Abstract
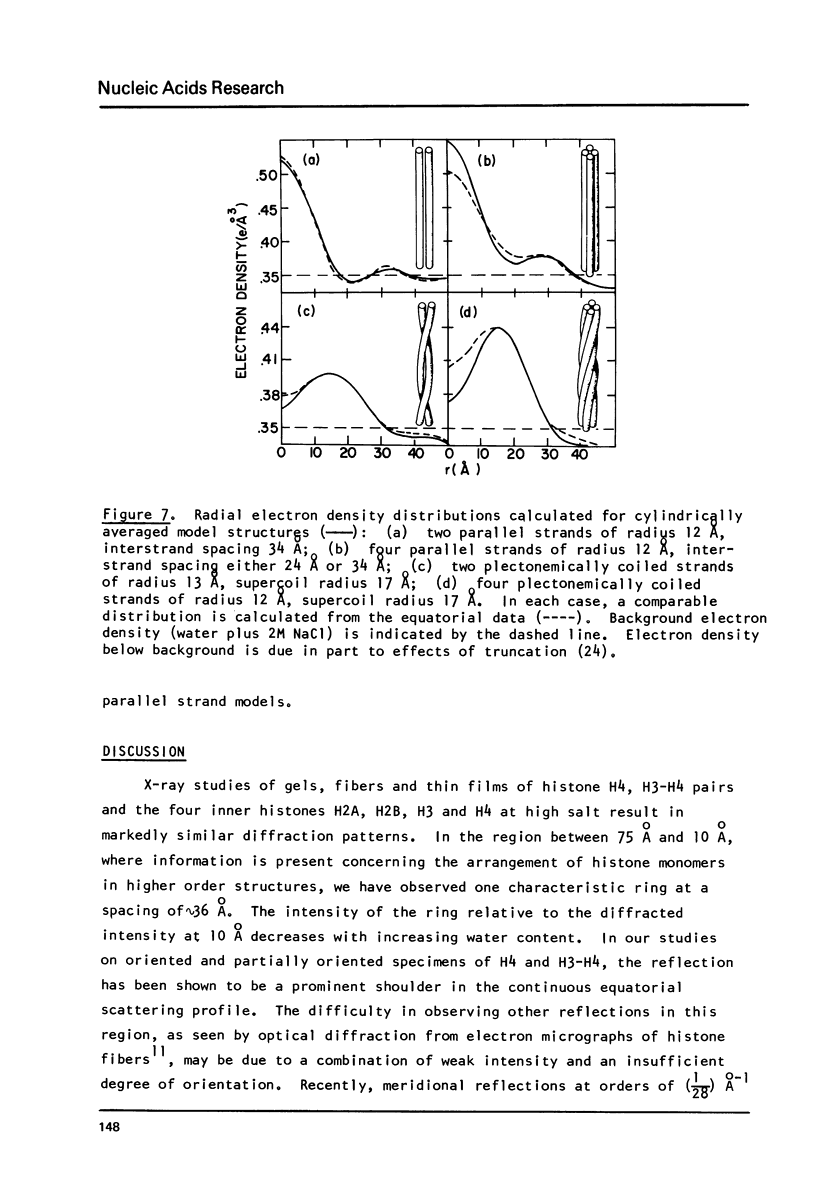
X-ray diffraction data from self-assembled histone fibers are presented for three systems: H4, H3-H4, and the four core histones H2A, H2B, H3 and H4. These data have been obtained under conditions of high ionic strength and high protein concentration which are thought to promote histone conformation similar to that found in intact chromatin. The low angle equatorial scattering (R less than .05 A-1) is analysed, and, with additional constraints imposed by electron microscopy data, four low resolution fibrillar models are derived. Two features common to all the possible models are a maximum outer diameter of approximately 60 A and a subfibril diameter of approximately 25 A. It is the interference of the protein subfibrils across a central region of low electron density - a 10 A "hole" - which gives rise to the characteristic diffraction peak at 36 A. Possible relationships of the models of the histone fibers to the structure of the histone component of chromatin are suggested.
