Abstract
Our understanding of the genetic basis of local adaptation has recently benefited from the increased power to identify functional variants associated with environmental variables at the genome scale. However, it often remains challenging to determine whether locally adaptive alleles are actively maintained at intermediate frequencies by spatially varying selection. Here, we evaluate the extent to which this particular type of balancing selection explains the retention of adaptive genetic variation in the extreme situation of perfect panmixia, using the American eel (Anguilla rostrata) as a model. We first conducted a genome scan between two samples from opposite ends of a latitudinal environmental gradient using 454 sequencing of individually tagged cDNA libraries. Candidate SNPs were then genotyped in 992 individuals from 16 sampling sites at different life stages of the same cohort (including larvae from the Sargasso Sea, glass eels, and 1-year-old individuals) as well as in glass eels of the following cohort. Evidence for spatially varying selection was found at 13 loci showing correlations between allele frequencies and environmental variables across the entire species range. Simulations under a multiple-niche Levene’s model using estimated relative fitness values among genotypes rarely predicted a stable polymorphic equilibrium at these loci. Our results suggest that some genetic-by-environment interactions detected in our study arise during the progress toward fixation of a globally advantageous allele with spatially variable effects on fitness.
VARIABLE environmental conditions across species’ ranges provide a basis for differential selection at polymorphic loci involved in local adaptation. In consequence, the level of locally adaptive genetic variation may be potentially increased, through a particular type of balancing selection whereby protected polymorphisms result from selection for different alleles in different environments. Depending on population structure, the degree of habitat choice, and the strength of selection, this process can lead to habitat specialization and eventually to ecological speciation (Maynard Smith 1966). However, when both dispersal across habitats and mating are random processes, local adaptation is impossible and polymorphism may be either lost by drift or, under special conditions, protected by selection (Yeaman and Otto 2011). This evolutionary mechanism was first investigated more than half a century ago (Levene 1953), through a local density regulation model integrating variation in fitness of genotypes across niches and differential contribution of the niches to a panmictic reproductive pool. Levene demonstrated that a sufficient condition for a locally adaptive polymorphism to be maintained by selection requires the harmonic mean fitness of the heterozygote genotype to be higher than that of each homozygote, a process called “harmonic mean overdominance”.
There is an increasing body of empirical evidence for cases of polymorphisms maintained by environmental heterogeneity (reviewed by Hedrick et al. 1976; Hedrick 1986, 2006).The most famous examples come from studies of allozyme variation (e.g., Kreitman 1983; Sezgin et al. 2004), color polymorphism (Nachman et al. 2003; Hoekstra et al. 2004), adaptation to climate (Hancock et al. 2008; Kolaczkowski et al. 2011), and soil type (Turner et al. 2010), as well as pathogen and insecticide resistance (Garrigan and Hedrick 2003; Weill et al. 2003; Pelz et al. 2005). Since habitat choice or reduced gene flow increases the opportunity for the maintenance of locally adaptive polymorphisms in subdivided populations (Felsenstein 1976), these cases are usually more fully understood under migration–selection models. However, the framework of Levene’s model remains highly relevant to the study of species for which random mating and dispersal exist over large (e.g., marine fishes and invertebrates) or local spatial scales (e.g., sympatric host races of insects).
To date, the principal limitation to evaluating the retention of locally adaptive alleles in such species has been the lack of genomic resources. Only two studies that focused on one or two selected genes have empirically tested the maintenance of polymorphism under Levene’s model, in the leafhopper (Prout and Savolainen 1996) and the acorn barnacle (Schmidt and Rand 2001). In practice, the discovery of locally adaptive polymorphisms in panmixia is not straightforward for several reasons. First, there might be a negative trade-off between the number of loci influenced by spatially varying selection and individual locus effects on fitness, such that the whole adaptive load due to selection on unlinked loci remains sustainable for the population. Second, environmental changes can shift the frequency of some protected variants out of their domain of stability, resulting in the loss of formerly stable polymorphisms. Third, recombination should rapidly erase the effects of selection on the chromosomal neighborhood of the selected sites (Charlesworth et al. 1997; Przeworski 2002). As such, partial selective sweeps provoked by the establishment of new protected variants should leave only transient genomic footprints, further reducing the chance to find them. Nevertheless, these difficulties can be partly overcome by tracking protected polymorphisms using a high-density genome-scan approach. Owing to the development of high-throughput sequencing techniques, this strategy is now achievable in most nonmodel species (Stapley et al. 2010).
The American eel Anguilla rostrata is one of the most appropriate organisms for studying the evolutionary effects of spatially varying selection within Levene’s model framework (Karlin 1977). Mating of the whole species occurs in the Sargasso Sea (Schmidt 1923), after which planktonic larvae are dispersed by the Antilles Current and the Gulf Stream to the eastern North American coast over a large continental habitat, extending from Florida to Quebec and Labrador (Tesch 2003). Studies based on neutral molecular markers have shown that in this textbook example of panmixia, random mating occurs at the species scale (Avise et al. 1986; Wirth and Bernatchez 2003). Evidence from the literature also supports that leptocephali larvae passively drift with the currents (Bonhommeau et al. 2010) and that, following metamorphosis, newly transformed unpigmented glass eels use a selective tidal stream transport mechanism to move landward (McCleave and Kleckner 1982). Thus, genotype-dependent habitat choice is unlikely to occur over a large geographical scale due to oriented horizontal swimming and newly recruited glass eels are exposed to highly unpredictable conditions with respect to environmental parameters (e.g., temperature, salinity, pathogens, and pollutants) during their early life history. Consistent with these observations, clinal variation attributed to single-generation footprints of spatially varying selection was found at three allozyme loci (Williams et al. 1973; Koehn and Williams 1978). However, allozyme studies focused on only a few metabolic genes and did not assess the retention of locally adaptive polymorphisms by spatially varying selection. Population genomics now offers powerful tools to bring empirical data to bear on this fundamental question in ecological genetics. Here, we discovered and typed annotated single-nucleotide polymorphisms (SNPs) in transcribed regions of the American eel genome to identify candidate genes potentially associated with environmental variables. An extensive spatiotemporal set of samples was then used to further test for selection at candidate loci, estimate niche-specific relative fitness among genotypes, and investigate conditions for the maintenance of polymorphism under a finite-population Levene’s model.
Materials and Methods
Preparation of cDNA libraries, contig assembly, and SNP discovery
We prepared cDNA libraries for 454 sequencing following the protocol described in Pierron et al. (2011). Two samples of 20 glass eels were collected just prior to settlement in freshwater at two river mouths located near the extreme ends of the species’ latitudinal range: the Grande Rivière Blanche in the lower St. Lawrence estuary (RB, 48°78′N, 67°70′W) and Florida (FL, 30°00′N, 81°19′W). Briefly, Poly(A) RNAs were individually extracted from entire glass eels and used as a template for cDNA amplification. Amplified cDNAs were then fragmented by sonication, and fragments from 300 to 800 bp were ligated to the standard 454 B primer and the standard 454 A primer, holding a 10-bp barcode extension at its 3′ end. Therefore, each individual could be identified by its unique barcode. For each sampling site, the 20 individually tagged libraries were pooled in equal amounts and sequenced on a half-plate of Roche GS-FLX DNA Sequencer at Genome Quebec Innovation Center (McGill University, Montreal, QC, Canada).
