Abstract
Lagging exposure information is often undertaken to allow for a latency period in cumulative exposure-disease analyses. The authors first consider bias and confidence interval coverage when using the standard approaches of fitting models under several lag assumptions and selecting the lag that maximizes either the effect estimate or model goodness of fit. Next, they consider bias that occurs when the assumption that the latency period is a fixed constant does not hold. Expressions were derived for bias due to misspecification of lag assumptions, and simulations were conducted. Finally, the authors describe a method for joint estimation of parameters describing an exposure-response association and the latency distribution. Analyses of associations between cumulative asbestos exposure and lung cancer mortality among textile workers illustrate this approach. Selecting the lag that maximizes the effect estimate may lead to bias away from the null; selecting the lag that maximizes model goodness of fit may lead to confidence intervals that are too narrow. These problems tend to increase as the within-person exposure variation diminishes. Lagging exposure assignment by a constant will lead to bias toward the null if the distribution of latency periods is not a fixed constant. Direct estimation of latency periods can minimize bias and improve confidence interval coverage.
Keywords: asbestos, cohort studies, latency, neoplasms, survival analysis
In an epidemiologic study of the association between a protracted or repeated exposure and disease, an epidemiologist may calculate a summary exposure metric, such as each person’s cumulative exposure. Under the premise that there is typically an induction and latency period between exposure and its observed impact on disease (1, 2), a summary exposure metric may be “lagged” by excluding exposures that occurred in the months or years immediately preceding the outcome (3). Often an epidemiologist will evaluate several exposure lags and select the lag that maximizes the magnitude of the resultant effect estimate or the model’s goodness of fit (4).
In this paper, we show that such approaches are liable to result in biased effect estimates or confidence intervals that are too narrow. For simplicity, we will use the term “latency period” to refer to the combined induction and latency periods. Using analytical results and simulations, we illustrate the impact of standard approaches to exposure lagging on effect estimates under various assumptions about the population distribution of latency periods, and we propose an approach that addresses these limitations.
MATERIALS AND METHODS
We focus on estimation of a cumulative exposure-disease association in a setting of protracted or repeated exposures. Repeated exposure measurements on the same person often are correlated over time. To allow for the possibility of simple exposure correlation over time, suppose that the exposure measure for person i at time j may be described by a model such as dij = μ + αi + ϵij, where μ is long-term overall mean exposure, αi is the deviation of person i’s exposure from μ, and ϵij is the variation in exposure at time j from person i’s mean. If αi and ϵij are independent, normally distributed variables with αi ∼ N(0, ) and ϵij ∼ N(0, ), the variance components and describe exposure variability within and between persons, respectively. A similar model of the form ln(dij) = μ + αi + ϵij often is used in environmental and occupational settings where the distribution of exposure tends to be lognormal (5), and simple extensions of this model allow for a systematic change of exposure over time.
The cumulative exposure accrued by a person up to age t is . We focus on the common situation in which exposure levels are nonnegative; therefore cumulative exposure level is only increasing, so that that the cumulative exposure level at one age must be at least as high as at any earlier age.
Latency period
We are interested in scenarios in which there is a latency period between exposure and resultant health outcome. Let L denote the length of the period (where L ≥ 0). Exposures that occur during the L years immediately prior to age t have no effect on the health outcome. Let Di(t) represent the cumulative exposure accrued up to age t, discounting those exposures that occurred during the L years immediately prior to t,
where IL(t − j) equals 1 if L ≤ (t − j), else 0.
With regard to exposure lags, an epidemiologist will often evaluate a range of exposure lag assumptions, for example, G = Gmin, …, Gmax, where G is a fixed constant (i.e., a value that is identical for all study members). We denote the measured cumulative exposure at age t for person i under a proposed lag, G, as
where IG(t − j) equals 1 if G ≤ (t − j), else 0. Note that Xi (t, G) denotes the cumulative exposure under a proposed lag G, while Di(t) denotes the cumulative exposure accounting for the true latency period.