Full text
PDF


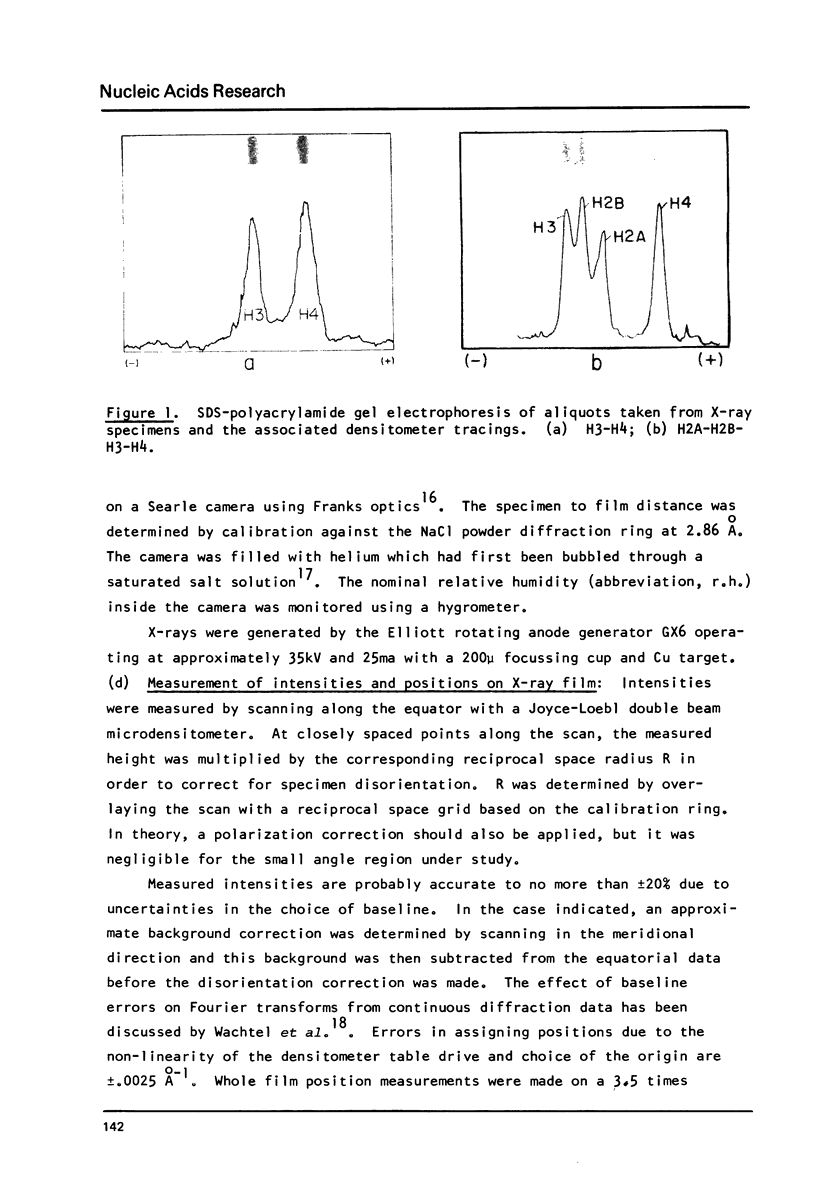



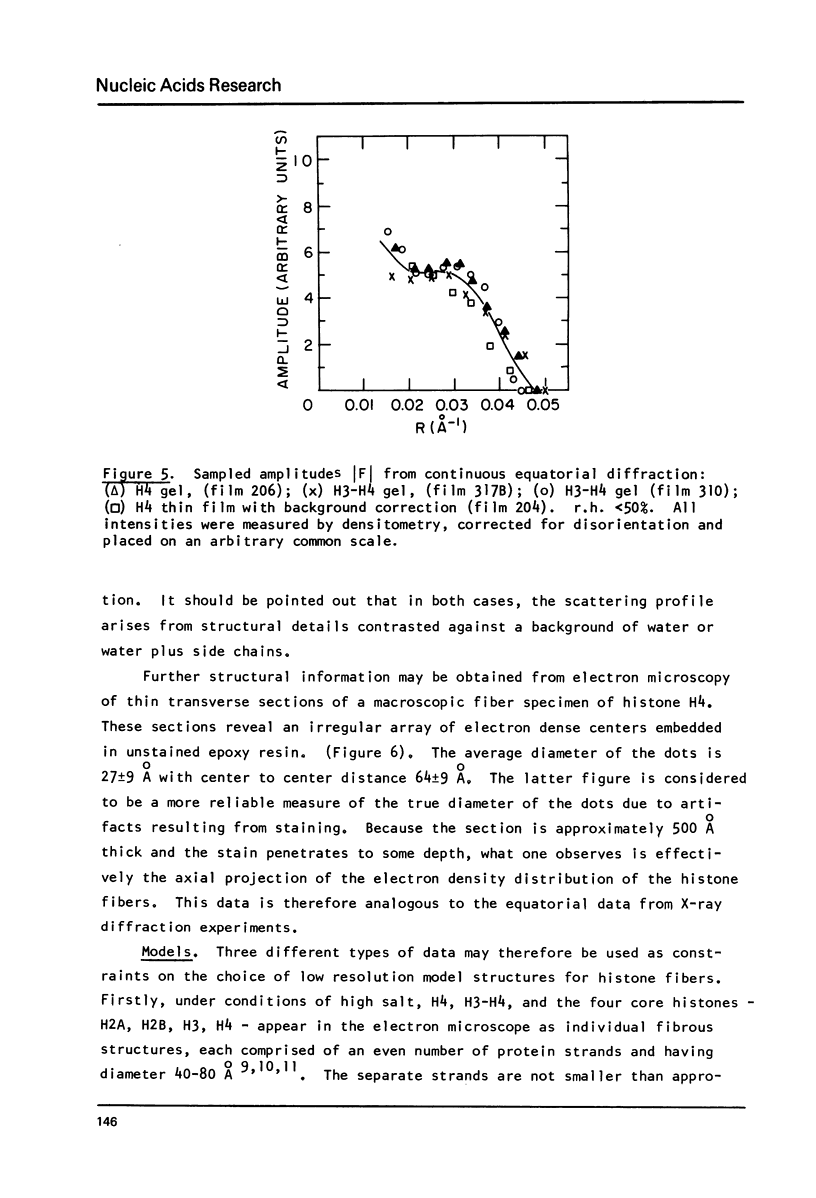





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bina-Stein M., Simpson R. T. Specific folding and contraction of DNA by histones H3 and H4. Cell. 1977 Jul;11(3):609–618. doi: 10.1016/0092-8674(77)90078-2. [DOI] [PubMed] [Google Scholar]
- Boseley P. G., Bradbury E. M., Butler-Browne G. S., Carpenter B. G., Stephens R. M. Physical studies of chromatin. The recombination of histones with DNA. Eur J Biochem. 1976 Feb 2;62(1):21–31. doi: 10.1111/j.1432-1033.1976.tb10093.x. [DOI] [PubMed] [Google Scholar]
- Camerini-Otero R. D., Sollner-Webb B., Felsenfeld G. The organization of histones and DNA in chromatin: evidence for an arginine-rich histone kernel. Cell. 1976 Jul;8(3):333–347. doi: 10.1016/0092-8674(76)90145-8. [DOI] [PubMed] [Google Scholar]
- D'Anna J. A., Jr, Isenberg I. A histone cross-complexing pattern. Biochemistry. 1974 Nov 19;13(24):4992–4997. doi: 10.1021/bi00721a019. [DOI] [PubMed] [Google Scholar]
- DeLange R. J., Smith E. L. Histones: structure and function. Annu Rev Biochem. 1971;40:279–314. doi: 10.1146/annurev.bi.40.070171.001431. [DOI] [PubMed] [Google Scholar]
- Fasman G. D., Chou P. Y., Adler A. J. Prediction of the conformation of the histones. Biophys J. 1976 Oct;16(10):1201–1238. doi: 10.1016/S0006-3495(76)85768-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finch J. T., Lutter L. C., Rhodes D., Brown R. S., Rushton B., Levitt M., Klug A. Structure of nucleosome core particles of chromatin. Nature. 1977 Sep 1;269(5623):29–36. doi: 10.1038/269029a0. [DOI] [PubMed] [Google Scholar]
- Fraser R. D., MacRae T. P., Miller A. X-ray diffraction patterns of alpha-fibrous proteins. J Mol Biol. 1965 Dec;14(2):432–442. doi: 10.1016/s0022-2836(65)80193-0. [DOI] [PubMed] [Google Scholar]
- Hjelm R. P., Kneale G. G., Sauau P., Baldwin J. P., Bradbury E. M., Ibel K. Small angle neutron scattering studies of chromatin subunits in solution. Cell. 1977 Jan;10(1):139–151. doi: 10.1016/0092-8674(77)90148-9. [DOI] [PubMed] [Google Scholar]
