Abstract
The molecular mechanisms governing PEPC expression in maize remain to be fully defined. Differential methylation of a region in the PEPC promoter has been shown to correlate with transcript accumulation, however, to date, investigations into the role of DNA methylation in maize PEPC expression have relied on the use of methylation-sensitive restriction enzymes. Bisulphite sequencing was used here to provide a single-base resolution methylation map of the maize PEPC promoter. It is shown that four cytosine residues in the PEPC promoter are heavily methylated in maize root tissue. In leaves, de-methylation of these cytosines is dependent on illumination and is coincident with elevated PEPC expression. Furthermore, light-regulated de-methylation of these cytosines occurs only in mesophyll cells. No methylation was discovered in the 0.6 kb promoter required for mesophyll-specific expression indicating that cytosine methylation is not required to direct the cell-specificity of PEPC expression. This raises interesting questions regarding the function of the cell-specific cytosine de-methylation observed in the upstream region of the PEPC promoter.
Keywords: Bundle sheath, C4 photosynthesis, maize, mesophyll, methylation, PEPC
Introduction
In plants, phoshoenolpyruvate carboxylase (PEPC) is a cytosolic enzyme that catalyses the conversion of phosphoenolpyruvate (PEP) and bicarbonate () to the four carbon acid oxaloacetate (OAA) and inorganic phosphate (Chollet et al., 1996). In all known C4 plants, PEPC operates as the primary carboxylase enzyme; in two-celled systems fixing CO2 as bicarbonate into OAA in mesophyll (M) cells prior to decarboxylation around Ribulose 1,5-Bisphosphate Carboxylase/Oxygenase (RuBisCO) in the parenchymatous bundle sheath (PBS) cells.
The compartmentation of proteins between M and PBS cells is considered a key characteristic of the C4 leaf (Brown et al., 2005), and numerous mechanisms underlying cell specificity have been reported (Hibberd and Covshoff, 2010; Brown et al., 2011). In the dicotyledonous C4 plants Flaveria trinervia (Spreng.) C. Mohr. and F. bidentis (L.) Kuntze, the control of PEPC expression is primarily exerted at the level of transcription. Fusion of 2 kb of the F. trinervia promoter to the β-glucuronidase (GUS) reporter gene is sufficient to generate M-specific GUS accumulation in F. bidentis (Stockhaus et al., 1997). A 41 nucleotide region of this promoter called the mesophyll enhancing module 1 (MEM1) containing a CACT tetranucleotide was shown to be capable of directing M-specific PEPC expression when integrated into the promoter of the C3 plant F. pringlei (Gowik et al., 2004; Akyildiz et al., 2007). To date, however, trans-acting factors associated with this element remain to be identified. In maize, GUS reporter experiments have shown that 0.6 kb of the PEPC promoter is capable of driving M-specific expression (Taniguchi et al., 2000; Kausch et al., 2001). ZmPEPC transcription occurs in all cell-types (except xylem tissue) in very young leaves and is subsequently repressed in all but M cells (Langdale et al., 1987, 1988; Kausch et al., 2001) suggesting that developmental signals are important in regulating the transcriptional activity of the promoter. The C4 PEPC promoter in maize has been shown to bind various protein complexes (Yanagisawa and Izui, 1990, 1992, 1993; Kano-Murakami et al., 1991; Yanagisawa, 1995; Yanagisawa and Sheen, 1998). However, the identity and specific function of these proteins remains to be defined.
Epigenetic modifications have also been shown to correlate with maize PEPC expression as both histone and DNA methylation have been implicated in its regulation. While light induces histone H4 acetylation in both M and PBS cells (Offermann et al., 2006) histone H3K4 tails are heavily tri-methylated in M compared with PBS cells. This pattern of histone modification remained unchanged in dark-grown leaves when PEPC expression was low, suggesting that this alone is not sufficient to account for the high amount of M-specific PEPC transcripts seen in maize leaves (Danker et al., 2008). Interestingly, the maize C4 NADP-ME gene that is expressed in PBS cells shows an inverse pattern in which tri-methylation of H3K4 occurs in PBS cells (Danker et al., 2008).
Although chromatin patterns are important in regulating PEPC expression in maize, during cell division chromatin structures are removed from DNA (Lucchini and Sogo, 1995) and therefore must subsequently be re-established following replication, implying a further level of regulation. The selective methylation of DNA provides a mechanism for regulating gene expression in plants (Spena et al., 1983; Hepburn et al., 1987; Bianchi and Viotti, 1988; Bucherna et al., 2001) and animals (Cedar, 1988) and has been shown to impact strongly on chromatin patterns (Lande-Diner et al., 2007) forming a basal template for chromatin arrangements (Weber and Schubeler, 2007; Suzuki and Bird, 2008). Indeed, previous work has linked DNA methylation with the expression of PEPC in plants. For example, methylation of four cytosines located in the promoter region and de-methylation of four cytosines in the 5' UTR of the McPPC1 gene from the facultative Crassulacean Acid Metabolism (CAM) plant Mesembryanthemum crystallinum is co-incident with an increase in expression associated with the switch from C3 to CAM metabolism (Huang et al., 2010). Furthermore, differential methylation of a PvuII restriction site 3.1 kb upstream from the maize PEPC transcription start site in response to illumination was correlated with changes to PEPC expression (Langdale et al., 1991). To provide additional insight into the extent to which DNA methylation of the maize PEPC promoter occurs, we used bisulphite sequencing. Treatment of DNA with sodium bisulphite results in deamination of unmethylated cytosines to uracil, however, 5-methylcytosines remain unconverted. Following sequencing of bisulphite-converted DNA unmethylated and methylated cytosines can be distinguished from one another because they appear as thymines and cytosines, respectively, in the amplified product (Frommer et al., 1992). Therefore, sequencing of bisulphite-treated DNA can determine the methylation status of a given DNA sequence at single-nucleotide resolution. In this paper, to assess the extent to which regulation of maize PEPC is related to the methylation status of the promoter, methylation was examined at single base resolution.
Materials and methods
Plant material
Maize (B73) plant material was germinated after overnight imbibition in molecular grade biology water. It was then planted in Levington M3 potting compost (Scotts, Ohio, USA) treated with intercept (200 mg l−1) (Scotts Miracle-Gro, Ohio, USA). All plant material was grown at a relative humidity (RH) of 50%, a constant temperature of 28 °C and an atmospheric CO2 concentration of 400 μl l−1. Etiolated seedling material was harvested above the mesocotyl under a dim green safelight after 7 d. The remaining dark-grown plants were then transferred to a 16/8 h light/dark regime (400 μmol m−2 s−1) 2 h into the light period and second leaves were harvested after 72 h. Leaves exposed to 72 h light were used for cell separation. In all cases, material was flash-frozen in liquid nitrogen and stored at –80 °C for subsequent DNA and RNA extraction.
