Abstract
The genetic basis of the phenotypic diversity of yeast is still poorly understood. Wine yeast strains have specific abilities to grow and ferment under stressful conditions compared with other strains, but the genetic basis underlying these traits is unknown. Understanding how sequence variation influences such phenotypes is a major challenge to address adaptation mechanisms of wine yeast. We aimed to identify the genetic basis of fermentation traits and gain insight into their relationships with variations in gene expression among yeast strains. We combined fermentation trait QTL mapping and expression profiling of fermenting cells in a segregating population from a cross between a wine yeast derivative and a laboratory strain. We report the identification of QTL for various fermentation traits (fermentation rates, nitrogen utilization, metabolites production) as well as expression QTL (eQTL). We found that many transcripts mapped to several eQTL hotspots and that two of them overlapped with QTL for fermentation traits. A QTL controlling the maximal fermentation rate and nitrogen utilization overlapping with an eQTL hotspot was dissected. We functionally demonstrated that an allele of the ABZ1 gene, localized in the hotspot and involved in p-aminobenzoate biosynthesis, controls the fermentation rate through modulation of nitrogen utilization. Our data suggest that the laboratory strain harbors a defective ABZ1 allele, which triggers strong metabolic and physiological alterations responsible for the generation of the eQTL hotspot. They also suggest that a number of gene expression differences result from some alleles that trigger major physiological disturbances.
Keywords: wine yeast, fermentation, QTL, transcriptome, p-aminobenzoate
Industrial wine yeast strains exhibit specific traits making them suitable for alcoholic fermentation. These yeasts are specifically adapted to stressful conditions and can ferment efficiently under conditions of high ethanol concentrations, low pH, nutrient starvation, while producing suitable metabolites, especially aroma compounds. Although a large phenotypic diversity has been described, the molecular basis remains largely unknown. Various molecular mechanisms may contribute to wine yeast adaptation, including gene deletions or amplifications, chromosome polyploidy (Dunham et al. 2002; Dunn et al. 2005; Infante et al. 2003; Perez-Ortin et al. 2002; Rachidi et al. 1999), and nucleotide polymorphisms (Borneman et al. 2008; Doniger et al. 2008; Goffeau et al. 1996; Kellis et al. 2003; Liti et al. 2009; Schacherer et al. 2009; Wei et al. 2007). Analysis of the complete genome sequence of a commercial wine yeast also revealed that new (non-Saccharomyces) genes arising from horizontal gene transfer were present in the wine strains (Novo et al. 2009).
Linking genetic variation with phenotypic diversity will help to increase our understanding of the adaptation of yeast to industrial stressful environments and will facilitate strain improvement through breeding and genetic engineering. Quantitative trait locus (QTL) mapping is a proven approach to map the genetic variation responsible for quantitative traits in S. cerevisiae. It was successfully applied to high-temperature growth (Sinha et al. 2008; Sinha et al. 2006; Steinmetz et al. 2002), sporulation (Ben-Ari et al. 2006; Deutschbauer and Davis 2005; Gerke et al. 2006; Katou et al. 2009), cell morphology (Nogami et al. 2007), drug sensitivity (Kim and Fay, 2007), ethanol tolerance and growth (Hu et al. 2007; Katou et al. 2009; Smith and Kruglyak 2008), and flocculation (Brauer et al. 2006). QTL approaches have also been used to dissect the molecular basis of several wine yeast metabolic traits such as acetic acid production, hydrogen sulphide production, and release of volatile phenol (Marullo et al. 2006).
Several studies have highlighted the role of expression variations on associated phenotypes in yeast (Brown et al. 2008; Cavalieri et al. 2000; Fay et al. 2004). Variations in expression between wine strains have been reported in various genome-wide analyses (Rossignol 2004; Zuzuarregui et al. 2006). However, their impact on strain properties is poorly understood. Genomic approaches to variations in gene expression have been used to establish relationships between differences in transcript abundance and genetic polymorphisms via the mapping of expression QTL (eQTL) (Jansen and Nap 2001; Brem et al. 2002; Yvert et al. 2003; Brem and Kruglyak 2005; Ronald and Akey 2007; Ansel et al. 2008)). Combining the search of eQTL with that of QTL of industrially relevant traits may be of interest to gain insight into the relationships between expression variations and wine yeast traits. Indeed when an eQTL colocalizes with a phenotypic QTL, one can hypothesize that a common polymorphism controls transcription differences and the physiological trait.
Here, we report an integrated approach where we searched for both phenotypic fermentation QTL and eQTL in the laboratory yeast strain S288c and a haploid derivative of the EC1118 industrial wine strain. We identified QTL for various fermentation traits, including kinetic traits, as well as eQTL for many transcripts. We show that eQTL display hotspot regions that control many transcripts and interestingly, two of them overlap with fermentation traits QTL. We functionally characterized a QTL that controlled the maximal fermentation rate and showed that a gene involved in p-aminobenzoate synthesis (ABZ1), probably defective in the strain S288C, modulates the fermentation rate by controlling nitrogen utilization. The relationships between structural alteration of ABZ1 and eQTL hotspot generation are discussed.
Materials and Methods
Strains, growth conditions, and fermentation conditions
The two parental strains compared in this study are the standard S288c (MATα; SUC2; gal2) strain and a haploid derivative of the industrial EC1118 (HO/ho) wine yeast strain, which is referred to as 59A (MATa; ho). This strain is phototrophic and has fermentation properties close to the diploid EC1118 strain (see Figure 1). The strains BY4742 (MATα; his3Δ1; leu2Δ0; lys2Δ0; ura3Δ0 and BY4742∆ABZ1 (Matα; his3Δ1; leu2Δ0; lys2Δ0; ura3Δ0; YNR033w::kanMX4) were used for hemizygous constructions.
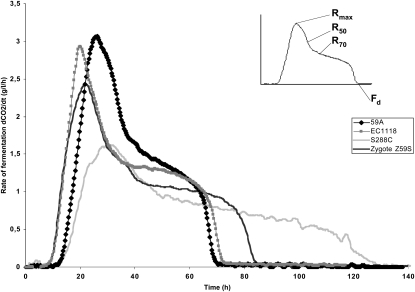
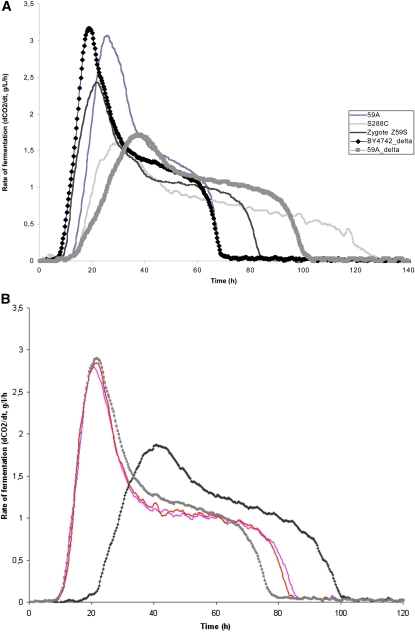
Figure 1 .
Fermentation profiles of industrial and laboratory strains. Fermentation rate profiles of the industrial wine yeast EC1118 (gray squares), its haploid derivative 59A (black diamonds), the laboratory strain S288C (gray line), and the hybrid Z59S (black line).
Fermentation experiments and precultures were carried out at 28°C on synthetic MS300 medium, which mimics a natural must described in Bely et al. (1990). After preculture in 125 ml shake flasks for 24 h, fermentations were performed in 1.2 l fermenters equipped with airlocks to maintain anaerobiosis and with constant stirring.
Construction of strains for functional analysis
Hybrid strains BY4742∆ABZ1/59A and BY4742/59A∆ABZ1 were constructed for the functional characterization of the ABZ1 candidate gene via a reciprocal hemizygosity analysis.
The BY4742∆ABZ1strain, which isogenic to S288c but with a copy of the ABZ1 gene deleted, was obtained from the Euroscarff society. The copy of ABZ1 in the 59A strain was deleted and replaced with a KanMX4 deletion module using the short-flanking homology PCR (SFH-PCR) technique (Schiestl and Gietz 1989). The two custom 60-mer primers used begin with 40 nucleotides that are identical to the upstream or the downstream region of ABZ1 followed by 20 nucleotides that can amplify the KanMX4 module, ABZ1 forward: ATGCTGTCCGATACAATTGACACAAAGCAACAACAGCAACTTCGTACGCTGCAGGTCGAC; and ABZ1 reverse: CTACATGAAAATTTGCAAGTTGCTCTCCAACTTGGTGTACGCATAGGCCACTAGTGGATCTG. After transformation using the lithium acetate transformation protocol of Schiestl and Gietz (1989), yeasts were grown with YEPD plus geneticin (200 µg/l) overnight at 28°C to select cells with the KanMX4 integrated module. Clones were finally tested for integration by PCR on genomic DNA with the primers ABZ1 forwardctrl: TTATCGGTGCGGCAATAAAG and ABZ1 reversectrl: TATGTGACCGTCAGGACG.
Measure of phenotypic parameters
To characterize the fermentation parameters, the fermenters were automatically weighed every 20 min and the rate of CO2 production was calculated from the weight loss by a method of polynomial smoothing. The number of cells was determined with an electronic particle counter (Beckman Coulter). At the end of fermentation, samples were analyzed by high-pressure liquid chromatography (HPLC) using an HPX-87H ion-exclusion column (Bio-Rad) to assess the sugars (glucose, fructose), the organic acids (succinic acid, acetic acid, pyruvic acid), and alcohols (glycerol, ethanol) according to the procedure described by Calull et al. (1992). Amino acids were separated by ion-exchange chromatography on an anionic Ultropac-8 lithium-form resin (Amersham Pharmacia Biotech) with a Chromakon 400 (Kontron) and a Biochrom 20 analyzer (Amersham Pharmacia Biotech). Amino acids were detected by reaction with nihydrin. Ammonia was measured using the enzymatic method from Bergmeyer and Beutler (1990) based on its conversion into glutamate by the glutamate dehydrogenase.
Fermentation parameters of the parental strains were determined from five independent fermentations and from duplicated fermentations for the other strains (segregants, hemizygous strains). Metabolites were measured in five biological replicates for the parental strains and in duplicate for the segregants, except the amino acids, which were determined from a single fermentation.
Gene expression analysis
Microarray hybridization and image analysis was performed at the Biochip Platform in Toulouse (http://biopuces.genotoul.fr). The microarrays were obtained by spotting in duplicate the 6308 oligonucleotides (70-mer) from the yeast genome Oligoset (Operon) onto UltraGAP chips. Gene expression was analyzed for five independent cultures of each parent and one culture of each segregant. RNA was isolated at the midtime of fermentation when half of the sugar present in the medium was consumed (equivalent to 45 g CO2), and then labeled with the Chipshot direct labeling and cleanup kit (Promega). Each sample labeled with the Cy3 dye was compared with a common reference pool labeled with Cy5 dye and composed of a mix of equivalent concentration of the RNA of each segregant of the population (star design). Hybridizations were done twice and thus technical replicates are available for each measurement. Data analysis was performed using the R 2.9.2 software and the limma package (R Development Core Team 2010; Smyth 2005, Smyth et al. 2005; Smyth and Speed 2003). Within-array normalization was performed using the print-tip-loess method followed by a quantile method for between-slide normalization. For each measurement, the two technical replicates were averaged. Gene expression between the two parental strains (2 × 5 microarrays) was compared using t-tests. Differentially expressed genes were defined by filtering on adjusted P = 0.01 to control the false discovery rate (FDR) (Benjamini et al. 2001). For linkage analysis in the population of segregants, transcripts with a low signal-to-noise ratio were filtered out: we required the mean red (Cy5) signal to be three times greater than the mean background signal (Cy5). The complete data set is available on the Gene Expression Omnibus database (accession number GSE26437) (Edgar et al. 2002). An analysis of the correlation between gene expression and phenotype parameters was carried out using the Spearman correlation coefficient and the associated statistical test, followed by a correction of the multiplicity using Benjamini and Hochberg procedure. We selected genes with correlation coefficients > 0.6 and adjusted P < 0.05 to control the FDR.
Genotyping and linkage analysis of the population of segregants
Genomic DNA was isolated, fragmented, labeled, and hybridized to Affymetrix YGS98 microarrays as previously described (Winzeler et al. 1999). This procedure was applied three times for the parental strains S288c and 59A and one time for each of the 30 segregants. Probes with a unique match on the S288c genome were then screened to identify biallelic markers using the same statistical procedure as Brem et al. (2002). First, probes identified as low perfect-match (PM) vs. mismatch (MM) hybridizers to S288c genomic DNA were discarded. All further analyses were performed on normalized log(PM/MM) values. We selected probe pairs that had high hybridization differences between S288c and 59A using the Z and z statistics described in Brem et al. (2002). A second test based on a clustering algorithm was used to select probe pairs for evidence of 2:2 segregation in the segregating population. Genotypes were then inferred from cluster assignments as described in Brem et al. (2002). This resulted in the selection of 2465 probes. The probes were further filtered using the genomic sequences of the parental strains S288c and 59A. For each probe, the presence of polymorphisms in the 59A genome was confirmed. We used Illumina high-throughput sequencing to obtain SNP information for 59A. Genomic DNA of the strain 59A was sequenced using Illumina Genome Analyzer II technology (Illumina, Inc.) with paired reads of 36 bp using standard manufacturer protocols. Read sequences (deposited in the NCBI Sequence Read Archive as study SRP004704) were aligned to the S288c genome with MAQ (ver. 0.7.1) (Li et al. 2008) using default parameters to detect SNP. This filtering step resulted in a map of 1834 markers used in the linkage analysis.