Base calling was performed using PyroBayes (Quinlan et al. 2008) after trimming adapters. Each read was then renamed according to its individual barcode, which was subsequently removed together with potential primers used for cDNA amplification. We performed a de novo assembly of the total sequencing data using CLC Genomic Workbench 3.7 (CLC bio), with a minimal read length fraction of 0.5 and a similarity parameter of 0.95. The consensus sequence of each de novo built contig was then used as a template for a reference assembly under the same parameters. This second round of assembly aimed at screening for additional reads that were not included into contigs during the step of de novo assembly and excluding poor-quality contigs that did not recruit any read during the reference assembly procedure.
SNP discovery was performed using the neighborhood quality standard (NQS) algorithm (Altshuler et al. 2000; Brockman et al. 2008) implemented in CLC Genomic Workbench 3.7 (CLC bio). This method takes into account the base quality values to distinguish sequencing errors from actual SNPs. We set a minimum coverage of 20× per SNP site and used either a frequency threshold of 5% or a count threshold of 5 for the rarest variant (when the coverage exceeded 100×) to avoid the detection of sequencing errors as SNPs. Only biallelic SNPs were considered.
Individual genotype inference
There is a significant risk to misscore a heterozygote genotype by repeatedly sampling the same allele when the individual coverage is <5× (see Supporting Information, Figure S1). We corrected such artifactual heterozygote deficiencies by supposing within-sample Hardy–Weinberg equilibrium (HWE) while taking into account the stochasticity induced by the binomial sampling process of homologous sequences at each locus for each individual. For each SNP having >10 individuals sequenced in each sample (i.e., total coverage ≥20×), allele counts were used to determine the observed genotype of each individual (AA, Aa, aa, or NA when no sequence data were available) to calculate the observed allelic frequencies. We then supposed within-sample HWE to estimate the number of expected individuals within each genotypic class in each sample given the number of individuals sequenced and the observed allelic frequencies in the sample. When the observed number of heterozygotes was below HWE predictions, new genotypes that were consistent with observed individual data were randomly drawn from a trinomial distribution with event probabilities (P(AA)i,j; P(Aa)i,j; P(aa)i,j) corresponding to the probabilities of each genotype, given the observed data for the jth individual in sample i. For each locus showing HW deficiency, a new array of individual genotypes was generated until HWE expectations were verified for the sample. The individual genotype probabilities used to parameterize the trinomial sampling process were obtained from the following equation giving the probabilities of real genotypes (GR) knowing the observed data (GO) at a given locus,
where Ni,j is the number of reads (i.e., individual coverage) of individual j in sample i, is its observed genotype
and pi is the frequency of the A allele in sample i. Under this procedure, the genotype of an observed heterozygote was never modified, whereas observed homozygotes could be probabilistically assigned to heterozygotes. Since the sequencing error rate was already taken into account by the SNP detection method, it was neglected at this step to simplify the approach. Methodological validation performed on simulated data sets showed that our correction efficiently restored up to 50% of the hidden heterozygotes (see Figure S1).
Outlier detection
Individual genotypes obtained after treating for the heterozygote deficiency bias were used to detect SNPs potentially affected by diversifying selection between the two samples RB and FL. The empirical distribution of pairwise FST as a function of within-samples heterozygosity was compared to a neutral distribution simulated under a symmetrical two-island model assuming near random mating (Beaumont and Nichols 1996), using ARLEQUIN ver. 3.5 (Excoffier and Lisher 2010). This approach is more conservative than drawing random samples from a single panmictic population to derive the neutral distribution. For each outlier locus (i.e., FST value located above the 99.5% quantile of the simulated distribution), the contig’s consensus sequence was blasted against the nonredundant NCBI protein database (nr), using BLASTX with an E-value threshold of 10−5 (Altschul et al. 1997).
SNP genotyping
Individual SNP assays were developed using the KBiosciences Competitive Allele-Specific PCR genotyping system (KASPar). For each candidate contig, we targeted the SNP showing the highest FST value when possible. We also developed assays for SNPs identified within contigs of allozyme coding genes showing clinal variation in Williams et al. (1973): the Sorbitol dehydrogenase gene (SDH), two Phosphoglucose isomerase isoforms (PGI-1 and PGI-2), and the Alcohol dehydrogenase gene (ADH-3). Our validation panel was finally completed with nonoutlier SNPs to 100 markers. All assays were tested with 80 individuals and only successfully genotyped SNPs were retained for subsequent genotyping.
A total of 992 individuals belonging to four distinct sample categories were genotyped (Table 1): (i) A reference sample of the 2007 cohort (before selection) consisting of 48 young leptocephali larvae collected in the Sargasso Sea soon after hatching in March and April 2007 (SAR7) during the Galathea III expedition (Munk et al. 2010); (ii) the first wave of recruiting glass eels belonging to the 2007 cohort, collected between January and July 2008 at 16 river mouths distributed from Florida to Quebec (GLASS8); (iii) 1-year-old individuals from the 2007 cohort, sampled between February and June 2009 from four localities previously sampled in 2008, ranging between South Carolina and Quebec (OYO9); and (iv) glass eels belonging to the 2008 cohort, collected in 2009 at 5 river mouths distributed from South Carolina to Quebec (GLASS9) and that were also sampled in 2008 for the 2007 cohort.
Table 1 . Sampling location and date, sample size, developmental stage, and river mouth temperature for each analyzed sample.
| Category | Sampling locality | Code | Development stage | N | Latitude | Longitude | Day | Month | Year | Temperature (°) |
|---|---|---|---|---|---|---|---|---|---|---|
| SAR7 | Sargasso Sea | SAR_7 | Leptocephali | 48 | — | — | — | — | 2007 | — |
| GLASS8 | Gaspésie, Grande Rivière Blanche | GAS_G8 | Glass eel | 40 | 48.78 | −67.70 | 14 | June | 2008 | 14.21a |
| Newfoundland, Codroy Bay | NF_G8 | Glass eel | 40 | 47.85 | −59.26 | 16 | July | 2008 | 15.86a | |
| Prince Edward Island, Rustico Bay | PEI_G8 | Glass eel | 40 | 46.43 | −63.24 | 3 | July | 2008 | 19.83a | |
| New Scotia, St. John’s River | NS_G8 | Glass eel | 40 | 45.54 | −64.70 | 25 | April | 2008 | 3.41a | |
| Maine, Boothbay Harbor | MAI_G8 | Glass eel | 40 | 43.85 | −69.65 | 1 | May | 2008 | 6.22a | |
| New Hampshire, Taylor River | NH_G8 | Glass eel | 40 | 42.91 | −70.84 | 23 | April | 2008 | 6.78a | |
| Massachusetts, Parker River | MA_G8 | Glass eel | 40 | 42.69 | −70.79 | 16 | April | 2008 | 7.04a | |
| Connecticut, Taylor River | CO_G8 | Glass eel | 40 | 41.41 | −70.55 | 5 | May | 2008 | 10.44a | |
| New Jersey, Patcong Creek | NJ_G8 | Glass eel | 40 | 41.17 | −72.23 | 4 | April | 2008 | 7.79a | |
| Pensylvania, Delaware River | PEN_G8 | Glass eel | 40 | 39.89 | −75.26 | 1 to 5 | May | 2008 | 9.50a | |
| Delaware, Millsboro Pond Spillway | DEL_G8 | Glass eel | 40 | 38.35 | −75.17 | 5 | February | 2008 | 7.00a | |
| Virginia, Wormley Creek | VIR_G8 | Glass eel | 40 | 37.21 | −76.49 | 28 | March | 2008 | 10.01a | |
| North Carolina, Black Creek | NC_G8 | Glass eel | 40 | 34.46 | −76.48 | 5 to 7 | February | 2008 | 10.44a | |
| South Carolina, Cooper River | SC_G8 | Glass eel | 40 | 32.55 | −80.00 | 13 | February | 2008 | 17.19a | |
| Georgia, Altamaha River | GEO_G8 | Glass eel | 22 | 31.18 | −81.28 | 8 to 23 | January | 2008 | 15.46a | |
| Florida, Guana River | FLO_G8 | Glass eel | 40 | 30.00 | −81.19 | 28 | January | 2008 | 18.09a | |
| OYO9 | Gaspésie, Grande Rivière Blanche | GAS_E9 | Elver 1+ | 35 | 48.79 | −67.70 | — | June | 2009 | 0.16b |
| Massachusetts, Parker River | MA_E9 | Elver 1+ | 39 | 42.69 | −70.79 | — | April | 2009 | 5.62b | |
| Pensylvania, Crum Creek | PEN_E9 | Elver 1+ | 40 | 39.86 | −75.32 | — | May | 2009 | 7.22b | |
| South Carolina, Cooper River | SC_E9 | Elver 1+ | 29 | 32.55 | −80.00 | — | February | 2009 | 14.68b | |
| GLASS9 | Gaspésie, Grande Rivière Blanche | GAS_G9 | Glass eel | 40 | 48.79 | −67.70 | — | June | 2009 | 10.57c |
| Nova Scotia, Caledonia | NS_G9 | Glass eel | 40 | 46.04 | −59.96 | — | June | 2009 | 9.55c | |
| Massachusetts, Parker River | MA_G9 | Glass eel | 39 | 42.69 | −70.79 | — | April | 2009 | 5.65c | |
| Pennsylvania, Crum Creek | PEN_G9 | Glass eel | 40 | 39.86 | −75.32 | — | May | 2009 | 13.57c | |
| South Carolina, Cooper River | SC_G9 | Glass eel | 20 | 32.55 | −80.00 | — | February | 2009 | 12.31c |
Ten-day average sea-surface temperature before sampling date (source NOAA: SST14NA).
