Abstract
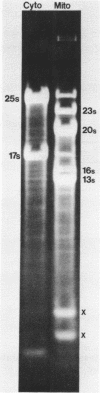
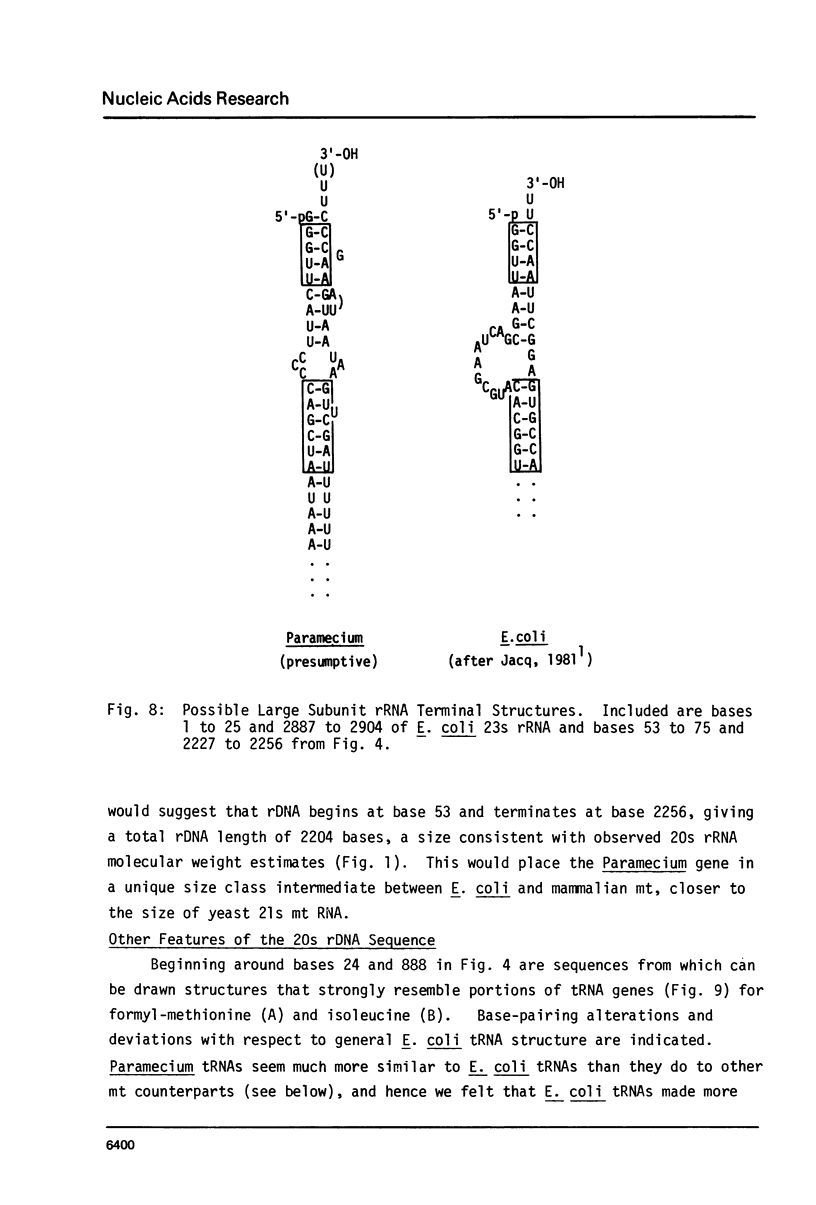
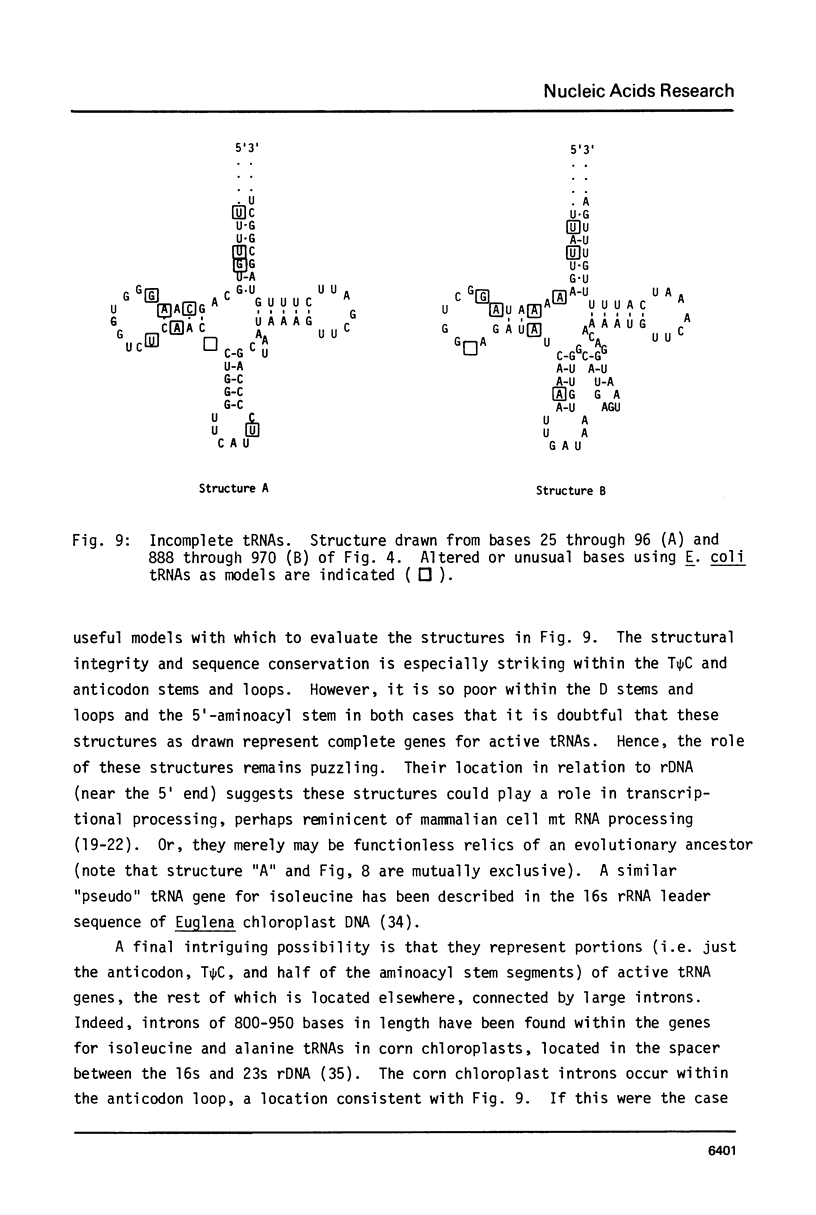
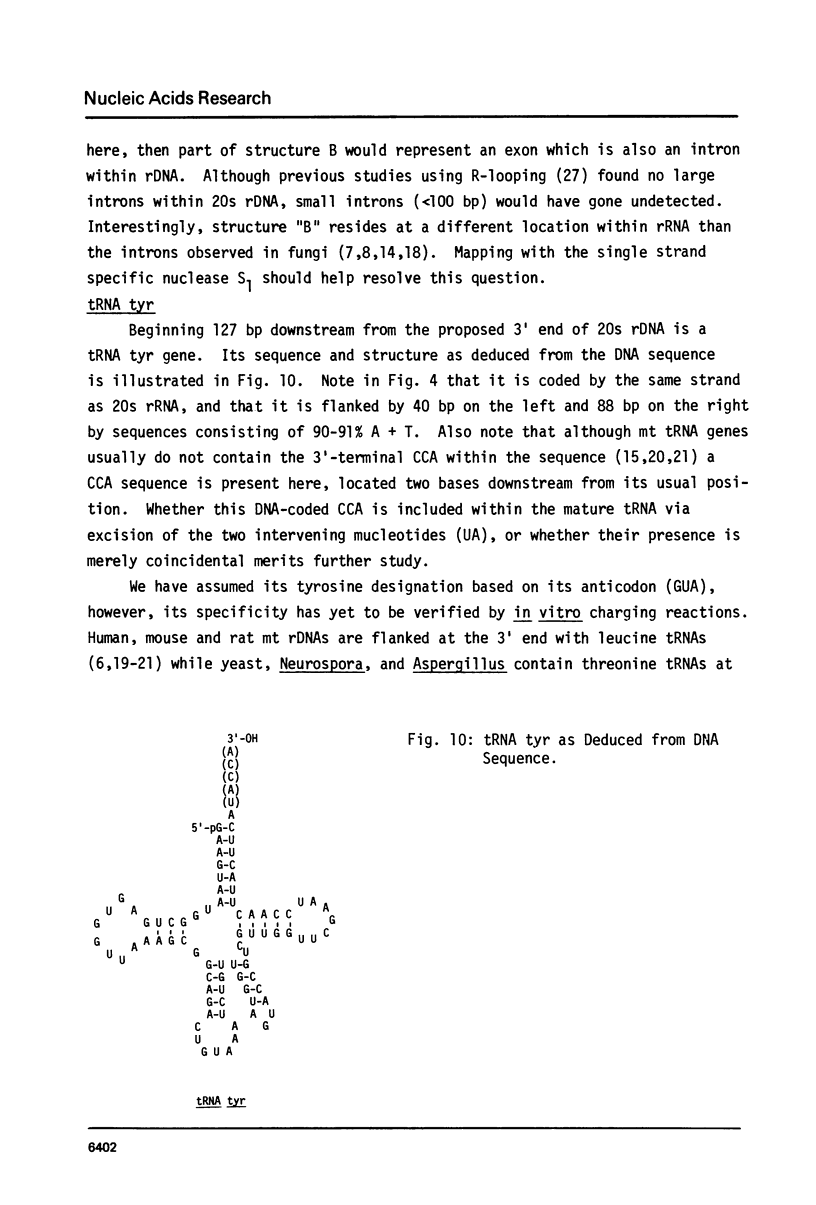
The sequence and structure of the large (20s) mitochondrial (mt) rRNA gene and flanking regions from Paramecium primaurelia have been determined. The gene contains two regions of strong homology with other large mt rRNAs: one 44-base region near the 5' end and a 321-base region near the 3' end. Another region of strong homology to both ends of E. coli 23s RNA exists at loci consistent with these regions. The Paramecium gene appears to be 2204 bases in length and contains slightly more homology to E. coli rRNA than its mammalian or fungal counterparts. The gene, located about 1200 bp from the replicative terminal end of the linear mt DNA, is transcribed in the same polarity as replication. Previous R-looping studies detected no large introns within the gene. Here we describe sequences resembling degenerate rRNAs, one of which could represent a small intron. A tRNA tyr gene was found on the same DNA strand, 127 bp downstream from the large rRNA presumptive 3' end. The tRNA is flanked on both sides by short DNA regions of approximately 90% A + T content.
Full text
PDF


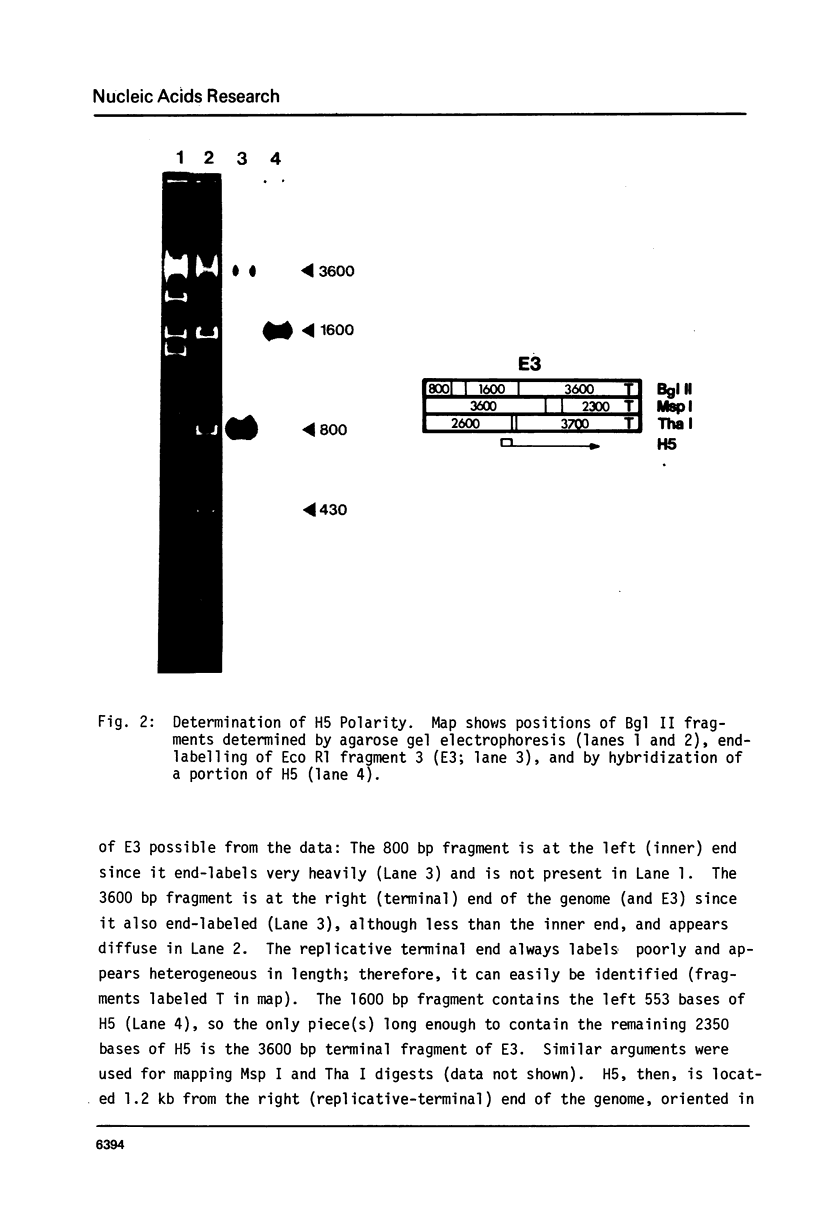




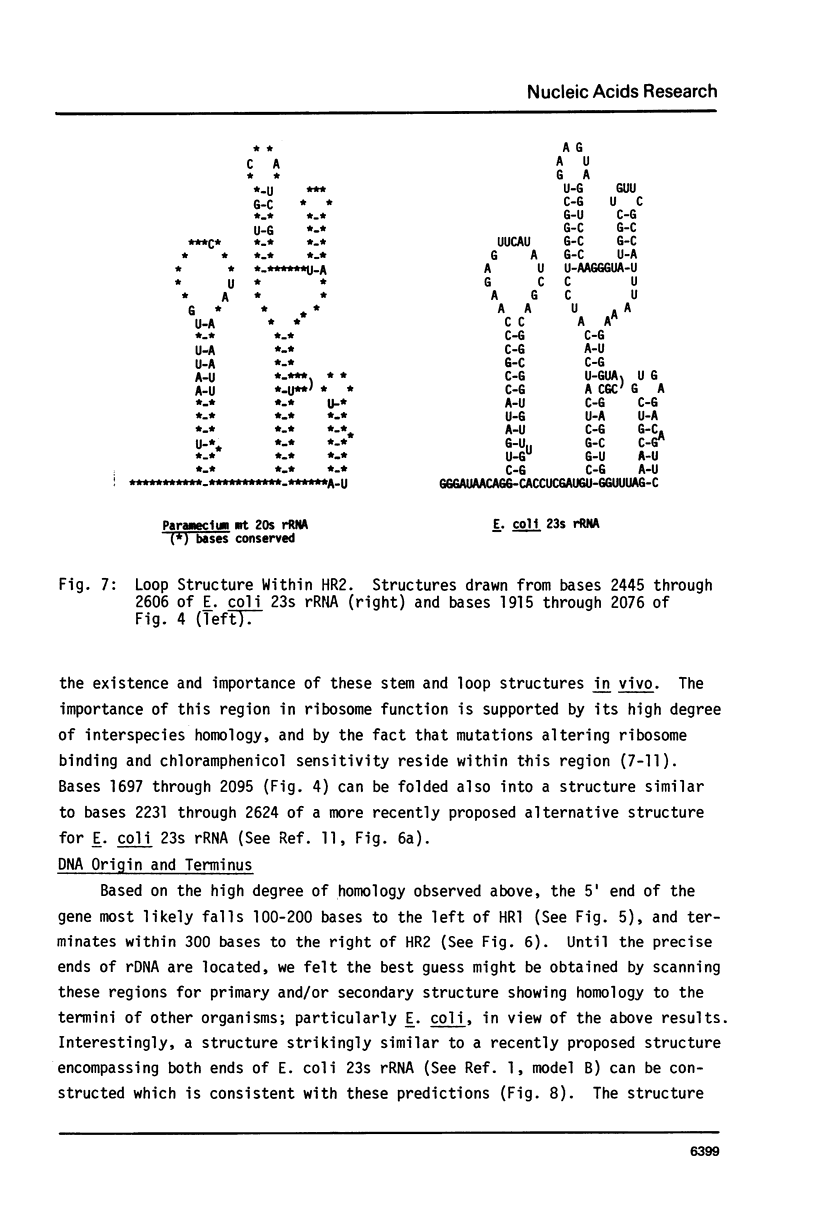




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson S., Bankier A. T., Barrell B. G., de Bruijn M. H., Coulson A. R., Drouin J., Eperon I. C., Nierlich D. P., Roe B. A., Sanger F. Sequence and organization of the human mitochondrial genome. Nature. 