Abstract
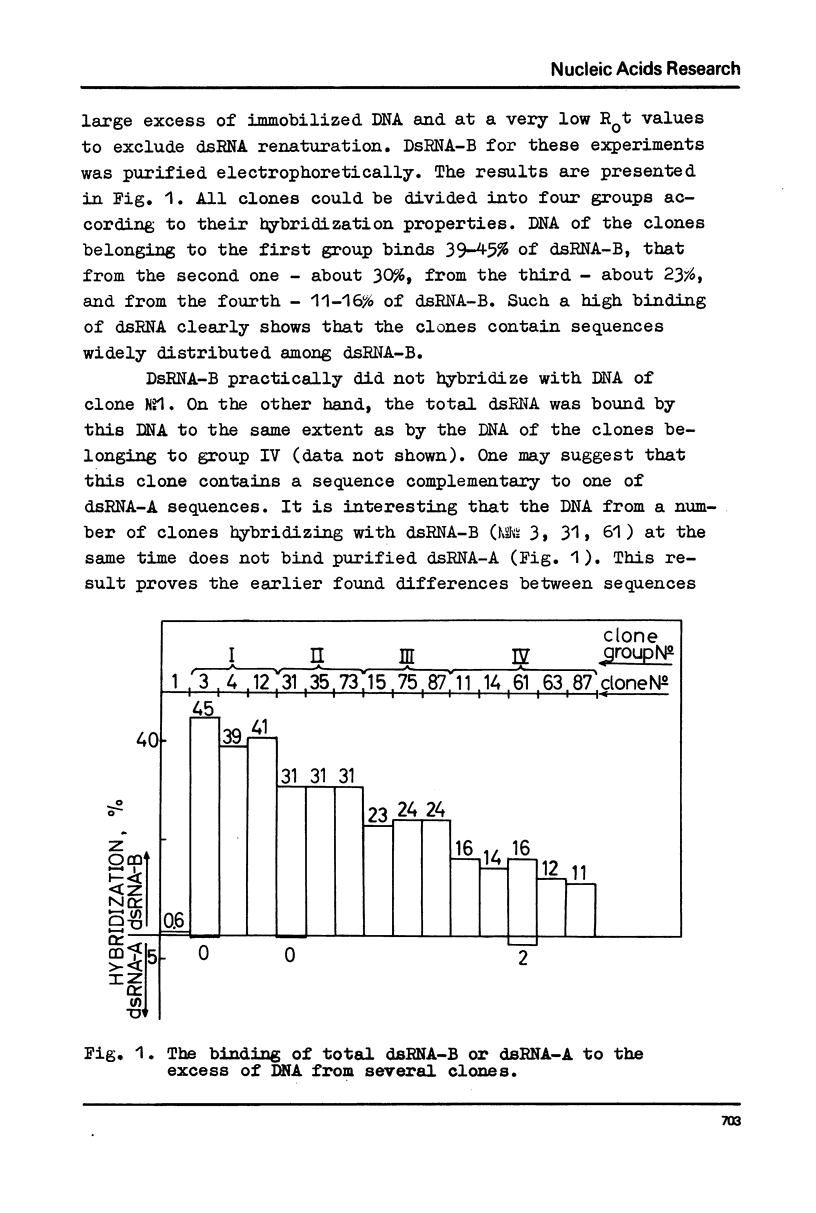
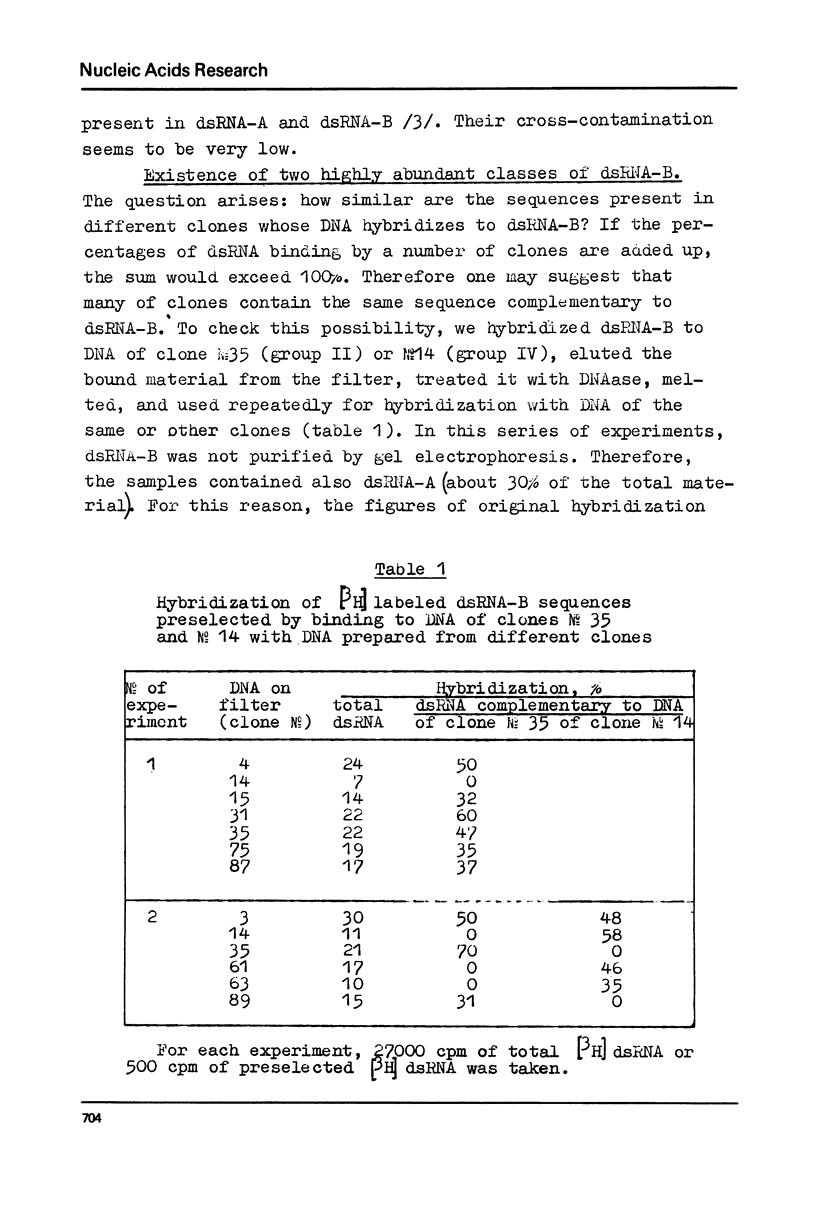
DNA preparations from about hundred randomly selected clones containing mouse DNA fragments were screened for the existence of sequences complementary to long double-stranded regions of pre-mRNA able to snap back after melting (dsRNA-B). Many clones containing such sequences were found. The cloned sequences can be subdivided into three groups: (1) those complementary to about a half (at least to 30-40%) of the total dsRNA, designated as sequences B1; (2) those complementary to a part of sequence B1; and (3) sequences complementary to about a quarter (at least to 15%) of the total dsRNA referred to as sequence B2. The size of DNA sequence complementary to dsRNA is about 400 base pairs.
