Abstract
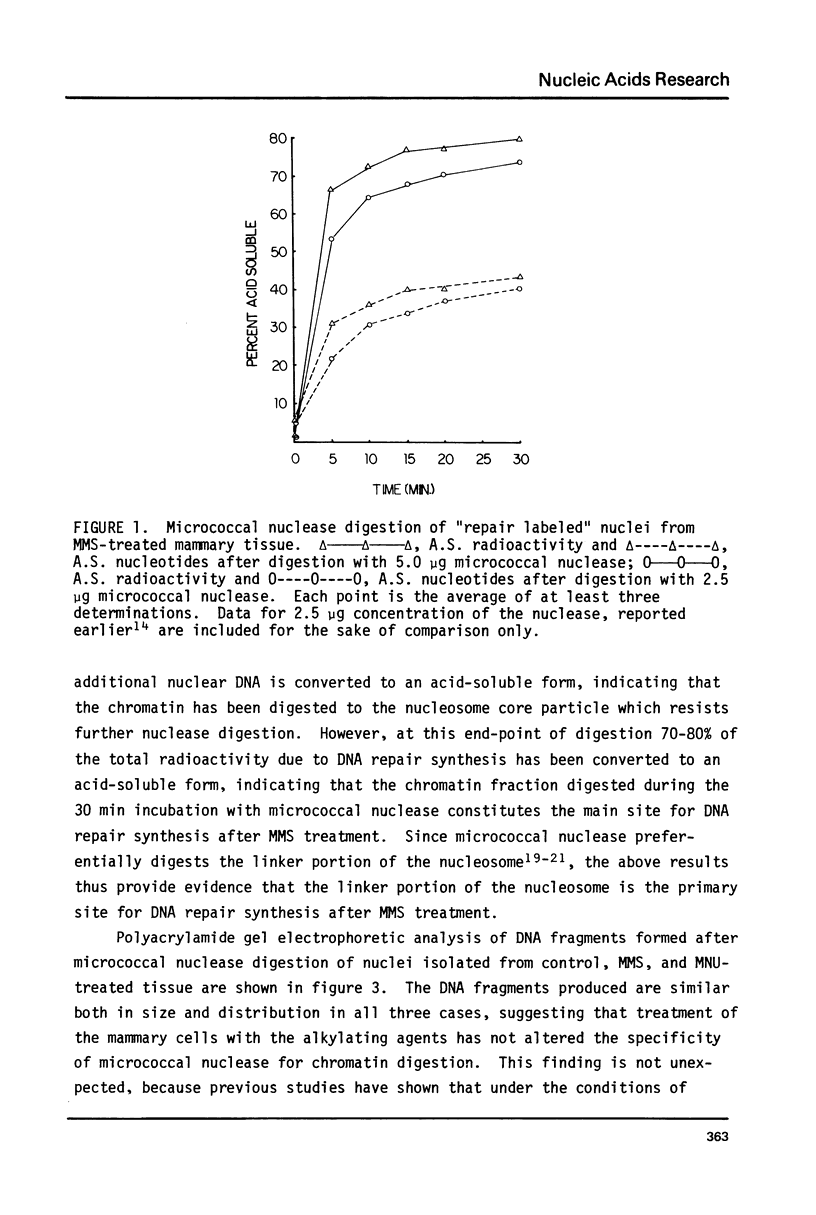
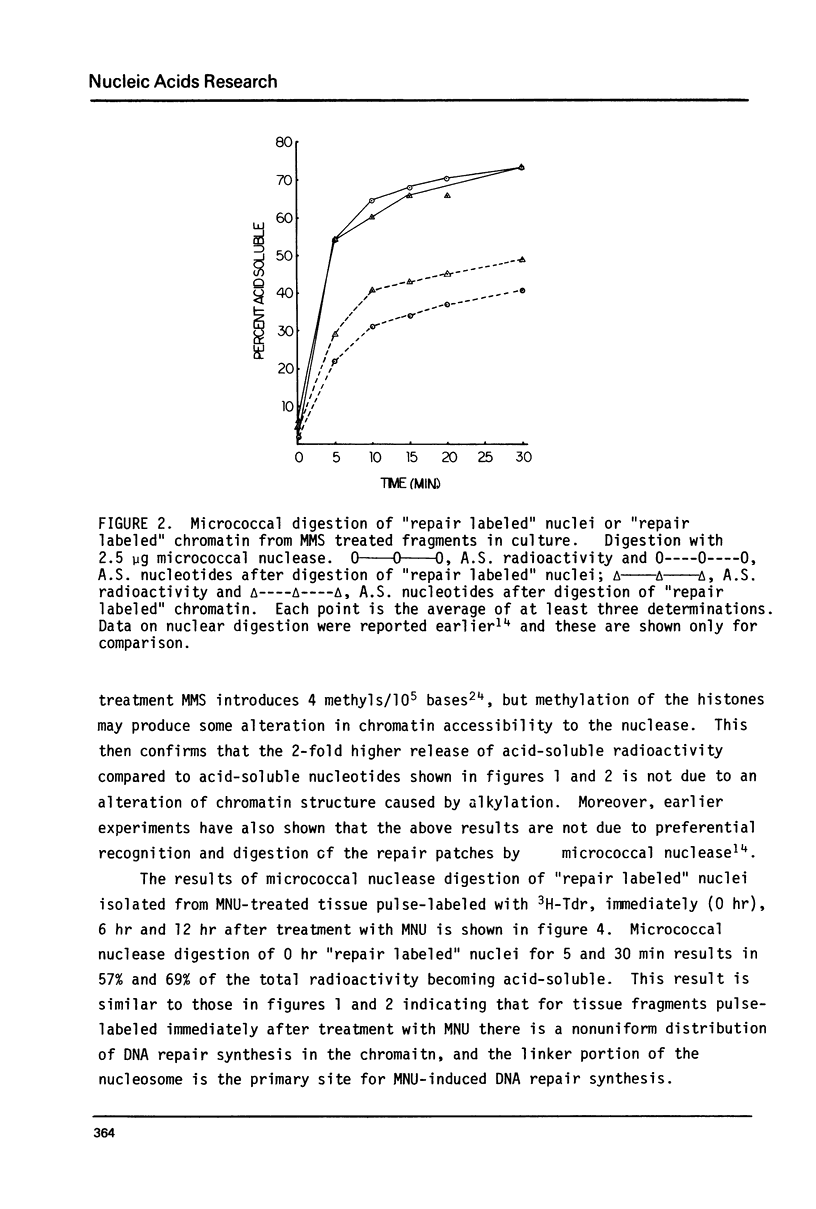
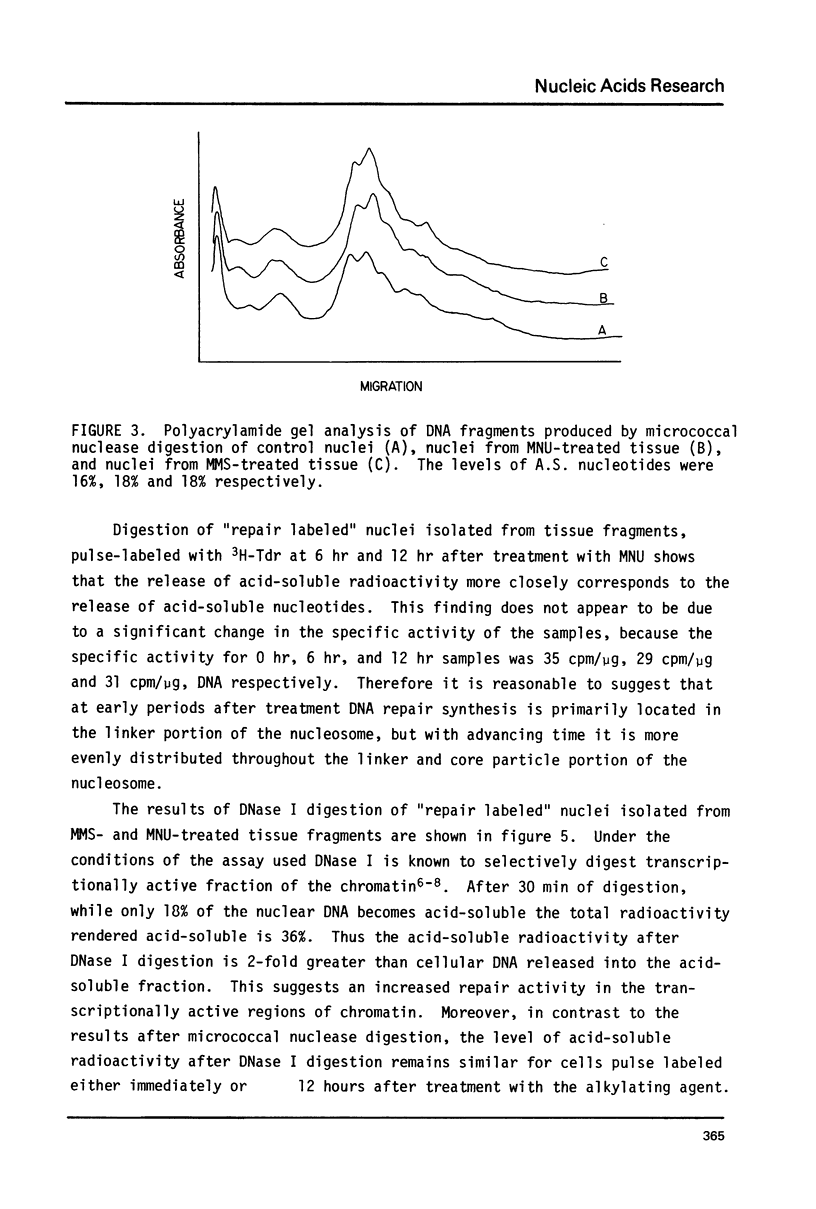
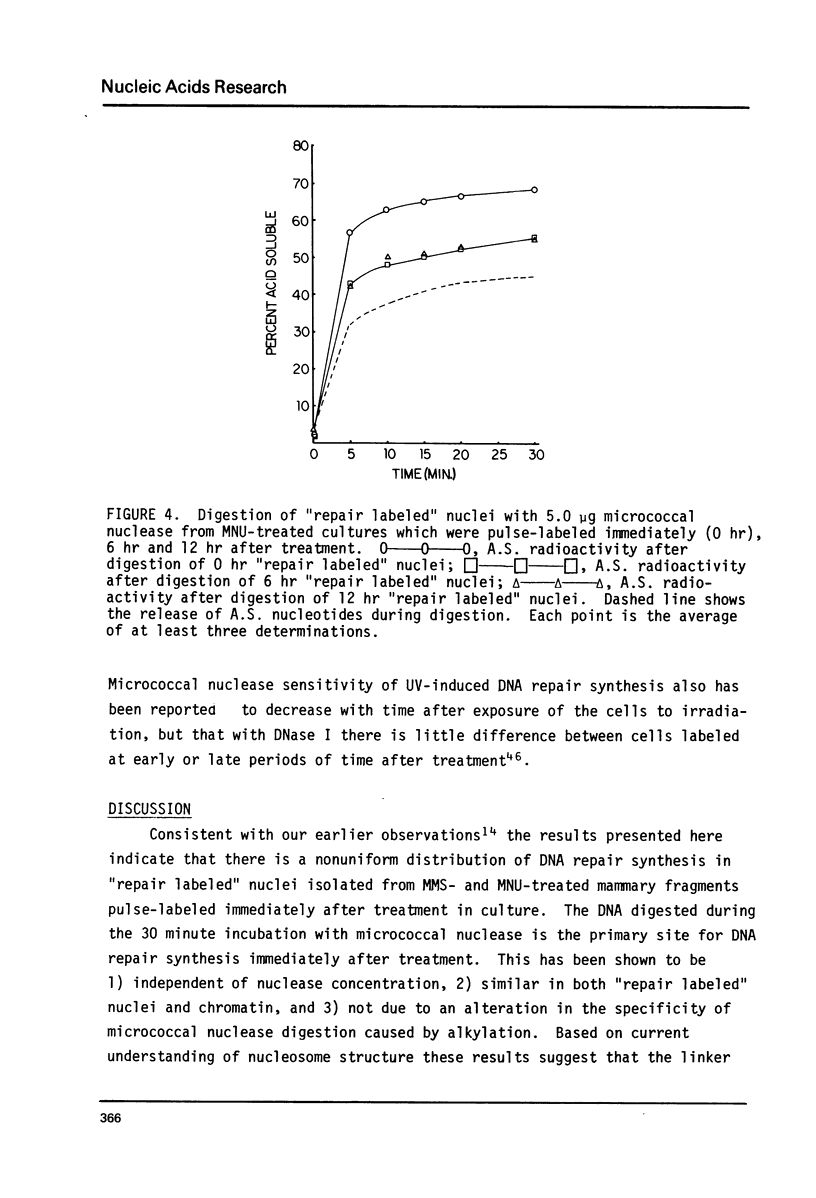
The influence of chromatin structure on the distribution of DNA repair synthesis was studied by enzymatic digestion of "repair labeled" nuclei of mouse mammary cells: "repair labeled" nuclei were isolated from pregnancy mammary tissue fragments, treated in vitro with methylmethanesulfonate (MMS) or methylnitrosourea (MNU), and pulse-labeled with 3H-thymidine in the presence of hydroxyurea in the culture medium. Micrococcal nuclease digestion of "repair labeled" nuclei indicates that at early hours after treatment with the alkylating agents 70-80% of the total repair synthesis is located in the linker portion of the nucleosome. However, 6-12 hours after treatment DNA repair synthesis is more evenly distributed throughout the core and linker portion of the nucleosome. "Repair labeled" mammary cell nuclei were also digested with DNase I under conditions selective for transcriptionally active chromatin. A two-fold higher level of repair synthesis was found in the transcriptionally active chromatin of "repair labeled" nuclei isolated from MMS or MNU treated mammary fragments, pulse-labeled at different times after treatment. The results indicate that structural constitution of the chromatin may influence the distribution of DNA repair synthesis both at the nucleosome level, and at higher levels of chromatin organization. This may be due to 1) nonrandom base alkylation in chromatin or 2) areas in chromatin with increased accessibility for the repair enzymes to the alkylated bases.
Full text
PDF






Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Axel R. Cleavage of DNA in nuclei and chromatin with staphylococcal nuclease. Biochemistry. 1975 Jul;14(13):2921–2925. doi: 10.1021/bi00684a020. [DOI] [PubMed] [Google Scholar]
- Biswas B. B., Ganguly A., Das A. Eukaryotic RNA polymerases and the factors that control them. Prog Nucleic Acid Res Mol Biol. 1975;15(0):145–184. doi: 10.1016/s0079-6603(08)60119-1. [DOI] [PubMed] [Google Scholar]
- Bodell W. J., Banerjee M. R. DNA repair in normal and preneoplastic mammary tissues. Cancer Res. 1978 Mar;38(3):736–740. [PubMed] [Google Scholar]
- Bodell W. J., Banerjee M. R. Reduced DNA repair in mouse satellite DNA after treatment with methylmethanesulfonate, and N-methyl-N-nitrosourea. Nucleic Acids Res. 1976 Jul;3(7):1689–1701. doi: 10.1093/nar/3.7.1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodell W. J. Nonuniform distribution of DNA repair in chromatin after treatment with methyl methanesulfonate. Nucleic Acids Res. 1977 Aug;4(8):2619–2628. doi: 10.1093/nar/4.8.2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhl S. N., Setlow R. B., Regan J. D. DNA repair in Potorous tridactylus. Biophys J. 1974 Oct;14(10):791–803. doi: 10.1016/S0006-3495(74)85949-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cech T., Pardue M. L. Cross-linking of DNA with trimethylpsoralen is a probe for chromatin structure. Cell. 1977 Jul;11(3):631–640. doi: 10.1016/0092-8674(77)90080-0. [DOI] [PubMed] [Google Scholar]
- Cleaver J. E. Defective repair replication of DNA in xeroderma pigmentosum. Nature. 1968 May 18;218(5142):652–656. doi: 10.1038/218652a0. [DOI] [PubMed] [Google Scholar]
- Cleaver J. E. Nucleosome structure controls rates of excision repair in DNA of human cells. Nature. 1977 Dec 1;270(5636):451–453. doi: 10.1038/270451a0. [DOI] [PubMed] [Google Scholar]