1981 Apr 9;290(5806):457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- Baer R. J., Dubin D. T. Methylated regions of hamster mitochondrial ribosomal RNA: structural and functional correlates. Nucleic Acids Res. 1981 Jan 24;9(2):323–337. doi: 10.1093/nar/9.2.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey J. M., Davidson N. Methylmercury as a reversible denaturing agent for agarose gel electrophoresis. Anal Biochem. 1976 Jan;70(1):75–85. doi: 10.1016/s0003-2697(76)80049-8. [DOI] [PubMed] [Google Scholar]
- Barrell B. G., Anderson S., Bankier A. T., de Bruijn M. H., Chen E., Coulson A. R., Drouin J., Eperon I. C., Nierlich D. P., Roe B. A. Different pattern of codon recognition by mammalian mitochondrial tRNAs. Proc Natl Acad Sci U S A. 1980 Jun;77(6):3164–3166. doi: 10.1073/pnas.77.6.3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battey J., Clayton D. A. The transcription map of human mitochondrial DNA implicates transfer RNA excision as a major processing event. J Biol Chem. 1980 Dec 10;255(23):11599–11606. [PubMed] [Google Scholar]
- Blanc H., Wright C. T., Bibb M. J., Wallace D. C., Clayton D. A. Mitochondrial DNA of chloramphenicol-resistant mouse cells contains a single nucleotide change in the region encoding the 3' end of the large ribosomal RNA. Proc Natl Acad Sci U S A. 1981 Jun;78(6):3789–3793. doi: 10.1073/pnas.78.6.3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohnert H. J., Gordon K. H., Crouse E. J. Homologies among ribosomal RNA and messenger RNA genes in chloroplasts, mitochondria and E. coli. Mol Gen Genet. 1980;179(3):539–545. doi: 10.1007/BF00271743. [DOI] [PubMed] [Google Scholar]
