Abstract
In the decade since Disrupted in Schizophrenia 1 (DISC1) was first identified it has become one of the most convincing risk genes for major mental illness. As a multi-functional scaffold protein, DISC1 has multiple identified protein interaction partners that highlight pathologically relevant molecular pathways with potential for pharmaceutical intervention. Amongst these are proteins involved in neuronal migration (e.g. APP, Dixdc1, LIS1, NDE1, NDEL1), neural progenitor proliferation (GSK3β), neurosignalling (Girdin, GSK3β, PDE4) and synaptic function (Kal7, TNIK). Furthermore, emerging evidence of genetic association (NDEL1, PCM1, PDE4B) and copy number variation (NDE1) implicate several DISC1-binding partners as risk factors for schizophrenia in their own right. Thus, a picture begins to emerge of DISC1 as a key hub for multiple critical developmental pathways within the brain, disruption of which can lead to a variety of psychiatric illness phenotypes.
This article is part of a Special Issue entitled ‘Schizophrenia’.
Keywords: DISC1, Schizophrenia, Neurodevelopment, Signalling, Synapse, Association studies
Abbreviations: APP, Amyloid precursor protein; ATF4, Activating transcription factor 4; BACE1, β-site APP-cleaving enzyme-1; BBS4, Bardet–Biedl syndrome 4; CEP290, Centrosomal protein 290 kDa; CNV, Copy number variation; CRE, cAMP response element; DBZ, DISC1-binding zinc finger; DISC1, Disrupted in schizophrenia 1; Dixdc1, Dishevelled-axin domain containing-1; FEZ1, Fasciculation and elongation protein zeta 1; GluR, Glutamate receptor; GSK3β, Glycogen synthase kinase 3β; Kal7, Kalirin-7; LEF/TCF, Lymphoid enhancer factor/T cell factor; LIS1, Lissencephaly 1; mTOR, Mammalian target of rapamycin; NDE1, Nuclear distribution factor E homologue 1 or Nuclear distribution element 1; NDEL1, NDE-like 1; NRG, Neuregulin; PACAP, Pituitary adenylate cyclase-activating polypeptide; PCM1, Pericentriolar material 1; PCNT, Pericentrin; PDE4, Phosphodiesterase 4; PI3 K, Phosphatidylinositiol 3-kinase; PSD, Post-synaptic density; Rac1, Ras-related C3 botulinum toxin substrate 1; TNIK, Traf2 and Nck interacting kinase
Highlights
► We review genetic data associating DISC1 with psychiatric illness. ► DISC1 binding partners include proteins involved in neuronal migration. ► Others are involved in neuronal signalling or synaptic function. ► These binding partners suggest putative disease-related molecular pathways. ► Several are now also implicated in psychiatric illness in their own right.
1. Introduction
A key objective of genetics and genomics research into psychiatric illness is to identify perturbed biological pathways and, as a consequence, potential targets for pharmacological intervention. The genetic entrée point need not itself explain a large fraction of the liability to schizophrenia – it is sufficient that genetic abrogation can cause schizophrenia. DISC1 and the extended DISC1 pathway illustrate this contention par excellence. DISC1 was identified through a unique family in which a chromosomal translocation event co-segregates strongly with major mental illness (Blackwood et al., 2001; St Clair et al., 1990). This translocation event directly disrupts both a protein coding gene, DISC1, and an antisense RNA only gene, DISC2 (Millar et al., 2000). In the intervening period, the DISC locus has been repeatedly implicated in psychiatric illness by genetic linkage, association and mutation detection (reviewed in Chubb et al., 2008, see Table 1 for references and summaries of recent studies). Some studies have also pointed to epistatic interactions between the DISC locus and other candidate genes (Burdick et al., 2008; Hennah et al., 2007; Nicodemus et al., 2010). Despite these many confirmatory studies, there are also negative studies (Chubb et al., 2008 and references in Table 1) and, as yet, no firm basis on which to estimate the proportion of genetic liability attributable to the DISC locus. The DISC locus has appeared as a gene-wide, but not a genome-wide finding in some (Sullivan et al., 2008) but not other (Sanders et al., 2008) studies. The critical issue is what we can learn from the identification of DISC1 regarding the specifics and generalities of the biological underpinning of schizophrenia and other major mental illness.
Table 1.
Studies investigating genetic links between the DISC locus or the adjacent TSNAX locus and major mental illness, an endophenotype thereof or, in one study, chronic fatigue syndrome published since those reviewed by Chubb et al. (2008). SNPs bracketed together indicate haplotypes. SNPs separated by a hyphen (−) indicate significance when alleles are considered together, but not independently.
| Study | Sample | Condition/phenotype | SNP, haplotype, marker or variant | Notes |
|---|---|---|---|---|
| Positive genetic association studies | ||||
| Palo et al. (2007) | Finnish families | Psychotic disorders | (rs1655285, rs751229) | Males only |
| (rs751229, rs3738401) | Males only | |||
| (rs751229, rs3738401, rs1538977) | Males only, principally those without bipolar spectrum disorder | |||
| (rs1655285, rs751229, rs3738401) | Males only, principally those with bipolar spectrum disorder | |||
| Bipolar spectrum disorders | rs1655285 | |||
| (rs1630250, rs1615409) | Principally those without psychotic disorder | |||
| (rs1655285, rs751229) | Females only | |||
| (rs1000731, rs821616) | ||||
| (rs821616, rs1411771) | ||||
| (rs821616, rs1411771, rs980898) | ||||
| Finnish families with bipolar disorder | General intellectual functioning | rs1615409 | Significant by two measures | |
| rs821616 | Significant by one measure | |||
| rs980989 | Significant by three measures | |||
| Attention/working memory | rs980989 | Significant by three measures | ||
| Verbal learning | rs751229 | Significant by two measures | ||
| rs1322784 | Significant by one measure | |||
| rs1000731 | Significant by one measure | |||
| rs980989 | Significant by two measures | |||
| Executive functions | rs821616 | Significant by one measure | ||
| Kilpinen et al. (2008) | Finnish families | Autism | D1S2709 | |
| Asperger’s syndrome | rs1322784 | Males only | ||
| (rs751229, rs3738401) | ||||
| (rs751229, rs3738401, rs1322784) | Males only | |||
| Kim et al. (2008a) | Korean | Schizophrenia with poor concentration | rs821616 | |
| Perlis et al. (2008) | American trios | Bipolar disorder | (rs10495308, rs2793091, rs2793085) | |
| Saetre et al. (2008) | Danish | Schizophrenia | rs3737597 | |
| Norwegian | Schizophrenia | rs3737597 | ||
| Swedish | Schizophrenia | rs3737597 | ||
| Hennah et al. (2009) | Finnish | Bipolar disorder | rs1538979 | Males only |
| English | Bipolar disorder | rs821577 | Females only | |
| British/Finnish | Schizophrenia | rs821633-rs1538979 | Females only. | |
| Rastogi et al. (2009) | Canadian families | Schizophrenia | (rs11122359, rs701158) | |
| (rs6675281, rs11122359) | ||||
| (rs701158, rs821597) | ||||
| Schumacher et al. (2009) | German | Schizophrenia and early onset schizophrenia | rs1015101 | Females only. |
| rs999710 | Females only. | |||
| rs4333837 | Females only. | |||
| Schizophrenia | 18x haplotyes | 5 in males only, 11 in females only | ||
| rs1538979 | Significant in males when stratified on rs821633 allele | |||
| Tomppo et al. (2009b) | Finnish | Social anhedonia | rs821577 | |
| rs11122381 | Females only. | |||
| rs821592 | Females only. | |||
| rs821633 | Significant when stratified on rs1538979 and rs821577 alleles | |||
| Fukuda et al. (2010) | Japanese | Chronic fatigue syndrome | rs821616 | Females only. |
| Harris et al. (2010) | Scottish, elderly | Anxiety scores | rs821577 | Lower in males, higher in females |
| rs821633 | Lower in males, higher in females | |||
| Depression scores | rs821577 | Females only | ||
| rs821633 | Females only | |||
| Emotional stability scores | rs821577 | Females only | ||
| rs821633 | Females only | |||
| Neuroticism scores | rs821577 | Females only | ||
| rs821633 | Females only | |||
| Lepagnol-Bestel et al. (2010) | French trios | Schizophrenia | rs6675281 | |
| Negative symptom scores | rs6675281 | |||
| Algerian trios | Schizophrenia | rs821616 | ||
| Negative symptom scores | rs6675281 | |||
| Mouaffak et al. (2010) | French | Ultra-resistant schizophrenia | rs3738401 | |
| Nicodemus et al. (2010) | American | Schizophrenia | rs10744743-rs1411771 | rs10744743 is an SNP in the CIT gene |
| Okuda et al. (2010) | Japanese | Major depressive disorder | rs766288 | Females only |
| Schosser et al. (2010) | English | Bipolar disorder | rs2492367 | |
| rs7546310 | ||||
| (rs7546310, rs821597) | ||||
| (rs766288, rs2492367) | ||||
| (rs1000731, rs7546310) | ||||
| British | Major depressive disorder | (rs7546310, rs821597) | ||
| Meta-analysis of association studies | ||||
| Schumacher et al. (2009) | European | Schizophrenia | rs17817356 | |
| Negative association studies | ||||
| Arai et al. (2007) | Japanese | Bipolar disorder | Negative | |
| Major depressive disorder | Negative | |||
| Sanders et al. (2008) | European ancesrtry | Schizophrenia | Negative | |
| Lim et al. (2009) | Korean | Autism spectrum disorders | ||
| Houlihan et al. (2009) | Scottish, elderly | Cognitive traits | Negative | |
| Okuda et al. (2010) | Japanese | Bipolar disorder | Negative | |
| Ultra-rare variants | ||||
| Song et al. (2008) | Single patients | Schizophrenia | Point mutations: G14A, R37 W, S90L, R418H, T603I | Not in 10,000 + sequenced controls. S90L seen in two patients |
| Crepel et al. (2010) | Two brothers | Autism | 2 Mb duplication including DISC locus | Not in 1577 controls |
| Williams et al. (2009) | Single patient | Autism spectrum disorder | 2 Mb deletion including TSNAX/DISC locus | |
| Song et al. (2010) | Single patients | Bipolar disorder | Point mutations: S209R R338Q, R418H, T754S | Not in 10,000 + sequenced controls |
The DISC1 protein has no known enzymatic activity; rather it exerts its effect on multiple proteins through interaction to modulate their functional states and biological activities in time and space. Many putative interacting proteins have been identified through extensive yeast-2-hybrid screening (Brandon et al., 2004; Camargo et al., 2007; Millar et al., 2003; Morris et al., 2003; Ozeki et al., 2003) and, where these have been examined, a large proportion have been validated by downstream experimentation (reviewed in Chubb et al., 2008). These multiple interactions, combined with the widespread subcellular distribution of DISC1 (reviewed in Chubb et al., 2008), a complex pattern of protein isoforms (James et al., 2004) and splice variants (Nakata et al., 2009), have led to the suggestion that DISC1 acts as a protein scaffold within the cell, dynamically interacting with and affecting the function of different proteins at different locations and developmental times (Brandon et al., 2009; Porteous, 2008; Porteous and Millar, 2009). DISC1-related psychiatric illness is therefore likely to arise through the simultaneous dysregulation of not just one, but more likely several, protein interactions, physiological states and activities, with a consequential complexity and pervasiveness of effect. Identifying the key DISC1 interactors is therefore of exceptional importance in coming to understand the nature of the devastating condition that is schizophrenia, and will facilitate the search for downstream elements which may be susceptible to pharmaceutical intervention.
In this review, we will focus on what is known about the biological functions of DISC1-interacting proteins, with particular attention to aspects of their biology which potentially relate to psychiatric illness, through effects on neurodevelopment, neurotransmission or neurosignalling, along with the emerging genetic evidence implicating many of these as schizophrenia-risk factors in their own right. Whereas a neurodevelopmental component to schizophrenia is well established, and pre-morbid features are recognised, it is typically not until adolescence or early adulthood that the debilitating symptoms emerge. That DISC1 and its pathway of interacting proteins affect both neurodevelopmental pathways and also signalling pathways in the adult brain, suggest that the study of DISC1 genetics and biology may help towards a more unified understanding of schizophrenia and with it the potential to develop rational interventions in the symptomatic adult and or even earlier.
2. Roles of DISC1-interacting proteins in neural development
2.1. DISC1-binding partners in cytoskeletal functions and neurite outgrowth
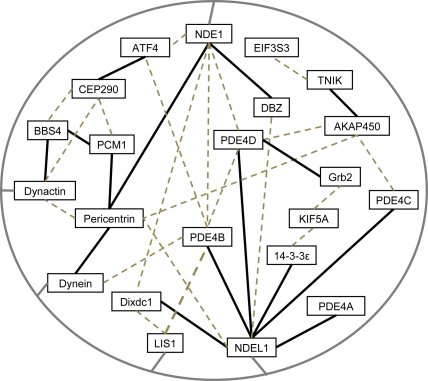
The complex and intricate task of co-ordinating the microtubule network of neurons, which is vital for maintaining correct development, morphology and migration, is performed in large part by the microtubule organising centre, or centrosome (Higginbotham and Gleeson, 2007). Multiple lines of evidence demonstrate that DISC1 is part of a protein complex at the centrosome (Fig. 1 and references in the legend) and is involved in cytoskeletal processes involved in neuronal migration, including nucleokinesis and neurite outgrowth. LIS1, NDE1, and NDEL1 are a trio of such centrosomal DISC1 interactors (Bradshaw et al., 2009; Brandon et al., 2004; Burdick et al., 2008; Millar et al., 2003; Morris et al., 2003; Ozeki et al., 2003) which play pivotal roles in the progression of the cell cycle, dynein-related transport along microtubules and nucleokinesis (reviewed in Chubb et al., 2008; Wynshaw-Boris et al., 2010). The localisation of NDEL1 and LIS1 in axons is known to be dependent on expression of DISC1 (Taya et al., 2007). Knock-down of either NDEL1 or LIS1 using RNAi in culture leads to reduced neurite outgrowth (Kamiya et al., 2006; Shim et al., 2008; Taya et al., 2007), while granule neurons from heterozygous NDEL1 or LIS1 knock-out mice show impaired migration in in vitro assays (Toyo-oka et al., 2005). NDEL1 is also known to play a role in axon regeneration after injury (Toth et al., 2008) and has an additional DISC1-modulated function as a cysteine endopeptidase (Hayashi et al., 2005) which appears to be important for regulation of neurite outgrowth (Hayashi et al., 2010). Although highly similar in amino-acid sequence to NDEL1, the role of NDE1 in many of these processes has yet to be determined.
Fig. 1.