RT-qPCR
Total RNA for RT-qPCR was extracted from 7-d-old maize seedlings using the Qiagen RNeasy® Plant Mini Kit (Qiagen, Alameda, CA) according to the manufacturer's instructions. To remove contaminating genomic DNA, samples were treated with 10 U μl−1 RNase-free DNase (Qiagen Alameda, CA) for 30 min at 20 °C and 15 min at 65 °C. RNA quality was analysed using an Agilent 2100 Bioanalyser and an RNA nano-chip. All samples had RNA integrity numbers (RINs) above 6.60 indicating the RNA was high quality (Fleige and Pfaffl, 2006). 1 μg RNA was reverse transcribed using an oligo(dT) primer and Superscript II (InVitrogen Life Technologies, USA). The total cDNA volume of 20 μl was stored at –20 °C overnight. Real-time quantitative PCR (RT-qPCR) was carried out using SYBR Green JumpStart Taq Ready Mix (Sigma-Aldrich, Germany) and 5-fold dilution of the template and primers at 0.2 μM final concentration. Primers were designed using Primer 3 software (http://frodo.wi.mit.edu/primer3) to have melting temperatures of 60 °C. Sequences of primers used to detect ZmPEPC and ZmMAZ95 (Lin et al., 2008) are listed in Supplementary Table S1 at JXB online. RT-qPCR was performed in a Rotor-Gene™ thermal cycler (Qiagen Alameda, CA). Cycling conditions were: 94 °C for 2 min, followed by 40 cycles of 94 °C for 20 s, 60 °C for 30 s, 72 °C for 30 s, and 75 °C for 5 s. The fluorescence threshold was set to a constant value of 0.04, which was manually determined to be as early as possible into the exponential phase of fluorescence for all transcripts. The CT values were calculated from means of three technical replicates for three independent biological replicates of each line. Relative abundance of transcripts (to ZmMAZ95) was calculated using the 2−ΔΔCT method after Livak and Schmittgen (2001). Standard errors were calculated from 2−ΔΔCT values of each combination of (biological and technical) replicates.
Parenchymatous bundle sheath/mesophyll cell extraction
Leaves from a minimum of 20 plants exposed to 72 h light were used for cell separation. M and PBS cells were separated after the method described by Markelz et al. (2003). Details of each preparation are given below. Protoplast and PBS strand integrity was assessed by light microscopy.
M cell preparation
Second and third maize leaves (corresponding to 5 g of leaf tissue) were cut perpendicularly to remove the midrib and subsequently transversely into 1–2 mm strips. The leaf samples were subjected to enzymatic digestion in enzyme buffer (20 mM MES (pH 5.5), 1 mM MgCl2, 0.6 M sorbitol, 2% (w/v) Cellulase Onazuka (Yakult Pharmaceuticals, Tokyo), and 0.1% (w/v) macerase (Calbiochem, San Diego) for 3 h at 21 °C. The strips were filtered through a 135 μm nylon mesh (Millipore, MA, USA) and resuspended in 50 ml wash buffer (50 mM TRIS-HCl (pH 7.5), 1 mM MgCl2, 0.6 M sorbitol, and 100 mM β-mercaptoethanol). Gentle pressure was applied with a stainless steel spoon for approximately 1.5 min to release the protoplasts. Removal of cellular debris was conducted by filtration through a 60 μm nylon mesh (Millipore, MA, USA). The filtrate containing protoplasts was subjected to centrifugation at 1200 g for 10 min, resuspended in wash buffer, and re-centrifuged. The pellet was resuspended in 500 μl of wash buffer solution and dropped into liquid nitrogen in peel-away cups (VWR Scientific, NJ, USA).
PBS cell preparation
To isolate PBS cells from maize leaves, second and third leaves were cut into 2×2 mm squares (4 g tissue) and disrupted by three 10 s pulses on ‘low’ setting in a blender (Waring Products, CT, USA) in 50 ml PBS buffer I (0.33 M sorbitol, 0.3 M NaCl, 0.01 M EGTA, 0.01 M dithiothreitol, 0.005 M diethyldithiocarbamic acid, and 0.2 M TRIS-HCl (pH 9.0)). The resulting buffer/tissue solution was filtered using 60 μm nylon mesh (Millipore, MA, USA) and subsequently blended for three 1 min pulses on ‘high’ setting, in PBS buffer II (0.35 M sorbitol, 0.005 M EDTA, 0.1% (v/v) β-mercaptoethanol, and 0.05 M TRIS (pH 8.0)) re-filtering through the mesh between each pulse. The blender was washed out with molecular biology grade water between each filtration. PBS strands retained on the nylon mesh were dried briefly by placing the mesh on paper towels to wick away excess moisture. PBS strands were then removed from the mesh and flash frozen in liquid nitrogen.
Bisulphite sequencing
Total cellular DNA was extracted from 100 mg fresh plant material (root, dark-grown and light-grown leaves, M, PBS) ground in liquid nitrogen. In the case of total leaf extractions three leaves from independent plants were ground together in liquid nitrogen at –80°C. DNA extraction was performed using the DNeasy© DNA extraction kit (Qiagen Ltd., West Sussex, UK) following the manufacturer's recommended protocol. The quantity of DNA was determined spectrophotometrically using a NanoDrop 1000™ spectrophotometer (Thermo Scientific, Wilmington, USA). The quality of the DNA sample was validated via agarose gel electrophoresis. 1 μg DNA and 5 μl DNA loading buffer (comprising 50% (v/v) glycerol, 1 mM ethylenediamine tetra-acetic acid disodium salt (EDTA), 0.4% (w/v) bromophenol blue (BPB), 0.005× TBE (TRIS-borate-EDTA) buffer, 48% (v/v) formamide) were heated to 60 °C for 10 min and subsequently run on a 1.5% (w/v) agarose gel containing 0.5 μg ml−1 ethidium bromide using 0.5× TBE buffer (44.5 mM tris-hydroxymethyl) aminomethane, 44.5 mM boric acid, and 1 mM EDTA at pH 8.0).
500 ng maize genomic DNA was treated with sodium bisulphite (NaHSO3) using the EZ DNA Methylation-Gold™ Kit (Zymo Research Corporation, CA, USA) according to the manufacturer's protocol. Treatment of DNA with sodium bisulphite results in the selective deamination of non-methylated cytosines to uracil, whereas 5' methylated cytosines remain unconverted during the treatment (Wang et al., 1980). The methylation status of the DNA can be determined by DNA sequencing of sodium bisulphite-treated and untreated controls following PCR amplification. Primers (see Supplementary Table S1 at JXB online) were used to amplify a region of the maize PEPC promoter (sense strand), 3.22–2.88 kb upstream of the transcription initiation site, surrounding the differentially-methylated PvuII (–3.029 kb) restriction site identified by Langdale et al. (1991) in order to validate the sensitivity of bisulphite sequencing in this context. Primers for bisulphite sequencing (see Supplementary Table S1 at JXB online) were designed after Henderson et al. (2010) and were biased to amplify from bisulphite-converted template DNA.