For each phenotype and each expression trait, linkage analysis was performed using a normal model with the Haley-Knott regression method implemented in the R/qtl package (Broman et al. 2003). Logarithm of odds (LOD) scores were computed for each marker every 2.5 cM. An interval estimate of the location of each QTL or eQTL was obtained as the 1-LOD support interval: the region in which the LOD score is within 1 unit of the peak LOD score. Statistical significance was assessed by random permutation of the phenotypes or expression levels relative to the genotype data. For eQTL, the permutation was performed 10 times, and the average number of transcripts showing linkage at a specific LOD score was used to calculate FDR. For each phenotypic QTL, 1000 permutations were used to calculate an individual LOD score threshold and FDR.
The GenYeasTrait resource
A database named GenYeasTrait, which is dedicated to the analysis of QTL and eQTL data, was developed using the Gmod/Gbrowse software version 2.15 (Stein et al. 2002). It includes the annotation of the S288c genome from the Saccharomyces Genome Database, SNPs detected from the comparison of the 59A strain sequence with the S288c reference genome, and the fermentation and expression QTL of the project. The database can be queried by using the Gbrowse interface, which allows user friendly graphical representations of features on chromosomes and facilitates data exploration. It is available at http://genome.jouy.inra.fr/genyeastrait/.
Results
Characterization of segregants from a cross between a derivative haploid of the wine yeast EC1118 and the laboratory strain S288C
To generate a population of segregants for QTL analysis, the S288C laboratory strain and the 59A strain (a haploid derivative of the EC1118 strain) were used for crossing (Figure 1). The fermentation profile of the haploid derivative 59A does not however perfectly overlap with the EC1118 pattern. Such variation is consistent with the known heterozygosity of the wine yeast EC1118 genome that was shown to contain 0.2% of heterozygous SNP (Novo et al. 2009). The hybrid (Z59S) obtained by crossing 59A with S288C displays a fermentation profile intermediate between the two parental strains (Figure 1). Fermentation rate profiles obtained in a synthetic medium (MS300) that mimics natural grape musts provided relevant criteria to characterize the fermentation capacity of the strains. As shown in Figure 1, the two parental strains exhibit very different fermentation profiles with a much higher maximum fermentation rate (Rmax) for 59A compared with S288c. Furthermore, the 59A strain also shows a higher fermentation rate at 50% of sugar utilization (R50) with a consequently shorter duration of fermentation duration (Fd). All of these data are consistent with a higher fermentation capacity of the 59A wine strain derivative compared vs. the laboratory strain. Under such fermentation conditions, the growth phase is restricted to the period of increase of the fermentation rate until Rmax, and then cells are subsequently fermented in a stationary phase (Rossignol et al. 2003).
The zygote Z59S was used to generated a population of haploid segregants. The spores obtained had a low viability with 54% viable. The proportion of asci with four viable spores was low (only 15%), and a majority (around 50%) had three viable spores. We kept only one complete tetrad in the analyzed population, and the other segregants were selected from 20 different asci. The 30 haploid progenies where characterized under the fermentation conditions previously described. To describe the fermentation capacity, we considered the fermentation rate at three different stages of fermentation: Rmax, R50 (fermentation rate at 50% fermentation), and R70 (fermentation rate at 70% fermentation). Other fermentation traits, such as fermentation duration (Fd) and cellular population (Cp), were taken into account. We also considered metabolites, such as the amount of assimilated nitrogen (Nass), glycerol (gly), acetic acid (ace), succinic acid (suc) and pyruvic acid (pyr) (Table S1, Table S2, and Table S3). We observed a high variability in the segregants fermentation profiles (supporting information, Figure S1). Several traits values (Rmax, R50, gly, pyr, ace) were roughly normally distributed within the population (P > 0.05, Shapiro-Wilk normality test), supporting a polygenic determinism of these phenotypes (Figure S2). The amount of assimilated nitrogen and, to some extent, R70 and Fd, displayed a bimodal distribution suggesting a possible control by a major locus. Furthermore, all of these parameters, except R50, displayed values outside the parental range. This transgressive segregation indicates that alleles with opposite effects were present in the parental strains.
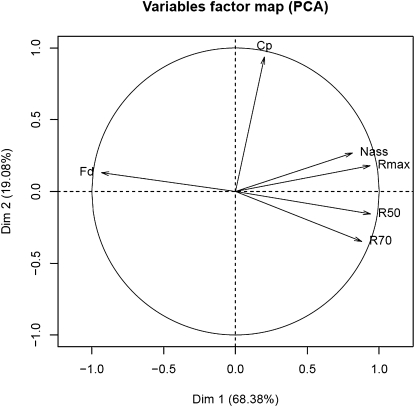
To investigate potential relationships among fermentation traits, principal component analysis (PCA) was performed (Figure 2). The projection on two principal axes preserves 87% of the information and explains 68% and 19% of this variation, respectively. This analysis shows that Fd, R70, and R50 are negatively correlated. This is consistent with the idea that the fermentation rate during the stationary phase strongly determines the overall duration of fermentation. The second axis also distinguishes the final cell population Cp and R70 variables (or Cp and Rmax) and the Cp and Fd variables which are not correlated. This suggests that the fermentation rate is independent from the total cell population. Finally, the fermentation rate at all stages (Rm, R50, and R70) was positively correlated with the assimilated nitrogen Nass. These data indicate that cell capacity to use nitrogen has a strong impact on the fermentation rate. This is consistent with previous reports on the role of nitrogen availability in fermentation rate (Bely et al. 1990, Varela et al. 2004). The laboratory strain has a poor ability to consume nitrogen, which was associated to a weak fermentation capacity (Figure S1 and Figure S2).
Figure 2 .
Principal component analysis (PCA) of kinetic and metabolic traits. This analysis shows that all kinetic parameters are correlated. The first component shows that Nass and fermentation rates (Rmax, R50, R70) are negatively correlated with Fd. The second component shows that Cp is not correlated with the other parameters.
Transcripts abundance in parental strains and in the population of segregants
To assess the gene expression level of the entire genome of the segregants and parental strains, we performed a transcriptome analysis. The physiological condition chosen for analyzing gene expression was the point of midfermentation (50% of fermentation process). At this stage, cells are in stationary phase and are experiencing a general stress due to nutrient starvation and alcohol accumulation (about 6% at this stage). Under these physiological conditions, the transcriptome is stable and expected to provide a relevant picture regarding the capacity to resist to such stressful conditions (Rossignol et al. 2003). The transcriptomes of both parents and each segregant were compared with the mean transcriptome of the population (see Materials and Methods). We first observed that 2184 genes were differentially expressed between the parental strains at P < 0.01 and 262 genes up- or downregulated with a ratio higher than 2 (see Materials and Methods). This indicates that a large fraction of the genes in the genome are differentially expressed, with only a small subset displaying strong variations in agreement with previous reports (Brem et al. 2002). Among the top 100 genes that were strongly overexpressed in the parental strains (Table S3), 59A exhibited a large set of stress- and anaerobiosis-inducible cell wall genes from the PAU and DAN families. These genes are known to be highly expressed during alcoholic fermentation (Rossignol et al. 2003); however, given that several of them can cross-hybridize, it is not possible to identify unambiguously the set of genes really overexpressed. In the laboratory strain, there was a massive overexpression of 76 retrotransposons in agreement with a reshaping of retrotransposons in S288c and an amplification of Ty1 in the laboratory strain compared with EC1118 (Novo et al. 2009). Another striking feature in the laboratory strain was the strong overexpression of several genes involved in the stress response (SPS100, HSP30, HSP12).
Correlations between gene expression and phenotypes
To address the relationships between fermentation phenotypes and gene expression, we carried out a correlation analysis (see Materials and Methods). The expression of a large number of genes exhibited significant correlation with kinetic parameters (Table 1 and Figure S4). Genes with the strongest correlation with Rmax were involved in nitrogen metabolism (MEP2, PUT1, MAE1), protein synthesis and degradation (MAP1, SUP35, PRE1), or Ty protein. Several of these genes were also correlated with the amount of assimilated nitrogen Nass. Expression of these genes is controlled by nitrogen catabolic repression (NCR), and they are induced at the beginning of the stationary phase in response to nitrogen depletion (Rossignol et al. 2003). Their correlation with Rmax and Nass is consistent with their control by nitrogen availability and the role of nitrogen in fermentation rate (Figure 2 and Table 1). The genes that correlated with the fermentation rate at midfermentation (R50) and at 70% fermentation (R70) were very similar. Several genes involved in thiamine metabolism (THI3, THI4, and PET18) displayed a high level of correlation as did the actin gene ACT1, and a cell wall gene (SCW4). Correlation was also seen, though to a lesser extent, for genes involved in nitrogen metabolism (MEP2, PUT1, MAP1). The genes correlated with fermentation duration (Fd) were mostly consistent with those that correlated with R70 or R50, but with the opposite relationship (these two traits are negatively correlated in PCA; see Figure 2). Peculiarly, this included various genes involved in the stress response (CUP1, HSP12, HSP30, CTT1, SSA4, ATC1) which were positively correlated with the fermentation duration Fd, indicating that a high stress response was associated with a low fermentation capacity. Several members of the PAU/DAN family previously observed to be overexpressed in the industrial strain were correlated with nitrogen assimilation, and to some extent, with Rmax and R50. For the other fermentation traits, such as cell population and amount of metabolites, weak or no correlation was found with gene expression. This was probably because the stage at which gene expression was analyzed (50% of fermentation progress) was not appropriate to address these parameters, which are mainly associated with the growth phase.
Table 1 . Correlations between fermentation phenotypes and gene expression.