22 known DISC1-interacting proteins, the majority of which are found in or around the centrosome. Proteins which are known to bind directly to each other are linked by thick black lines, while proteins which are known to co-exist in the same complex, but for which a direct-interaction has, to our knowledge, yet to be demonstrated are linked by grey dashed lines. The circle linking dynein, dynactin, LIS1, NDE1 and NDEL1 signifies that these five proteins complex with each other. Note that the DISC1-CEP290, DISC1-AKAP450 and TNIK-AKAP450 interactions have only been shown by yeast-2-hybrid screening and remain to be confirmed, while Grb2 binds only to a single isoform of PDE4D. Data on interactions with DISC1 and between DISC1-binding partners were taken from the following papers: (Beard et al., 1999; Bradshaw et al., 2008, 2009; Brandon et al., 2004; Burdick et al., 2008; Camargo et al., 2007; Chang et al., 2006; Collins et al., 2008; Ewing et al., 2007; Faulkner et al., 2000; Feng et al., 2000; Guo et al., 2006; Hattori et al., 2007; Hirohashi et al., 2006; Hutchins et al., 2010; Kim et al., 2004, 2008b; Kitagawa et al., 2000; McCahill et al., 2005; Millar et al., 2003, 2005; Miyoshi et al., 2004; Morris et al., 2003; Murdoch et al., 2007; Niethammer et al., 2000; Ogawa et al., 2005; Ozeki et al., 2003; Purohit et al., 1999; Sasaki et al., 2000; Sawamura et al., 2008; Sayer et al., 2006; Shinoda et al., 2007; Singh et al., 2010; Smith et al., 2000; Stehman et al., 2007; Sweeney et al., 2001; Tai et al., 2002; Takahashi et al., 2002; Taya et al., 2007; Toyo-oka et al., 2005; Wang et al., 2010).
DISC1 is also known to be involved in transport along microtubules to the distal parts of axons as part of a ternary complex with kinesin-1 and the adaptor protein Grb2 (Shinoda et al., 2007; Taya et al., 2007). Cargo transported in this manner includes the DISC1 interactors LIS1, NDEL1 and 14-3-3ɛ (Taya et al., 2007). The interaction of DISC1 with Grb2 may also be required for neutrophin-3-related axon elongation (Shinoda et al., 2007). Of potential therapeutic relevance, expression of GRB2 mRNA is known to be upregulated following electroconvulsive seizure, an established anti-depression therapy (Newton et al., 2004). FEZ1, another DISC1-interacting protein (Miyoshi et al., 2003), is known to be involved in the activation of the kinesin-1 motor protein (Blasius et al., 2007; Fujita et al., 2007), regulation of neurite outgrowth (Fujita et al., 2007) and the establishment of neuronal polarity (Ikuta et al., 2007). DISC1-FEZ1 interaction is enhanced during neurodifferentiation, and expression of the FEZ1-binding domain of DISC1 has a dominant negative effect on neurite outgrowth in a cellular model (Miyoshi et al., 2003), implying co-operation of DISC1 and FEZ1 in this signalling pathway.
At the centrosome, DISC1 also interacts with the scaffold protein pericentrin (also known as kendrin, Miyoshi et al., 2004), a molecule that is known to play important roles in microtubule nucleation and aster formation (reviewed in Delaval and Doxsey, 2010) in a seemingly DISC1-dependent manner (Shimizu et al., 2008). Intriguingly, mutations in the PCNT gene, which encodes pericentrin, are heavily implicated in a form of dwarfism associated with reduced brain size, suggesting it to be important for neurodevelopment (Griffith et al., 2008; Rauch et al., 2008). Recruitment of pericentrin to the centrosome is essential for correct microtubule organisation and is facilitated by another large scaffold protein PCM1 (Dammermann and Merdes, 2002). PCM1 in turn is recruited co-operatively by interacting proteins DISC1 and BBS4 (Kamiya et al., 2008). Centrosomal PCM1 is known to be required for correct axon morphology (Calderon de Anda et al., 2010) and embryonic neurogenesis (Ge et al., 2010). Intriguingly, the level of localisation of PCM1 to the centrosome in human glial cells is altered by two common DISC1 amino-acid substitutions, Ser704Cys and Leu607Phe (Eastwood et al., 2010), one possible mechanism by which these alleles lead to DISC1 dysfunction, as measured by brain imaging (Callicott et al., 2005; Di Giorgio et al., 2008; Hashimoto et al., 2006; Prata et al., 2008; Szeszko et al., 2008; Takahashi et al., 2009) and elevated risk of psychiatric illness (Table 1 and references therein).
Other centrosomal interactors of DISC1 include the DISC1-Binding Zinc finger protein (DBZ, also known as Su48 or ZNF365). DBZ is a brain expressed protein which binds to DISC1, NDE1 and NDEL1 (Camargo et al., 2007; Hattori et al., 2007; Hirohashi et al., 2006; Wang et al., 2006). Co-expression of DISC1 and DBZ results in a reduction in the number of PC12 cells bearing neurites, while expression of the DISC1-binding domain of DBZ lead to reduced neurite outgrowth in mouse primary hippocampal neurons (Hattori et al., 2007).
2.2. DISC1-binding proteins in neuronal migration and differentiation within the mouse brain
Results from cell-based models such as those described above beg the question as to how binding partners of DISC1 might be involved in regulating neurodevelopment. Important insights have come from various in vivo mouse studies. In the hippocampus, knock-down of DISC1 using shRNA methods have been shown to lead to aberrant positioning and dendritic structure of adult-born neurons (Duan et al., 2007). Intriguingly, the defects caused by one DISC1 shRNA of mild effect were greatly enhanced by co-expression of an shRNA to knock-down levels of NDEL1 (Duan et al., 2007), strongly implying that these two proteins co-operate together, and consistent with the role of NDEL1 established in cultured cells. Deficiencies in the migration of neurons in the developing cortex can be seen following silencing of DISC1, PCM1 or BBS4 (Calderon de Anda et al., 2010; Kamiya et al., 2005, 2008) and in NDE1 and NDEL1 knock-out mice (Feng and Walsh, 2004; Sasaki et al., 2005) implying that the various DISC1-containing complexes involved in microtubule regulation are critical for cortical development.
Migration defects in cortical neurons can also be caused by silencing the DISC1-interacting protein Dixdc1 or by inhibiting DISC1-Dixdc1 interaction using an interfering peptide (Singh et al., 2010). Intriguingly, Dixdc1 is also an interactor of NDEL1, and a phosphorylation site key to this interaction is required to rescue migration defects caused by suppression of DIXDC1 expression (Singh et al., 2010). Thus, DISC1, Dixdc1 and NDEL1 appear to co-operate in regulating migration in the developing cortex. Another potential member of this pathway is the Alzheimer’s disease-related Amyloid Precursor Protein (APP, Young-Pearse et al., 2010). Knock-down of APP levels by RNAi in the developing cortex leads to migration defects reminiscent of DISC1 knock-down which can be largely reversed by DISC1 over-expression (Young-Pearse et al., 2007, 2010). There is also evidence to suggest that APP is involved in the localisation of DISC1 to the centrosome in the cortex (Young-Pearse et al., 2010).
Both Dixdc1 and DISC1 impact on the Wnt-signalling pathway. Silencing of DISC1 or Dixdc1 reduces lymphoid enhancer factor/T cell factor (LEF/TCF) mediated transcription and thus differentiation of neural progenitors (Mao et al., 2009; Singh et al., 2010). These effects caused by down-regulation of DISC1 expression can be rescued by expression of Dixdc1 and vice versa. The key linking molecules are Glycogen Synthase Kinase 3β (GSK3β) and β-catenin. The kinase activity of GSK3β is inhibited on binding to DISC1 (Mao et al., 2009), preventing degradation of β-catenin and allowing it to translocate to the nucleus where it stimulates transcription of neurogenesis-related genes. These effects of DISC1/Dixdc1 silencing can be rescued by expression of β-catenin or by inhibiting GSK3β (Mao et al., 2009; Singh et al., 2010). Also, and of clinical relevance, GSK3β is a well-established target for lithium chloride, widely used in the management of bipolar disorder (Ross and Margolis, 2009). GSK3β-specific inhibitors can also rescue hyperlocomotion in open field tests observed in mice expressing the DISC1-L100P mutant or in which DISC1 has been silenced in the hippocampus (Lipina et al., 2010a; Mao et al., 2009) as well as pre-pulse and latent inhibition deficits in the L100P mouse (Lipina et al., 2010a). Intriguingly, GSK3 activity is also regulated by the APP-derived β-amyloid peptide (reviewed in Hernández et al., 2010).
Girdin (also known as KIAA1212, APE, GIV and HkRP1) is another DISC1-interacting protein (Camargo et al., 2007; Enomoto et al., 2009; Kim et al., 2009), over-expression of which leads to adult-born neurons of the dentate gyrus displaying enhanced dendritic growth, increased dendritic number and over-extended migration into the outer granule cell layer and molecular layer (Kim et al., 2009), mirroring the effects of DISC1 depletion (Duan et al., 2007). Incorrect neuronal localisation and impaired mossy fibre development are also seen in girdin knock-out mice (Enomoto et al., 2009). These effects of girdin appear to be mediated via its ability to bind to and increase the activity of the serine/threonine kinase Akt (Anai et al., 2005). DISC1 depletion increases Akt activity (Hashimoto et al., 2006) at least in part through binding to girdin and preventing its Akt-stimulating activity (Kim et al., 2009). In agreement with this, use of rapamycin to inhibit mTOR, which lies downstream of Akt signalling, can rescue the neuronal abnormalities caused by Girdin over-expression or DISC1 knock-down (Kim et al., 2009). Akt is also a negative modulator of GSK3β, although an inhibitor of GSK3β was not seen to rescue these girdin-related developmental defects (Kim et al., 2009).
2.3. DISC1-binding partners at the post-synaptic density
In addition to modulating the proliferation, migration and integration of neurons, it can also be postulated that proteins of the DISC1 complex might impact upon major mental illness by modulation of synaptic transmission. In support of this, DISC1 and several of its binding partners, including citron, PDE4B, LIS1, NDE1 and NDEL1, have been found to localise to the post-synaptic density (PSD, Bradshaw et al., 2008; Clapcote et al., 2007; Furuyashiki et al., 1999; Kirkpatrick et al., 2006; Niethammer et al., 2000; Zhang et al., 1999). To date however, relatively little is understood of the synaptic functions of these proteins.
In contrast, more is known about the role of the PSD-localised DISC1 interactor TNIK (Camargo et al., 2007), a kinase expressed in the mouse hippocampus (Wang et al., 2010) whose activity is involved in regulation of the cytoskeleton (Fu et al., 1999). DISC1-binding inhibits the kinase activity of TNIK, leading to the degradation of several key PSD proteins, including the important structural protein PSD95, and modulating the surface expression of glutamate receptor 1 (GluR1, Wang et al., 2010). More generally, knock-down of DISC1 expression leads to an increase in the formation of spines and GluR1-expessing synapses in mature rat neurons, a process dependent on its interaction with kalirin-7 (Kal7, Hayashi-Takagi et al., 2010). Kal7-dependent regulation of spine formation occurs through its activity as a GDP/GTP exchange factor for Rac1 (Xie et al., 2007), and DISC1 appears to inhibit its activity by binding to Kal7 and PSD95 (Hayashi-Takagi et al., 2010). Activation of NMDA receptors causes dissociation of DISC1, Kal7 and PSD95, making Kal7 available to modulate Rac1 and thus spine structure (Hayashi-Takagi et al., 2010). Thus DISC1 appears to modulate the formation of PSD complexes and dendritic spines through regulation of TNIK, Kal7 and Rac1.
Other DISC1 interactors of potential importance at the synapse include APP (Young-Pearce et al., 2010), given its involvement in spine formation (Lee et al., 2009) and enhancement of NMDA receptor activity (Hoe et al., 2009). Another DISC1 interactor, Activating Transcription Factor 4 (ATF4 or CREB2, Millar et al., 2003; Morris et al., 2003; Sawamura et al., 2008) is known to bind to GABAB receptors in the synapse (Nehring et al., 2000; Vernon et al., 2001; White et al., 2000), and its transport from the synapse to the nucleus, where it acts as a transcriptional repressor, is implicated in long-term depression and memory (Lai et al., 2008).
3. Roles of DISC1-interacting proteins in signalling
Another important theme in DISC1 biology is its role in a wide variety of signalling pathways, including the GSK3β and Akt pathways discussed above in Section 2.2. A third such pathway involves signalling by the ubiquitous secondary messenger molecule cAMP. The phosphodiesterase 4 family of enzymes degrade cAMP (reviewed in Houslay and Adams, 2003) and isoforms from all four PDE4 subtypes (PDE4A-D) have been demonstrated to interact with DISC1 (Millar et al., 2005; Murdoch et al., 2007). DISC1 binds PDE4 in a low-activity conformation (Millar et al., 2005; Murdoch et al., 2007) and PDE4 activity is diminished in mice with a mutation, Q31L, in a PDE4-binding site on DISC1 (Clapcote et al., 2007). Downstream effects of DISC1–PDE4 interaction remain to be determined, but are likely to include regulation of the activity of cAMP-dependent Protein Kinase A (PKA), substrates of which include the DISC1-interactors NDE1 (Bradshaw et al., 2008) and ATF4 (Elefteriou et al., 2005). In support of this, over-expression of DISC1 exaggerates the repression of CRE-dependent gene transcription caused by ATF4 in response to PKA (Karpinski et al., 1992; Sawamura et al., 2008). PDE4 is the known target for rolipram and other small molecules which have anti-depressant and anti-psychotic activity in rodent models (Kanes et al., 2007; Maxwell et al., 2004; O’Donnell and Zhang, 2004).
Another family of proteins heavily implicated in major mental illness are the neuregulins and the ErbB family of receptors for which cleaved NRG domains act as ligands (reviewed in Schmitt et al., 2008). Intriguingly, NRG1 and NRG2 signalling is seen to increase the expression of a specific DISC1 isoform in a process dependant on the activity of BACE to cleave neuregulins, forming extracellular peptide ligands (Seshadri et al., 2010). This pathway appears to be mediated by PI3 K/Akt signalling and is transcription-dependent (Seshadri et al., 2010). Downstream effects of NRG1 signalling include inducing the expression and activity of ATF4 (Talukder et al., 2000). Expression of DISC1 also appears to be upregulated following signalling by the neuropeptide PACAP, which additionally stimulates interaction of DISC1 with DBZ (Hattori et al., 2007). DISC1 is also implicated in dopamine signalling, which is altered in several DISC1 mouse models (Ayhan et al., 2010; Lipina et al., 2010b; Niwa et al., 2010). Silencing of DISC1 in rat striatal neurons leads to loss of dopamine receptor-expressing cilia (Marley and von Zastrow, 2010).
In summary, it is increasingly apparent that DISC1 is not simply a scaffold for the formation of protein complexes, but more an active hub for regulating divergent signalling pathways, including PDE4/cAMP, Akt/mTOR and GSK3β/β-catenin that are each well known to impact upon neurodevelopmental and/or psychiatric illness. An interesting side point is the apparent convergence of the DISC1 pathway with proteins involved in Alzheimer’s disease. DISC1 is now known to interact with APP (Young-Pearse et al., 2010), from which the β-amyloid peptide is derived, along with the related APLP1 protein (Millar et al., 2003). DISC1 also inhibits the activity of GSK3β (Mao et al., 2009), which is modulated by and may also modulate β-amyloid peptides (Hernández et al., 2010), and DISC1 levels are indirectly regulated by BACE (Seshadri et al., 2010), the APP-cleaving enzyme. By implication, DISC1 plays a critical role in integrating these otherwise independent pathways, elaborating the details of which represents a key future challenge.