Three winter months (December through February) average sea-surface temperature (source NOAA: SST14NA).
Sampling month average sea-surface temperature (source NOAA: SST14NA).
Statistical analyses
We tested for HWE at each diploid locus within each of the four eel sample categories, using ARLEQUIN ver. 3.5 (Excoffier and Lisher 2010). We corrected for multiple independent tests using the false discovery rate correction (α = 0.05). Multilocus global FST values among localities within sample categories were estimated and tested through 10,000 permutations. Outlier SNPs were searched on the basis of their level of genetic differentiation among localities within categories as well as between pairs of localities, using coalescent simulations under a symmetrical island model assuming near random mating.
For each locus, statistical associations between allelic frequencies and a set of four explanatory variables (sample category, latitude, longitude, and temperature) were assessed through logistic regressions using the R package glmulti (Calcagno and De Mazancourt 2010). Temperature data were obtained from a National Oceanic and Atmospheric Administration (NOAA) database containing geo-referenced sea-surface temperatures along North America’s coastlines (SST14NA), with a nominal spatial resolution of 14 km and a 48-hr update frequency. More precisely, we took the sea-surface temperature at river mouth averaged across the 10 days preceding the sampling date in each locality, which corresponded to recruitment at river mouths for the GLASS8 category. Because the exact date of arrival at river mouths was not known for the two other categories of samples, we used different temperature criteria: the three winter months (December to February) average river mouth temperature was used for the OYO9 category, and the sampling month average river mouth temperature was used for the GLASS9 category (Table 1). All possible models involving the four explanatory variables (including pairwise interactions) were fitted using samples from the three continental categories (GLASS8, OYO9, and GLASS9), and the best model was identified using a Bayesian information criterion (BIC). Because the best geographical coverage was achieved for the 2008 glass eels, the same approach was also performed using samples from the GLASS8 category only. For each SNP found in association with explanatory variables, individual haplotype information was retrieved from 454 sequencing data and used to evaluate between-sites linkage disequilibrium (LD), using the method for partially phased haplotypes in Haploview v4.2 (Barrett et al. 2005).
The multilocus spatial component of genetic variability at loci inferred to be influenced by spatially varying selection was determined using the spatial principal component analysis method (sPCA) (Jombart et al. 2008) implemented in the R package adegenet_1.2-2 (Jombart 2008). The sPCA includes spatial information in the analysis of genetic data, which helps to reveal subtle global spatial structures such as geographic clines. The spatial proximity network among localities was built using the neighborhood-by-distance method. An abrupt decrease of the eigenvalues obtained by decomposing the genetic diversity from the spatial autocorrelation was used as a criterion to choose the principal component to interpret.
Evolution under Levene’s model
The classical one-locus–two-allele model of Levene (1953) was extended by the addition of a genetic drift component. At each generation, mating occurs in panmixia, followed by random dispersal of genotypes across niches. Selection is a niche-specific process in which the frequency of allele A before selection in the ith niche, noted qi, passes to after selection following the equation
where Wi and Vi, respectively, denote the fitness of the homozygote genotypes AA and aa relative to that of the heterozygote genotype in the ith niche. Genetic drift is then modeled by randomly drawing Ne × Ci genotypes in each niche from a trinomial distribution with event probabilities
The new frequency of allele A after selection and drift in the ith niche is noted , and since Ci corresponds to the relative contribution of niche i to the global reproductive pool of effective size Ne, the frequency of allele A equals in the next mating pool.
To test for equilibrium under Levene’s model, empirical values of Wi and Vi were estimated from the observed genotypic frequencies in the SAR7 larval pool (fAA; fAa; faa) and the modeled niche-specific genotypic frequencies after selection following
where the ratios and were predicted by the regression models of and , using the observed genotypic frequencies in the GLASS8 samples and the explanatory variables previously selected for this category (see Results). For each locus inferred to be influenced by spatially varying selection, the 16 estimated pairs of (Wi; Vi) were used to parameterize a 16-niche Levene’s model in which the allelic frequencies observed in SAR7 were used as starting values. Different distributions of the Ci were explored, from uniform to normally distributed outputs among niches, and the population effective size parameter was set between 104 and 106 to assess genetic drift effects.
Results
Sequence assembly and SNP discovery
A total of 292.6 Mb of sequences were obtained from the two half-runs of 454 GS-FLX pyrosequencing, among which were 482,322 reads from the St. Lawrence estuary sample (RB, mean read length of 296 bp) and 495,482 reads from the Florida sample (FL, mean read length of 303 bp). These sequences were deposited in the NCBI sequence read archive SRA045712. Trimming adapters and individual barcodes and then filtering for sequence quality removed 5.3% of the reads from the RB data set and 5.1% from the FL data set. Processed reads were assembled into 22,093 contigs with an average length of 464 bp. In silico SNP detection allowed identifying 70,912 putative SNPs, 13,293 of which were retained after filtering for a minimal coverage of 10 reads from at least 10 different individuals in both samples RB and FL (i.e., total coverage ≥20×). This filtering step allowed inferring 78.1% of the 265,860 genotypes (20 individuals × 13,293 SNPs) in sample RB and 79.7% in sample FL.
Candidate SNP detection and genotyping
A total of 163 outlier SNPs with estimated FST values ranging from 0.167 to 0.637 were detected (see Figure S2). However, for most candidate SNPs exhibiting the highest FST values, BLAST searches revealed the presence of reads matching alternative copies of duplicated genes within contigs. These false SNPs, which probably reflected differential expression patterns of paralogous genes (principally myosin isoforms) between samples RB and FL, were removed from subsequent analyses.
