Abstract
The light-harvesting chlorophyll a/b binding proteins (LHCB) are perhaps the most abundant membrane proteins in nature. It is reported here that the down-regulation or disruption of any member of the LHCB family, LHCB1, LHCB2, LHCB3, LHCB4, LHCB5, or LHCB6, reduces responsiveness of stomatal movement to ABA, and therefore results in a decrease in plant tolerance to drought stress in Arabidopsis thaliana. By contrast, over-expression of a LHCB member, LHCB6, enhances stomatal sensitivity to ABA. In addition, the reactive oxygen species (ROS) homeostasis and a set of ABA-responsive genes are altered in the lhcb mutants. These data demonstrate that LHCBs play a positive role in guard cell signalling in response to ABA and suggest that they may be involved in ABA signalling partly by modulating ROS homeostasis.
Keywords: Abscisic acid signalling, Arabidopsis thaliana, light-harvesting chlorophyll a/b binding protein, reactive oxygen species, stomatal movement
Introduction
The light-harvesting chlorophyll a/b-binding (LHCB) proteins are the apoproteins of the light-harvesting complex of photosystem II (PSII), which are normally complexed with chlorophyll and xanthophylls and serve as the antenna complex (Jansson, 1994, 1999). As important components of the major light-harvesting complex, the PSII outer antenna proteins LHCBs are perhaps the most abundant membrane proteins in nature. Expression of the LHCB genes is regulated by multiple environmental and developmental cues, including mainly light (Silverthorne and Tobin, 1984; Sun and Tobin, 1990; Peer et al., 1996; Millar and Kay, 1996; Weatherwax et al., 1996; Yang et al., 1998; Humbeck and Krupinska, 2003), oxidative stress (for reviews, see Nott et al., 2006; Staneloni et al., 2008), chloroplast retrograde signal (for review, see Nott et al., 2006), circadian clock (Paulsen and Bogorad, 1988; Strayer et al., 2000; Alabadi et al., 2001; Thain et al., 2002; Andronis et al., 2008), and the phytohormone abscisic acid (ABA) (Bartholomew et al., 1991; Chang and Walling, 1991; Weatherwax et al., 1996; Staneloni et al., 2008). Previous studies showed that exogenously-applied ABA down-regulates LHCB gene expression in tomato leaves (Bartholomew et al., 1991), Arabidopsis seedlings (Staneloni et al., 2008), Lemna gibba cells grown on liquid medium (Weatherwax et al., 1996), and developing seeds of soybean (Chang and Walling, 1991), whereas a recent report showed that the treatments of the 6-d-old Arabidopsis seedlings with low levels of ABA (from 0.125 to 1 μM) enhanced LHCB1.2 mRNA levels (Voigt et al., 2010). The regulation of the LHCB expression is considered to be one of the important mechanisms for plants to modulate chloroplast functions (Nott et al., 2006; De Montaigu et al., 2010; Pruneda-Paz and Kay, 2010; Thines and Harmon, 2010).
ABA is a vital phytohormone to regulate many aspects of plant growth and development, and especially to modulate the plant response to stressful conditions (Finkelstein et al., 2002; Adie et al., 2007). ABA signal transduction has been extensively studied, and numerous signalling components have been identified, which include plasma membrane and intracellular ABA receptors (Shen et al., 2006; Fujii et al., 2009; Ma et al., 2009; Pandey et al., 2009; Park et al., 2009; Wu et al., 2009; Cutler et al., 2010; Shang et al., 2010). Previous reports showed that the members of the LHCB family play an important role in plant adaptation to environmental stresses (Andersson et al., 2001, 2003; Ganeteg et al., 2004; Kovacs et al., 2006), as well as their expression being regulated by ABA (Bartholomew et al., 1991; Chang and Walling, 1991; Weatherwax et al., 1996; Staneloni et al., 2008). However, it remains unknown whether the decline of plant stress tolerance due to a lack of the LHCB proteins is associated with the plant response to ABA under environmental stresses. It is reported here that the Arabidopsis LHCBs are positively involved in guard cell signalling in response to ABA, and they may affect ABA signalling partly by modulating ROS homeostasis. These findings help understand the complex mechanism of ABA signalling and the positive role of LHCB proteins in plant stress tolerance.
Materials and methods
Plant materials
Arabidopsis thaliana ecotype Columbia (Col-0) was used in the generation of transgenic plants. The open reading frame (ORF) cDNA of the LHCB6 gene (At1g15820) was introduced into Col plants as a green fluorescence protein (GFP)-fusion protein to generate LHCB6-over-expressing transgenic lines. The cDNA was isolated by polymerase chain reaction (PCR) using the forward primer 5′-GCTCTAGAATGGCGATGGCGGTCTCC-3′ and reverse primer 5′-CGGTCGACTCACAAACCAAGAGCACCGAG-3′. The cauliflower mosaic virus (CaMV) 35S::LHCB6 chimeric gene construct was generated by ligating the ORF (777 bp) of the LHCB6 gene into the pCAMBIA1300 vector by XbaI and SalI sites. The construct was confirmed by sequencing, and introduced into the GV3101 strain Agrobacterium tumefaciens and transformed into plants by floral infiltration. The homozygous T3 seeds of the transgenic plants were used for analysis. More than 20 LHCB6-over-expressing transgenic lines were screened, all of which showed ABA hypersensitivity in stomatal movement, and four representative lines have been shown (see Supplementary Fig. S2 at JXB online).
The T-DNA insertion mutants lhcb1.1 (SALK-134810) in the LHCB1.1 gene (At1g29920; referred to as LHCB1 and representative of LHCB1.1, LHCB1.2, LHCB1.3, LHCB1., and LHCB1.5), lhcb2.2 (SALK-005614) in the LHCB2.2 gene (At2g05070; referred to as LHCB2 and representative of LHCB2.1, LHCB2.2, LHCB2.3, and LHCB2.4), lhcb3 (SALK-036200) in the LHCB3 gene (At5g54270), lhcb4.3 (SALK-032779) in the LHCB4.3 gene (At2g40100; referred to as LHCB4 and representative of LHCB4.1, LHCB4.2, and LHCB4.3), lhcb5 (SALK-139667) in the LHCB5 gene (At4g10340), and lhcb6 (SALK-074622) in the LHCB6 gene (At1g15820) were used in this study and the seeds of these mutants were obtained from the Arabidopsis Biological Resource Center (ABRC). The screening for the knockout or knockdown mutants was done following the recommended procedures. The sequences of the primers for the screening are presented in Supplementary Table S1 at JXB online. The T-DNA insertion in the mutants was identified by PCR and DNA gel-blot analysis and the exact position was determined by sequencing. The mutants lhcb1.1 (SALK-134810), lhcb2.2 (SALK-005614), lhcb4.3 (SALK-032779), lhcb5 (SALK-139667), and lhcb6 (SALK-074622) are also knockdown mutants in their corresponding genes except for the mutant lhcb3 (SALK-036200) that is a knockout mutant in the LHCB3 gene. DNA gel-blot analysis showed that all the mutants have one single copy of T-DNA in their genome.
