membrane-based polyspermy block in other organisms, the connection between CG exocytosis and polyspermy in C. elegans is not well understood (Wessel, Brooks et al. 2001). In C. elegans, polyspermy has been observed after the depletion of chs-1, gna-2, or egg-4/5 (Parry, Velarde et al. 2009; Johnston, Krizus et al. 2010). gna-2 encodes a GLD-regulated glucosamine-6-P N acetyltransferase that supplies UDP-N-acteyl glucosamine for chitin biosynthesis (Johnston, Krizus et al. 2006). The deposition of chitin is independent of CG exocytosis and the presence of chitin seems to play a role in the membrane-based polyspermy block (Sato, Sato et al. 2006; Sato, Grant et al. 2008; Parry, Velarde et al. 2009; Johnston, Krizus et al. 2010). Potential defects in the polyspermy block can be assessed by DAPI staining intact hermaphrodites or dissected embryos and looking for evidence of multiple sperm chromatin masses within recently fertilized, meiotic stage oocytes (Parry, Velarde et al. 2009; Johnston, Krizus et al. 2010). However, in intact or poorly dissected animals, it can be difficult to distinguish nuclei within the embryo from those in the surrounding periphery (Parry, Velarde et al. 2009; Johnston, Krizus et al. 2010). When staining dissected embryos that do not have eggshells, a grouping of embryos can lack clear demarcation of each embryo boundary. The use of GFP:PH (Plextrin Homology) construct can be used to help visualize the plasma membrane (Audhya, Hyndman et al. 2005) and minimize this problem.
XII. Analysis of Fertilization-Specific Gene Products
Antibody-based approaches are useful for determining the sub-cellular localization of fertilization-specific gene products. The standard approach is to generate polyclonal anti-peptide antibodies against two or more regions of the candidate fertility protein since not all regions will be useful for generating antibodies (Zannoni, L’Hernault et al. 2003; Chatterjee, Richmond et al. 2005; Parry, Velarde et al. 2009). For these fertility proteins, polyclonal, anti-peptide antibodies have two distinct advantages over monoclonal antibodies: 1) they do not require the isolation of pure sperm and oocytes, 2) polyclonal antibodies typically have higher binding efficiencies, which is useful when attempting to detect cell-surface proteins that are typically expressed at low levels. It is also important always to pre-screen the animals used for antibody production since many express unrelated nematode antibodies stemming from previous nematode infections.
It is useful to understand the localization pattern of SPE proteins in both spermatids and spermatozoa. Sperm squashes are used for staining spermatids, and spermatozoa preparations can be obtained either from mated hermaphrodites or from males dissected in non-protease-based sperm activators. Samples should be fixed separately in both paraformaldehyde and cold methanol as membrane proteins can differ in their response to each. C. elegans spermatids are small and possess only four major organelles, (a central chromatin mass, an associated inactive centriole, numerous mitochondria, and multiple membranous organelles [MOs]) simplifying the analysis of localization patterns. MO localization may be confirmed using the monoclonal antibody 1CB4 (Okamoto and Thomson 1985) as was done for the proteins SPE-9 and SPE-38 (Zannoni, L’Hernault et al. 2003; Chatterjee, Richmond et al. 2005) (Fig. 8A–F).
Live cell staining (with antibodies added before fixation) can also be used to assess protein localization on the external surface of the plasma membrane or in fused MOs (Chatterjee, Richmond et al. 2005). fer-1 mutants can be used to confirm an initial restriction to the MO since the fer-1 spermatozoa are specifically defective in MO fusion despite their ability to form a small motile pseudopod (Achanzar and Ward 1997; Xu and Sternberg 2003; Chatterjee, Richmond et al. 2005).
To date, all fertilization-defective SPE proteins have at least partially localized to the pseudopods of spermatozoa, now presumed to be the point of oocyte-sperm contact (Xu and Sternberg 2003; Zannoni, L’Hernault et al. 2003; Chatterjee, Richmond et al. 2005). In spermatids, some fertilization-defective SPE proteins like SPE-9 localize to the plasma membrane (Zannoni, L’Hernault et al. 2003) while others, like SPE-38 and TRP-3/SPE-41, localize to the unfused MOs of spermatids. Any patterns of localization common to multiple members of the SPE-9 class remain to be determined (Fig. 8) (Chatterjee, Richmond et al. 2005).
Fusion proteins (e.g. GFP fusions) can be convenient tools for determining a protein’s distribution and dynamics in fixed or live cells. However, the expression of oocyte and sperm fusion proteins is often repressed in the germline (Kelly, Xu et al. 1997; Seydoux and Schedl 2001; Putiri, Zannoni et al. 2004). Oocyte fusion protein expression (Fig. 8G) can be enhanced using complex arrays (Kelly, Xu et al. 1997), integrated low copy number microparticle bombardment transformation strategies (Praitis, Casey et al. 2001), or Mos1 transposon single copy insertion (MosSCI) based methods (Robert and Bessereau 2010). Reliable expression of sperm fusion proteins has not yet been achieved. For a more in depth discussion of protein localization studies see the chapter by Hutter.