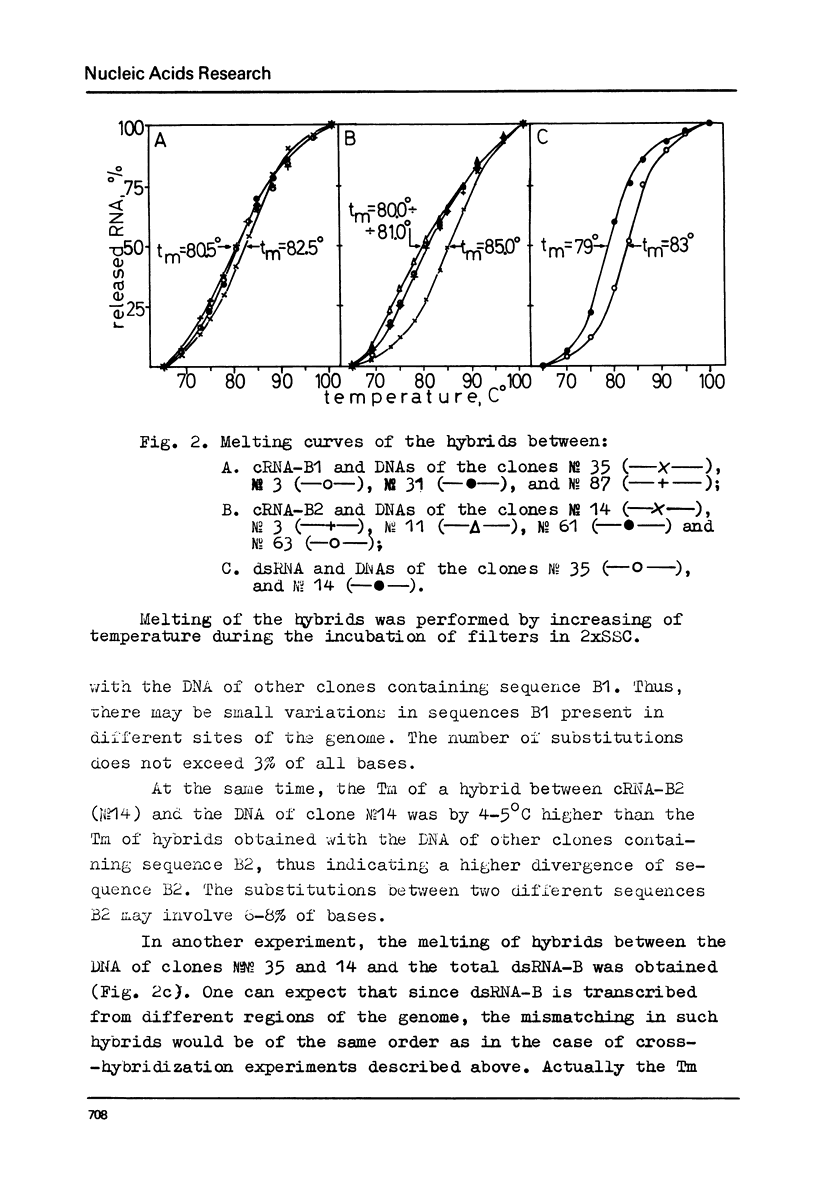
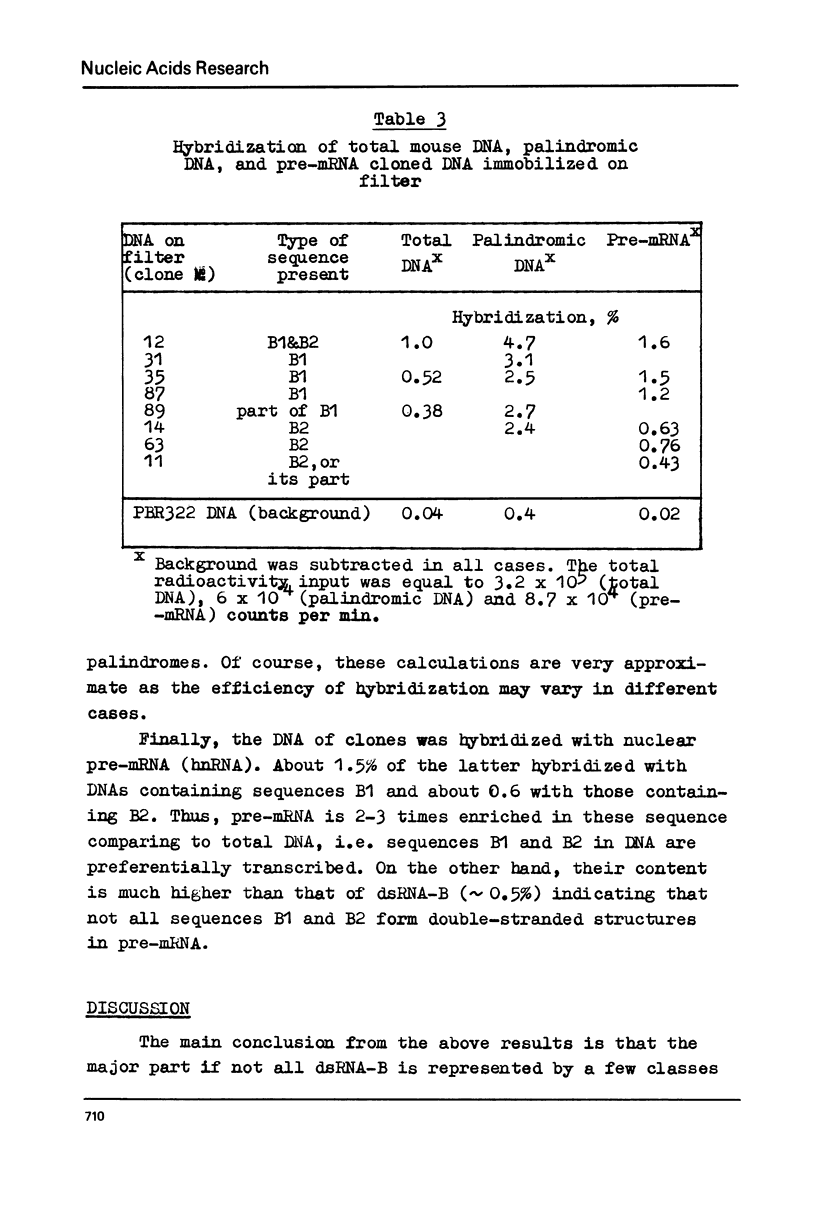
Melting experiments with hybrids show that the members of B1 family are very similar if not identical, while the divergence among B2 sequences is higher, but still the number of substitutions does not exceed 9% of bases. Thus, the major part of dsRNA-B consists of a small number of highly abundant sequences as was suggested earlier on the basis of renaturation kinetics /1-3/. Sequences B1 and B2 are represented by many copies in the mouse genome and in pre-mRNA, and many of them probably do not form hairpin-like structures.
Full text
PDF












Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bolivar F., Rodriguez R. L., Greene P. J., Betlach M. C., Heyneker H. L., Boyer H. W., Crosa J. H., Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. [PubMed] [Google Scholar]
- Cech T. R., Hearst J. E. An electron microscopic study of mouse foldback DNA. Cell. 1975 Aug;5(4):429–446. doi: 10.1016/0092-8674(75)90062-8. [DOI] [PubMed] [Google Scholar]
- Denhardt D. T. A membrane-filter technique for the detection of complementary DNA. Biochem Biophys Res Commun. 1966 Jun 13;23(5):641–646. doi: 10.1016/0006-291x(66)90447-5. [DOI] [PubMed] [Google Scholar]
- GEORGIEV G. P., MANT'EVA V. L. [Messenger and ribosomal ribonucleic acid in the chromosomal-nucleolar apparatus, methods of isolation and nuceotide structure]. Biokhimiia. 1962 Sep-Oct;27:949–957. [PubMed] [Google Scholar]
- Georgiev G. P., Ryskov A. P., Coutelle C., Mantieva V. L., Avakyan E. R. On the structure of transcriptional unit in mammalian cells. Biochim Biophys Acta. 1972 Jan 31;259(2):259–283. doi: 10.1016/0005-2787(72)90066-4. [DOI] [PubMed] [Google Scholar]
- Georgiev G. P., Varshavsky A. J., Ryskov A. P., Church R. B. On the structural organization of the transcriptional unit in animal chromosomes. Cold Spring Harb Symp Quant Biol. 1974;38:869–884. doi: 10.1101/sqb.1974.038.01.089. [DOI] [PubMed] [Google Scholar]