- Bonitz S. G., Berlani R., Coruzzi G., Li M., Macino G., Nobrega F. G., Nobrega M. P., Thalenfeld B. E., Tzagoloff A. Codon recognition rules in yeast mitochondria. Proc Natl Acad Sci U S A. 1980 Jun;77(6):3167–3170. doi: 10.1073/pnas.77.6.3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borst P., Grivell L. A. The mitochondrial genome of yeast. Cell. 1978 Nov;15(3):705–723. doi: 10.1016/0092-8674(78)90257-x. [DOI] [PubMed] [Google Scholar]
- Borst P. Mitochondrial nucleic acids. Annu Rev Biochem. 1972;41:333–376. doi: 10.1146/annurev.bi.41.070172.002001. [DOI] [PubMed] [Google Scholar]
- Bos J. L., Osinga K. A., Van der Horst G., Hecht N. B., Tabak H. F., Van Ommen G. J., Borst P. Splice point sequence and transcripts of the intervening sequence in the mitochondrial 21S ribosomal RNA gene of yeast. Cell. 1980 May;20(1):207–214. doi: 10.1016/0092-8674(80)90248-2. [DOI] [PubMed] [Google Scholar]
- Brosius J., Dull T. J., Noller H. F. Complete nucleotide sequence of a 23S ribosomal RNA gene from Escherichia coli. Proc Natl Acad Sci U S A. 1980 Jan;77(1):201–204. doi: 10.1073/pnas.77.1.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummings D. J., Maki R. A., Conlon P. J., Laping J. Anatomy of mitochondrial DNA from Paramecium aurelia. Mol Gen Genet. 1980;178(3):499–510. doi: 10.1007/BF00337854. [DOI] [PubMed] [Google Scholar]
- Delius H., Koller B. Sequence homologies between Escherichia coli and chloroplast ribosomal DNA as seen by heteroduplex analysis. J Mol Biol. 1980 Sep 15;142(2):247–261. doi: 10.1016/0022-2836(80)90048-0. [DOI] [PubMed] [Google Scholar]