PCR was performed in a total volume of 20 μl (9.2 μl molecular biology grade water, 4 μl 5× BioTaq buffer, 0.2 μl dNTPs, 1 μM Forward primer, 1 μM Reverse primer, 200 ng DNA template, 0.2 μl BioTaq polymerase). PCRs were carried out using a Techne™ thermal cycler and BioTaq™ high-fidelity DNA polymerase (TaKaRa, Shiga, Japan). Taq polymerase was selected for non-proofreading activity since proofreading polymerases stall after incorporation of deoxyuracil, a base which is efficiently incorporated into amplified products by BioTaq. PCR began with a hot start before BioTaq polymerase was added (3 min at 95 °C) to reduce non-specific binding, followed by 35 cycles of; further denaturation (20 s at 95 °C), annealing (30 s at 50–55 °C), and extension (60 s at 62 °C). A final extension was carried out at 62 °C for 10 min. PCR products were examined on 1.5% agarose gel, by loading 5 μl PCR product and 5 μl loading buffer. A Hyperladder IV™ (Bioline, Ltd., London, UK) size marker was used to determine the molecular weight of the products. Amplified products were size-excluded and purified using the QIAquick™ gel extraction kit (Qiagen, Alameda, CA) according to the manufacturer's protocol. PCR products amplified this way were cloned into PJet 1.2 plasmid vector using the CloneJET™ PCR Cloning Kit (Fermentas, Germany) as per the manufacturer's instructions and transformed into Escherichia coli DH5α competent cells. Bacteria were plated on LB agar selective media containing 100 mg ml−1 ampicillin. Successfully transformed colonies were screened by PCR.
Cycle sequencing of cleaned PCR products was performed in a Techne™ thermal cycler (initial denaturation at 96 °C, followed by 25 cycles of 96 °C for 10 s; 50 °C for 5 s, and, finally, 60 °C for 4 min) using the following reagents: 200 ng PCR product (plasmid), PJet 1.2 Forward Primer (10 μM) 0.5 μl, ddH2O (to 10 μl) BigDye v3.1 5X cycle sequencing buffer 2 μl and BigDye v3.1 1 μl (Applied Biosystems, Foster City, CA). DNA precipitation and removal of unincorporated terminators and sequencing was performed by the sequencing facility at the Department of Biochemistry, University of Cambridge, UK. Sequence analysis was performed using BioEdit™ v7.0.5 sequence alignment software for Windows™ on a Dell™ Optiplex 740 computer. Since all cytosines are converted to uracil as a consequence of bisulphite treatment, cytosine residues in amplified sequences (excluding primer sequences) were interpreted as methylated bases. Thymine residues occurring at the equivalent positions as cytosines in untreated controls were classified as unmethylated. Initially, the vector inserts of at least 10 independent clones were sequenced using forward primers against the PJet 1.2 vector backbone, followed by a further 10 independent clones per treatment in regions where cytosine methylation was detected. Care was taken to ensure that all sequences analysed varied at a minimum of one C/T base in order that the same amplicon was not sequenced multiple times. Results are expressed as percentages of clones with cytosine residues at the nucleotide position indicated.
Results
PEPC transcripts accumulate in maize leaves in response to illumination
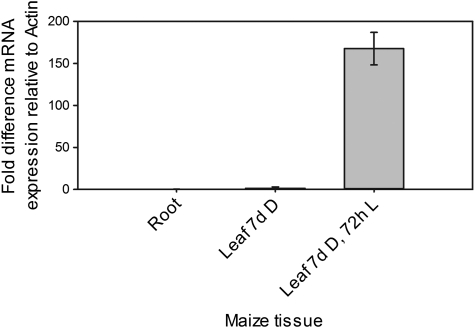
To establish the amount of PEPC transcripts in each tissue type in maize, RNA was extracted from root tissue, leaves from 7-d-old plants grown in the dark or leaves transferred to the light for 72 h. After production of cDNA, Real-time quantitative Polymerase Chain Reaction (RT-qPCR) was used to quantify PEPC transcripts relative to maize actin (ZmMaz95). Very little PEPC expression was detected in roots and dark-grown leaves (Fig. 1). However, significant transcript accumulation was observed in light-grown leaves indicating that PEPC expression was responsive to illumination and that PEPC transcripts were more abundant in light-grown leaves relative to roots and dark-grown leaves (Fig. 1).
Fig. 1.
PEPC transcripts accumulate in maize leaves in response to illumination. The 2−ΔΔCT method was used to quantify the relative abundance of transcripts (Livak and Schmittgen, 2001). ZmMaz95 (Lin et al., 2008) was used as a reference. Results are expressed as mean CT values calculated from a minimum of three biological and three technical replicates. Error bars represent one standard error of the mean calculated from 2−ΔΔCT values of each combination of biological replicates.
Four cytosine residues in the PEPC promoter are heavily methylated in maize root tissue
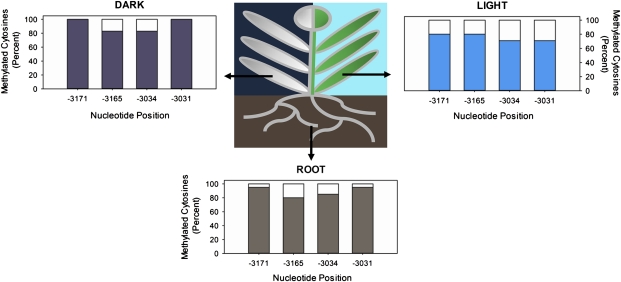
In order to define precisely the methylation status of the differentially methylated PvuII restriction site (CAGCTG) identified in the maize PEPC promoter by Langdale et al. (1991), DNA extracted from 7-d-old root tissue was subjected to modification with sodium bisulphite to convert unmethylated cytosines to uracil. A 270 bp region of the sense strand DNA (–3178 to –2908) surrounding the PvuII site was then amplified from this template via PCR, sequenced, and compared with an untreated control. The majority of cytosines in this region appeared as thymines in the amplified product validating this method in converting unmethylated cytosines to uracil. By contrast, four cytosine residues (CHG context) on the sense strand of the 270 bp region showed high methylation frequencies in root tissue samples taken from 7-d-old maize seedlings. Methylation of both cytosines in the PvuII site (positions –3034 and –3031) was detected and at two previously unidentified cytosines further upstream (–3171 and –3165) (Fig. 2). 95% and 80% of cytosines (both CAG context) at positions –3171 and –3165 were found to be methylated in root samples (Fig. 2). Similarly, both cytosines in the PvuII site –3034 and –3031 kb upstream from the transcription start site were methylated at a frequency of 85% and 95%, respectively (Fig. 2).
Fig. 2.