XIII. Future Prospects/Issues
This chapter attempts to provide a useful overview of how C. elegans can be used as a model system for addressing questions of fertility. A clear understanding of any biological process as complex as fertilization is impossible without a complete inventory of its cellular and molecular components; consequently, each new gene identified adds significantly to our knowledge and insight into fertilization mechanisms.
Although much has already been elucidated via C. elegans fertility research, many fundamental questions remain unanswered. For example, how are the events surrounding fertilization (e.g. meiotic maturation, ovulation, fertilization, sperm migratory behavior and the nature of signaling events) coordinated? What is the mechanism of the block to polyspermy? How are the events of fertilization and egg activation related to early development and patterning of the embryo?
The fundamental question of what happens to sperm and oocyte fertility proteins during the physical joining of the gametes is being actively studied in our laboratories. The investigation of this deceptively simple question has proven challenging since there are at most only two recently fertilized oocytes present in any hermaphrodite at one time, and only those in the very earliest stage of post-fertilization meiosis are informative (A. Richmond and D. Shakes, unpublished data). Although difficult, such studies are feasible and will be ultimately useful in analyzing the interactions between sperm and oocyte fertility proteins.
Exciting progress continues to be made and the field is still developing new experimental tools. Calcium imaging, for instance, has been applied to C. elegans sperm and oocytes (Samuel, Murthy et al. 2001; Xu and Sternberg 2003). Using Calcium Green-1 dextran as an indicator, an increase in cytoplasmic calcium is observed at the same time as fertilization, but the trigger for the calcium release and its functional consequence are unknown (Samuel, Murthy et al. 2001). Voltage-sensitive reagents or other physiological approaches could be used to test whether the fast block to polyspermy is dependent on depolarization of the oocyte plasma membrane (Yanagimachi 1994) as in other organisms. New transgenic approaches are helping to overcome the germline expression repression that currently hampers research in this area (Robert and Bessereau 2010). In vitro fertilization systems in C. elegans could be useful for analyzing the function of newly discovered molecules; however, the important physical interactions between gametes and the somatic germline suggest that this approach may not be feasible.
Insights gained from the study of fertilization in C. elegans will also increase our understanding of diverse reproductive strategies and those mechanisms relevant to molecular evolution and speciation. The recent sequencing projects of other nematode species (Bird, Blaxter et al. 2005; Mitreva, Blaxter et al. 2005; Ghedin, Wang et al. 2007) and baseline spe gene phenotypic data (Geldziler, Chatterjee et al. 2006) should enable this work to progress quickly.
C. elegans remains an extremely useful organism with which to study the nature of fertilization. An integrated approach to the analysis of fertilization that combines molecular genetic and cell biological worm techniques with the more traditional antibody-based and biochemical methods will continue to further our understanding of this most fascinating and fundamental process.
References
- Achanzar WE, Ward S. A nematode gene required for sperm vesicle fusion. Journal of Cell Science. 1997;110:1073–1081. doi: 10.1242/jcs.110.9.1073. [DOI] [PubMed] [Google Scholar]
- Albertson DG, Thomson JN. Segregation of holocentric chromosomes at meiosis in the nematode, Caenorhabditis elegans. Chromosome Res. 1993;1(1):15–26. doi: 10.1007/BF00710603. [DOI] [PubMed] [Google Scholar]
- Aroian RV, Field C, et al. Isolation of actin-associated proteins from Caenorhabditis elegans oocytes and their localization in the early embryo. Embo J. 1997;16(7):1541–9. doi: 10.1093/emboj/16.7.1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artal-Sanz M, Tsang WY, et al. The mitochondrial prohibitin complex is essential for embryonic viability and germline function in Caenorhabditis elegans. J Biol Chem. 2003;278(34):32091–9. doi: 10.1074/jbc.M304877200. [DOI] [PubMed] [Google Scholar]