- Dubin D. T., Timko K. D., Baer R. J. The 3' terminus of the large ribosomal subunit ("17S") RNA from hamster mitochondria is ragged and oligoadenylated. Cell. 1981 Jan;23(1):271–278. doi: 10.1016/0092-8674(81)90291-9. [DOI] [PubMed] [Google Scholar]
- Dujon B. Sequence of the intron and flanking exons of the mitochondrial 21S rRNA gene of yeast strains having different alleles at the omega and rib-1 loci. Cell. 1980 May;20(1):185–197. doi: 10.1016/0092-8674(80)90246-9. [DOI] [PubMed] [Google Scholar]
- Edwards K., Kössel H. The rRNA operon from Zea mays chloroplasts: nucleotide sequence of 23S rDNA and its homology with E.coli 23S rDNA. Nucleic Acids Res. 1981 Jun 25;9(12):2853–2869. doi: 10.1093/nar/9.12.2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eperon I. C., Anderson S., Nierlich D. P. Distinctive sequence of human mitochondrial ribosomal RNA genes. Nature. 1980 Jul 31;286(5772):460–467. doi: 10.1038/286460a0. [DOI] [PubMed] [Google Scholar]
- Glotz C., Zwieb C., Brimacombe R., Edwards K., Kössel H. Secondary structure of the large subunit ribosomal RNA from Escherichia coli, Zea mays chloroplast, and human and mouse mitochondrial ribosomes. Nucleic Acids Res. 1981 Jul 24;9(14):3287–3306. doi: 10.1093/nar/9.14.3287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard J. M., Cummings D. J. Mitochondrial DNA replication in Paramecium aurelia. Cross-linking of the initiation end. J Mol Biol. 1977 Jan 15;109(2):327–344. doi: 10.1016/s0022-2836(77)80037-5. [DOI] [PubMed] [Google Scholar]