| Name | Correlation | P | Adjusted P | Function |
|---|---|---|---|---|
| Spearman correlations relative to Rmax | ||||
| MEP2 | 0.7456 | 2.27E−06 | 0.002 | Ammonium permease involved in regulation of pseudohyphal growth |
| MAE1 | 0.6902 | 2.44E−05 | 0.01 | Mitochondrial malic enzyme |
| PUT1 | 0.6833 | 3.16E−05 | 0.01 | Proline oxidase |
| SNG1 | 0.6824 | 3.28E−05 | 0.01 | Protein involved in nitrosoguanidine (MNNG) resistance |
| SUP35 | 0.6795 | 3.64E−05 | 0.01 | Translation termination factor eRF3 |
| YLR410W-B | 0.6598 | 7.30E−05 | 0.013 | Retrotransposon TYA Gag and TYB Pol genes |
| YGR038C-B | 0.6452 | 1.18E−04 | 0.017 | Retrotransposon TYA Gag and TYB Pol genes |
| YFL002W-A | 0.6357 | 1.60E−04 | 0.018 | Retrotransposon TYA Gag and TYB Pol genes |
| YBR016W | 0.6272 | 2.08E−04 | 0.02 | Plasma membrane protein of unknown function |
| YPR137C-B | 0.6241 | 2.28E−04 | 0.02 | Retrotransposon TYA Gag and TYB Pol genes |
| YCL076W | 0.6239 | 2.30E−04 | 0.02 | Dubious open reading frame unlikely to encode a protein |
| YLR035C-A | 0.6208 | 2.52E−04 | 0.022 | Retrotransposon TYA Gag and TYB Pol genes |
| RPN6 | 0.6117 | 3.29E−04 | 0.025 | Essential |
| YML131W | 0.6094 | 3.51E−04 | 0.026 | Putative protein of unknown function with similarity to medium chain dehydrogenase/reductases |
| YDR261W-B | 0.6038 | 4.11E−04 | 0.029 | Retrotransposon TYA Gag and TYB Pol genes |
| YBL113C | −0.7505 | 1.79E−06 | 0.002 | Helicase-like protein encoded within the telomeric Y element |
| YHR219W | −0.7487 | 1.95E−06 | 0.002 | Putative protein of unknown function with similarity to helicases |
| YKL050C | −0.7429 | 2.58E−06 | 0.002 | Protein of unknown function |
| YHR097C | −0.7414 | 2.78E−06 | 0.002 | Putative protein of unknown function |
| YLR326W | −0.7409 | 2.84E−06 | 0.002 | Putative protein of unknown function |
| YRF1-3 | −0.7209 | 7.01E−06 | 0.004 | Helicase encoded by the Y element of subtelomeric regions |
| YOR289W | −0.716 | 8.64E−06 | 0.005 | Putative protein of unknown function |
| YJL225C | −0.6931 | 2.18E−05 | 0.01 | Putative protein of unknown function |
| GFD1 | −0.6902 | 2.44E−05 | 0.01 | Coiled-coiled protein of unknown function |
| APM1 | −0.6864 | 2.82E−05 | 0.01 | Mu1-like medium subunit of the clathrin-associated protein complex (AP-1) |
| PHM7 | −0.6848 | 2.99E−05 | 0.01 | Protein of unknown function |
| YMR244C-A | −0.6775 | 3.91E−05 | 0.01 | Putative protein of unknown function |
| MSC3 | −0.6742 | 4.42E−05 | 0.011 | Protein of unknown function |
| MSO1 | −0.6713 | 4.90E−05 | 0.011 | Probable component of the secretory vesicle docking complex |
| TMA23 | −0.671 | 4.94E−05 | 0.011 | Nucleolar protein of unknown function implicated in ribosome biogenesis |
| MRS1 | −0.6693 | 5.26E−05 | 0.011 | Protein required for the splicing of two mitochondrial group I introns (BI3 in COB and AI5beta in COX1) |
| CUP1-2 | −0.6659 | 5.91E−05 | 0.012 | Metallothionein |
| BNA1 | −0.6591 | 7.47E−05 | 0.013 | 3-hydroxyanthranilic acid dioxygenase |
| IES5 | −0.6589 | 7.53E−05 | 0.013 | Protein that associates with the INO80 chromatin remodeling complex under low-salt conditions |
| VOA1 | −0.6515 | 9.65E−05 | 0.015 | Putative protein of unknown function |
| NSA1 | −0.6512 | 9.72E−05 | 0.015 | Constituent of 66S pre-ribosomal particles |
| YGR251W | −0.6486 | 1.06E−04 | 0.016 | Essential protein required for maturation of 18S rRNA |
| CUP1-1 | −0.6475 | 1.10E−04 | 0.016 | Metallothionein |
| YPL080C | −0.6437 | 1.24E−04 | 0.017 | Dubious open reading frame unlikely to encode a protein |
| PIN2 | −0.6419 | 1.32E−04 | 0.017 | Protein that induces appearance of [PIN+] prion when overproduced |
| BSC4 | −0.6406 | 1.37E−04 | 0.017 | Protein of unknown function |
| CSM2 | −0.6397 | 1.41E−04 | 0.017 | Protein required for accurate chromosome segregation during meiosis |
| YMR086W | −0.6392 | 1.43E−04 | 0.017 | Protein of unknown function that may interact with ribosomes |
| YEL077C | −0.6386 | 1.46E−04 | 0.017 | Helicase-like protein encoded within the telomeric Y element |
| COX16 | −0.6357 | 1.60E−04 | 0.018 | Mitochondrial inner membrane protein |
| YLL066C | −0.6352 | 1.62E−04 | 0.018 | Putative protein of unknown function with similarity to helicases |
| OXR1 | −0.6348 | 1.64E−04 | 0.018 | Protein of unknown function required for normal levels of resistance to oxidative damage |
| YRF1-6 | −0.6328 | 1.75E−04 | 0.018 | Helicase encoded by the Y element of subtelomeric regions |
| YML133C | −0.629 | 1.97E−04 | 0.02 | Putative protein of unknown function with similarity to helicases |
| DAL81 | −0.6285 | 2.00E−04 | 0.02 | Positive regulator of genes in multiple nitrogen degradation pathways |
| TOA1 | −0.6268 | 2.11E−04 | 0.02 | TFIIA large subunit |
| MDM12 | −0.6254 | 2.19E−04 | 0.02 | Mitochondrial outer membrane protein |
| CDC12 | −0.6248 | 2.24E−04 | 0.02 | Component of the septin ring of the mother-bud neck that is required for cytokinesis |
| YMR306C-A | −0.619 | 2.66E−04 | 0.022 | Dubious open reading frame unlikely to encode a functional protein |
| VPS24 | −0.6174 | 2.78E−04 | 0.023 | One of four subunits of the endosomal sorting complex required for transport III (ESCRT-III) |
| DDC1 | −0.6154 | 2.95E−04 | 0.024 | DNA damage checkpoint protein |
| ATH1 | −0.6139 | 3.09E−04 | 0.025 | Acid trehalase required for utilization of extracellular trehalose |
| KES1 | −0.6127 | 3.19E−04 | 0.025 | Member of the oxysterol binding protein family |
| MEC3 | −0.611 | 3.36E−04 | 0.025 | DNA damage and meiotic pachytene checkpoint protein |
| PUP1 | −0.6075 | 3.71E−04 | 0.027 | Endopeptidase with trypsin-like activity that cleaves after basic residues |
| YDL173W | −0.6038 | 4.11E−04 | 0.029 | Putative protein of unknown function |
| SPG1 | −0.6028 | 4.23E−04 | 0.03 | Protein required for survival at high temperature during stationary phase |
| Spearman correlations relative to R50 | ||||
| THI3 | 0.7442 | 5.30E−06 | 0.003 | Probable alpha-ketoisocaproate decarboxylase |
| THI4 | 0.7228 | 1.18E−05 | 0.006 | Thiazole synthase |
| ACT1 | 0.7201 | 1.31E−05 | 0.006 | Actin |
| SCW4 | 0.7152 | 1.57E−05 | 0.006 | Cell wall protein with similarity to glucanases |
| YLR444C | 0.7063 | 2.19E−05 | 0.007 | Dubious open reading frame unlikely to encode a functional protein |
| HPA3 | 0.7032 | 2.45E−05 | 0.007 | D-Amino acid N-acetyltransferase |
| SNG1 | 0.6866 | 2.79E−05 | 0.007 | Protein involved in nitrosoguanidine (MNNG) resistance |
| TIM21 | 0.6845 | 4.75E−05 | 0.01 | Constituent of the mitochondrial inner membrane presequence translocase (TIM23 complex) |
| PRE1 | 0.6783 | 5.87E−05 | 0.011 | Beta 4 subunit of the 20S proteasome |
| PET18 | 0.6747 | 6.61E−05 | 0.012 | Protein required for respiratory growth and stability of the mitochondrial genome |
| MAP1 | 0.6556 | 1.22E−04 | 0.018 | Methionine aminopeptidase |
| MAE1 | 0.6534 | 1.31E−04 | 0.018 | Mitochondrial malic enzyme |
| MEP2 | 0.6525 | 1.34E−04 | 0.018 | Ammonium permease involved in regulation of pseudohyphal growth |
| YNR048W | 0.6463 | 1.62E−04 | 0.021 | Protein proposed to interact with phospholipid translocases |
| ERG20 | 0.6449 | 1.69E−04 | 0.021 | Farnesyl pyrophosphate synthetase |
| LYS9 | 0.6387 | 2.03E−04 | 0.024 | Saccharopine dehydrogenase (NADP+) |
| TPK1 | 0.6356 | 2.22E−04 | 0.025 | cAMP-dependent protein kinase catalytic subunit |
| YAR069C | 0.6343 | 2.31E−04 | 0.025 | Dubious open reading frame unlikely to encode a protein |
| BUD31 | 0.6343 | 2.31E−04 | 0.025 | Protein involved in bud-site selection |
| ARC1 | 0.6307 | 2.56E−04 | 0.025 | Protein that binds tRNA and methionyl- and glutamyl-tRNA synthetases (Mes1p and Gus1p) |
| PAU1 | 0.6294 | 2.65E−04 | 0.025 | Part of 23-member seripauperin multigene family encoded mainly in subtelomeric regions |
| YLR179C | 0.6294 | 2.65E−04 | 0.025 | Protein of unknown function |
| CDC45 | 0.6249 | 3.01E−04 | 0.027 | DNA replication initiation factor |
| RPS21A | 0.6236 | 3.12E−04 | 0.027 | Protein component of the small (40S) ribosomal subunit |
| SOL2 | 0.6218 | 3.28E−04 | 0.028 | Protein with a possible role in tRNA export |
| SSZ1 | 0.6151 | 3.93E−04 | 0.032 | Hsp70 protein that interacts with Zuo1p (a DnaJ homolog) to form a ribosome-associated complex that binds the ribosome via the Zuo1p subunit |
| CDC7 | 0.6036 | 5.35E−04 | 0.038 | DDK (Dbf4-dependent kinase) catalytic subunit required for firing origins and replication fork progression in mitosis through phosphorylation of Mcm2-7p complexes and Cdc45p |
| DRN1 | 0.6031 | 5.41E−04 | 0.038 | Putative debranching enzyme associated ribonuclease |
| JSN1 | 0.6013 | 5.66E−04 | 0.04 | Member of the Puf family of RNA-binding proteins |
| TVP23 | −0.7913 | 1.42E−06 | 0.002 | Integral membrane protein localized to late Golgi vesicles along with the v-SNARE Tlg2p |
| SPC1 | −0.7878 | 1.51E−06 | 0.002 | Subunit of the signal peptidase complex (SPC) |
| GCN3 | −0.7709 | 2.21E−06 | 0.002 | Alpha subunit of the translation initiation factor eIF2B |
| CUP1-2 | −0.7677 | 2.42E−06 | 0.002 | Metallothionein |
| YLL032C | −0.7602 | 3.04E−06 | 0.002 | Protein of unknown function that may interact with ribosomes |
| HSP30 | −0.7595 | 1.14E−06 | 0.002 | Hydrophobic plasma membrane localized |
| CUP1-1 | −0.7179 | 1.42E−05 | 0.006 | Metallothionein |
| APM1 | −0.7126 | 1.74E−05 | 0.006 | Mu1-like medium subunit of the clathrin-associated protein complex (AP-1) |
| Q0182 | −0.7006 | 2.70E−05 | 0.007 | Dubious open reading frame unlikely to encode a protein |
| HSP12 | −0.6988 | 2.88E−05 | 0.007 | Plasma membrane localized protein that protects membranes from desiccation |
| ATG1 | −0.693 | 3.54E−05 | 0.008 | Protein ser/thr kinase required for vesicle formation in autophagy and the cytoplasm-to-vacuole targeting (Cvt) pathway |
| GND2 | −0.693 | 3.54E−05 | 0.008 | 6-phosphogluconate dehydrogenase (decarboxylating) |
| ATC1 | −0.6814 | 5.28E−05 | 0.011 | Nuclear protein |
| YGR026W | −0.6814 | 5.28E−05 | 0.011 | Putative protein of unknown function |
| YEL077C | −0.6756 | 6.41E−05 | 0.012 | Helicase-like protein encoded within the telomeric Y element |
| CRS5 | −0.6659 | 8.83E−05 | 0.014 | Copper-binding metallothionein |
| RHO5 | −0.6641 | 9.35E−05 | 0.015 | Nonessential small GTPase of the Rho/Rac subfamily of Ras-like proteins |
| SPG1 | −0.659 | 7.49E−05 | 0.013 | Protein required for survival at high temperature during stationary phase |
| SPS100 | −0.6512 | 1.40E−04 | 0.019 | Protein required for spore wall maturation |
| YOR277C | −0.6412 | 1.34E−04 | 0.018 | Dubious open reading frame unlikely to encode a protein |
| YOL014W | −0.6409 | 1.90E−04 | 0.023 | Putative protein of unknown function |
| SSA4 | −0.6343 | 2.31E−04 | 0.025 | Heat shock protein that is highly induced upon stress |
| ADA2 | −0.632 | 2.46E−04 | 0.025 | Transcription coactivator |
| PIB2 | −0.6298 | 2.62E−04 | 0.025 | Protein binding phosphatidylinositol 3-phosphate |
| YNR047W | −0.6249 | 3.01E−04 | 0.027 | Putative protein kinase that |
| INP53 | −0.6249 | 3.01E−04 | 0.027 | Polyphosphatidylinositol phosphatase |
| YPC1 | −0.6218 | 3.28E−04 | 0.028 | Alkaline ceramidase that also has reverse (CoA-independent) ceramide synthase activity |
| YJL114W | −0.6165 | 3.79E−04 | 0.031 | Retrotransposon TYA Gag gene cotranscribed with TYB Pol |
| HXT8 | −0.6102 | 4.49E−04 | 0.035 | Protein of unknown function with similarity to hexose transporter family members |
| CYC1 | −0.6085 | 4.70E−04 | 0.036 | Cytochrome c |
| CTT1 | −0.6067 | 4.93E−04 | 0.037 | Cytosolic catalase T |
| ATP6 | −0.605 | 3.98E−04 | 0.032 | Mitochondrially encoded subunit a of the F0 sector of mitochondrial F1F0 ATP synthase |