- Audhya A, Hyndman F, et al. A complex containing the Sm protein CAR-1 and the RNA helicase CGH-1 is required for embryonic cytokinesis in Caenorhabditis elegans. J Cell Biol. 2005;171(2):267–79. doi: 10.1083/jcb.200506124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bembenek JN, Richie CT, et al. Cortical granule exocytosis in C. elegans is regulated by cell cycle components including separase. Development. 2007;134(21):3837–48. doi: 10.1242/dev.011361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird DM, Blaxter ML, et al. A white paper on nematode comparative genomics. J Nematol. 2005;37(4):408–16. [PMC free article] [PubMed] [Google Scholar]
- Browning H, Strome S. A sperm-supplied factor required for embryogenesis in C. elegans. Development. 1996;122:391–404. doi: 10.1242/dev.122.1.391. [DOI] [PubMed] [Google Scholar]
- Burrows AE, Sceurman BK, et al. The C. elegans Myt1 ortholog is required for the proper timing of oocyte maturation. Development. 2006;133(4):697–709. doi: 10.1242/dev.02241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee I, Richmond A, et al. The Caenorhabditis elegans spe-38 gene encodes a novel four-pass integral membrane protein required for sperm function at fertilization. Development. 2005;132:2795–2808. doi: 10.1242/dev.01868. [DOI] [PubMed] [Google Scholar]
- Cheng KC, Klancer R, et al. Regulation of MBK-2/DYRK by CDK-1 and the pseudophosphatases EGG-4 and EGG-5 during the oocyte-to-embryo transition. Cell. 2009;139(3):560–72. doi: 10.1016/j.cell.2009.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church DL, Guan KL, et al. Three genes of the MAP kinase cascade, mek-2, mpk-1/sur-1 and let-60 ras, are required for meiotic cell cycle progression in Caenorhabditis elegans. Development. 1995;121(8):2525–35. doi: 10.1242/dev.121.8.2525. [DOI] [PubMed] [Google Scholar]
- Davis ES, Wille L, et al. Multiple subunits of the Caenorhabditis elegans anaphase-promoting complex are required for chromosome segregation during meiosis I. Genetics. 2002;160(2):805–13. doi: 10.1093/genetics/160.2.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Detwiler MR, Reuben M, et al. Two zinc finger proteins, OMA-1 and OMA-2, are redundantly required for oocyte maturation in C. elegans. Dev Cell. 2001;1(2):187–99. doi: 10.1016/s1534-5807(01)00026-0. [DOI] [PubMed] [Google Scholar]
- Doniach T, Hodgkin J. A sex-determining gene, fem-1, required for both male and hermaphrodite development in Caenorhabditis elegans. Dev Biol. 1984;106(1):223–35. doi: 10.1016/0012-1606(84)90077-0. [DOI] [PubMed] [Google Scholar]
- Doree M, Hunt T. From Cdc2 to Cdk1: when did the cell cycle kinase join its cyclin partner? J Cell Sci. 2002;115(Pt 12):2461–4. doi: 10.1242/jcs.115.12.2461. [DOI] [PubMed] [Google Scholar]
- Edgar LG. Blastomere culture and analysis. Methods Cell Biol. 1995;48:303–21. doi: 10.1016/s0091-679x(08)61393-x. [DOI] [PubMed] [Google Scholar]
- Edgley ML, Baillie DL, et al. Genetic balancers. Methods in Cell Biology. 1995;48:147–184. [PubMed] [Google Scholar]
- Edgley ML, Baillie DL, et al. Genetic balancers. WormBook. 2006:1–32. doi: 10.1895/wormbook.1.89.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ensslin MA, Shur BD. Identification of mouse sperm SED1, a bimotif EGF repeat and discoidin-domain protein involved in sperm-egg binding. Cell. 2003;114(4):405–17. doi: 10.1016/s0092-8674(03)00643-3. [DOI] [PubMed] [Google Scholar]
- Fay D. Genetic mapping and manipulation: chapter 1--Introduction and basics. WormBook. 2006:1–12. doi: 10.1895/wormbook.1.90.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire A, Xu S, et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Furuta T, Tuck S, et al. EMB-30: an APC4 homologue required for metaphase-to-anaphase transitions during meiosis and mitosis in Caenorhabditis elegans. Mol Biol Cell. 2000;11(4):1401–19. doi: 10.1091/mbc.11.4.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geldziler B, Chatterjee I, et al. A comparative study of sperm morphology, cytology and activation in Caenorhabditis elegans, Caenorhabditis remanei and Caenorhabditis briggsae. Dev Genes Evol. 2006;216(4):198–208. doi: 10.1007/s00427-005-0045-4. [DOI] [PubMed] [Google Scholar]
- Geldziler B, Chatterjee I, et al. The genetic and molecular analysis of spe-19, a gene required for sperm activation in Caenorhabditis elegans. Dev Biol. 2005;283(2):424–36. doi: 10.1016/j.ydbio.2005.04.036. [DOI] [PubMed] [Google Scholar]
- Geldziler B, Kadandale P, et al. Molecular genetic approaches to studying fertilization in model systems. Reproduction. 2004;127(4):409–16. doi: 10.1530/rep.1.00009. [DOI] [PubMed] [Google Scholar]
- Ghedin E, Wang S, et al. Draft genome of the filarial nematode parasite Brugia malayi. Science. 2007;317(5845):1756–60. doi: 10.1126/science.1145406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golden A. Cytoplasmic flow and the establishment of polarity in C. elegans 1-cell embryos. Curr Opin Genet Dev. 2000;10(4):414–20. doi: 10.1016/s0959-437x(00)00106-4. [DOI] [PubMed] [Google Scholar]