All the double mutants were generated by genetic crosses and identified by PCR genotyping.
Plants were grown in a growth chamber at 19–20 °C on Murashige-Skoog (MS) medium (Sigma) at about 80 μmol photons m−2 s−1, or in compost soil at about 120 μmol photons m−2 s−1 over a 16 h photoperiod.
Complementation of the lhcb mutants
The ABA-related phenotypes of the lhcb1, lhcb2, lhcb3, lhcb4, lhcb5, and lhcb6 mutants were complemented by introducing into the mutant plants, respectively, the LHCB1, LHCB2, LHCB3, LHCB4, LHCB5, and LHCB6 ORF cDNAs driven by the 35S promoter integrated into the pCAMBIA1300-221 vector. The primers for cloning the LHCB1, LHCB2, LHCB3, LHCB4, LHCB5, and LHCB6 ORF cDNAs are listed in Supplementary Table S1 at JXB online.
Protein extraction and immunoblotting
The extraction of the Arabidopsis total proteins was performed essentially according to procedures proposed by the LHCB-antibody supplier Agrisera. The plant tissues were frozen in liquid N2, ground in a pre-chilled mortar with a pestle to a fine powder and transferred to a 1.5 ml tube. The extraction buffer consists of 50 mM TRIS-HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 0.1% (v/v)Triton X-100, 10% (v/v) glycerol, and 5 μg ml−1 protein inhibitor cocktail. The extraction buffer was added to the tube (buffer:sample=4:1), which was immediately frozen in liquid N2. The mixture was carefully subjected to sonication just until the sample was thawed, and was re-frozen immediately in liquid N2 to avoid heating. The sonication step was repeated three times. The mixture was centrifuged for 3 min at 10 000 g to remove insoluble material and unbroken cells and the supernatant was transferred to a new tube for use. The SDS-PAGE and immunoblotting assays were done essentially according to our previously described procedures (Wu et al., 2009; Shang et al., 2010). The specific antibodies against, respectively, LHCB1, LHCB2, LHCB3, LHCB4, LHCB5, and LHCB6 were purchased from Agrisera (Stockholm, Sweden; website:www.agrisera.com; product No. AS08300).
Real-time PCR analysis
Total RNA was isolated from leaves of 2-week-old Arabidopsis seedlings using a Total RNA Rapid Extraction Kit (BioTeke), treated with RNase-free DNase I (TAKARA) at 37 °C for 30 min to degrade genomic DNA and purified by using an RNA Purification Kit (BioTeke). A 2 μg aliquot of RNA was subjected to first-strand cDNA synthesis using M-MLV reverse transcriptase (Promega), and an oligo (dT21) primer. The primers used for real-time PCR are listed in Supplementary Table S1 at JXB online. Analysis was performed using the Bio-Rad Real-Time System CFX96TM C1000 Thermal Cycler (Singapore). All experiments were repeated at least three times along with three independent repetitions of the biological experiments.
Chlorophyll measurements
The contents of chlorophyll were assayed essentially by the previously described procedures (Shen et al., 2006).
ROS measurements
ROS detection in whole leaves was conducted by nitroblue tetrazolium (NBT) staining, essentially as described previously by Lee et al. (2002). Leaves were sampled from 3-week-old plants and preincubated in a medium composed of 50 mM KCl, 10 mM MES-TRIS (pH 6.15) supplemented with different concentrations of (±)-ABA (as indicated) under light at 200 μmol m−2 s−1 for 1 h, and then the leaves were vacuum-infiltrated with 0.1 mg ml−1 NBT (Amresco, Solon, OH, USA) in 100 mM potassium phosphate buffer, pH 7.6. Samples were incubated at room temperature in the dark for 2 h. To remove chlorophylls, the stained samples were transferred to boiling 80% ethanol for 10 min.
ROS production from guard cells was examined by loading epidermal peels with H2DCF-DA (Molecular Probes) essentially as previously described by Miao et al. (2006). The epidermal strips were preincubated for 2 h under conditions promoting stomatal opening in the MES-TRIS buffer (the same as mentioned above) supplemented with 0 (ethanol, as a control) or 5 μM (±)-ABA, and were incubated in the loading buffer with 50 mM TRIS-KCl (pH 7.2) containing 50 mM H2DCF-DA in the dark for 20 min, and then the epidermal tissues were washed with the same MES-TRIS preincubation buffer to remove excess dye. Examinations of peel fluorescence were performed using fluorescence microscopy (Olympus, BX51, Japan). Fluorescent optical sections were collected from dye-loaded guard cells with the following settings: excitation, 488 nm; emission, 525 nm.
Stomatal aperture assay
Stomatal aperture was assayed as previously described (Wu et al., 2009; Shang et al., 2010). Leaves sampled from 3-week old plants were used. To observe ABA-induced stomatal closure, leaves were floated in the buffer containing 50 mM KCl and 10 mM MES-TRIS (pH 6.15) under a halogen cold-light source (Colo-Parmer) at 200 μmol m−2 s−1 for 2.5 h followed by the addition of different concentrations of (±)-ABA. Apertures were recorded on epidermal strips after 2.5 h of further incubation to estimate ABA-induced closure. To study ABA-inhibited stomatal opening, leaves were floated on the same buffer in the dark for 2.5 h before they were transferred to the cold-light for 2.5 h in the presence of ABA, and then apertures were determined.
Accession numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative database under the following accession numbers: At5g13630 (ABAR/CHLH), At1g29920 (LHCB1), At2g05070 (LHCB2), At5g54270 (LHCB3), At2g40100 (LHCB4), At4g10340 (LHCB5), and At1g15820 (LHCB6). Germplasm identification numbers for mutant lines and SALK lines: lhcb1.1 (SALK-134810), lhcb2.2 (SALK-005614), lhcb3 (SALK-036200), lhcb4.4 (SALK-032779), lhcb5 (SALK-139667), and lhcb6 (SALK-074622).
Results
Down-regulation or disruption of LHCB genes reduces, but up-regulation of LHCB6 gene enhances, ABA sensitivity in stomatal movement
AT-DNA insertion mutant for each of the LHCB genes was isolated (Fig. 1A–F). The lhcb1, lhcb2, lhcb4, lhcb5, and lhcb6 are knockdown alleles and the lhcb3 is a knockout allele (Fig. 1G). It was observed that down-regulation of one LHCB gene altered the expression of other LHCB members (Fig. 1G). This may be due to a feedback effect in the LHCB family, where a decrease or the removal of one protein in a multiple protein complex can result in the decreased stability of the others (Andersson et al., 2001; Ganeteg et al., 2004; Kovacs et al., 2006). The levels of chlorophyll, ABA, and dry substances and the growth of these lhcb mutants were not significantly affected in any of these mutants (Fig. 1H; see Supplementary Fig. S1at JXB online). It is noteworthy, however, that a knockout lhcb6 mutation significantly affected seedling growth (Kovacs et al., 2006), while the present lhcb6 knockdown-mutant allele did not significantly alter plant growth, probably because the LHCB6 level in this knockdown mutant is still sufficient for normal plant growth.