- Hyde J. E., Walker I. O. A model for chromatin sub-structure incorporating symmetry considerations of histone oligomers. Nucleic Acids Res. 1975 Mar;2(3):405–421. doi: 10.1093/nar/2.3.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johns E. W. Studies on histones. 7. Preparative methods for histone fractions from calf thymus. Biochem J. 1964 Jul;92(1):55–59. doi: 10.1042/bj0920055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg R. D. Structure of chromatin. Annu Rev Biochem. 1977;46:931–954. doi: 10.1146/annurev.bi.46.070177.004435. [DOI] [PubMed] [Google Scholar]
- Kornberg R. D. Structure of chromatin. Annu Rev Biochem. 1977;46:931–954. doi: 10.1146/annurev.bi.46.070177.004435. [DOI] [PubMed] [Google Scholar]
- LUFT J. H. Improvements in epoxy resin embedding methods. J Biophys Biochem Cytol. 1961 Feb;9:409–414. doi: 10.1083/jcb.9.2.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Marvin D. A., Wiseman R. L., Wachtel E. J. Filamentous bacterial viruses. XI. Molecular architecture of the class II (Pf1, Xf) virion. J Mol Biol. 1974 Jan 15;82(2):121–138. doi: 10.1016/0022-2836(74)90336-2. [DOI] [PubMed] [Google Scholar]
- Marvin D. A. X-ray diffraction and electron microscope studies on the structure of the small filamentous bacteriophage fd. J Mol Biol. 1966 Jan;15(1):8–17. doi: 10.1016/s0022-2836(66)80205-x. [DOI] [PubMed] [Google Scholar]
- Pardon J. F., Richards B. M., Cotter R. I. X-ray diffraction studies on oriented nucleohistone gels. Cold Spring Harb Symp Quant Biol. 1974;38:75–81. doi: 10.1101/sqb.1974.038.01.010. [DOI] [PubMed] [Google Scholar]
- Pardon J. F., Worcester D. L., Wooley J. C., Cotter R. I., Lilley D. M., Richards R. M. The structure of the chromatin core particle in solution. Nucleic Acids Res. 1977 Sep;4(9):3199–3214. doi: 10.1093/nar/4.9.3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sollner-Webb B., Camerini-Otero R. D., Felsenfeld G. Chromatin structure as probed by nucleases and proteases: evidence for the central role of histones H3 and H4. Cell. 1976 Sep;9(1):179–193. doi: 10.1016/0092-8674(76)90063-5. [DOI] [PubMed] [Google Scholar]