- Golden A, Sadler PL, et al. Metaphase to anaphase (mat) transition-defective mutants in Caenorhabditis elegans. J Cell Biol. 2000;151(7):1469–82. doi: 10.1083/jcb.151.7.1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein B, Hird SN. Specification of the anteroposterior axis in Caenorhabditis elegans. Development. 1996;122:1467–1474. doi: 10.1242/dev.122.5.1467. [DOI] [PubMed] [Google Scholar]
- Govindan JA, Greenstein D. Embryogenesis: anchors away! Curr Biol. 2007;17(20):R890–2. doi: 10.1016/j.cub.2007.08.019. [DOI] [PubMed] [Google Scholar]
- Grant B, Hirsh D. Receptor-mediated endocytosis in the Caenorhabditis elegans oocyte. Mol Biol Cell. 1999;10(12):4311–26. doi: 10.1091/mbc.10.12.4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumienny TL, Lambie E, et al. Genetic control of programmed cell death in the Caenorhabditis elegans hermaphrodite germline. Development. 1999;126(5):1011–22. doi: 10.1242/dev.126.5.1011. [DOI] [PubMed] [Google Scholar]
- Guven-Ozkan T, Nishi Y, et al. Global transcriptional repression in C. elegans germline precursors by regulated sequestration of TAF-4. Cell. 2008;135(1):149–60. doi: 10.1016/j.cell.2008.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill DP, Shakes DC, et al. A sperm-supplied product essential for initiation of normal embryogenesis in Caenorhabditis elegans is encoded by the paternal-effect embryonic-lethal gene, spe-11 [published erratum appears in Dev Biol 1990 May;139(1):230] Dev Biol. 1989;136(1):154–66. doi: 10.1016/0012-1606(89)90138-3. [DOI] [PubMed] [Google Scholar]
- Hill KL, L’Hernault SW. Analyses of reproductive interactions that occur after heterospecific matings within the genus Caenorhabditis. Dev Biol. 2001;232(1):105–114. doi: 10.1006/dbio.2000.0136. [DOI] [PubMed] [Google Scholar]
- Hodgkin J. In: Appendix 1, Genetics C. elegans II. Riddle DL, Blumenthal T, Meyer BJ, Priess JR, editors. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 1997. pp. 881–1048. [Google Scholar]
- Hodgkin J. Conventional genetics C. elegans: A Practical Approach. I. Hope. New York: Oxford University Press; 1999. pp. 245–270. [Google Scholar]
- Horner VL, Wolfner MF. Transitioning from egg to embryo: triggers and mechanisms of egg activation. Dev Dyn. 2008;237(3):527–44. doi: 10.1002/dvdy.21454. [DOI] [PubMed] [Google Scholar]
- Hsu JY, Sun ZW, et al. Mitotic phosphorylation of histone H3 is governed by Ipl1/aurora kinase and Glc7/PP1 phosphatase in budding yeast and nematodes. Cell. 2000;102(3):279–91. doi: 10.1016/s0092-8674(00)00034-9. [DOI] [PubMed] [Google Scholar]
- Iwasaki K, McCarter J, et al. emo-1, a Caenorhabditis elegans Sec61p gamma homologue, is required for oocyte development and ovulation. J Cell Biol. 1996;134(3):699–714. doi: 10.1083/jcb.134.3.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen G, Hazendonk E, et al. Reverse genetics by chemical mutagenesis in Caenorhabditis elegans. Nat Genet. 1997;17(1):119–21. doi: 10.1038/ng0997-119. [DOI] [PubMed] [Google Scholar]
- Jaramillo-Lambert A, Ellefson M, et al. Differential timing of S phases, X chromosome replication, and meiotic prophase in the C. elegans germ line. Dev Biol. 2007;308(1):206–21. doi: 10.1016/j.ydbio.2007.05.019. [DOI] [PubMed] [Google Scholar]
- Johnston WL, Krizus A, et al. The eggshell is required for meiotic fidelity, polar-body extrusion and polarization of the C. elegans embryo. BMC Biol. 2006;4:35. doi: 10.1186/1741-7007-4-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston WL, Krizus A, et al. Eggshell chitin and chitin-interacting proteins prevent polyspermy in C. elegans. Curr Biol. 2010;20(21):1932–7. doi: 10.1016/j.cub.2010.09.059. [DOI] [PubMed] [Google Scholar]
- Jorgensen EM, Mango SE. The art and design of genetic screens: Caenorhabditis elegans. Nat Rev Genet. 2002;3(5):356–69. doi: 10.1038/nrg794. [DOI] [PubMed] [Google Scholar]
- Kadandale P, Geldziler B, et al. Use of SNPs to determine the breakpoints of complex deficiencies, facilitating gene mapping in Caenorhabditis elegans. BMC Genet. 2005;6(1):28. doi: 10.1186/1471-2156-6-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadandale P, Singson A. Oocyte production and sperm utilization patterns in semi-fertile strains of Caenorhabditis elegans. BMC Dev Biol. 2004;4(1):3. doi: 10.1186/1471-213X-4-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadandale P, Stewart-Michaelis A, et al. The Egg Surface LDL Receptor Repeat-Containing Proteins EGG-1 and EGG-2 Are Required for Fertilization in Caenorhabditis elegans. Curr Biol. 2005;15(24):2222–9. doi: 10.1016/j.cub.2005.10.043. [DOI] [PubMed] [Google Scholar]