- Gilbert W. Why genes in pieces? Nature. 1978 Feb 9;271(5645):501–501. doi: 10.1038/271501a0. [DOI] [PubMed] [Google Scholar]
- Gross-Bellard M., Oudet P., Chambon P. Isolation of high-molecular-weight DNA from mammalian cells. Eur J Biochem. 1973 Jul 2;36(1):32–38. doi: 10.1111/j.1432-1033.1973.tb02881.x. [DOI] [PubMed] [Google Scholar]
- Jelinek W. R. Inverted repeated DNA from Chinese hamster ovary cells studied with cloned DNA fragments. Proc Natl Acad Sci U S A. 1978 Jun;75(6):2679–2683. doi: 10.1073/pnas.75.6.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelinek W. R. Specific nucleotide sequences in HeLa cell inverted repeated DNA: enrichment for sequences found in double-stranded regions of heterogeneous nuclear RNA. J Mol Biol. 1977 Oct 5;115(4):591–601. doi: 10.1016/0022-2836(77)90104-8. [DOI] [PubMed] [Google Scholar]
- Jelinek W., Darnell J. E. Double-stranded regions in heterogeneous nuclear RNA from Hela cells. Proc Natl Acad Sci U S A. 1972 Sep;69(9):2537–2541. doi: 10.1073/pnas.69.9.2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramerov D. A., Ryskov A. P., Georgiev G. P. The structural organization of nuclear pre-mRNA. II. Very long double-stranded structures in nuclear pre-mRNA. Biochim Biophys Acta. 1977 Apr 4;475(3):461–475. doi: 10.1016/0005-2787(77)90062-4. [DOI] [PubMed] [Google Scholar]
- Maniatis T., Jeffrey A., Kleid D. G. Nucleotide sequence of the rightward operator of phage lambda. Proc Natl Acad Sci U S A. 1975 Mar;72(3):1184–1188. doi: 10.1073/pnas.72.3.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prensky W., Steffensen D. M., Hughes W. L. The use of iodinated RNA for gene localization. Proc Natl Acad Sci U S A. 1973 Jun;70(6):1860–1864. doi: 10.1073/pnas.70.6.1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryskov A. P., Farashyan V. R., Georgiev G. P. Ribonuclease-stable base sequences specific exclusively for giant dRNA. Biochim Biophys Acta. 1972 Apr 12;262(4):568–572. doi: 10.1016/0005-2787(72)90502-3. [DOI] [PubMed] [Google Scholar]
