Abstract
amylose extender (ae−) starches characteristically have modified starch granule morphology resulting from amylopectin with reduced branch frequency and longer glucan chains in clusters, caused by the loss of activity of the major starch branching enzyme (SBE), which in maize endosperm is SBEIIb. A recent study with ae− maize lacking the SBEIIb protein (termed ae1.1 herein) showed that novel protein–protein interactions between enzymes of starch biosynthesis in the amyloplast could explain the starch phenotype of the ae1.1 mutant. The present study examined an allelic variant of the ae− mutation, ae1.2, which expresses a catalytically inactive form of SBEIIb. The catalytically inactive SBEIIb in ae1.2 lacks a 28 amino acid peptide (Val272–Pro299) and is unable to bind to amylopectin. Analysis of starch from ae1.2 revealed altered granule morphology and physicochemical characteristics distinct from those of the ae1.1 mutant as well as the wild-type, including altered apparent amylose content and gelatinization properties. Starch from ae1.2 had fewer intermediate length glucan chains (degree of polymerization 16–20) than ae1.1. Biochemical analysis of ae1.2 showed that there were differences in the organization and assembly of protein complexes of starch biosynthetic enzymes in comparison with ae1.1 (and wild-type) amyloplasts, which were also reflected in the composition of starch granule-bound proteins. The formation of stromal protein complexes in the wild-type and ae1.2 was strongly enhanced by ATP, and broken by phosphatase treatment, indicating a role for protein phosphorylation in their assembly. Labelling experiments with [γ-32P]ATP showed that the inactive form of SBEIIb in ae1.2 was phosphorylated, both in the monomeric form and in association with starch synthase isoforms. Although the inactive SBEIIb was unable to bind starch directly, it was strongly associated with the starch granule, reinforcing the conclusion that its presence in the granules is a result of physical association with other enzymes of starch synthesis. In addition, an Mn2+-based affinity ligand, specific for phosphoproteins, was used to show that the granule-bound forms of SBEIIb in the wild-type and ae1.2 were phosphorylated, as was the granule-bound form of SBEI found in ae1.2 starch. The data strongly support the hypothesis that the complement of heteromeric complexes of proteins involved in amylopectin synthesis contributes to the fine structure and architecture of the starch granule.
Keywords: Amylopectin, amyloplasts, amylose extender, high-amylose, protein phosphorylation, protein–protein interactions, starch synthase, starch branching enzyme, starch phosphorylase, starch granule-bound proteins, starch synthesis
Introduction
A key feature of starch granules is their water insolubility, which is of enormous biological significance, allowing cells to store large quantities of glucose while avoiding adverse osmotic consequences. Most starches contain ∼25% by mass of a sparsely branched polymer called amylose, the remainder being amylopectin, a highly ordered branched polymer which essentially defines granule structure and is responsible for crystallinity and associated physical properties (French, 1984). An important factor underpinning glucan crystallinity is the clustering of α-(1→6) branch points in amylopectin, achieving close packing of adjacent chains which spontaneously form double helices with each other, or, potentially, other malto-oligosaccharides with a minimum chain length of degree of polymerization (DP) 6–10 (Gidley and Bulpin, 1987). Helix formation is also modulated by other factors such as the presence of salts of high lyotropic number and free fatty acids (Hizukuri et al., 1960, 1980). Three types of higher ordered crystalline organization (allomorphs) are found in starch granules (termed A-, B-, and C-type; Katz, 1930) representing differences in helix packing; A-type allomorphs characteristic of cereal storage starches are the most tightly packed due to smaller intercluster chain lengths (Hizukuri et al., 1983). B-type starch allomorphs (e.g. potato tuber) have a less packed, open structure with more water between helices, and C-types appear to be a mixture of A- and B-types (Wu and Sarko, 1978).
The enzyme classes responsible for the construction of this complex glucan matrix and its associated variations are starch (glucan) synthases (SSs; EC 2.4.1.21) and starch branching enzymes (SBEs; EC 2.4.1.18), and multiple isoforms of each class are expressed in cereals. Studies with plant and algal mutants of SS and SBE have been important in assigning a role for the different isoforms, and have also demonstrated overlap of function in many cases (Ball and Morell, 2003; Tetlow, 2011). A third class of enzymes appears to be critical for the formation of starch; these are the debranching enzymes [DBEs, including isoamylases (Isos), EC 3.2.1.41, and pullulanase, EC 3.2.1.68] which probably modify the glucan structure (pre-amylopectin) produced by SSs and SBEs by debranching α-(1→6) branch points, and allowing clusters of glucan chains to form crystalline water-insoluble amylopectin (Morris and Morris, 1939; James et al., 1995; Mouille et al., 1996; Zeeman et al., 1998; Wattebled et al., 2005, 2008; Fujita et al., 2009).
The relative catalytic activities and kinetic characteristics of individual SS and SBE isoforms operating in the amyloplast clearly play a critical role in developing the final structure of the starch granule. The effects of loss of SS and SBE isoforms on starch structure are highly variable, and dependent on the species/tissue involved and the relative contribution of different isoforms to the overall structure of amylopectin. A complicating factor in attempting to understand the effects of different mutations in the starch biosynthetic pathway is the fact that many of the enzymes have been shown to operate in multimeric protein complexes (Tetlow et al., 2004, 2008; Hennen-Bierwagen et al., 2008, 2009).
One of the most dramatic, and widely exploited, alterations in starch structure occurs when the catalytic activity of the major form of SBE is lost in cereal endosperms. In maize this is termed the amylose extender (ae−) mutant which has a characteristic kernel and starch phenotype and is caused by loss of SBEIIb activity (Hedman and Boyer, 1982, 1983; Stinard et al., 1993). ae− starches are characterized by longer internal chain lengths in amylopectin compared with normal starches, and less frequently branched outer chains (Hilbert and MacMasters, 1946; Banks et al., 1974; Klucinec and Thompson, 2002). ae− starches have also been shown to demonstrate B-type crystallinity, indicating alterations in amylopectin cluster and blocklet architecture (Nishi et al., 2001; Gérard et al., 2002; Kubo et al., 2008; Saibene and Seetharaman, 2011) as distinct from the A-type associated with wild-type cereal endosperms. The higher proportion of relatively long-chain branch points in ae− starches leads to them being termed ‘high-amylose’ starches, although this term is misleading, since studies with ae−/waxy double mutants (i.e. ae− in an amylose-free background) demonstrate that these starches have modified amylopectin with some amylose-like properties (Nishi et al., 2001; Yao et al., 2004), and recent studies with SBEIIb-deficient rice demonstrated no alteration in the amylose content (Butardo et al., 2011). The relative proportions of the different classes of SBEII activity in different cereals appear to be important in determining the effects of loss of a particular SBEII form on starch structure. The major SBEII isoform expressed in maize and rice endosperm is SBEIIb, and its loss in the ae− mutant causes the alterations in amylopectin structure noted above. However, lesions in SBEIIa in maize (expressed at low levels in the endosperm, but the dominant form of SBEII in leaf tissue) lead to unaltered endosperm starch, but leaf starch with increased apparent amylose (Blauth et al., 2001). In wheat and barley, ae−-like starches are only produced by suppression of both genes encoding the SBEIIa and SBEIIb forms, resulting in starches containing >70% apparent amylose (Regina et al., 2006, 2010), and this probably reflects the higher levels of expression of SBEIIa isoforms in these tissues compared with maize (Gao et al., 1997; Sun et al., 1998).
A detailed biochemical analysis of the maize ae− mutant, lacking expression of the SBEIIb protein (which is now termed ae1.1), was recently reported by Liu et al. (2009). This study showed that isolated amyloplasts from the ae1.1 mutant possessed modified forms of heteromeric protein complexes normally found in the stroma of wild-type cereals. In particular, a complex consisting of SSI, SSIIa, and SBEIIb found in wild-type amyloplasts was replaced by one or more protein complexes in which SBEIIb (absent from ae1.1) was substituted by SBEI, starch phosphorylase (SP; EC 2.4.1.1), and SBEIIa (Liu et al., 2009). Liu et al. argued that recruitment of SBEI and SBEIIa as replacement branching enzymes for SBEIIb in the mutant protein complex contributes to the starch phenotype of ae1.1, a view supported by the known biochemical characteristics of these branching enzymes. SBEI has a higher affinity for branching long glucan chains (including amylose) and results in lower branching frequency with amylopectin compared with SBEII forms (Guan and Preiss, 1993; Takeda et al., 1993; Rydberg et al., 2001). Its association in the mutant protein complex was postulated as leading to a lower frequency of branch points and branched chains, with longer glucan chains relative to wild-type starch.
The present study analyzed an allelic variant of the ae− mutant of maize which expresses an inactive form of SBEIIb, which is termed ae1.2. In particular, the aim was to determine the basis of the mutation in SBEIIb, and whether the presence of the inactive protein impacts protein–protein interactions with SSs with consequences for the phenotype of starch. The data show that the mutant protein lacks a 28 amino acid peptide, lacks glucan binding ability, and that novel protein complexes are formed in ae1.2 amyloplasts, resulting in starch with properties distinct from both the wild-type and ae1.1. The significance of these observations lies in the distinct nature of the starch phenotypes arising from the different alleles of ae− mutants, suggesting that the complement of enzymes present in amylopectin-synthesizing protein complexes in amyloplasts influences starch structure.
Materials and methods
Plant material
Two recessive ae− alleles, ae1-ref (herein referred to as ae1.1) and ae1-Elmore (herein referred to as ae1.2), were examined in a common maize inbred line background, CG102. The ae− alleles were obtained from the Maize Genetics Cooperation Stock Centre and were introgressed into CG102 by backcrossing for four generations followed by two generations of self-pollinations. The resulting BC3S2 lines are homozygous for the ae− mutant alleles and on average possess 93.75% of the CG102 genome. Wild-type CG102 and ae− mutant maize plants were grown at 25–27 °C in the glasshouse at the University of Guelph under conditions previously described for growing wheat (Tetlow et al., 2008). Self-pollinated kernels, obtained through controlled pollinations, were collected at 9–12 days after pollination (DAP), 20–25 DAP, and 29–35 DAP, and used to prepare endosperm amyloplasts, starch, and whole cell soluble extracts. Plant materials were flash frozen in liquid nitrogen and stored at –80 °C until future use.
Preparation of whole cell extracts from developing endosperm
Whole cell extracts were prepared as described previously (Tetlow et al., 2003) with some modification. Approximately 10 g of endosperm tissue was quickly frozen in liquid nitrogen and immediately ground into a fine powder using a chilled mortar and pestle under liquid nitrogen. The frozen powder was mixed with ice-cold rupturing buffer containing 100 mM Tricine/KOH, pH 7.8, 1 mM Na2-EDTA, 1 mM dithiothreitol (DTT), 5 mM MgCl2, and a protease inhibitor cocktail (Sigma-Aldrich, catalogue no. P 9599, used at 10 μl ml−1). The mixture was further gently ground and allowed to stand on ice for 5 min, followed by centrifugation at 13 500 g for 5 min at 4 °C. The supernatant was subjected to ultracentrifugation at 120 000 g for 15 min in a Beckman Optima Max-XP Ultracentrifuge to remove membranes and particulate material. The supernatant obtained following ultracentrifugation was used for experiments.
Amyloplast isolation
Maize endosperm amyloplasts were isolated using a modification of the methods described by Tetlow et al. (2008). Fresh endosperm tissue was washed and chopped with a razor blade in ice-cold amyloplast extraction buffer (50 mM HEPES/KOH, pH 7.5, containing 0.8 M sorbitol, 1 mM KCl, 2 mM MgCl2, and 1 mM Na2-EDTA). The resulting whole cell extract was then filtered through four layers of Miracloth (CalBiochem, catalogue no. 475855) wetted in the same buffer. Approximately 25 ml of the filtrate was then carefully layered onto 15 ml of 3% (w/v) Histodenz (Sigma, catalogue no. D2158) in amyloplast extraction buffer followed by centrifugation at 100 g at 4 °C for 20 min, and the supernatant was carefully decanted. Intact amyloplasts appeared as a yellow ring on top of the starch in the pellet and were lysed osmotically by the addition of ice-cold rupturing buffer (see above). The plastid lysate was then centrifuged at 13 500 g for 2 min at 4 °C to remove starch granules, followed by ultracentrifugation at 120 000 g for 15 min to remove plastidial membranes. The supernatant from the ultracentrifugation step, termed plastid stroma (0.5–1.1 mg protein ml−1), was flash frozen in liquid nitrogen and stored at –80 °C until future use.
