Abstract
Thiol enzymes have single- or double-catalytic site cysteine residues and are redox active. Oxidoreductases and isomerases contain double-catalytic site cysteine residues, which are oxidized to a disulfide via a sulfenyl intermediate and reduced to a thiol or a thiolate. The redox changes of these enzymes are involved in their catalytic processes. On the other hand, transferases, and also some phosphatases and hydrolases, have a single-catalytic site cysteine residue. The cysteines are redox active, but their sulfenyl forms, which are inactive, are not well explained biologically. In particular, oxidized forms of sulfurtransferases, such as mercaptopyruvate sulfurtransferase and thiosulfate sulfurtransferase, are not reduced by reduced glutathione but by reduced thioredoxin. This paper focuses on why the catalytic site cysteine of sulfurtransferase is redox active.
1. Introduction
Cysteine residues in proteins maintain the protein conformation, coordinate metal(s), and regulate protein function [1–3]. Enzymes with catalytic site cysteines (Table 1) [4–42] have critical roles in biologic processes such as cell cycle regulation, apoptosis, and signal transduction [43].
Table 1.
Typical thiol enzymes.
| Enzyme | Oxidative inactivation | ||
|---|---|---|---|
| Classification | name defined as a thiol enzyme | ||
| Oxidoreductase | Glutathione family [4] | Not defined | |
| Glutaredoxin family [5] | Not defined | ||
| Glyceraldehyde-3-phosphate dehydrogenase [6] | Yes [7, 8] | ||
| Peptide-methionine (S)-S-oxide reductase [9] | Not defined | ||
| Peroxiredoxin [10, 11] | Yes [12] | ||
| Sulphiredoxin [13] | Not defined | ||
| Thioredoxin family [14] | Not defined | ||
| Transferase | Desulfurase | Cysteine desulfurase1 [15] | Not defined |
| Phosphatase | Cdc2 25 family [16] | Yes [17, 18] | |
| Protein-tyrosine phosphatases [19] | Yes [20] | ||
| Sulfurtransferase | Mercaptopyruvate sulfurtransferase [21, 22] | Yes [23] | |
| Thiosulfate sulfurtransferase [24] | Yes [25, 26] | ||
| Hydrolase | Cysteine protease | Actinidain family [27] | Not defined |
| Bromelain family [28] | Yes [29] | ||
| Calpain family [30] | Yes [31] | ||
| Caspase family [32] | Yes [33] | ||
| Cathepsin family [34] | Yes [35] | ||
| Chymopapain family [36] | Yes [37] | ||
| Ficin family [38] | Not defined | ||
| Mir1-CP3 [39] | Not defined | ||
| Papain family [40] | Yes [41] | ||
| Isomerase | Protein disulfide isomerase [42] | Not defined | |
1pyridoxal 5′-phosphate-dependent enzyme
2cdc, cell division cycle
3Mir1-CP. Maize insect resistance-cysteine protease
A cysteine residue that easily accepts and donates (an) electron(s) is referred to as a redox-active cysteine, and has a lower pK a value than an unperturbed cysteine residue. Clairborne and colleagues extensively and successfully studied redox changes of cysteine residues and reviewed the biologic importance of redox-active cysteine [44, 45]; a redox-active cysteine is generally a thiolate at physiologic pH and is easily oxidized to a sulfenic acid. Cysteine-related enzymes are generally inhibited by mild oxidation and are reversibly reduced by thioredoxin or glutathione. The sulfenyl form is a reaction intermediate for peroxiredoxin to form disulfide [46] or protein tyrosine phosphatase 1B to form sulfenyl amide [47, 48].
The sulfenyl form is further oxidized to the sulfinyl form and/or sulfonyl form. It is noteworthy that cysteine sulfinate desulfinase catalyzes the desulfination of cysteine sulfinic acid [49, 50], which is not a reversible reaction. On the other hand, cysteine sulfinic acid reductase (sulfiredoxin) catalyzes the reduction of cysteine sulfinic acid [51, 52], although neither thioredoxin nor glutathione can reduce sulfinic acid. Thus, sulfination of cysteine residues is a reversible oxidative process under the conditions that cysteine sulfinic acid reductase can access the catalytic site cysteine of an enzyme. When the reductase cannot access the catalytic site cysteine, sulfination is as irreversible as sulfonation. Recent studies in redox biology indicate that sulfenic acid is a molecular switch [53].
2. The Catalytic Site Cysteine Residue of Sulfurtransferase Is Redox Active
The catalytic site cysteine of a thiol enzyme is generally redox active: a cysteine residue with a low pK a value easily accepts and donates (an) electron(s). The catalytic site cysteine is essential for oxidoreductase to form a (intramolecular) disulfide and/or sulfenyl intermediate, and its high reactivity of the nucleophilic cysteine is advantageous for the catalysis of transferase (desulfurase, phosphatase, and sulfurtransferase), hydrolase (cysteine protease), and isomerase (protein disulfide isomerase) (Table 1).
The effects of perturbing the pK a of a cysteine residue in a protein are not well explained. It is generally considered that a decrease in the pK a of a cysteine residue is caused by positively charged groups of neighboring amino acid residues and/or strengthening of electrostatic interactions between the group and the sulfur atom due to an increase in the electron density of the sulfur atom of the cysteine residue. Further, hydrogen bonding stabilizes the proton-dissociated state of the cysteine residue to maintain the pK a perturbation. Hol and colleagues proposed the interesting notion that the alpha-helix macropole in a protein structure contributes to lowering the pK a of a cysteine residue [54, 55].