- Ryskov A. P., Kramerov D. A., Georgiev G. P. The structural organization of nuclear messenger RNA precursor. I. Reassociation and hybridization properties of double-stranded hairpin-like loops in messenger RNA precursor. Biochim Biophys Acta. 1976 Oct 4;447(2):214–229. doi: 10.1016/0005-2787(76)90344-0. [DOI] [PubMed] [Google Scholar]
- Ryskov A. P., Saunders G. F., Farashyan V. R., Georgiev G. P. Double-helical regions in nuclear precursor of mRNA (pre-mRNA). Biochim Biophys Acta. 1973 Jun 8;312(1):152–164. doi: 10.1016/0005-2787(73)90060-9. [DOI] [PubMed] [Google Scholar]
- Ryskov A. P., Tokarskaya O. V., Georgiev G. P., Coutelle C., Thiele B. Globin mRNA contains a sequence complementary to double-stranded region of nuclear pre-mRNA. Nucleic Acids Res. 1976 Jun;3(6):1487–1498. doi: 10.1093/nar/3.6.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SCHERRER K., DARNELL J. E. Sedimentation characteristics of rapidly labelled RNA from HeLa cells. Biochem Biophys Res Commun. 1962 Jun 4;7:486–490. doi: 10.1016/0006-291x(62)90341-8. [DOI] [PubMed] [Google Scholar]
- Schmid C. W., Manning J. E., Davidson N. Inverted repeat sequences in the Drosophila genome. Cell. 1975 Jun;5(2):159–172. doi: 10.1016/0092-8674(75)90024-0. [DOI] [PubMed] [Google Scholar]
- Vogt V. M. Purification and further properties of single-strand-specific nuclease from Aspergillus oryzae. Eur J Biochem. 1973 Feb 15;33(1):192–200. doi: 10.1111/j.1432-1033.1973.tb02669.x. [DOI] [PubMed] [Google Scholar]
- Weiss B., Live T. R., Richardson C. C. Enzymatic breakage and joining of deoxyribonucleic acid. V. End group labeling and analysis of deoxyribonucleic acid containing single straned breaks. J Biol Chem. 1968 Sep 10;243(17):4530–4542. [PubMed] [Google Scholar]