Fig. 1.
Molecular and biochemical characterization of the lhcb mutants (from lhcb1 to lhcb6). (A–F) T-DNA insertion sites in lhcb1.1 (SALK_134810) (A), lhcb2.2 (SALK_005614) (B), lhcb3 (SALK_036200) (C), lhcb4.3 (SALK_032779) (D), lhcb5 (SALK_139667) (E), and lhcb6 (SALK_074622) (F). LP, left genomic primer, and RP, right genomic primer with the suffix numbers corresponding to the numbers of the LHCB genes (1 to 6). LBa1, left border primer, and RBa1, right border primer for the flanking sequences of the T-DNA. Boxes and lines represent exons and introns, respectively. The locations of the primers for the identification of the mutants are indicated by arrows. LB and RB represent the left and right border of the T-DNA insertion, respectively; T-DNA1 and T-DNAn, first and last copy of the inserted T-DNAs, respectively, noting that the two or more than two copies were inserted in an inverted manner. (A) One single copy of the T-DNA was inserted into the promoter region at nt –592 to –523 in the 5′-upstream region of the translation start codon (ATG) of the LHCB1.1 gene with a 70 bp fragment deleted in the lhcb1-1 mutant. (B) Tandem T-DNA of two copies (or more than two copies) was inserted into the promoter region in an inverted fashion at the same locus for the lhcb2 mutant, which generates a 24 bp deletion from nt –126 to –102 in the 5′-upstream region of the translation start codon (ATG) of the LHCB2.2 gene. (C) One single copy of T-DNA was inserted into the first exon at nt 120 to 125 of the LHCB3 gene with a 6 bp fragment deleted in the lhcb3 mutant. (D-F) Tandem T-DNA of two copies (or more than two copies) was inserted into the promoter region in an inverted fashion at the same locus for lhcb4.3 (D), lhcb5 (E), and lhcb6 (F) mutants, which generates a 17 bp deletion from nt –1263 to –1247 for lhcb4.3, a 7 bp deletion from nt –483 to –477 for lhcb5, and a 46–bp deletion from nt –391 to –346 in the 5′-upstream region of the translation start codon (ATG) of the corresponding genes LHCB4.3, LHCB5, and LHCB6, respectively. (G) Quantitative real-time PCR analysis (columns) and immunoblotting (protein bands below the columns) for LHCB gene expression in the mutants (from lhcb1 to lhcb6). Actin was used as a loading control for immunoblotting. Relative protein band intensities, normalized relative to the intensity of Col, are indicated by numbers in the box below the bands. Expression of all the six numbers of LHCBs (from LHCB1 to LHCB6) was assessed in each mutant, and the red arrow indicates the level of the corresponding mutated LHCB gene. Note that lhcb1-1, lhcb2, lhcb4, lhcb5, and lhcb6 are knockdown mutants in their corresponding genes, while lhcb3 is a knockout mutant in the LHCB3 gene. The immunoblotting assays were repeated three times with independent biological experiments which gave similar results. Each value for real-time PCR is the mean ±SE of three independent biological determinations. (H) The chlorophyll a/b contents are not significantly affected in the mutants (from lhcb1 to lhcb6). Left panel, the concentrations of chloroplast a (Chla) and b (Chlb) and total chlorophyll in the different mutants. Each value is the mean ±SE of three independent biological determinations. Right panel, the status of the seedlings of the different mutants, showing that no chlorophyll-deficient phenotype can be seen for these mutants.
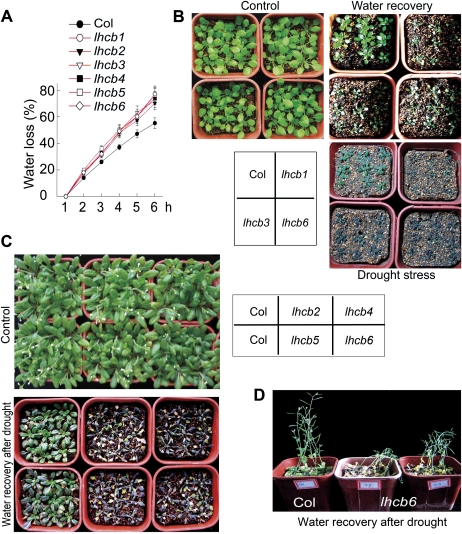
All the T-DNA insertion lhcb mutants showed ABA-insensitive phenotypes, but the over-expression lines of the LHCB6 gene showed ABA-hypersensitive phenotypes in the ABA-induced promotion of stomatal closure and in the inhibition of stomatal opening (Fig. 2A, B; see Supplementary Fig. S2Aat JXB online). The stomatal apertures of the mutants were only slightly reduced at 20 μM or 30 μM (±)ABA that reduced dramatically stomatal apertures of the wild-type plants (Fig. 2A, B). The double mutant lhcb1 lhcb6 showed substantially the same extent of ABA-insensitive phenotypes in stomatal movement (Fig. 2D). This enhanced resistance of stomatal closure to ABA suggests that the lhcb mutants should be more susceptible to drought. Indeed, it was also observed that the detached leaves of the lhcb mutants lost more water than those of wild-type plants under dehydration conditions (Fig. 3A), and that both young seedlings and mature plants of the lhcb mutants had a lower capacity to conserve their water during drought stress compared with wild-type plants (Fig. 3B–D).
Fig. 2.
Down- or up-regulation of members of the LHCB family alters ABA sensitivities in stomatal movement. (A) ABA-induced stomatal closure (top) and inhibition of stomatal opening (bottom) in wild-type Col, ch1-1, cch, lhcb1, lhcb2, and lhcb3 mutants and a complemented line of the lhcb3 mutant (lhcb3/LHCB3). (B) ABA-induced stomatal closure (top) and inhibition of stomatal opening (bottom) in wild-type Col, ch1-1, cch, lhcb4, lhcb5, and lhcb6 mutants and a transgenic LHCB6-over-expressor (LHCB6, line OE5 as described in Supplementary Fig. S2at JXB online). (C) ABA-induced stomatal closure (top) and inhibition of stomatal opening (bottom) in wild-type Col, cch and lhcb6 mutants and a transgenic LHCB6-over-expressiing line in the cch mutant (cch/LHCB6). (D) ABA-induced stomatal closure (top) and inhibition of stomatal opening (bottom) in wild-type Col, cch and lhcb6 single mutants, and lhcb1 lhcb6, and lhcb6 cch double mutants. Values presented in (A) to (D) are the means ±SE from three independent experiments; n=60 apertures per experiment.