Comparative studies of primary structures of sulfurtransferases (mercaptopyruvate sulfurtransferase [MST] and evolutionarily related rhodanese [TST] [22, 55, 56]) revealed that the consensus sequences around the catalytic cysteine of MST and TST are CG(S/T)G and C(R/Y)(K/H)G, respectively (Figure 1) [22, 55, 56].
Figure 1.

Comparison of the amino acid sequences around a catalytic site cysteine residue between MST and TST Sequence identity was analyzed using GENETYX (GENETYX CORPORATION). Box, a catalytic site. E, E. coli (D10496 for MST, NP_417883 for TST); H, Homo sapiens (BC009450 for MST, D87292 for TST); L. Leishmania (CAC85741); P1 and P2, Arabidopsis thaliana (AB032864 and AB032865 for MSTs); R, Rattus norvegicus (D50564 for MST, BC088449 for TST). Underlined amino acids, consensus sequences for MST or TST.
The tertiary structures of MST and TST are persulfurated enzymes and stable catalytic intermediates (and also free-TST) [57–60]. In X-ray structural studies of bovine TST by Ploegman and colleagues [58–60] and Hol et al. [61], persulfide was stabilized by a ring of persulfide-stabilizing NH groups; Arg248, Lys249, Val251, and Thr252 (Figure 1) contributed to hydrogen bonding with an outer sulfur atom of a persulfide at the catalytic site Cys247, and in addition, Gly254 and Ser274 with the Sγ of Cys247. Further, two helix-dipoles (α9 and α10) (Figure 2(a)) contribute to lowering the pK a of the catalytic cysteine residue to approximately 6.5 [54, 62].
Figure 2.
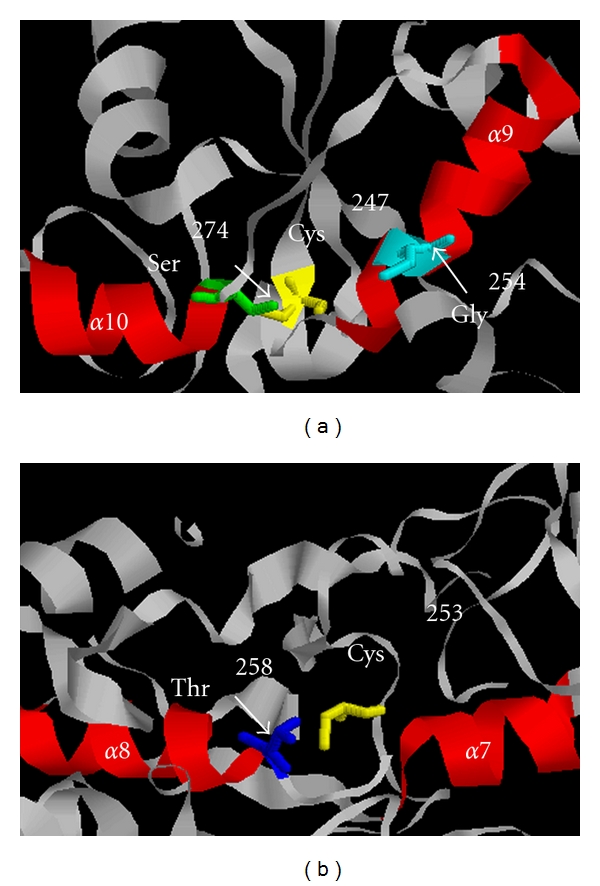
Model for the two α-helix dipoles of TST and MST, each structure is represented using RasMol. (a) bovine liver TST from 1DP2, red ribbon structure represents two helix-dipoles (α9 and α10) and ball-and-stick model in yellow represents a catalytic site Cys247. (b) Leishmania major MST from 1CKG red ribbon structure represents two helix-dipoles (α8 and α9), red ribbon structure represents two helix-dipoles (α8 and α9), and ball-and-stick model in yellow represents a catalytic site Cys253.
Similar to TST, an X-ray structural study of Leishmania major persulfurated MST by Alphey et al. [57] revealed that Gly254, Ser255, Gly256, Val257, Thr258, and Ala259 (Figure 1) contribute to hydrogen binding with an outer sulfur atom of a persulfide at the catalytic site Cys253, and further, Thr258 with the Sγ of Cys253. Two helix-dipoles (α8 and α9) (Figure 2(b)) also contribute to lowering the pK a of the catalytic cysteine.
The Cdc25 phosphatase family is a rhodanese superfamily [64, 65], and the catalytic subunit contains an alpha-helix macropole like MST and TST [66], which could contribute to lowering the pK a of the catalytic cysteine. A member of the pyridoxal 5′-phosphate-dependent enzyme family, cysteine desulfurase (E. coli NifS CsdB), has an alpha-helix macropole like MST and TST [63, 67].
In sulfurtransferases, alpha-helix macropoles surrounding a catalytic cysteine characterize the cysteine as redox active, indicating that hydrogen bonding between an outer sulfur atom of a persulfide at the catalytic site cysteine with surrounding amino acids is important for stabilizing catalytic intermediates.