4. Genetic evidence implicating DISC1 interactors in schizophrenia and related disorders
Several positive genetic association studies have been reported for genes encoding DISC1 interactors (summarised in Table 2), implying that multiple DISC1-related pathways need to be considered as relevant to risk of psychopathology. ATF4, CIT (encoding citron), FEZ1, NDE1, PAFAH1B1 (encoding LIS1), PCNT (encoding pericentrin), PDE4D, TNIK and YWHAE (encoding 14-3-3ɛ) are thus all implicated in schizophrenia, although some of these are single studies or report modest associations that await firm replication, with some studies failing to replicate (Table 3 and references therein). Replication of genetic association between one or more SNPs and major mental illness supports PDE4B, NDEL1 and PCM1 in their own right (Table 2 and references therein).
Table 2.
Studies which have found positive evidence of association between variants in genes encoding DISC1-interacting proteins and major mental illness. SNPs grouped together in brackets indicate haplotypes. SNPs separated by a hyphen (−) indicate significance when alleles are considered together, but not independently.
| Gene | Study | Condition | Sample | SNP, haplotype or marker | Notes |
|---|---|---|---|---|---|
| ATF4 | Qu et al. (2008) | Schizophrenia | Han Chinese | (rs17001266, rs4894) | Males only |
| CIT | Lyons-Warren et al. (2005) | Bipolar disorder | American | rs203368 | |
| rs435136 | |||||
| (rs435136, hCV3259834) | |||||
| (rs203368, rs435136) | |||||
| (rs203368, rs435136, hCV3259834) | |||||
| (rs278109, rs203368) | |||||
| (rs2285595, rs278109, rs203368) | |||||
| (rs2285595, rs278109, rs203368, rs435136) | |||||
| Nicodemus et al. (2010) | Schizophrenia | American | rs10744743 | ||
| rs3847960-rs203332 | |||||
| rs3847960-rs440299 | |||||
| CIT-DISC1 | Nicodemus et al. (2010) | Schizophrenia | American | rs10744743-rs1411771 | |
| CIT-NDEL1 | Nicodemus et al. (2010) | Schizophrenia | American | rs10744743-rs4791707 | |
| FEZ1 | Rastogi et al. (2009) | Schizophrenia | Canadian | (rs2845846, rs2849222) | |
| NDE1 | Hennah et al. (2007) | Schizophrenia | Finnish families | (rs4781678, rs2242549, rs881803, rs2075512) | Females only, conditioned on DISC1 HEP3 haplotype |
| Burdick et al. (2008) | Schizophrenia | American Caucasian | (rs8061376, rs4781679, rs3784859, rs12934645) | Amongst DISC1 C704 carriers | |
| NDEL1 | Burdick et al. (2008) | Schizophrenia | American Caucasian | (rs1391768, rs1391766, rs931672, rs35261231) | Not amongst DISC1 C704 carriers |
| Tomppo et al. (2009a) | Schizophrenia | Finnish families | rs17806986 | ||
| (rs17806986, rs1391768, rs1391766, rs3817003) | |||||
| Nicodemus et al. (2010) | Schizophrenia | American | rs4791707 | ||
| PAFAH1B1 | Rastogi et al. (2009) | Schizophrenia | Canadian families | (rs8081803, rs12938775) | |
| Nicodemus et al. (2010) | Schizophrenia | American | rs12938775 | ||
| PCM1 | Gurling et al. (2006) | Schizophrenia | British & Icelandic families | D8S261 | |
| Brisith | D8S261 | ||||
| (rs445422, 87366_66, rs370429) | |||||
| (rs454755, rs3780103, rs6991775) | |||||
| (rs454755, 87366_66, rs3780103, rs6991775) | |||||
| Scottish | (rs454755, rs3780103, rs6991775) | ||||
| American trios | D8S261 | ||||
| Datta et al. (2010) | Schizophrenia | British | rs208747 | ||
| rs445422 | |||||
| rs13276297 | |||||
| rs370429 | |||||
| 14 haplotypes | |||||
| Scottish | rs445422 | ||||
| Moens et al. (2010) | Schizophrenia | Swedish | rs13276297 | ||
| European | rs445422 | Meta-analysis of populations in Datta & Moens studies | |||
| rs208747 | |||||
| PCNT | Anitha et al. (2009) | Schizophrenia | Japanese | rs2249057 | |
| (rs9981892, rs2249057) | |||||
| (rs9981892, rs2249057, rs2839222) | |||||
| Numata et al. (2009b) | Major depression | Japanese | rs3788265 | ||
| rs2073376 | |||||
| PDE4B | Pickard et al. (2007) | Schizophrenia | Scottish | (rs2503166, rs583018, rs526772) | Females only |
| Fatemi et al. (2008) | Schizophrenia | American Caucasian | rs1354064 | ||
| rs4320761 | |||||
| rs1040716 | |||||
| rs910694 | |||||
| rs1321177 | |||||
| rs2144719 | |||||
| rs783038 | |||||
| African American | rs599381 | ||||
| rs1040716 | |||||
| rs910694 | |||||
| Numata et al. (2008a) | Schizophrenia | Japanese | rs2180335 | ||
| rs910694 | |||||
| rs472952 | |||||
| Numata et al. (2009a) | Major depressive disorder | Japanese | rs472952 | Not replicated in second sample | |
| Rastogi et al. (2009) | Schizophrenia | Canadian | (rs614350, rs2503174) | ||
| (rs12068439, rs12743648) | |||||
| (rs2503174, rs1577844) | |||||
| Tomppo et al. (2009a) | Schizophrenia | Finnish families | rs7412571 | ||
| (rs10158178, rs7412571, rs5999235, rs2069278) | |||||
| (rs4503327, rs2503222, rs6588186) | |||||
| PDE4D | Tomppo et al. (2009a) | Schizophrenia | Finnish families | rs1120303 | |
| (rs13190249, rs1120303, rs921942, rs10805515, rs10514862) | |||||
| TNIK | Potkin et al. (2009) | Schizophrenia associated with a quantitative trait | American | rs2088885 | |
| rs7627954 | |||||
| YWHAE | Ikeda et al. (2008) | Schizophrenia | Japanese | rs34041110 | |
| rs7224258 | |||||
| rs3752826 | |||||
| rs11655548 | |||||
| rs2131431 | |||||
| rs1873827 | |||||
| rs28365859 |
Table 3.
Studies which failed to find evidence of association of mental illness with any SNP examined of a gene encoding a DISC1-interacting protein.
| Gene | Study | Condition | Population |
|---|---|---|---|
| ATF4 | Kakiuchi et al. (2007) | Bipolar disordera | Japanese |
| DBZ | Anitha et al. (2009) | Schizophrenia | Japanese |
| Bipolar disorder | Japanese | ||
| FEZ1 | Yamada et al. (2004) | Bipolar disorder | Japanese |
| Schizophreniaa | Japanese | ||
| Hodgkinson et al. (2007) | Schizophrenia | American Caucasian | |
| African American | |||
| Koga et al. (2007) | Schizophrenia | Japanese | |
| Nicodemus et al. (2010) | Schizophreniab | American | |
| GRB2 | Ikeda et al. (2008) | Schizophrenia | Japanese |
| KIF5A | Ikeda et al. (2008) | Schizophrenia | Japanese |
| NDE1 | Numata et al. (2008b) | Schizophrenia | Japanese |
| Nicodemus et al. (2010) | Schizophreniab | American | |
| NDEL1 | Kähler et al. (2008) | Schizophrenia | Scandanavian |
| Ikeda et al. (2008) | Schizophrenia | Japanese | |
| Rastogi et al. (2009) | Schizophrenia | Canadian | |
| PAFAH1B1 | Ikeda et al. (2008) | Schizophrenia | Japanese |
| Kähler et al. (2008) | Schizophrenia | Scandanavian | |
| PCNT | Numata et al. (2010) | Schizophreniaa | Japanese |
| Anitha et al. (2008) | Bipolar disorder | Japanese | |
| PDE4B | Holliday et al. (2009) | Schizophreniac | Tamil Nadu, India |
| Rastogi et al. (2009) | Schizophreniaa | Canadian | |
| Kähler et al. (2009) | Schizophreniaa | Scandinavian | |
| Bipolar disorder | Scandinavian | ||
| YWHAE | Kähler et al. (2008) | Schizophrenia | Scandanavian |
These studies found nominal association with one or more SNPs, but these did not survive correction for multiple testing.
Nicodemus et al. were principally looking for evidence of genetic epistasis between genes rather than evidence that individual SNPs were associated with schizophrenia.
Holliday et al. found a risk locus proximal to PDE4B in a ethnically homogenous sample, but found no evidence of association to PDE4B itself.
It is important here also to distinguish between the strict statistical tests for significance that must be applied to gene-wide or genome-wide test of association and the insight which can be gained from specific mutations in individual cases and families (Mitchell and Porteous, 2009; Porteous, 2008). Thus, in much the same way as DISC1 was discovered at a translocation breakpoint (Blackwood et al., 2001; Millar et al., 2000; St Clair et al., 1990), PDE4B was found to be directly disrupted by a t(1:16) translocation in a proband with schizophrenia, who also had an affected cousin (Millar et al., 2005). Similarly, both deletions and duplications at chromosomal locus 16p13.1, containing the NDE1 gene, are significantly over-represented in schizophrenia patients in Scottish and other European populations, with a similar deletion also seen in an African–American individual with the condition (Ingason et al., 2011; Need et al., 2009). Ultra-rare missense mutations in patients with schizophrenia have been reported for APP (Jones et al., 1992) PCM1 (Kamiya et al., 2008), and indeed DISC1 (Song et al., 2008). These rare mutations point the finger directly at these genes and associated pathways, and further demonstrate their biological relevance.
Evidence has also been reported of transcripts encoding several of these proteins being either up- or down-regulated in brain tissue of individuals with psychiatric illness, compared to that from healthy controls (Table 4 and references therein). Such differences in mRNA expression could be the result of direct mutation in those genes, or indirectly, due to dysregulation of transcription factors or other modulatory proteins. Additionally, certain DISC1 SNPs are associated with reduced levels of FEZ1, LIS1 and NDEL1 transcripts in the hippocampus (Lipska et al., 2006).
Table 4.
Studies which have found levels of transcripts encoding DISC1-interacting proteins to be significantly altered in RNA from individuals with major mental illness compared to healthy controls.
| Gene | Study | Condition | Associated expression profile |
|---|---|---|---|
| FEZ1 | Lipska et al. (2006) | Schizophrenia | Reduced in the hippocampus and dorsolateral prefrontal cortex |
| NDE1 | Fatemi et al. (2010) | Schizophrenia | Increased in cerebellum |
| Bipolar disorder | Increased in cerebellum | ||
| Major depression | Increased in cerebellum | ||
| NDEL1 | Lipska et al. (2006) | Schizophrenia | Reduced in the hippocampus |
| PAFAH1B1 | Lipska et al. (2006) | Schizophrenia | Reduced in the hippocampus and dorsolateral prefrontal cortex |
| PDE4B | Numata et al. (2009a) | Major depression | Increased in peripheral leukocytes |
| TNIK | Glatt et al. (2005) | Schizophrenia | Increased in dorsolateral prefrontal cortex |
| Matigian et al. (2007) | Bipolar disorder | Increased in lymphoblastoids (relative to healthy monozygotic twin) |
Additionally, as one would predict biologically, evidence for epistatic interaction is emerging: three-way interaction between specific SNPs in CIT, DISC1 and NDEL1 has been reported for schizophrenia (Nicodemus et al., 2010); there is strong statistical interplay between the HEP3 haplotype and NDE1 in the Finnish population (Hennah et al., 2007) and haplotypes of NDE1 and NDEL1 show association that is dependent on the Ser or Cys variant at position 704 in DISC1 (Burdick et al., 2008).
Although the emphasis of genetic studies to date has been on their potential pathological impact, it is emerging that common variants of DISC1 and its interactors impact on normal variation and intermediate phenotypes, for example memory tasks (Burdick et al., 2005; Cannon et al., 2005; Hennah et al., 2005) and in quantitative measures of personality and mood (Harris et al., 2010). We have recently reported (Hennah and Porteous, 2009) that common cis-acting variants of DISC1 modulate expression within normal subjects by up to 20%. Moreover, variants in DISC1, PDE4 and NDE1 impact on the expression of genes involved in the cytoskeleton, neurosignalling and sensory perception, and are significantly enriched for current drug development targets in psychiatry.
In a similar vein, the recent demonstration of at least fifty different DISC1 transcripts including an abundance that are specific to foetal development and some for which expression is altered in the hippocampus of those suffering from schizophrenia or carrying DISC1 schizophrenia-risk alleles (Nakata et al., 2009), raised a whole new series of questions about how DISC1 expression is regulated, and with what effect on neurodevelopment and signalling.
5. Caveats and limitations
Whereas the growing literature on DISC1 and the DISC1 pathway, as summarised here, provides multiple avenues of promising research to pursue that is relevant to neurodevelopment, signalling and psychopathology, there are gaps and limitations. For example, with respect to the DISC1 interactome derived from yeast-two-hybrid analysis, only a minority of putative DISC1 interactors have been formally tested and confirmed by co-immunoprecipitation or co-localisation in native tissue. Although multiple transcripts and protein isoforms of DISC1 have been described, the functional role of the former and the amino-acid sequence of the later remain to be determined. Evidence from the original family from which DISC1 was identified is consistent with a simple haploinsufficiency model, but in the absence of patient tissue other than lymphoblastoid cell lines, it is not possible to rule out a dominant negative effect of hypothetical truncated or fusion DISC1 protein expression during development or in the adult brain. This family is an example of an ultra rare, in this case unique, genetic event revealing a more general genetic contribution though other genetic variants at the locus. A number of amino-acid substitutions in DISC1 been described and regulatory mutations hypothecated from association evidence, but a comprehensive analysis of all possible mutations awaits the results of large scale resequencing studies. Our understanding of the biological consequences of S704C and L607F, the best studied to date, remain partial. Regarding the cell and animal models used to test for biological effects, these do not as yet model known clinical variants, nor, for obvious reasons, do the models allow testing of psychiatric phenotypes, only at best surrogates and proxies. That said, a still modest, but growing body of evidence is making links between mouse models and human studies through comparative brain histology and imaging (reviewed in Johnstone et al., 2010). Thus, despite the remarkable progress, much remains to be done not just in vitro or in model systems, but in clinical studies too.