After these filtering procedures, our validation panel included 57 outlier SNPs and was completed to 100 markers with nonoutlier SNPs selected across the full range of heterozygosity. Successful genotyping was obtained for 73 of these 100 SNPs (see File S1), 70 of which were functionally annotated using BLASTX (see Table S1), and 44 were outliers from the initial screen. The genotyping success rate across all samples and loci was >98%; KASPar primers used for genotyping are provided Table S2.
SNP variation patterns
Only one locus departed significantly from HWE expectations within the SAR7 category, whereas 15, 7, and 10 loci showed significant HW disequilibrium within the continental categories GLASS8, OYO9, and GLASS9, respectively (see Table S3). Overall multilocus FST values calculated among locality samples were not significantly different from zero within each of the three continental categories (GLASS8, FST = 0.0003, P = 0.318; OYO9, FST = 0.0015, P = 0.171; GLASS9, FST = 0.0022, P = 0.060).
Logistic regressions between allelic frequencies and explanatory variables based on the data set containing the three continental categories (GLASS8, OYO9, and GLASS9) revealed contrasting patterns across loci. For 61 of 73 SNPs, all possible models involving the explanatory variables and their pairwise interactions were rejected. However, significant associations were detected for 10 loci and marginally significant associations for 2 loci (Table 2). The same approach performed within the GLASS8 category alone revealed statistical associations for 8 loci, all but MDH being already detected in the analysis including all continental samples (Table 2). Among the 13 loci for which significant associations were found, 11 were also detected in outlier tests due to atypically high FST values ranging from 0.052 to 0.175 between some pairs of localities. After removing these 13 loci from the data set, the global multilocus FST values calculated among locality samples became null in two continental categories (GLASS8, FST = −0.0015, P = 0.981; OYO9, FST = −0.0007, P = 0.647) and was reduced in the third one (GLASS9, FST = 0.0016, P = 0.151).
Table 2 . Models selected for 13 loci associated with explanatory variables, for both the GLASS8 data set and the three continental categories.
| Locus | Gene | GLASS8 | Slope P-value | Three continental categories | Slope P-value |
|---|---|---|---|---|---|
| ACP_13914 | Acyl carrier protein | TEMP | 0.0013 | TEMP | 0.0001 |
| ANX_2_249 | Annexin A2-A | TEMP:LAT | 0.0385 | TEMP+TEMP:LONG+TEMP:LAT+LAT:LONG | 0.0017a |
| CST_21113 | Cystatin | Null | — | TEMP:COHORT | 0.0021 |
| EIF_3F_341 | Translation initiation factor 3 subunit F | Null | — | TEMP | 0.0075 |
| GPX_4_19607 | Glutathione peroxidase 4 | TEMP:LONG | 0.0013 | TEMP:LONG | 0.0008 |
| HSP_90A_15666 | Heat-shock protein 90 alpha | TEMP:LONG | 0.0355 | TEMP+LONG+TEMP:LONG | 0.0058a |
| MDH_1393 | Malate dehydrogenase | TEMP:LONG | 0.0403 | Null | 0.0874b |
| NCP_2_15547 | Nucleolar complex protein 2 | Null | — | TEMP | 0.0539 |
| NRAP_1541 | Nebulin-related anchoring protein | TEMP:LAT | 0.0425 | TEMP:LONG | 0.0590 |
| PRP_40_16504 | Pre-mRNA–processing factor 40 homolog A | LAT | 0.0044 | TEMP+LAT+TEMP:LAT | 0.0202a |
| SN4_TDR_374 | Staphylococcal muclease domain-containing protein 1 | Null | — | TEMP:LONG | 0.0281 |
| TENT_02_11046 | — | Null | — | TEMP | 0.0082 |
| UGP_2_2128 | UDP-glucose pyrophosphorylase 2 | TEMP:LONG | 0.0177 | TEMP+COHORT+TEMP:COHORT | 0.0318c |
LAT, latitude; LONG, longitude; TEMP, temperature.
P-value associated to the term identified as best model for GLASS8, in the best model for all three continental categories.
P-value associated to the best model for GLASS8.
P-value associated to the term TEMP, in the best model for all 3 continental categories.
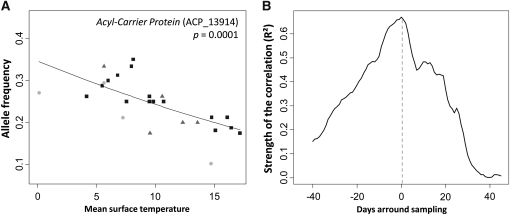
Most selected regression models revealed significant interactions between spatial variables and river mouth temperature (Table 2), which was measured at different timescales for the three continental categories (Table 1). River mouth temperature at recruitment was selected as the best model for one locus in the GLASS8 category (Acyl-Carrier Protein, ACP; Figure 1A). To identify more precisely when this parameter was the most biologically relevant for this locus, we used a sliding-window analysis to test whether the correlation could be improved, using temperature data from different time periods surrounding glass eels recruitment. We found that the correlation was rapidly lost when the 10-days window used to calculate the average sea-surface temperature was shifted around the period corresponding to recruitment (Figure 1B). Furthermore, because the timing of glass eels’ recruitment varied considerably across sampling locations, river mouth temperatures were neither correlated with latitude (R2 = 0.113, P = 0.203) nor correlated with longitude (R2 = 0.036, P = 0.484) during the recruitment period (Table 1).
Figure 1 .
Correlation between river mouth temperature and allele frequencies at locus ACP. Logistic regression is based on all three continental categories. (A) Allele frequencies in the GLASS8 category are represented by solid squares, OYO9 by circles with light shading, and GLASS9 by triangles with dark shading. (B) Sliding-window analysis of the coefficient of determination (R2) between allele frequencies at locus ACP in the GLASS8 category and the values predicted using river mouth temperature data. For each day within a 3-month period centered on the sampling date (which also corresponded approximately to the date of arrival at river mouths), surface temperature was taken as the mean value across the 10 previous days.
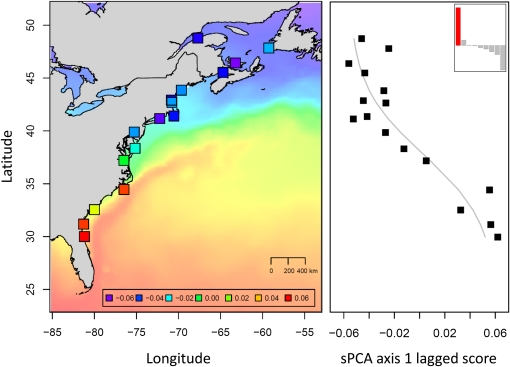
Multivariate analysis of the eight loci significantly associated with explanatory variables in the GLASS8 category showed that most of the variability was explained by the first principal component, since the first eigenvalue of the sPCA was highly positive (Figure 2, top right). The global structure illustrated by individual lagged scores on the first principal component showed a synthetic latitudinal cline (Figure 2), corresponding to the multilocus spatial component of genetic variation at the eight loci inferred to be under spatially varying selection. A logistic regression of locality scores against latitude (R2 = 0.76, P < 0.0001) showed that the center of the cline coincides with the coastal zone where the latitudinal gradient of nearshore sea-surface temperature is the strongest over the sampling period. Moreover, river mouth temperature averaged across the whole sampling period was a better predictor of locality scores than latitude (R2 = 0.88, P < 0.0001). A highly similar synthetic latitudinal cline was obtained when analyzing the three continental categories together (see Figure S3), supporting the temporal stability of the observed pattern.
Figure 2 .