3. Sulfenate Formation at a Catalytic Site in Sulfurtransferase
When MST and TST are oxidized, catalytic site cysteines are reversibly sulfenated [23, 68] and are stable, probably due to hydrogen bonding. Sulfenyl TST was confirmed by the observation of thioredoxin oxidase activity and was reduced by reduced thioredoxin (Figure 3(a)) [68]. On the other hand, sulfenyl MST was confirmed by the observation of thioredoxin peroxidase activity (Figure 3(b)) and mass spectrometric data, and was reduced not by reduced glutathione but rather by reduced thioredoxin [23]. These findings indicate that the half-redox potential of sulfenate is lower than that of glutathione and higher than that of thioredoxin (“low redox potential sulfenate” [23]). The redox potential of the cysteine residue is pH-dependent due to pH-dependent perturbation of the electric field strength surrounding the cysteine residue via interactions of the cysteine residue with basic amino acids. In fact, the pH-dependent perturbation of the redox potential of the cysteine residue was demonstrated in the thioredoxin superfamily [69]. The active-site loop of TSTs contains two basic residues whereas no charged residues are observed in MSTs [65], suggesting that the electric field strength surrounding cysteine residue of mitochondrial TST is larger than that of MST. This hypothesis, however, has not been tested experimentally.
Figure 3.
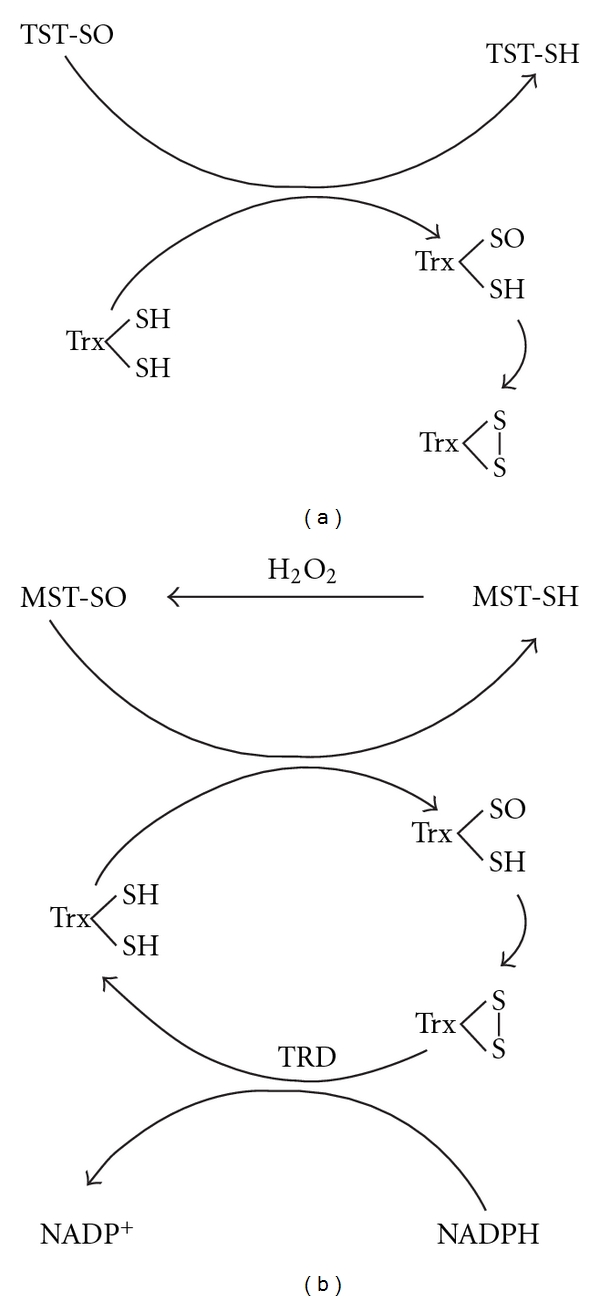
Thioredoxin oxidase activity of TST and thioredoxin peroxidase activity of MST, Proposed thioredoxin oxidase activity of TST (a) reported by Nandi and colleagues [63], which is same as thioredoxin peroxidase activity of MST (b) (from Figure 10 of Nagahara et al. Current Medical Chemistry 2009. 16: 4422). Trx: thioredoxin; TRD: thioredoxin reductase.
4. Possible Biologic Function of Catalytic SiteSulfenate of Sulfurtransferase
Sulfenyl sulfurtransferase is neither a reaction intermediate nor an active form whereas the sulfenyl form is a reaction intermediate of a thiol-oxidoreductase. Therefore, the biologic relevance of a conversion between sulfenate and thiolate at a catalytic cysteine is not clear. There are two possibilities: first, the molecular feature was accidentally acquired during the molecular evolution of the thiol enzyme family, and second, some molecular entity, such as an antioxidant protein, has evolved under oxidizing atmospheric conditions.
MST and TST are widely distributed in eukaryotes and prokaryotes [56, 70], and in eukaryotic cells, MST is distributed in the cytoplasm, mitochondria, and in chloroplasts (in plants) [71, 72]. On the other hand, TST distribution is restricted to the mitochondria and chloroplasts (in plants) [72–74]. Thus, both MST and TST are located in mitochondria and chloroplasts (in plants). Based on the minor catalytic contributions, the latter possibility is likely: MST and TST could locally serve as antioxidant proteins.