Choosing a lag based on magnitude of association
One approach to choosing a lag assumption is to estimate the exposure-disease association under a range of lag assumptions and to select the lag that yields the largest effect estimate. This approach is premised on the notion that the maximal effect estimate will be the one that is least biased by exposure misclassification (1).
We represent the expected estimate of association given the cumulative exposure under the guessed lag by E(φi|Xi (t, G)). Suppose we assume that E(φi|Di (t), Xi (t, G)) = E(φi|Di (t)); that is, once the true exposure is known, the measured exposure under the proposed lag provides no additional information regarding the association of interest. If the excess relative rate of the outcome of interest increases as a linear function of Di (t), denoted by the function E(φi|Di (t)) = βDi (t), where β is the parameter of interest to be estimated, then it follows that E(φi|Xi (t, G)) = E(E(φi|Di (t), Xi (t, G))|Xi (t, G)) = E(E(φi|Di (t))|Xi (t, G)) = E(βDi (t)|Xi (t, G)) = βE(Di (t)|Xi (t, G)). Thus, the exposure-response association is a function of the conditional expectation of the true exposure given the measured exposure under the guessed lag.
We express the conditional expectation of the true exposure given a measured exposure, x, as follows: E(Di (t)|Xi (t, G) = x) = x + E[Δ], where E[Δ] denotes the expected difference between the true and measured dose,
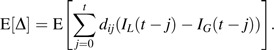 |
If we let η = [(x + E[Δ])/x], then E[Di (t)|Xi (t, G) = x] = ηx, where η equals 1 if E[Δ] = 0, as occurs when the proposed lag equals the true latency period, G = L. Therefore, the exposure-response association under a proposed lag is given by E(φi|Xi (t, G) = x) = βηx = β*x, where β* = βη is the cumulative exposure-response estimate that would be obtained under the proposed lag.
If G = L, then β* = β because η equals 1, confirming that an investigator will obtain an unbiased estimate of association if the proposed lag equals the true latency period (it is also true that if β = 0 then β* = β). However, will the proposed lag that yields the largest estimated value for β* be the one that is least biased (i.e., the value of β* closest to β)? Not necessarily. To illustrate the potential for bias to occur if an investigator selects the proposed lag that yields the largest effect estimate, consider the situation in which exposure is constant over time, such that exposure levels differ between people but not within persons. If G > L, then η will take a value greater than 1; and if L > G, then η will take a value less than 1. Therefore, β* will be biased away from the null if G > L and will be biased toward the null if L > G. Consequently, the approach of selecting the lag that yields the largest magnitude of association is susceptible to bias in settings where there is constant exposure. More generally, as the within-person variation in exposure diminishes, the estimate of a cumulative exposure-response association under the proposed lag that yields the largest estimate of association may be biased away from the true association.
Choosing a lag based on model goodness of fit
Another approach to selection of a lag is to choose the lag that maximizes the model goodness of fit (4). This approach is premised on the idea that if the model is correctly specified, the proposed lag that yields the best fit will yield an unbiased estimate of association.
This approach also depends upon the degree of within-person exposure variability over the period during which exposure histories will be summarized. As the within-person variation in exposure diminishes, the correlation of values for Xi (t, G) obtained under various values for G increases, and the ability to compare lags based on model goodness of fit diminishes. Consider the scenario in which exposure is constant over time. The likelihood for the regression function, E(φi|Xi (t, G) = x) = βηx, depends upon β and G. If Xi (t, G) values derived under a range of proposed lags are perfectly correlated, the model likelihood will be identical across that range. More generally, as σw → 0, values of Xi (t, G) derived under different values of G become increasingly correlated, and the likelihood surface becomes increasingly flat with respect to G.