Four cytosine residues in the PEPC promoter are de-methylated in maize leaves in response to illumination. The methylation status of PEPC promoter region –3178 to –2906 kb upstream of the transcription start site in maize roots, dark-grown leaves, and leaves transferred to light for 72 h. The extent of DNA methylation was determined by bisulphite sequencing; cytosine residues in amplified products indicated methylated bases, whereas thymines in the place of cytosines were classified as unmethylated. Percentages refer to the percentage of clones with cytosine residues at the nucleotide position indicated. At least 20 independent clones were sequenced per treatment and all sequences varied at a minimum of one C/T base.
De-methylation of the PEPC promoter in maize leaves is dependent on illumination
To determine whether the methylation status of the 270 bp region (–3178 to –2908) within the PEPC promoter was preserved in leaf as well as root tissue, it was amplified via PCR from leaves of 7-d-old etiolated seedlings. The four cytosines methylated in roots were found to be methylated at similar frequencies in leaves of etiolated seedlings, with 100% of cytosines at positions –3171 and –3031 and 83% at positions –3165 and –3034 methylated, respectively. This indicated that methylation of these cytosines is maintained in roots and dark-grown leaf tissue (Fig. 2).
However, in DNA from leaves of 7-d-old etiolated seedlings which were subsequently transferred to light for 72 h, these cytosines were less heavily methylated. Although cytosines at position –3165 were methylated at a similar level to those in roots, the methylation of cytosines at –3171 was reduced to 80% (Fig. 2). Both cytosines in the PvuII site (positions –3034 and –3031) were de-methylated upon illumination in 29% of amplified products (Fig. 2). De-methylation of these cytosine residues in the PEPC promoter therefore coincides with an increase in PEPC expression in maize leaves in response to illumination.
Light induced de-methylation of the PEPC promoter occurs predominantly in M cells
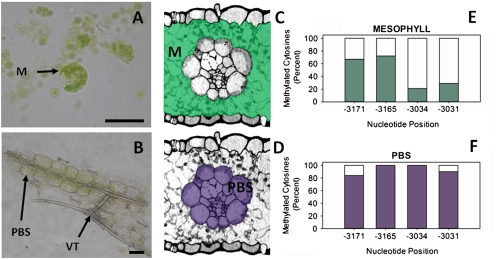
In order to determine whether de-methylation of the cytosine residues was spatially coincident with PEPC expression in the maize leaf, the 270 bp region of DNA (sense strand –3178 to –-2908) surrounding the four sites was amplified and sequenced from bisulphite-treated DNA extracted from M and PBS cells of 3-week-old light-grown leaf tissue. PBS strands were assessed for contamination by M cells and we estimate this contamination as being less than 5% M. This agrees with previous work in which contamination of PBS or M cell preparations by the other cell type is typically lower than 5% (Sawers et al., 2007). Representative images of the M and PBS preparations are shown in Fig. 3A and B. In M cells all four cytosines were de-methylated in response to light, while methylation of these residues was retained in PBS cells. Whilst only 67% and 72% of cytosines at positions –3171 and –3165 remained methylated in M cell extracts (Fig. 3C, E), 84% and 100% of cytosines at these positions were found to be methylated in PBS cells, respectively (Fig. 3D, F). De-methylation in cytosines at positions –3034 and –3031 was more pronounced with only 21% and 29% methylated in M cells (Fig. 3C, E), whereas 100% and 90% of cytosines, respectively, were methylated in PBS cells (Fig. 3D, F) corresponding to a 79% and 61% change in methylation at these nucleotides between the two cell types. The changes in methylation status of cytosine residues observed upon illumination of whole leaves can thus be attributed to a M-specific de-methylation of four cytosine residues, coincident with PEPC transcript accumulation.
Fig. 3.
Light induced de-methylation of the PEPC promoter occurs only in M cells. Representative pictures of two cell extracts from maize leaf tissue used for DNA extraction and subsequent bisulphite sequencing (A, B). The position of these extracts within the leaf is shown on transverse sections of maize leaf tissue in (C) and (D). The methylation status of PEPC promoter region –3178 to –2908 kb upstream of the transcription start site in M and PBS cells of light-grown maize leaves determined by bisulphite sequencing is shown in (E) and (F). The occurrence of cytosine residues in amplified products indicated methylated bases, whereas thymines in the place of cytosines were classified as unmethylated. Percentages refer to the percentage of clones with cytosine residues at the nucleotide position indicated. At least 20 independent clones were sequenced per treatment and all sequences varied at a minimum of one C/T base. Shaded areas indicate tissue types in each extract used for DNA extraction. Abbreviations are as follows: M, mesophyll; PBS, parenchyma bundle sheath; VT, vascular tissue. Scale bars=25 μm.
DNA methylation in 1 kb upstream of the transcription initiation site does not regulate PEPC expression
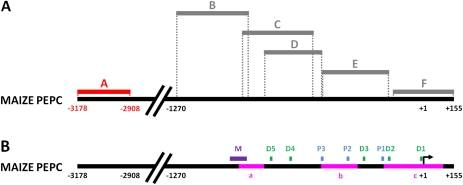
Despite the discovery of four cytosine residues ∼3.1 kb upstream from the transcription start site (TSS) which are specifically de-methylated in M cells in response to light, several studies have shown that only –1212 to +88 of the PEPC gene is required for M-specific expression in maize (Taniguchi et al., 2000; Kausch et al., 2001). Indeed, maize PEPC and PPDK promoters also generate GUS expression in the appropriate cell-types in a light-dependent manner when placed in rice (Matsuoka et al., 1993, 1994; Ku et al., 1999; Nomura et al., 2000) indicating that the relevant trans-acting factors required for the recognition of these genes are not only conserved in these species but also operate in the correct cell-types. Strong GUS accumulation in maize M cells when the uidA gene encoding β-glucoronidase are fused to 1.7 kb or 0.6 kb of the maize PEPC promoter indicates that the site ∼3.1 kb upstream of the TSS is not necessary for M-specific expression in maize (Taniguchi et al., 2000; Kausch et al., 2001). Although studies using methylation-sensitive restriction enzymes have not shown evidence of methylation in the ‘core’ PEPC promoter (–1212 to +88) (Langdale et al., 1991), it was hypothesized that methylated cytosines in this region may have gone undetected as a consequence of the limited resolution inherent in experiments predicated on the occurrence of appropriate restriction sites. To discover whether methylation of this region is involved in directing tissue- and/or cell-specificity of PEPC transcript accumulation, the sense strand of this region of genomic DNA (–158 to +155) of PEPC in roots and dark/light-grown leaves as well as in M and PBS of light-grown leaves was interrogated using bisulphite sequencing as described previously. No cytosine residues in either CG or CHG contexts in the cluster were found to be methylated/de-methylated in response to light or in a cell-specific manner (Fig. 4).
Fig. 4.