Unlike sulfurtransferases, a sulfenyl amide is found at the catalytic site cysteine in protein tyrosine phosphatase IB in an unusual oxidized form. This enzyme is oxidized to form sulfenate at the catalytic site cysteine, and the Sγ atom of the cysteine covalently binds to the main chain nitrogen atom of an adjacent serine to form sulfenyl amide [47, 48]. This sulfenyl amide enzyme is inactive. Reduced glutathione cleaves (reduces) the ring structure of sulfenyl amide to completely restore activity [44, 45]. The redox regulation of the enzymatic activity correlates with signal transduction [75–77] via the regulation of protein dephosphorylation [78–81].
Cdc25C, a member of the phosphatase family, has two redox active cysteines (Cys330 and Cys377). Mild oxidation forms sulfenate at one of the two redoxactive cysteines (Cys377) resulting in the formation of an intramolecular disulfide between them, which produces an inactive form of the enzyme [17, 18]. Further, the oxidized form is reduced not by reduced glutathione but rather by reduced thioredoxin [18], meaning that the cdc25 family forms a low redox potential disulfide. The redox regulation of the enzymatic activity correlates with the regulation of the cell cycle via the regulation of protein dephosphorylation [17, 18]. Further oxidation forms sulfinate at Cys377, which is an inactivated form, resulting in degradation of the protein [17].
The cysteine protease caspase, which regulates apoptosis, is also inactivated by mild oxidation, probably due to sulfenate formation at the catalytic site cysteine, and can be reduced by reduced glutathione in vitro [33]. Physiologic levels of glutathione, however, are unable to restore activity [33], and other cellular reductants such as thioredoxin have not been examined. The biologic importance of redox regulation of the caspase activity remains unknown.
5. Summary
Both MST and TST are localized in mitochondria and chloroplasts, and probably serve as antioxidant proteins.
The catalytic site cysteine residue of MST and TST is redox active, probably due to helix dipoles.
Stable and low redox sulfenate is formed at the catalytic site cysteine of MST and TST, and is reduced by thioredoxin.
References
- 1.Nagahara N, Matsumura T, Okamoto R, Kajihara Y. Protein cysteine modifications: (1) medicinal chemistry for proteomics. Current Medicinal Chemistry. 2009;16(33):4419–4444. doi: 10.2174/092986709789712880. [DOI] [PubMed] [Google Scholar]
- 2.Nagahara N, Matsumura T, Okamoto R, Kajihara Y. Protein cysteine modifications: (2) reactivity specificity and topics of medicinal chemistry and protein engineering. Current Medicinal Chemistry. 2009;16(34):4490–4501. doi: 10.2174/092986709789760643. [DOI] [PubMed] [Google Scholar]
- 3.Nagahara N. Intermolecular disulfide bond to modulate protein function as a redox-sensing switch. doi: 10.1007/s00726-010-0508-4. Amino Acids. In press. [DOI] [PubMed] [Google Scholar]
- 4.Quastel JH, Stewart CP, Tunnicliffe HE. On glutathione. IV. Constitution. Biochemical Journal. 1923;17:586–592. doi: 10.1042/bj0170586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Holmgren A. Glutathione-dependent synthesis of deoxyribonucleotides. Purification and characterization of glutaredoxin from Escherichia coli . Journal of Biological Chemistry. 1979;254(9):3664–3671. [PubMed] [Google Scholar]
- 6.Perham RN, Harris JL. Amino acid sequences around the reactive cysteine residues in glyceraldehyde-3-phosphate dehydrogenases. Journal of Molecular Biology. 1963;7:316–320. doi: 10.1016/s0022-2836(63)80011-x. [DOI] [PubMed] [Google Scholar]
- 7.Mohr S, Stamler J, Brune B. Mechanism of covalent modification of glyceraldehyde-3- phosphate dehydrogenase at its active site thiol by nitric oxide, peroxynitrite and related nitrosating agents. FEBS Letters. 1994;348(3):223–227. doi: 10.1016/0014-5793(94)00596-6. [DOI] [PubMed] [Google Scholar]
- 8.Ishii T, Sunami O, Nakajima H, Nishio H, Takeuchi T, Hata F. Critical role of sulfenic acid formation of thiols in the inactivation of glyceraldehyde-3-phosphate dehydrogenase by nitric oxide. Biochemical Pharmacology. 1999;58(1):133–143. doi: 10.1016/s0006-2952(99)00060-x. [DOI] [PubMed] [Google Scholar]