A flat likelihood with respect to a range of lags suggests that the data in hand do not contain enough information for one to choose between lags within that range. Indeed, at the extreme, the parameters in the likelihood are not identifiable. The epidemiologist may find that a range of models under alternative lag assumptions yield similar goodness of fit but imply markedly different estimates of association.
Under the assumption that there is enough information to yield a nonflat likelihood surface, the approach of evaluating a range of proposed lags and selecting a single lag value on the basis of model goodness of fit will not lead to biased estimates of the cumulative exposure-response association. However, this approach will tend to yield confidence intervals for the estimated exposure effect that are too narrow. If, a priori, the investigator assumed that a range of values for the latency period were equally plausible, then the appropriate confidence interval should reflect the range of values for the exposure-response parameter that are statistically compatible with the observed data given the joint estimation of the lag assumption. If a range of lags are consistent with the data and the estimated exposure-disease association varies across this range of lags, then the appropriate confidence interval obtained by the joint estimation of β and G will tend to be wider than the confidence interval obtained had G been chosen a priori. We discuss joint estimation of β and G in the Web Appendix, which appears on the Journal’s Web site (http://aje.oxfordjournals.org).
Random versus fixed latency periods
The standard practice of lagging exposure treats the latency period as a fixed constant. Suppose, instead, that L depends on unmeasured individual characteristics, such that Li may be viewed as a random variable. Given a random variable Li, the expected exposure-disease association may be obtained by averaging over the population distribution of L, denoted fL(l), as follows:
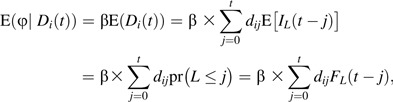 |
where which is the cumulative density function for the random variable L.
The expected cumulative exposure-disease association obtained under a proposed fixed constant lag assumption, G, is given by E(φi|Xi (t, G)) = βE(Di (t)|X(t, G)), where E(Di (t)|Xi (t, G)) = Xi (t, G) + E[Δ]. For the scenario in which dij is constant over j,
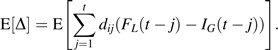 |
Again, letting η denote [(x + E[Δ])/x], the observed exposure-response is given by E(φi|Xi (t, G) = x) = βE(Di (t)|Xi (t, G) = x) = βηx = β*x. If FL approximates a step function that transitions from 0 to 1 at L and if G = L, then little or no bias will be observed. However, if the cumulative density function, FL, does not approximate a step function (e.g., it is more gradual than a step function), then even if an investigator selects the lag, G, that conforms to the mode (or mean) of the underlying distribution of latency intervals, L, bias may arise.
The guessed lag, G, which yields a function IG that best approximates the underlying cumulative density function of the latency periods, FL, will be the best fitting model. However, misspecification of the underlying distribution of latency periods may lead to bias.
Regression models for latency periods
The estimation problems arising from the unobserved latency periods described above may be addressed by using a regression model that allows estimation of parameters describing the distribution of latency periods along with the other regression model parameters. The Web Appendix provides SAS code (SAS Institute, Inc., Cary, North Carolina) illustrating how point estimates and confidence intervals are derived for the parameters describing the distribution of latency periods along with other regression model parameters for linear excess relative rate models fit by maximum likelihood, as well as for analyses based on a log-linear model where the excess relative rate of the outcome of interest increases as an exponential function of Di (t), φi(Di (t); β) = exp(βDi(t)) − 1.