Isolation of starch granule-bound proteins
Isolation of starch granule-bound proteins (i.e. proteins trapped inside the granule matrix as opposed to proteins attached to the granule surface) was performed as described previously (Tetlow et al., 2004; Liu et al., 2009). Starch granules from plastid lysates (see above) were resuspended in cold aqueous washing buffer (50 mM TRIS-acetate, pH 7.5, 1 mM Na2-EDTA, and 1 mM DTT) and centrifuged at 3000 g for 1 min at 4 °C. This washing step was repeated five times. The pellet was then washed three times with acetone followed by three washes with 2% (w/v) sodium dodecyl sulphate (SDS). Starch granule-bound proteins were extracted by boiling the washed starch in SDS loading buffer [62.5 mM TRIS-HCl, pH 6.8, 2% (w/v) SDS, 10% (w/v) glycerol, 5% (v/v) β-mercaptoethanol, 0.001% (w/v) bromophenol blue]. Boiled samples were centrifuged at 13 000 g for 5 min and the supernatant was used for SDS–PAGE and immunoblotting analysis of granule-bound proteins.
In addition to the acetone/SDS washing methods described above, granule-bound proteins were also prepared using protease treatment employing a modification of the method described by Rahman et al. (1995). Starch granules (15 mg) from plastid lysates were washed in buffer as described above and then incubated with 12.5 U of trypsin (Promega) in 470 μl of a buffer containing 50 mM TRIS-HCl (pH 7.6) and 1 mM CaCl2 for 30 min at 37 °C on a rotator. After incubation, the starch granules were washed in buffer, acetone, and SDS as described above.
Phosphorylation of amyloplast proteins in vitro
Plastid lysates (containing 0.5–1.1 mg protein ml−1) prepared from wild-type and ae− amyloplasts isolated from endosperm at 20–25 DAP were incubated with 1 mM ATP with 600–1200 Ci mmol−1 [γ-32P]ATP (Perkin-Elmer, Boston, MA, USA) in a total volume of 0.5 ml for 30 min at 25 °C with gentle rocking. The phosphorylation reaction was terminated by the addition of 10 mM Na2-EDTA made up in 100 mM Tricine-KOH (pH 7.8). Phosphorylated stromal proteins were used in immunoprecipitation experiments (see below). Radiolabelled proteins were visualized by autoradiography and identified by immunoblotting and mass spectrometry (MS).
Expression and purification of recombinant SBEIIb in Escherichia coli
The SBEIIb cDNA sequences from CG102 and ae1.2/CG102 were ligated to pET29a vectors (Novagen) with restriction sites NcoI and XhoI after removing sequences encoding transit peptides. The recombinant plasmids were transformed to ArcticExpress competent cells (Stratagene) and protein expression was induced by 1 mM isopropyl-β-D-thiogalactopyranoside (IPTG) at 10 °C, 250 rpm for 24 h. Escherichia coli (ArcticExpress) cells were collected by centrifugation and lysed using ‘BugBuster Protein Extraction Reagent’ (Novagen). Recombinant SSI, SBEI, and SBEIIa were purified from inclusion bodies. A Protein Refolding Kit (Novagen catalogue no. 70123-3) was employed for the purification of inclusion bodies and refolding of recombinant proteins following the manufacturer’s manual. Soluble recombinant SBEIIb proteins (CG102 and ae1.2/CG102 origin sequences) were precipitated by 35% (w/v) ammonium sulphate followed by dialysis for 4 h at 4 °C. The dialysates were centrifuged for 30 min at 27 000 g at 4 °C, and the supernatant applied to a 1 ml ResourceQ column (connected to an AKTA Explorer FPLC, Amersham Biosciences at 4 °C) pre-equilibrated with 50 mM TRIS-acetate buffer, pH 7.5, containing 0.05% (v/v) Triton X-100. The columns were initially washed with the equilibration buffer, followed by elution with a linear gradient of 0-0.6 M KCl in the equilibration buffer. The flow rates were 1 ml min−1, and 0.5 ml per fraction was collected. Eluates were tested by SDS–PAGE and Coomassie Blue staining for recombinant SBEIIb. Isolated SBEIIb was concentrated on a Centricon YM50 filter (Millipore) and then applied to a Superdex 200 10/300GL gel permeation column connected to an AKTA Explorer FPLC (Amersham Biosciences) at 4 °C. Purified recombinant protein was collected from fractions corresponding to a molecular mass of ∼85–90 kDa (identified by immunoblotting) for both CG102 and ae1.2/CG102. The biochemical functions of SBEIIb were measured using 14C-labelled substrate assays, and native gel zymogram assays (see below). Recombinant proteins were stored at –20 °C in 40% (v/v) glycerol and catalytic activities were checked every 2–3 months.
Enzyme assays
Starch branching enzymes
SBE was assayed indirectly by stimulation of incorporation of 14C from [U-14C]α-D-glucose 1-phosphate (Glc1P) into glucan by phosphorylase a according to methods previously described (Smith, 1990), and modified by Tetlow et al. (2008). The reaction contained, in a total volume of 200 μl, 100 mM sodium citrate, pH 7.0, 1 mM Na2-EDTA, 1 mM DTT, 2.5 mM AMP, and 0.2 U of rabbit muscle phosphorylase a (from rabbit muscle; product no. P-1261, Sigma-Aldrich). Protein extract (60 μl) was added immediately before initiation of the reaction with 20 μl of 50 mM [U-14C]Glc 1-P (3.7–7.4 kBq per assay; Amersham Biosciences), incubated at 25 °C for 90 min, and terminated by heating at 95 °C for 5 min. Prior to washing with 75% (v/v) methanol–1% (w/v) KCl (MeOH/KCl), an 8% (w/v) aqueous solution of rabbit liver glycogen (type III, Sigma-Aldrich) was added to precipitate the newly synthesized 14C-labelled glucan.
The modified SBE assays for maize were each optimized with respect to substrate concentration and glucan primer used for each genotype, and reactions were all shown to be linear with respect to protein concentration and reaction time prior to experimentation.
Starch phosphorylase
SP activity could be detected on SBE zymograms, producing a dark blue coloured band of glucan product (the position of this activity band aligned with an immunoreactive band with anti-SP antibodies on corresponding native gels). A quantitative assay for SP was also employed measuring glucan-synthesizing activity as described by Chang and Su (1986) with some modifications. SP activity was assayed in a final volume of 200 μl containing 50 mM MES-NaOH buffer (pH 6), 1 mM DTT, 10 mM Glc-1P (Sigma, catalogue no. G7000-25G), 2.5% (w/v) rabbit liver glycogen (type III, Sigma-Aldrich), and 50 μg of amyloplast stromal proteins. The assay system was pre-incubated at 37 °C for 2 min. Protein extracts (60 μl) were added immediately before initiation of the reaction, followed by the addition of 20 μl of 50 mM [U-14C]Glc 1-P (3.7–7.4 kBq per assay; Amersham Biosciences). The mixture was incubated at 37 °C for 30 min, and terminated by heating at 95 °C for 5 min. An 8% (w/v) aqueous solution of rabbit liver glycogen (type III, Sigma-Aldrich) was added to the mixture and precipitated together with newly elongated glucan by adding 1 ml of MeOH/KCl followed by centrifugation at 13 500 g for 5 min. The precipitated glucan was resuspended with 0.3 ml of H2O on a disruptor for 5 min; glucan was again precipitated with 1 ml of MeOH/KCl and centrifuged at 13 500 g for 5 min; the pellet was resuspended again with 0.5 ml of H2O, and washed glucan was added to a vial for counting 14C radioactivity in a Beckman LS6500 liquid scintillation counter.
Glucan binding assays with recombinant SBEIIb
Approximately 100 μg of recombinant SBEIIb (wild-type or ae1.2) containing an N-terminal S-tag was incubated with 15 mg of purified maize starch (wild-type and ae1.2, purified using the protocol described above for granule-bound protein analysis) at 4 °C for 1 h on a rotator. Following incubation, starch was washed five times with 1.5 ml of 20 mM TRIS-acetate buffer (pH 7.5) containing 1 mM DTT, followed by a single wash in 1.5 ml of 1% (w/v) SDS. Washed starch was boiled in 500 μl of SDS loading buffer, centrifuged at 14 000 g for 15 min, and 45 μl of the supernatant was loaded onto 10% (w/v) discontinuous polyacrylamide gels. Up to 150 μg of bovine serum albumin (BSA) or the E. coli chaperone protein Cpn60 were used as control proteins in the glucan binding assays; none of these control proteins showed binding to starch granules (data not shown).
Zymograms and native PAGE
Zymograms for measuring SS and SBE activity were run according to methods modified previously (Tetlow et al., 2004, 2008). Zymograms were in-gel assays employing native 5% (w/v) polyacrylamide gels in 375 mM TRIS-HCl, pH 8.8, and 10 mg of the α-amylase inhibitor Acarbose (‘Prandase’, Bayer). SS zymograms contained 0.3% (w/v) rabbit liver glycogen (type III, Sigma-Aldrich) as primer in the gel and were incubated for 48–72 h in a buffer containing 50 mM glycylglycine, pH 9.0, 100 mM (NH4)2SO4, 20 mM DTT, 5 mM MgCl2, 0.5 mg ml−1 BSA, and 4 mM ADP-Glc. The native gel for the SBE zymogram contained 0.2% (w/v) maltoheptaose (Sigma-M7755) and 1.4 U of phosphorylase a (from rabbit muscle; Sigma-Aldrich, catalogue no. P-1261), and was incubated in a buffer containing 20 mM MES-NaOH, pH 6.6, 100 mM Na-citrate, 45 mM Glc-1-P, 2.5 mM AMP, 1 mM DTT, 1 mM Na2-EDTA for 2–3 h at 28 °C. SS and SBE zymograms were developed with Lugol’s solution and visualized immediately. Native gels were prepared as for zymograms but with substrates and inhibitors omitted. Following electrophoresis, native gels were electroblotted onto nitrocellulose (see below). Protein samples used for zymograms and native PAGE were mixed with a buffer containing 62.5 mM TRIS-HCl, pH 6.8, 10% (w/v) glycerol, and 0.001% (w/v) bromophenol blue prior to loading into wells.
Denaturing SBE zymograms
Samples prepared for denaturing SBE zymograms were boiled in loading buffer according to the protocols used for SDS–PAGE. Proteins were separated in a gel identical to that used for the native SBE zymogram (above) except for the addition of 1% (w/v) SDS in a running buffer identical to that used for SDS–PAGE. Following electrophoresis, the gel was washed overnight in a renaturing buffer containing 20 mM TRIS-HCl (pH 8.5) and 1 mM DTT. Following this washing step, the gel was incubated for 24 h in the incubation buffer used for native SBE zymograms and developed in Lugol’s solution.
Size exclusion chromatography
Recombinant proteins were separated by size exclusion chromatography in order to separate monomer recombinant SBEs from inactive aggregates prior to experiments. A Superdex 200 10/300GL gel permeation column was connected to an AKTA Explorer FPLC (Amersham Biosciences) at 4 °C. The column was routinely calibrated using commercial protein standards from 13.7 kDa to 669 kDa (Amersham Biosciences). The column was pre-equilibrated with two column volumes of running buffer containing 10 mM HEPES-NaOH, pH 7.5, 100 mM NaCl, 1 mM DTT, and 0.5 mM phenylmethylsulphonyl fluoride (PMSF), at a flow rate of 0.25 ml min−1. The protein samples (∼1 mg ml−1) were loaded onto the column in a final volume of 0.5 ml, and fractions of 0.5 ml were collected.