- Goddard J. M., Cummings D. J. Structure and replication of mitochondrial DNA from Paramecium aurelia. J Mol Biol. 1975 Oct 5;97(4):593–609. doi: 10.1016/s0022-2836(75)80061-1. [DOI] [PubMed] [Google Scholar]
- Goodman H. M., Abelson J., Landy A., Brenner S., Smith J. D. Amber suppression: a nucleotide change in the anticodon of a tyrosine transfer RNA. Nature. 1968 Mar 16;217(5133):1019–1024. doi: 10.1038/2171019a0. [DOI] [PubMed] [Google Scholar]
- Green M. R., Grimm M. F., Goewert R. R., Collins R. A., Cole M. D., Lambowitz A. M., Heckman J. E., Yin S., RajBhandary U. L. Transcripts and processing patterns for the ribosomal RNA and transfer RNA region of Neurospora crassa mitochondrial DNA. J Biol Chem. 1981 Feb 25;256(4):2027–2034. [PubMed] [Google Scholar]
- Heckman J. E., Alzner-Deweerd B., RajBhandary U. L. Interesting and unusual features in the sequence of Neurospora crassa mitochondrial tyrosine transfer RNA. Proc Natl Acad Sci U S A. 1979 Feb;76(2):717–721. doi: 10.1073/pnas.76.2.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckman J. E., RajBhandary U. L. Organization of tRNA and rRNA genes in N. crassa mitochondria: intervening sequence in the large rRNA gene and strand distribution of the RNA genes. Cell. 1979 Jul;17(3):583–595. doi: 10.1016/0092-8674(79)90266-6. [DOI] [PubMed] [Google Scholar]
- Heckman J. E., Sarnoff J., Alzner-DeWeerd B., Yin S., RajBhandary U. L. Novel features in the genetic code and codon reading patterns in Neurospora crassa mitochondria based on sequences of six mitochondrial tRNAs. Proc Natl Acad Sci U S A. 1980 Jun;77(6):3159–3163. doi: 