| FCP1 | −0.6049 | 5.16E−04 | 0.038 | Carboxy-terminal domain (CTD) phosphatase |
| Spearman correlations relative to R70 | ||||
| THI3 | 0.8427 | 5.18E−09 | 0 | Probable alpha-ketoisocaproate decarboxylase |
| YLR444C | 0.8028 | 9.39E−08 | 0 | Dubious open reading frame unlikely to encode a functional protein |
| THI4 | 0.7817 | 3.40E−07 | 0 | Thiazole synthase |
| RPS21A | 0.7759 | 4.71E−07 | 0 | Protein component of the small (40S) ribosomal subunit |
| PET18 | 0.7483 | 1.99E−06 | 0.001 | Protein required for respiratory growth and stability of the mitochondrial genome |
| TPK1 | 0.7468 | 2.14E−06 | 0.001 | cAMP-dependent protein kinase catalytic subunit |
| SCW4 | 0.7452 | 2.31E−06 | 0.001 | Cell wall protein with similarity to glucanases |
| ACT1 | 0.741 | 2.83E−06 | 0.001 | Actin |
| HPA3 | 0.7243 | 6.04E−06 | 0.001 | D-Amino acid N-acetyltransferase |
| ERG20 | 0.7227 | 6.47E−06 | 0.001 | Farnesyl pyrophosphate synthetase |
| TIM21 | 0.7165 | 8.45E−06 | 0.001 | Constituent of the mitochondrial inner membrane presequence translocase (TIM23 complex) |
| DRN1 | 0.7112 | 1.06E−05 | 0.001 | Putative debranching enzyme associated ribonuclease |
| PRE1 | 0.7109 | 1.07E−05 | 0.001 | Beta 4 subunit of the 20S proteasome |
| YAR069C | 0.6963 | 1.93E−05 | 0.002 | Dubious open reading frame unlikely to encode a protein |
| ARR1 | 0.6951 | 2.02E−05 | 0.002 | Transcriptional activator of the basic leucine zipper (bZIP) family |
| RPL14B | 0.694 | 2.10E−05 | 0.002 | Protein component of the large (60S) ribosomal subunit |
| CCT7 | 0.6931 | 2.18E−05 | 0.002 | Subunit of the cytosolic chaperonin Cct ring complex |
| MAP1 | 0.6931 | 2.18E−05 | 0.002 | Methionine aminopeptidase |
| RPL14A | 0.6916 | 2.31E−05 | 0.002 | N-terminally acetylated protein component of the large (60S) ribosomal subunit |
| CDC7 | 0.6871 | 2.74E−05 | 0.003 | DDK (Dbf4-dependent kinase) catalytic subunit required for firing origins and replication fork progression in mitosis through phosphorylation of Mcm2-7p complexes and Cdc45p |
| BUD31 | 0.6862 | 2.83E−05 | 0.003 | Protein involved in bud-site selection |
| LYS9 | 0.6851 | 2.95E−05 | 0.003 | Saccharopine dehydrogenase (NADP+) |
| PDC5 | 0.6836 | 3.13E−05 | 0.003 | Minor isoform of pyruvate decarboxylase |
| YLR179C | 0.6836 | 3.13E−05 | 0.003 | Protein of unknown function |
| SSZ1 | 0.6785 | 3.78E−05 | 0.003 | Hsp70 protein that interacts with Zuo1p (a DnaJ homolog) to form a ribosome-associated complex that binds the ribosome via the Zuo1p subunit |
| CTS1 | 0.6707 | 5.00E−05 | 0.004 | Endochitinase |
| YHB1 | 0.6575 | 7.87E−05 | 0.006 | Nitric oxide oxidoreductase |
| HTA1 | 0.6569 | 8.05E−05 | 0.006 | Histone H2A |
| POL30 | 0.6551 | 8.55E−05 | 0.006 | Proliferating cell nuclear antigen (PCNA) |
| STR3 | 0.6547 | 8.68E−05 | 0.006 | Cystathionine beta-lyase |
| VPS66 | 0.6493 | 1.04E−04 | 0.007 | Cytoplasmic protein of unknown function involved in vacuolar protein sorting. |
| RPS10A | 0.6455 | 1.17E−04 | 0.008 | Protein component of the small (40S) ribosomal subunit |
| CYS4 | 0.6429 | 1.28E−04 | 0.008 | Cystathionine beta-synthase |
| ARC19 | 0.6391 | 1.44E−04 | 0.009 | Subunit of the ARP2/3 complex |
| HYP2 | 0.6375 | 1.51E−04 | 0.009 | Translation initiation factor eIF-5A |
| RPL25 | 0.6373 | 1.52E−04 | 0.009 | Primary rRNA-binding ribosomal protein component of the large (60S) ribosomal subunit |
| YNR048W | 0.6326 | 1.76E−04 | 0.01 | Protein proposed to interact with phospholipid translocases |
| CHO2 | 0.6315 | 1.82E−04 | 0.01 | Phosphatidylethanolamine methyltransferase (PEMT) |
| THI21 | 0.6271 | 2.09E−04 | 0.011 | Hydroxymethylpyrimidine phosphate kinase |
| RSC58 | 0.6242 | 2.27E−04 | 0.012 | Component of the RSC chromatin remodeling complex |
| GFA1 | 0.6228 | 2.37E−04 | 0.012 | Glutamine-fructose-6-phosphate amidotransferase |
| GEA1 | 0.619 | 2.65E−04 | 0.014 | Guanine nucleotide exchange factor for ADP ribosylation factors (ARF) |
| SMF3 | 0.6177 | 2.76E−04 | 0.014 | Putative divalent metal ion transporter involved in iron homeostasis |
| ZEO1 | 0.6153 | 2.96E−04 | 0.015 | Peripheral membrane protein of the plasma membrane that interacts with Mid2p |
| DAL7 | 0.6128 | 3.18E−04 | 0.015 | Malate synthase |
| ADH7 | 0.6117 | 3.29E−04 | 0.016 | NADPH-dependent medium chain alcohol dehydrogenase with broad substrate specificity |
| LYS12 | 0.6095 | 3.50E−04 | 0.016 | Homo-isocitrate dehydrogenase |
| JSN1 | 0.6075 | 3.71E−04 | 0.017 | Member of the Puf family of RNA-binding proteins |
| YOX1 | 0.605 | 3.97E−04 | 0.018 | Homeodomain-containing transcriptional repressor |
| CTA1 | 0.6046 | 4.02E−04 | 0.018 | Catalase A |
| SPC1 | −0.8714 | 3.72E−10 | 0 | Subunit of the signal peptidase complex (SPC) |
| HSP30 | −0.8505 | 2.66E−09 | 0 | Hydrophobic plasma membrane localized |
| GCN3 | −0.844 | 4.64E−09 | 0 | Alpha subunit of the translation initiation factor eIF2B |
| TVP23 | −0.824 | 2.21E−08 | 0 | Integral membrane protein localized to late Golgi vesicles along with the v-SNARE Tlg2p |
| HSP12 | −0.8238 | 2.24E−08 | 0 | Plasma membrane localized protein that protects membranes from desiccation |
| ATG1 | −0.8129 | 4.84E−08 | 0 | Protein ser/thr kinase required for vesicle formation in autophagy and the cytoplasm-to-vacuole targeting (Cvt) pathway |
| GND2 | −0.8033 | 9.13E−08 | 0 | 6-phosphogluconate dehydrogenase (decarboxylating) |
| SPS100 | −0.7748 | 5.01E−07 | 0 | Protein required for spore wall maturation |
| CRS5 | −0.7639 | 9.03E−07 | 0 | Copper-binding metallothionein |
| CUP1-2 | −0.7632 | 9.35E−07 | 0 | Metallothionein |
| Q0182 | −0.7597 | 1.12E−06 | 0 | Dubious open reading frame unlikely to encode a protein |
| CTT1 | −0.7352 | 3.70E−06 | 0.001 | Cytosolic catalase T |
| YLL032C | −0.7308 | 4.53E−06 | 0.001 | Protein of unknown function that may interact with ribosomes |
| MSC1 | −0.7295 | 4.80E−06 | 0.001 | Protein of unknown function |
| SSA4 | −0.7232 | 6.34E−06 | 0.001 | Heat shock protein that is highly induced upon stress |
| INP53 | −0.7212 | 6.92E−06 | 0.001 | Polyphosphatidylinositol phosphatase |
| RHO5 | −0.7172 | 8.22E−06 | 0.001 | Nonessential small GTPase of the Rho/Rac subfamily of Ras-like proteins |
| ATP3 | −0.7136 | 9.55E−06 | 0.001 | Gamma subunit of the F1 sector of mitochondrial F1F0 ATP synthase |
| CUP1-1 | −0.7127 | 9.91E−06 | 0.001 | Metallothionein |
| ADA2 | −0.7094 | 1.14E−05 | 0.002 | Transcription coactivator |
| YPC1 | −0.6983 | 1.78E−05 | 0.002 | Alkaline ceramidase that also has reverse (CoA-independent) ceramide synthase activity |
| NCE102 | −0.694 | 2.10E−05 | 0.002 | Protein of unknown function |
| DDR2 | −0.6931 | 2.18E−05 | 0.002 | Multistress response protein |
| YJL114W | −0.6905 | 2.41E−05 | 0.002 | Retrotransposon TYA Gag gene cotranscribed with TYB Pol |
| UFD1 | −0.6891 | 2.54E−05 | 0.003 | Protein that interacts with Cdc48p and Npl4p |
| ATP6 | −0.6831 | 3.19E−05 | 0.003 | Mitochondrially encoded subunit a of the F0 sector of mitochondrial F1F0 ATP synthase |
| APM1 | −0.6814 | 3.40E−05 | 0.003 | Mu1-like medium subunit of the clathrin-associated protein complex (AP-1) |
| HXT8 | −0.672 | 4.77E−05 | 0.004 | Protein of unknown function with similarity to hexose transporter family members |
| PMP3 | −0.668 | 5.49E−05 | 0.004 | Small plasma membrane protein related to a family of plant polypeptides that are overexpressed under high salt concentration or low temperature |
| FCP1 | −0.6535 | 9.01E−05 | 0.007 | Carboxy-terminal domain (CTD) phosphatase |
| YNR047W | −0.6529 | 9.21E−05 | 0.007 | Putative protein kinase that |
| ATC1 | −0.6509 | 9.84E−05 | 0.007 | Nuclear protein |
| NDD1 | −0.6504 | 9.98E−05 | 0.007 | Transcriptional activator essential for nuclear division |
| SIP18 | −0.6498 | 1.02E−04 | 0.007 | Protein of unknown function whose expression is induced by osmotic stress |
| HXT13 | −0.646 | 1.15E−04 | 0.008 | Hexose transporter |
| CYC1 | −0.6455 | 1.17E−04 | 0.008 | Cytochrome c |
| YGR026W | −0.6449 | 1.20E−04 | 0.008 | Putative protein of unknown function |
| SME1 | −0.6386 | 1.46E−04 | 0.009 | Core Sm protein Sm E |
| YLR132C | −0.6353 | 1.62E−04 | 0.01 | Essential protein of unknown function |
| TOM7 | −0.634 | 1.69E−04 | 0.01 | Component of the TOM (translocase of outer membrane) complex responsible for recognition and initial import steps for all mitochondrially directed proteins |
| YNL190W | −0.63 | 1.91E−04 | 0.011 | Cell wall protein of unknown function |
| TRP4 | −0.6282 | 2.02E−04 | 0.011 | Anthranilate phosphoribosyl transferase of the tryptophan biosynthetic pathway |
| SFT1 | −0.6226 | 2.39E−04 | 0.012 | Intra-Golgi v-SNARE |
| COB | −0.6199 | 2.58E−04 | 0.013 | Cytochrome b, mitochondrially encoded subunit of the ubiquinol-cytochrome c reductase complex which includes Cobp |
| PGD1 | −0.6155 | 2.94E−04 | 0.015 | Subunit of the RNA polymerase II mediator complex |
| DCS2 | −0.6142 | 3.06E−04 | 0.015 | Nonessential |
| GIS3 | −0.6121 | 3.24E−04 | 0.016 | Protein of unknown function |
| YBR013C | −0.6086 | 3.59E−04 | 0.016 | Putative protein of unknown function |
| NSR1 | −0.6086 | 3.59E−04 | 0.016 | Nucleolar protein that binds nuclear localization sequences |
| YOR277C | −0.6029 | 4.22E−04 | 0.019 | Dubious open reading frame unlikely to encode a protein |
| FCY1 | −0.6026 | 4.25E−04 | 0.019 | Cytosine deaminase |
| JJJ2 | −0.6012 | 4.42E−04 | 0.019 | Protein of unknown function |
| Spearman correlations relative to Fd | ||||
| CUP1-2 | 0.8062 | 7.56E−08 | 0.0003 | Metallothionein |
| CUP1-1 | 0.7866 | 2.56E−07 | 0.0004 | Metallothionein |
| YLL032C | 0.7857 | 2.69E−07 | 0.0004 | Protein of unknown function that may interact with ribosomes |
| APM1 | 0.777 | 4.43E−07 | 0.0005 | Mu1-like medium subunit of the clathrin-associated protein complex (AP-1) |
| SPC1 | 0.7372 | 3.38E−06 | 0.0028 | Subunit of the signal peptidase complex (SPC) |
| TVP23 | 0.7336 | 3.98E−06 | 0.0028 | Integral membrane protein localized to late Golgi vesicles along with the v-SNARE Tlg2p |
| GCN3 | 0.7309 | 4.49E−06 | 0.0028 | Alpha subunit of the translation initiation factor eIF2B |
| ATG1 | 0.7051 | 1.36E−05 | 0.006 | Protein ser/thr kinase required for vesicle formation in autophagy and the cytoplasm-to-vacuole targeting (Cvt) pathway |
| HSP12 | 0.696 | 1.95E−05 | 0.006 | Plasma membrane localized protein that protects membranes from desiccation |
| HSP30 | 0.695 | 2.02E−05 | 0.006 | Hydrophobic plasma membrane localized |
| SPG1 | 0.6903 | 2.43E−05 | 0.0065 | Protein required for survival at high temperature during stationary phase |
| ADA2 | 0.6884 | 2.61E−05 | 0.0065 | Transcription coactivator |
| GND2 | 0.6797 | 3.61E−05 | 0.0076 | 6-phosphogluconate dehydrogenase (decarboxylating) |
| SSA4 | 0.6626 | 6.63E−05 | 0.0127 | Heat shock protein that is highly induced upon stress |
| CYC1 | 0.6566 | 8.14E−05 | 0.0133 | Cytochrome c |
| SPT7 | 0.6535 | 9.03E−05 | 0.0137 | Subunit of the SAGA transcriptional regulatory complex |
| YHR219W | 0.651 | 9.79E−05 | 0.0137 | Putative protein of unknown function with similarity to helicases |
| YNL108C | 0.6492 | 1.04E−04 | 0.0138 | Putative protein of unknown function with similarity to Tfc7p and prokaryotic phosphotransfer enzymes |
| YPL108W | 0.6466 | 1.13E−04 | 0.0147 | Cytoplasmic protein of unknown function |
| YOR277C | 0.6453 | 1.18E−04 | 0.0147 | Dubious open reading frame unlikely to encode a protein |
| YGR026W | 0.6434 | 1.25E−04 | 0.0147 | Putative protein of unknown function |