Synthetic multilocus spatial variation component in the 2008 glass eels. The spatial component analysis was based on genetic variation at the eight loci significantly associated with explanatory variables in the GLASS8 category. The 16 sampling sites are represented on the map by squares colored according to each locality’s lagged score on the first principal component, as indicated in the inset. Sea-surface temperatures averaged across the whole sampling period (from January 8 to July 16, 2008) are represented on the same color scale for indication (purple, 0.2°; red, 27.3°). The plot on the right shows the shape of the synthetic multilocus cline, as well as the decomposition of the product of the variance and the spatial autocorrelation into positive, null, and negative components (top right corner). The clinal structure corresponds to the highly positive eigenvalue in red.
One of the 13 SNPs associated with explanatory variables was a nonsynonymous polymorphism (Nucleolar Complex Protein 2, NCP-2), whereas for 9 of the 12 other SNPs, at least one nonsynonymous segregating site was identified within a 1-kb region (see Figure S4). Heterozygosity within the 13 contigs usually followed a monotonical trend and substantial levels of linkage disequilibrium (r2 > 0.5) were sometimes found between remote SNPs. These results suggest that the indirect influence of selection through linkage with a nearby functional mutation was more likely than direct selection at the focal SNPs. However, this should not introduce any bias in the estimation of the relative fitness values used in the following simulations.
Assessment of polymorphism stability under Levene’s model
Simulating the evolution of allelic frequencies for the eight loci significantly associated with explanatory variables in the GLASS8 category led to two different predictions. Under the hypothesis of uniform contribution among niches and an effective population size (Ne) of 105, simulations predicted polymorphism stability for two SNPs and allele fixation for the remaining six loci (Table 3). The simulated evolution of allelic frequencies over generations as generated by the model can be found for each of the eight loci in Figure S5. Frequency at equilibrium was fairly close to that measured in the SAR7 sample for the two SNPs predicted to be protected by spatially varying selection. Concerning the six transient polymorphisms, allele fixation was generally reached within <80 generations. Most importantly, the invading allele was always the derived state after identifying the ancestral allele through BLASTN search.
Table 3 . Simulated evolution of allelic frequencies under Levene’s model for the eight loci statistically associated with explanatory variables in the GLASS8 category.
| Locus | Predictive model | 1/∑(Ci/Wi) | 1/∑(Ci/Vi) | Frequency Sargasso | Levene’s model prediction |
|---|---|---|---|---|---|
| ACP_13914 | TEMP | 0.7661 | 1.3688 | 0.6875 | Fixation of the derived allele |
| ANX_2_249 | TEMP:LAT | 0.8325 | 0.2900 | 0.9565 | Equilibrium at 0.81 |
| GPX_4_19607 | TEMP:LONG | 1.0031 | 1.8379 | 0.9149 | Fixation of the derived allele |
| HSP_90A_15666 | TEMP:LONG | 2.1926 | 1.0759 | 0.1170 | Fixation of the derived allele |
| MDH_1393 | TEMP:LONG | 0.6397 | 0.7761 | 0.3913 | Equilibrium at 0.375 |
| NRAP_1541 | TEMP:LAT | 0.6065 | 1.1201 | 0.5426 | Fixation of the derived allele |
| PRP_40_16504 | LAT | 4.7856 | 1.1736 | 0.1277 | Fixation of the derived allele |
| UGP_2_2128 | TEMP:LONG | 0.7568 | 1.4254 | 0.4468 | Fixation of the derived allele |
Uniform contribution among niches and a population effective size of 105 were assumed in these simulations.
The results obtained under different assumptions on the relative contributions among niches and population size did not radically change these predictions, as the same two protected polymorphisms were repeatedly inferred across scenarios. However, estimating the niche-specific relative fitness directly from the observed genotype frequencies in the GLASS8 category (i.e., instead of using values predicted by the regression models) increased the number of protected polymorphisms from two to four (see Table S4).
Discussion
Evidence for single-generation footprints of spatially varying selection
Our results provide strong indications that young glass eels colonizing different areas of the species range are exposed to differential patterns of selection, resulting in significant shifts in allele frequencies within a short timescale. The alternative hypothesis of a subtle neutral population genetic structure in the American eel was not supported by previous works, since panmixia has never been rejected using neutral markers (Avise et al. 1986; Wirth and Bernatchez 2003). This conclusion was reiterated here on the basis of 60 neutral SNPs genotyped over 944 individuals distributed across three temporal categories. Moreover, a neutral pattern imposed by spatially restricted gene flow is not consistent with the finding that 85% (11 of 13) of the loci associated with explanatory variables were also detected as outliers and that different regression models were selected across these markers. Consequently, our data do not support the existence of a spatial population structure due to deviation from panmixia in A. rostrata. Alternatively, passive genotype-specific habitat choice could possibly occur if the genes associated with environmental variables underlie differences in leptocephalus stage duration. However, the observation that early-metamorphosing leptocephali preferentially recruit to the center of the continental distribution range, whereas late-metamorphosing larvae mostly settle in northern and southern locations (Wang and Tzeng 1998), is inconsistent with the clinal multilocus spatial component detected at these loci (Figure 2). Moreover, half of these loci displayed HWE deviations in continental samples but not in the larval sample, which is incompatible with the habitat choice hypothesis. Therefore, we propose that spatially varying selection is the most parsimonious mechanism underlying the observed patterns of genetic variation.
The diversity of the statistical models retained to explain genetic variation across selected loci may suggest the implication of different locus-specific selective factors. Without detailed information on such agents, the choice of spatial and temperature variables as proxies for the ecological conditions experienced by eels was justified, as, for instance, several environmental factors covary with latitude along the North Atlantic coasts (Schmidt et al. 2008). Covariation between latitude and spatially varying selective factors has previously been used to illustrate multilocus spatial patterns attributed to selection in heterogeneous environments in Drosophila melanogaster (Sezgin et al. 2004). Here, we additionally used this synthetic multilocus signal to demonstrate the overall temporal stability of the observed patterns (Figure S3). Owing to the apparent panmixia and the lack of evidence for large-scale habitat choice in the American eel, any genetic pattern left by spatially varying selection at a given generation will be inevitably erased at the next generation. Consequently, the temporal stability of the observed genetic patterns between the GLASS8 and GLASS9 categories probably reflects the repeated action of similar natural selection pressures in the two consecutive year cohorts covered by this study. Given that only a very small proportion (<0.5%) of the larvae survive until glass eels reach the coasts and that the glass eel survival rate is ∼10% (Bonhommeau et al. 2009), our sampling scheme was designed with the intent to detect changes in allelic frequencies occurring during the early stages of eels’ life cycle. Moreover, we sampled the first wave of early-recruiting glass eels before potential settlement cues may affect up-estuary migration depending on individual condition and temperature (Sullivan et al. 2009).
Although we used variables that mirror continental factors better than open-ocean factors, a decoupling between river mouth and nearshore continental shelf sea-surface temperatures in the northeastern part of the species range [localities Gaspésie (GAS), Newfoundland (NF), and Prince Edward Island (PEI); Table 1] may have influenced our results. Glass eels recruiting in this region during early summer first face cold water temperatures while crossing the continental shelf before entering warmer estuary waters influenced by river outflows. Differential mortality during the cross-shelf transport may thus have resulted in the selection of explanatory models involving interactions between river mouth temperature and latitude or longitude, as observed for six loci of eight in the GLASS8 category. This interpretation is further supported by the close correspondence between the multilocus spatial variation component and the nearshore averaged sea-surface temperature pattern in Figure 2. Although it suggests that the continental shelf sea-surface temperature should have been used in the regression analyses, this variable remains too difficult to measure without knowing the trajectories of eels during the cross-shelf transport. Using hydrodynamic models for backtracking larval transport may thus help in selecting additional meaningful variables in future studies. Admittedly, as in any other study of this type, our approach cannot fully capture the signal of all spatially varying selection pressures and probably underestimates the number of genes under spatially varying selection. On the other hand, the strong association found at locus Acyl-Carrier Protein (ACP) between allele frequencies and temperature at recruitment (Figure 1) shows that the river mouth temperature is a relevant variable. Indeed, settlement in estuaries is a critical period during which glass eels do not feed (Sullivan et al. 2009) and probably live on their fatty acids reserves.