- Kamath RS, Ahringer J. Genome-wide RNAi screening in Caenorhabditis elegans. Methods. 2003;30(4):313–21. doi: 10.1016/s1046-2023(03)00050-1. [DOI] [PubMed] [Google Scholar]
- Kelly WG, Xu S, et al. Distinct requirements for somatic and germline expression of a generally expressed Caenorhabditis elegans gene. Genetics. 1997;146:227–238. doi: 10.1093/genetics/146.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimble J, Ward S. Germ-line development and fertilization. In: Wood WB, editor. The Nematode Caenorhabditis elegans. Cold Spring Harbor: Cold Spring Harbor Laboratory; 1988. pp. 191–213. [Google Scholar]
- Kosinski M, McDonald K, et al. C. elegans sperm bud vesicles to deliver a meiotic maturation signal to distant oocytes. Development. 2005;132(15):3357–69. doi: 10.1242/dev.01916. [DOI] [PubMed] [Google Scholar]
- Kroft TL, Gleason EJ, et al. The spe-42 gene is required for sperm-egg interactions during C. elegans fertilization and encodes a sperm-specific transmembrane protein. Dev Biol. 2005;286:169–181. doi: 10.1016/j.ydbio.2005.07.020. [DOI] [PubMed] [Google Scholar]
- Kubagawa HM, Watts JL, et al. Oocyte signals derived from polyunsaturated fatty acids control sperm recruitment in vivo. Nat Cell Biol. 2006;8(10):1143–8. doi: 10.1038/ncb1476. [DOI] [PubMed] [Google Scholar]
- L’Hernault SW. In: Spermatogenesis C. Elegans II. Riddle DL, Blumenthal T, Meyer BJ, Priess JR, editors. Cold Spring Harbor: Cold Spring Harbor Laboratory; 1997. pp. 271–294. [PubMed] [Google Scholar]
- L’Hernault SW T. C. e. R. Community, . Spermatogenesis. WormBook: online review of C. elegans biology. 2006 doi: 10.1895/wormbook.1.85.1. http://www.wormbook.org. [DOI] [PMC free article] [PubMed]
- L’Hernault SW, Roberts TM. Cell biology of nematode sperm. Methods in Cell Biology. 1995;48:273–301. doi: 10.1016/s0091-679x(08)61392-8. [DOI] [PubMed] [Google Scholar]
- L’Hernault SW, Shakes DC, et al. Developmental genetics of chromosome I spermatogenesis-defective mutants in the nematode Caenorhabditis elegans. Genetics. 1988;120:435–452. doi: 10.1093/genetics/120.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaMunyon CW, Ward S. Sperm precedence in a hermaphroditic nematode Caenorhabditis elegans is due to competitive superiority of male sperm. Experientia. 1995;51(8):817–23. doi: 10.1007/BF01922436. [DOI] [PubMed] [Google Scholar]
- LaMunyon CW, Ward S. Increased competitiveness of nematode sperm bearing the male X chromosome. Proc Natl Acad Sci U S A. 1997;94(1):185–9. doi: 10.1073/pnas.94.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaMunyon CW, Ward S. Larger sperm outcompete smaller sperm in the nematode Caenorhabditis elegans. Proceedings of the Royal Society, Series B. 1998;265:1997–2002. doi: 10.1098/rspb.1998.0531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaMunyon CW, Ward S. Evolution of sperm size in nematodes: sperm competition favours larger sperm. Proceedings of the Royal Society, Series B. 1999;266(1416):263–7. doi: 10.1098/rspb.1999.0631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MH, Schedl T. Identification of in vivo mRNA targets of GLD-1, a maxi-KH motif containing protein required for C. elegans germ cell development. Genes Dev. 2001;15(18):2408–20. doi: 10.1101/gad.915901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessard C, Pendola JK, et al. New mouse genetic models for human contraceptive development. Cytogenet Genome Res. 2004;105(2–4):222–7. doi: 10.1159/000078192. [DOI] [PubMed] [Google Scholar]
- Lin R. A gain-of-function mutation in oma-1, a C. elegans gene required for oocyte maturation, results in delayed degradation of maternal proteins and embryonic lethality. Dev Biol. 2003;258(1):226–39. doi: 10.1016/s0012-1606(03)00119-2. [DOI] [PubMed] [Google Scholar]
- Liu J, Vasudevan S, et al. CUL-2 and ZYG-11 promote meiotic anaphase II and the proper placement of the anterior-posterior axis in C. elegans. Development. 2004;131(15):3513–25. doi: 10.1242/dev.01245. [DOI] [PubMed] [Google Scholar]
- Liu LX, Spoerke JM, et al. High-throughput isolation of Caenorhabditis elegans deletion mutants. Genome Res. 1999;9(9):859–67. doi: 10.1101/gr.9.9.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda I, Kohara Y, et al. Large-scale analysis of gene function in Caenorhabditis elegans by high-throughput RNAi. Curr Biol. 2001;11(3):171–6. doi: 10.1016/s0960-9822(01)00052-5. [DOI] [PubMed] [Google Scholar]
- Marcello MR, Singson A. Fertilization and the oocyte-to-embryo transition in C. elegans. BMB Rep. 2010;43(6):389–99. doi: 10.5483/bmbrep.2010.43.6.389. [DOI] [PubMed] [Google Scholar]