Empirical example
To illustrate the proposed method for joint estimation of parameters that describe an exposure-response association and a latency distribution, we use empirical data analyses of cumulative asbestos exposure-lung cancer mortality associations among 3,072 South Carolina asbestos textile workers employed in production for at least 1 month between January 1, 1940, and December 31, 1965 (6). Vital status was ascertained through December 31, 2001. The outcome of interest, lung cancer mortality, was defined on the basis of underlying cause of death. Cumulative asbestos exposure, expressed in fiber-years per milliliter, was computed for each worker as the product of the length of employment in each job in a year by the estimated asbestos exposure rate for that job. For each lung cancer death, a risk set was formed that included all workers who were alive and eligible to be in the study at the age of death of the index case; controls were also matched to cases on sex, race, and calendar year of birth (defined in 5-year categories from before 1960 to 1990 and later). Up to 40 controls were selected for each lung cancer death by random sampling without replacement from all controls from the risk set. As in previous analyses of these data, we fitted a linear excess rate ratio (ERR) model for the association between cumulative asbestos exposure and lung cancer mortality (7). Estimates of the ERR per 100 fiber-years/mL, as well as associated regression model goodness of fit, were derived under proposed lags of 1, 2, 3, …, 19, and 20 years. We identified the lag that yielded the largest estimated exposure-outcome coefficient, and we identified the lag that yielded the best model fit (as determined by the −2 log likelihood). In addition, we directly estimated the length of the latency period under the assumption that this period is a fixed constant, and we estimated the mode and coefficient of variation of an assumed underlying lognormal distribution of latency periods.
The analysis included 198 lung cancer cases and 7,505 controls. The magnitude of the estimated association between cumulative asbestos exposure and lung cancer mortality increased with increasing duration of the exposure lag assumption, from an estimated ERR per 100 fiber-years/mL = 1.07 under a 1-year exposure lag assumption to an estimated ERR per 100 fiber-years/mL = 1.32 under a 20-year exposure lag assumption.
If an investigator selected the exposure lag based on the largest magnitude of association over the range of proposed lags, this would yield an estimated ERR per 100 fiber-years/mL = 1.32 (95% confidence interval (CI): 0.45, 2.62) under the 20-year exposure lag. In contrast, if an investigator selected the exposure lag based upon the best model goodness of fit over the range of examined lags, this would yield an estimated ERR per 100 fiber-years/mL = 1.15 obtained under a 9-year lag assumption. Now, treating the 9-year lag as if it had been chosen a priori yields a 95% confidence interval of 0.43, 2.22. However, the 9-year lag was arrived at through a “best fit search,” so, as expected, the 95% confidence interval of 0.42, 2.33 that takes into account the joint estimation of lag and exposure effect is slightly wider.
A regression model was fitted in which the latency periods were assumed to arise from a lognormal distribution, resulting in an estimated ERR = 1.15 (95% CI: 0.43, 2.25) with the mode of the lognormal distribution at 8.47 years (95% CI: 0.66, 24.53) and coefficient of variation = 0.148.
Simulation example
Simulations were used to illustrate bias and confidence interval coverage due to misspecification of lag assumptions. Data were simulated for 10,000 hypothetical studies of the association between cumulative radon exposure and lung cancer mortality; the simulation method follows an approach described previously for simulating nested case-control data by using empirical data from the Colorado Plateau uranium miners study (8). Letting i denote worker and j denote year of observation, radon exposure histories were assigned such that the exposure intensity for each worker-year, dij, conforms to the model ln(dij) = μ + αi + ϵij, where μ = 0.1, ai ∼ N(0, σ2B), and ϵij ∼ N(0, σ2w). Data were simulated for scenarios in which σB = 1 and σw = 0.1, 0.5, or 1.0. Risk sets were then formed from the cohort at each of the ages of the 258 lung cancer deaths. First, we randomly pick one member of the risk set, k, as the index case by specifying the rate of lung cancer death for each worker, i, at age t, λi(t), as a function of the person’s exposure history up to that age from the multinomial distribution with probabilities λi(tk)/Σj λj(tk), where the sum is over the risk set members, and then randomly sampled up to 40 controls from the risk set; and λi(t) = exp(βD), where β = 1.0 and D is the cumulative exposure accrued L years prior to the index date. Two series of simulations were