6. Summary and conclusions
The complexities of schizophrenia and related psychiatric illness were never likely to yield to single methodologies or models, but a combined genetic and biological approach offers promise. From what might have been viewed as an unlikely start point, the molecular genetic characterisation of a single family with a high loading for psychiatric illness, the insights from the discovery of DISC1 have been manifold and far-reaching, a paradigm for future work. It is now not so much a question of the role of DISC1 per se, but much more about the DISC1 pathway in neurodevelopment and signalling, brought to light through the multiplicity of DISC1-interacting proteins. Mechanistic details remain to be filled in and potential therapeutic targets evaluated. But in the decade since DISC1 was cloned, much progress towards these goals has been made by a combination of genetics, biochemistry, neurobiology and animal models. The next decade promises further exciting prospects to enhance our understanding of the DISC1 pathway to the benefit of patients.
Acknowledgements
This work was funded by Wellcome Trust grant 088179/A/09/Z. The funding source had no involvement in the planning or preparation of this review. The authors thank Shaun Mackie, Kirsty Millar and Dinesh Soares for critical reading of this manuscript and regret that, for reasons of brevity, much interesting research into individual DISC1-interacting partners had to be excluded or described only in a summary form.
Contributor Information
Nicholas J. Bradshaw, Email: nicholas.bradshaw@uni-duesseldorf.de.
David J. Porteous, Email: David.Porteous@ed.ac.uk.
References
- Anai M., Shojima N., Katagiri H., Ogihara T., Sakoda H., Onishi Y., Ono H., Fujishiro M., Fukushima Y., Horike N., Viana A., Kikuchi M., Noguchi N., Takahashi S., Takata K., Oka Y., Uchijima Y., Kurihara H., Asano T. A novel protein kinase B (PKB)/AKT-binding protein enhances PKB kinase activity and regulates DNA synthesis. J. Biol. Chem. 2005;280:18525–18535. doi: 10.1074/jbc.M500586200. [DOI] [PubMed] [Google Scholar]
- Anitha A., Nakamura K., Yamada K., Iwayama Y., Toyota T., Takei N., Iwata Y., Suzuki K., Sekine Y., Matsuzaki H., Kawai M., Miyoshi K., Katayama T., Matsuzaki S., Baba K., Honda A., Hattori T., Shimizu S., Kumamoto N., Tohyama M., Yoshikawa T., Mori N. Gene and expression analyses reveal enhanced expression of pericentrin 2 (PCNT2) in bipolar disorder. Biol. Psychiatry. 2008;63:678–685. doi: 10.1016/j.biopsych.2007.07.010. [DOI] [PubMed] [Google Scholar]
- Anitha A., Nakamura K., Yamada K., Iwayama Y., Toyota T., Takei N., Iwata Y., Suzuki K., Sekine Y., Matsuzaki H., Kawai M., Thanseem I., Miyoshi K., Katayama T., Matsuzaki S., Baba K., Honda A., Hattori T., Shimizu S., Kumamoto N., Kikuchi M., Tohyama M., Yoshikawa T., Mori N. Association studies and gene expression analyses of the DISC1-interacting molecules, pericentrin 2 (PCNT2) and DISC1-binding zinc finger protein (DBZ), with schizophrenia and with bipolar disorder. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 2009;150B:967–976. doi: 10.1002/ajmg.b.30926. [DOI] [PubMed] [Google Scholar]
- Arai M., Obata N., Kockelkorn T.T.J.P., Yamada K., Toyota T., Haga S., Yoshida Y., Ujike H., Sora I., Ikeda K., Yoshikawa T., Itokawa M. Lack of association between polymorphisms in the 5′ upstream region of the DISC1 gene and mood disorders. Psychiatr. Genet. 2007;17:357. doi: 10.1097/YPG.0b013e3281c8f275. [DOI] [PubMed] [Google Scholar]
- Ayhan Y., Abazyan B., Nomura J., Kim R., Ladenheim B., Krasnova I.N., Sawa A., Margolis R.L., Cadet J.L., Mori S., Vogel M.W., Ross C.A., Pletnikov M.V. Differential effects of prenatal and postnatal expressions of mutant human DISC1 on neurobehavioral phenotypes in transgenic mice: evidence for neurodevelopmental origin of major psychiatric disorders. Mol. Psychiatry. 2010 doi: 10.1038/mp.2009.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beard M.B., O’Connell J.C., Bolger G.B., Houslay M.D. The unique N-terminal domain of the cAMP phosphodiesterase PDE4D4 allows for interaction with specific SH3 domains. FEBS Lett. 1999;460:173–177. doi: 10.1016/s0014-5793(99)01335-6. [DOI] [PubMed] [Google Scholar]
- Blackwood D.H.R., Fordyce A., Walker M.T., St.Clair D.M., Porteous D.J., Muir W.J. Schizophrenia and affective disorders – cosegregation with a translocation at chromosome 1q42 that directly disrupts brain-expressed genes: clinical and P300 findings in a family. Am. J. Hum. Genet. 2001;69:428–433. doi: 10.1086/321969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasius T.L., Cai D., Jih G.T., Toret C.P., Verhey K.J. Two binding partners cooperate to activate the molecular motor kinesin-1. J. Cell Biol. 2007;176:11–17. doi: 10.1083/jcb.200605099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradshaw N.J., Ogawa F., Antolin-Fontes B., Chubb J.E., Carlyle B.C., Christie S., Claessens A., Porteous D.J., Millar J.K. DISC1, PDE4B, and NDE1 at the centrosome and synapse. Biochem. Biophysical Res. Commun. 2008;377:1091–1096. doi: 10.1016/j.bbrc.2008.10.120. [DOI] [PubMed] [Google Scholar]
- Bradshaw N.J., Christie S., Soares D.C., Carlyle B.C., Porteous D.J., Millar J.K. NDE1 and NDEL1: multimerisation, alternate splicing and DISC1 interaction. Neurosci. Lett. 2009;449:228–233. doi: 10.1016/j.neulet.2008.10.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandon N.J., Handford E.J., Schurov I., Rain J.-C., Pelling M., Duran-Jimeriz B., Camargo L.M., Oliver K.R., Beher D., Shearman M.S., Whiting P.J. Disrupted in schizophrenia 1 and nudel form a neurodevelopmentally regulated protein complex: implications for schizophrenia and other major neurological disorders. Mol. Cell. Neurosci. 2004;25:42–55. doi: 10.1016/j.mcn.2003.09.009. [DOI] [PubMed] [Google Scholar]
- Brandon N.J., Millar J.K., Korth C., Sive H., Singh K.K., Sawa A. Understanding the role of DISC1 in psychiatric disease and during normal development. J. Neurosci. 2009;29:12768–12775. doi: 10.1523/JNEUROSCI.3355-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdick K.E., Hodgkinson C.A., Szeszko P.R., Lencz T., Ekholm J.M., Kane J.M., Goldman D., Malhotra A.K. DISC1 and neurocognitive function in schizophrenia. Neuroreport. 2005;16:1399–1402. doi: 10.1097/01.wnr.0000175248.25535.f6. [DOI] [PubMed] [Google Scholar]
- Burdick K.E., Kamiya A., Hodgkinson C.A., Lencz T., DeRosse P., Ishizuka K., Elashvili S., Arai H., Goldman D., Sawa A., Malhotra A.K. Elucidating the relationship between DISC1, NDEL1, and NDE1 and the risk for schizophrenia: evidence of epistasis and competitive binding. Hum. Mol. Genet. 2008;17:2462–2473. doi: 10.1093/hmg/ddn146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderon de Anda F., Meletis K., Ge X., Rei D., Tsai L.-H. Centrosome motility is essential for initial axon formation in the neocortex. J. Neurosci. 2010;30:10391–10406. doi: 10.1523/JNEUROSCI.0381-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callicott J.H., Straub R.E., Pezawas L., Egan M.F., Mattay V.S., Hariri A.R., Verchinski B.A., Meyer-Lindenberg A., Balkissoon R., Kolachana B., Goldberg T.E., Weinberger D.R. Variation in DISC1 affects hippocampal structure and function and increases risk for schizophrenia. Prot. Natl. Acad. Sci. USA. 2005;102:8627–8632. doi: 10.1073/pnas.0500515102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camargo L.M., Collura V., Rain J.-C., Mizuguchi K., Hermjakob H., Kerrien S., Bonnert T.P., Whiting P.J., Brandon N.J. Disrupted in schizophrenia 1 interactome: evidence for the close connectivity of risk genes and a potential synaptic basis for schizophrenia. Mol. Psychiatry. 2007;12:74–86. doi: 10.1038/sj.mp.4001880. [DOI] [PubMed] [Google Scholar]
- Cannon T.D., Hennah W., van Erp T.G.M., Thompson P.M., Lonnqvist J., Huttunen M., Gasperoni T., Tuulio-Henriksson A., Pirkola T., Toga A.W., Kaprio J., Mazziotta J., Peltonen L. Association of DISC1/TRAX haplotypes with schizophrenia, reduced prefrontal gray matter, and impaired short- and long-term memory. Arch. Gen. Psychiatry. 2005;62:1205–1213. doi: 10.1001/archpsyc.62.11.1205. [DOI] [PubMed] [Google Scholar]
- Chang B., Khanna H., Hawes N., Jimeno D., He S., Lillo C., Parapuram S.K., Cheng H., Scott A., Hurd R.E., Sayer J.A., Otto E.A., Attanasio M., O’Toole J.F., Jin G., Shou C., Hildebrandt F., Williams D.S., Heckenlively J.R., Swaroop A. In-frame deletion in a novel centrosomal/ciliary protein CEP290/NPHP6 perturbs its interaction with RPGR and results in early-onset retinal degeneration in the rd16 mouse. Hum. Mol. Genet. 2006;15:1847–1857. doi: 10.1093/hmg/ddl107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chubb J.E., Bradshaw N.J., Soares D.C., Porteous D.J., Millar J.K. The DISC locus in psychiatric illness. Mol. Psychiatry. 2008;13:36–64. doi: 10.1038/sj.mp.4002106. [DOI] [PubMed] [Google Scholar]
- Clapcote S.J., Lipina T.V., Millar J.K., Mackie S., Christie S., Ogawa F., Lerch J.P., Trimble K., Uchiyama M., Sakuraba Y., Kaneda H., Shiroishi T., Houslay M.D., Henkelman R.M., Sled J.G., Gondo Y., Porteous D.J., Roder J.C. Behavioral phenotypes of Disc1 missense mutations in mice. Neuron. 2007;54:387–402. doi: 10.1016/j.neuron.2007.04.015. [DOI] [PubMed] [Google Scholar]
- Collins D.M., Murdoch H., Dunlop A.J., Charych E., Baillie G.S., Wang Q., Herberg F.W., Brandon N., Prinz A., Houslay M.D. Ndel1 alters its conformation by sequestering cAMP-specific phosphodiesterase-4D3 (PDE4D3) in a manner that is dynamically regulated through Protein Kinase A (PKA) Cell. Signal. 2008;20:2356–2369. doi: 10.1016/j.cellsig.2008.09.017. [DOI] [PubMed] [Google Scholar]
- Crepel A., Breckpot J., Fryns J.-P., De la Marche W., Steyaert J., Devriendt K., Peeters H. DISC1 duplication in two brothers with autism and mild mental retardation. Clin. Genet. 2010;77:389–394. doi: 10.1111/j.1399-0004.2009.01318.x. [DOI] [PubMed] [Google Scholar]
- Dammermann A., Merdes A. Assembly of centrosomal proteins and microtubule organization depends on PCM-1. J. Cell Biol. 2002;159:255–266. doi: 10.1083/jcb.200204023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta S.R., McQuillin A., Rizig M., Blaveri E., Thirumalai S., Kalsi G., Lawrence J., Bass N.J., Puri V., Choudhury K., Pimm J., Crombie C., Fraser G., Walker N., Curtis D., Zvelebil M., Pereira A., Kandaswamy R., St Clair D., Gurling H.M.D. A threonine to isoleucine missense mutation in the pericentriolar material 1 gene is strongly associated with schizophrenia. Mol. Psychiatry. 2010;15:615–628. doi: 10.1038/mp.2008.128. [DOI] [PubMed] [Google Scholar]
- Delaval B., Doxsey S.J. Pericentrin in cellular function and disease. J. Cell Biol. 2010;188:181–190. doi: 10.1083/jcb.200908114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Giorgio A., Blasi G., Sambataro F., Rampino A., Papazacharias A., Gambi F., Romano R., Caforio G., Rizzo M., Latorre V., Popolizio T., Kolachana B., Callicott J.H., Nardini M., Weinberger D.R., Bertolino A. Association of the Ser704Cys DISC1 polymorphism with human hippocampal formation gray matter and function during memory encoding. Eur. J. Neurosci. 2008;28:2129–2136. doi: 10.1111/j.1460-9568.2008.06482.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan X., Chang J.H., Ge S., Faulkner R.L., Kim J.Y., Kitabatake Y., Liu X.-b., Yang C.-H., Jordan J.D., Ma D.K., Liu C.Y., Ganesan S., Cheng H.-J., Ming G.-l., Lu B., Song H. Disrupted-in-schizophrenia 1 regulates integration of newly generated neurons in the adult brain. Cell. 2007;130:1146–1158. doi: 10.1016/j.cell.2007.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eastwood S.L., Walker M., Hyde T.M., Kleinman J.E., Harrison P.J. The DISC1 Ser704Cys substitution affects centrosomal localization of its binding partner PCM1 in glia in human brain. Hum. Mol. Genet. 2010;19:2487–2496. doi: 10.1093/hmg/ddq130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elefteriou F., Ahn J.D., Takeda S., Starbuck M., Yang X., Liu X., Kondo H., Richards W.G., Bannon T.W., Noda M., Clement K., Vaisse C., Karsenty G. Leptin regulation of bone resorption by the sympathetic nervous system and CART. Nature. 2005;434:514–520. doi: 10.1038/nature03398. [DOI] [PubMed] [Google Scholar]
- Enomoto A., Asai N., Namba T., Wang Y., Kato T., Tanaka M., Tatsumi H., Taya S., Tsuboi D., Kuroda K., Kaneko N., Sawamoto K., Miyamoto R., Jijiwa M., Murakumo Y., Sokabe M., Seki T., Kaibuchi K., Takahashi M. Roles of disrupted-in-schizophrenia 1-interacting protein girdin in postnatal development of the dentate gyrus. Neuron. 2009;63:774–787. doi: 10.1016/j.neuron.2009.08.015. [DOI] [PubMed] [Google Scholar]