- Maruyama R, Velarde NV, et al. EGG-3 regulates cell-surface and cortex rearrangements during egg activation in Caenorhabditis elegans. Curr Biol. 2007;17(18):1555–60. doi: 10.1016/j.cub.2007.08.011. [DOI] [PubMed] [Google Scholar]
- McCarter J, Bartlett B, et al. On the control of oocyte meiotic maturation and ovulation in C. elegans. Dev Biol. 1999;205:111–128. doi: 10.1006/dbio.1998.9109. [DOI] [PubMed] [Google Scholar]
- McNally K, Audhya A, et al. Katanin controls mitotic and meiotic spindle length. J Cell Biol. 2006;175(6):881–91. doi: 10.1083/jcb.200608117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally KL, McNally FJ. Fertilization initiates the transition from anaphase I to metaphase II during female meiosis in C. elegans. Dev Biol. 2005;282(1):218–30. doi: 10.1016/j.ydbio.2005.03.009. [DOI] [PubMed] [Google Scholar]
- Mello C, Fire A. DNA transformation. Methods in Cell Biology. 1995;48:451–82. [PubMed] [Google Scholar]
- Miller MA. Sperm and oocyte isolation methods for biochemical and proteomic analysis. Methods Mol Biol. 2006;351:193–201. doi: 10.1385/1-59745-151-7:193. [DOI] [PubMed] [Google Scholar]
- Miller MA, V, Nguyen Q, et al. A sperm cytoskeletal protein that signals oocyte meiotic maturation and ovulation. Science. 2001;291(5511):2144–7. doi: 10.1126/science.1057586. [DOI] [PubMed] [Google Scholar]
- Miller MA, Ruest PJ, et al. An Eph receptor sperm-sensing control mechanism for oocyte meiotic maturation in Caenorhabditis elegans. Genes Dev. 2003;17(2):187–200. doi: 10.1101/gad.1028303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitreva M, Blaxter ML, et al. Comparative genomics of nematodes. Trends Genet. 2005;21(10):573–81. doi: 10.1016/j.tig.2005.08.003. [DOI] [PubMed] [Google Scholar]
- Nelson GA, Ward S. Vesicle fusion, pseudopod extension and amoeboid motility are induced in nematode spermatids by the ionophore monensin. Cell. 1980;19(2):457–64. doi: 10.1016/0092-8674(80)90520-6. [DOI] [PubMed] [Google Scholar]
- Nishi Y, Lin R. DYRK2 and GSK-3 phosphorylate and promote the timely degradation of OMA-1, a key regulator of the oocyte-to-embryo transition in C. elegans. Dev Biol. 2005;288(1):139–49. doi: 10.1016/j.ydbio.2005.09.053. [DOI] [PubMed] [Google Scholar]
- Nishimura H, L’Hernault SW. Spermatogenesis-defective (spe) mutants of the nematode Caenorhabditis elegans provide clues to solve the puzzle of male germline functions during reproduction. Dev Dyn. 2010;239(5):1502–14. doi: 10.1002/dvdy.22271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nykjaer A, Willnow TE. The low-density lipoprotein receptor gene family: a cellular Swiss army knife? Trends Cell Biol. 2002;12(6):273–80. doi: 10.1016/s0962-8924(02)02282-1. [DOI] [PubMed] [Google Scholar]
- O’Connell KF, Leys CM, et al. A genetic screen for temperature-sensitive cell-division mutants of Caenorhabditis elegans. Genetics. 1998;149(3):1303–21. doi: 10.1093/genetics/149.3.1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto H, Thomson JN. Monoclonal antibodies which distinguish certain classes of neuronal and supporting cells in the nervous tissue of the nematode Caenorhabditis elegans. J Neurosci. 1985;5(3):643–53. doi: 10.1523/JNEUROSCI.05-03-00643.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang KM, Ishidate T, et al. The minibrain kinase homolog, mbk-2, is required for spindle positioning and asymmetric cell division in early C. elegans embryos. Dev Biol. 2004;265(1):127–39. doi: 10.1016/j.ydbio.2003.09.024. [DOI] [PubMed] [Google Scholar]
- Parry JM, Singson A. EGG Molecules Couple the Oocyte-to-Embryo Transition with Cell Cycle Progression. Results Probl Cell Differ. 2011;53:135–51. doi: 10.1007/978-3-642-19065-0_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry JM, Velarde NV, et al. EGG-4 and EGG-5 Link Events of the Oocyte-to-Embryo Transition with Meiotic Progression in C. elegans. Curr Biol. 2009;19(20):1752–7. doi: 10.1016/j.cub.2009.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellettieri J, Reinke V, et al. Coordinate activation of maternal protein degradation during the egg-to-embryo transition in C. elegans. Dev Cell. 2003;5(3):451–62. doi: 10.1016/s1534-5807(03)00231-4. [DOI] [PubMed] [Google Scholar]
- Praitis V, Casey E, et al. Creation of low-copy integrated transgenic lines in Caenorhabditis elegans. Genetics. 2001;157(3):1217–26. doi: 10.1093/genetics/157.3.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Primakoff P, Myles DG. Penetration, adhesion, and fusion in mammalian sperm-egg interaction. Science. 2002;296(5576):2183–5. doi: 10.1126/science.1072029. [DOI] [PubMed] [Google Scholar]