- Cleaver J. E. Xeroderma pigmentosum: a human disease in which an initial stage of DNA repair is defective. Proc Natl Acad Sci U S A. 1969 Jun;63(2):428–435. doi: 10.1073/pnas.63.2.428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLange R. J., Smith E. L. Histones: structure and function. Annu Rev Biochem. 1971;40:279–314. doi: 10.1146/annurev.bi.40.070171.001431. [DOI] [PubMed] [Google Scholar]
- Edenberg H. J., Huberman J. A. Eukaryotic chromosome replication. Annu Rev Genet. 1975;9:245–284. doi: 10.1146/annurev.ge.09.120175.001333. [DOI] [PubMed] [Google Scholar]
- Felsenfeld G. Chromatin. Nature. 1978 Jan 12;271(5641):115–122. doi: 10.1038/271115a0. [DOI] [PubMed] [Google Scholar]
- Finch J. T., Klug A. Solenoidal model for superstructure in chromatin. Proc Natl Acad Sci U S A. 1976 Jun;73(6):1897–1901. doi: 10.1073/pnas.73.6.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garel A., Axel R. Selective digestion of transcriptionally active ovalbumin genes from oviduct nuclei. Proc Natl Acad Sci U S A. 1976 Nov;73(11):3966–3970. doi: 10.1073/pnas.73.11.3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goth R., Rajewsky M. F. Persistence of O6-ethylguanine in rat-brain DNA: correlation with nervous system-specific carcinogenesis by ethylnitrosourea. Proc Natl Acad Sci U S A. 1974 Mar;71(3):639–643. doi: 10.1073/pnas.71.3.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewish D. Features of the structure of replicating and non-replicating chromatin in chicken erythroblasts. Nucleic Acids Res. 1977 Jun;4(6):1881–1890. doi: 10.1093/nar/4.6.1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrand C. E., Walters R. A. Rapid assembly of newly synthesized DNA into chromatin subunits prior to joining to small DNA replication intermediates. Biochem Biophys Res Commun. 1976 Nov 8;73(1):157–163. doi: 10.1016/0006-291x(76)90510-6. [DOI] [PubMed] [Google Scholar]