Preparation and analysis of polyclonal maize antibodies
Polyclonal rabbit antisera targeted to maize SSI, SSIIa, SBEI, SBEIIa, SBEIIb, SP, GBSSI, SSIV, pullulanase, Iso-1, and Iso-2 were raised against synthetic peptides prepared commercially (http://www.anaspec.com/services/antibody.asp). The specific peptide sequences used for SSI, SSIIa, GBSSI, SBEI, SBEIIa, SBEIIb, SP, Iso-1, and Iso-2 were as described in Liu et al. (2009). Peptide sequences for the other antibodies were as follows: SSIV, ANHRNRASIQRDRASASI, corresponding to residues 55–72 of the full-length protein (GenBank accession no. ACC63897); and pullulanase, DEILRSKSLDRDSYDSGDW, corresponding to residues 764–782 of the full-length protein (GenBank accession no. AAD11599.1).
Crude antisera were further purified using peptide affinity columns according to methods described by Liu et al. (2009). Pre-immune sera for each of the antibodies used above were employed as negative controls, and showed no cross-reaction with proteins from plastid lysates and co-immunoprecipitation experiments (data not shown).
Co-immunoprecipitation
Immunoprecipitation and co-immunoprecipitation experiments were conducted using the methods described by Liu et al. (2009). Purified SSI, SSIIa, SBEI, and SBEIIb antibodies (each ∼10 μg) were individually used for the co-immunoprecipitation experiments with both wild-type and amylose extender mutant amyloplast lysates (1 ml, 0.5–1 mg ml−1 proteins). The mixture of antibody and amyloplast lysate was incubated at room temperature on a rotator for 50 min and the immunoprecipitation of the antibody was performed by adding 50 μl of protein A–Sepharose (Sigma-Aldrich) made up as a 50% (w/v) slurry with phosphate-buffered saline (PBS; 137 mM NaCl, 10 mM Na2HPO4, 2.7 mM KCl, 1.8 mM KH2PO4, pH 7.4) at room temperature for 40 min. The protein A–Sepharose–antibody–protein complex was centrifuged at 2000 g for 5 min at 4 °C in a refrigerated micro-centrifuge, and the supernatant discarded. The pellet was washed five times (1.3 ml each) with PBS, followed by washing five times with a buffer containing 10 mM HEPES-NaOH, pH 7.5, and 150 mM NaCl. Washed pellets were boiled in SDS loading buffer and separated by SDS–PAGE, followed by immunoblot analysis (see below). In order to exclude the possibility that the co-immunoprecipitation of the proteins observed in the immunoprecipitation pellet was a result of SS, SBE, or SP binding to the same glucan chain, plastid lysates used for immunoprecipitation were pre-incubated with glucan-degrading enzymes as follows. Wild-type and ae− plastid lysates were incubated with 5 U each of amyloglucosidase (EC 3.2.1.3, Sigma product number A7255, from Rhizopus) and α-amylase (EC 3.2.1.1, Sigma product number A2643, from porcine pancreas) for 20 min at 25 °C. Glucose released as a result of amyloglucosidase/α-amylase digestion of glucans was measured spectrophotometrically using a hexokinase/glucose 6-phosphate dehydrogenase-linked assay as described previously (Tetlow et al., 1994). Control experiments indicated that 0.1 mg of glucan (glycogen, starch, or amylopectin) could be completely digested under these conditions (data not shown).
SDS–PAGE and immunoblotting
Protein samples for SDS–PAGE and immunoblotting were mixed with SDS loading buffer and boiled for 5 min. SDS gels used were either 12% (w/v) acrylamide gels or pre-cast NUPAGE Novex 4–12% BISTRIS acrylamide gradient gels (Invitrogen Canada, catalogue no. NP0335BOX), following the manufacturer’s instructions. Gradient gels were run at room temperature in a MOPS-based running buffer prepared according to the manufacturer’s (Invitrogen) instructions.
For immunoblotting, separated proteins in gels were transblotted onto nitrocellulose membranes (Pall Life Science), and blocked in 1.5% (w/v) BSA for 15 min at room temperature with shaking. The purified maize SSI, SSIIa, SSIII, SBEI, SBEIIa, SBEIIb, pullulanase, and Iso-1 antibodies (mentioned above) were used at a dilution of 1:1000 in 1.5% (w/v) BSA; the purified SP and SSIV antibodies were diluted 1:2000 and 1:500, respectively, in 1.5% (w/v) BSA. Alkaline phosphatase (APase)-conjugated goat anti-rabbit IgG (Sigma) was used as a secondary antibody.
Separation and detection of phosphoproteins using Phos-tag SDS–PAGE
SBEIIb and SBEI proteins with different phosphorylation status were separated by electrophoresis using a general 8% SDS–PAGE system, containing 25 mM Mn2+–Phos-tag (Wako Pure Chemical Industries Ltd, VA, USA) at a constant current of 35 mA per gel for 5 h at room temperature. Following electrophoresis, Mn2+ was removed from the gel by soaking in 1.0 l of transfer buffer (20% of methanol in 25 mM TRIS and 192 mM glycine running buffer) containing 1 mM Na2-EDTA for 10 min with gentle agitation, and followed by further soaking in a general transfer buffer without Na2-EDTA for 10 min with gentle agitation before immunoblotting.
Molecular characterization of SBEIIb sequences
Total mRNA was extracted with an RNeasy Plant Mini Kit (Qiagen) from endosperms of CG102 and ae1.2/CG102 at 22–25 DAP grown in a glasshouse. First-strand cDNA was synthesized by reverse transcription of total RNA using oligo d(T) and a Qiagen kit. The SBEIIb sequences were amplified using primers covering the full-length of coding regions plus parts of the 5′- and 3′-untranslated regions (5′-CCCAGAGCAGACCCGGATTTCGCTC and 3′-GAGAGGACAACGCAGCGAGATGCATG). PCR products were sequenced by the Advanced Analysis Centre at University of Guelph using the above primers and two more primers (GGGATTTATTATGATCCTCCTGAAGAG and GCATACCATGATACCTAAGATAGTCTG). SBEIIb cDNA sequences from different genotypes were analyzed using GeneRunner3.05, and polypeptide sequences were multialigned online (http://mbs.cbrc.jp/papia-cgi/mul_menu.pl).
Morphology of starch granules by scanning electron microscopy (SEM)
The morphology of purified starch granules was observed by SEM (Hiachi Tabletop Microscope TM-1000) based on variable pressure technology combined with the high sensitivity backscattered electron detector (Goldstein et al., 1981).
Measurement of endosperm starch content
The amount of starch present in the endosperms from the maize kernels from the different genotypes was determined as previously described for wheat endosperm (Tetlow et al., 1994).
Estimation of apparent amylose content by iodometry
Apparent amylose content was estimated following the colorimetric method of Morrison and Laignelet (1983) with slight modifications as described by Regina et al. (2004). The analysis was conducted on 2 mg of purified starch. Accurately weighed starch was defatted by incubation in 85% methanol at 65 °C. Following defatting, the dried starch was dissolved in urea dimethylsulphoxide (UDMSO) solution (nine parts of DMSO and one part of 6 M urea). A 50 μl aliquot of starch–UDMSO solution was treated with I2–KI reagent and the absorbance was read at 620 nm. Standard samples containing an amylose content ranging from 0 to 100% were used to generate a standard curve. The absorbance of the test samples was converted into amylose content using a regression equation derived from the standard curve (Morrison and Laignelet, 1983).
Chain length distribution (CLD) analysis by capillary electrophoresis
The CLD of starch was analyzed following the method of O’Shea et al. (1998) using a P/ACE 5510 capillary electrophoresis system (Beckman) with argon-LIF detection. Debranching of native starch and β-amylase-treated starch was performed using Iso (Megazyme, Co., Wicklow, Ireland). Debranched starch was labelled with 8-amino-1,3,6,-pyrenetrisulphonic acid before conducting the capillary electrophoresis.
Isoamylase-debranched reducing end assay
The method used was a modification of the reducing end assay developed by Bernfeld (1955). Iso-debranched starch solution (50 μl) was mixed with 150 μl of dinistrosalicylic acid solution and incubated at 100 °C for 10 min. DNS (3, 5-dinitrosalicylate) solution was prepared by dissolving 5 g of DNS in 100 ml of 2 M NaOH followed by adding di-sodium tartrate solution (150 g dissolved in 250 ml of water) and making up the final volume to 500 ml. After incubation of the starch–DNS solution for 10 min at 100 °C, 180 μl of water was added and the absorbance was measured at 540 nm. A calibration curve was generated using malto-triose solution for a concentration ranging from 0 to 200 nmol. The reducing end of the starch sample was estimated as units of equivalent malto-triose.
Differential scanning calorimetry (DSC)
Thermal properties of starches were analyzed using a differential scanning calorimeter (2920 Modulated DSC; TA Instruments, New Castle, DE, USA) equipped with a refrigerated cooling system for gelatinization and retrogradation of the starches. Samples of starch granules from different genotypes were weighed into high-volume pans. Distilled water was added using a micropipette to produce suspensions with 70% moisture content. Sample weights were ∼11 mg. Pans were sealed and equilibrated overnight at room temperature before heating in the DSC. The measurements were carried out at a heating rate of 100 °C min−1 from 5 °C to 2000 °C. The instrument was calibrated using indium and an empty pan as reference. The enthalpy (ΔH) of phase transitions was measured from the endotherm of DSC thermograms using software (Universal Analysis, Version 2.6D, TA Instruments) based on the mass of dry solid. Transition temperatures (onset, To; peak, Tp; and completion, Tc) of endotherms were also measured from DSC thermograms. The reported values are the means of duplicate measurements.
Estimation of protein content
Protein was determined using the Bio-Rad protein assay (Bio-Rad Laboratories, Canada) according to the manufacturer’s instructions and using thyroglobulin as a standard.
Mass spectrometry
In-gel digestion with trypsin and preparation of peptides for MS were as described previously (Tetlow et al., 2008). Tandem electrospray mass spectra were recorded using a hybrid Q-TOF spectrometer (Micromass) interfaced to a Micromass CapLC capillary chromatograph as previously described (Tetlow et al., 2004).
Results
Biochemical characterization of the amylose extender mutants
Amyloplast stromal proteins
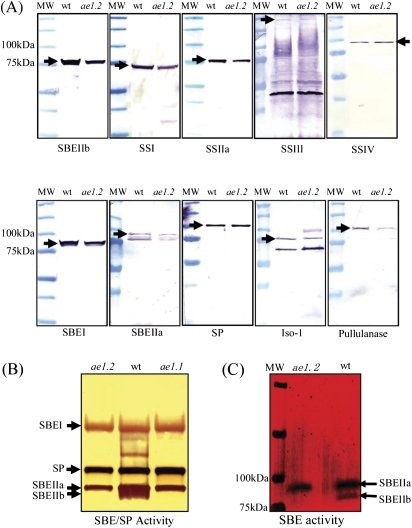
Amyloplasts were isolated from developing maize endosperms at 20–25 DAP, during maximal rates of starch deposition, and used to prepare lysates for immunoblot analysis of enzymes of starch synthesis. Figure 1A shows that all of the major enzymes of starch synthesis, including SBEIIb, are present in the wild-type and the ae1.2 mutant of the same cultivar in comparable amounts as judged by their immunoreactivity with their respective antibodies. Zymogram analyses were performed in order to determine the relative catalytic activities of the different SBE isoforms (given that the ae− phenotype is associated with alterations in SBE activity). Figure 1B shows that amyloplast lysates from ae− endosperm lack a band of activity on the zymogram which corresponds to SBEIIb (based on immunoblots of wild-type material, data not shown). The lack of SBEIIb activity in the mutant was also demonstrated following denaturation and renaturation in SDS-containing gels (Fig, 1C).
Fig. 1.