Fig. 3.
Down-regulation of members of the LHCB family reduces the ability of plants to conserve water. (A) Water loss rates during a 6 h period from the detached leaves of wild-type Col and different lhcb mutants. Values are the means ±SE of five individual plants per genotype. (B, C) Water loss assays with young seedlings for wild-type Col, lhcb1, lhcb3, and lhcb6 mutants (B) or for wild-type Col, lhcb2, lhcb4, lhcb5, and lhcb6 mutants (C). Plants were well watered (Control) or drought-stressed by withholding water for 18 d and then the drought-stressed plants were rewatered (Water recovery) and growth status was recorded 2 d later. The entire experiment was replicated three times with similar results. (D) Assays with mature plants for wild-type Col and lhcb6 mutants. Plants were drought-stressed by withholding water for 21 d and then the plants were rewatered and growth status was recorded 2 d later. The entire experiment was replicated three times with similar results.
A chlorophyll b-deficient mutant ch1-1, which results in low expression of the LHCB genes (Espineda et al., 1999), was used to assess the relationships between chlorophyll-deficiency-caused LHCB decrease and ABA responsiveness. The ch1-1 mutant did not show any ABA insensitive phenotype (Fig. 2A, B).
The transgenic complementation lines of all the lhcb mutants displayed the wild-type ABA phenotypes in the ABA-induced promotion of stomatal closure and the inhibition of stomatal opening (see Supplementary Fig. S3at JXB online), showing that the phenotypes of the lhcb mutants did indeed result from the down-regulation or disruption of the LHCB genes. The LHCBs are expressed ubiquitously in different tissues/organs except for dry seeds and the LHCB mRNA is detectable even in roots, although the expression levels are low in roots (see Supplementary Fig. S4 at JXB online). This suggests that the LHCB members can function at the whole plant level. It is noteworthy, however, that the function of LHCB members in roots remains to be determined.
Double mutation in LHCB and ABAR genes confers ABA-insensitivity, and over-expression of LHCB6 partly restores the ABA sensitivity of the cch mutant
It was previously reported that the Mg-chelatase H subunit (CHLH/ABAR) functions as a chloroplast/plastid ABA receptor (Shen et al., 2006; Wu et al., 2009; Shang et al., 2010). A possible relationship between CHLH/ABAR- and LHCB in guard cell signalling in response to ABA was assessed. Using a strong stomata-ABA-insensitive mutant allele of the CHLH/ABAR gene, cch (Shen et al., 2006; Wu et al., 2009; Fig. 2A–D), the double mutant lhcb6 cch was generated and it was observed that the lhcb6 cch double mutant showed ABA-insensitive phenotypes in the ABA-induced promotion of stomatal closure and the inhibition of stomatal opening, and the strength of the ABA-insensitive phenotypes was comparable with that of the cch mutant, stronger than that of the lhcb6 single mutant and lhcb1 lhcb6 double mutant (Fig. 2C, D). Interestingly, it was observed that over-expression of the LHCB6 gene partly restored the wild-type phenotype of the cch mutant in the stomatal responses to ABA (Fig. 2C; see Supplementary Fig. S2B at JXB online), suggesting that LHCBs function downstream of the ABAR-mediated ABA signalling pathway.
Down-regulation or disruption of LHCB members affects homeostasis of reactive oxygen species
It has been well known that reactive oxygen species (ROS) are involved in ABA signalling (Pei et al., 2000; Murata et al., 2001; Mustilli et al., 2002; Kwak et al., 2006; Miao et al., 2006; Zhang et al., 2009), and chloroplasts are major sites of ROS production (Kwak et al., 2006; Nott et al., 2006; Galvez-Valdivieso and Mullineaux, 2010) where LHCBs play a key role (Jansson, 1994, 1999; Galvez-Valdivieso and Mullineaux, 2010). Thus, ROS production was investigated in the lhcb mutants using techniques of NBT-leaf-staining and CFDA-guard-cell staining. The results showed that the ROS levels increased in all the lhcb mutants compared with wild-type plants, which was observed in both the whole leaves and in guard cells (Fig. 4A–C). It was found that ABA treatments at relatively low concentrations (1–5 μM) stimulated ROS levels in the wild-type plants (Fig. 4A, B) which is consistent with previous observations (Pei et al., 2000; Murata et al., 2001; Mustilli et al., 2002; Kwak et al., 2006; Miao et al., 2006; Zhang et al., 2009), but high levels of ABA had no significant stimulating effect (10 μM) or, inversely, reduced ROS levels (50 μM) in these wild-type plants (Fig. 4A). In all the lhcb mutants, however, ABA treatments at low concentrations reduced ROS levels in both whole leaves (1–10 μM ABA application; Fig. 4A, B) and guard cells (5 μM ABA application; Fig. 4C). The stomatal apertures of the lhcb mutants were not significantly affected by the higher levels of ROS in the absence of ABA, but showed resistance to ABA when exogenous ABA was applied (Fig. 2; see Supplementary Fig. 5at JXB online) which, by contrast, reduced ROS levels in these mutants (Fig. 4A–C). In addition, experiments were conducted with 3,5-diaminobenzidine (DAB) staining which detects H2O2 production and substantially the same results were obtained as those with NBT staining (see Supplementary Fig. S6 at JXB online). These data demonstrate that down-regulation or disruption of the LHCB members alters the homeostasis of ROS and ABA responsiveness of ROS in plant cells.
Fig. 4.
ROS homeostasis is altered in lhcb mutants. (A) ROS production in leaves in response to different concentrations of ABA (from 0–50 μM for Col and 0–10 μM for lhcb mutants), detected by nitroblue tetrazolium staining in wild-type Col and different lhcb mutants. The entire experiment was replicated five times with similar results. (B) Quantitative estimation of the ROS production described in (A). Relative ROS-staining intensities estimated by scanning the staining profiles, are normalized relative to the ROS-staining intensity of Col (taken as 100%). Each value is the mean ±SE of five independent biological determinations. (C) ROS production from guard cells in response to ABA (5 μM), examined by H2DCF-DA imaging in wild-type Col and different lhcb mutants. The entire experiment was replicated three times with similar results. For the stomatal apertures of the treated plants, see Supplementary Fig. S5 at JXB online.