- Jahn C. L., Litman G. W. Distribution of covalently bound benzo(a)pyrene in chromatin. Biochem Biophys Res Commun. 1976 May 23;76(2):534–540. doi: 10.1016/0006-291x(77)90757-4. [DOI] [PubMed] [Google Scholar]
- Kornberg R. D. Structure of chromatin. Annu Rev Biochem. 1977;46:931–954. doi: 10.1146/annurev.bi.46.070177.004435. [DOI] [PubMed] [Google Scholar]
- Kriek E. Persistent binding of a new reaction product of the carcinogen N-hydroxy-N-2-acetylaminofluorene with guanine in rat liver DNA in vivo. Cancer Res. 1972 Oct;32(10):2042–2048. [PubMed] [Google Scholar]
- Lacy E., Axel R. Analysis of DNA of isolated chromatin subunits. Proc Natl Acad Sci U S A. 1975 Oct;72(10):3978–3982. doi: 10.1073/pnas.72.10.3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy B., Dixon G. H. Renaturation kinetics of cDNA complementary to cytoplamic polyadenylated RNA from rainbow trout testis. Accessibility of transcribed genes to pancreatic DNase. Nucleic Acids Res. 1977 Apr;4(4):883–898. doi: 10.1093/nar/4.4.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman M. W., Dipple A. Removal of bound carcinogen during DNA repair in nondividing human lymphocytes. Cancer Res. 1972 Sep;32(9):1855–1860. [PubMed] [Google Scholar]
- Maher V. M., McCormick J. J., Grover P. L., Sims P. Effect of DNA repair on the cytotoxicity and mutagenicity of polycyclic hydrocarbon derivatives in normal and xeroderma pigmentosum human fibroblasts. Mutat Res. 1977 Apr;43(1):117–138. doi: 10.1016/0027-5107(77)90137-3. [DOI] [PubMed] [Google Scholar]
- Metzger G., Wilhelm F. X., Wilhelm M. L. Non-random binding of a chemical carcinogen to the DNA in chromatin. Biochem Biophys Res Commun. 1977 Apr 11;75(3):703–710. doi: 10.1016/0006-291x(77)91529-7. [DOI] [PubMed] [Google Scholar]