Immunological characterization of amyloplast lysates from wild-type and ae1.2 maize endosperm. Amyloplast lysates (∼0.8 mg ml−1) were prepared from developing wild-type (wt) and ae− endosperms at 20–25 DAP (individual kernel fresh weight ∼300 mg). Aliquots (12–15 μg) of soluble (stromal) proteins were loaded onto each gel lane and separated on 12% acrylamide gels, electroblotted onto nitrocellulose membranes, and developed with various peptide-specific anti-maize antibodies as shown in (A). Arrows indicate cross-reactions of each of the antibodies with its corresponding target protein. The approximate molecular mass is given for each protein based on its amino acid sequence and, in cases where the migration of the protein on SDS–PAGE does not correspond to its predicted molecular mass based on amino acid sequence, both the mass based on migration in SDS–PAGE and the predicted value based on amino acid sequence are given; SBEIIb (85 kDa), SSI (74 kDa), SSIIa (76 kDa, but with a predicted mass of 85 kDa), SSIII (250 kDa), SSIV (95 kDa), SBEI (80 kDa), SBEIIa (90 kDa), SP (112 kDa), Iso-1 (80 kDa, but with a predicted mass of 90 kDa), and pullulanase (105 kDa). MW, molecular mass markers. (B) Native activity gel (zymogram) analysis of SBE isoforms and SP from whole cell extracts of wild-type and ae1.2 endosperms (20–25 DAP). Approximately 100 μg of soluble proteins per lane were separated on a 7 cm native 5% acrylamide gel containing 0.2% (w/v) maltoheptaose, and then developed for 3 h at 28 °C. SBE and SP activities were visualized by staining with I2–KI. (C) Denaturing SBE zymogram analysis of whole cell extracts from wild-type and ae1.2 endosperms. Samples were prepared as for SDS–PAGE, and the 0.2% (w/v) maltoheptaose-containing gels also included 1% (w/v) SDS.
Fig. 2.
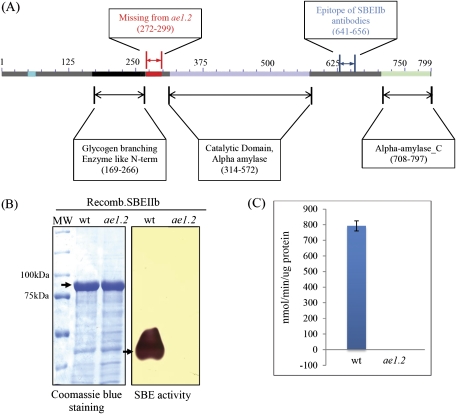
(A) Schematic representation of the catalytically inactive SBEIIb from ae1.2 maize endosperm showing the 28 amino acid region missing in the mutant protein, the epitope recognized by the anti-maize SBEIIb antibodies, and putative glucan-binding and catalytic domains determined from previous structural studies with E. coli branching enzyme (Abad et al., 2002). (B) Measurement of SBE activity in recombinant wild-type and ae1.2 SBEIIb proteins using zymogram analysis; a Coomassie-stained SDS gel of recombinant wild-type and ae1.2 SBEIIb proteins shows equivalent amounts of protein that were loaded onto the zymogram. (C) SBE activity was also determined using the 14C-based phosphorylase a stimulation assay; values given are following subtraction of boiled controls. The recombinant ae1.2 SBEIIb protein was catalytically inactive. (This figure is available in colour at JXB online.)
Amino acid sequence analysis of SBEIIb from ae1.2 endosperm and enzyme characteristics
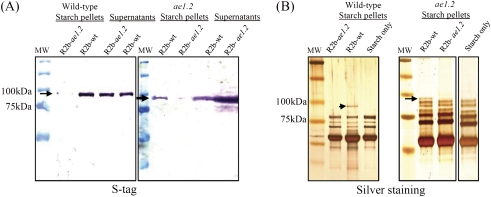
The sequence of the catalytically inactive SBEIIb in ae1.2 was determined following RT-PCR. Structural analysis of SBEIIb from the ae1.2 mutant indicates that the inactive enzyme lacks a region of 28 amino acids (Val272–Pro299 of the full-length sequence) in the N-terminal region of the protein (Fig. 2A). In contrast to the catalytically active wild-type recombinant maize SBEIIb, the mutant SBEIIb produced in E. coli showed no catalytic activity on zymograms, or with the phosphorylase a stimulation assay (Fig.2B and 2C). Despite the 28 amino acid deletion in the mutant SBEIIb, no difference in mobility between the mutant and wild-type SBEIIb was observed when the proteins were separated by SDS–PAGE (Figs 1A, 2B). Since the 28 amino acid deletion in the mutant was proximal to a putative glucan-binding domain (within a region termed the glycogen branching enzyme-like N-terminus; Fig.2A) and an α-amylase-like catalytic domain, the ability of the recombinant mutant protein to bind starch was tested. Glucan binding assays with recombinant wild-type and ae1.2 SBEIIb enzymes show that the mutant protein is unable to bind to glucan (purified maize starch), irrespective of whether the starch was derived from wild-type or ae1.2 endosperm (Fig. 3). Recombinant wild-type SBEIIb was able to bind to both wild-type and ae1.2 starch and was detected using anti-S-tag antibodies (Fig. 3A) and also on silver-stained gels (identified by MS) (Fig. 3B). The data therefore confirm that the ae−genotype expresses a catalytically inactive form of SBEIIb, which is termed ae1.2 to distinguish it from other ae− mutants which lack the SBEIIb protein (ae1.1).
Fig. 3.
Starch binding assays using recombinant wild-type and ae1.2 SBEIIb proteins. Approximately 100 μg of recombinant wild-type SBEIIb (R2b-wt) and recombinant ae1.2 SBEIIb (R2b-ae2) were incubated with 15 mg of purified maize starch (wild-type or ae1.2) for 1 h at 4 °C. Following incubation, starch granules were separated from the incubation buffer by centrifugation and the proteins in the supernatant and starch pellet analyzed by SDS–PAGE and immunoblotting. (A) Immunoblot analysis of washed starch pellet and supernatants of samples incubated with R2b-wt and R2b-ae2 using anti-S-tag antibodies to detect the recombinant proteins. (B) Silver-stained gel of granule-bound proteins in washed starch pellets following incubation with either R2b-wt or R2b-ae2. Wild-type and ae1.2 starch granule proteins extracted from untreated starch are included as controls. Arrows indicate the position of the bound recombinant protein. (This figure is available in colour at JXB online.)
Granule-bound proteins
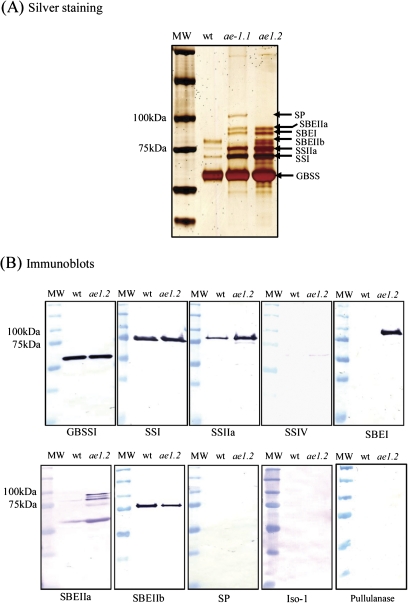
A specific group of proteins consistently remain bound within the starch granule matrix following extensive washing with buffers, acetone, and SDS; these are termed granule-bound proteins (Denyer et al., 1995; Liu et al., 2009) and are distinct from proteins which adhere to the surface of the granules (Rahman et al., 1995). No difference was observed between the granule-bound protein profiles of starches washed using the buffer/acetone/SDS method and the protease treatment method (data not shown). All data presented for granule-bound proteins were obtained from starch samples washed with buffer, acetone, and SDS. The granule-bound proteins of wild-type and ae1.2 endosperms were analyzed by SDS–PAGE and immunoblotting (Fig. 4). The major granule-bound proteins were identified by quadrupole time of flight-MS (Q-TOF-MS) analysis (Fig. 4A, and MS data not shown). Proteins previously identified as being granule-bound were observed in wild-type and ae1.2 starch granules; granule-bound starch synthase (GBSS), SSI, SSIIa, and SBEIIb. In addition to the granule-bound proteins found in wild-type starch granules, ae1.2 granules also contained SBEI and SBEIIa. The catalytically inactive form of SBEIIb was detected on SDS–PAGE as two closely migrating bands, possibly suggesting differential post-translational modification, since SBEIIb is known to be phosphorylated (Tetlow et al., 2004; Grimaud et al., 2008). SP was not detected in ae1.2 granules, although it was detected in ae1.1 starch granules (Liu et al., 2009, and Fig. 4A). Immunoblot experiments confirmed the presence and identity of the proteins observed in silver-stained gels, and also indicated that Iso-1, pullulanase, SSIV, and SP were not present as granule-bound proteins in wild-type or ae1.2 endosperm (Fig. 4B).
Fig. 4.
Analysis of starch granule-bound proteins from wild-type and ae− amyloplasts. Starch granules were isolated from amyloplasts at 20–25 DAP and washed extensively to remove proteins loosely bound to the granule surface. A 40 mg aliquot of purified starch was boiled in 600 μl of SDS loading buffer, and 35 μl of the supernatant from the boiled sample loaded onto gels. (A) Granule-bound proteins from wild-type, ae1.1, and ae1.2 plastids were separated by SDS–PAGE using 4–12% acrylamide gradient gels and visualized by silver staining. Horizontal arrows indicate the major polypeptides which were excised from wild-type and ae− starch granules and identified by Q-TOF-MS analysis; SBEI (found only in ae1.1 and ae1.2), SBEIIb (absent in ae1.1), SSIIa, SSI, SBEIIa and SP (only detected in ae1.1), and GBSSI. MW indicates molecular mass markers with their molecular masses shown on the left of the gel. (B) Starch granule proteins from wild-type and ae1.2 starch were separated by SDS–PAGE and subjected to immunoblot analysis using various peptide-specific anti-maize antibodies. MW indicates molecular mass markers with their molecular masses shown on the left of the blot. (This figure is available in colour at JXB online.)
Starch composition and physicochemical characteristics
Starch from wild-type endosperm was compared with starches extracted from the two allelic variants of amylose extender, ae1.1 (lacking SBEIIb protein) and ae1.2 (expressing an inactive SBEIIb).
Apparent amylose content was determined using a colorimetric assay based on differential iodine binding. The data presented in Table 1 show significant differences in apparent amylose content between the different lines, both amylose extender varieties having significantly higher apparent amylose (P < 0.0005) than the wild-type of ∼25%. There was notably a significant difference (P < 0.0002) between the apparent amylose values of the different amylose extender starches, ae1.1 having an apparent amylose content of 65.6%, whereas ae1.2 starch possessed ∼45% apparent amylose.
Table 1.
Comparison of physicochemical properties of wild-type and amylose extender starches isolated from developing maize endosperm at 20–25 DAP. Values given are means ±SEM of 3–5 biological replicates.
| Genotype | Granule size (μm) | Starch content (%) | Amylose content (%) | Branching frequencya | Gelatinization temperature (°C) |
|||
| Onset | Peak | Completion | ΔH (J g−1) | |||||
| Wild-type | 8.37±1.33 | 41.02±0.83 | 24.9±2.2 | 863.2±71.0 | 68.1±0.1 | 73.3±0.1 | 85.9±0.5 | 15.3±1.5 |
| ae-1.1 | 6.04±0.56 | 38.12±1.22 | 65.6±0.4 | 667.7±37.8 | 67.9±0.1 | 80.6±0.4 | 101.7±0.1 | 14.7±0.1 |
| ae1.2 | 4.32±0.94 | 35.82±0.51 | 49.3±2.2 | 652.8±25.1 | 69.9±0.0 | 78.7±0.0 | 102.7±1.9 | 13.3±0.2 |
nmol malto-triose.
The frequency of α-(1→6) branch linkages in the different starches was estimated by debranching with Iso and measuring the reducing ends created, with the results expressed as malto-triose equivalents (Table 1). The results show a significant reduction in branching of starches of both amylose extender alleles (P < 0.0136) compared with the wild-type starch. However, there was no difference observed in branching frequency between ae1.1 and ae1.2 starches.