- Sperling R., Amos L. A. Arrangement of subunits in assembled histone H4 fibers. Proc Natl Acad Sci U S A. 1977 Sep;74(9):3772–3776. doi: 10.1073/pnas.74.9.3772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperling R., Bustin M. Dynamic equilibrium in histone assembly: self-assembly of single histones and histone pairs. Biochemistry. 1975 Jul 29;14(15):3322–3331. doi: 10.1021/bi00686a006. [DOI] [PubMed] [Google Scholar]
- Sperling R., Bustin M. Self assembly of histone F2a1. Proc Natl Acad Sci U S A. 1974 Nov;71(11):4625–4629. doi: 10.1073/pnas.71.11.4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suau P., Kneale G. G., Braddock G. W., Baldwin J. P., Bradbury E. M. A low resolution model for the chromatin core particle by neutron scattering. Nucleic Acids Res. 1977 Nov;4(11):3769–3786. doi: 10.1093/nar/4.11.3769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas G. J., Jr, Prescott B., Olins D. E. Secondary structure of histones and DNA in chromatin. Science. 1977 Jul 22;197(4301):385–388. doi: 10.1126/science.560060. [DOI] [PubMed] [Google Scholar]
- Thomas J. O., Kornberg R. D. An octamer of histones in chromatin and free in solution. Proc Natl Acad Sci U S A. 1975 Jul;72(7):2626–2630. doi: 10.1073/pnas.72.7.2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas J. O., Kornberg R. D. Cleavable cross-links in the analysis of histone-histone associations. FEBS Lett. 1975 Oct 15;58(1):353–358. doi: 10.1016/0014-5793(75)80296-1. [DOI] [PubMed] [Google Scholar]
- Wachtel E. J., Wiseman R. L., Pigram W. J., Marvin D. A. Filamentous bacterial viruses. XIII. Molecular structure of the virion in projection. J Mol Biol. 1974 Sep 25;88(3):601–618. doi: 10.1016/0022-2836(74)90411-2. [DOI] [PubMed] [Google Scholar]
- Weintraub H., Palter K., Van Lente F. Histones H2a, H2b, H3, and H4 form a tetrameric complex in solutions of high salt. Cell. 1975 Sep;6(1):85–110. doi: 10.1016/0092-8674(75)90077-x. [DOI] [PubMed] [Google Scholar]
- Worcel A., Benyajati C. Higher order coiling of DNA in chromatin. Cell. 1977 Sep;12(1):83–100. doi: 10.1016/0092-8674(77)90187-8. [DOI] [PubMed] [Google Scholar]