- Mortelmans K., Friedberg E. C., Slor H., Thomas G., Cleaver J. E. Defective thymine dimer excision by cell-free extracts of xeroderma pigmentosum cells. Proc Natl Acad Sci U S A. 1976 Aug;73(8):2757–2761. doi: 10.1073/pnas.73.8.2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicoll J. W., Swann P. F., Pegg A. E. Effect of dimethylnitrosamine on persistence of methylated guanines in rat liver and kidney DNA. Nature. 1975 Mar 20;254(5497):261–262. doi: 10.1038/254261a0. [DOI] [PubMed] [Google Scholar]
- Noll M. Subunit structure of chromatin. Nature. 1974 Sep 20;251(5472):249–251. doi: 10.1038/251249a0. [DOI] [PubMed] [Google Scholar]
- Paterson M. C., Lohman P. H., Sluyter M. L. Use of UV endonuclease from Micrococcus luteus to monitor the progress of DNA repair in UV-irradiated human cells. Mutat Res. 1973 Aug;19(2):245–256. doi: 10.1016/0027-5107(73)90083-3. [DOI] [PubMed] [Google Scholar]
- Peacock A. C., Dingman C. W. Molecular weight estimation and separation of ribonucleic acid by electrophoresis in agarose-acrylamide composite gels. Biochemistry. 1968 Feb;7(2):668–674. doi: 10.1021/bi00842a023. [DOI] [PubMed] [Google Scholar]
- Regan J. D., Trosko J. E., Carrier W. L. Evidence for excision of ultraviolet-induced pyrimidine dimers from the DNA of human cells in vitro. Biophys J. 1968 Mar;8(3):319–325. doi: 10.1016/S0006-3495(68)86490-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seale R. L. Assembly of DNA and protein during replication in HeLa cells. Nature. 1975 May 15;255(5505):247–249. doi: 10.1038/255247a0. [DOI] [PubMed] [Google Scholar]