- Ewing R.M., Chu P., Elisma F., Li H., Taylor P., Climie S., McBroom-Cerajewski L., Robinson M.D., O’Connor L., Li M., Taylor R., Dharsee M., Ho Y., Heilbut A., Moore L., Zhang S., Ornatsky O., Bukhman Y.V., Ethier M., Sheng Y., Vasilescu J., Abu-Farha M., Lambert J.-P., Duewel H.S., Stewart I.I., Kuehl B., Hogue K., Colwill K., Gladwish K., Muskat B., Kinach R., Adams S.-L., Moran M.F., Morin G.B., Topaloglou T., Figeys D. Large-scale mapping of human protein–protein interactions by mass spectrometry. Mol. Syst. Biol. 2007;3:89. doi: 10.1038/msb4100134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatemi S.H., King D.P., Reutiman T.J., Folsom T.D., Laurence J.A., Lee S., Fan Y.-T., Paciga S.A., Conti M., Menniti F.S. PDE4B polymorphisms and decreased PDE4B expression are associated with schizophrenia. Schizophr. Res. 2008;101:36–49. doi: 10.1016/j.schres.2008.01.029. [DOI] [PubMed] [Google Scholar]
- Fatemi S.H., Folsom T.D., Reutiman T.J., Vazquez G. Phosphodiesterase signaling system is disrupted in the cerebella of subjects with schizophrenia, bipolar disorder, and major depression. Schizophr. Res. 2010;119:266–267. doi: 10.1016/j.schres.2010.02.1055. [DOI] [PubMed] [Google Scholar]
- Faulkner N.E., Dujardin D.L., Tai C.-Y., Vaughan K.T., O’Connell C.B., Wang Y.-l., Vallee R.B. A role for the lissencephaly gene LIS1 in mitosis and cytoplasmic dynein function. Nat. Cell Biol. 2000;2:784–791. doi: 10.1038/35041020. [DOI] [PubMed] [Google Scholar]
- Feng Y., Walsh C.A. Mitotic spindle regulation by Nde1 controls cerebral cortical size. Neuron. 2004;44:279–293. doi: 10.1016/j.neuron.2004.09.023. [DOI] [PubMed] [Google Scholar]
- Feng Y., Olson E.C., Stukenberg P.T., Flanagan L.A., Kirschner M.W., Walsh C.A. LIS1 regulates CNS lamination by interacting with mNudE, a central component of the centrosome. Neuron. 2000;28:665–679. doi: 10.1016/s0896-6273(00)00145-8. [DOI] [PubMed] [Google Scholar]
- Fu C.A., Shen M., Huang B.C.B., Lasaga J., Payan D.G., Luo Y. TNIK, a novel member of the germinal center kinase family that activates the c-Jun N-terminal kinase pathway and regulates the cytoskeleton. J. Biol. Chem. 1999;274:30729–30737. doi: 10.1074/jbc.274.43.30729. [DOI] [PubMed] [Google Scholar]
- Fujita T., Maturana A.D., Ikuta J., Hamada J., Walchli S., Suzuki T., Sawa H., Wooten M.W., Okajima T., Tatematsu K., Tanizawa K., Kuroda i S. Axonal guidance protein FEZ1 associates with tubulin and kinesin motor protein to transport mitochondria in neurites of NGF-stimulated PC12 cells. Biochem. Biophysical Res. Commun. 2007;361:605–610. doi: 10.1016/j.bbrc.2007.07.050. [DOI] [PubMed] [Google Scholar]
- Fukuda S., Hashimoto R., Ohi K., Yamaguti K., Nakatomi Y., Yasuda Y., Kamino K., Takeda M., Tajima S., Kuratsune H., Nishizawa Y., Watanabe Y. A functional polymorphism in the disrupted-in schizophrenia 1 gene is associated with chronic fatigue syndrome. Life Sci. 2010;86:722–725. doi: 10.1016/j.lfs.2010.03.007. [DOI] [PubMed] [Google Scholar]
- Furuyashiki T., Fujisawa K., Fujita A., Madaule P., Uchino S., Mishina M., Bito H., Narumiya S. Citron, a rho-target, interacts with PSD-95/SAP-90 at glutamatergic synapses in the thalamus. J. Neurosci. 1999;19:109–118. doi: 10.1523/JNEUROSCI.19-01-00109.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge X., Frank C.L., Calderon de Anda F., Tsai L.-H. Hook3 interacts with PCM1 to regulate pericentriolar material assembly and the timing of neurogenesis. Neuron. 2010;65:191–203. doi: 10.1016/j.neuron.2010.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glatt S.J., Everall I.P., Kremen W.S., Corbeil J., Sasik R., Khanlou N., Han M., Liew C.-C., Tsuang M.T. Comparative gene expression analysis of blood and brain provides concurrent validation of SELENBP1 up-regulation in schizophrenia. Proc. Natl. Acad. Sci. USA. 2005;102:15533–15538. doi: 10.1073/pnas.0507666102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith E., Walker S., Martin C.-A., Vagnarelli P., Stiff T., Vernay B., Al Sanna N., Saggar A., Hamel B., Earnshaw W.C., Jeggo P.A., Jackson A.P., O’Driscoll M. Mutations in pericentrin cause Seckel syndrome with defective ATR-dependent DNA damage signaling. Nat. Genet. 2008;40:232–236. doi: 10.1038/ng.2007.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J., Yang Z., Song W., Chen Q., Wang F., Zhang Q., Zhu X. Nudel contributes to microtubule anchoring at the mother centriole and is involved in both dynein-dependent and -independent centrosomal protein assembly. Mol. Biol. Cell. 2006;17:680–689. doi: 10.1091/mbc.E05-04-0360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurling H.M.D., Critchley H., Datta S.R., McQuillin A., Blaveri E., Thirumalai S., Pimm J., Krasucki R., Kalsi G., Quested D., Lawrence J., Bass N., Choudhury K., Puri V., O’Daly O., Curtis D., Blackwood D., Muir W., Malhotra A.K., Buchanan R.W., Good C.D., Frackowiak R.S.J., Dolan R.J. Genetic association and brain morphology studies and the chromosome 8p22 pericentriolar material 1 (PCM1) gene in susceptibility to schizophrenia. Arch. Gen. Psychiatry. 2006;63:844–854. doi: 10.1001/archpsyc.63.8.844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris S.E., Hennah W., Thomson P.A., Luciano M., Starr J.M., Porteous D.J., Deary I.J. Variation in DISC1 is associated with anxiety, depression and emotional stability in elderly women. Mol. Psychiatry. 2010;15:232–234. doi: 10.1038/mp.2009.88. [DOI] [PubMed] [Google Scholar]
- Hashimoto R., Numakawa T., Ohnishi T., Kumamaru E., Yagasaki Y., Ishimoto T., Mori T., Nemoto K., Adachi N., Izumi A., Chiba S., Noguchi H., Suzuki T., Iwata N., Ozaki N., Taguchi T., Kamiya A., Kosuga A., Tatsumi M., Kamijima K., Weinberger D.R., Sawa A., Kunugi H. Impact of the DISC1 Ser704Cys polymorphism on risk for major depression, brain morphology, and ERK signaling. Hum. Mol. Genet. 2006;15:3024–3033. doi: 10.1093/hmg/ddl244. [DOI] [PubMed] [Google Scholar]
- Hattori T., Baba K., Matsuzaki S., Honda A., Miyoshi K., Inoue K., Taniguchi M., Hashimoto H., Shintani N., Baba A., Shimizu S., Yukioka F., Kumamoto N., Yamaguchi A., Tohyama M., Katayama T. A novel DISC1-interacting partner DISC1-binding zinc-finger protein: implication in the modulation of DISC1-dependent neurite outgrowth. Mol. Psychiatry. 2007;12:398–407. doi: 10.1038/sj.mp.4001945. [DOI] [PubMed] [Google Scholar]
- Hayashi M.A.F., Portaro F.C.V., Bastos M.F., Guerreiro J.R., Oliveira V., Gorrao S.S., Tambourgi D.V., Sant’Anna O.A., Whiting P.J., Camargo L.M., Konno K., Brandon N.J., Camargo A.C.M. Inhibition of NUDEL (nuclear distribution element-like)-oligopeptidase activity by disrupted-in-schizophrenia 1. Proc. Natl. Acad. Sci. USA. 2005;102:3828–3833. doi: 10.1073/pnas.0500330102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi M.A.F., Guerreiro J.R., Charych E., Kamiya A., Barbosa R.L., Machado M.F., Campeiro J.D., Oliveira V., Sawa A., Camargo A.C.M., Brandon N.J. Assessing the role of endooligopeptidase activity of Ndel1 (nuclear-distribution gene E homolog like-1) in neurite outgrowth. Mol. Cell. Neurosci. 2010;44:353–361. doi: 10.1016/j.mcn.2010.04.006. [DOI] [PubMed] [Google Scholar]
- Hayashi-Takagi A., Takaki M., Graziane N., Seshadri S., Murdoch H., Dunlop A.J., Makino Y., Seshadri A.J., Ishizuka K., Srivastava D.P., Xie Z., Baraban J.M., Houslay M.D., Tomoda T., Brandon N.J., Kamiya A., Yan Z., Penzes P., Sawa A. Disrupted-in-schizophrenia 1 (DISC1) regulates spines of the glutamate synapse via Rac1. Nat. Neurosci. 2010;13:327–332. doi: 10.1038/nn.2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennah W., Porteous D. The DISC1 pathway modulates expression of neurodevelopmental, synaptogenic and sensory perception genes. PLOS One. 2009;4:e4906. doi: 10.1371/journal.pone.0004906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennah W., Tuulio-Henriksson A., Paunio T., Ekelund J., Varilo T., Partonen T., Cannon T.D., Lonnqvist J., Peltonen L. A haplotype within the DISC1 gene is associated with visual memory functions in families with a high density of schizophrenia. Mol. Psychiatry. 2005;10:1097–1103. doi: 10.1038/sj.mp.4001731. [DOI] [PubMed] [Google Scholar]
- Hennah W., Tomppo L., Hiekkalinna T., Palo O.M., Kilpinen H., Ekelund J., Tuulio-Henriksson A., Silander K., Partonen T., Paunio T., Terwilliger J.D., Lonnqvist J., Peltonen L. Families with the risk allele of DISC1 reveal a link between schizophrenia and another component of the same molecular pathway, NDE1. Hum. Mol.Genet. 2007;6:453–462. doi: 10.1093/hmg/ddl462. [DOI] [PubMed] [Google Scholar]
- Hennah W., Thomson P., McQuillin A., Bass N., Loukola A., Anjorin A., Blackwood D., Curtis D., Deary I.J., Harris S.E., Isometsa E.T., Lawrence J., Lonnqvist J., Muir W., Palotie A., Partonen T., Paunio T., Pylkko E., Robinson M., Soronen P., Suominen K., Suvisaari J., Thirumalai S., St Clair D., Gurling H., Peltonen L., Porteous D. DISC1 association, heterogeneity and interplay in schizophrenia and bipolar disorder. Mol. Psychiatry. 2009;14:865–873. doi: 10.1038/mp.2008.22. [DOI] [PubMed] [Google Scholar]
- Hernández F., Gómez de Barreda E., Fuster-Matanzo A., Lucas J.J., Avila J. GSK3: a possible link between beta amyloid peptide and tau protein. Exp. Neurol. 2010;223:322–325. doi: 10.1016/j.expneurol.2009.09.011. [DOI] [PubMed] [Google Scholar]
- Higginbotham H.R., Gleeson J.G. The centrosome in neuronal development. Trends Neurosci. 2007;30:276–283. doi: 10.1016/j.tins.2007.04.001. [DOI] [PubMed] [Google Scholar]
- Hirohashi Y., Wang Q., Liu Q., Li B., Du X., Zhang H., Furuuchi K., Masuda K., Sato N., Greene M.I. Centrosomal proteins Nde1 and Su48 form a complex regulated by phosphorylation. Oncogene. 2006;25:6048–6055. doi: 10.1038/sj.onc.1209637. [DOI] [PubMed] [Google Scholar]
- Hodgkinson C.A., Goldman D., Ducci F., DeRosse P., Caycedo D.A., Newman E.R., Kane J.M., Roy A., Malhotra A.K. The FEZ1 gene shows no association to schizophrenia in Caucasian or African American populations. Neuropsychopharmacology. 2007;32:190–196. doi: 10.1038/sj.npp.1301177. [DOI] [PubMed] [Google Scholar]
- Hoe H.-S., Fu Z., Makarova A., Lee J.-Y., Lu C., Feng L., Pajoohesh-Ganji A., Matsuoka Y., Hyman B.T., Ehlers M.D., Vicini S., Pak D.T.S., Rebeck G.W. The effects of amyloid precursor protein on postsynaptic composition and activity. J. Biol. Chem. 2009;284:8495–8506. doi: 10.1074/jbc.M900141200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holliday E.G., Nyholt D.R., Tirupati S., John S., Ramachandran P., Ramamurti M., Ramadoss A.J., Jeyagurunathan A., Kottiswaran S., Smith H.J., Filippich C., Nertney D.A., Nancarrow D.J., Hayward N.K., Watkins W.S., Jorde L.B., Thara R., Mowry B.J. Strong evidence for a novel schizophrenia risk locus on chromosome 1p31.1 in homogeneous pedigrees from Tamil Nadu, India. Am. J. Psychiatry. 2009;166:206–215. doi: 10.1176/appi.ajp.2008.08030442. [DOI] [PubMed] [Google Scholar]
- Houlihan L.M., Harris S.E., Luciano M., Gow A.J., Starr J.M., Visscher P.M., Deary I.J. Replication study of candidate genes for cognitive abilities: the Lothian Birth Cohort 1936. Genes Brain Behav. 2009;8:238–247. doi: 10.1111/j.1601-183X.2008.00470.x. [DOI] [PubMed] [Google Scholar]
- Houslay M.D., Adams D.R. PDE4 cAMP phosphodiesterases: modular enzymes that orchestrate signalling cross-talk, desensitization and compartmentalization. Biochem. J. 2003;370:1–18. doi: 10.1042/BJ20021698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchins J.R.A., Toyoda Y., Hegemann B., Poser I., Heriche J.-K., Sykora M.M., Augsburg M., Hudecz O., Buschhorn B.A., Bulkescher J., Conrad C., Comartin D., Schleiffer A., Sarov M., Pozniakovsky A., Slabicki M.M., Schloissnig S., Steinmacher I., Leuschner M., Ssykor A., Lawo S., Pelletier L., Stark H., Nasmyth K., Ellenberg J., Durbin R., Buchholz F., Mechtler K., Hyman A.A., Peters J.-M. Systematic analysis of human protein complexes identifies chromosome segregation proteins. Science. 2010;328:593–599. doi: 10.1126/science.1181348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda M., Hikita T., Taya S., Uraguchi-Asaki J., Toyo-oka K., Wynshaw-Boris A., Ujike H., Inada T., Takao K., Miyakawa T., Ozaki N., Kaibuchi K., Iwata N. Identification of YWHAE, a gene encoding 14-3-3epsilon, as a possible susceptibility gene for schizophrenia. Hum. Mol. Genet. 2008;17:3212–3222. doi: 10.1093/hmg/ddn217. [DOI] [PubMed] [Google Scholar]