10.1073/pnas.77.6.3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacq B. Sequence homologies between eukaryotic 5.8S rRNA and the 5' end of prokaryotic 23S rRNa: evidences for a common evolutionary origin. Nucleic Acids Res. 1981 Jun 25;9(12):2913–2932. doi: 10.1093/nar/9.12.2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearsey S. E., Craig I. W. Altered ribosomal RNA genes in mitochondria from mammalian cells with chloramphenicol resistance. Nature. 1981 Apr 16;290(5807):607–608. doi: 10.1038/290607a0. [DOI] [PubMed] [Google Scholar]
- Koch W., Edwards K., Kössel H. Sequencing of the 16S-23S spacer in a ribosomal RNA operon of Zea mays chloroplast DNA reveals two split tRNA genes. Cell. 1981 Jul;25(1):203–213. doi: 10.1016/0092-8674(81)90245-2. [DOI] [PubMed] [Google Scholar]
- Köchel H. G., Lazarus C. M., Basak N., Küntzel H. Mitochondrial tRNA gene clusters in Aspergillus nidulans: organization and nucleotide sequence. Cell. 1981 Feb;23(2):625–633. doi: 10.1016/0092-8674(81)90158-6. [DOI] [PubMed] [Google Scholar]
- Lazarus C. M., Lünsdorf H., Hahn U., Stepień P. P., Küntzel H. Physical map of Aspergillus nidulans mitochondrial genes coding for ribosomal RNA: an intervening sequence in the large rRNA cistron. Mol Gen Genet. 1980 Feb;177(3):389–397. doi: 10.1007/BF00271477. [DOI] [PubMed] [Google Scholar]
- Levens D., Ticho B., Ackerman E., Rabinowitz M. Transcriptional initiation and 5' termini of yeast mitochondrial RNA. J Biol Chem. 1981 May 25;256(10):5226–5232. [PubMed] [Google Scholar]