Cytosine methylation in the ∼1 kb upstream region of the TSS does not correlate with PEPC expression. The methylation status of PEPC promoter region +155 to –1270 in roots, dark-grown leaves, and M and PBS cells of maize leaves transferred to light for 72 h is shown in panel (A). Grey bars indicate amplicons where no methylation was detected. Red bars indicate differentially-methylated regions. Dashed lines show the position of amplified sequences on the maize PEPC promoter. Regions amplified from bisulphite treated DNA are as follows: (A) –3178/–2908; (B) –1270/–888; (C) –902/–551; (D) –817/–518; (E) –521/–176; (F) –158/+155. The sites of protein complex binding relative to the maize PEPC promoter sequence are indicated in panel (B). Abbreviations are as follows: M, MNF1; D, DOF; P, PEP-I. The positions of CpG islands in the PEPC promoter predicted by Methprimer™ software are depicted as pink bars in panel (B) (a, –946 to –817; b, –782/–595; c, –200/+105). The program settings were as follows: 100 bp segments, observed/expected CpG dinucleotides >0.6 and 50% GC content.
The regulatory protein complex PEP-I has been identified as an important regulator of PEPC expression in maize (Kano-Murakami et al., 1991). PEP-I binding is sensitive to methylation interference at two guanine residues in the consensus binding sequence (Kano-Murakami et al., 1991). It was therefore hypothesized that differential methylation of cytosines in the PEP-I binding sites could influence the binding affinity of PEP-I to this region of the promoter. To test this, after treatment with sodium bisulphite, a 328 bp region (–521 to –176) of sense strand DNA including the PEP-I binding sites was amplified and sequenced. None of the cytosines in root, dark-grown leaves or illuminated leaves were found to be methylated and no differences were found between M and PBS sequences for this region. Overall, this indicates that cytosine methylation is unlikely to regulate differential binding of PEP-I in maize roots or leaves. Similarly, when the remainder of the maize PEPC promoter region up to –1270 downstream of the TSS (regions B, C, and D) was screened for changes in cytosine methylation by bisulphite sequencing, no cytosine methylation was detected. These regions include binding sites for MNF1 and Dof, indicating that cytosine methylation at these sites does not regulate the binding of these proteins to the PEPC promoter in maize nor is it required for tissue- or cell-specificity of maize PEPC expression. The lack of methylation in this region of the promoter led to an examination of the distribution of localized concentrations of CpG dinucleotides (cytsoine and guanine nucleotides separated by a single phosphate) in the maize PEPC promoter. The –1270 to +155 bp region was screened for CpG clusters using the Methprimer™ software (urogene.com) (Li and Dahiya, 2002). Using the programme default settings (100 bp segments with 50% GC content, observed/expected ratio of CpG dinucleotides >0.6), three CpG islands were predicted within the PEPC promoter (–946 to –817; –782/–595; –200/+105) (Fig. 4). Increasing the stringency of the search parameters to 300 bp segments, observed/expected ratio >0.6 and >70% GC revealed one large CG cluster (–200 to +105) spanning the PEPC TATA box, TSS and ATG sequences.
Discussion
Regulatory protein binding is unlikely to be regulated by cytosine methylation
Several regulatory protein complexes have been found to bind the PEPC promoter. One in particular, PEP-I, interacts with the promoter in a tissue-specific manner, binding in maize leaves but not roots. It has also been shown that PEP-I binding in vitro is sensitive to methylation interference at two guanine residues in the consensus binding sequence (Kano-Murakami et al., 1991). However, differential methylation of cytosines between –521 and –176 did not differ between roots and dark- or light-grown leaves. Similarly, no differences in the methylation status of cytosine residues were observed between tissue- or cell types in the region from –905 to –818 corresponding to the binding site of MNF1 (Yanagisawa and Izui, 1992) or the DOF proteins (Yanagisawa and Izui, 1993; Yanagisawa, 1995; Yanagisawa and Sheen, 1998), indicating that cytosine methylation is unlikely to play a role in regulating the binding affinity of these proteins in vivo.
Cytosine methylation is not required for cell-specific expression of PEPC
Two cytosine residues in the upstream region of PEPC which undergo M-specific de-methylation in response to illumination have been identified. Two more cytosines are de-methylated at an adjacent PvuII restriction site, previously discovered by Langdale et al. (1991); the extent of cytosine de-methylation at this site in different maize tissues and cell-types has now been quantified. As GUS accumulates in maize M cells when the uidA gene encoding β-glucoronidase is fused to 0.6 kb of the ZmPEPC promoter (Taniguchi et al., 2000; Kausch et al., 2001), neither of these sites at 3.1 kb that undergo de-methylation appear necessary for M-specific expression in maize. In fact, the PEPC promoter sequence from –389 upstream to the first ATG generated 75% reporter gene expression in transient assays of isolated maize M cells (Shäffner and Sheen, 1992). Similarly, when maize PEPC is transformed into rice under the control of maize 1.2 kb promoter sequence, PEPC transcripts accumulate faithfully in M cells (Matsuoka et al., 1994). It is shown here that there is no cytosine methylation in the first 1.3 kb of the maize PEPC promoter in roots, etiolated leaves or M and PBS cells of light-grown leaves. This indicates that cytosine methylation in the minimal promoter of maize PEPC is not directly involved in regulating its cell-specific expression. Due to a high GC content, the availability of appropriate sites for bisulphite primer design was limited (Henderson et al., 2010) and so a stretch of 16 nt (–175 to –157) was not analysed in this study. Although this region could contain differentially-methylated sites, it represents 1.1% of the total promoter sequence interrogated and contains only two CpG sites.
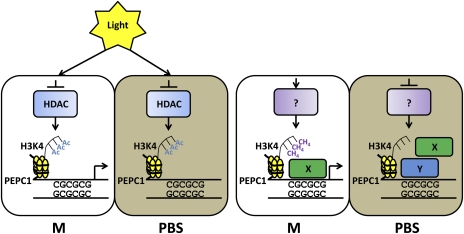
The discovery that ∼1.3 kb of the PEPC promoter is not methylated is rather unexpected, given the cell-specific chromatin patterns observed in the promoter associated with its transcription in M cells. The discovery of three clusters of CG dinucleotides within the promoter sequence is also unusual. These regions, called CpG islands are typically found to be unmethylated, but, in animals, are normally associated with constitutive expression observed in genes with housekeeping functions (Cedar, 1988) rather than cell-specific gene expression. However, there is at least one example in maize where this is not the case. The expression of the maize gene encoding alcohol dehydrogenase (Adh1) is repressed in leaves (Okimoto et al., 1980), yet a 900 bp CpG island in the promoter of the Adh1 gene is not methylated in this tissue (Nick et al., 1986). It is not known how CpG islands are maintained in an unmethylated state, although it has been hypothesized that proteins may bind these regions to protect them from methylation (Voo et al., 2000; Bader et al., 2003) and it has been shown that CXXC finger protein 1 (Cfp1) binds to over 80% of unmethylated CpG islands in mammals and directly influences local chromatin structures (Blackledge et al., 2010; Thomson et al., 2010). In the case of maize PEPC it could be that protein complexes binding the region between –0.6 kb and the TSS, containing two CpG islands, perform this function. It is particularly interesting in this context that some of the proteins that have been identified as binding the maize PEPC promoter also bind other promoters and that Cfp1 was shown to be closely associated with tri-methylation at H3K4 (Thompson et al., 2010), which was shown to occur in the PEPC promoter in M cells (Danker et al., 2008). With this in mind, a model is proposed whereby a protein with similar properties to Cfp1 binds unmethylated CpGs in the PEPC promoter in M cells and directs H3K4 trimethylation, maintaining an open chromatin conformation and permitting transcription to occur. Either the absence of this protein or competition for binding sites in PBS cells prevents H3K4 methylation and therefore transcription (Figure 5).