- Ikuta J., Maturana A., Fujita T., Okajima T., Tatematsu K., Tanizawa K., Kuroda i S. Fasciculation and elongation protein zeta-1 (FEZ1) participates in the polarization of hippocampal neuron by controlling the mitochondrial motility. Biochem. Biophysical Res. Commun. 2007;353:127–132. doi: 10.1016/j.bbrc.2006.11.142. [DOI] [PubMed] [Google Scholar]
- Ingason A., Rujescu D., Cichon S., Sigurdsson E., Sigmundsson T., Pietilainen O.P.H., Buizer-Voskamp J.E., Strengman E., Francks C., Muglia P., Gylfason A., Gustafsson O., Olason P.I., Steinberg S., Hansen T., Jakobsen K.D., Rasmussen H.B., Giegling I., Moller H.-J., Hartmann A., Crombie C., Fraser G., Walker N., Lonnqvist J., Suvisaari J., Tuulio-Henriksson A., Bramon E., Kiemeney L.A., Franke B., Murray R., Vassos E., Toulopoulou T., Muhleisen T.W., Tosato S., Ruggeri M., Djurovic S., Andreassen O.A., Zhang Z., Werge T., Ophoff R.A., GROUP Invesitgators, Rietschel M., Nothen M.M., Petursson H., Stefansson H., Peltonen L., Collier D., Stefansson K., St. Clair D.M. Copy number variations of chromosome 16p13.1 region associated with schizophrenia. Mol Psychiatry. 2011;16:17–25. doi: 10.1038/mp.2009.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James R., Adams R.R., Christie S., Buchanan S.R., Porteous D.J., Millar J.K. Disrupted in schizophrenia 1 (DISC1) is a multicompartmentalized protein that predominantly localizes to mitochondria. Mol. Cell. Neurosci. 2004;16:112–122. doi: 10.1016/j.mcn.2004.01.013. [DOI] [PubMed] [Google Scholar]
- Johnstone M., Thomson P.A., Hall J., McIntosh A.M., Lawrie S.M., Porteous D.J. DISC1 in schizophrenia: genetic mouse models and human genomic imaging. Schizophr. Bull. 2010 doi: 10.1093/schbul/sbq135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones C.T., Morris S., Yates C.M., Moffoot A., Sharpe C., Brock D.J.H., St. Clair D. Mutation in codon 713 of the b amyloid precursor protein gene presenting with schizophrenia. Nat. Genet. 1992;1:306–309. doi: 10.1038/ng0792-306. [DOI] [PubMed] [Google Scholar]
- Kähler A.K., Djurovic S., Kulle B., Jönsson E.G., Agartz I., Hall H., Opjordsmoen S., Jakobsen K.D., Hansen T., Melle I., Werge T., Steen V.M., Andreassen O.A. Association analysis of schizophrenia on 18 genes involved in neuronal migration: MDGA1 as a new susceptibility gene. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 2008;147B:1089–1100. doi: 10.1002/ajmg.b.30726. [DOI] [PubMed] [Google Scholar]
- Kähler A.K., Otnæss M.K., Wirgenes K.V., Hansen T., Jönsson E.G., Agartz I., Hall H., Werge T., Morken G., Mors O., Mellerup E., Dam H., Koefod P., Melle I., Steen V.M., Andreassen O.A., Djurovic S. Association study of PDE4B gene variants in Scandinavian schizophrenia and bipolar disorder multicenter case–control samples. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 2009;153B:86–96. doi: 10.1002/ajmg.b.30958. [DOI] [PubMed] [Google Scholar]
- Kakiuchi C., Ishiwata M., Nanko S., Kunugi H., Minabe Y., Nakamura K., Mori N., Fujii K., Yamada K., Yoshikawa T., Kato T. Association analysis of ATF4 and ATF5, genes for interacting-proteins of DISC1, in bipolar disorder. Neurosci. Lett. 2007;417:316–321. doi: 10.1016/j.neulet.2007.02.054. [DOI] [PubMed] [Google Scholar]
- Kamiya A., Kubo K.-i., Tomoda T., Takaki M., Youn R., Ozeki Y., Sawamura N., Park U., Kudo C., Okawa M., Ross C.A., Hatten M.E., Nakajima K., Sawa A. A schizophrenia-associated mutation of DISC1 perturbs cerebral cortex development. Nat. Cell Biol. 2005;7:1167–1178. doi: 10.1038/ncb1328. [DOI] [PubMed] [Google Scholar]
- Kamiya A., Tomoda T., Chang J., Takaki M., Zhan C., Morita M., Cascio M.B., Elashvili S., Koizumi H., Takanezawa Y., Dickerson F., Yolken R., Arai H., Sawa A. DISC1-NDEL1/NUDEL protein interaction, an essential component for neurite outgrowth, is modulated by genetic variations of DISC1. Hum. Mol. Genet. 2006;15:3313–3323. doi: 10.1093/hmg/ddl407. [DOI] [PubMed] [Google Scholar]
- Kamiya A., Tan P.L., Kubo K., Engelhard C., Ishizuka K., Kubo A., Tsukita S., Pulver A.E., Nakajima K., Cascella N.G., Katsanis N., Sawa A. Recruitment of PCM1 to the centrosome by the cooperative action of DISC1 and BBS4: a candidate for psychiatric illnesses. Arch. Gen. Psychiatry. 2008;65:996–1006. doi: 10.1001/archpsyc.65.9.996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanes S.J., Tokarczyk J., Siegel S.J., Bilker W., Abel T., Kelly M.P. Rolipram: a specific phosphodiesterase 4 inhibitor with potential antipsychotic activity. Neuroscience. 2007;144:239–246. doi: 10.1016/j.neuroscience.2006.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpinski B.A., Morle G.D., Huggenvik J., Uhler M.D., Leiden J.M. Molecular cloning of human CREB-2: an ATF/CREB transcription factor that can negatively regulate transcription from the cAMP response element. Proc. Natl. Acad. Sci. U.S.A. 1992;89:4820–4824. doi: 10.1073/pnas.89.11.4820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilpinen H., Ylisaukko-oja T., Hennah W., Palo O.M., Varilo T., Vanhala R., Nieminen-von Wendt T., von Wendt L., Paunio T., Peltonen L. Association of DISC1 with autism and Asperger syndrome. Mol. Psychiatry. 2008;13:187–196. doi: 10.1038/sj.mp.4002031. [DOI] [PubMed] [Google Scholar]
- Kim J.C., Badano J.L., Sibold S., Esmail M.A., Hill J., Hoskins B.E., Leitch C.C., Venner K., Ansley S.J., Ross A.J., Leroux M.R., Katsanis N., Beales P.L. The Bardet–Biedl protein BBS4 targets cargo to the pericentriolar region and is required for microtubule anchoring and cell cycle progression. Nat. Genet. 2004;36:462–470. doi: 10.1038/ng1352. [DOI] [PubMed] [Google Scholar]
- Kim H.-J., Park H.J., Jung K.H., Ban J.Y., Ra J., Kim J.W., Park J.K., Choe B.-K., Yim S.V., Kwon Y.K., Chung J.-H. Association study of polymorphisms between DISC1 and schizophrenia in a Korean population. Neurosci. Lett. 2008;430:60–63. doi: 10.1016/j.neulet.2007.10.010. [DOI] [PubMed] [Google Scholar]
- Kim J., Krishnaswami S.R., Gleeson J.G. CEP290 interacts with the centriolar satellite component PCM-1 and is required for Rab8 localization to the primary cilium. Hum. Mol. Genet. 2008;17:3796–3805. doi: 10.1093/hmg/ddn277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.Y., Duan X., Liu C.Y., Jang M.-H., Guo J.U., Pow-anpongkul N., Kang E., Song H., Ming G.-l. DISC1 regulates new neuron development in the adult brain via modulation of AKT-mTOR signaling through KIAA1212. Neuron. 2009;63:761–773. doi: 10.1016/j.neuron.2009.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick B., Xu L., Cascella N., Ozeki Y., Sawa A., Roberts R.C. DISC1 immunoreactivity at the light and ultrastructural level in the human neocortex. J. Comp. Neurol. 2006;497:436–450. doi: 10.1002/cne.21007. [DOI] [PubMed] [Google Scholar]
- Kitagawa M., Umezu M., Aoki J., Koizumi H., Arai H., Inoue K. Direct association of LIS1, the lissencephaly gene product, with a mammalian homologue of a fungal nuclear distribution protein, rNUDE. FEBS Lett. 2000;479:57–62. doi: 10.1016/s0014-5793(00)01856-1. [DOI] [PubMed] [Google Scholar]
- Koga M., Ishiguro H., Horiuchi Y., Albalushi T., Inada T., Iwata N., Ozaki N., Ujike H., Muratake T., Someya T., Arinami T. Failure to confirm the association between the FEZ1 gene and schizophrenia in a Japanese population. Neurosci. Lett. 2007;417:326–329. doi: 10.1016/j.neulet.2007.02.055. [DOI] [PubMed] [Google Scholar]
- Lai K.-O., Zhao Y., Ch’ng T.H., Martin K.C. Importin-mediated retrograde transport of CREB2 from distal processes to the nucleus in neurons. Proc. Natl. Acad. Sci. U.S.A. 2008;105:17175–17180. doi: 10.1073/pnas.0803906105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K.J., Moussa C.E.H., Lee Y., Sung Y., Howell B.W., Turner R.S., Pak D.T.S., Hoe H.S. Beta amyloid-independent role of amyloid precursor protein in generation and maintenance of dendritic spines. Neuroscience. 2009;169:344–356. doi: 10.1016/j.neuroscience.2010.04.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepagnol-Bestel A.-M., Dubertret C., Benmessaoud D., Simonneau M., Adès J., Kacha F., Hamdani N., Gorwood P., Ramoz N. Association of DISC1 gene with schizophrenia in families from two distinct French and Algerian populations. Psychiatr. Genet. 2010;20:298–303. doi: 10.1097/YPG.0b013e32833aa5c4. [DOI] [PubMed] [Google Scholar]
- Lim S.M., Kim H.-J., Nam M., Chung J.-H., Park Y.H. Association study of DISC1 in Korean population with autism spectrum disorders. Psychiatr. Genet. 2009;19:160. doi: 10.1097/YPG.0b013e32832a9bd1. [DOI] [PubMed] [Google Scholar]
- Lipina T.V., Kaidanovich-Beilin O., Patel S., Wang M., Clapcote S.J., Liu F., Woodgett J.R., Roder J.C. Genetic and pharmacological evidence for schizophrenia-related Disc1 interaction with GSK-3. Synapse. 2010 doi: 10.1002/syn.20839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipina T.V., Niwa M., Jaaro-Peled H., Fletcher P.J., Seeman P., Sawa A., Roder J.C. Enhanced dopamine function in DISC1-L100P mutant mice: implications for schizophrenia. Genes Brain Behav. 2010;9:777–789. doi: 10.1111/j.1601-183X.2010.00615.x. [DOI] [PubMed] [Google Scholar]
- Lipska B.K., Peters T., Hyde T.M., Halim N., Horowitz C., Mitkus S., Weickert C.S., Matsumoto M., Sawa A., Straub R.E., Vakkalanka R., Herman M.M., Weinberger D.R., Kleinman J.E. Expression of DISC1 binding partners is reduced in schizophrenia and associated with DISC1 SNPs. Hum. Mol. Genet. 2006;15:1245–1258. doi: 10.1093/hmg/ddl040. [DOI] [PubMed] [Google Scholar]
- Lyons-Warren A., Chang J.J., Balkissoon R., Kamiya A., Garant M., Nurnberger J., Scheftner W., Reich T., McMahon F., Kelsoe J., Gershon E., Coryell W., Byerley W., Berrettini W., DePaulo R., McInnis M., Sawa A. Evidence of association between bipolar disorder and Citron on chromosome 12q24. Mol. Psychiatry. 2005;10:807–809. doi: 10.1038/sj.mp.4001703. [DOI] [PubMed] [Google Scholar]
- Mao Y., Ge X., Frank C.L., Madison J.M., Koehler A.N., Doud M.K., Tassa C., Berry E.M., Soda T., Singh K.K., Biechele T., Petryshen T.L., Moon R.T., Haggarty S.J., Tsai L.-H. Disrupted in schizophrenia 1 regulates neuronal progenitor proliferation via modulation of GSK3b/b-catenin signaling. Cell. 2009;136:1017–1031. doi: 10.1016/j.cell.2008.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marley A., von Zastrow M. DISC1 regulates primary cilia that display specific dopamine receptors. PLOS One. 2010;5:e10902. doi: 10.1371/journal.pone.0010902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matigian N., Windus L., Smith H., Filippich C., Pantelis C., McGrath J., Mowry B., Hayward N. Expression profiling in monozygotic twins discordant for bipolar disorder reveals dysregulation of the WNT signalling pathway. Mol. Psychiatry. 2007;12:815–825. doi: 10.1038/sj.mp.4001998. [DOI] [PubMed] [Google Scholar]
- Maxwell C.R., Kanes S.J., Abel T., Siegel S.J. Phosphodiesterase inhibitors: a novel mechanism for receptor-independent antipsychotic medications. Neuroscience. 2004;129:101–107. doi: 10.1016/j.neuroscience.2004.07.038. [DOI] [PubMed] [Google Scholar]
- McCahill A., McSorley T., Huston E., Hill E.V., Lynch M.J., Gall I., Keryer G., Lygren B., Tasken K., van Heeke G., Houslay M.D. In resting COS1 cells a dominant negative approach shows that specific, anchored PDE4 cAMP phosphodiesterase isoforms gate the activation, by basal cyclic AMP production, of AKAP-tethered protein kinase A type II located in the centrosomal region. Cell. Sig. 2005;17:1158–1173. doi: 10.1016/j.cellsig.2005.04.003. [DOI] [PubMed] [Google Scholar]
- Millar J.K., Wilson-Annan J.C., Anderson S., Christie S., Taylor M.S., Semple C.A.M., Devon R.S., St Clair D.M., Muir W.J., Blackwood D.H.R., Porteous D.J. Disruption of two novel genes by a translocation co-segregating with schizophrenia. Hum. Mol. Genet. 2000;9:1415–1425. doi: 10.1093/hmg/9.9.1415. [DOI] [PubMed] [Google Scholar]
- Millar J.K., Christie S., Porteous D.J. Yeast two-hybrid screens implicate DISC1 in brain development and function. Biochem. Biophysical Res. Commun. 2003;311:1019–1025. doi: 10.1016/j.bbrc.2003.10.101. [DOI] [PubMed] [Google Scholar]
- Millar J.K., Pickard B.S., Mackie S., James R., Christie S., Buchanan S.R., Malloy M.P., Chubb J.E., Huston E., Baille G.S., Hill E.V., Houslay M.D., Brandon N.J., Rain J.-C., Camargo L.M., Whiting P.J., Blackwood D.H.R., Muir W.J., Porteous D.J. DISC1 and PDE4B are interacting genetic factors in schizophrenia that regulate cAMP signalling. Science. 2005;310:1187–1191. doi: 10.1126/science.1112915. [DOI] [PubMed] [Google Scholar]
- Mitchell K.J., Porteous D.J. GWAS for psychiatric disease: is the framework built on a solid foundation? Mol. Psychiatry. 2009;14:740–741. doi: 10.1038/mp.2009.17. [DOI] [PubMed] [Google Scholar]