| SIP18 | 0.6434 | 1.25E−04 | 0.0147 | Protein of unknown function whose expression is induced by osmotic stress |
| Q0182 | 0.6417 | 1.33E−04 | 0.0148 | Dubious open reading frame unlikely to encode a protein |
| OXR1 | 0.6358 | 1.59E−04 | 0.0171 | Protein of unknown function required for normal levels of resistance to oxidative damage |
| SMD2 | 0.6343 | 1.67E−04 | 0.0175 | Core Sm protein Sm D2 |
| FCP1 | 0.6301 | 1.90E−04 | 0.0192 | Carboxy-terminal domain (CTD) phosphatase |
| SMA1 | 0.6294 | 1.94E−04 | 0.0192 | Protein of unknown function involved in the assembly of the prospore membrane during sporulation |
| YBL113C | 0.6287 | 1.98E−04 | 0.0192 | Helicase-like protein encoded within the telomeric Y element |
| YJL114W | 0.6274 | 2.07E−04 | 0.0193 | Retrotransposon TYA Gag gene cotranscribed with TYB Pol |
| INP53 | 0.621 | 2.51E−04 | 0.0221 | Polyphosphatidylinositol phosphatase |
| CRS5 | 0.6198 | 2.59E−04 | 0.0221 | Copper-binding metallothionein |
| SKS1 | 0.6196 | 2.61E−04 | 0.0221 | Putative serine/threonine protein kinase |
| POP6 | 0.6161 | 2.90E−04 | 0.0221 | Subunit of both RNase MRP |
| GIS3 | 0.6147 | 3.01E−04 | 0.0221 | Protein of unknown function |
| FMP48 | 0.6143 | 3.05E−04 | 0.0221 | Putative protein of unknown function |
| YHR097C | 0.6143 | 3.05E−04 | 0.0221 | Putative protein of unknown function |
| RHO5 | 0.6143 | 3.05E−04 | 0.0221 | Nonessential small GTPase of the Rho/Rac subfamily of Ras-like proteins |
| TMA23 | 0.614 | 3.07E−04 | 0.0221 | Nucleolar protein of unknown function implicated in ribosome biogenesis |
| YGL117W | 0.6103 | 3.42E−04 | 0.0227 | Putative protein of unknown function |
| SFT1 | 0.6103 | 3.42E−04 | 0.0227 | Intra-Golgi v-SNARE |
| SPS100 | 0.6089 | 3.56E−04 | 0.0227 | Protein required for spore wall maturation |
| MSC1 | 0.6079 | 3.66E−04 | 0.0227 | Protein of unknown function |
| ALG11 | 0.602 | 4.32E−04 | 0.024 | Alpha-1 |
| SME1 | 0.602 | 4.32E−04 | 0.024 | Core Sm protein Sm E |
| YDL144C | 0.6018 | 4.35E−04 | 0.024 | Putative protein of unknown function |
| DCS2 | 0.6016 | 4.37E−04 | 0.024 | Nonessential |
| YOL014W | 0.6009 | 4.46E−04 | 0.0241 | Putative protein of unknown function |
| YJL156W-A | 0.6005 | 4.51E−04 | 0.0241 | Dubious open reading frame unlikely to encode a protein |
| THI3 | −0.714 | 9.39E−06 | 0.0052 | Probable alpha-ketoisocaproate decarboxylase |
| SCW4 | −0.7056 | 1.33E−05 | 0.006 | Cell wall protein with similarity to glucanases |
| TIM21 | −0.6984 | 1.77E−05 | 0.006 | Constituent of the mitochondrial inner membrane presequence translocase (TIM23 complex) |
| TPK1 | −0.6953 | 2.00E−05 | 0.006 | cAMP-dependent protein kinase catalytic subunit |
| SNG1 | −0.6947 | 2.05E−05 | 0.006 | Protein involved in nitrosoguanidine (MNNG) resistance |
| THI4 | −0.6877 | 2.68E−05 | 0.0065 | Thiazole synthase |
| YLR444C | −0.6862 | 2.84E−05 | 0.0066 | Dubious open reading frame unlikely to encode a functional protein |
| PUT1 | −0.6815 | 3.38E−05 | 0.0074 | Proline oxidase |
| ACT1 | −0.6739 | 4.45E−05 | 0.0089 | Actin |
| MEP2 | −0.661 | 7.00E−05 | 0.0128 | Ammonium permease involved in regulation of pseudohyphal growth |
| HPA3 | −0.6588 | 7.55E−05 | 0.0133 | D-Amino acid N-acetyltransferase |
| RPS21A | −0.6577 | 7.84E−05 | 0.0133 | Protein component of the small (40S) ribosomal subunit |
| DAN2 | −0.6543 | 8.77E−05 | 0.0137 | Cell wall mannoprotein with similarity to Tir1p |
| BUD31 | −0.6515 | 9.65E−05 | 0.0137 | Protein involved in bud-site selection |
| YLR179C | −0.6506 | 9.94E−05 | 0.0137 | Protein of unknown function |
| PET18 | −0.643 | 1.27E−04 | 0.0147 | Protein required for respiratory growth and stability of the mitochondrial genome |
| ARC1 | −0.6412 | 1.35E−04 | 0.0148 | Protein that binds tRNA and methionyl- and glutamyl-tRNA synthetases (Mes1p and Gus1p) |
| PAU1 | −0.6283 | 2.01E−04 | 0.0192 | Part of 23-member seripauperin multigene family encoded mainly in subtelomeric regions |
| HRT3 | −0.6221 | 2.43E−04 | 0.0221 | Putative SCF-ubiquitin ligase F-box protein |
| CDC7 | −0.6192 | 2.64E−04 | 0.0221 | DDK (Dbf4-dependent kinase) catalytic subunit required for firing origins and replication fork progression in mitosis through phosphorylation of Mcm2-7p complexes and Cdc45p |
| RNR2 | −0.6187 | 2.68E−04 | 0.0221 | Ribonucleotide-diphosphate reductase (RNR) |
| YIL067C | −0.6183 | 2.71E−04 | 0.0221 | Uncharacterized protein of unknown function |
| PRE1 | −0.614 | 3.07E−04 | 0.0221 | Beta 4 subunit of the 20S proteasome |
| URA7 | −0.6118 | 3.27E−04 | 0.0227 | Major CTP synthase isozyme (see also URA8) |
| DBP5 | −0.6105 | 3.40E−04 | 0.0227 | Cytoplasmic ATP-dependent RNA helicase of the DEAD-box family involved in mRNA export from the nucleus |
| PAU7 | −0.6092 | 3.53E−04 | 0.0227 | Part of 23-member seripauperin multigene family |
| ERG20 | −0.6089 | 3.56E−04 | 0.0227 | Farnesyl pyrophosphate synthetase |
| YAR069C | −0.6087 | 3.58E−04 | 0.0227 | Dubious open reading frame unlikely to encode a protein |
| JSN1 | −0.6083 | 3.63E−04 | 0.0227 | Member of the Puf family of RNA-binding proteins |
| RPL23B | −0.6072 | 3.73E−04 | 0.0228 | Protein component of the large (60S) ribosomal subunit |
| CHO2 | −0.6045 | 4.03E−04 | 0.024 | Phosphatidylethanolamine methyltransferase (PEMT) |
| CDC45 | −0.6034 | 4.16E−04 | 0.024 | DNA replication initiation factor |
| YNR048W | −0.6031 | 4.19E−04 | 0.024 | Protein proposed to interact with phospholipid translocases |
| SOL2 | −0.6016 | 4.37E−04 | 0.024 | Protein with a possible role in tRNA export |
| GLN1 | −0.6 | 4.57E−04 | 0.0241 | Glutamine synthetase (GS) |
| Spearman correlations relative to Nass | ||||
| PAU7 | 0.6761 | 6.32E−05 | 0.019 | Part of 23-member seripauperin multigene family |
| SNG1 | 0.6568 | 8.07E−05 | 0.019 | Protein involved in nitrosoguanidine (MNNG) resistance |
| PUT1 | 0.6556 | 1.22E−04 | 0.023 | Proline oxidase |
| DAN2 | 0.6547 | 1.25E−04 | 0.023 | Cell wall mannoprotein with similarity to Tir1p |
| PAU8 | 0.6383 | 2.06E−04 | 0.027 | Hypothetical protein |
| MEP2 | 0.6383 | 2.06E−04 | 0.027 | Ammonium permease involved in regulation of pseudohyphal growth |
| PLB2 | 0.6245 | 3.05E−04 | 0.033 | Phospholipase B (lysophospholipase) involved in phospholipid metabolism |
| LHP1 | 0.6214 | 3.32E−04 | 0.034 | RNA binding protein required for maturation of tRNA and U6 snRNA precursors |
| PAU13 | 0.6182 | 3.62E−04 | 0.034 | Putative protein of unknown function |
| PAU1 | 0.6182 | 3.62E−04 | 0.034 | Part of 23-member seripauperin multigene family encoded mainly in subtelomeric regions |
| MOG1 | 0.6151 | 3.93E−04 | 0.034 | Conserved nuclear protein that interacts with GTP-Gsp1p |
| PAU14 | 0.6142 | 4.03E−04 | 0.034 | Hypothetical protein |
| PAU4 | 0.6133 | 4.13E−04 | 0.034 | Part of 23-member seripauperin multigene family encoded mainly in subtelomeric regions |
| SRB7 | 0.6125 | 4.23E−04 | 0.034 | Subunit of the RNA polymerase II mediator complex |
| PAU10 | 0.6071 | 4.87E−04 | 0.038 | Hypothetical protein |
| CBR1 | 0.6027 | 5.47E−04 | 0.038 | Microsomal cytochrome b reductase |
| YBR016W | 0.6018 | 5.60E−04 | 0.038 | Plasma membrane protein of unknown function |
| PEX29 | 0.6009 | 5.73E−04 | 0.039 | Peroxisomal integral membrane peroxin |
| YKL050C | −0.8091 | 1.11E−06 | 0.005 | Protein of unknown function |
| MRS1 | −0.7499 | 4.30E−06 | 0.009 | Protein required for the splicing of two mitochondrial group I introns (BI3 in COB and AI5beta in COX1) |
| YBL113C | −0.7344 | 7.64E−06 | 0.01 | Helicase-like protein encoded within the telomeric Y element |
| YHR097C | −0.7286 | 9.51E−06 | 0.01 | Putative protein of unknown function |
| YJL225C | −0.7152 | 1.57E−05 | 0.014 | Putative protein of unknown function |
| YMR086W | −0.709 | 1.98E−05 | 0.015 | Protein of unknown function that may interact with ribosomes |
| YGR251W | −0.6974 | 3.02E−05 | 0.018 | Essential protein required for maturation of 18S rRNA |
| COX16 | −0.6948 | 3.32E−05 | 0.018 | Mitochondrial inner membrane protein |
| TOA1 | −0.6881 | 4.20E−05 | 0.019 | TFIIA large subunit |
| YPL080C | −0.6823 | 5.12E−05 | 0.019 | Dubious open reading frame unlikely to encode a protein |
| PEX14 | −0.6796 | 5.61E−05 | 0.019 | Peroxisomal membrane peroxin that is a central component of the peroxisomal protein import machinery |
| DDC1 | −0.6739 | 6.80E−05 | 0.019 | DNA damage checkpoint protein |
| YRF1-3 | −0.6734 | 6.90E−05 | 0.019 | Helicase encoded by the Y element of subtelomeric regions |
| YLL066C | −0.6707 | 7.54E−05 | 0.019 | Putative protein of unknown function with similarity to helicases |
| YLR326W | −0.669 | 7.99E−05 | 0.019 | Putative protein of unknown function |
| TMA23 | −0.6672 | 8.46E−05 | 0.019 | Nucleolar protein of unknown function implicated in ribosome biogenesis |
| PHM7 | −0.6659 | 8.83E−05 | 0.019 | Protein of unknown function |
| IES5 | −0.6637 | 6.38E−05 | 0.019 | Protein that associates with the INO80 chromatin remodeling complex under low-salt conditions |
| NSA1 | −0.6636 | 9.48E−05 | 0.019 | Constituent of 66S pre-ribosomal particles |
| MTF1 | −0.6515 | 9.65E−05 | 0.019 | Mitochondrial RNA polymerase |
| YMR244C-A | −0.6507 | 1.42E−04 | 0.023 | Putative protein of unknown function |
| MDM12 | −0.6503 | 1.44E−04 | 0.023 | Mitochondrial outer membrane protein |
| YMR306C-A | −0.6489 | 1.50E−04 | 0.023 | Dubious open reading frame unlikely to encode a functional protein |
| ERG24 | −0.6489 | 1.50E−04 | 0.023 | C-14 sterol reductase |
| ENP2 | −0.6485 | 1.52E−04 | 0.023 | Essential nucleolar protein of unknown function |
| AIM23 | −0.6458 | 1.64E−04 | 0.024 | Putative protein of unknown function |
| YHR219W | −0.6392 | 2.00E−04 | 0.027 | Putative protein of unknown function with similarity to helicases |
| YDL173W | −0.6374 | 2.11E−04 | 0.027 | Putative protein of unknown function |
| VOA1 | −0.6352 | 2.25E−04 | 0.027 | Putative protein of unknown function |
| CUE5 | −0.6352 | 2.25E−04 | 0.027 | Protein containing a CUE domain that binds ubiquitin |
| YOR021C | −0.6316 | 2.49E−04 | 0.029 | Putative protein of unknown function |
| YAR028W | −0.6298 | 2.62E−04 | 0.03 | Putative integral membrane protein |
| GFD1 | −0.6267 | 2.86E−04 | 0.031 | Coiled-coiled protein of unknown function |
| JID1 | −0.6263 | 2.14E−04 | 0.027 | Probable Hsp40p cochaperone |
| SCM3 | −0.6209 | 3.36E−04 | 0.034 | Nonhistone component of centromeric chromatin that binds stoichiometrically to CenH3-H4 histones |
| YLR363W-A | −0.6205 | 3.40E−04 | 0.034 | Putative protein of unknown function |
| YOR289W | −0.616 | 3.84E−04 | 0.034 | Putative protein of unknown function |
| IRC25 | −0.6147 | 3.98E−04 | 0.034 | Component of a heterodimeric Poc4p-Irc25p chaperone involved in assembly of alpha subunits into the 20S proteasome |
| TPK3 | −0.6142 | 4.03E−04 | 0.034 | cAMP-dependent protein kinase catalytic subunit |
| HCA4 | −0.6133 | 4.13E−04 | 0.034 | Putative nucleolar DEAD box RNA helicase |
| SGF11 | −0.6102 | 4.49E−04 | 0.036 | Integral subunit of SAGA histone acetyltransferase complex |
| YRF1-8 | −0.608 | 4.76E−04 | 0.037 | One of several telomeric Y element-encoded DNA helicases |
| CUS1 | −0.6044 | 5.22E−04 | 0.038 | Protein required for assembly of U2 snRNP into the spliceosome |
| GLC8 | −0.6031 | 5.41E−04 | 0.038 | Regulatory subunit of protein phosphatase 1 (Glc7p) |
| YML133C | −0.6027 | 5.47E−04 | 0.038 | Putative protein of unknown function with similarity to helicases |
| SPO13 | −0.6018 | 5.60E−04 | 0.038 | Meiosis-specific protein |
| ELA1 | −0.6004 | 5.79E−04 | 0.039 | Elongin A |
For each gene, the P value, the adjusted P value (Benjamini and Hochberg adjustment), and function are indicated.