Down-regulation or disruption of LHCB genes alters the expression of a set of ABA responsive genes
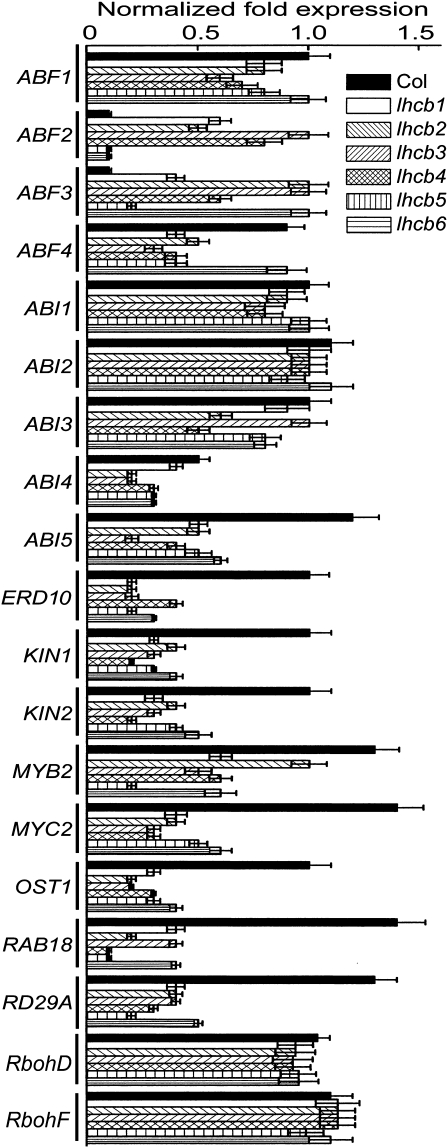
The expression of the following ABA responsive genes was assayed in the lhcb mutants: ABF1, ABF2/AREB1, ABF3, and ABF4/AREB2 (Choi et al., 2000; Uno et al., 2000), ABI1 (Leung et al., 1994; Meyer et al., 1994; Gosti et al., 1999), ABI2 (Leung et al., 1997), ABI3 (Giraudat et al., 1992), ABI4 (Finkelstein et al., 1998), ABI5 (Finkelstein and Lynch, 2000), ERD10 (Kiyosue et al., 1994), KIN1 and KIN2 (Kurkela and Borg-Franck, 1992), MYB2 and MYC2 (Abe et al., 2003), OST1 (Mustilli et al., 2002), RAB18 (Lang and Palva, 1992), and RD29A (Yamaguchi-Shinozaki and Shinozaki, 1994). Expression of ten ABA-positively-responsive genes, ABI4, ABI5, ERD10, KIN1, KIN2, MYB2, MYC2, OST1, RAB18, and RD29A, was significantly repressed in all the lhcb mutants (from lhcb1 to lhcb6, Fig. 5). Expression of three genes encoding important transcription factors that positively regulate ABA signalling, ABF1, ABF4, and ABI3, was also significantly repressed in the lhcb mutants except for lhcb1, lhcb3, and lhcb6 (Fig. 5). However, expression of two genes that encode negative regulators of ABA signalling acting directly downstream of the ABA receptor PYR/PYL/RCAR (Fujii et al., 2009), ABI1 and AB12, was not altered in any of the lhcb mutants (Fig. 5). By contrast, expression of two genes coding for two transcription factors positively involved in ABA signalling, ABF2 and ABF3, was up-regulated in the lhcb mutants except for lhcb5 and lhcb6 (Fig. 5). This expression profile of the ABA-responsive genes is essentially consistent with the idea that LHCBs are positively involved in ABA signalling but with a complex underlying mechanism.
Fig. 5.
Expression of a set of ABA-responsive genes is altered in lhcb mutants. Gene expression was assayed by real-time PCR. Each value is the mean ±SE of three independent biological determinations.
Gene expression of two major members of the NADPH oxidases was also analysed, termed respiratory burst oxidase homologues (Rbohs), RbohD and RbohF, which are plasma membrane-associated proteins and involved in ABA-induced stomatal closure (Kwak et al., 2003; Bright et al., 2006). The results showed that expression of RbohD and RbohF genes was not significantly affected in the lhcb mutants in comparison with wild-type plants (Fig. 5).
Discussion
LHCB members are positively involved in guard cell signalling in response to ABA
Genetic evidence is provided here that the members of the LHCB family are positively involved in guard cell signalling in response to ABA and so LHCB members have been identified as new players in ABA signalling in stomatal movement. Consistently, previous studies showed that down-regulation of the LHCB members reduced plant tolerance to environmental stresses with lowered seed production (Andersson et al., 2001, 2003; Ganeteg et al., 2004; Kovacs et al., 2006) except for the lhcb3 mutant that showed comparable seed production with that of wild-type plants (Damkjaer et al., 2009). The differences observed in the lhcb3 mutants between our present observations and the previously reported data (Damkjaer et al., 2009) may be due to the characteristics of the different lhcb3 mutants: in the lhcb3 mutant used here (SALK_036200 or N536200), the protein levels of the other LHCB members were not affected (Fig. 1) while in the lhcb3 mutant used by Damkjaer et al (2009) (N520342 or SALK_020342), the protein levels of LHCB1 and LHCB2 were significantly up-regulated (Damkjaer et al., 2009), which may partly compensate for the disruption of the LHCB3 protein. Nevertheless, the defects in ABA signalling in stomatal movement in the lhcb mutants, among other defects in the photosynthesis apparatus (Andersson et al., 2001, 2003; Ganeteg et al., 2004; Kovacs et al., 2006; Damkjaer et al., 2009), are at least partly responsible for the previously-observed decline of the plants’ ability to adapt to environmental stresses (Andersson et al., 2001, 2003; Ganeteg et al., 2004; Kovacs et al., 2006).
The concentrations of ABA, dry substances and chlorophyll a/b were not affected in the lhcb mutants used in the present experiments (Fig. 1; see Supplementary Fig. S1 at JXB online), which shows that the ABA-insensitive phenotypes of these mutants in stomatal movement were associated neither with ABA biosynthesis nor with photoassimilate accumulation and, in particular, that down-regulation of the LHCB members could affect ABA signalling without altering chlorophyll homeostasis. However, the stability of the LHCB proteins is associated with chlorophyll a/b, of which the deficiency may result in ac decrease of the LHCB proteins (Adam, 1996; Espineda et al., 1999). The chlorophyll b-deficient mutants ch1-1 and ch1-2, which results in low expression of the LHCB genes (Espineda et al., 1999), showed slight or no ABA insensitive phenotype in the stomatal response to ABA (Fig. 2; Shen et al., 2006). The possible explanation of this phenomenon is that chlorophyll deficiency may induce more complex consequences than degradation of the LHCB proteins, which may compensate for the effects of the LHCB protein deficiency.
The lhcb double mutants showed ABA-insensitive phenotypes similar to the lhcb single mutants (Fig. 2), suggesting that a compensatory feed-back mechanism to maintain the LHCB homeostasis may function in the LHCB-related ABA signalling in guard cells. Each of the six lhcb single mutants showed similar ABA-insensitive phenotypes in stomatal movement and similar drought-hypersensitive phenotypes (Figs 2, 3), suggesting that each of the LHCB members is necessary for building the antenna complex and keeping the complex intact, which functions as a whole both in photosynthesis and ABA signalling in guard cells. So, deficiency of any of the LHCB members may damage this core molecular complex of the PSII antenna machinery, and thus affect ABA signalling in stomatal movement. This point of view is consistent with the previous reports, which showed that each member of the LHCB family plays a specific role in the regulation of the photosynthetic machinery (Andersson et al., 2001, 2003; Ganeteg et al., 2004; Kovacs et al., 2006; Damkjaer et al., 2009).