- 9.Levine RL, Mosoni L, Berlett BS, Stadtman ER. Methionine residues as endogenous antioxidants in proteins. Proceedings of the National Academy of Sciences of the United States of America. 1996;93(26):15036–15040. doi: 10.1073/pnas.93.26.15036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheong NAE, Choi YO, Lee KO, et al. Molecular cloning, expression, and functional characterization of a 2Cys-peroxiredoxin in Chinese cabbage. Plant Molecular Biology. 1999;40(5):825–834. doi: 10.1023/a:1006271823973. [DOI] [PubMed] [Google Scholar]
- 11.Stacy RAP, Munthe E, Steinum T, Sharma B, Aalen RB. A peroxiredoxin antioxidant is encoded by a dormancy-related gene, Per1, expressed during late development in the aleurone and embryo of barley grains. Plant Molecular Biology. 1996;31(6):1205–1216. doi: 10.1007/BF00040837. [DOI] [PubMed] [Google Scholar]
- 12.Yang KS, Kang SW, Woo HA, et al. Inactivation of human peroxiredoxin I during catalysis as the result of the oxidation of the catalytic site cysteine to cysteine-sulfinic acid. Journal of Biological Chemistry. 2002;277(41):38029–38036. doi: 10.1074/jbc.M206626200. [DOI] [PubMed] [Google Scholar]
- 13.Biteau B, Labarre J, Toledano MB. ATP-dependent reduction of cysteine-sulphinic acid by S. cerevisiae sulphiredoxin. Nature. 2003;425(6961):980–984. doi: 10.1038/nature02075. [DOI] [PubMed] [Google Scholar]
- 14.Hatch MD, Turner JF. A protein disulphide reductase from pea seeds. The Biochemical journal. 1960;76:556–562. doi: 10.1042/bj0760556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zheng L, White RH, Cash VL, Dean DR. Mechanism for the desulfurization of L-cysteine catalyzed by the NIFs gene product. Biochemistry. 1994;33(15):4714–4720. doi: 10.1021/bi00181a031. [DOI] [PubMed] [Google Scholar]
- 16.Dunphy WG, Kumagai A. The cdc25 protein contains an intrinsic phosphatase activity. Cell. 1991;67(1):189–196. doi: 10.1016/0092-8674(91)90582-j. [DOI] [PubMed] [Google Scholar]
- 17.Savitsky PA, Finkel T. Redox regulation of Cdc25C. Journal of Biological Chemistry. 2002;277(23):20535–20540. doi: 10.1074/jbc.M201589200. [DOI] [PubMed] [Google Scholar]
- 18.Sohn J, Rudolph J. Catalytic and chemical competence of regulation of Cdc25 phosphatase by oxidation/reduction. Biochemistry. 2003;42(34):10060–10070. doi: 10.1021/bi0345081. [DOI] [PubMed] [Google Scholar]
- 19.Guan KL, Dixon JE. Evidence for protein-tyrosine-phosphatase catalysis proceeding via a cysteine-phosphate intermediate. Journal of Biological Chemistry. 1991;266(26):17026–17030. [PubMed] [Google Scholar]
- 20.Heffetz D, Bushkin I, Dror R, Zick Y. The insulinomimetic agents H2O2 and vanadate stimulate protein tyrosine phosphorylation in intact cells. Journal of Biological Chemistry. 1990;265(5):2896–2902. [PubMed] [Google Scholar]
- 21.Pallini R, Guazzi GC, Cannella C, Cacace MG. Cloning and sequence analysis of the human liver rhodanese: comparison with the bovine and chicken enzymes. Biochemical and Biophysical Research Communications. 1991;180(2):887–893. doi: 10.1016/s0006-291x(05)81148-9. [DOI] [PubMed] [Google Scholar]
- 22.Nagahara N, Nishino T. Role of amino acid residues in the active site of rat liver mercaptopyruvate sulfurtransferase: cDNA cloning, overexpression, and site- directed mutagenesis. Journal of Biological Chemistry. 1996;271(44):27395–27401. doi: 10.1074/jbc.271.44.27395. [DOI] [PubMed] [Google Scholar]
- 23.Nagahara N, Katayama A. Post-translational regulation of mercaptopyruvate sulfurtransferase via a low redox potential cysteine-sulfenate in the maintenance of redox homeostasis. Journal of Biological Chemistry. 2005;280(41):34569–34576. doi: 10.1074/jbc.M505643200. [DOI] [PubMed] [Google Scholar]
- 24.Sörbo BH. Crystalline rhodanese. II. The enzyme catalyzed reaction. Acta Chemica Scandinavica. 1953;7:1137–1145. [Google Scholar]
- 25.Horowitz PM, Falksen K. Oxidative inactivation of the enzyme rhodanese by reduced nicotinamide adenine dinucleotide. Journal of Biological Chemistry. 1986;261(36):16953–16956. [PubMed] [Google Scholar]
- 26.Horowitz PM, Bowman S. Conformational changes accompany the oxidative inactivation of rhodanese by a variety of reagents. Journal of Biological Chemistry. 1987;262(18):8728–8733. [PubMed] [Google Scholar]
- 27.Baker EN. Structure of actinidin, after refinement at 1.7 θ resolution. Journal of Molecular Biology. 1980;141(4):441–484. doi: 10.1016/0022-2836(80)90255-7. [DOI] [PubMed] [Google Scholar]
- 28.Heinicke RM, Mori R. Effect of diisopropylfluorophosphate on sulfhydryl proteases. Science. 1959;129(3364):p. 1678. doi: 10.1126/science.129.3364.1678. [DOI] [PubMed] [Google Scholar]