Marker map construction and identification of QTL for fermentation parameters
To genotype the entire population of this study, we hybridized the genomic DNA of the 30 segregants and parental strains on high-density olignonucleotide microarrays Affymetrix YGS98 (Winzeler et al. 1998). Using the genome sequences of the two strains, we obtained a dense coverage of the genome with 1834 physical markers and an average spacing of 6.6 kbp between each marker (Figure S5). The markers are expected to be reliable as we checked that each marker corresponded to a SNP in the genome sequence of the strain 59A. The coverage of the genome was rather homogeneous, with only few gaps on chromosome III and on the right arm of chromosome XIV. These regions of low coverage also have low SNP density that may explain the reduced frequency of markers (see Gbrowse at http://genome.jouy.inra.fr/genyeastrait/). The distribution of the markers in the 30 segregants was used to build a recombination map (Figure S6). Only very few chromosomes did not display crossover. We used the single tetrad to estimate the number of crossovers and found a frequency of 70 crossovers per meiosis, a number close to previous reports (Brem et al. 2002; Cubillos et al. 2011). The parental genomes markers were evenly distributed in the population, with 49.8% originating from the strain 59A and 50.2% from S288C.
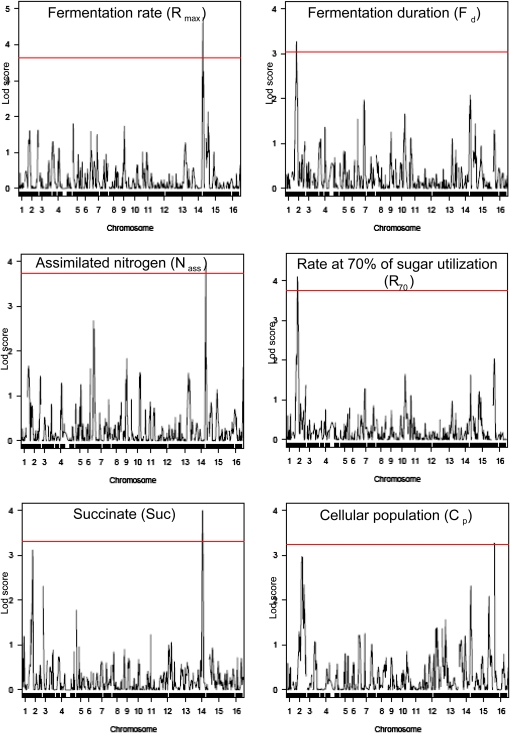
We then used these markers to map QTL for the different fermentation and metabolic parameters using an interval mapping approach (see Materials and Methods). We obtained a significant LOD score threshold for six different phenotypes. QTL were detected for Rmax on chromosome XIV, for the cell population Cp on chromosome XVI, for the assimilated nitrogen Nass on chromosome XIV, and for the fermentation duration Fd and R70 on chromosome II (Figure 3). A QTL for metabolite levels was found only for succinate production with a clear QTL located on chromosome XIV and one additional potential QTL, with a LOD score below the threshold, on chromosome II (Figure 3). We observed that QTL for various fermentation traits overlapped (Table 2). This concerned a QTL for the maximum fermentation rate and a QTL for the assimilated nitrogen that overlapped in a 37.5 kb region on chromosome XIV (664875 kb to 702375 kb). These results are consistent with the previous PCA results that highlighted relationships between nitrogen utilization Nass and Rmax (Figure 2). Two other parameters, R70 and Fd, mapped to a common region on chromosome II, whereas R50 and succinate production had a potential linkage on the same region with a LOD score just below the threshold. Careful examination of the LOD score profiles suggests that the QTL for succinate is distinct from the R70 QTL (Figure S7). The overlap of QTL for R70, R50, and Fd is consistent with the known correlations between these parameters.
Figure 3 .
QTL mapping of kinetics traits and succinate production. The concatenated chromosomes are displayed on the X-axis and LOD score values on the Y-axis. The significant LOD score thresholds are indicated by a red line. Each peak of the LOD curve corresponds to the LOD value of linkage between a marker located in the X position and the value of each trait. Further details of QTL are in Table 2.
Table 2 . Chromosomal location and size of the different phenotypic QTL.
| Traits | Chromosome | Size (kb) | Start Position (pb) | End Position (pb) | LOD Threshold | LOD Score |
|---|---|---|---|---|---|---|
| Rmax | XIV | 37.50 | 664875 | 702375 | 3.64 | 4.76 |
| Nass | XIV | 37.50 | 664875 | 702375 | 3.73 | 3.91 |
| Fd | II | 69.74 | 246162 | 315903 | 3.05 | 3.27 |
| R50 | II | 69.74 | 246162 | 315903 | 3.74 | 3.11 |
| Succinate | II | 43.58 | 227328 | 270903 | 3.31 | 3.12 |
| R70 | II | 60.00 | 263403 | 323403 | 3.75 | 4.1 |
| Cp | XVI | 37.16 | 25617 | 62772 | 3.24 | 3.27 |
| Succinate | XIV | 60.00 | 447375 | 507375 | 3.31 | 4.01 |
The gray colors in the table indicate QTL that are overlapping. The first group of overlapping QTL, which is located on chromosome XIV, involved the maximum fermentation rate variation (Rmax) and nitrogen assimilation (Nass). The other group, which is localized on chromosome II, involves four traits as the succinate production, the fermentation duration, and the fermentation rate at 50% and 70% of sugar consumption. Two QTL mentioned (R50 and succinate on chromosome II) have a maximum LOD score just below the threshold but are conserved because they overlap with Fd and R70 QTL on chromosome II.
Identification of expression QTL
In parallel, to identify loci controlling gene expression, the yeast genome was scanned for markers by gene expression linkages. We treated the expression signals from the whole data set of 4398 genes as quantitative traits without additional filtering and subjected them to linkage analysis using the set of markers. In doing so, a test statistic evaluating each marker-by-gene association was computed to detect the number of eQTL with different LOD score thresholds. For each significance threshold we then estimated a corresponding FDR using a permutation approach (see Materials and Methods). Depending on the LOD score threshold used, we could detect from 92 to 897 eQTL. Ninety-two eQTL were detected with a LOD score threshold of 4.5 (FDR of 0.26). A less stringent LOD score threshold of 3.5 (FDR of 0.50) was associated with 409 eQTL, and a LOD score threshold of 3.0 (FDR of 0.63) was associated with 897 eQTL. The high FDR associated with LOD scores was not surprising given the small population size of segregants used in our analysis. Indeed, all previous eQTL studies in yeast were performed using larger population sizes (Brem and Kruglyak 2005).
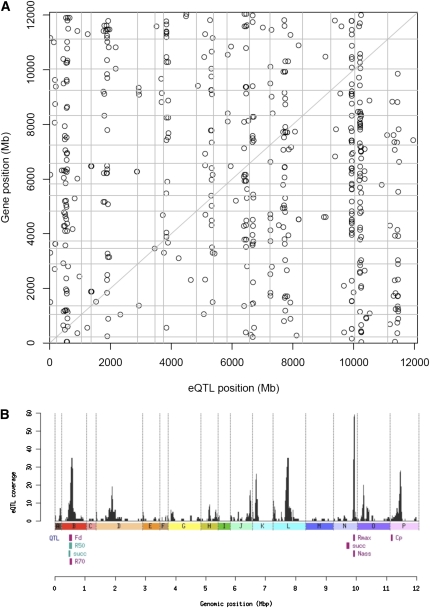
The main interest in this study was to use the QTL analysis to gain an initial insight into the eQTL architecture and its relationship with fermentation QTL. We therefore decided to analyze the data set using a permissive LOD score threshold of 3.5. The physical location of eQTL across the genome was plotted against the physical location of genes that were differentially deregulated and possessed an eQTL using this LOD score (Figure 4). The point in the diagonal indicates cis-linkages, i.e., meaning that the QTL linkage peak coincides with the physical location of the open reading frame for the expression trait. All of the eQTL located outside this diagonal represent trans-linkages with a linkage peaks at loci distant from the physical location of the open reading frames. We detected only a limited number, eight, of cis-associations visible on the diagonal line in Figure 4A. Among them, four ASP3 genes (ASP3-1, ASP3-2, ASP3-3, and ASP3-4) displayed a local regulation consistent with the known deletion of these genes in the industrial strain (Novo et al. 2009). Surprisingly, by observing within trans-eQTL distribution, we noticed the linkage of many transcripts to the same chromosomal region, which gave rise to vertical alignments of eQTL (Figure 4A). Seven such eQTL trans-bands could be observed and localized on chromosomes II, IV, XI, XII, XIV, XV and XVI. This distribution of eQTL corresponds to hotspot regions, which contain loci exerting a trans-control on the expression of a large set of genes (Figure 4A). These trans-regulatory bands were also visible at more stringent threshold levels (Figure S8), and this strengthens the notion of a role of these loci in the variation of expression of a large set of transcripts. The number of eQTL linked to these regions ranged from 12 on chromosome XVI to 42 on chromosome XIV (Figure 4 and Table S4). The position of individual eQTL can be examined using the Gbrowse interface at http://genome.jouy.inra.fr/genyeastrait/.
Figure 4 .
Genomic view of eQTL distribution and relationships with QTL for fermentation traits. A) Genomic view of eQTL mapping. The X-axis represents the genome location of markers, and the Y-axis represents the genome location of the regulated linked genes on concatenated chromosomes. EQTL values with LOD scores greater than 3.5 are displayed. The diagonal pattern is called “cis-eQTL band” and represents an association between the expression level of a gene and the genotype at the gene’s locus. Multiple vertical bands, called “trans-eQTL bands,” illustrate associations between the expression of many genes and a single locus. B) Overlapping of eQTL and QTL for fermentation traits. The scale below the figure indicates the genomic position across the genome in mega base pairs (Mpb). We can observe the overlapping of the hotspot on chromosome II with Fd, R70, R50, and succinate (succ) parameters and the overlap of the hotspot located on chromosome XIV with Rmax and Nass. QTL for traits with LOD scores just below the threshold are shown in blue.
Overlap between eQTL and QTL for fermentation traits
To determine whether the chromosomal loci detected in the QTL/eQTL analysis have an impact both on fermentation phenotypes and the massive deregulation of gene expression we examined the possible relationships between eQTL and QTL of fermentation phenotypes. Figure 4B displays the positions of fermentation QTL and the number of eQTL detected across the genome to visualize the hotspots in parallel. Among the seven detected eQTL hotspot regions, two overlap with fermentation QTL. The hotspot located on chromosome II overlaps with QTL involved in the variations of Fd, R70, and succinate production. The 63 kb hotspot on chromosome XIV overlaps with QTL associated with variations in Rmax and Nass (Figure 4B). The colocalization of the two fermentation QTL with the eQTL hotspots suggests that a common regulator affects both the expression of the set of genes and the fermentation traits. As variations in parameters such as nitrogen assimilation are expected to be associated with strong physiological changes, their coupling with massive changes in gene expression is unsurprising. In addition, some genes, whose expression was previously found to be correlated with the fermentation parameters, are linked to the same hotspot (e.g., the stress-responsive genes HSP12, HSP30, and SPS100 are correlated with Fd and R70 and have a linkage on the chromosome-II hotspot).