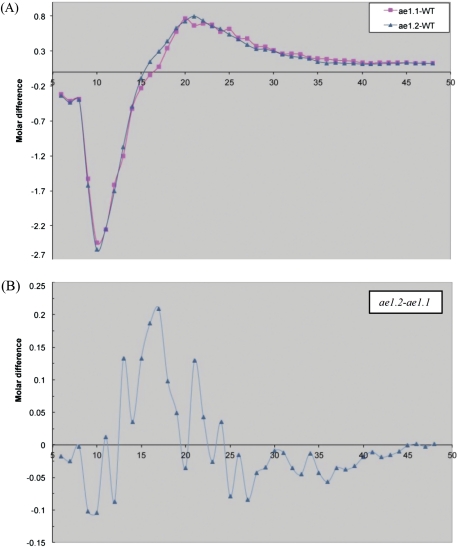
The glucan CLD of starch following de-branching with Iso was determined using fluorophore-assisted carbohydrate electrophoresis (FACE). The results summarized in Fig. 5 show normalized glucan chain distribution of wild-type starch as a reference to ae− starches where wild-type values are subtracted from the ae− values. Results show a reduction in the proportion of chains of DP 9–15, and a significant increase in the proportion of DP16–35 in both ae− mutants compared with the wild-type. Comparative CLD analysis of the two ae− mutants showed a measureable reduction in the proportion of glucan chains of DP 16–20 in ae1.1 starch compared with ae1.2. While small, the same reduction in CLD was observed between the two allelic variants crossed into a second maize genotype (cgr04, data not shown).
Fig. 5.
Glucan chain length distribution (CLD) profiles of debranched wild-type (circles), ae1.1 (squares), and ae1.2 (triangles) starches expressed as mol% difference of ae1.1 and ae1.2 compared with the wild-type. Both ae− starches show reductions in DP 9–15, and significant increases in the proportion of DP 16–35 chains compared with the wild-type. Data presented are representative of three replicate analyses. (This figure is available in colour at JXB online.)
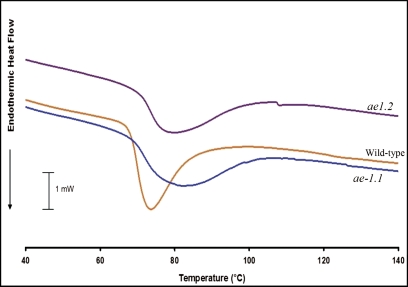
Differential scanning calorimetry
The gelatinization temperature of starch is a measure of granule order, and represents the temperature at which the granule loses internal order following heating in water. The gelatinization temperatures of purified wild-type and ae− starches were analyzed by DSC, and the data are summarized in Fig. 6. Both ae− starches had significantly higher gelatinization temperatures than the wild-type (P < 0.0001 for ae1.2 versus the wild-type). It is notable that ae1.1 starches exhibited a significantly higher gelatinization temperature than the ae1.2 starch (P=0.0012). There was no statistically significant difference between the onset temperatures of gelatinization for the wild-type and ae1.1, but both exhibited significantly lower onset temperatures than ae1.2 starch (P-values of <0.0001 for ae1.2 versus the wild-type and <0.0001 for ae1.2 versus ae1.1).
Fig. 6.
Maize starch thermal properties measured by differential scanning calorimetry (DSC). (This figure is available in colour at JXB online.)
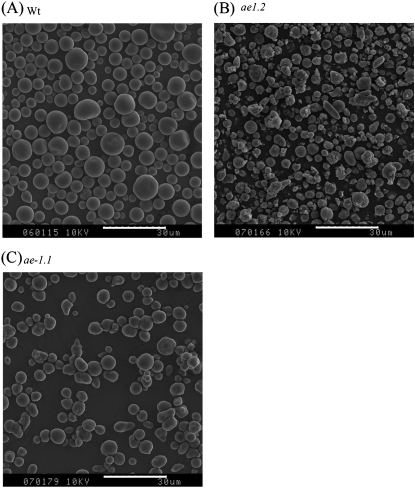
Granule morphology
Starch granule morphology was investigated using SEM (Fig. 7). The starch granules of wild-type starch were of the size and morphology described previously. Starches of both ae− alleles showed a significant reduction in average diameter (P < 0.001, Table 1) and altered morphologies compared with the wild-type, consistent with previous observations (Shannon et al., 2009). ae− starches were less regular in shape compared with the wild-type but, importantly, ae1.2 starches were highly irregular, and smaller than ae1.1 starches (Fig. 7, Table 1).
Fig. 7.
Scanning electron micrographs of purified starch granules from developing maize endosperms (20–25 DAP). (a) Wild-type, (b) ae1.2, and (c) ae1.1. White bars indicating 30 μm are shown on each micrograph.
Co-immunoprecipitation of enzymes of amylopectin synthesis in wild-type and ae1.2 amyloplast lysates
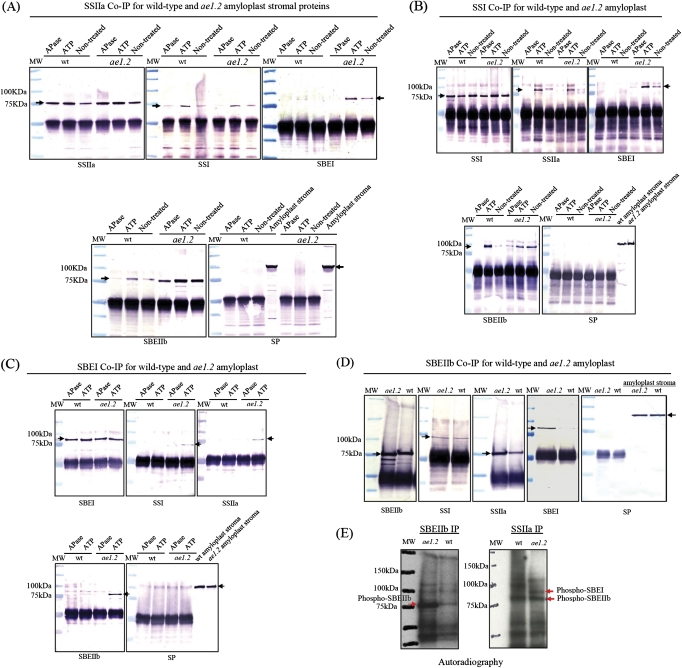
Peptide-specific antibodies were used in reciprocal co-immunoprecipitation experiments to analyse protein–protein interactions among starch-synthesizing enzymes in wild-type and ae1.2 maize amyloplasts at 20–25 DAP. Previous experimental work has shown that protein phosphorylation governs the assembly of protein complexes involving amylopectin-synthesizing enzymes in cereal amyloplasts. In the present study, samples were treated with either 1 mM ATP to promote phosphorylation, or 30 U of E. coli APase to dephosphorylate proteins. All of the antibodies used in co-immunoprecipitation experiments (anti-SSI, anti-SSIIa, SBEIIb, and anti-SBEI) were able to recognize, and precipitate, their respective target protein irrespective of prior treatment with ATP or APase (Fig. 8). Figure 8A and B shows that antibodies to either SSI or SSIIa co-precipitated with each other in all cases, and that co-precipitation of SSI and SSIIa is increased in the presence of ATP, and abolished following dephosphorylation with APase. In both wild-type and ae1.2 maize, SBEIIb was clearly co-immunoprecipated by either SSI or SSIIa antibodies, whereas neither SBEIIa, SBEI nor SP was detected (Fig. 8A, B). Notably, in the ae1.2 mutant, antibodies to either SSI or SSIIa also co-precipitated SBEI in a phosphorylation-dependent manner (Fig. 8A, B). In wild-type and ae1.2 amyloplast lysates, no interactions were observed between SSs, SBEs, and Iso-1, or Iso-2 (data not shown). Anti-SBEI co-immunoprecipitation experiments demonstrated the reciprocal associations predicted from the above (Fig. 8C). In wild-type maize amyloplast extracts, neither SSI, SSIIa, SBEIIb, SBEIIa, nor SP was immunoprecipitated with antibodies to SBEI. In the case of the ae1.2 mutant, antibodies to SBEI were clearly able to co-precipitate SSI, SSIIa, and SBEIIb, but not SP. Addition of a protein phosphatase inhibitor cocktail (Sigma) to ATP-treated plastid lysates caused no increase in co-immunoprecipitation of the interacting proteins (data not shown). Pre-incubation of wild-type and ae1.2 plastid lysates with glucan-degrading enzymes (amyloglucosidase and α-amylase) did not prevent co-immunoprecipitation of SS, SBE, and SP isoforms, indicating that their association in each of the genotypes tested is due to specific protein–protein interactions, and not a result of SSs, SBEs, or SP binding to a common glucan chain.
Fig. 8.
Co-immunoprecipitation of stromal proteins from wild-type (wt) and ae1.2 amyloplasts. Aliquots (0.5–1 ml) of amyloplast lysates (0.8–1 mg protein ml−1) prepared from wild-type and ae1.2 endosperm 20–25 DAP were incubated with peptide-specific anti-SS and anti-SBE antibodies at 25 °C for 50 min, and then immunoprecipitated with protein A–Sepharose. For the analysis of phosphorylation-dependent protein–protein interactions in maize endosperm amyloplasts, wild-type and ae1.2 amyloplasts (20–25 DAP) were pre-incubated with either 1 mM ATP or 30 U of E. coli APase for 30 min at 25 °C. Non-treated samples were incubated in an equivalent volume of rupturing buffer. The washed protein A–Sepharose–antibody–antigen complexes were boiled in 200 μl of SDS loading buffer and 30 μl was loaded onto 4–12% pre-cast polyacrylamide gradient gels, electroblotted onto nitrocellulose, and developed with various anti-maize antisera as shown. Co-immunoprecipitation using (A) anti-SSIIa antibodies, (B) anti-SSI antibodies, (C) anti-SBEI antibodies, and (D) anti-SBEIIb antibodies. Horizontal arrows indicate cross-reactions with the various antisera used: SSI at 74 kDa, SSIIa at 76 kDa, SBEI at 80 kDa, SBEIIa at 90 kDa, SBEIIb at 85 kDa, and SP at 112 kDa. The large band observed at ∼50 kDa in all lanes is due to autorecognition of the IgG heavy chain. MW indicates pre-stained molecular mass markers with molecular masses shown on the left of the immunoblot. (E) Autoradiography of 32P-labelled phosphoproteins immunoprecipitated with anti-maize anti-SBEIIb and anti-SSIIa antibodies from wild-type and ae1.2 amyloplast stroma. Amyloplasts were incubated with 1 mM [γ-32P]ATP for 30 min at 25 °C, and then incubated with anti-maize SBEIIb or SSIIa antibodies as described in the Materials and methods. Arrows shown on the autoradiographs indicate phosphorylated forms of SBEIIb being immunoprecipitated with anti-SBEIIb antibodies in both genotypes, and phosphorylated SBEIIb co-precipitated with anti-SSIIa antibodies. MW indicates the positions of molecular mass markers (shown as bands) with their corresponding masses in kDa. (This figure is available in colour at JXB online.)
Phosphorylation of SBEs
Active SBEIIb in wild-type maize is post-translationally modified by protein phosphorylation (Liu et al., 2009). In order to determine whether the catalytically inactive SBEIIb from ae1.2 endosperm is phosphorylated like the wild-type SBEIIb, wild-type and ae− amyloplast lysates were incubated with 1 mM [γ-32P]ATP, and anti-SBEIIb and anti-SSIIa antibodies, respectively, were used to immunoprecipitate the SBEIIb, and protein complexes containing SBEIIb. Figure 8E shows that following incubation with [γ-32P]ATP, phosphorylated forms of SBEIIb could be immunoprecipitated using anti-SBEIIb antibodies, and that the SBEIIb from ae1.2 amyloplasts appears to be more strongly phosphorylated. In both wild-type and ae1.2 amyloplasts, γ-32P-labelled SBEIIb co-precipitated with SSIIa, indicating that SBEIIb is phosphorylated in both wild-type and mutant protein complexes (Fig. 8E). A number of non-specifically γ-32P-labelled proteins were also observed within the immunocomplexes (Fig. 8E). Previous work suggests that many of these proteins are components of the immunoglobulins and serum proteins, as well as unidentified plastidial phosphoproteins associating with the immunocomplexes or their target proteins. No further analysis or identification of these proteins was undertaken in the present study.