Fig. 5.
Epigenetic interactions influencing PEPC transcription in maize leaves. Abbreviations: A, acetylation; , methylation; HDAC, histone deacetylase; M, mesophyll; PBS, parenchymatous bundle sheath; H3K4, histone 3 lysine 4; X, putative positive regulatory protein; Y, Putative negative regulatory protein.
Our study demonstrates that methylation of the PEPC promoter is unlikely to be involved in directing cell-specific expression, and the question of how the differential methylation of four cytosines 3.1 kb upstream relates to PEPC expression remains unresolved. Preliminary in silico analysis of maize chromosome 9 suggests that the nearest predicted gene upstream (5') of this region on the same strand is approximately 12 kb away (see Supplementary Figs S1–S4 at JXB online), indicating that the site is unlikely to be regulating a gene upstream of PEPC in the opposite orientation. Another possibility is that the differentially-methylated region operates as an enhancer element to PEPC expression. Studies introducing maize PEPC into rice with a minimal promoter (–1212 to +88) show that the amount of expression was not identical to PEPC transcript abundance in maize (Matsuoka et al., 1994) supporting this hypothesis. To test the effect of methylation at the PvuII site more directly, we attempted to grow maize on methylation inhibitors. However, preliminary analysis indicates that in maize plants grown on media containing 5’-azacytidine or zebularine, cytosine analogues which inhibit methylation in actively dividing cells (Jones et al., 1985; Christman, 2002; Baubec et al., 2009), the amount of methylation in leaves remain unaffected (H Woodfield, unpublished results).
Recent developments in nanopore sequencing technology (Kasianowicz et al., 1996; Astier et al., 2006; Clarke et al., 2009) include the ability to distinguish 5-methylcytosine from unmethylated nucleotide bases due to differences in its ionic conductivity (Clarke et al., 2009) and promise to provide a high-throughput, affordable method of analysing the methylome of species with large genomes such as maize, to single base resolution. This will enable the methylation status of M and PBS cell genomes to be interrogated simultaneously and promises to greatly improve our understanding of the role of DNA methylation in regulating C4 gene expression.
In summary, methylation marks within the PvuII site identified by Langdale et al. (1991) have been defined and two additional cytosine residues have been identified in the PEPC promoter that are methylated in maize root tissue. In leaves, de-methylation of these cytosines is dependent on illumination and is coincident with elevated PEPC expression. Furthermore, light-regulated de-methylation of these cytosines occurs only in M cells. No evidence of cytosine methylation was found in the 0.6 kb promoter required for M-specific expression indicating that cytosine methylation does not play a direct role in directing cell-specificity. However, the abundance of unmethylated CpG sites in the PEPC promoter suggests that the epigenetic status of the PEPC promoter may be important in maintaining an open chromatin structure for transcription factor binding. The function of the four differentially-methylated cytosines in the upstream region of the PEPC promoter remains unclear: however, the possibility remains that this site exerts some distant regulatory control over the cell-specific expression of the gene. The identification of proteins binding to the 5' flanking region of maize PEPC gene is a priority and together with developments in nanopore sequencing, should provide a clearer picture of the regulatory mechanisms governing PEPC expression in maize.
Supplementary data
Supplementary data can be found at JXB online.
Supplementary Fig. S1. Gene prediction in the region 15 kb upstream of ZmPEPC1.
Supplementary Fig. S2. Predicted protein coding sequences in the region 15 kb upstream of ZmPEPC1.
Supplementary Fig. S3. Gene prediction in the region 50 kb upstream of ZmPEPC1.
Supplementary Fig. S4. Predicted protein coding sequences in the region 50 kb upstream of ZmPEPC1.
Supplementary Table S1. Primer sequences used in bisulphite sequencing PCR to amplify regions of the maize PEPC1 promoter.
Acknowledgments
BJT was supported by a PhD award from The Gatsby Charitable Foundation. We thank Dr Ian Henderson and Dr Andrew Bassett for useful conversations and suggestions, Professor Jane Langdale for critical assessment, and Dr Sarah Covshoff, Susan Stanley, and Julie Bull for practical help. Authors’ contributions: BJT, HW, and JMH designed the research; BJT, HW, and SW performed the research; BJT, HW, and JMH analysed data; and BT and JMH wrote the paper.