- Shinohara K., Cerutti P. A. Excision repair of benzo[a]pyrene-deoxyguanosine adducts in baby hamster kidney 21/C13 cells and in secondary mouse embryo fibroblasts C57BL/6J. Proc Natl Acad Sci U S A. 1977 Mar;74(3):979–983. doi: 10.1073/pnas.74.3.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson R. T., Whitlock J. P., Jr Chemical evidence that chromatin DNA exists as 160 base pair beads interspersed with 40 base pair bridges. Nucleic Acids Res. 1976 Jan;3(1):117–127. doi: 10.1093/nar/3.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smerdon M. J., Tlsty T. D., Lieberman M. W. Distribution of ultraviolet-induced DNA repair synthesis in nuclease sensitive and resistant regions of human chromatin. Biochemistry. 1978 Jun 13;17(12):2377–2386. doi: 10.1021/bi00605a020. [DOI] [PubMed] [Google Scholar]
- Stein G., Stein J., Kleinsmith L., Park W., Jansing R., Thomson J. Nonhistone chromosomal proteins and histone gene transcription. Prog Nucleic Acid Res Mol Biol. 1976;19:421–445. doi: 10.1016/s0079-6603(08)60935-6. [DOI] [PubMed] [Google Scholar]
- Takebe H., Nii S., Ishii M. I., Utsumi H. Comparative studies of host-cell reactivation, colony forming ability and excision repair after UV irradiation of xeroderma pigmentosum, normal human and some other mammalian cells. Mutat Res. 1974 Dec;25(3):383–390. doi: 10.1016/0027-5107(74)90067-0. [DOI] [PubMed] [Google Scholar]
- Weintraub H., Groudine M. Chromosomal subunits in active genes have an altered conformation. Science. 1976 Sep 3;193(4256):848–856. doi: 10.1126/science.948749. [DOI] [PubMed] [Google Scholar]
- Wieshahn G. P., Hyde J. E., Hearst J. E. The photoaddition of trimethylpsoralen to Drosophila melanogaster nuclei: a probe for chromatin substructure. Biochemistry. 1977 Mar 8;16(5):925–932. doi: 10.1021/bi00624a018. [DOI] [PubMed] [Google Scholar]
- Wilkins R. J., Hart R. W. Preferential DNA repair in human cells. Nature. 1974 Jan 4;247(5435):35–36. doi: 10.1038/247035a0. [DOI] [PubMed] [Google Scholar]