- Putiri E, Zannoni S, et al. Functional domains and temperature-sensitive mutations in SPE-9, an EGF repeat-containing protein required for fertility in Caenorhabditis elegans. Dev Biol. 2004;272(2):448–59. doi: 10.1016/j.ydbio.2004.05.014. [DOI] [PubMed] [Google Scholar]
- Quintin S, Mains PE, et al. The mbk-2 kinase is required for inactivation of MEI-1/katanin in the one-cell Caenorhabditis elegans embryo. EMBO Rep. 2003;4(12):1175–81. doi: 10.1038/sj.embor.7400029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman MM, Kipreos ET. The specific roles of mitotic cyclins revealed. Cell Cycle. 2010;9(1):22–3. doi: 10.4161/cc.9.1.10577. [DOI] [PubMed] [Google Scholar]
- Rappleye CA, Paredez AR, et al. The coronin-like protein POD-1 is required for anterior-posterior axis formation and cellular architecture in the nematode Caenorhabditis elegans. Genes Dev. 1999;13(21):2838–51. doi: 10.1101/gad.13.21.2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinke V, Cutter AD. Germline expression influences operon organization in the Caenorhabditis elegans genome. Genetics. 2009;181(4):1219–28. doi: 10.1534/genetics.108.099283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinke V, I, Gil S, et al. Genome-wide germline-enriched and sex-biased expression profiles in Caenorhabditis elegans. Development. 2004;131(2):311–23. doi: 10.1242/dev.00914. [DOI] [PubMed] [Google Scholar]
- Ringstad N, Horvitz HR. FMRFamide neuropeptides and acetylcholine synergistically inhibit egg-laying by C. elegans. Nat Neurosci. 2008;11(10):1168–76. doi: 10.1038/nn.2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert VJ, Bessereau JL. Manipulating the Caenorhabditis elegans genome using mariner transposons. Genetica. 2010;138(5):541–9. doi: 10.1007/s10709-009-9362-2. [DOI] [PubMed] [Google Scholar]
- Roberts TM, Pavalko FM, et al. Membrane and cytoplasmic proteins are transported in the same organell complex during nematode spermatogenesis. Journal of Cell Biology. 1986;102:1787–1796. doi: 10.1083/jcb.102.5.1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadler PL, Shakes DC. Anucleate Caenorhabditis elegans sperm can crawl, fertilize oocytes and direct anterior-posterior polarization of the 1-cell embryo. Development. 2000;127(2):355–66. doi: 10.1242/dev.127.2.355. [DOI] [PubMed] [Google Scholar]
- Samuel AD, Murthy VN, et al. Calcium dynamics during fertilization in C. elegans. BMC Dev Biol. 2001;1:8. doi: 10.1186/1471-213X-1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K, Sato M, et al. Dynamic regulation of caveolin-1 trafficking in the germ line and embryo of Caenorhabditis elegans. Mol Biol Cell. 2006;17(7):3085–94. doi: 10.1091/mbc.E06-03-0211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato M, Grant BD, et al. Rab11 is required for synchronous secretion of chondroitin proteoglycans after fertilization in Caenorhabditis elegans. J Cell Sci. 2008;121(Pt 19):3177–86. doi: 10.1242/jcs.034678. [DOI] [PubMed] [Google Scholar]
- Sawada H, Tanaka E, et al. Self/nonself recognition in ascidian fertilization: vitelline coat protein HrVC70 is a candidate allorecognition molecule. Proc Natl Acad Sci U S A. 2004;101(44):15615–20. doi: 10.1073/pnas.0401928101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt A, Nebreda AR. Signalling pathways in oocyte meiotic maturation. J Cell Sci. 2002;115(Pt 12):2457–9. doi: 10.1242/jcs.115.12.2457. [DOI] [PubMed] [Google Scholar]
- Seydoux G, Schedl T. The germline in C. elegans: origins, proliferation, and silencing. Int Rev Cytol. 2001;203:139–85. doi: 10.1016/s0074-7696(01)03006-6. [DOI] [PubMed] [Google Scholar]
- Shakes DC, Ward S. Initiation of spermiogenesis in C. elegans: a pharmacological and genetic analysis. Dev Biol. 1989;134:189–200. doi: 10.1016/0012-1606(89)90088-2. [DOI] [PubMed] [Google Scholar]
- Shakes DC, Wu JC, et al. Spermatogenesis-specific features of the meiotic program in Caenorhabditis elegans. PLoS Genet. 2009;5(8):e1000611. doi: 10.1371/journal.pgen.1000611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singaravelu G, Chatterjee I, et al. Isolation and In vitro Activation of Caenorhabditis elegans Sperm. J Vis Exp. 2011;(47) doi: 10.3791/2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singson A. Every sperm is sacred: fertilization in Caenorhabditis elegans. Dev Biol. 2001;230(2):101–9. doi: 10.1006/dbio.2000.0118. [DOI] [PubMed] [Google Scholar]
- Singson A. Sperm activation: time and tide wait for no sperm. Curr Biol. 2006;16(5):R160–2. doi: 10.1016/j.cub.2006.02.039. [DOI] [PubMed] [Google Scholar]
- Singson A, Hang JS, et al. Genes required for the common miracle of fertilization in Caenorhabditis elegans. Int J Dev Biol. 2008;52(5–6):647–56. doi: 10.1387/ijdb.072512as. [DOI] [PubMed] [Google Scholar]