conducted: In the first, the latency period, L, was a fixed constant equal to 5 years; in the second, the latency period was assigned to each person by random sampling from a lognormal distribution with mode equal to 5 years and coefficient of variation = 0.1, 0.2, or 0.3. Each simulated data set was analyzed by using conditional logistic regression. Estimates of the cumulative exposure-outcome association were derived under proposed lags of 1, 2, …, 9, and 10 years. For each simulation, we identified the proposed lag that yielded the largest estimated exposure-outcome coefficient, and we identified the proposed lag that yielded the best model fit (as determined by the −2 log likelihood). For simulations in which the assigned latency period was a fixed constant, an estimate of the cumulative exposure-outcome association was derived by using a model that jointly estimated the exposure effect estimate and latency period (Web Appendix). For simulations in which a latency period was assigned to each person by random sampling from a lognormal distribution, an estimate of the cumulative exposure-outcome association was derived by using a model that simultaneously estimated the exposure effect and parameters for a lognormal distribution of latency periods (Web Appendix). From 10,000 trials, we computed the mean log rate ratio (“estimated β”), empirical standard deviation of the estimated log rate ratio (“empirical SE”), average of the estimated standard error of the log rate ratio (“estimated SE”), and proportion of estimated 95% Wald-type confidence intervals that covered the specified true value for β (“CI coverage”). We note that the framework laid out in the paper is more general than the parametric model used in the simulations.
We first simulated data specifying a 5-year fixed latency period under scenarios in which σw was 0.1, 0.5, or 1.0 (Table 1). For each iteration of the scenario, models were fitted under a range of proposed lags (1–10 years). As in our motivating example, as the proposed lag increased so did the magnitude of the estimated association. Therefore, selecting the proposed lag that yielded the largest estimate of the magnitude of association consistently corresponded to the longest lag evaluated (i.e., a 10-year lag). The average estimate of association was 1.33, 1.31, or 1.24 under scenarios in which σw was 0.1, 0.5, or 1.0. Selecting the lag that yielded the best model fit for each iteration of the simulation resulted in estimates of association that were close to the true value specified for the simulations. The average estimates of association were 1.04, 1.03, and 1.02 under scenarios in which σw was 0.1, 0.5, and 1.0; 95% confidence interval coverage was estimated to be 62%, 68%, and 85% under these scenarios, suggesting that the practice of comparing the fit of a series a models under various lag assumptions and selecting the lag that yields the best model fit results in confidence intervals for the effect estimate that are too narrow. Using the same simulation data, we directly estimated the latency period simultaneously with the parameter describing the exposure-response association. The average estimates of association were log(relative rate/100 units) = 1.01 (CI coverage, 88%), 0.99 (CI coverage, 88%), and 1.01 (CI coverage, 92%) under scenarios in which σw was 0.1, 0.5, and 1.0, indicating little bias and confidence interval coverage closer to the nominal 95% level. Of course, if the true latency period was known a priori and the investigator specified a 5-year lag for each iteration of the simulation, the confidence interval will be narrower. In our simulations, the average estimates of association under these scenarios equal log(relative rate/100 units) = 1.00 (CI coverage, 95%) (not shown).
Table 1.
Estimated Log Relative Rate per 100 WLMs Under Various Approaches to Selecting the Exposure Lag Assumptiona
| Three Scenarios Regarding σw | Estimated β | Empirical SE | Estimated SE | CI Coverage |
| σw = 0.1 | ||||
| Select “guessed” lag that yields largest estimate of association | 1.33 | 0.126 | 0.120 | 0.218 |
| Select “guessed” lag that yields best model fit | 1.04 | 0.195 | 0.092 | 0.623 |
| Simultaneously estimate lag and estimate of association | 1.01 | 0.190 | 0.134 | 0.878 |
| σw = 0.5 | ||||
| Select “guessed” lag that yields largest estimate of association | 1.31 | 0.095 | 0.093 | 0.083 |
| Select “guessed” lag that yields best model fit | 1.03 | 0.147 | 0.072 | 0.684 |
| Simultaneously estimate lag and estimate of association | 0.99 | 0.114 | 0.077 | 0.880 |
| σw = 1.0 | ||||
| Select “guessed” lag that yields largest estimate of association | 1.24 | 0.076 | 0.070 | 0.050 |
| Select “guessed” lag that yields best model fit | 1.02 | 0.078 | 0.056 | 0.851 |
| Simultaneously estimate lag and estimate of association | 1.01 | 0.071 | 0.055 | 0.915 |
Abbreviations: CI, confidence interval; SE, standard error; WLM, working level month.