Three of the five allozymes previously studied (Williams et al. 1973) were included in our analysis to assess whether clines observed at the protein level could be detected at the DNA level. The SNP developed for the Malate dehydrogenase gene (MDH), which was also detected as an outlier in our initial 454 transcriptome scan, was the only one to show a significant association with environmental variables. In the allozyme study, however, genetic heterogeneity at this locus was observed only among samples of adults and not at the glass eel stage. The lack of a significant pattern for the SNPs developed at the ADH and PGI loci may be due to problems of paralogy or to a lack of LD with the SNPs under selection.
The fate of selected polymorphisms under Levene’s model
Covariation between environmental variables such as temperature and the direction and strength of selection has been suspected for a long time to actively maintain polymorphisms in heterogeneous environments (reviewed by Hedrick et al. 1976; Karlin 1977). Depending on the overall sum of local selective effects, spatially varying selection can, however, lead to two different outcomes: (i) balanced selection for different alleles in different environments can maintain polymorphism over generations, while (ii) globally unbalanced local effects of directional selection may lead to allelic fixation (Levene 1953). Recent theoretical developments have shown that substantial multilocus polymorphism can be maintained under Levene’s model, in particular when locally advantageous alleles are partially dominant (Bürger 2010). This includes cases of local dominance that specifically arise with enzymes when fitness is a concave function of the activity level while the heterozygote’s enzymatic activity is intermediate to that of both homozygotes (Gillespie and Langley 1974).
Here, equilibrium was tested through simulations under Levene’s model to account for combinations of parameters leading to nontrivial evolution of allelic frequencies within a finite population. This approach is relevant since the underlying conditions of Levene’s model perfectly fit the American eel’s life cycle, which is characterized by random mating and dispersal, and a local density regulation (Vollestad and Jonsson 1988). While exploring a realistic range of parameter values, stable equilibrium was predicted at only two of eight tested loci. This relatively low proportion may be partly explained by uncertainties due to the methodological approach. For instance, a lack of precision in the estimation of fitness parameters, but also in the relative outputs among niches, will obviously affect the realism of the simulations (Schmidt and Rand 2001). Here, we considered only 16 river mouths among a much greater number of existing rivers harboring the American eel along the North Atlantic coast. Yet, those sampling locations are distributed evenly across the species range and are therefore representative of the variation in selection direction and intensity potentially encountered by glass eels. By considering only the earliest life stages, we may fail to catch differential selection acting later in the life cycle. While we cannot rule it out, the fact that most of the mortality occurs before entering freshwater (Bonhommeau et al. 2009) reduces this possibility. Moreover, it is likely that selection acting at later life stages plays on different sets of genes. Finally, successive waves of recruiting glass eels can face different conditions depending on their date of arrival (i.e., sea-surface temperature), and interannual global variations in the selective parameters may also exist. The natural settings are thus likely more complex than considered in the model. However, it has been shown that temporal variation in selection is less efficient in maintaining locally adaptive polymorphisms compared to spatially varying selection (Ewing 1979).
Thus, it appears plausible that the low proportion of protected polymorphisms truly reflects the relatively restrictive conditions required for equilibrium (Levene 1953). When a locally advantageous allele appears by mutation and successfully escapes random loss when rare, it has more chance to invade the panmictic gene pool and to become fixed than to stabilize at an intermediate, stable frequency. Because the transitory phase to fixation will often last for a few hundred generations, loci that are undergoing incomplete selective sweeps may not be easily discovered unless they are frequent enough to be detected with a genome-scan approach. In populations with a large census size, however, new adaptive mutations can frequently occur (Karasov et al. 2010) and may result in selective sweeps if the overall effects of spatially varying selection are unbalanced. The finding that, in our simulations, the invading allele was always a derived state for each of the six predicted unstable SNPs supports the hypothesis of linkage with such a globally advantageous mutation that has not already reached fixation.
Incomplete sweeps are expected to leave a specific pattern in the haplotype structure. Because the derived allele increasing in frequency has an atypically long-range LD compared to neutral ancestral variants segregating at the same frequency (Sabeti 2006; Voight et al. 2006), the measure of LD can be used to detect ongoing directional selection. Here, sequence information retrieved from 454 sequencing data was insufficient to perform such tests on the basis of the haplotype structure, which require phased haplotypes extending outside the selected gene. However, measuring LD on the basis of available information within contigs revealed the existence of substantial linkage (r2 > 0.5) between some sites that are likely separated by a few kilobases if the presence of introns is taken into account.
Implications for adaptation and conservation of American eel
The Gene Ontology (GO) molecular functions of the genes inferred to be involved in G × E interactions mostly encompassed major metabolic functions, among which are lipid metabolism (ANX-2, inhibition of phospholipase A2; ACP, acyl carrier activity; GPX-4, phospholipid–hydroperoxide glutathione peroxidase activity), saccharide metabolism (MDH, malate dehydrogenase activity; UGP-2, UDP-glucose pyrophosphorylase activity), and protein biosynthesis (EIF-3F, translation initiation factor; PRP-40, pre-mRNA–processing activity). The best predictive models of all these genes included temperature, a factor known to have a strong influence on the level of metabolism in the American eel (Walsh et al. 1983). Moreover, the center of the synthetic multilocus latitudinal cline coincided with the region where the warm waters of the Gulf Stream drift away from the coasts. Although the American eel occupies a wide latitudinal range, its thermal preferendum is rather elevated for the temperate zone, since glass eels have a highly reduced swimming ability below 7° (Wuenschel and Able 2008), elvers optimally grow at 28° (Tzeng et al. 1998), and yellow eels stop feeding and become metabolically depressed below 10° (Walsh et al. 1983). Therefore, selective effects are logically expected at the relatively low temperatures locally encountered between metamorphosis and recruitment to estuaries, although phenotypic plasticity may also account for the wide range of temperature tolerance in A. rostrata (Daverat et al. 2006).
Two genes involved in defense response were also detected (CST, cysteine endopeptidase inhibitor activity; SN4-TDR, nuclease activity). Since selective factors related to pathogen exposure do not always correlate with temperature or geographic coordinates, other genes whose variation patterns could not be explained with our set of explanatory variables may also play a role in resistance to pathogens in A. rostrata. For instance, the innate immune response gene TRIM-35 showed strong departure from HWE in glass eels and atypically high levels of genetic differentiation between some localities (FST values up to 0.174). In parallel, simulating the evolution of allelic frequencies at this locus on the basis of observed genotype frequencies predicted a stable equilibrium (results not shown). This observation warrants further investigation, especially since the TRIM-35 gene cluster, which is located in a region of significantly elevated nucleotide diversity in the threespine stickleback (Gasterosteus aculeatus), is also a candidate target of balancing selection in this species (Hohenlohe et al. 2010).