Dissection of the eQTL hotspot associated to the variation of Rm and Nass
We explored the hotspot covering the region from 635 kbp to 732 kbp on chromosome XIV to identify putative candidate genes that could be involved in the control of Rmax and Nass. We used the genome sequence of the 59A strain to search for genes with either nonsynonymous SNP leading to amino acid changes in the coding region or with SNPs in the regulatory region and a self-eQTL linkage in the hotspot. Genes potentially connected to nitrogen metabolism were preferentially considered. We found a relevant candidate gene, ABZ1, which codes for a p-aminobenzoate synthase. This gene harbors 13 SNPs in the coding sequence and 5 of these are nonsynonymous substitutions (see http://genome.jouy.inra.fr/genyeastrait). ABZ1 is also involved in methyl donor synthesis required for biosynthesis of methionine and nucleotides. The ABZ1 gene is therefore connected to the nitrogen biosynthetic pathways and was considered to be worth for further investigations. We used a reciprocal-hemizygosity analysis described by Steinmetz et al. (2002) to investigate the impact of the ABZ1 allele on the fermentation phenotype. We constructed an isogenic pair of strains by crossing 59A with BY4742Δabz1 on one side and a 59A strain, in which the ABZ1 gene was deleted (59AΔabz1), with BY4742 on the other side. The two diploid strains differ only in their ABZ1 alleles. In each strain, only one allele of ABZ1 is functional, originating from either BY4742 or 59A. The fermentation profiles obtained with these two strains revealed that the ABZ1 allele has a strong impact on fermentation behavior (Figure 5A). Rmax was strongly decreased whereas Fd was increased when the hemizygous strain carried the ABZ1 allele originating from BY4742. These results are a first confirmation of the implication of ABZ1 in the control of the fermentation rate. The lower fermentation rate with the ABZ1 allele from the BY4742 strain suggests that this allele could be associated with a defect in p-aminobenzoate synthesis. Actually, the synthetic medium used (MS300) (see Materials and Methods) did not contain p-aminobenzoate. We therefore examined how the fermentation kinetics of the hemizygous strains were altered following supplementing the medium with p-aminobenzoate. Indeed, when p-aminobenzoate was added (1 mg/l) to the fermentation medium, the hemizygous strain carrying the BY4742 ABZ1 allele recovered a high fermentation rate (Figure 5B). In contrast, supplementation with p-aminobenzoate had no effect on the reciprocal strain carrying the 59A allele. These data are consistent with the fermentation rate being modulated by a limitation in p-aminobenzoate biosynthesis due to the presence of ABZ1 allele of the laboratory strain. Since the QTL for Nass, overlapped with the QTL for Rmax, we checked whether Nass was modulated by the ABZ1 allele. An amino-acid analysis of the fermentation medium revealed that residual assimilable nitrogen was 30 mg/l with the strain carrying the ABZ1 allele from BY4742 vs. 11 mg/l with the hemizygous strain containing the 59A form. These results indicate that nitrogen assimilation is also modulated by the ABZ1 allele.
Figure 5 .
Reciprocal hemizygous analysis of ABZ1 and impact of p-aminobenzoate on the fermentation profiles. A) Fermentation rate profiles of the two reciprocal hemizygous strains carrying the ABZ1 allele from BY4742 or 59A. The hemizygous strains harbor either an active ABZ1 BY allele (strain BY4742/ABZ1-59A∆, thick gray line) or the 59A allele (strain 59A/ABZ1- BY4742∆, black line with diamonds). Profiles of the strains 59A (dark-blue line), the laboratory strain S288C (thin gray line), and the hybrid Z59S (black line) are shown. B) Impact of p-aminobenzoate on the fermentation profiles of hemizygous strains. Fermentation kinetics of two hemizygous strains in MS300 supplemented or not with 1 mg/l of p-aminobenzoic acid. Hemizygous strain carrying S288c ABZ1 allele in MS300 without (black line) or supplemented with p-aminobenzoic acid (gray kinetic line). Hemizygous strain carrying 59A ABZ1 allele in MS300, without (red kinetic line) or supplemented with p-aminobenzoic acid (pink kinetic line). The supplementation with p-aminobenzoic acid improves the fermentation capacity of the hemizygous strain carrying the S288c ABZ1 allele.
Discussion
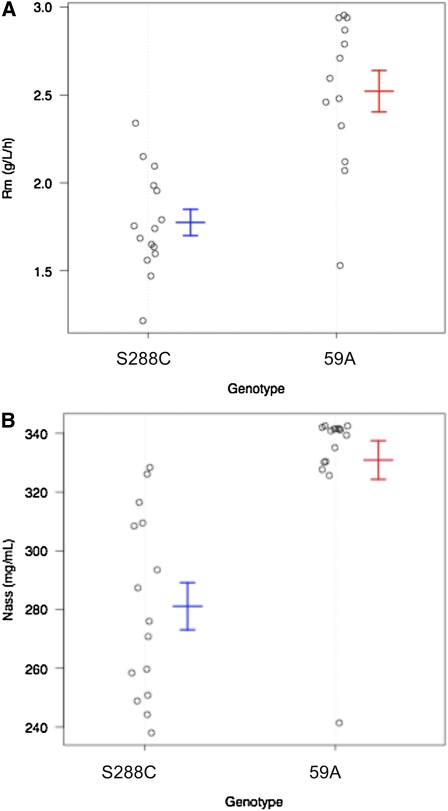
The genetic determinants of variation for most of the fermentation parameters in yeast are still unknown. In this study, we addressed the genetic basis of several fermentation traits and combined this with gene-expression analysis and an eQTL approach. Using a segregating population of a limited size we identified QTL for several fermentation traits. Accurate measures of kinetic parameters—specifically fermentation rates—allowed us to show that depending on the progress of fermentation, different QTL are associated with different fermentation rates; one for Rmax, and one for R70. This is consistent with the notion that, depending on fermentation progress, growth, or starvation, different cellular mechanisms are critical in controlling fermentation flux. On the other hand, these parameters share other components of cell metabolism, which is indicated by their high correlation and strong association with nitrogen utilization. Indeed, we found that Rmax and Nass shared a common QTL. This QTL was dissected, and we provided functional evidence that a gene involved in p-aminobenzoate synthesis plays a role in controlling Rmax and Nass. The ABZ1 allele has a strong effect on the fermentation rate and on nitrogen utilization as visualized in Figure 6. We calculated that ABZ1 could explain 51% of the variance of Rmax and 45% of the variance of Nass in the segregants population.
Figure 6 .
Impact of the ABZ1 genotype on the fermentation phenotypes Rmax and Nass of the 30 segregants. A) Fermentation rate Rmax of the segregants that inherited the ABZ1 locus from S288C or 59A. B) Nitrogen assimilation (Nass) phenotype of the segregants that inherited the ABZ1 locus from S288C or 59A. The average phenotype and standard deviation are indicated for each genotype group.
The association of a transcriptomic analysis of the segregants population to a classical QTL approach has provided an important value of the study. Given the small size of the population, these data did not intend to decipher the regulatory variations globally. However, they could help at elucidating fermentation QTL and offer new insights into their relationships with gene expression. An outstanding observation was the evidence of overlaps among several fermentation QTL and eQTL hotspots. Two out of the seven identified hotspots overlapped with phenotypes. The other hotspots were not associated with phenotypic QTL. However, only a very small number of phenotypes were investigated, and examination of additional phenotypes is required to address potential associations with other hotspots. Similar associations between eQTL hotspots and phenotypes have been reported in yeast (Yvert et al. 2003) and other organisms (Fu et al. 2009). They are thought to originate from sequence variations that have broad pleiotropic effects. In the model plant, Arabidopsis thaliana, variation of complex quantitative traits could be explained by a few hotspot genomic regions that controlled various parameters (Fu et al. 2009). In the present study, we show that the ABZ1 allele has a strong impact on the fermentation capacity of the strain. Indeed, such physiological changes are expected to be coupled to massive changes in gene expressions, which can explain the hotspot. In this case, the hotspot results from the effects of a strong metabolic alteration and not directly from modification of a general regulator.
Our data show that the ABZ1 allele from S288c is unable to support an Rmax that is similar to the industrial form. Because this phenomenon is abolished by the addition of p-aminobenzoate to the fermentation medium, we can infer that a limitation in the flux of this metabolite is responsible for the lower fermentation rate. p-aminobenzoate is an intermediate of the tetrahydrofolate biosynthetic pathway, which leads to a family of cofactors required for one-carbon transfer reactions. These methyl-donor compounds are involved in the synthesis of methionine, serine, and purines (Thomas and Surdin-Kerjan 1997). A limiting flux in p-aminobenzoate can therefore trigger a limitation in the availability of one of these metabolites. Given the pivotal role of methionine in translation, this may limit the rate of protein synthesis and in turn the incorporation of nitrogen sources. This mechanism is consistent with the observed correlation between Rmax and Nass, as well as with the overlap of their QTL. Our results highlight the critical role of methyl-donor biosynthesis in the control of nitrogen utilization during alcoholic fermentation. Interestingly, we did not observe any significant effect of the ABZ1 allele on the growth rate of the hemizygous strains (data not shown). This suggests that the methyl-donor pathway is more critical for sugar flux then for cell growth. The difference in nitrogen utilization triggered by the ABZ1 alleles is also clearly responsible for the differential expression of genes regulated by nitrogen sources. This explains the correlations of the genes under NCR control, such as MEP2 and PUT1, with Rmax. Most of the genes linked to the hotspot are however not involved in nitrogen metabolism but respond to various environmental changes including nitrogen depletion (Gasch et al. 2000). This suggests that they correspond to both direct and indirect effects of the ABZ1 allele. Unexpectedly, we did not observe any deregulation of the ABZ1 gene itself; it was not differentially expressed in the parental strains and displayed no change in the segregants. A similar lack of differential expression was also noticed for genes downstream ABZ1 in the biosynthetic pathway, ABZ2 and FOL1 (data not shown). This suggests that the genes of the pathway are not or poorly regulated by the Abz1 product, 4-amino-4-deoxychorismate, or by downstream metabolites such as p-aminobenzoate.
The Abz1 protein from the strain 59A contains five amino acids changes compared with the S288c form. Comparison with the Abz1sequence of Saccharomyces cerevisiae strains from various origins available at the Sanger Institute (Liti et al. 2009) or in Genbank shows that the same changes are found in other wine yeasts, such as VIN13 (Borneman et al. 2011) and RM11-1a (Ruderfer et al. 2006) (Figure S9). Three of these amino acid changes (T313R, Q650E, N777T) are found in all Saccharomyces cerevisiae strains (except laboratory ones) and a fourth mutation (N475D) in all except 3 of the 39 strains sequenced at the Sanger Institute. Only the L559 was found in strains from different origins [e.g, wild (UWOPS87.2421, UWOPS05.217) or sake (Y12)]. The four common amino acids changes were also found in ABZ1 orthologous of other Saccharomyces species, indicating that they correspond to an ancestral form of the gene. This situation is consistent with a divergence of the ABZ1 gene of S288c. This evolution has been likely associated to a partial loss of function of the p-aminobenzoate synthase probably due to a cultivation of the strain in rich laboratory media (Kvitek et al. 2008). This idea is strengthened by additional information obtained from a large-scale analysis of fermentation phenotypes of Saccharomyces strains that included those from the Sanger list (Liti et al. 2009; Camarasa et al.; paper in preparation). It was observed that alteration of Rmax in the absence of PABA was associated to Abz1 amino acids that are shared by laboratory strains (Camarasa, personal communication). This is consistent with an evolution ABZ1 toward a defective form in the laboratory strains. The laboratory allele of ABZ1 is probably not heavily defective since it does not lead to a true PABA auxotrophy. The strain S288c grows normally in a minimal medium without PABA with no change observed by comparison with a medium supplemented with 1 mg/l PABA) (data not shown). Such a situation may have facilitated the conservation of an ABZ1 form that triggers a phenotype only under specific conditions.
More information based on variation of fermentation parameters and their relationships with expression will certainly be gained from the dissection of the QTL corresponding to the chromosome II hotspot that controls the fermentation rate in the late phase at R70 and consistently at R50. Several genes involved in stress responses (HSP30, HSP12, SPS100) whose level of expression correlated negatively with R70 have a linkage in the hotspot. These stress genes displayed a high level of expression in the laboratory strain and in the progeny that inherited the laboratory-strain form of the region (data not shown). This indicates that the locus from S288c leads to a specific stress response and a low fermentation rate. The potential mechanisms for this observation remain unknown. Our study did not address the expression of the genes acquired by the wine strain EC1118 from a horizontal transfer (Novo et al. 2009) as these genes were unknown at the beginning of the study. However, we have checked for linkage to makers flanking these regions and did not find any with fermentation traits and only one weak with an eQTL, suggesting that they have no strong impact on the parameters considered here.
The biological device used in the present study with a small population of segregants was able to detect fermentation QTL, but it had a lower power to detect eQTL. An additional difficulty for eQTL detection may originate from the specific physiological conditions under which we analyzed gene expression (i.e., nongrowing cells under conditions of stress). Until now, yeast eQTL studies have been performed with growing cells. Under starvation and conditions of stress, many genes that respond to environmental changes have an expression more noisy than average (Gasch 2007; Razer et al. 2004). However, because most critical phenomena in industrial alcoholic fermentations (reduction in carbon flux, ethanol inhibition, cell death, etc.) take place during this phase, an assessment of the relationships between variations in gene expression and fermentation traits is required under such conditions. The findings of our study are the first step toward understanding this process. We demonstrated the role of a QTL relevant for alcoholic fermentation performance and provided an initial basis to address the relationships among fermentation QTL and variations in gene expression.
Supplementary Material
Acknowledgments
This work was supported by Agence Nationale de la Recherche grant Genyeastrait ANR-07-BLAN-0205. C. Ambroset was supported by a fellowship from the INRA Département Microbiologie et Chaîne Alimentaire (MICA) and the region Languedoc-Roussillon.
Footnotes
Supporting information is available online at http://www.g3journal.org/lookup/suppl/doi:10.1534/g3.111.000422/-/DC1SI.
Sequence data from this article have been deposited in the NCBI Sequence Read Archive as study SRP004704. Gene expression data from this article have been submitted to the Gene Expression Omnibus database under accession no. GSE26437.