- 29.Ota S, Umi H, Muta E, Okamoto Y. Chemical modification of stem bromelain I-1 and fruit bromelain A with 2 hydroxy 5 nitrobenzyl bromide, tetranitromethane, and hydrogen peroxide. Journal of Biochemistry. 1975;78(3):627–635. doi: 10.1093/oxfordjournals.jbchem.a130949. [DOI] [PubMed] [Google Scholar]
- 30.Murachi T, Tanaka K, Hatanaka M, Murakami T. Intracellular Ca2+-dependent protease (CALPAIN) and its high-molecular-weight endogenous inhibitor (CALPASTATIN) Session VII Complex interactions and biologically active proteins. In: Weber G, editor. Advance in Enzyme Regulation 19. New York, NY, USA: Pergamon Press; 1981. pp. 407–424. [DOI] [PubMed] [Google Scholar]
- 31.Morelli A, Grasso M, De Flora A. Oxidative inactivation of the calcium-stimulated neutral proteinase from human red blood cells by divicine and intracellular protection by reduced glutathione. Archives of Biochemistry and Biophysics. 1986;251(1):1–8. doi: 10.1016/0003-9861(86)90044-5. [DOI] [PubMed] [Google Scholar]
- 32.Wilson KP, Black JAF, Thomson JA, et al. Structure and mechanism of interleukin-1β converting enzyme. Nature. 1994;370(6487):270–275. doi: 10.1038/370270a0. [DOI] [PubMed] [Google Scholar]
- 33.Borutaite V, Brown GC. Caspases are reversibly inactivated by hydrogen peroxide. FEBS Letters. 2001;500(3):114–118. doi: 10.1016/s0014-5793(01)02593-5. [DOI] [PubMed] [Google Scholar]
- 34.Elliott SD. A proteolytic enzyme produced by group a streptococci with special reference to its effect on the type-specific m antigen. The Journal of Experimental Medicine. 1945;81:573–592. doi: 10.1084/jem.81.6.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vissers MC, Winterbourn CC. Myeloperoxidase-dependent oxidative inactivation of neutrophil neutral proteinases and microbicidal enzymes. Biochemical Journal. 1987;245(1):277–280. doi: 10.1042/bj2450277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ebata M, Yasunobu KT. Chymopapain. I. Isolation, crystallization, and preliminary characterization. The Journal of Biological Chemistry. 1962;237:1086–1094. [PubMed] [Google Scholar]
- 37.Vissers MC, Winterbourn CC. Myeloperoxidase-dependent oxidative inactivation of neutrophil neutral proteinases and microbicidal enzymes. Biochemical Journal. 1987;245(1):277–280. doi: 10.1042/bj2450277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Metrione RM, Johnston RB, Seng R. Purification, partial characterization, and sequence around a reactive sulfhydryl of ficin. Archives of Biochemistry and Biophysics. 1967;122(1):137–143. doi: 10.1016/0003-9861(67)90132-4. [DOI] [PubMed] [Google Scholar]
- 39.Pechan T, Jiang B, Steckler D, et al. Characterization of three distinct cDNA clones encoding cysteine proteinases from maize (Zea mays L.) callus. Plant Molecular Biology. 1999;40(1):111–119. doi: 10.1023/a:1026494813936. [DOI] [PubMed] [Google Scholar]
- 40.Elliott SD. A proteolytic enzyme produced by group a streptococci with special reference to its effect on the type-specific m antigen. The Journal of Experimental Medicine. 1945;81:573–592. doi: 10.1084/jem.81.6.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lin WS, Armstrong DA, Gaucher GM. Formation and repair of papain sulfenic acid. Canadian Journal of Biochemistry. 1975;53(3):298–307. doi: 10.1139/o75-042. [DOI] [PubMed] [Google Scholar]
- 42.Edman JC, Ellis L, Blacher RW, Roth RA, Rutter WJ. Sequence of protein disulphide isomerase and implications of its relationship to thioredoxin. Nature. 1985;317(6034):267–270. doi: 10.1038/317267a0. [DOI] [PubMed] [Google Scholar]
- 43.Jacob C, Knight I, Winyard PG. Aspects of the biological redox chemistry of cysteine: from simple redox responses to sophisticated signalling pathways. Biological Chemistry. 2006;387(10-11):1385–1397. doi: 10.1515/BC.2006.174. [DOI] [PubMed] [Google Scholar]
- 44.Claiborne AL, Yeh JI, Mallett TC, et al. Protein-sulfenic acids: diverse roles for an unlikely player in enzyme catalysis and redox regulation. Biochemistry. 1999;38(47):15407–15416. doi: 10.1021/bi992025k. [DOI] [PubMed] [Google Scholar]
- 45.Poole LB, Karplus PA, Claiborne AL. Protein sulfenic acids in redox signaling. Annual Review of Pharmacology and Toxicology. 2004;44:325–347. doi: 10.1146/annurev.pharmtox.44.101802.121735. [DOI] [PubMed] [Google Scholar]
- 46.Hofmann B, Hecht HJ, Flohé L. Peroxiredoxins. Biological Chemistry. 2002;383(3-4):347–364. doi: 10.1515/BC.2002.040. [DOI] [PubMed] [Google Scholar]