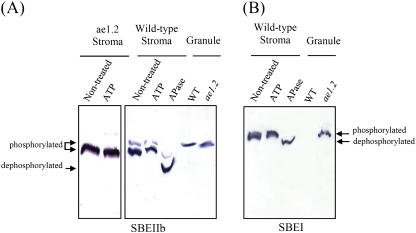
In addition to direct labelling using [γ-32P]ATP, the phosphorylation status of SBEs was also determined by measuring the differential mobility of phosphorylated and dephosphorylated SBEs in polyacrylamide gels containing the Mn2+–Phos-tag ligand (Fig. 9). This technique was particularly useful in determining the phosphorylation status of granule-bound proteins. When proteins are phosphorylated at multiple sites, phosphoproteins show differential retardation in the Mn2+–Phos-tag ligand-containing gel corresponding to the number of phosphorylated residues; for example, 0 (dephosphorylated) showing the greatest mobility, and increased numbers of phosphorylated residues causing greater retardation. Results with the Mn2+–Phos-tag ligand clearly showed that SBEIIb and SBEI isoforms in the amyloplast stroma were retarded relative to the APase-treated control, indicating that these SBE isoforms are phosphorylated in both the wild-type and ae1.2 plastid stroma (Fig. 9A and B, respectively). The non-treated and ATP-treated samples of stromal SBEIIb and SBEI showed two strong bands on immunoblots, both of which exhibited significantly lower mobility than the protein which had prior treatment with APase, suggestive of multiple (at least two) phosphorylation sites. Analysis of the granule-bound proteins from the two genotypes showed that phosphorylated forms of wild-type and ae1.2 SBEIIb were present in isolated starch granules, whereas a phosphorylated form of SBEI was present as a granule-bound protein only in ae1.2 starch granules (Fig. 9B). The data for SBEIIb suggest that this protein may be subject to phosphorylation on more than one site, but that only a multiple phosphorylated form is present in the granule.
Fig. 9.
Detection of phosphorylated forms of SBEs in amyloplast stroma and in starch granules from wild-type and ae1.2 endosperm using an affinity-based ligand (Mn2+–Phos-tag) polyacrylamide electrophoresis. The mobility of specific SBE isoforms was detected using immunoblotting and peptide-specific antisera. Stromal proteins were treated with 1 mM ATP or 30 U of APase for 25 min and 15–20 μg of protein was loaded onto each lane to show the differential mobility of phosphorylated and dephosphorylated forms of SBEs (marked with arrows). The mobility of the granule-bound forms of SBE suggests that they are phosphorylated. (This figure is available in colour at JXB online.)
Discussion
This paper describes an allelic variant of the amylose extender mutation of maize which possesses a catalytically inactive form of the expressed protein, and compares it with both the wild-type and a null mutant of the same gene. Classical Mendelian genetics predict that the loss of function arising from a mutation in a single gene should produce a single distinct phenotype. The present results show clear differences in starch structure and the organization and regulation of amylopectin-synthesizing enzymes between the two ae− mutants, and these are discussed in detail below.
Western blot and zymogram analyses of native and SDS–polyacrylamide gels of soluble proteins from amyloplast lysates show that the ae1.2 mutant used in this study expressed a catalytically inactive form of the SBEIIb protein. This was confirmed by expression of the recombinant mutant protein which was demonstrated to be unable to bind starch and showed no detectable catalytic activity (Figs 2, 3). The protein levels of SSI, SSIIa, Iso-1, Iso-2, pullulanase, plastidial SP, and the other forms of SBE were unaltered compared with the wild-type. Unlike the previously characterized ae1.1 mutant which showed a reduction in measurable SBEI activity (Colleoni et al., 2003; Liu et al., 2009), no changes in SBEI activity in the ae1.2 mutant were apparent from the zymogram (Fig.1B) or the phosphorylase a stimulation assay (data not shown). Other related enzyme activities, determined by zymogram analysis (SBEIIa and SP), showed no change in the ae1.2 mutant when compared with the wild-type, whereas in the ae1.1 mutant SP was shown to decrease in measurable catalytic activity (Liu et al., 2009).
Comparative analysis of the physicochemical properties of ae− starches with the wild-type showed that both ae1.1 and ae1.2 mutants display characteristics typical of other ae− starches analyzed previously. Both ae− variants showed a modest reduction in total endosperm starch content typical of this mutation in other maize backgrounds and in rice (Boyer et al., 1976; Garwood et al., 1976; Mizuno et al., 1993), and also observed in other cereals (Regina et al., 2010). In maize, SBEIIb is responsible for most of the branching of amylopectin in the endosperm, so a reduction in branching frequency (observed in both ae− starches in relation to the wild-type; see Table 1) may be expected to reduce the overall rate of amylopectin biosynthesis through a reduction in the number of available non-reducing termini of glucan chains available for SSs. Indeed, the ‘high-amylose’ starch phenotype of the ae− mutants probably results from a combination of factors. This includes reduced amylopectin synthesis, coupled with unaltered rates of amylose synthesis (no difference was observed in the amount of GBSS protein in ae− mutants and wild-type starches, but amylose synthesis per se was not measured), and also the formation of modified amylopectin with reduced branching frequency, and branches with relatively longer DP (Jane et al., 1999; Klucinec and Thompson, 2002). The increase in apparent amylose content of both ae− starches compared with the wild-type is consistent with this interpretation. Other ‘high-amylose’ phenotypes such as the barley sex6 mutation (resulting from loss of SSIIa activity) are probably caused solely by a reduction in the rate of amylopectin synthesis relative to amylose biosynthesis, making the structural properties of sex6 and ae− ‘high-amylose’ starches quite distinct (Morell et al., 2003). Both maize ae− mutants analyzed showed increased starch gelatinization temperature compared with the wild-type starch (DSC analysis, Table 1), a characteristic associated with ae− starches (Krueger et al., 1987; Fuwa et al., 1999), since the longer amylopectin chains of ae− starches require more energy to disrupt during the gelatinization process. Previous studies have shown that the length of the glucan double helix is correlated to the gelatinization temperature (Gidley and Bulpin, 1987; Cooke and Gidley, 1992; Gidley et al., 1995; Moates et al., 1997; Safford et al., 1998), and more recent studies with ae− rice concluded that the gelatinization temperature of rice starch granules is determined by the proportion of short to long glucan chains within amylopectin clusters (Nishi et al., 2001; Tanaka et al., 2004).
An important component of this work was to determine the effect of expression of a non-catalytic protein (SBEIIb) on the ability to form protein complexes, and the consequences for the biosynthesis and structure of starch. There is now abundant evidence for the operation of functional protein complexes between specific enzymes of amylopectin synthesis during starch biosynthesis in cereal endosperms (Tetlow et al., 2004, 2008; Hennen-Bierwagen et al., 2008). Associations between SS and SBE forms in wheat and maize endosperms have been shown to be phosphorylation dependent (Tetlow et al., 2008; Hennen-Bierwagen et al., 2009; Liu et al., 2009), and the SBEII forms within these protein complexes are phosphorylated at one or more serine residues (Tetlow et al., 2004, 2008; Liu et al., 2009). It has been postulated that a trimeric protein complex containing SSI, SSIIa, and SBEII is involved in the assembly of the short and intermediate sized glucan clusters of amylopectin at the surface of the granule, and that these proteins become entrapped and/or tightly bound to the amylopectin as the granule grows, eventually becoming the granule-bound proteins (Liu et al., 2009). There is strong experimental evidence supporting the idea that the granule-bound amylopectin-synthesizing proteins are the components of stromal protein complexes. In the ae1.1 mutant of maize, a novel protein complex (or complexes) is formed in which SBEI, SBEIIa, and SP interact with SSI and SSIIa and appear to substitute for the loss of the SBEIIb protein. Importantly, it was determined that the soluble proteins present in the new complex(es) are also found as granule-bound proteins (Liu et al., 2009). In the present study, analysis of the protein–protein interactions and granule-bound proteins occurring in ae1.2 amyloplasts supports the idea that the amylopectin-synthesizing proteins found in starch granules are a result of their physical association in one or more protein complexes (see Table 2). The catalytically inactive form of SBEIIb in the ae1.2 mutant was found to be associated with SSI, SSIIa, and SBEI, and all of these enzymes were found as granule-bound proteins in ae1.2 starch (Fig. 4). The SBEIIb expressed in the ae1.2 mutant lacks a peptide of 28 amino acids which is close to a putative glucan-binding domain (see Fig. 2A), although not affecting the four conserved catalytic domains associated with branching enzymes (Kuriki et al., 1996, 1997). The deletion clearly affects the interaction of the protein with glucans, as the mutant protein was unable to bind to starch or amylopectin, unlike its wild-type counterpart (Fig. 3), thus indirectly affecting the enzyme’s catalytic capacity. Given the mutant SBEIIb’s inability to bind glucan (Fig. 3), it must be concluded that its association with ae1.2 starch granules in vivo is mediated through interaction with other proteins in a complex whose other components have strong glucan binding affinity.
Table 2.
Summary of possible protein complexes and their relationship to granule-bound proteins in different maize genotypes
| Genotype | Possible protein complexes | Starch granule-bound proteins |
| Wild-type | SSI, SSIIa, SBEIIb | GBSSI |
| SBEIa, SBEIIb | SSI, SSIIa, SBEIIb | |
| ae-1.1 | SSI, SSIIa, SBEI, SBEIIa, SP | GBSSI |
| SSI, SSIIa, SBEI | SSI, SSIIa, SBEI, SBEIIa | |
| SSI, SSIIa, SBEI, SBEIIa | SP | |
| SSI, SSIIa, SBEI, SP | ||
| SSI, SSIIa, SBEIIa | ||
| SSI, SSIIa, SBEIIa, SP | ||
| SBEI, SBEIIa, SP | ||
| ae1.2 | SSI, SSIIa, SBEI, SBEIIa, SBEIIb (inactive) | GBSSI SSI, SSIIa, SBEI, SBEIIa |
| SSI, SSIIa, SBEI, SBEIIb (inactive) | SBEIIb (inactive) | |
| SSI, SSIIa, SBEI, SBEIIa | ||
| SSI, SSIIa, SBEIIb (inactive) | ||
| SSI, SSIIa, SBEIIa | ||
| SSI, SSIIa, SBEI | ||
| SBEI, SBEIIb |
SBEI–SBEIIb protein complex observed by Liu et al. (2009).
Since both ae− mutants lack SBEIIb activity, it is argued that differences in starch phenotype between the two mutants are explained by alterations in protein–protein interactions between enzymes of amylopectin synthesis. Both ae− mutants recruit SBEI into one or more protein complexes with SS isoforms, as well as small amounts of SBEIIa [a minor SBE in maize endosperm, mutation of which produces no phenotype in this tissue (Blauth et al., 2001)]. It has been argued that the presence of SBEI in the protein complex with SS isoforms contributes to the observed ae− starch phenotype (long glucan chains in clusters and relatively lower branching frequency within clusters) as SBEI has a low branching frequency with amylopectin, and preferentially branches longer glucan chains (Guan and Preiss, 1993; Takeda et al., 1993). The properties of ae1.2 starch are noticeably different from those of ae1.1; in particular, a smaller granule size with altered morphology, a much lower proportion of apparent amylose (intermediate between the wild-type and ae1.1, resulting in a lower gelatinization temperature compared with ae1.1 starch), and a higher proportion of DP 16–20 chains.
Previous studies have shown that interactions between SSs and SBEs in cereal amyloplasts are controlled by protein phosphorylation (Tetlow et al., 2004, 2008; Hennen-Bierwagen et al., 2009; Liu et al., 2009). Work with the ae1.1 mutant by Liu et al. (2009) showed that the stability of the protein–protein interactions observed in this mutant was unaffected by pre-treatment with either ATP or phosphatase. Interestingly, assembly and disassembly of the novel protein complex(es) formed in the ae1.2 mutant were clearly shown to be dependent on phosphorylation, in common with the wild-type complexes. The observable differences between the protein complexes found in the two ae− mutants are the presence of the catalytically inactive SBEIIb in the ae1.2 amyloplasts, and the presence of SP in the enzyme complexes observed in ae1.1, which was not detected in the ae1.2 complex(es). Presently it has not been possible to determine whether the presence of SBEIIb (active or inactive) or the absence of SP in the protein complex is the basis for a requirement for the control of complex assembly by protein phosphorylation. In both the wild-type and ae1.2, the SBEIIb present in the protein complex(es) was clearly shown to be phosphorylated, and phosphorylated forms of SBEIIb were also detected in isolated starch granules (Figs 8E, 9).