In all simulations, the specified true association is β = 1. The natural log of annual exposure is distributed with μ = 0.1 and σB = 1.
Data were also simulated for scenarios in which the latency period was a random variable arising from a lognormal distribution (Table 2). We first consider the approach of selecting the lag that yields the largest magnitude of association; this estimate is obtained under the longest proposed lag value and is a biased estimate of association. In the majority of simulations, this corresponded to the longest lag assumption considered. The average estimates of association were 1.36, 1.33, and 1.29 under scenarios in which the population variations in the latency period were 0.1, 0.2, and 0.3. Next, we considered the approach of selecting the lag that yielded the best model fit. Bias increased as the population variation in the latency period increased. The average estimates of association were 0.99, 0.97, and 0.94 under scenarios in which the population variations in the latency period were 0.1, 0.2, and 0.3. The 95% confidence interval coverages were estimated to be 81%, 78%, and 65% under these scenarios. We fit a model to directly estimate the coefficient of variation of a lognormal latency period distribution, as would be estimated if the mode of the distribution of latency period was known a priori to be equal to 5 years (or, as would be estimated if the investigator specified a preferred lag assumption and evaluated the sensitivity of results to the assumption that the latency period was lognormally distributed rather than a fixed constant). The average estimates of association were 1.05 (CI coverage, 95%), 1.04 (CI coverage, 94%), and 1.04 (CI coverage, 90%) under scenarios in which the population variations in the latency interval were 0.1, 0.2, and 0.3. We also fitted a model to directly estimate the mode and coefficient of variation of a lognormal distribution of latency periods. The average estimates of association were 1.05 (CI coverage, 92%), 1.05 (CI coverage, 91%), and 1.04 (CI coverage, 87%) under scenarios in which the population variations in the latency period were 0.1, 0.2, and 0.3.
Table 2.
Estimated Log Relative Rate per 100 WLMs Under Various Approaches to Selecting the Exposure Lag Assumption or Modeling the Population Distribution of Latency Intervalsa
| Three Scenarios Regarding the CV of This Distribution | Estimated β | Empirical SE | Estimated SE | CI coverage |
| CV = 0.1 | ||||
| Select “guessed” lag that yields largest estimate of association | 1.36 | 0.076 | 0.075 | 0.000 |
| Select “guessed” lag that yields best model fit | 0.99 | 0.079 | 0.053 | 0.814 |
| Simultaneously estimate CV and estimate of association | 1.05 | 0.148 | 0.122 | 0.947 |
| Simultaneously estimate mode, CV, and estimate of association | 1.05 | 0.182 | 0.141 | 0.923 |
| CV = 0.2 | ||||
| Select “guessed” lag that yields largest estimate of association | 1.33 | 0.076 | 0.075 | 0.002 |
| Select “guessed” lag that yields best model fit | 0.97 | 0.081 | 0.053 | 0.779 |
| Simultaneously estimate CV and estimate of association | 1.04 | 0.161 | 0.130 | 0.930 |
| Simultaneously estimate mode, CV, and estimate of association | 1.04 | 0.187 | 0.152 | 0.900 |
| CV = 0.3 | ||||
| Select “guessed” lag that yields largest estimate of association | 1.29 | 0.075 | 0.074 | 0.006 |
| Select “guessed” lag that yields best model fit | 0.94 | 0.087 | 0.051 | 0.654 |
| Simultaneously estimate CV and estimate of association | 1.04 | 0.182 | 0.148 | 0.897 |
| Simultaneously estimate mode, CV, and estimate of association | 1.04 | 0.214 | 0.176 | 0.870 |
Abbreviations: CI, confidence interval; CV, coefficient of variation; SE, standard error; WLM, working level month.