- 47.Salmeen A, Andersen JN, Myers MP, et al. Redox regulation of protein tyrosine phosphatase 1B involves a sulphenyl-amide intermediate. Nature. 2003;423(6941):769–773. doi: 10.1038/nature01680. [DOI] [PubMed] [Google Scholar]
- 48.Van Montfort RLM, Congreve M, Tisi D, Carr R, Jhoti H. Oxidation state of the active-site cysteine in protein tyrosine phosphatase 1B. Nature. 2003;423(6941):773–777. doi: 10.1038/nature01681. [DOI] [PubMed] [Google Scholar]
- 49.Soda K, Novogrodsky A, Meister A. Enzymatic desulfination of cysteine sulfinic acid. Biochemistry. 1964;3(10):1450–1454. doi: 10.1021/bi00898a010. [DOI] [PubMed] [Google Scholar]
- 50.Mihara H, Kurihara T, Yoshimura T, Soda K, Esaki N. Cysteine sulfinate desulfinase, a NIFS-like protein of Escherichia coli with selenocysteine lyase and cysteine desulfurase activities. Gene cloning, purification, and characterization of a novel pyridoxal enzyme. Journal of Biological Chemistry. 1997;272(36):22417–22424. doi: 10.1074/jbc.272.36.22417. [DOI] [PubMed] [Google Scholar]
- 51.Biteau B, Labarre J, Toledano MB. ATP-dependent reduction of cysteine-sulphinic acid by S. cerevisiae sulphiredoxin. Nature. 2003;425(6961):980–984. doi: 10.1038/nature02075. [DOI] [PubMed] [Google Scholar]
- 52.Georgiou G, Masip L. An overoxidation journey with a return ticket. Science. 2003;300(5619):592–594. doi: 10.1126/science.1084976. [DOI] [PubMed] [Google Scholar]
- 53.Jacob C, Holme AL, Fry FH. The sulfinic acid switch in proteins. Organic and Biomolecular Chemistry. 2004;2(14):1953–1956. doi: 10.1039/b406180b. [DOI] [PubMed] [Google Scholar]
- 54.Hol WGJ. The role of the α-helix dipole in protein function and structure. Progress in Biophysics and Molecular Biology. 1985;45(3):149–195. doi: 10.1016/0079-6107(85)90001-x. [DOI] [PubMed] [Google Scholar]
- 55.Kortemme T, Creighton TE. Ionisation of cysteine residues at the termini of model α-helical peptides. Relevance to unusual thiol pK a values in proteins of the thioredoxin family. Journal of Molecular Biology. 1995;253(5):799–812. doi: 10.1006/jmbi.1995.0592. [DOI] [PubMed] [Google Scholar]
- 56.Nagahara N. Molecular evolution of thioredoxin-dependent redox-sensing switch in mercaptopyruvate sulfurtransferase. In: Mohan RM, editor. Research Advances in Biological Chemistry. Vol. 1. Kerala, India: Global Research Network; 2007. pp. 19–26. [Google Scholar]
- 57.Alphey MS, Williams RAM, Mottram JC, Coombs GH, Hunter WN. The crystal structure of leishmania major 3-mercaptopyruvate sulfurtransferase. A three-domain architecture with a serine protease-like triad at the active site. Journal of Biological Chemistry. 2003;278(48):48219–48227. doi: 10.1074/jbc.M307187200. [DOI] [PubMed] [Google Scholar]
- 58.Ploegman JH, Drent G, Kalk KH, Hol WGJ. Structure of bovine liver rhodanese. I. Structure determination at 2.5 Å resolution and a comparison of the conformation and sequence of its two domains. Journal of Molecular Biology. 1978;123(4):557–594. [PubMed] [Google Scholar]
- 59.Ploegman JH, Drent GÉ, Kalk KH, Hol WGJ. The structure of bovine liver rhodanese. II. The active site in the sulfur-substituted and the sulfur-free enzyme. Journal of Molecular Biology. 1979;127(2):149–162. doi: 10.1016/0022-2836(79)90236-5. [DOI] [PubMed] [Google Scholar]
- 60.Ploegman JH, Drent G, Kalk KH. The covalent and tertiary structure of bovine liver rhodanese. Nature. 1978;273(5658):124–129. doi: 10.1038/273124a0. [DOI] [PubMed] [Google Scholar]
- 61.Hol WGJ, Lijk LJ, Kalk KH. The high resolution three-dimensional structure of bovine liver rhodanese. Fundamental and Applied Toxicology. 1983;3(5):370–376. doi: 10.1016/s0272-0590(83)80007-4. [DOI] [PubMed] [Google Scholar]
- 62.Schlesinger P, Westley J. An expanded mechanism for rhodanese catalysis. Journal of Biological Chemistry. 1974;249(3):780–788. [PubMed] [Google Scholar]
- 63.Fujii T, Maeda M, Mihara H, Kurihara T, Esaki N, Hata Y. Structure of a NifS homologue: X-ray structure analysis of CsdB, an Escherichia coli counterpart of mammalian selenocysteine lyase. Biochemistry. 2000;39(6):1263–1273. doi: 10.1021/bi991732a. [DOI] [PubMed] [Google Scholar]
- 64.Bordo D, Deriu D, Colnaghi R, Carpen A, Pagani S, Bolognesi M. The crystal structure of a sulfurtransferase from Azotobacter vinelandii highlights the evolutionary relationship between the rhodanese and phosphatase enzyme families. Journal of Molecular Biology. 2000;298(4):691–704. doi: 10.1006/jmbi.2000.3651. [DOI] [PubMed] [Google Scholar]