- Ojala D., Merkel C., Gelfand R., Attardi G. The tRNA genes punctuate the reading of genetic information in human mitochondrial DNA. Cell. 1980 Nov;22(2 Pt 2):393–403. doi: 10.1016/0092-8674(80)90350-5. [DOI] [PubMed] [Google Scholar]
- Orozco E. M., Jr, Rushlow K. E., Dodd J. R., Hallick R. B. Euglena gracilis chloroplast ribosomal RNA transcription units. II. Nucleotide sequence homology between the 16 S--23 S ribosomal RNA spacer and the 16 S ribosomal RNA leader regions. J Biol Chem. 1980 Nov 25;255(22):10997–11003. [PubMed] [Google Scholar]
- Parish J. H., Kirby K. S. Reagents which reduce interactions between ribosomal RNA and rapidly labelled RNA from rat liver. Biochim Biophys Acta. 1966 Dec 21;129(3):554–562. doi: 10.1016/0005-2787(66)90070-0. [DOI] [PubMed] [Google Scholar]
- Saccone C., Cantatore P., Gadaleta G., Gallerani R., Lanave C., Pepe G., Kroon A. M. The nucleotide sequence of the large ribosomal RNA gene and the adjacent tRNA genes from rat mitochondria. Nucleic Acids Res. 1981 Aug 25;9(16):4139–4148. doi: 10.1093/nar/9.16.4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Van Etten R. A., Walberg M. W., Clayton D. A. Precise localization and nucleotide sequence of the two mouse mitochondrial rRNA genes and three immediately adjacent novel tRNA genes. Cell. 1980 Nov;22(1 Pt 1):157–170. doi: 10.1016/0092-8674(80)90164-6. [DOI] [PubMed] [Google Scholar]
- Veldman G. M., Klootwijk J., de Jonge P., Leer R. J., Planta R. J. The transcription termination site of the ribosomal RNA operon in yeast. Nucleic Acids Res. 1980 Nov 25;8(22):5179–5192. doi: 10.1093/nar/8.22.5179. [DOI] [PMC free article] [PubMed] [Google Scholar]