In all simulations, the specified true association is β = 1. The natural log of annual exposure is distributed with μ = 0.1, σw = 1, and σB = 1. The population distribution of the latency interval, L, is lognormally distributed with a mode equal to 5.
RESULTS AND DISCUSSION
Lagging of exposure assignment is a widely used method to account for a latency period between exposure and disease (3, 4, 9). Common analytical approaches to account for the latency period are to impose a number of lag periods and to choose the one that either 1) maximizes the effect estimate or 2) has the best fit. Both of these approaches implicitly assume that the latency period is approximately the same across persons. If this is the case, we showed that the former approach can lead to bias in the effect estimate and that the latter approach, while unbiased, will result in confidence intervals that are narrower than they should be. If the equal latency period assumption is true and the lagging method is used, we showed that joint likelihood estimation of lag and effect yields unbiased estimation of the exposure effect and that confidence intervals provide reasonably accurate coverage. We then showed that, when the equal latency period assumption is not true, that is, when there is (moderate) variability in the latency periods over persons, selecting the lag that yields the best model fit produces a biased estimate of exposure effect; the degree of bias increases with the coefficient of variation of the latency periods (Table 2). Finally, because the equal latency period assumption is implausible, we proposed a likelihood approach that accommodates variable latency intervals across persons and showed that this approach performed reasonably well.
Of course, one way to avoid the problems that arise from an iterative search for the best lag assumption is to specify a single lag assumption a priori based on biologic knowledge or expert opinion. However, rarely do investigators have a strong prior basis for selecting a specific value for the latency period (or a basis for specifying the mode and variance for a population distribution of such values). If the investigator does have a strong prior basis regarding the latency period, then this could be used to inform the joint estimation of the exposure effect and latency period, suggesting extensions of the approach described in this paper to explicitly incorporate prior knowledge in a Bayesian framework. As we illustrate, in the absence of a strong prior basis, the data in hand often may be consistent with a range of latency periods, and this may impact confidence intervals for the estimate of the exposure effect.
As illustrated in our simulations, if there is substantial population variation in the latency period, then standard approaches to lagging exposure assignment may lead to biased effect estimates. We propose an estimation technique, implemented under the assumption that the distribution of latency periods, L, conforms to a lognormal distribution, denoted fL(l). If this assumption is wrong, bias may occur; however, it should be noted that, because fL(l) asymptotically approaches the density function for a step function, the proposed approach can necessarily perform at least as well as a standard exposure lagging approach. Of course, other distributions, such as exponential or gamma, could replace the lognormal in our approach. However, a lognormal distribution is a reasonable choice, as it precludes values for L at or below 0; in addition, it offers a cumulative density function that may approximate those of many other reasonable population distributions of latency periods (such as normal). Use of a lognormal model for induction and latency periods was discussed for cancer by Breslow and Day in 1987 and infectious diseases by Sartwell in 1950 on the basis of the observations that latency (incubation) time distributions in several malignant (infectious) diseases are skew with a marked tail and that the logarithms of the times are approximately normally distributed (10, 11). As illustrated by our empirical example, even in a moderately sized study, estimates of these parameters may be obtained.
Models with additional parameters to describe variation over time in the effect of exposure have been proposed (12–14). However, the simple models discussed in the current paper offer a useful connection to the standard approach of lagging exposure assignment while overcoming some important limitations.