Literature Cited
- Ansel J., Bottin H., Rodriguez-Beltran C., Damon C., Nagarajan M., et al. , 2008. Cell-to-cell stochastic variation in gene expression is a complex genetic trait. PLoS Genet. 4: e1000049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bely M., Sablayrolles J.-M., Barre P., 1990. Automatic detection of assimilable nitrogen deficiencies during alcoholic fermentation in oenological conditions. J. Ferment. Bioeng. 70: 246–252 [Google Scholar]
- Ben-Ari G., Zenvirth D., Sherman A., David L., Klutstein M., et al. , 2006. Four linked genes participate in controlling sporulation efficiency in budding yeast. PLoS Genet. 2: e195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y., Drai D., Elmer G., Kafkafi N., Golani I., 2001. Controlling the false discovery rate in behavior genetics research. Behav. Brain Res. 125: 279–284 [DOI] [PubMed] [Google Scholar]
- Bergmeyer H. U., Beutler H. O., 1990. Ammonia, pp. 454–461 in Methods of Enzymatic Analysis, 3rd Ed, Vol. III, edited by Bergmeyer H. U. VCH Publishers Ltd., Cambridge, UK [Google Scholar]
- Borneman A. R., Forgan A. H., Pretorius I. S., Chambers P. J., 2008. Comparative genome analysis of a Saccharomyces cerevisiae wine strain. FEMS Yeast Res. 8: 1185–1195 [DOI] [PubMed] [Google Scholar]
- Borneman A. R., Desany B. A, Riches D., Affourtit J. P., Forgan A. H., Pretorius I. S., et al. , 2011. Whole-genome comparison reveals novel genetic elements that characterize the genome of industrial strains of Saccharomyces cerevisiae. PLoS Genet. 7: e1001287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brauer M. J., Christianson C. M., Pai D. A., Dunham M. J., 2006. Mapping novel traits by array-assisted bulk segregant analysis in Saccharomyces cerevisiae. Genetics 173: 1813–1816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brem R. B., Yvert G., Clinton R., Kruglyak L., 2002. Genetic dissection of transcriptional regulation in budding yeast. Science 296: 752–755 [DOI] [PubMed] [Google Scholar]
- Brem R. B., Kruglyak L., 2005. The landscape of genetic complexity across 5,700 gene expression traits in yeast. Proc. Natl. Acad. Sci. USA 102: 1572–1577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broman K. W., Wu H., Sen S., Churchill G. A., 2003. R/qtl: QTL mapping in experimental crosses. Bioinformatics 19: 889–890 [DOI] [PubMed] [Google Scholar]
- Brown K. M., Landry C. R., Hartl D. L., Cavalieri D., 2008. Cascading transcriptional effects of a naturally occurring frameshift mutation in Saccharomyces cerevisiae. Mol. Ecol. 17: 2985–2997 [DOI] [PubMed] [Google Scholar]
- Calull M., Lopez E., Marce R. M., Olucha J. C., Borrull F., 1992. Optimization of an ion exchange high-performance liquid chromatographic method for determination of carboxylic acids, sugars, glycerol, and ethanol in wines. J. Chromatogr. A 589: 151–158 [DOI] [PubMed] [Google Scholar]
- Cavalieri D., Townsend J. P., Hartl D. L., 2000. Manifold anomalies in gene expression in a vineyard isolate of Saccharomyces cerevisiae revealed by DNA microarray analysis. Proc. Natl. Acad. Sci. USA 97: 12369–12374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubillos F. A., Billi E., Zorgo E., Parts L., Fargier P., et al. , 2011. Assessing the complex architecture of complex traits in diverged yeast populations. Mol. Ecol. 20: 1401–1413 [DOI] [PubMed] [Google Scholar]
- Deutschbauer A. M., Davis R. W., 2005. Quantitative trait loci mapped to single-nucleotide resolution in yeast. Nat. Genet. 37: 1333–1340 [DOI] [PubMed] [Google Scholar]
- Doniger S. W., Kim H. S., Swain D., Corcuera D., Williams M., et al. , 2008. A catalog of neutral and deleterious polymorphism in yeast. PLoS Genet. 4: e1000183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunham M. J., Badrane H., Ferea T., Adams J., Brown P. O., et al. , 2002. Characteristic genome rearrangements in experimental evolution of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 99: 16144–16149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn B., Levine R. P., Sherlock G., 2005. Microarray karyotyping of commercial wine yeast strains reveals shared, as well as unique, genomic signatures. BMC Genomics 6: 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R., Domrachev M., Lash A. E., 2002. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30: 207–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay J. C., McCullough H. L., Sniegowski P. D., Eisen M. B., 2004. Population genetic variation in gene expression is associated with phenotypic variation in Saccharomyces cerevisiae. Genome Biol. 5: R26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J., Keurentjes J. J., Bouwmeester H., America T., Verstappen F. W., et al. , 2009. System-wide molecular evidence for phenotypic buffering in Arabidopsis. Nat. Genet. 41: 166–167 [DOI] [PubMed] [Google Scholar]
- Gasch A. P., Spelman P. T., Kao C. M., Carmel-Harel O., Eisen M. B., et al. , 2000. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell 11: 4241–4257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasch A. P., 2007. Comparative genomics of the environmental stress response in ascomycete fungi. Yeast 24: 961–976 [DOI] [PubMed] [Google Scholar]
- Gerke J. P., Chen C. T., Cohen B. A., 2006. Natural isolates of Saccharomyces cerevisiae display complex genetic variation in sporulation efficiency. Genetics 174: 985–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goffeau A., Barrell B., Bussey H., Davis R., Dujon B., et al. , 1996. Life with 6000 genes. Science 274: 546, 563–567 [DOI] [PubMed] [Google Scholar]
- Hu X. H., Wang M. H., Tan T., Li J. R., Yang H., et al. , 2007. Genetic dissection of ethanol tolerance in the budding yeast Saccharomyces cerevisiae. Genetics 175: 1479–1487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Infante J. J., Dombek K. M., Rebordinos L., Cantoral J. M., Young E. T., 2003. Genome-wide amplifications caused by chromosomal rearrangements play a major role in the adaptive evolution of natural yeast. Genetics 165: 1745–1759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Instituto Nacional de la Investigación en Agronomía (INRA). GenYeasTrait Genome Browser. Available at: http://genome.jouy.inra.fr/genyeastrait/. Accessed: March 01, 2011
- Institut National des Sciences Appliquées. Plateforme Biopuces. Available at: http://biopuces.genotoul.fr. Accessed: January 1, 2011
- Jansen R. C., Nap J. P., 2001. Genetical genomics: the added value from segregation. Trends Genet. 17: 388–391 [DOI] [PubMed] [Google Scholar]
- Katou T., Namise M., Kitagaki H., Akao T., Shimoi H., 2009. QTL mapping of sake brewing characteristics of yeast. J. Biosci. Bioeng. 107: 383–393 [DOI] [PubMed] [Google Scholar]
- Kellis M., Patterson N., Endrizzi M., Birren B., Lander E. S., 2003. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature 423: 241–254 [DOI] [PubMed] [Google Scholar]
- Kim H. S., Fay J. C., 2007. Genetic variation in the cysteine biosynthesis pathway causes sensitivity to pharmacological compounds. Proc. Natl. Acad. Sci. USA 104: 19387–19391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvitek D. J., Will J. L., Gasch A. P., 2008. Variations in stress sensitivity and genomic expression in diverse S. cerevisiae isolates. PLoS Genet. 4: e1000223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Ruan J., Durbin R., 2008. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 18: 1851–1858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liti G., Carter D. M., Moses A. M., Warringer J., Parts L., et al. , 2009. Population genomics of domestic and wild yeasts. Nature 458: 337–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marullo P., Bely M., Masneuf-Pomarede I., Pons M., Aigle M., et al. , 2006. Breeding strategies for combining fermentative qualities and reducing off-flavor production in a wine yeast model. FEM. Yeast Res. 6: 268–279 [DOI] [PubMed] [Google Scholar]
- Nogami S., Ohya Y., Yvert G., 2007. Genetic complexity and quantitative trait loci mapping of yeast morphological traits. PLoS Genet. 3: e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novo M., Bigey F., Beyne E., Galeote V., Gavory F., et al. , 2009. Eukaryote-to-eukaryote gene transfer events revealed by the genome sequence of the wine yeast Saccharomyces cerevisiae EC1118. Proc. Natl. Acad. Sci. USA 106: 16333–16338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Ortin J. E., Querol A., Puig S., Barrio E., 2002. Molecular characterization of a chromosomal rearrangement involved in the adaptive evolution of yeast strains. Genome Res. 12: 1533–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team, 2010. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria. Available at: www.R-project.org
- Rachidi N., Barre P., Blondin B., 1999. Multiple Ty-mediated chromosomal translocations lead to karyotype changes in a wine strain of Saccharomyces cerevisiae. Mol. Gen. Genet. 261: 841–850 [DOI] [PubMed] [Google Scholar]
- Razer J. M., O’Shea E. K., 2004. Control of stochasticity in eukaryotic gene expression. Science 304: 1811–1814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronald J., Akey J. M., 2007. The evolution of gene expression QTL in Saccharomyces cerevisiae. PLoS ONE 2: e678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossignol T., Dulau L., Julien A., Blondin B., 2003. Genome-wide monitoring of wine yeast gene expression during alcoholic fermentation. Yeast 20: 1369–1385 [DOI] [PubMed] [Google Scholar]
- Rossignol T., 2004. Analyse de l'expression du génome des levures œnologiques en fermentation alcoolique par des approches post-génomiques. Ph.D. Thesis. University Montpellier II. Montpellier, France [Google Scholar]
- Ruderfer D. M., Pratt S. C., Seidel H. S., Kruglyak L., 2006. Population genomic analysis of outcrossing and recombination in yeast. Nat. Genet. 38: 1077–1081 [DOI] [PubMed] [Google Scholar]
- Saccharomyces Genome Database, 2008. Saccharomyces Genome Database. http://www.yeastgenome.org. Accessed: February 1, 2011 [Google Scholar]
- Schacherer J., Shapiro J. A., Ruderfer D. M., Kruglyak L., 2009. Comprehensive polymorphism survey elucidates population structure of Saccharomyces cerevisiae. Nature 458: 342–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiestl R. H., Gietz R. D., 1989. High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr. Genet. 16: 339–346 [DOI] [PubMed] [Google Scholar]
- Sinha H., Nicholson B. P., Steinmetz L. M., McCusker J. H., 2006. Complex genetic interactions in a quantitative trait locus. PLoS Genet. 2: e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha H., David L., Pascon R. C., Clauder-Munster S., Krishnakumar S., et al. , 2008. Sequential elimination of major-effect contributors identifies additional quantitative trait loci conditioning high-temperature growth in yeast. Genetics 180: 1661–1670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth G. K., 2005. Limma: linear models for microarray data, pp. 397–420 Bioinformatics and Computational Biology Solutions using R and Bioconductor, edited by Gentleman R., Carey V., Dudoit S., Irizarry R., Huber W. Springer, New York [Google Scholar]
- Smyth G. K., Speed T. P., 2003. Normalization of cDNA microarray data. Methods 31: 265–273 [DOI] [PubMed] [Google Scholar]
- Smyth G. K., Michaud J., Scott H., 2005. The use of within-array replicate spots for assessing differential expression in microarrays experiments. Bioinformatics 21: 2067–2075 [DOI] [PubMed] [Google Scholar]
- Smith E. N., Kruglyak L., 2008. Gene-environment interaction in yeast gene expression. PLoS Biol. 6: e83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein L. D., Mungall C., Shu S., Caudy M., Mangone M., et al. , 2002. The generic genome browser: a building block for a model organism system database. Genome Res. 12: 1599–1610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz L. M., Sinha H., Richards D. R., Spiegelman J. I., Oefner P. J., et al. , 2002. Dissecting the architecture of a quantitative trait locus in yeast. Nature 416: 326–330 [DOI] [PubMed] [Google Scholar]
- Thomas D., Surdin-Kerjan Y., 1997. Metabolism of sulfur amino acids in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 61: 503–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varela C., Pizarro F., Agosin E., 2004. Biomass content governs fermentation rate in nitrogen-deficient wine musts. Appl. Environ. Microbiol. 70: 3392–3400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W., McCusker J. H., Hyman R. W., Jones T., Ning Y., et al. , 2007. Genome sequencing and comparative analysis of Saccharomyces cerevisiae strain YJM789. Proc. Natl. Acad. Sci. USA 104: 12825–12830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winzeler E. A., Richards D. R., Conway A. R., Goldstein A. L., Kalman S., et al. , 1998. Direct allelic variation scanning of the yeast genome. Science 281: 1194–1197 [DOI] [PubMed] [Google Scholar]
- Winzeler E. A., Lee B., McCusker J. H., Davis R. W., 1999. Whole genome genetic-typing in yeast using high-density oligonucleotide arrays. Parasitology 118 Suppl: S73–S80 [DOI] [PubMed] [Google Scholar]
- Yvert G., Brem R. B., Whittle J., Akey J. M., Foss E., et al. , 2003. Trans-acting regulatory variation in Saccharomyces cerevisiae and the role of transcription factors. Nat. Genet. 35: 57–64 [DOI] [PubMed] [Google Scholar]
- Zuzuarregui A., Monteoliva L., Gil C., Del Olmo M., 2006. Transcriptomic and proteomic approach for understanding the molecular basis of adaptation of Saccharomyces cerevisiae to wine fermentation. Appl. Environ. Microbiol. 72: 836–847 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.