In conclusion, we have screened >13,000 SNPs in transcribed regions of the American eel genome and identified several genes undergoing spatially varying selection associated with the highly heterogeneous habitat used by this species. Due to our methodological approach, however, the number of genes involved in G × E interactions has likely been underestimated, and the causative agents of selection remain partially unknown. Nevertheless, the higher proportion of transient vs. stable polymorphisms suggests that locally adaptive polymorphisms are not easily maintained by spatially varying selection when local adaptation is impossible. Under such conditions, theory predicts that phenotypic plasticity, by broadening the environmental tolerance of individual genotypes, provides a more functionally adaptive response to spatial environmental variation (Sultan and Spencer 2002). Indeed, the costs induced by selection on locally adaptive traits are particularly severe in the case of random mating and in the absence of habitat choice (Lenormand 2002). For eels, as for other highly fecund marine species facing huge mortality rates during larval stages, phenotypic plasticity may represent the main mechanism for coping with habitat heterogeneity (Edeline 2007), and our results suggest that differential expression of paralogous genes may be involved in this regulation. Nevertheless, the finding of locally selected mutations spreading to fixation in A. rostrata suggests that this high census size species may be regularly subject to new locally adaptive mutations. How the recent population decline of Atlantic eels (Wirth and Bernatchez 2003) affects their adaptability to changing environments is still poorly understood and will be a matter of further investigations.
Acknowledgments
We are grateful to M. Castonguay for his help in sample collection and to R. MacGregor as well as anonymous referees and Associate Editor D. Begun for their constructive and helpful comments on the manuscript. We also thank F. Pierron, M. H. Perreault, V. Bourret, and G. Côté for their precious help with molecular experiments and C. Sauvage for help with bioinformatic analyses. L.B. and P.A.G. designed research; P.A.G. performed research; C.C., M.M.H., E.N., and P.A.G. contributed reagents, materials, and analysis tools; P.A.G. analyzed data; and P.A.G., E.N., M.M.H. and L.B. wrote the paper. This research was funded by a grant from Natural Science and Engineering Research Council of Canada (Discovery grant program) as well as by a Canadian Research Chair in Genomics and Conservation of Aquatic Resources (to L.B.). P.A.G. was supported by a Government of Canada Postdoctoral Fellowship. M.M.H. was supported by the Danish Council for Independent Research|Natural Sciences (grant 09-072120).
Footnotes
Communicating editor: D. Begun
Literature Cited
- Altschul S. F., Madden T. L., Schaffer A. A., Zhang J., Zhang Z., et al. , 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altshuler D., Pollara V., Cowles C., Van Etten W., Baldwin J., et al. , 2000. An SNP map of the human genome generated by reduced representation shotgun sequencing. Nature 407: 513–516 [DOI] [PubMed] [Google Scholar]
- Avise J. C., Helfmant G. S., Saunders N. C., Hales L. S., 1986. Mitochondrial DNA differentiation in North Atlantic eels: population genetic consequences of an unusual life history pattern. Proc. Natl. Acad. Sci. USA 83: 4350–4354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett J. C., Fry B., Maller J., Daly M. J., 2005. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21: 263–265 [DOI] [PubMed] [Google Scholar]
- Beaumont M. A., Nichols R. A., 1996. Evaluating loci for use in the genetic analysis of population structure. Proc. R. Soc. Lond. B Biol. Sci. 263: 1619–1626 [Google Scholar]
- Bonhommeau S., Le Pape O., Gascuel D., Blanke B., Tréguier A. M., et al. , 2009. Estimates of the mortality and the duration of the trans-Atlantic migration of European eel Anguilla anguilla leptocephali using a particle tracking model. J. Fish Biol. 74: 1891–1914 [DOI] [PubMed] [Google Scholar]
- Bonhommeau S., Castonguay M., Rivot E., Sabatié R., Le Pape O., 2010. The duration of migration of Atlantic Anguilla larvae. Fish Fish. 11: 289–306 [Google Scholar]
- Brockman W., Alvarez P., Young S., Garber M., Giannoukos G., et al. , 2008. Quality scores and SNP detection in sequencing-by-synthesis systems. Genome Res. 18: 763–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bürger R., 2010. Evolution and polymorphism in the multilocus Levene model with no or weak epistasis. Theor. Popul. Biol. 78: 123–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calcagno V., de Mazancourt C., 2010. glmulti: an R package for easy automated model selection with (generalized) linear models. J. Stat. Softw. 34: i12 [Google Scholar]
- Charlesworth B., Nordborg M., Charlesworth D., 1997. The effects of local selection, balanced polymorphism and background selection on equilibrium patterns of genetic diversity in subdivided populations. Genet. Res. 70: 155–174 [DOI] [PubMed] [Google Scholar]
- Daverat F., Limburg K. E., Thibault I., Shiao J. C., Dodson J. J., et al. , 2006. Phenotypic plasticity of habitat use by three temperate eel species Anguilla anguilla, A. japonica and A. rostrata. Mar. Ecol. Prog. Ser. 308: 231–241 [Google Scholar]
- Edeline E., 2007. Adaptive phenotypic plasticity of eel diadromy. Mar. Ecol. Prog. Ser. 341: 229–232 [Google Scholar]
- Ewing E. P., 1979. Genetic variation in heterogeneous environment VII. Temporal and spatial heterogeneity in infinite populations. Am. Nat. 114: 197–212 [Google Scholar]
- Excoffier L., Lisher H. E. L., 2010. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Res. 10: 564–567 [DOI] [PubMed] [Google Scholar]
- Felsenstein, J., 1976 The theoretical population genetics of variable selection and migration. Annu. Rev. Genet. 10: 253–280. [DOI] [PubMed]
- Garrigan D., Hedrick P. W., 2003. Perspective: detecting adaptive molecular evolution, lessons from the MHC. Evolution 57: 1707–1722 [DOI] [PubMed] [Google Scholar]
- Gillespie J. H., Langley C. H., 1974. A general model to account for enzyme variation in natural populations. Genetics 76: 837–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock A. M., Witonsky D. B., Gordon A. S., Eshel G., Pritchard J. K., et al. , 2008. Adaptations to climate in candidate genes for common metabolic disorders. PLoS Genet. 4: e32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedrick P. W., 1986. Genetic polymorphism in heterogeneous environments: a decade later. Annu. Rev. Ecol. Syst. 17: 535–566 [Google Scholar]
- Hedrick P. W., 2006. Genetic polymorphism in heterogeneous environments: the age of genomics. Annu. Rev. Ecol. Evol. Syst. 37: 67–93 [Google Scholar]
- Hedrick P. W., Ginevan M. E., Ewing E. P., 1976. Genetic polymorphism in heterogeneous environments. Annu. Rev. Ecol. Syst. 7: 1–32 [Google Scholar]
- Hoekstra H. E., Drumm K. E., Nachman M. W., 2004. Ecological genetics of adaptive color polymorphism in pocket mice: geographic variation in selected and neutral genes. Evolution 58: 1329–1341 [DOI] [PubMed] [Google Scholar]