- Singson A, Hill KL, et al. Sperm competition in the absence of fertilization in Caenorhabditis elegans. Genetics. 1999;152(1):201–8. doi: 10.1093/genetics/152.1.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singson A, Mercer KB, et al. The C. elegans spe-9 gene encodes a sperm transmembrane protein that contains EGF-like repeats and is required for fertilization. Cell. 1998;93:71–79. doi: 10.1016/s0092-8674(00)81147-2. [DOI] [PubMed] [Google Scholar]
- Singson A, Zannoni S, et al. Molecules that function in the steps of fertilization. Cytokine Growth Factor Rev. 2001;12(4):299–304. doi: 10.1016/s1359-6101(01)00013-2. [DOI] [PubMed] [Google Scholar]
- Sonneville R, Gonczy P. Zyg-11 and cul-2 regulate progression through meiosis II and polarity establishment in C. elegans. Development. 2004;131(15):3527–43. doi: 10.1242/dev.01244. [DOI] [PubMed] [Google Scholar]
- Stanfield GM, Villeneuve AM. Regulation of sperm activation by SWM-1 is required for reproductive success of C. elegans males. Current Biology. 2006 doi: 10.1016/j.cub.2005.12.041. [DOI] [PubMed] [Google Scholar]
- Stitzel ML, Cheng KC, et al. Regulation of MBK-2/Dyrk kinase by dynamic cortical anchoring during the oocyte-to-zygote transition. Curr Biol. 2007;17(18):1545–54. doi: 10.1016/j.cub.2007.08.049. [DOI] [PubMed] [Google Scholar]
- Stitzel ML, Seydoux G. Regulation of the oocyte-to-zygote transition. Science. 2007;316(5823):407–8. doi: 10.1126/science.1138236. [DOI] [PubMed] [Google Scholar]
- Strome S, Powers J, et al. Spindle dynamics and the role of gamma-tubulin in early Caenorhabditis elegans embryos. Mol Biol Cell. 2001;12(6):1751–64. doi: 10.1091/mbc.12.6.1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumiyoshi E, Sugimoto A, et al. Protein phosphatase 4 is required for centrosome maturation in mitosis and sperm meiosis in C. elegans. J Cell Sci. 2002;115(Pt 7):1403–10. doi: 10.1242/jcs.115.7.1403. [DOI] [PubMed] [Google Scholar]
- Tenenhaus C, Subramaniam K, et al. PIE-1 is a bifunctional protein that regulates maternal and zygotic gene expression in the embryonic germ line of Caenorhabditis elegans. Genes Dev. 2001;15(8):1031–40. doi: 10.1101/gad.876201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmons L, Court DL, et al. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene. 2001;263(1–2):103–12. doi: 10.1016/s0378-1119(00)00579-5. [DOI] [PubMed] [Google Scholar]
- Timmons L, Fire A. Specific interference by ingested dsRNA. Nature. 1998;395(6705):854. doi: 10.1038/27579. [DOI] [PubMed] [Google Scholar]
- Wakimoto BT, Lindsley DL, et al. Toward a comprehensive genetic analysis of male fertility in Drosophila melanogaster. Genetics. 2004;167(1):207–16. doi: 10.1534/genetics.167.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward S, Argon Y, et al. Sperm morphogenesis in wild-type and fertilization-defective mutants of Caenorhabditis elegans. J Cell Biol. 1981;91(1):26–44. doi: 10.1083/jcb.91.1.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward S, Carrel JS. Fertilization and sperm competition in the nematode Caenorhabditis elegans. Dev Biol. 1979;73:304–321. doi: 10.1016/0012-1606(79)90069-1. [DOI] [PubMed] [Google Scholar]
- Wassarman PM. Mammalian fertilization: molecular aspects of gamete adhesion, exocytosis, and fusion. Cell. 1999;96:175–183. doi: 10.1016/s0092-8674(00)80558-9. [DOI] [PubMed] [Google Scholar]
- Wessel GM, Brooks JM, et al. The biology of cortical granules. Int Rev Cytol. 2001;209:117–206. doi: 10.1016/s0074-7696(01)09012-x. [DOI] [PubMed] [Google Scholar]
- Wu TF, Nera B, et al. Elucidating gene regulatory mechanisms for sperm function through the integration of classical and systems approaches in C. elegans. Syst Biol Reprod Med. 2010;56(3):222–35. doi: 10.3109/19396361003749986. [DOI] [PubMed] [Google Scholar]
- Xu XZ, Sternberg PW. A C. elegans sperm TRP protein required for sperm-egg interactions during fertilization. Cell. 2003;114(3):285–97. doi: 10.1016/s0092-8674(03)00565-8. [DOI] [PubMed] [Google Scholar]
- Yamamoto I, Kosinski ME, et al. Start me up: cell signaling and the journey from oocyte to embryo in C. elegans. Dev Dyn. 2006;235(3):571–85. doi: 10.1002/dvdy.20662. [DOI] [PubMed] [Google Scholar]
- Yanagimachi R. In: Mammalian fertilization. The Physiology of Reproduction. Knobil E, Neill JD, editors. New York: Raven Press; 1994. pp. 189–317. [Google Scholar]
- Zannoni S, L’Hernault SW, et al. Dynamic localization of SPE-9 in sperm: a protein required for sperm-oocyte interactions in Caenorhabditis elegans. BMC Dev Biol. 2003;3(1):10. doi: 10.1186/1471-213X-3-10. [DOI] [PMC free article] [PubMed] [Google Scholar]