References
- Akyildiz M, Gowik U, Engelmann S, Koczor M, Streubel M, Westhoff P. Evolution and function of a cis-regulatory module for mesophyll-specific gene expression in the C4 dicot Flaveria trinervia. The Plant Cell. 2007;19:3391–3402. doi: 10.1105/tpc.107.053322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astier Y, Braha O, Bayley H. Toward single molecule DNA sequencing: direct identification of ribonucleoside and deoxyribonucleoside 5'-monophosphates by using an engineered protein nanopore equipped with a molecular adapter. Journal of the American Chemical Society. 2006;128:1705–1710. doi: 10.1021/ja057123+. [DOI] [PubMed] [Google Scholar]
- Bader S, Walker M, McQueen HA, Sellar R, Oei E, Wopereis S, Zhu Y, Peter A, Bird AP, Harrison DJ. MBD1, MBD2, and CGBP genes at chromosome 18q21 are infrequently mutated in human colon and lung cancers. Oncogene. 2003;22:3506–3510. doi: 10.1038/sj.onc.1206574. [DOI] [PubMed] [Google Scholar]
- Baubec T, Pecinka A, Rozhon W, Scheid OM. Effective, homogeneous and transient interference with cytosine methylation in plant genomic DNA by zebularine. The Plant Journal. 2009;57:542–554. doi: 10.1111/j.1365-313X.2008.03699.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi MW, Viotti A. DNA Methylation and tissue-specific transcription of the storage protein genes of maize. Plant Molecular Biology. 1988;11:203–214. doi: 10.1007/BF00015672. [DOI] [PubMed] [Google Scholar]
- Blackledge NP, Zhou JC, Tolstorukov MY, Farcas AM, Park PJ, Klose RJ. CpG islands recruit a histone H3 lysine 36 demethylase. Molecular Cell. 2010;38:179–190. doi: 10.1016/j.molcel.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown NJ, Newell CA, Stanley S, Chen JE, Perrin AJ, Kajala K, Hibberd JM. Independent and parallel recruitment of pre-existing mechanisms underlying C4 photosynthesis. Science. 2011;331:1436–1439. doi: 10.1126/science.1201248. [DOI] [PubMed] [Google Scholar]
- Brown NJ, Parsley K, Hibberd JM. The future of C4 research: maize, Flaveria orCleome? Trends in Plant Science. 2005;10:215–221. doi: 10.1016/j.tplants.2005.03.003. [DOI] [PubMed] [Google Scholar]
- Bucherna N, Szabo E, Heszky LS, Nagy I. DNA methylation and gene expression differences during alternative in vitro morphogenic processes in eggplant (Solanum melongena L.) In vitro Cellular and Developmental Biology-Plant. 2001;37:672–677. [Google Scholar]
- Cedar H. DNA methylation and gene activity. Cell. 1988;53:3–4. doi: 10.1016/0092-8674(88)90479-5. [DOI] [PubMed] [Google Scholar]
- Chollet R, Vidal J, O'Leary MH. Phospho enolpyruvate carboxylase: a ubiquitous, highly regulated enzyme in plants. Annual Review of Plant Physiology and Plant Molecular Biology. 1996;47:273–298. doi: 10.1146/annurev.arplant.47.1.273. [DOI] [PubMed] [Google Scholar]
- Christman JK. 5-Azacytidine and 5-aza-2'-deoxycytidine as inhibitors of DNA methylation: mechanistic studies and their implications for cancer therapy. Oncogene. 2002;21:5483–5495. doi: 10.1038/sj.onc.1205699. [DOI] [PubMed] [Google Scholar]
- Clarke J, Wu HC, Jayasinghe L, Patel A, Reid A, Bayley H. Continuous base identification for single-molecule nanopore DNA sequencing. Nature Nanotechnology. 2009;4:265–270. doi: 10.1038/nnano.2009.12. [DOI] [PubMed] [Google Scholar]
- Danker T, Dreesen B, Offermann S, Horst I, Peterhaensel C. Developmental information but not promoter activity controls the methylation state of histone H3 lysine 4 on two photosynthetic genes in maize. The Plant Journal. 2008;53:465–474. doi: 10.1111/j.1365-313X.2007.03352.x. [DOI] [PubMed] [Google Scholar]
- Fleige S, Pfaffl MW. RNA integrity and the effect on the real-time qRT-PCR performance. Molecular Aspects of Medicine. 2006;27:126–139. doi: 10.1016/j.mam.2005.12.003. [DOI] [PubMed] [Google Scholar]
- Frommer M, McDonald LE, Millar DS, Collis CM, Watt F, Grigg GW, Molloy PL, Paul CL. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proceedings of the National Academy of Sciences, USA. 1992;89:1827–1831. doi: 10.1073/pnas.89.5.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowik U, Burscheidt J, Akyildiz M, Schlue U, Koczor M, Streubel M, Westhoff P. cis-Regulatory elements for mesophyll-specific gene expression in the C4 plant Flaveria trinervia, the promoter of the C4 phospho enolpyruvate carboxylase gene. The Plant Cell. 2004;16:1077–1090. doi: 10.1105/tpc.019729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson IR, Chan SR, Cao X, Johnson L, Jacobsen SE. Accurate sodium bisulfite sequencing in plants. Epigenetics. 2010;5:47–49. doi: 10.4161/epi.5.1.10560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepburn AG, Belanger FC, Mattheis JR. DNA methylation in plants. Developmental Genetics. 1987;8:475–493. [Google Scholar]
- Hibberd JM, Covshoff S. The regulation of gene expression required for C4 photosynthesis. Annual Review of Plant Biology. 2010;61:181–207. doi: 10.1146/annurev-arplant-042809-112238. [DOI] [PubMed] [Google Scholar]
- Huang N-C, Li C-H, Lee J-Y, Yen HE. Cytosine methylation changes in the ice plant Ppc1 promoter during transition from C3 to Crassulacean acid metabolism. Plant Science. 2010;178:41–46. [Google Scholar]
- Jones PA. Altering gene expression with 5-azacytidine. Cell. 1985;40:485–486. doi: 10.1016/0092-8674(85)90192-8. [DOI] [PubMed] [Google Scholar]
- Kano-Murakami Y, Suzuki I, Sugiyama T, Matsuoka M. Sequence-specific interactions of a maize factor with a GC-rich repeat in the phosphoenolpyruvate carboxylase gene. Molecular and General Genetics. 1991;225:203–208. doi: 10.1007/BF00269849. [DOI] [PubMed] [Google Scholar]
- Kasianowicz JJ, Brandin E, Branton D, Deamer DW. Characterization of individual polynucleotide molecules using a membrane channel. Proceedings of the National Academy of Sciences, USA. 1996;93:13770–13773. doi: 10.1073/pnas.93.24.13770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kausch AP, Owen TP, Zachwieja SJ, Flynn AR, Sheen J. Mesophyll-specific, light and metabolic regulation of the C4 PPCZm1 promoter in transgenic maize. Plant Molecular Biology. 2001;45:1–15. doi: 10.1023/a:1006487326533. [DOI] [PubMed] [Google Scholar]
- Ku MSB, Agarie S, Nomura M, et al. High-level expression of maize phosphoenolpyruvate carboxylase in transgenic rice plants. Nature Biotechnology. 1999;17:76–80. doi: 10.1038/5256. [DOI] [PubMed] [Google Scholar]
- Lande-Diner L, Zhang J, Ben-Porath I, Amariglio N, Keshet I, Hecht M, Azuara V, Fisher AG, Rechavi G, Cedar H. Role of DNA methylation in stable gene repression. Journal of Biological Chemistry. 2007;282:12194–12200. doi: 10.1074/jbc.M607838200. [DOI] [PubMed] [Google Scholar]
- Langdale JA, Metzler MC, Nelson T. The argentia mutation delays normal development of photosynthetic cell-types in. Zea mays. Developmental Biology. 1987;122:243–255. doi: 10.1016/0012-1606(87)90349-6. [DOI] [PubMed] [Google Scholar]