It would appear that SBEIIb, whether catalytically active or not, is phosphorylated at one or more serine residues, and is required for phosphorylation-dependent assembly of SS-containing protein complexes. Whether regulation of complex formation by protein phosphorylation in itself dictates glucan structure is an open question. It is more likely, however, that the presence of phosphorylated SBEIIb governs the regulation of protein complex assembly, and that this somehow prevents association of SP with the resulting complex. The lack of SP in the ae1.2 complex (in common with the wild-type SSI, SSIIa, and SBEIIb trimeric assembly) is associated with alterations in glucan structure of ae1.2 (see above). In addition, the finding of a phosphorylated form of the catalytically inactive SBEIIb in ae1.2, and which lacks glucan binding capability, is strong evidence of its association with other proteins within a protein complex prior to entrapment within the starch granule.
It was previously speculated that if SP plays a catalytic role in the protein complex found in ae1.1, then it would be in modifying glucan chain length appropriate for catalysis by SBEI and SBEIIa, and maintaining constant cluster size (Liu et al., 2009). If the loss of SP from the protein complex in ae1.2 is impacting starch structure, it may be through a reduction of longer glucan chains; that is, branched chains are not being elongated further by SP, thus contributing to a reduced amylose:amylopectin ratio and gelatinization temperature with respect to ae1.1. The mode of catalytic action by SP in the plastid, either as the free enzyme (dimer) or in a complex with other proteins, is not known, but operation in a predominantly glucan-synthetic direction in cereal amyloplasts appears likely. Much of the carbon for starch synthesis in cereal endosperm amyloplasts derives from the import of ADP-Glc produced by a cytosolic ADP-Glc pyrophosphorylase (Thorbjørnsen et al., 1996; Beckles et al., 2001; Tetlow et al., 2003; Bowsher et al., 2007) meaning that no plastidial Pi is generated as a result of starch biosynthesis via this route. Data from amyloplasts of wheat endosperm suggest that even during conditions promoting starch biosynthesis mediated via hexose phosphates, plastidial Pi levels remain low (Tetlow et al., 1998). Such conditions within the plastid would be expected to promote the glucan-synthetic activity of SP, assuming no other factors influence reaction equilibrium. The experimental techniques used to identify protein–protein interactions in this study do not allow the determination of the precise stoichiometry of the protein complexes. Consequently, it is not known whether the presence of the catalytically inactive SBEIIb prevents association of other proteins, for example SP.
In summary, this study has shown that two allelic maize mutants lacking SBEIIb activity display distinct starch phenotypes, and altered regulatory properties of the amylopectin-synthesizing machinery, strongly supporting the case for the involvement of multienzyme protein complexes in defining amylopectin structure. Analysis of the starch granule-bound proteins in the ae1.2 mutant support earlier work indicating that amylopectin-synthesizing enzymes found as granule-bound proteins are present as a result of their associations in multienzyme protein complexes working at the periphery of the growing starch granule. That two mutants, both lacking activity of the major form of SBEII, show such distinct biochemical properties and starch structure from each other is significant, and indicative of pleiotropic effects, which it is argued are mediated through different protein–protein interactions in the amyloplasts of the two ae− mutants. The corollary of these observations is that the specific organization of protein complex assemblies in amyloplasts is responsible for modulating the fine structure of amylopectin.
Acknowledgments
This work was supported by the Ontario Ministry of Agriculture and Food Bio-Products Research Grants (project no. 026262 to MJE and IJT, and project no. 200172 to IJT), Natural Sciences and Engineering Research Council (NSERC) Discovery Grants (no. 262209 to MJE and no. 400167 to IJT), a Canada Foundation for Innovation Leader's Opportunity Fund grant (no. 12949 to IJT), and an NSERC Strategic Grant (no. 048237 to MJE and IJT). The authors gratefully acknowledge Drs Alan Myers and Martha James (Iowa State University, Ames, Iowa, USA) for providing cDNAs for the mature sequences of maize SSI, SBEI, SBEIIa, and SBEIIb. We thank Dr Dyanne Brewer (University of Guelph Advanced Analysis Center) for Q-TOF-MS analyses.
Glossary
Abbreviations
- ADP-Glc
ADP-glucose
- ae−
amylose extender
- APase
alkaline phosphatase
- BSA
bovine serum albumin
- CLD
chain length distribution
- DAP
days after pollination
- DBE
debranching enzyme
- DP
degree of polymerization
- DTT
dithiothreitol
- GBSSI
granule-bound starch synthase
- Glc1P
α-D-glucose 1-phosphate
- GT
gelatinization temperature
- Iso
isoamylase
- MS
mass spectrometry
- Pi
inorganic orthophosphate
- PMSF
phenylmethylsulphonyl fluoride
- SBE
starch branching enzyme
- SDS-PAGE
sodium dodecyl sulphate-polyacrylamide gel electrophoresis
- SP
starch phosphorylase
- SS
starch synthase
- UDMSO
urea dimethylsulphoxide
References
- Abad M, Binderup K, Rios-Steiner K, Arni R, Preiss J, Geiger J. The X-ray crystallographic structure of Escherichia coli branching enzyme. Journal of Biological Chemistry. 2002;277:42164–42170. doi: 10.1074/jbc.M205746200. [DOI] [PubMed] [Google Scholar]
- Ball SG, Morell MK. From bacterial glycogen to starch: understanding the biogenesis of the plant starch granule. Annual Review of Plant Biology. 2003;54:207–233. doi: 10.1146/annurev.arplant.54.031902.134927. [DOI] [PubMed] [Google Scholar]
- Banks W, Greenwood CT, Muir DD. Studies on starches of high amylose content: part 17. A review of current concepts. Starch. 1974;26:289–300. [Google Scholar]
- Beckles DM, Smith AM, ap Rees T. A cytosolic ADP-glucose pyrophosphorylase is a feature of graminaceous endosperms, but not of other starch storing organs. Plant Physiology. 2001;125:818–827. doi: 10.1104/pp.125.2.818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernfeld P. Amylases, alpha and beta. Methods in Enzymology. 1955;1:149–158. [Google Scholar]
- Blauth SL, Yao Y, Klucinec JD, Shannon JC, Thompson DB, Guiltinan M. Identification of Mutator insertional mutants of starch-branching enzyme 2a in corn. Plant Physiology. 2001;125:1396–1405. doi: 10.1104/pp.125.3.1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowsher CG, Scrase-Field EFAL, Esposito S, Emes MJ, Tetlow IJ. Characterization of ADP-glucose transport across the cereal endosperm amyloplast envelope. Journal of Experimental Botany. 2008;58:1321–1332. doi: 10.1093/jxb/erl297. [DOI] [PubMed] [Google Scholar]
- Boyer CD, Daniels RR, Shannon JC. Abnormal starch granule formation in Zea mays L. endosperms possessing the amylose-extender mutant. Crop Science. 1976;16:298–301. [Google Scholar]
- Butardo VM, Fitzgerald MA, Bird AR, et al. Impact of down-regulation of starch branching enzyme IIb in rice by artificial microRNA and hairpin RNA-mediated RNA silencing. Journal of Experimental Botany. 2011 doi: 10.1093/jxb/err188. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang T, Su JC. Starch phosphorylase inhibitor from sweet potato. Plant Physiology. 1986;80:534–538. doi: 10.1104/pp.80.2.534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colleoni C, Myers AM, James MG. One- and two-dimensional native PAGE activity gel analyses of maize endosperm proteins reveal functional interactions between specific starch metabolizing enzymes. Journal of Applied Glycoscience. 2003;50:207–212. [Google Scholar]
- Cooke D, Gidley MJ. Loss of crystalline and molecular order during starch gelatinisation: origin of the enthalpic transition. Carbohydrate Research. 1992;227:103–112. [Google Scholar]
- Denyer K, Hylton CM, Jenner CF, Smith AM. Identification of multiple isoforms of soluble and granule-bound starch synthase in developing wheat endosperm. Planta. 1995;196:256–265. [Google Scholar]
- French D. Organisation of starch granules. In: Whistler RL, BeMiller JN, Paschall EF, editors. Starch: chemistry and technology. Orlando, FL: Academic Press; 1984. pp. 183–237. [Google Scholar]
- Fujita N, Toyosawa Y, Utsumi Y, et al. Characterization of pullulanase (PUL)-deficient mutants of rice (Oryza sativa L.) and the function of PUL on starch biosynthesis in the developing rice endosperm. Journal of Experimental Botany. 2009;60:1009–1023. doi: 10.1093/jxb/ern349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuwa H, Glover DV, Fujita S, Sugimoto Y, Inouchi N, Sugihara M, Yoshioka S, Yamada K. Structural and physicochemical properties of endosperm starches possessing different alleles at the amylose extender and waxy locus in maize (Zea mays L.) Starch. 1999;51:147–151. [Google Scholar]
- Gao M, Fisher DK, Kim K-N, Boyer CD, Guiltinan MJ. Independent genetic control of maize starch-branching enzymes IIa and IIb: isolation and characterization of Sbe2a cDNA. Plant Physiology. 1997;114:69–78. doi: 10.1104/pp.114.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garwood DL, Shannon JC, Creech RG. Starches of endosperms possessing different alleles of the amylose-extender locus in Zea mays. Cereal Chemistry. 1976;53:355–364. [Google Scholar]
- Gérard C, Colonna P, Buléon A, Planchot V. Order in maize mutant starches revealed by mild acid hydrolysis. Carbohydrate Polymers. 2002;48:131–141. [Google Scholar]
- Gidley MJ, Bulpin PV. Crystallisation of malto-oligosaccharides as models of the crystalline forms of starch: minimum chain-length requirement for the formation of double helices. Carbohydrate Research. 1987;161:291–300. [Google Scholar]
- Gidley MJ, Cooke D, Darke AH, Hoffmann RA, Russell AL, Greenwell P. Molecular order and structure in enzyme-resistant retrograded starch. Carbohydrate Polymers. 1995;28:23–31. [Google Scholar]
- Goldstein JI, Newbury DE, Echlin P, Joy DC, Fiori C, Lifshin E. Scanning electron microscopy and X-ray microanalysis. New York: Plenum Press; 1981. [Google Scholar]
- Grimaud F, Rogniaux H, James MG, Myers AM, Planchot V. Proteome and phosphoproteome analysis of starch granule-associated proteins from normal maize and mutants affected in starch biosynthesis. Journal of Experimental Botany. 2008;59:3395–3406. doi: 10.1093/jxb/ern198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan H-P, Preiss J. Differentiation of the properties of the branching isozymes from maize (Zea mays) Plant Physiology. 1993;102:1269–1273. doi: 10.1104/pp.102.4.1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedman KD, Boyer CD. Gene dosage at the Amylose-Extender locus of maize: effects on the levels of starch branching enzymes. Biochemical Genetics. 1982;20:483–492. doi: 10.1007/BF00484699. [DOI] [PubMed] [Google Scholar]
- Hedman KD, Boyer CD. Allelic studies of the Amylose-Extender locus of Zea mays L.: levels of the starch branching enzymes. Biochemical Genetics. 1983;21:1217–1222. doi: 10.1007/BF00488473. [DOI] [PubMed] [Google Scholar]