- Miyoshi K., Honda A., Baba K., Taniguchi M., Oono K., Fujita T., Kuroda S., Katayama T., Tohyama M. Disrupted-in-Schizophrenia 1, a candidate gene for schizophrenia, participates in neurite outgrowth. Mol. Psychiatry. 2003;8:685–694. doi: 10.1038/sj.mp.4001352. [DOI] [PubMed] [Google Scholar]
- Miyoshi K., Asanuma M., Miyazaki I., Diaz-Corrales F.J., Katayama T., Tohyama M., Ogawa N. DISC1 localizes to the centrosome by binding to kendrin. Biochem. Biophysical Res. Commun. 2004;317:1195–1199. doi: 10.1016/j.bbrc.2004.03.163. [DOI] [PubMed] [Google Scholar]
- Moens L.N., Ceulemans S., Alaerts M., Van Den Bossche M.J., Lenaerts A.-S., De Zutter S., Norrback K.-F., Adolfsson R., Del-Favero J. PCM1 and schizophrenia: a replication study in the Northern Swedish population. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 2010;153B:1240–1243. doi: 10.1002/ajmg.b.31088. [DOI] [PubMed] [Google Scholar]
- Morris J.A., Kandpal G., Ma L., Austin C.P. DISC1 (Disrupted-In-Schizophrenia 1) is a centrosome-associated protein that interacts with MAP1A, MIPT3, ATF4/5 and NUDEL: regulation and loss of interaction with mutation. Hum. Mol. Genet. 2003;12:1591–1608. doi: 10.1093/hmg/ddg162. [DOI] [PubMed] [Google Scholar]
- Mouaffak F., Kebir O., Chayet M., Tordjman S., Vacheron M.N., Millet B., Jaafari N., Bellon A., Olie J.P., Krebs M.-O. Association of disrupted in schizophrenia 1 (DISC1) missense variants with ultra-resistant schizophrenia. Pharmacogenomics J. 2010 doi: 10.1038/tpj.2010.40. [DOI] [PubMed] [Google Scholar]
- Murdoch H., Mackie S., Collins D.M., Hill E.V., Bolger G.B., Klussmann E., Porteous D.J., Millar J.K., Houslay M.D. Isoform-selective susceptibility of DISC1/phosphodiesterase-4 complexes to dissociation by elevated intracellular cAMP levels. J. Neurosci. 2007;27:9513–9524. doi: 10.1523/JNEUROSCI.1493-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakata K., Lipska B.K., Hyde T.M., Ye T., Newburn E.N., Morita Y., Vakkalanka R., Barenboim M., Sei Y., Weinberger D.R., Kleinman J.E. DISC1 splice variants are upregulated in schizophrenia and associated with risk polymorphisms. Proc. Natl. Acad. Sci. USA. 2009;106:15873–15878. doi: 10.1073/pnas.0903413106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Need A.C., Ge D., Weale M.E., Maia J., Feng S., Heinzen E.L., Shianna K.V., Yoon W., Kasperavičiūtė D., Gennarelli M., Strittmatter W.J., Bonvicini C., Rossi G., Jayathilake K., Cola P.A., McEvoy J.P., Keefe R.S.E., Fisher E.M.C., St.Jean P.L., Giegling I., Hartmann A.M., Möller H.-J., Ruppert A., Fraser G., Crombie C., Middleton L.T., St. Clair D., Roses A.D., Muglia P., Francks C., Rujescu D., Meltzer H.Y., Goldstein D.B. A genome-wide investigation of SNPs and CNVs in schizophrenia. PLoS Genet. 2009;5:e1000373. doi: 10.1371/journal.pgen.1000373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nehring R.B., Horikawa H.P.M., El Far O., Kneussel M., Brandstatter J.H., Stamm S., Wischmeyer E., Betz H., Karschin A. The metabotropic GABAB receptor directly interacts with the activating transcription factor 4. J. Biol. Chem. 2000;275:35185–35191. doi: 10.1074/jbc.M002727200. [DOI] [PubMed] [Google Scholar]
- Newton S.S., Collier E.F., Bennett A.H., Russell D.S., Duman R.S. Regulation of growth factor receptor bound 2 by electroconvulsive seizure. Mol. Brain Res. 2004;129:185–188. doi: 10.1016/j.molbrainres.2004.06.032. [DOI] [PubMed] [Google Scholar]
- Nicodemus K.K., Callicott J.H., Higier R.G., Luna A., Nixon D.C., Lipska B.K., Vakkalanka R., Giegling I., Rujescu D., St Clair D., Muglia P., Shugart Y.Y., Weinberger D.R. Evidence of statistical epistasis between DISC1, CIT and NDEL1 impacting risk for schizophrenia: biological validation with functional neuroimaging. Hum. Genet. 2010 doi: 10.1007/s00439-009-0782-y. [DOI] [PubMed] [Google Scholar]
- Niethammer M., Smith D.S., Ayala R., Peng J., Ko J., Lee M.-S., Morabito M., Tsai L.-H. NUDEL is a novel Cdk5 substrate that associates with LIS1 and cytoplasmic dynein. Neuron. 2000;28:697–711. doi: 10.1016/s0896-6273(00)00147-1. [DOI] [PubMed] [Google Scholar]
- Niwa M., Kamiya A., Murai R., Kubo K.-i., Gruber A.J., Tomita K., Lu L., Tomisato S., Jaaro-Peled H., Seshadri S., Hiyama H., Huang B., Kohda K., Noda Y., O’Donnell P., Nakajima K., Sawa A., Nabeshima T. Knockdown of DISC1 by in utero gene transfer disturbs postnatal dopaminergic maturation in the frontal cortex and leads to adult behavioral deficits. Neuron. 2010;65:480–489. doi: 10.1016/j.neuron.2010.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Numata S., Ueno S.-i., Iga J.-i., Hongwei S., Nakataki M., Tayoshi S.Y., Sumitani S., Tomotake M., Itakura M., Sano A., Ohmori T. Positive association of the PDE4B (phosphodiesterase 4B) gene with schizophrenia in the Japanese population. J. Psychiatr. Res. 2008;43:7–12. doi: 10.1016/j.jpsychires.2008.01.013. [DOI] [PubMed] [Google Scholar]
- Numata S., Ueno S.-i., Iga J.-i., Nakataki M., Tanahashi T., Itakura M., Sano A., Ohi K., Hashimoto R., Takeda M., Ohmori T. No association between the NDE1 gene and schizophrenia in the Japanese population. Schizophr. Res. 2008;99:367–369. doi: 10.1016/j.schres.2007.10.032. [DOI] [PubMed] [Google Scholar]
- Numata S., Iga J.-I., Nakataki M., Tayoshi S., Taniguchi K., Sumitani S., Tomotake M., Tanahashi T., Itakura M., Kamegaya Y., Tatsumi M., Sano A., Asada T., Kunugi H., Ueno S.-I., Ohmori T. Gene expression and association analyses of the phosphodiesterase 4B (PDE4B) gene in major depressive disorder in the Japanese population. Am. J. Med. Genet. B. 2009;150B:527–534. doi: 10.1002/ajmg.b.30852. [DOI] [PubMed] [Google Scholar]
- Numata S., Iga J., Nakataki M., Tayoshi S., Tanahashi T., Itakura M., Ueno S., Ohmori T. Positive association of the pericentrin (PCNT) gene with major depressive disorder in the Japanese population. J. Psychiatry Neurosci. 2009;34:195–198. [PMC free article] [PubMed] [Google Scholar]
- Numata S., Nakataki M., Iga J.-i., Tanahashi T., Nakadoi Y., Ohi K., Hashimoto R., Takeda M., Itakura M., Ueno S.-i., Ohmori T. Association study between the pericentrin (PCNT) gene and schizophrenia. Neuromolecular Med. 2010;12:243–247. doi: 10.1007/s12017-009-8106-x. [DOI] [PubMed] [Google Scholar]
- O’Donnell J.M., Zhang H.-T. Antidepressant effects of inhibitors of cAMP phosphodiesterase (PDE4) Trends Pharmacol. Sci. 2004;25:158–163. doi: 10.1016/j.tips.2004.01.003. [DOI] [PubMed] [Google Scholar]
- Ogawa F., Kasai M., Akiyama T. A functional link between disrupted-in-schizophrenia 1 and the eukaryotic translation initiation factor 3. Biochem. Biophysical Res. Commun. 2005;338:771–776. doi: 10.1016/j.bbrc.2005.10.013. [DOI] [PubMed] [Google Scholar]
- Okuda A., Kishi T., Okochi T., Ikeda M., Kitajima T., Tsunoka T., Okumukura T., Fukuo Y., Kinoshita Y., Kawashima K., Yamanouchi Y., Inada T., Ozaki N., Iwata N. Translin-associated factor X gene (TSNAX) may be associated with female major depressive disorder in the Japanese population. Neuromol. Med. 2010;12:78–85. doi: 10.1007/s12017-009-8090-1. [DOI] [PubMed] [Google Scholar]
- Ozeki Y., Tomoda T., Kleiderlein J., Kamiya A., Bord L., Fujii K., Okawa M., Yamada N., Hatten M.E., Snyder S.H., Ross C.A., Sawa A. Disrupted-in-schizophrenia-1 (DISC-1): mutant truncation prevents binding to NudE-like (NUDEL) and inhibits neurite outgrowth. Proc. Natl. Acad. Sci. USA. 2003;100:289–294. doi: 10.1073/pnas.0136913100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palo O.M., Antila M., Silander K., Hennah W., Kilpinen H., Soronen P., Tuulio-Henriksson A., Kieseppa T., Partonen T., Lonnqvist J., Peltonen L., Paunio T. Association of distinct allelic haplotypes of DISC1 with psychotic and bipolar spectrum disorders and with underlying cognitive impairments. Hum. Mol.Genet. 2007;16:2517–2528. doi: 10.1093/hmg/ddm207. [DOI] [PubMed] [Google Scholar]
- Perlis R.H., Purcell S., Fagerness J., Kirby A., Petryshen T.L., Fan J., Sklar P. Family-based association study of lithium-related and other candidate genes in bipolar disorder. Arch. Gen. Psychiatry. 2008;65:53–61. doi: 10.1001/archgenpsychiatry.2007.15. [DOI] [PubMed] [Google Scholar]
- Pickard B.S., Thomson P.A., Christoforou A., Evans K.L., Morris S.W., Porteous D.J., Blackwood D.H.R., Muir W.J. The PDE4B gene confers sex-specific protection against schizophrenia. Psychiatr. Genet. 2007;17:129–133. doi: 10.1097/YPG.0b013e328014492b. [DOI] [PubMed] [Google Scholar]
- Porteous D., Millar K. How DISC1 regulates postnatal brain development: girdin gets in on the AKT. Neuron. 2009;63:711–713. doi: 10.1016/j.neuron.2009.09.017. [DOI] [PubMed] [Google Scholar]
- Porteous D. Genetic causality in schizophrenia and bipolar disorder: out with the old and in with the new. Curr. Opin. Genet. Dev. 2008;18:229–234. doi: 10.1016/j.gde.2008.07.005. [DOI] [PubMed] [Google Scholar]
- Potkin S.G., Turner J.A., Guffanti G., Lakatos A., Fallon J.H., Nguyen D.D., Mathalon D., Ford J., Lauriello J., Macciardi F., FBIRN A genome-wide association study of schizophrenia using brain activation as a quantitative phenotype. Schizophr. Bull. 2009;35:96–108. doi: 10.1093/schbul/sbn155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prata D.P., Mechelli A., Fu C.H.Y., Picchioni M., Kane F., Kalidindi S., McDonald C., Kravariti E., Toulopoulou T., Miorelli A., Murray R., Collier D.A., McGuire P.K. Effect of disrupted-in-schizophrenia-1 on pre-frontal cortical function. Mol. Psychiatry. 2008;13:915–917. doi: 10.1038/mp.2008.76. [DOI] [PubMed] [Google Scholar]
- Purohit A., Tynan S.H., Vallee R., Doxsey S.J. Direct interaction of pericentrin with cytoplasmic dynein light intermediate chain contributes to mitotic spindle organization. J. Cell Biol. 1999;147:481–492. doi: 10.1083/jcb.147.3.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu M., Tang F., Wang L., Yan H., Han Y., Yan J., Yue W., Zhang D. Associations of ATF4 gene polymorphisms with schizophrenia in male patients. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 2008;147B:732–736. doi: 10.1002/ajmg.b.30675. [DOI] [PubMed] [Google Scholar]
- Rastogi A., Zai C., Likhodi O., Kennedy J.L., Wong A.H. Genetic association and post-mortem brain mRNA analysis of DISC1 and related genes in schizophrenia. Schizophr. Res. 2009;114:39–49. doi: 10.1016/j.schres.2009.06.019. [DOI] [PubMed] [Google Scholar]
- Rauch A., Thiel C.T., Schindler D., Wick U., Crow Y.J., Ekici A.B., van Essen A.J., Goecke T.O., Al-Gazali L., Chrzanowska K.H., Zweier C., Brunner H.G., Becker K., Curry C.J., Dallapiccola B., Devriendt K., Dorfler A., Kinning E., Megarbane A., Meinecke P., Semple R.K., Spranger S., Toutain A., Trembath R.C., Voss E., Wilson L., Hennekam R., de Zegher F., Dorr H.-G., Reis A. Mutations in the pericentrin (PCNT) gene cause primordial dwarfism. Science. 2008;319:816–819. doi: 10.1126/science.1151174. [DOI] [PubMed] [Google Scholar]
- Ross C.A., Margolis R.L. Schizophrenia: a point of disruption. Nature. 2009;458:976–977. doi: 10.1038/458976a. [DOI] [PubMed] [Google Scholar]
- Saetre P., Agartz I., De Franciscis A., Lundmark P., Djurovic S., Kähler A., Andreassen O.A., Jakobsen K.D., Rasmussen H.B., Werge T., Hall H., Terenius L., Jönsson E.G. Association between a disrupted-in-schizophrenia 1 (DISC1) single nucleotide polymorphism and schizophrenia in a combined Scandinavian case–control sample. Schizophr. Res. 2008;106:237–241. doi: 10.1016/j.schres.2008.08.024. [DOI] [PubMed] [Google Scholar]
- Sanders A.R., Duan J., Levinson D.F., Shi J., He D., Hou C., Burrell G.J., Rice J.P., Nertney D.A., Olincy A., Rozic P., Vinogradov S., Buccola N.G., Mowry B.J., Freedman R., Amin F., Black D.W., Silverman J.M., Byerley W.F., Crowe R.R., Cloninger C.R., Martinez M., Gejman P.V. No significant association of 14 candidate genes with schizophrenia in a large european ancestry sample: implications for psychiatric genetics. Am. J. Psychiatry. 2008;165:497–506. doi: 10.1176/appi.ajp.2007.07101573. [DOI] [PubMed] [Google Scholar]
- Sasaki S., Shionoya A., Ishida M., Gambello M.J., Yingling J., Wynshaw-Boris A., Hirotsune S. A LIS1/NUDEL/cytoplasmic dyenin heavy chain complex in the developing and adult nervous system. Neuron. 2000;28:681–696. doi: 10.1016/s0896-6273(00)00146-x. [DOI] [PubMed] [Google Scholar]