- Langdale JA, Rothermel BA, Nelson T. Cellular pattern of photosynthetic gene expression in developing maize leaves. Genes and Development. 1988;2:106–115. doi: 10.1101/gad.2.1.106. [DOI] [PubMed] [Google Scholar]
- Langdale JA, Taylor WC, Nelson T. Cell-specific accumulation of maize phospho enolpyruvate carboxylase is correlated with demethylation at a specific site >3 kb upstream of the gene. Molecular and General Genetics. 1991;225:49–55. doi: 10.1007/BF00282641. [DOI] [PubMed] [Google Scholar]
- Li L-C, Dahiya R. MethPrimer: designing primers for methylation PCRs. Bioinformatics. 2002;18:1427–1431. doi: 10.1093/bioinformatics/18.11.1427. [DOI] [PubMed] [Google Scholar]
- Lin C, Shen B, Xu Z, Köllner TG, Degenhardt J, Dooner HK. Characterization of the monoterpene synthase gene tps26, the ortholog of a gene induced by insect herbivory in maize. Plant Physiology. 2008;146:940–951. doi: 10.1104/pp.107.109553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔ CT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lucchini R, Sogo JM. Replication of transcriptionally active chromatin. Nature. 1995;374:276–280. doi: 10.1038/374276a0. [DOI] [PubMed] [Google Scholar]
- Markelz NH, Costich DE, Brutnell TP. Photomorphogenic responses in maize seedling development. Plant Physiology. 2003;133:1578–1591. doi: 10.1104/pp.103.029694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka M, Ichikawa H, Saito A, Tada Y, Fujimura T, Kano-Murakami Y. Expression of a rice homeobox gene causes altered morphology of transgenic plants. The Plant Cell. 1993;5:1039–1048. doi: 10.1105/tpc.5.9.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka M, Kyozuka J, Shimamoto K, Kano-Murakami Y. The promoters of two carboxylases in a C4 plant (maize) direct cell-specific, light-regulated expression in a C3 plant (rice) The Plant Journal. 1994;6:311–319. doi: 10.1046/j.1365-313x.1994.06030311.x. [DOI] [PubMed] [Google Scholar]
- Nick H, Bowen B, Ferl RJ, Gilbert W. Detection of cytosine methylation in the maize alcohol dehydrogenase gene by genomic sequencing. Nature. 1986;319:243–246. [Google Scholar]
- Nomura M, Sentoku N, Nishimura A, et al. The evolution of C4 plants: acquisition of cis-regulatory sequences in the promoter of C4-type pyruvate, orthophosphate dikinase gene. The Plant Journal. 2000;22:211–221. doi: 10.1046/j.1365-313x.2000.00726.x. [DOI] [PubMed] [Google Scholar]
- Offermann S, Danker T, Dreymüller D, Kalamajka R, Töpsch S, Weyand K, Peterhänsel C. Illumination is necessary and sufficient to induce histone acetylation independent of transcriptional activity at the C4-specific phosphoenolpyruvate carboxylase promoter in maize. Plant Physiology. 2006;141:1078–1088. doi: 10.1104/pp.106.080457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okimoto R, Sachs MM, Porter EK, Freeling M. Patterns of polypeptide synthesis in various maize organs under anaerobiosis. Planta. 1980;150:89–94. doi: 10.1007/BF00385619. [DOI] [PubMed] [Google Scholar]
- Sawers RJ, Liu P, Anufrikova K, Hwang JT, Brutnell TP. A multi-treatment experimental system to examine photosynthetic differentiation in the maize leaf. BMC Genomics. 2007;8:12. doi: 10.1186/1471-2164-8-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäffner AR, Sheen J-Y. Maize C4 photosynthesis involves differential regulation of phosphoenol pyruvate carboxylase genes. The Plant Journal. 1992;2:221–232. doi: 10.1046/j.1365-313x.1992.t01-44-00999.x. [DOI] [PubMed] [Google Scholar]
- Spena A, Viotti A, Pirotta V. Two adjacent genomic zein sequences: structure, organization and tissue-specific restriction pattern. Journal of Molecular Biology. 1983;169:799–811. doi: 10.1016/s0022-2836(83)80137-5. [DOI] [PubMed] [Google Scholar]
- Stockhaus J, Schlue U, Koczor M, Chitty JA, Taylor WC, Westhoff P. The promoter of the gene encoding the C4 form of phospho enolpyruvate carboxylase directs mesophyll-specific expression in transgenic C4 Flaveria spp. The Plant Cell. 1997;9:479–89. doi: 10.1105/tpc.9.4.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nature Reviews Genetics. 2008;9:465–476. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- Taniguchi M, Izawa K, Ku MSB, Lin JH, Saito H, Ishida Y, Ohta S, Komari T, Matsuoka M, Sugiyama T. Binding of cell type-specific nuclear proteins to the 5′-flanking region of maize C4 phospho enolpyruvate carboxylase gene confers its differential transcription in mesophyll cells. Plant Molecular Biology. 2000;44:543–57. doi: 10.1023/a:1026565027772. [DOI] [PubMed] [Google Scholar]
- Thomson JP, Skene PJ, Selfridge J, et al. CpG islands influence chromatin structure via the CpG-binding protein Cfp1. Nature. 2010;464:1082–1086. doi: 10.1038/nature08924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voo KS, Carlone DL, Jacobsen BM, Flodin A, Skalnik DG. Cloning of a mammalian transcriptional activator that binds unmethylated CpG motifs and shares a CXXC domain with DNA methyltransferase, human trithorax, and methyl-CpG binding domain protein 1. Molecular Cell Biology. 2000;20:2108–2121. doi: 10.1128/mcb.20.6.2108-2121.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang RYH, Gehrke CW, Ehrlich M. Comparison of bisulphite modification of 5-methyldeoxycytidine and deoxycytidine residues. Nucleic Acids Research. 1980;8:4777–4790. doi: 10.1093/nar/8.20.4777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber M, Schubeler D. Genomic patterns of DNA methylation: targets and function of an epigenetic mark. Current Opinion in Cell Biology. 2007;19:273–280. doi: 10.1016/j.ceb.2007.04.011. [DOI] [PubMed] [Google Scholar]
- Yanagisawa S. A novel DNA-binding domain that may form a single zinc finger motif. Nucleic Acids Research. 1995;23:3403–3410. doi: 10.1093/nar/23.17.3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagisawa S, Izui K. Multiple interactions between tissue-specific nuclear proteins and the promoter of the phosphoenol pyruvate carboxylase gene for C4 photosynthesis in. Zea mays. Molecular and General Genetics. 1990;224:325–332. doi: 10.1007/BF00262425. [DOI] [PubMed] [Google Scholar]
- Yanagisawa S, Izui K. MNF1, a leaf tissue-specific DNA-binding protein of maize, interacts with the cauliflower mosaic virus 35S promoter as well as the C4 photosynthetic phosphoenol pyruvate carboxylase gene promoter. Plant Molecular Biology. 1992;19:545–553. doi: 10.1007/BF00026781. [DOI] [PubMed] [Google Scholar]
- Yanagisawa S, Izui K. Molecular cloning of two DNA-binding proteins of maize that are structurally different but interact with the same sequence motif. Journal of Biological Chemistry. 1993;268:16028–16036. [PubMed] [Google Scholar]
- Yanagisawa S, Sheen J-Y. Involvement of maize Dof zinc finger proteins in tissue-specific and light-regulated gene expression. The Plant Cell. 1998;10:75–89. doi: 10.1105/tpc.10.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.