Supplementary Material
Acknowledgments
Author affiliations: Department of Epidemiology, School of Public Health, University of North Carolina at Chapel Hill, Chapel Hill, North Carolina (David B. Richardson, Stephen R. Cole); Division of Biostatistics, School of Public Health, University of Minnesota, Minneapolis, Minnesota (Haitao Chu); and Division of Biostatistics, Department of Preventive Medicine, Keck School of Medicine, University of Southern California, Los Angeles, California (Bryan Langholz).
This project was supported by grant R01-CA117841 from the National Cancer Institute, National Institutes of Health.
Conflict of interest: none declared.
Glossary
Abbreviations
- CI
confidence interval
- ERR
excess rate ratio
References
- 1.Rothman KJ. Induction and latent periods. Am J Epidemiol. 1981;114(2):253–259. doi: 10.1093/oxfordjournals.aje.a113189. [DOI] [PubMed] [Google Scholar]
- 2.Armenian HK. Incubation periods of cancer: old and new. J Chronic Dis. 1987;40(suppl 2) doi: 10.1016/s0021-9681(87)80004-8. 9S–15S. [DOI] [PubMed] [Google Scholar]
- 3.Checkoway H, Pearce N, Hickey JL, et al. Latency analysis in occupational epidemiology. Arch Environ Health. 1990;45(2):95–100. doi: 10.1080/00039896.1990.9935932. [DOI] [PubMed] [Google Scholar]
- 4.Salvan A, Stayner L, Steenland K, et al. Selecting an exposure lag period. Epidemiology. 1995;6(4):387–390. doi: 10.1097/00001648-199507000-00010. [DOI] [PubMed] [Google Scholar]
- 5.Loomis D, Kromhout H. Exposure variability: concepts and applications in occupational epidemiology. Am J Ind Med. 2004;45(1):113–122. doi: 10.1002/ajim.10324. [DOI] [PubMed] [Google Scholar]
- 6.Hein MJ, Stayner LT, Lehman E, et al. Follow-up study of chrysotile textile workers: cohort mortality and exposure-response. Occup Environ Med. 2007;64(9):616–625. doi: 10.1136/oem.2006.031005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Langholz B, Goldstein L. Risk set sampling in epidemiologic cohort studies. Stat Sci. 1996;11(1):35–53. [Google Scholar]
- 8.Langholz B, Richardson D. Are nested case-control studies biased? Epidemiology. 2009;20(3):321–329. doi: 10.1097/EDE.0b013e31819e370b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Breslow NE, Day NE. Statistical Methods in Cancer Research. Vol II. The Design and Analysis of Cohort Studies. Lyon, France: International Agency for Research on Cancer; 1987. (IARC scientific publication no. 82) [PubMed] [Google Scholar]
- 10.Boag JW. The presentation and analysis of the results of radiotherapy. Br J Radiol. 1948;21(243):128–138. doi: 10.1259/0007-1285-21-243-128. [DOI] [PubMed] [Google Scholar]
- 11.Sartwell PE. The distribution of incubation periods of infectious disease. Am J Hyg. 1950;51(3):310–318. doi: 10.1093/oxfordjournals.aje.a119397. [DOI] [PubMed] [Google Scholar]
- 12.Langholz B, Thomas D, Xiang A, et al. Latency analysis in epidemiologic studies of occupational exposures: application to the Colorado Plateau uranium miners cohort. Am J Ind Med. 1999;35(3):246–256. doi: 10.1002/(sici)1097-0274(199903)35:3<246::aid-ajim4>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 13.Hauptmann M, Wellmann J, Lubin JH, et al. Analysis of exposure-time-response relationships using a spline weight function. Biometrics. 2000;56(4):1105–1108. doi: 10.1111/j.0006-341x.2000.01105.x. [DOI] [PubMed] [Google Scholar]
- 14.Richardson DB, Ashmore JP. Investigating time patterns of variation in radiation cancer associations. Occup Environ Med. 2005;62(8):551–558. doi: 10.1136/oem.2004.017368. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