How do the LHCB proteins work in guard cell signalling in response to ABA?
The mechanism by which the LHCB members are involved in ABA signalling in guard cells may be highly complex. The ABA insensitivity of the cch mutant in stomatal movement was partly suppressed by LHCB6-over-expression, suggesting that CHLH/ABAR may function upstream of the LHCB members. It was observed that down-regulation of the LHCB members altered both the ROS homeostasis and the ABA responsiveness of ROS in plant leaves (Fig. 4). Expression of two major plasma membrane-associated NADPH oxidase genes, RbohD and RbohF, which are involved in ABA-induced stomatal closure (Kwak et al., 2003; Bright et al., 2006), was not affected in the lhcb mutants (Fig. 5), suggesting that the alteration in ROS levels in these lhcb mutants is mainly caused by a deficiency of the LHCB members, but may not involve the plasma-membrane NADPH oxidases. Under normal conditions, higher amounts of ROS accumulated in the lhcb mutants compared with wild-type plants (Fig. 4), which suggests that an imbalanced antenna complex reduces its efficiency, thus leading to ROS accumulation. In the presence of ABA, however, ROS levels in all the mutants decreased compared with the wild type in which ABA stimulates ROS production (Fig. 4), suggesting that ABA probably enhances the already activated ROS-detoxifying systems, thus lowering ROS levels that are abnormally enhanced by lhcb mutation. It is possible that LHCBs are involved in ABA signalling in guard cells partly by modulating ROS homeostasis. It will be of importance to elucidate upstream- and downstream-events of LHCBs to understand the complex mechanism of ABA signalling in guard cells and the positive role of LHCB proteins in plant stress tolerance.
Supplementary data
Supplementary data can be found at JXB online.
Supplementary Fig. S1. Concentrations of endogenous ABA and accumulation of dry substances of the different lhcb mutant plants.
Supplementary Fig. S2. Real-time PCR analysis of the LHCB6-RNAi and over-expression lines.
Supplementary Fig. S3. Expression of the 35S-promoter-driven LHCBs rescues ABA sensitivity of the lhcb mutants.
Supplementary Fig. S4. Different members of LHCBs are expressed ubiquitously in different tissues/organs except for dry seeds.
Supplementary Fig. S5. ROS homeostasis is altered in lhcb mutants.
Supplementary Fig. S6. Stomatal aperture of Col plants and lhcb mutants when assaying ROS levels in stomata.
Supplementary Table S1. Primers used in this study.
Acknowledgments
The mutant seeds were provided by the Arabidopsis Biological Resource Center (ABRC). This work was supported by grants from National Natural Science Foundation of China (grant nos. 90817104 and 30700053) and Ministry of Agriculture of China (grant no. 2008ZX08009-003).
Footnotes
This is an Open Access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/2.0/uk/) which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
These authors contributed equally to this work.
References
- Abe H, Urao T, Ito T, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcription activators in abscisic acid signaling. The Plant Cell. 2003;15:63–78. doi: 10.1105/tpc.006130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adam Z. Protein stability and degradation in chloroplasts. Plant Molecular Biology. 1996;32:773–783. doi: 10.1007/BF00020476. [DOI] [PubMed] [Google Scholar]
- Adie BAT, Perez-Perez J, Perez-Perez MM, Godoy M, Sanchez-Serrano JJ, Schmelz EA, Solanoa R. ABA is an essential signal for plant resistance to pathogens affecting JA biosynthesis and the activation of defenses in Arabidopsis. The Plant Cell. 2007;19:1665–1681. doi: 10.1105/tpc.106.048041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alabadi D, Oyama T, Yanovsky MJ, Harmon FG, Mas P, Kay SA. Reciprocal regulation between TOC1 and LHY/ CCA1 within the Arabidopsis circadian clock. Science. 2001;293:880–883. doi: 10.1126/science.1061320. [DOI] [PubMed] [Google Scholar]
- Andersson J, Walters RG, Horton P, Jansson S. Antisense inhibition of the photosynthetic antenna proteins CP29 and CP26: Implications for the mechanism of protective energy dissipation. The Plant Cell. 2001;13:1193–1204. doi: 10.1105/tpc.13.5.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson J, Wentworth M, Walters RG, Howard CA, Ruban AV, Horton P, Jansson S. Absence of the Lhcb1 and Lhcb2 proteins of the light-harvesting complex of the photosystem II: effects on photosynthesis, grana stacking and fitness. The Plant Journal. 2003;35:350–361. doi: 10.1046/j.1365-313x.2003.01811.x. [DOI] [PubMed] [Google Scholar]
- Andronis C, Barak S, Knowles SM, Sugano S, Tobin EM. The clock protein CCA1 and the bZIP transcription factor HY5 physically interact to regulate gene expression in Arabidopsis. Molecular Plant. 2008;1:58–67. doi: 10.1093/mp/ssm005. [DOI] [PubMed] [Google Scholar]
- Bartholomew DM, Bartley GE, Scolnik PA. Abscisic acid control of rbcS and cab transcription in tomato leaves. Plant Physiology. 1991;96:291–296. doi: 10.1104/pp.96.1.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bright J, Desikan R, Hancock JT, Weir IS, Neill SJ. ABA-induced NO generation and stomatal closure in Arabidopsis are dependent on H2O2 synthesis. The Plant Journal. 2006;45:113–122. doi: 10.1111/j.1365-313X.2005.02615.x. [DOI] [PubMed] [Google Scholar]
- Chang YC, Walling LL. Abscisic acid negatively regulates expression of chlorophyll a/ b binding protein genes during soybean embryogeny. Plant Physiology. 1991;97:1260–1264. doi: 10.1104/pp.97.3.1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi H, Hong J, Ha J, Kang J, Kim SY. ABFs, a family of ABA-responsive element binding factors. Journal of Biological Chemistry. 2000;275:1723–1730. doi: 10.1074/jbc.275.3.1723. [DOI] [PubMed] [Google Scholar]
- Cutler SR, Rodriguez PL, Finkelstein RR, Abrams SR. Abscisic acid: emergence of a core signaling network. Annual Review of Plant Biology. 2010;61:651–679. doi: 10.1146/annurev-arplant-042809-112122. [DOI] [PubMed] [Google Scholar]
- Damkjaer JT, Kereiche S, Johnson MP, Kovacs L, Kiss AZ, Boekema EJ, Ruban AV, Horton P, Jansson S. The photosystem II light-harvesting protein Lhcb3 affects the macrostructure of photosystem II and the rate of state transitions in. Arabidopsis. The Plant Cell. 2009;21:3245–3256. doi: 10.1105/tpc.108.064006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Montaigu A, Toth R, Coupland G. Plant development goes like clockwork. Trends in Genetics. 2010;26:296–308. doi: 10.1016/j.tig.2010.04.003. [DOI] [PubMed] [Google Scholar]