- Sasaki S., Mori D., Toyo-oka K., Chen A., Garrett-Beal L., Muramatsu M., Miyagawa S., Hiraiwa N., Yoshiki A., Wynshaw-Boris A., Hirotsune S. Complete loss of Ndel1 results in neuronal migration defects and early embryonic lethality. Mol. Cell. Biol. 2005;25:7812–7827. doi: 10.1128/MCB.25.17.7812-7827.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawamura N., Ando T., Maruyama Y., Fujimuro M., Mochizuki H., Honjo K., Shimoda M., Toda H., Sawamura-Yamamoto T., Makuch L.A., Hayashi A., Ishizuka K., Cascella N.G., Kamiya A., Ishida N., Tomoda T., Hai T., Furukubo-Tokunaga K., Sawa A. Nuclear DISC1 regulates CRE-mediated gene transcription and sleep homeostasis in the fruit fly. Mol. Psychiatry. 2008;13:1138–1148. doi: 10.1038/mp.2008.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayer J.A., Otto E.A., O’Toole J.F., Nurnberg G., Kennedy M.A., Becker C., Hennies H.C., Helou J., Attanasio M., Fausett B.V., Utsch B., Khanna H., Liu Y., Drummond I., Kawakami I., Kusakabe T., Tsuda M., Ma L., Lee H., Larson R.G., Allen S.J., Wilkinson C.J., Nigg E.A., Shou C., Lillo C., Williams D.S., Hoppe B., Kemper M.J., Neuhaus T., Parisi M.A., Glass I.A., Petry M., Kispert A., Gloy J., Ganner A., Walz G., Zhu X., Goldman D., Nurnberg P., Swaroop A., Leroux M.R., Hildebrandt F. The centrosomal protein nephrocystin-6 is mutated in Joubert syndrome and activates transcription factor ATF4. Nat. Genet. 2006;38:674–681. doi: 10.1038/ng1786. [DOI] [PubMed] [Google Scholar]
- Schmitt A., Parlapani E., Gruber O., Wobrock T., Falkai P. Impact of neuregulin-1 on the pathophysiology of schizophrenia in human post-mortem studies. Eur. Arch. Psychiatry Clin. Neurosci. 2008;258:35–39. doi: 10.1007/s00406-008-5019-x. [DOI] [PubMed] [Google Scholar]
- Schosser A., Gaysina D., Cohen-Woods S., Chow P.C., Martucci L., Craddock N., Farmer A., Korszun A., Gunasinghe C., Gray J., Jones L., Tozzi F., Perry J., Muglia P., Owen M.J., Craig I.W., McGuffin P. Association of DISC1 and TSNAX genes and affective disorders in the depression case-control (DeCC) and bipolar affective case-control (BACCS) studies. Mol. Psychiatry. 2010;15:844–849. doi: 10.1038/mp.2009.21. [DOI] [PubMed] [Google Scholar]
- Schumacher J., Laje G., Abou Jamra R., Becker T., Mühleisen T.W., Vasilescu C., Mattheisen M., Herms S., Hoffmann P., Hillmer A.M., Georgi A., Herold C., Schulze T.G., Propping P., Rietschel M., McMahon F.J., Nöthen M.M., Cichon S. The DISC locus and schizophrenia: evidence from an association study in a central European sample and from a meta-analysis across different European populations. Hum. Mol. Genet. 2009;18:2719–2727. doi: 10.1093/hmg/ddp204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seshadri S., Kamiya A., Yokota Y., Prikulis I., Kano S.-I., Hayashi-Takagi A., Stanco A., Eom T.-Y., Rao S., Ishizuka K., Wong P., Korth C., Anton E.S., Sawa A. Disrupted-in-schizophrenia-1 expression is regulated by b-site amyloid precursor protein cleaving enzyme-1-neuregulin cascade. Prot. Natl. Acad. Sci. USA. 2010;107:5622–5627. doi: 10.1073/pnas.0909284107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim S.Y., Samuels B.A., Wang J., Neumayer G., Belzil C., Ayala R., Shi Y., Shi Y., Tsai L.-H., Nguyen M.D. Ndel1 controls the dynein-mediated transport of vimentin during neurite outgrowth. J. Biol. Chem. 2008;283:12232–12240. doi: 10.1074/jbc.M710200200. [DOI] [PubMed] [Google Scholar]
- Shimizu S., Matsuzaki S., Hattori T., Kumamoto N., Miyoshi K., Katayama T., Tohyama M. DISC1-kendrin interaction is involved in centrosomal microtubule network formation. Biochem. Biophysical Res. Commun. 2008;377:1051–1056. doi: 10.1016/j.bbrc.2008.10.100. [DOI] [PubMed] [Google Scholar]
- Shinoda T., Taya S., Tsuboi D., Hikita T., Matsuzawa R., Kuroda S., Iwamatsu A., Kaibuchi K. DISC1 regulates neurotrophin-induced axon elongation via interaction with Grb2. J. Neurosci. 2007;27:4–14. doi: 10.1523/JNEUROSCI.3825-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh K.K., Ge X., Mao Y., Drane L., Meletis K., Samuels B.A., Tsai L.-H. Dixdc1 is a critical regulator of DISC1 and embryonic cortical development. Neuron. 2010;67:33–48. doi: 10.1016/j.neuron.2010.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith D.S., Niethammer M., Ayala R., Zhou Y., Gambello M.J., Wynshaw-Boris A., Tsai L.-H. Regulation of cytoplasmic dynein behaviour and microtubule organization by mammalian Lis1. Nat. Cell Biol. 2000;2:767–775. doi: 10.1038/35041000. [DOI] [PubMed] [Google Scholar]
- Song W., Li W., Feng J., Heston L.L., Scaringe W.A., Sommer S.S. Identification of high risk DISC1 structural variants with a 2% attributable risk for schizophrenia. Biochem. Biophysical Res. Commun. 2008;367:700–706. doi: 10.1016/j.bbrc.2007.12.117. [DOI] [PubMed] [Google Scholar]
- Song W., Li W., Noltner K., Yan J., Green E., Grozeva D., Jones I.R., Craddock N., Longmate J., Feng J., Sommer S.S. Identification of high risk DISC1 protein structural variants in patients with bipolar spectrum disorder. Neurosci. Lett. 2010 doi: 10.1016/j.neulet.2010.09.027. [DOI] [PubMed] [Google Scholar]
- St Clair D., Blackwood D., Muir W., Carothers A., Walker M., Spowart G., Gosden C., Evans H.J. Association within a family of a balanced autosomal translocation with major mental illness. Lancet. 1990;336:13–16. doi: 10.1016/0140-6736(90)91520-k. [DOI] [PubMed] [Google Scholar]
- Stehman S.A., Chen Y., McKenney R.J., Vallee R.B. NudE and NudEL are r. PLoS Genet. 2009;5:e1000373. [Google Scholar]
- Sullivan P.F., Lin D., Tzeng J.-Y., van den Oord E., Perkins D., Stroup T.S., Wagner M., Lee S., Wright F.A., Zou F., Liu W., Downing A.M., Lieberman J., Close S.L. Genomewide association for schizophrenia in the CATIE study: results of stage 1. Mol. Psychiatry. 2008;13:570–584. doi: 10.1038/mp.2008.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney K.J., Prokscha A., Eichele G. NudE-L, a novel Lis1-interacting protein, belongs to a family of vertebrate coiled-coil proteins. Mech. Dev. 2001;101:21–33. doi: 10.1016/s0925-4773(00)00543-8. [DOI] [PubMed] [Google Scholar]
- Szeszko P.R., Hodgkinson C.A., Robinson D.G., DeRosse P., Bilder R.M., Lencz T., Burdick K.E., Napolitano B., Betensky J.D., Kane J.M., Goldman D., Malhotra A.K. DISC1 is associated with prefrontal cortical gray matter and positive symptoms in schizophrenia. Biol. Psychol. 2008;79:103–110. doi: 10.1016/j.biopsycho.2007.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai C.-Y., Dujardin D.L., Faulkner N.E., Vallee R.B. Role of dynein, dynactin, and CLIP-170 interactions in LIS1 kinetochore function. J. Cell Biol. 2002;156:959–968. doi: 10.1083/jcb.200109046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi M., Yamagiwa A., Nishimura T., Mukai H., Ono Y. Centrosomal proteins CG-NAP and kendrin provide microtubule nucleation sites by anchoring g-tubulin ring complex. Mol. Biol. Cell. 2002;13:3235–3245. doi: 10.1091/mbc.E02-02-0112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi T., Suzuki M., Tsunoda M., Maeno N., Kawasaki Y., Zhou S.-Y., Hagino H., Niu L., Tsuneki H., Kobayashi S., Sasaoka T., Seto H., Kurachi M., Ozaki N. The disrupted-in-schizophrenia-1 Ser704Cys polymorphism and brain morphology in schizophrenia. Psychiatry Res. Neuroimaging. 2009;172:128–135. doi: 10.1016/j.pscychresns.2009.01.005. [DOI] [PubMed] [Google Scholar]
- Talukder A.H., Vadlamudi R., Mandal M., Kumar R. Heregulin induces expression, DNA binding activity, and transactivating functions of basic leucine zipper activating transcription factor 4. Cancer Res. 2000;60:276–281. [PubMed] [Google Scholar]
- Taya S., Shinoda T., Tsuboi D., Asaki J., Nagai K., Hikita T., Kuroda S., Kuroda K., Shimizu M., Hirotsune S., Iwamatsu A., Kaibuchi K. DISC1 regulates the transport of the NUDEL/LIS1/14-3-3epsilon complex through kinesin-1. J. Neurosci. 2007;27:15–26. doi: 10.1523/JNEUROSCI.3826-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomppo L., Hennah W., Lahermo P., Loukola A., Tuulio-Henriksson A., Suvisaari J., Partonen T., Ekelund J., Lönnqvist J., Peltonen L. Association between genes of disrupted in schizophrenia 1 (DISC1) interactors and schizophrenia supports the role of the DISC1 pathway in the etiology of major mental illnesses. Biol. Psychiatry. 2009;65:1055–1062. doi: 10.1016/j.biopsych.2009.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomppo L., Hennah W., Miettunen J., Jarvelin M.-R., Veijola J., Ripatti S., Lahermo P., Lichtermann D., Peltonen L., Ekelund J. Association of variants in DISC1 with psychosis-related traits in a large population cohort. Arch. Gen. Psychiatry. 2009;66:134–141. doi: 10.1001/archgenpsychiatry.2008.524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth C., Shim S.Y., Wang J., Jiang Y., Neumayer G., Belzil C., Liu W.-Q., Martinez J., Zochodne D., Nguyen M.D. Ndel1 promotes axon regeneration via intermediate filaments. PLOS One. 2008;3:e2014. doi: 10.1371/journal.pone.0002014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyo-oka K., Sasaki S., Yano Y., Mori D., Kobayashi T., Toyoshima Y.Y., Tokuoka S.M., Ishii S., Shimizu T., Muramatsu M., Hiraiwa N., Yoshiki A., Wynshaw-Boris A., Hirotsune S. Recruitment of katanin p60 by phosphorylated NDEL1, an LIS1 interacting protein, is essential for mitotic cell division and neuronal migration. Hum. Mol. Genet. 2005;14:3113–3128. doi: 10.1093/hmg/ddi339. [DOI] [PubMed] [Google Scholar]
- Vernon E., Meyer G., Pickard L., Dev K., Molnar E., Collingridge G.L., Henley J.M. GABAB receptors couple directly to the transcription factor ATF4. Mol. Cell. Neurosci. 2001;17:637–645. doi: 10.1006/mcne.2000.0960. [DOI] [PubMed] [Google Scholar]
- Wang Q., Du X., Meinkoth J., Hirohashi Y., Zhang H., Liu Q., Richter M., Greene M.I. Characterization of Su48, a centrosome protein essential for cell division. Proc. Natl. Acad. Sci. USA. 2006;103:6512–6517. doi: 10.1073/pnas.0601682103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Charych E.I., Pulito V.L., Lee J.B., Graziane N.M., Crozier R.A., Revilla-Sanchez R., Kelly M.P., Dunlop A.J., Murdoch H., Taylor N., Xie Y., Pausch M., Hayashi-Takagi A., Ishizuka K., Seshadri S., Bates B., Kariya K., Sawa A., Weinberg R.J., Moss S.J., Houslay M.D., Yan Z., Brandon N.J. The psychiatric disease risk factors DISC1 and TNIK interact to regulate synapse composition and function. Mol. Psychiatry. 2010 doi: 10.1038/mp.2010.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White J.H., McIllhinney R.A.J., Wise A., Ciruela F., Chan W.-Y., Emson P.C., Billinton A., Marshall F.H. The GABAB receptor interacts directly with the related transcription factors CREB2 and ATFx. Proc. Natl. Acad. Sci. USA. 2000;97:13967–13972. doi: 10.1073/pnas.240452197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams J.M., Beck T.F., Pearson D.M., Proud M.B., Cheung S.W., Scott D.A. A 1q42 deletion involving DISC1, DISC2, and TSNAX in an autism spectrum disorder. Am. J. Med. Genet. A. 2009;149A:1758–1762. doi: 10.1002/ajmg.a.32941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wynshaw-Boris A., Pramparo T., Youn Y.H., Hirotsune S. Lissencephaly: mechanistic insights from animal models and potential therapeutic strategies. Semin. Cell Dev. Biol. 2010 doi: 10.1016/j.semcdb.2010.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z., Srivastava D.P., Photowala H., Kai L., Cahill M.E., Woolfrey K.M., Shum C.Y., Surmeier D.J., Penzes P. Kalirin-7 controls activity-dependent structural and functional plasticity of dendritic spines. Neuron. 2007;56:640–656. doi: 10.1016/j.neuron.2007.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K., Nakamura K., Minabe Y., Iwayama-Shigeno Y., Takao H., Toyota T., Hattori E., Takei N., Sekine Y., Suzuki K., Iwata Y., Miyoshi K., Honda A., Baba K., Katayama T., Tohyama M., Mori N., Yoshikawa T. Association analysis of FEZ1 variants with schizophrenia in Japanese cohorts. Biol. Psychiatry. 2004;56:683–690. doi: 10.1016/j.biopsych.2004.08.015. [DOI] [PubMed] [Google Scholar]
- Young-Pearse T.L., Bai J., Chang R., Zheng J.B., LoTurco J.J., Selkoe D.J. A critical function for b-amyloid precursor protein in neuronal nigration revealed by in utero RNA interference. J. Neurosci. 2007;27:14459–14469. doi: 10.1523/JNEUROSCI.4701-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young-Pearse T.L., Suth S., Luth E.S., Sawa A., Selkoe D.J. Biochemical and functional interaction of disrupted-in-schizophrenia 1 and amyloid precursor protein regulates neuronal migration during mammalian cortical development. J. Neurosci. 2010;30:10431–10440. doi: 10.1523/JNEUROSCI.1445-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Vazquez L., Apperson M., Kennedy M.B. Citron binds to PSD-95 at glutamatergic synapses on inhibitory neurons in the hippocampus. J. Neurosci. 1999;19:96–108. doi: 10.1523/JNEUROSCI.19-01-00096.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]