- 65.Bordo D, Bork P. The rhodanese/Cdc25 phosphatase superfamily. Sequence-structure-function relations. EMBO Reports. 2002;3(8):741–746. doi: 10.1093/embo-reports/kvf150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Reynolds RA, Yem AW, Wolfe CL, Deibel MR, Chidester CG, Watenpaugh KD. Crystal structure of the catalytic subunit of Cdc25B required for G/M phase transition of the cell cycle. Journal of Molecular Biology. 1999;293(3):559–568. doi: 10.1006/jmbi.1999.3168. [DOI] [PubMed] [Google Scholar]
- 67.Lima CD. Analysis of the E. coli NifS CsdB protein at 2.0 Å reveals the structural basis for perselenide and persulfide intermediate formation. Journal of Molecular Biology. 2002;315(5):1199–1208. doi: 10.1006/jmbi.2001.5308. [DOI] [PubMed] [Google Scholar]
- 68.Nandi DL, Horowitz PM, Westley J. Rhodanese as a thioredoxin oxidase. International Journal of Biochemistry and Cell Biology. 2000;32(4):465–473. doi: 10.1016/s1357-2725(99)00035-7. [DOI] [PubMed] [Google Scholar]
- 69.Moutevelis E, Warwicker J. Prediction of pK a and redox properties in the thioredoxin superfamily. Protein Science. 2004;13(10):2744–2752. doi: 10.1110/ps.04804504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nagahara N, Okazaki T, Nishino T. Cytosolic mercaptopyruvate sulfurtransferase is evolutionarily related to mitochondrial rhodanese. Striking similarity in active site amino acid sequence and the increase in the mercaptopyruvate sulfurtransferase activity of rhodanese by site-directed mutagenesis. Journal of Biological Chemistry. 1995;270(27):16230–16235. doi: 10.1074/jbc.270.27.16230. [DOI] [PubMed] [Google Scholar]
- 71.Nakamura T, Yamaguchi Y, Sano H. Plant mercaptopyruvate sulfurtransferases molecular cloning, subcellular localization and enzymatic activities. European Journal of Biochemistry. 2000;267(17):5621–5630. doi: 10.1046/j.1432-1327.2000.01633.x. [DOI] [PubMed] [Google Scholar]
- 72.de Duve C, Pressman BC, Gianetto R, Wattiaux R, Appehmans F. Tissue fractionation studies. 6. Intracellular distribution patterns of enzymes in rat-liver tissue. The Biochemical journal. 1955;60(4):604–617. doi: 10.1042/bj0600604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Koj A, Frendo J, Wojtczak L. Subcellular distribution and intramitochondrial localization of three sulfurtransferases in rat liver. FEBS Letters. 1975;57(1):42–46. doi: 10.1016/0014-5793(75)80148-7. [DOI] [PubMed] [Google Scholar]
- 74.Papenbrock J, Schmidt A. Characterization of a sulfurtransferase from Arabidopsis thaliana. European Journal of Biochemistry. 2000;267(1):145–154. doi: 10.1046/j.1432-1327.2000.00980.x. [DOI] [PubMed] [Google Scholar]
- 75.Elchebly M, Payette P, Michaliszyn E, et al. Increased insulin sensitivity and obesity resistance in mice lacking the protein tyrosine phosphatase-1B gene. Science. 1999;283(5407):1544–1548. doi: 10.1126/science.283.5407.1544. [DOI] [PubMed] [Google Scholar]
- 76.Klaman LD, Boss O, Peroni OD, et al. Increased energy expenditure, decreased adiposity, and tissue-specific insulin sensitivity in protein-tyrosine phosphatase 1B-deficient mice. Molecular and Cellular Biology. 2000;20(15):5479–5489. doi: 10.1128/mcb.20.15.5479-5489.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Neel BG, Tonks NK. Protein tyrosine phosphatases in signal transduction. Current Opinion in Cell Biology. 1997;9(2):193–204. doi: 10.1016/s0955-0674(97)80063-4. [DOI] [PubMed] [Google Scholar]
- 78.Denu JM, Dixon JE. Protein tyrosine phosphatases: Mechanisms of catalysis and regulation. Current Opinion in Chemical Biology. 1998;2(5):633–641. doi: 10.1016/s1367-5931(98)80095-1. [DOI] [PubMed] [Google Scholar]
- 79.Lee S-R, Kwon K-S, Kim S-R, Rhee SG. Reversible inactivation of protein-tyrosine phosphatase 1B in A431 cells stimulated with epidermal growth factor. Journal of Biological Chemistry. 1998;273(25):15366–15372. doi: 10.1074/jbc.273.25.15366. [DOI] [PubMed] [Google Scholar]
- 80.Meng TC, Fukada T, Tonks NK. Reversible oxidation and inactivation of protein tyrosine phosphatases in vivo. Molecular Cell. 2002;9(2):387–399. doi: 10.1016/s1097-2765(02)00445-8. [DOI] [PubMed] [Google Scholar]
- 81.Narhi LO, Arakawa T, Aoki KH, et al. The effect of carbohydrate on the structure and stability of erythropoietin. Journal of Biological Chemistry. 1991;266(34):23022–23026. [PubMed] [Google Scholar]