- Espineda CE, Linford AS, Devine D, Brusslan JA. The AtCAO gene, encoding chlorophyll a oxygenase, is required for chlorophyll b synthesis in. Arabidopsis thaliana. Proceedings of the National Academy of Sciences, USA. 1999;96:10507–10511. doi: 10.1073/pnas.96.18.10507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein RR, Gampala S, Rock C. Abscisic acid signaling in seeds and seedlings. The Plant Cell. 2002;14(Supplement):S15–S45. doi: 10.1105/tpc.010441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein RR, Lynch TJ. The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. The Plant Cell. 2000;12:599–609. doi: 10.1105/tpc.12.4.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein RR, Wang ML, Lynch TJ, Rao S, Goodman HM. The Arabidopsis abscisic acid response locus ABI4 encodes an APETALA2 domain protein. The Plant Cell. 1998;10:1043–1054. doi: 10.1105/tpc.10.6.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii H, Chinnusamy V, Rodrigues A, Rubio S, Antoni R, Park SY, Cutler SR, Sheen J, Rodriguez PL, Zhu JK. In vitro reconstitution of an abscisic acid signaling pathway. Nature. 2009;462:660–664. doi: 10.1038/nature08599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galvez-Valdivieso G, Mullineaux PM. The role of reactive oxygen species in signaling from chloroplasts to the nucleus. Physiologia Plantarum. 2010;138:430–439. doi: 10.1111/j.1399-3054.2009.01331.x. [DOI] [PubMed] [Google Scholar]
- Ganeteg U, Kulheim C, Andersson J, Jansson S. Is each light-harvesting complex protein important for plant fitness? Plant Physiology. 2004;134:502–509. doi: 10.1104/pp.103.033324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraudat J, Hauge BM, Valon C, Smalle J, Parcy F, Goodman HM. Isolation of Arabidopsis ABI3 gene by positional cloning. The Plant Cell. 1992;4:1251–1261. doi: 10.1105/tpc.4.10.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosti F, Beaudoin N, Serizet C, Webb AAR, Vartanian N, Giraudata J. ABI1 protein phosphatase 2C is a negative regulator of abscisic acid signaling. The Plant Cell. 1999;11:1897–1909. doi: 10.1105/tpc.11.10.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humbeck K, Krupinska K. The abundance of minor chlorophyll a/b-binding proteins CP29 and LHCI of barley (Hordeum vulgare L.) during leaf senescence is controlled by light. Journal of Experimental Botany. 2003;54:375–383. doi: 10.1093/jxb/erg012. [DOI] [PubMed] [Google Scholar]
- Jansson S. The light-harvesting chlorophyll a/ b-binding proteins. Biochimica et Biophysica Acta. 1994;1184:1–19. doi: 10.1016/0005-2728(94)90148-1. [DOI] [PubMed] [Google Scholar]
- Jansson S. A guide to the Lhc genes and their relatives in. Arabidopsis. Trends in Plant Sciences. 1999;4:236–240. doi: 10.1016/s1360-1385(99)01419-3. [DOI] [PubMed] [Google Scholar]
- Kiyosue T, Yamaguchi-Shinozaki K, Shinozaki K. Characterization of two cDNAs (ERD10 and ERD14) corresponding to genes that respond rapidly to dehydration stress in. Arabidopsis thaliana. Plant and Cell Physiology. 1994;35:225–231. [PubMed] [Google Scholar]
- Kovacs L, Damkjær J, Kereiche S, Ilioaia C, Ruban AV, Boekema EJ, Jansson S, Horton P. Lack of the light-harvesting complex CP24 affects the structure and function of the grana membranes of higher plant chloroplasts. The Plant Cell. 2006;18:3106–3120. doi: 10.1105/tpc.106.045641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurkela S, Borg-Franck M. Structure and expression of kin2, one of two cold- and ABA-induced genes of. Arabidopsis thaliana. Plant Molecular Biology. 1992;19:689–692. doi: 10.1007/BF00026794. [DOI] [PubMed] [Google Scholar]
- Kwak JM, Mori IC, Pei ZM, Leonhardt N, Torres MA, Dang JL, Bloom RE, Boddle S, Jones JDG, Schroeder JI. NADPH oxidase AtrbohD and AtrbohF genes function in ROS dependent ABA signaling in Arabidopsis. EMBO Journal. 2003;22:2623–2633. doi: 10.1093/emboj/cdg277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak JM, Nguyen V, Schroeder JI. The role of reactive oxygen species in hormonal responses. Plant Physiology. 2006;141:323–329. doi: 10.1104/pp.106.079004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang V, Palva ET. The expression of a tab-related gene, rab18, is induced by abscisic acid during the cold acclimation process of Arabidopsis thaliana (L.) Heynh. Plant Molecular Biology. 1992;20:951–962. doi: 10.1007/BF00027165. [DOI] [PubMed] [Google Scholar]
- Lee B, Lee H, Xiong L, Zhu JK. A mitochondrial complex I defect impairs cold-regulated nuclear gene expression. The Plant Cell. 2002;14:1235–1251. doi: 10.1105/tpc.010433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung J, Bouvier-Durand M, Morris PC, Guerrier D, Chefdor F, Giraudat J. Arabidopsis ABA response gene ABI1: features of a calcium-modulated protein phosphatase. Science. 1994;264:1448–1452. doi: 10.1126/science.7910981. [DOI] [PubMed] [Google Scholar]
- Leung J, Merlot S, Giraudat J. The Arabidopsis ABSCISIC ACD-INSENSITIVE2 (ABI2) and ABI1 encode homologous protein phosphatase 2C involved in abscisic acid signal transduction. The Plant Cell. 1997;9:759–771. doi: 10.1105/tpc.9.5.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Szostkiewicz I, Korte A, Moes D, Yang Y, Christman A, Grill E. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science. 2009;324:1064–1068. doi: 10.1126/science.1172408. [DOI] [PubMed] [Google Scholar]
- Meyer K, Leube MP, Grill E. A protein phosphatase 2C involved in ABA signal transduction in Arabidopsis thaliana. Science. 1994;264:1452–1455. doi: 10.1126/science.8197457. [DOI] [PubMed] [Google Scholar]
