Summary
Gene expression signatures relating mammary stem cell populations to breast cancers have focused on adult tissue. Here, we identify, isolate and characterize the fetal mammary stem cell (fMaSC) state since the invasive and proliferative processes of mammogenesis resemble phases of cancer progression. fMaSC frequency peaks late in embryogenesis, enabling more extensive stem cell purification than achieved with adult tissue. fMaSCs are self-renewing, multipotent, and co-express multiple mammary lineage markers. Gene expression, transplantation, and in vitro analyses reveal putative autocrine and paracrine regulatory mechanisms including ErbB and FGF signaling pathways impinging on fMaSC growth. Expression profiles from fMaSCs and associated stroma exhibit significant similarities to basal-like and Her2+ intrinsic breast cancer subtypes. Our results reveal significant links between development and cancer and provide resources to identify new candidates for diagnosis, prognosis and therapy.
Introduction
Breast cancers are a heterogeneous group of diseases distinguishable by histopathology and molecular profiling. Expression profiling of patient samples enabled categorization into molecular subtypes referred to as luminal A, luminal B, Her2 positive, basal-like and claudin-low (Herschkowitz et al., 2007; Perou et al., 2000). These divisions identify critical differences in cellular composition and molecular pathways suggesting treatment options and correlating with patient survival (Prat and Perou, 2011). Prognostic expression signatures, refined by related approaches, are being tested or used clinically (Fan et al., 2011; Paik et al., 2006; van ’t Veer et al., 2002; van de Vijver et al., 2002). Previously reported prognostic signatures and subtype designations identify a limited set of biologic programs correlating with hormone receptor status (Estrogen and Progesterone Receptors, ER and PR), Her2 expression and proliferation (Desmedt et al., 2008; Fan et al., 2006; Haibe-Kains et al., 2008; Prat and Perou, 2011; Sotiriou and Piccart, 2007). While hormone receptors and Her2 are therapeutic targets, many breast cancers, including most basal-like subtypes, lack ER, PR and Her2 expression and associated targeted treatment options (Pal et al., 2011).
Stem cell biology offers promise for understanding the origins and progression of breast and other cancers, and may also reveal the next generation of molecular targets for breast cancers not susceptible to current agents. For example, basal-like breast cancers are poorly differentiated, and exhibit gene expression similarities to embryonic and induced pluripotent stem cells (Ben-Porath et al., 2008; Mizuno et al., 2010). Expression profiles derived from adult mammary cells of different differentiation stages have also been used to designate cancers as stem like or non-stem like (Lim et al., 2009; Lim et al., 2010; Perou et al., 2010). Breast cancer cells that generate xenografted tumors with high efficiency, regenerate the cellular complexity of the originiating tumor, and that self-renew as defined by secondary transplantation exhibit properties attributed to stem cells, and have consequently been called breast “cancer stem cells” (Al-Hajj et al., 2003). However, defining potential relationships between stem-like cells in breast cancer, breast cancer stem cells, and normal mammary stem cells (MaSC) requires MaSC isolation and characterization.
Adult MaSCs (aMaSCs) have been enriched using stem cell isolation methods, and their gene expression signatures have been reported (Lim et al., 2009; Lim et al., 2010; Pece et al., 2010; Raouf et al., 2008; Shackleton et al., 2006; Stingl et al., 2006). However, aMaSC rarity combined with the cellular complexity of the adult gland make purification challenging (Shackleton et al., 2006; Stingl et al., 2006), and co-purifying stroma and differentiated mammary cells complicate elucidation of their core self-renewal and differentiation programs.
The developing mammary gland is less complex than the adult gland, suggesting that it may facilitate stem cell identification and purification. Furthermore, while the extensive proliferation, migration, and invasion required for mammogenesis do not occur in the resting adult mammary gland, they do resemble processes mediating breast cancer progression (Veltmaat et al., 2003). These observations suggest that stem cells present in fetal mammary rudiments (i.e., fMaSCs) might express genes comprising pathways overlooked by analyses focused on the adult mammary gland, and that fMaSCs may reveal new targets to aide detection, prognosis, and treatment of breast cancers. Consistent with this idea, gene expression profiling of bulk epithelium from early mammogenesis revealed significant differences with the adult (Wansbury et al., 2011). Importantly, this study did not assess whether the profiled cells exhibited stem cell activity, so the relevance of these signatures to fMaSCs remains to be determined.
Mouse mammary gland development begins at approximately embryonic day 11.5 (E11.5) with a thickening of the ventral ectoderm to generate five pairs of mammary placodes. The placodes become spherical buds by E12.5, which elongate into ductal sprouts in females by E16 (Veltmaat et al., 2003). Invasion of the extending rudiment into the fat pad precursor begins by E16.5, and by E18.5 the mammary rudiment constitutes a primitive branched network within the mammary fat pad (Veltmaat et al., 2003).
Classic rudiment transplantation studies suggest that mammary stem cells may arise coincident with the morphologic specification of the mammaray gland (Sakakura et al., 1979). For example, transplanting multiple intact E13 to E17 epithelial rudiments generated full mammary outgrowths (Sakakura et al., 1979). Heterotypic recombination experiments involving salivary mesenchyme demonstrated that the mammary epithelium is committed to develop into a mammary phenotype by E12.5 (Cunha and Hom, 1996).
Here, we quantify fMaSCs, characterize fMaSC activity during fetal mammogenesis, and analyze the relationship of their gene expression programs to those found in human breast cancer. Our data reveal the unexpected finding that fMaSCs are extremely rare in early embryogenesis, but increase rapidly as the mammary rudiment invades into the fat pad precursor. We also show that fMaSCs and their associated stroma exhibit gene expression programs related to those found in specific forms of human breast cancer.