- Hennen-Bierwagen TA, Lin Q, Grimaud F, Planchot V, Keeling PL, James MG, Myers AM. Proteins from multiple metabolic pathways associate with starch biosynthetic enzymes in high molecular weight complexes: a model for regulation of carbon allocation in maize amyloplasts. Plant Physiology. 2009;149:1541–1559. doi: 10.1104/pp.109.135293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennen-Bierwagen TA, Liu F, Marsh R, Kim S, Gan Q, Tetlow IJ, Emes MJ, James MG, Myers AM. Multiple starch biosynthetic enzymes from developing Zea mays endosperm associate in multisubunit complexes. Plant Physiology. 2008;146:1892–1908. doi: 10.1104/pp.108.116285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilbert GE, MacMasters MM. Pea starch, a starch of high amylose content. Journal of Biological Chemistry. 1946;162:229–238. [PubMed] [Google Scholar]
- Hizukuri S, Fuji M, Nikuni Z. The effect of inorganic ions on the crystallization of amylodextrin. Biochimica et Biophysica Acta. 1960;40:346–348. doi: 10.1016/0006-3002(60)91360-3. [DOI] [PubMed] [Google Scholar]
- Hizukuri S, Kaneko T, Takeda Y. Measurement of the chain length of AMP and its relevance to the origin of crystalline polymorphism of starch granules. Biochimica et Biophysica Acta. 1983;760:188–191. [Google Scholar]
- Hizukuri S, Takeda Y, Usami S, Takase Y. Effect of aliphatic hydrocarbon groups on the crystallization of amylodextrin: model experiments for starch crystallization. Carbohydrate Research. 1980;83:193–199. [Google Scholar]
- James MG, Robertson DS, Myers AM. Characterization of the maize gene sugary1, a determinant of starch composition in kernels. The Plant Cell. 1995;7:417–429. doi: 10.1105/tpc.7.4.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jane J, Chen YY, Lee LF, McPherson AE, Wong KS, Radoslavljevic M, Kasemsuwan T. Effects of amylopectin branch chain length and amylose content on the gelatinisation and pasting properties of starch. Cereal Chemistry. 1999;76:629–637. [Google Scholar]
- Katz JR. Über die anderungen in Rontgenspektrum der stärke brim baker and beam altbacken warden des brutes. Zeitschrift fur Physikalische Chemie. 1930;150:37–59. [Google Scholar]
- Kubo A, Yuguchi Y, Takeniasa M, Susuki S, Satoh H, Kitamura S. The use of micro-beam X-ray diffraction for the characterization of starch crystal structure in rice mutant kernels of waxy, amylose extender, and sugary1. Journal of Cereal Science. 2008;48:92–97. [Google Scholar]
- Kuriki T, Guan HP, Sivak M, Preiss J. Analysis of the active center of branching enzyme II from maize endosperm. Journal of Protein Chemistry. 1996;15:305–313. doi: 10.1007/BF01887119. [DOI] [PubMed] [Google Scholar]
- Kuriki T, Stewart DC, Preiss J. Construction of chimeric enzymes out of maize endosperm branching enzymes I and II: activity and properties. Journal of Biological Chemistry. 1997;272:28999–29004. doi: 10.1074/jbc.272.46.28999. [DOI] [PubMed] [Google Scholar]
- Klucinec JD, Thompson DB. Structure of amylopectins from ae-containing maize starches. Cereal Chemistry. 2002;79:19–23. [Google Scholar]
- Krueger BR, Walker CE, Knutson CA, Inglett GE. Differential scanning calorimetry of raw and annealed starch isolated from normal and mutant maize genotypes. Cereal Chemistry. 1987;64:187–190. [Google Scholar]
- Liu F, Makhmoudova A, Lee EA, Wait R, Emes MJ, Tetlow IJ. The amylose extender mutant of maize conditions novel protein–protein interactions between starch biosynthetic enzymes in amyloplasts. Journal of Experimental Botany. 2009;60:4423–4440. doi: 10.1093/jxb/erp297. [DOI] [PubMed] [Google Scholar]
- Mizuno K, Kawasaki K, Shimada H, Satoh H, Kobayashi E, Okumura S, Arai Y, Baba T. Alteration of the structural properties of starch components by the lack of an isoform of starch branching enzyme in rice seeds. Journal of Biological Chemistry. 1993;268:19084–19091. [PubMed] [Google Scholar]
- Moates GK, Noel TR, Parker R, Ring SG. The effect of chain length and solvent interactions on the dissolution of the B-type crystalline polymorph of amylose in water. Carbohydrate Research. 1997;298:327–333. [Google Scholar]
- Morell MK, Kosar-Hashemi B, Cmiel M, Samuel MS, Chandler P, Rahman S, Buleon A, Batey IL, Li Z. Barley sex6 mutants lack starch synthase IIa activity and contain a starch with novel properties. The Plant Journal. 2003;34:173–185. doi: 10.1046/j.1365-313x.2003.01712.x. [DOI] [PubMed] [Google Scholar]
- Morris DL, Morris CT. Glycogen in the seed of Zea mays. Journal of Biological Chemistry. 1939;130:535–544. [Google Scholar]
- Morrison WR, Laignelet B. An improved colorimetric procedure for determining apparent and total amylose in cereal and other starches. Journal of Cereal Science. 1983;1:9–20. [Google Scholar]
- Mouille G, Maddelein M-L, Libessart N, Talaga P, Decq A, Delrue B, Ball S. Phytoglycogen processing: a mandatory step for starch biosynthesis in plants. The Plant Cell. 1996;8:1353–1366. doi: 10.1105/tpc.8.8.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishi A, Nakamura Y, Tanaka N, Satoh H. Biochemical and genetic effects of amylose-extender mutation in rice endosperm. Plant Physiology. 2001;127:459–472. [PMC free article] [PubMed] [Google Scholar]
- O’Shea MG, Samuel MS, Konik CM, Morell MK. Fluorophore assisted carbohydrate electrophoresis (FACE) of oligosaccharides: efficiency of labelling and high-resolution separation. Carbohydrate Research. 1998;307:1–12. [Google Scholar]
- Rahman S, Kosar-Hashemi B, Samuel MS, Hill A, Abbott DC, Skerritt JH, Preiss J, Appels R, Morell MK. The major proteins of wheat endosperm starch granules. Australian Journal of Plant Physiology. 1995;22:793–803. [Google Scholar]
- Regina A, Bird A, Topping D, Bowden S, Freeman J, Barsby T, Kosar-Hashemi B, Li ZY, Rahman S, Morell MK. High-amylose wheat generated by RNA interference improves indices of large-bowel health in rats. Proceedings of the National Academy of Sciences, USA. 2006;103:3546–3551. doi: 10.1073/pnas.0510737103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regina A, Kosar-Hashemi B, Li Z, Rahman S, Morell MK. Control of starch branching in barley defined through differential RNAi suppression of starch branching enzyme IIa and IIb. Journal of Experimental Botany. 2010;61:1469–1482. doi: 10.1093/jxb/erq011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regina A, Kosar-Hashemi B, Li Z, Rampling L, Cmiel M, Gianibelli MC, Konik-Rose C, Larroque O, Rahman S, Morell MK. Multiple isoforms of starch branching enzyme-1 in wheat: lack of the major SBE-I isoform does not alter starch phenotype. Functional Plant Biology. 2004;31:591–601. doi: 10.1071/FP03193. [DOI] [PubMed] [Google Scholar]
- Rydberg U, Andersson L, Andersson R, Åman P, Larsson H. Comparison of starch branching enzyme I and II from potato. European Journal of Biochemistry. 2001;268:6140–6145. doi: 10.1046/j.0014-2956.2001.02568.x. [DOI] [PubMed] [Google Scholar]
- Safford R, Jobling SA, Sidebottom CM, Westcott RJ, Cooke D, Tober KJ, Strongitharm BH, Russel AL, Gidley MJ. Consequences of antisense RNA inhibition of starch branching enzyme activity on properties of potato starch. Carbohydrate Polymers. 1997;35:155–168. [Google Scholar]
- Saibene D, Seetharaman K. Segmental mobility of polymers in starch granules at low moisture contents. Carbohydrate Polymers. 2011 (in press) [Google Scholar]
- Shannon JC, Garwood DL, Boyer CD. Genetics and physiology of starch development. In: James B, Roy W, editors. Starch. 3rd edn. San Diego: Academic Press; 2009. pp. 23–82. [Google Scholar]
- Smith AM. Enzymes of starch synthesis. Methods in Plant Biochemistry. 1990;3:93–102. [Google Scholar]
- Stinard PS, Robertson DS, Schnable PS. Genetic isolation, cloning, and analysis of a mutator-induced, dominant antimorph of the maize amylose extender1 locus. The Plant Cell. 1993;5:1555–1566. doi: 10.1105/tpc.5.11.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C, Sathish P, Ahlandsberg S, Jansson C. The two genes encoding starch-branching enzymes IIa and IIb are differentially expressed in barley. Plant Physiology. 1998;118:37–49. doi: 10.1104/pp.118.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda Y, Guan H-P, Preiss J. Branching of amylose by the branching isoenzymes of maize endosperm. Carbohydrate Research. 1993;240:253–263. [Google Scholar]
- Tanaka N, Fujita N, Nishi A, Satoh H, Hosaka Y, Ugaki M, Kawasaki S, Nakamura Y. The structure of starch can be manipulated by changing the expression levels of starch branching enzyme IIb in rice endosperm. Plant Biotechnology Journal. 2004;2:507–516. doi: 10.1111/j.1467-7652.2004.00097.x. [DOI] [PubMed] [Google Scholar]
- Tetlow IJ. Starch biosynthesis in developing seeds. Seed Science Research. 2011;21:5–32. [Google Scholar]
- Tetlow IJ, Beisel KG, Cameron S, Makhmoudova A, Liu F, Bresolin NS, Wait R, Morell MK, Emes MJ. Analysis of protein complexes in amyloplasts reveals functional interactions among starch biosynthetic enzymes. Plant Physiology. 2008;146:1878–1891. doi: 10.1104/pp.108.116244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tetlow IJ, Blissett KJ, Emes MJ. Metabolite pools during starch synthesis and carbohydrate oxidation in amyloplasts isolated from wheat endosperm. Planta. 1998;204:100–108. [Google Scholar]
- Tetlow IJ, Blissett KJ, Emes MJ. Starch synthesis and carbohydrate oxidation in amyloplasts from developing wheat endosperm. Planta. 1994;194:454–460. [Google Scholar]
- Tetlow IJ, Davies EJ, Vardy KA, Bowsher CG, Burrell MM, Emes MJ. Subcellular localization of ADPglucose pyrophosphorylase in developing wheat endosperm and analysis of a plastidial isoform. Journal of Experimental Botany. 2003;54:715–725. doi: 10.1093/jxb/erg088. [DOI] [PubMed] [Google Scholar]
- Tetlow IJ, Wait R, Lu Z, Akkasaeng R, Bowsher CG, Esposito S, Kosar-Hashemi B, Morell MK, Emes MJ. Protein phosphorylation in amyloplasts regulates starch branching enzyme activity and protein–protein interactions. The Plant Cell. 2004;16:694–708. doi: 10.1105/tpc.017400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorbjørnsen T, Villand P, Denyer K, Olsen OA, Smith AM. Distinct isoforms of ADPglucose pyrophosphorylase occur inside and outside the amyloplasts in barley endosperm. The Plant Journal. 1996;10:243–250. [Google Scholar]
- Wattebled F, Dong Y, Dumez S, et al. Mutants of Arabidopsis lacking a chloroplastic isoamylase accumulate phytoglycogen and an abnormal form of amylopectin. Plant Physiology. 2005;138:184–195. doi: 10.1104/pp.105.059295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wattebled F, Planchot V, Dong Y, Szydlowski N, Pontoire B, Devin A, Ball S, D’Hulst C. Further evidence for the mandatory nature of polysaccharide debranching for the aggregation of semicrystalline starch and for overlapping functions of debranching enzymes in Arabidopsis leaves. Plant Physiology. 2008;148:1309–1323. doi: 10.1104/pp.108.129379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H-CH, Sarko A. The double-helical molecular structure of crystalline A-amylose. Carbohydrate Research. 1978;61:27–40. [Google Scholar]
- Yao Y, Thompson DB, Guiltinan MJ. Maize starch-branching enzyme isoforms and amylopectin structure. In the absence of starch-branching enzyme IIb, the further absence of starch-branching enzyme Ia leads to increased branching. Plant Physiology. 2004;136:3515–3523. doi: 10.1104/pp.104.043315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeeman SC, Umemoto T, Lue WL, Au-Yeung P, Martin C, Smith AM, Chen J. A mutant of Arabidopsis lacking a chloroplastic isoamylase accumulates both starch and phytoglycogen. The Plant Cell. 1998;10:1699–1712. doi: 10.1105/tpc.10.10.1699. [DOI] [PMC free article] [PubMed] [Google Scholar]