- Miao Y, Lv D, Wang P, Wang XC, Chen J, Miao C, Song CP. An Arabidopsis glutathione peroxidase functions as both a redox transducer and a scavenger in abscisic acid and drought stress responses. The Plant Cell. 2006;18:2749–2766. doi: 10.1105/tpc.106.044230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar AJ, Kay S. Integration of circadian and phototransduction pathways in the network controlling CAB gene transcription in Arabidopsis. Proceedings of the National Academy of Sciences, USA. 1996;93:15491–15494. doi: 10.1073/pnas.93.26.15491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata Y, Pei ZM, Mori IC, Schroeder JI. Abscisic acid activation of plasma membrane Ca2+ channels in guard cells requires cytosolic NAD(P)H and is differentially disrupted upstream and downstream of reactive oxygen species production in abi1-1 and abi2-1 protein phosphatase 2C mutants. The Plant Cell. 2001;13:2513–2523. doi: 10.1105/tpc.010210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustilli AC, Merlot S, Vavasseur A, Fenzi F, Giraudat J. Arabidopsis OST1 protein kinase mediates the regulation of stomatal aperture by abscisic acid and acts upstream of reactive oxygen species production. The Plant Cell. 2002;14:3089–3099. doi: 10.1105/tpc.007906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nott A, Jung HS, Koussevitzky S, Chory J. Plastid-to-nucleus retrograde signaling. Annual Review of Plant Biology. 2006;57:739–759. doi: 10.1146/annurev.arplant.57.032905.105310. [DOI] [PubMed] [Google Scholar]
- Pandey S, Nelson DC, Assmann SM. Two novel GPCR-type G proteins are abscisic acid receptors in Arabidopsis. Cell. 2009;136:136–148. doi: 10.1016/j.cell.2008.12.026. [DOI] [PubMed] [Google Scholar]
- Park S-Y, Fung P, Nishimura N, et al. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science. 2009;324:1068–1071. doi: 10.1126/science.1173041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen H, Bogorad L. Diurnal and circadian rhythms in the accumulation and synthesis of mRNA for the light-harvesting chlorophyll a/ b-binding protein in tobacco. Plant Physiology. 1988;88:1104–1109. doi: 10.1104/pp.88.4.1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peer W, Silverthorne J, Peters J. Developmental and light-regulated expression of individual members of the light-harvesting complex b gene family in. Pinus palustris. Plant Physiology. 1996;111:627–634. doi: 10.1104/pp.111.2.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei ZM, Murata Y, Benning G, Thomine S, Klusener B, Allen GJ, Grill E, Schroeder JI. Calcium channels activated by hydrogen peroxide mediate abscisic acid signalling in guard cells. Nature. 2000;406:731–734. doi: 10.1038/35021067. [DOI] [PubMed] [Google Scholar]
- Pruneda-Paz JL, Kay SA. An expanding universe of circadian networks in high plants. Trends in Plant Sciences. 2010;15:259–265. doi: 10.1016/j.tplants.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang Y, Yan L, Liu ZQ, et al. The Mg-chelatase H subunit antagonizes a group of WRKY transcription repressors to relieve ABA-responsive genes of inhibition. The Plant Cell. 2010;22:1909–1935. doi: 10.1105/tpc.110.073874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen YY, Wang XF, Wu FQ, et al. The Mg-chelatase H subunit is an abscisic acid receptor. Nature. 2006;443:823–826. doi: 10.1038/nature05176. [DOI] [PubMed] [Google Scholar]
- Silverthorne J, Tobin EM. Demonstration of transcriptional regulation of specific genes by phytochrome action. Proceedings of the National Academy of Sciences, USA. 1984;81:1112–1116. doi: 10.1073/pnas.81.4.1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staneloni RT, Rodriguez-Batiller MJ, Casal JJ. Abscisic acid, high-light, and oxidative stress down-regulate a photosynthetic gene via a promoter motif not involved in phytochrome-mediated transcriptional regulation. Molecular Plant. 2008;1:75–83. doi: 10.1093/mp/ssm007. [DOI] [PubMed] [Google Scholar]
- Strayer C, Oyama T, Schultz TF, Raman R, Somers DE, Mas P, Panda S, Kreps JA, Kay SA. Cloning of the Arabidopsis clock gene TOC1, an autoregulatory response regulator homolog. Science. 2000;289:768–771. doi: 10.1126/science.289.5480.768. [DOI] [PubMed] [Google Scholar]
- Sun L, Tobin EM. Phytochrome-regulated expression of genes encoding light-harvesting chlorophyll a/ b-binding protein in two long hypocotyls mutants and wild type plants of. Arabidopsis thaliana. Photochemistry and Photobiology. 1990;52:51–56. doi: 10.1111/j.1751-1097.1990.tb01754.x. [DOI] [PubMed] [Google Scholar]
- Thain SC, Murtas G, Lynn JR, McGrath RB, Millar AJ. The circadian clock that controls gene expression in Arabidopsis is tissue specific. Plant Physiology. 2002;130:102–110. doi: 10.1104/pp.005405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thines B, Harmon FG. Four easy pieces: mechanisms underlying circadian regulation of growth and development. Current Opinion in Plant Biology. 2010;14:1–7. doi: 10.1016/j.pbi.2010.09.009. [DOI] [PubMed] [Google Scholar]
- Uno Y, Furihata T, Abe H, Yoshida R, Shinozaki K, Yamagushi-Shinozaki K. Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. Proceedings of the National Academy of Sciences, USA. 2000;97:11632–11637. doi: 10.1073/pnas.190309197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voigt C, Oster U, Bornke F, Jahns P, Dietze KJ, Leister D, Kleine T. In-depth analysis of the distinctive effects of norflurazon implies that tetrapyrrole biosynthesis, organellar gene expression and ABA cooperate in the GUN-type of plastid signaling. Physiologia Plantarum. 2010;138:503–519. doi: 10.1111/j.1399-3054.2009.01343.x. [DOI] [PubMed] [Google Scholar]
- Weatherwax SC, Ong MS, Degenhardt J, Bray EA, Tobin EM. The interaction of light and abscisic acid in the regulation of plant gene expression. Plant Physiology. 1996;111:363–370. doi: 10.1104/pp.111.2.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu FQ, Xin Q, Cao Z, et al. The Mg-chelatase H subunit binds abscisic acid and functions in abscisic acid signaling: new evidence in Arabidopsis. Plant Physiology. 2009;150:1940–1954. doi: 10.1104/pp.109.140731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K. A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. The Plant Cell. 1994;6:251–264. doi: 10.1105/tpc.6.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang DH, Webster J, Adam Z, Lindahl M, Andersson B. Induction of acclimative proteolysis of the light-harvesting chlorophyll a/ b protein of photosystem II in response to elevated light intensities. Plant Physiology. 1998;118:827–834. doi: 10.1104/pp.118.3.827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Zhu H, Zhang Q, Li M, Yan M, Wang R, Wang L, Welti R, Zhang W, Wang X. Phospholipase Dα1 and phosphatidic acid regulate NADPH oxidase activity and production of reactive oxygen species in ABA-mediated stomatal closure in Arabidopsis. The Plant Cell. 2009;21:2357–2377. doi: 10.1105/tpc.108.062992. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.