- Hohenlohe P. A., Bassham S., Etter P. D., Stiffler N., Johnson E. A., et al. , 2010. Population genomics of parallel adaptation in threespine stickleback using sequenced RAD tags. PLoS Genet. 6: e1000862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jombart T., 2008. adegenet: an R package for the multivariate analysis of genetic markers. Bioinformatics 24: 1403–1405 [DOI] [PubMed] [Google Scholar]
- Jombart T., Devillard S., Dufour A. B., Pontier D., 2008. Revealing cryptic spatial patterns in genetic variability by a new multivariate method. Heredity 101: 92–103 [DOI] [PubMed] [Google Scholar]
- Karasov T., Messer S. W., Petrov D. A., 2010. Evidence that adaptation in Drosophila is not limited by mutation at single sites. PLoS Genet. 6: e1000924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlin S., 1977. Gene frequency patterns in the Levene subdivided population model. Theor. Popul. Biol. 11: 356–385 [DOI] [PubMed] [Google Scholar]
- Koehn R. K., Williams G. C., 1978. Genetic differentiation without isolation in the American eel, Anguilla rostrata. II. Temporal stability of geographic patterns. Evolution 32: 624–637 [DOI] [PubMed] [Google Scholar]
- Kolaczkowski B., Kern A. D., Holloway A. K., Begun D. J., 2011. Genomic differentiation between temperate and tropical Australian populations of Drosophila melanogaster. Genetics 187: 245–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreitman M., 1983. Nucleotide polymorphism at the alcohol dehydrogenase locus of Drosophila melanogaster. Nature 304: 411–417 [DOI] [PubMed] [Google Scholar]
- Lenormand T., 2002. Gene flow and the limits to natural selection. Trends Ecol. Evol. 17: 183–189 [Google Scholar]
- Levene H., 1953. Genetic equilibrium when more than one ecological niche is available. Am. Nat. 87: 331–333 [Google Scholar]
- Maynard Smith J., 1966. Sympatric speciation. Am. Nat. 100: 637–650 [Google Scholar]
- McCleave J. D., Kleckner R. C., 1982. Selective tidal stream transport in the estuarine migration of glass eels of the American eel (Anguilla rostrata). J. Conseil 40: 262–271 [Google Scholar]
- Munk P., Hansen M. M., Maes G. E., Nielsen T. G., Castonguay M., et al. , 2010. Oceanic fronts in the Sargasso Sea control the early life and drift of Atlantic eels. Proc. Biol. Sci. 277: 3593–3599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachman M. W., Hoekstra H. E., D’Agostino S. L., 2003. The genetic basis of adaptive melanism in pocket mice. Proc. Natl. Acad. Sci. USA 100: 5268–5273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelz H. J., Rost S., Hünerberg M., Fregin C., Heiberg A. C., et al. , 2005. The genetic basis of resistance to anticoagulants in rodents. Genetics 170: 1839–1847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierron F., Normandeau E., Defo M., Campbell P., Bernatchez L., et al. , 2011. Effects of chronic metal exposure on wild fish populations revealed by high-throughput cDNA sequencing. Ecotoxicology 20: 1388–1399 [DOI] [PubMed] [Google Scholar]
- Prout T., Savolainen O., 1996. Genotype-by-environment interaction is not sufficient to maintain variation: levene and the leafhopper. Am. Nat. 148: 930–936 [Google Scholar]
- Przeworski M., 2002. The signature of positive selection at randomly chosen loci. Genetics 160: 1179–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan A. R., Stewart D. A., Strömberg M. P., Marth G. T., 2008. Pyrobayes: an improved base caller for SNP discovery in pyrosequences. Nat. Methods 5: 179–181 [DOI] [PubMed] [Google Scholar]
- Sabeti P. C., 2006. Positive natural selection in the human lineage. Science 312: 1614–1620 [DOI] [PubMed] [Google Scholar]
- Schmidt J., 1923. The breeding places of the eel. Philos. Trans. R. Soc. B Biol. Sci. 211: 179–208 [Google Scholar]
- Schmidt P. S., Rand D. M., 2001. Adaptive maintenance of genetic polymorphism in an intertidal barnacle: habitat-and-life-stage specific survivorship of MPI genotypes. Evolution 55: 1336–1344 [DOI] [PubMed] [Google Scholar]
- Schmidt P. S., Serrao E. A., Pearson G. A., Riginos C., Rawson P. D., et al. , 2008. Ecological genetics in the North Atlantic: environmental gradients and adaptation at specific loci. Ecology 89: S91–S107 [DOI] [PubMed] [Google Scholar]
- Sezgin E., Duvernell D. D., Matzkin L. M., Duan Y., Zhu C.-T., et al. , 2004. Single-locus latitudinal clines and their relationship to temperate adaptation in metabolic genes and derived alleles in Drosophila melanogaster. Genetics 168: 923–931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stapley J., Reger J., Feulner P. G. D., Smadja C., Galindo J., et al. , 2010. Adaptation genomics: the next generation. Trends Ecol. Evol. 25: 705–712 [DOI] [PubMed] [Google Scholar]
- Sullivan M. C., Wuenschel M. J., Able K. W., 2009. Inter and intra-estuary variability in ingress, condition and settlement of the American eel Anguilla rostrata: implications for estimating and understanding recruitment. J. Fish Biol. 74: 1949–1969 [DOI] [PubMed] [Google Scholar]
- Sultan S. E., Spencer H. G., 2002. Metapopulation structure favors plasticity over local adaptation. Am. Nat. 160: 271–283 [DOI] [PubMed] [Google Scholar]
- Tesch F., 2003. The Eel. Blackwell Science, Oxford [Google Scholar]
- Turner T. L., Bourne E. C., Von Wettberg E. J., Hu T. T., Nuzhdin S. V., 2010. Population resequencing reveals local adaptation of Arabidopsis lyrata to serpentine soils. Nat. Genet. 42: 260–263 [DOI] [PubMed] [Google Scholar]
- Tzeng W. N., Wang Y. T., Wang C. H., 1998. Optimal growth temperature of the American eel, Anguilla rostrata (Le Sueur). J. Fish Soc. Taiwan 25: 111–115 [Google Scholar]
- Voight B. F., Kudaravalli S., Wen X., Pritchard J. K., 2006. A map of recent positive selection in the human genome. PLoS Biol. 4: e72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollestad L. A., Jonsson B., 1988. A 13-year study of the population dynamics and growth of the European eel Anguilla anguilla in a Norwegian river: evidence for density-dependent mortality, and development of a model for predicting yield. J. Anim. Ecol. 57: 983–997 [Google Scholar]
- Walsh P. J., Foster G. D., Moon T. W., 1983. The effects of temperature on metabolism of the American eel Anguilla rostrata (Le Sueur): compensation in the summer and torpor in the winter. Physiol. Zool. 56: 532–540 [Google Scholar]
- Wang C.-H., Tzeng W.-N., 1998. Interpretation of geographic variation in size of American eel Anguilla rostrata elvers on the Atlantic coast of North America using their life history and otolith ageing. Mar. Ecol. Prog. Ser. 168: 35–43 [Google Scholar]
- Weill R., Lutfalla G., Mogensen K., Chandre F., Berthomieu A., et al. , 2003. Insecticide resistance in mosquito vectors. Nature 423: 136–137 [DOI] [PubMed] [Google Scholar]
- Williams G. C., Koehn R. K., Mitton J. B., 1973. Genetic differentiation without isolation in the American Eel, Anguilla rostrata. Evolution 27: 192–204 [DOI] [PubMed] [Google Scholar]
- Wirth T., Bernatchez L., 2003. Decline of North Atlantic eels: A fatal synergy? Proc. Biol. Sci. 270: 681–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuenschel M., Able K., 2008. Swimming ability of eels (Anguilla rostrata, Conger oceanicus) at estuarine ingress: contrasting patterns of cross-shelf transport? Mar. Biol. 154: 775–786 [Google Scholar]
- Yeaman S., Otto S. P., 2011. Establishment and maintenance of adaptive genetic divergence under migration, selection, and drift. Evolution 65: 2123–2129 [DOI] [PubMed] [Google Scholar]