Results
Mammary stem cells are rare early, but increase dramatically during late fetal mammogenesis
We used limiting dilution transplantation analyses (LDTA) to measure mammary repopulating unit (MRU) frequency using single-hit Poisson statistical analyses (Bonnefoix et al., 1996; Hu and Smyth, 2009; Stingl et al., 2006). Accordingly, fMRU must contain an fMaSC but may additionally require other cells or components for mammary outgrowth. However, as a single stem cell can generate a mammary gland (Shackleton et al., 2006), we will refer to fMaSC frequency when quantifying fMRUs.
We transplanted limiting dilutions of rudiment-derived cell suspensions obtained at different developmental stages of CD1 embryos into immune compromised CB17-SCID recipients (Figure 1A; Table 1 and S1) and in parallel transplanted intact mammary rudiments including surrounding mesenchyme (Figure S1A and S1D). Single intact mammary rudiments from as early as E12.5 transplanted at frequencies >70%. In contrast, measurable fMaSC activity was not evident in dissociated bulk mammary cell populations prior to E15.5 (Figure 1A and S1D; Table 1 column 2, S1 column 3, and S2). This was surprising as we transplanted 10,000 viable cells (epithelial and mesenchymal) per fat pad, which is more than we estimate to be present in a single rudiment (Figure 1A; Table 1 and S1, column 3). While rudiment dissociation could have reduced repopulation efficiency, especially if contextual cues analogous to niche interactions (Spradling et al., 2001) are required for stem cell function, identically dispersed E18.5 rudiment cells generated mammary outgrowths from 100 cells (Table S1, column 3). Thus, fMaSC concentration increases dramatically during fetal development (Figure 1A; Table 1, column 2 and S1, column 3).
Figure 1.
fMaSCs identified in late embryogenesis express high levels of CD24 and CD49f. (A) Mammary stem cell frequency estimates at various stages of fetal development and in the adult in the presence or absence of Matrigel. Gross morphological appearance of the gland at various stages is illustrated. e=epithelium, m=mesenchyme and fp=fat pad. *p < 0.001, pairwise group difference. Error bars, 95% confidence interval. (B) Confocal images showing CD49f (i-iii) and CD24 (iv-vi) expression in whole mounts at E13.5, E15.5 and E18.5. (C) Histogram and FACS contour plot showing the distribution of cells expressing CD24 and CD49f in the LIN− population (DAPI−CD31−CD45−TER119−) in mammary glands from a nulliparous adult mouse (black) and actin-eGFP E18.5 female embryos (green). Adult eGFP− mammary and eGFP+ E18.5 fetal mammary cell suspensions were mixed, co-stained and analyzed together. (D) Immunofluorescence analysis of paraffin sections of a regenerated mammary gland from a parous recipient showing Casein/K8 (i) and K14/K8 (ii). Inset, secondary antibody control. (E) Representative FACS dot plots showing very similar patterns of expression of CD24 and CD49f in viable lineage depleted mammary cells from a nulliparous adult mouse (top panel) and from a mammary gland regenerated by the fMaSCs (bottom panel). (F) Representative whole mount of actin-eGFP mammary outgrowth arising from transplantation of the fMaSC population (Lin−CD24highCD49fhigh) isolated from E18.5 embryos. The mammary glands were harvested from primary (i) and secondary (ii) recipients 12 weeks after transplantation. See also Table 1, S1 and Figure S1.
Table 1.
Fetal mammary rudiments in late embryogenesis are found to be highly enriched with mammary stem cells

|
The importance of extracellular cues in stem cell function (Spradling et al., 2001) prompted us to determine whether Matrigel would increase fMaSC transplantation efficiency as it does aMaSCs (Lim et al., 2009). While Matrigel significantly increased the sensitivity of fMaSC detection, it still equated to one or fewer stem cells per E13.5 rudiment (1/12,000 cells; Figure 1A; Table 1, column 3 and S1, column 4). fMaSC activity peaked at E18.5, increasing to 1 in 60 cells with Matrigel, a 14-fold increase compared to transplantation in its absence (Figure 1A; Table 1 and S1). Thus, fMaSC concentration increases at least 200-fold between E13.5 to E18.5 when measured in the presence of Matrigel (Figure 1A; Table 1, column 3). A striking nine-fold increase in fMaSC abundance occurred during the narrow developmental window between E15.5-E16.5 (1 in 1,800 to 1 in 200, p < 0.001) (Figure 1A; Table 1 and S1).
These quantitative measurements of stem cell frequency during embryogenesis reveal for the first time the surprising finding that transplantable fMaSCs are rare early in mammogenesis but are abundant late. Furthermore, the fMaSC frequency in unfractionated E18.5 embryonic mammary rudiments is ~5-fold higher than that obtained using bulk adult mammary cell populations measured under identical conditions (1 in 60 versus 1 in 300, p<0.001) (Table 1 and S1). These differences may, in part, reflect different contaminating epithelial and stromal components within the cell populations obtained at different stages. Nevertheless, the data indicate that mammary stem cell frequency is higher in the E18.5 mammary rudiment than in the adult gland. This facilitates their purification for subsequent molecular analyses (see below).
Stem activity is restricted to a unique fetal population expressing high levels of CD24 and CD49f
aMaSCs can be enriched using fluorescence activated cell sorting for surface markers such as CD24 (heat stable antigen, HSA) and CD49f (α6 integrin) (Stingl et al., 2006). Both proteins were expressed in the stromal and epithelial compartments of E13.5 and E15.5 mammary rudiments (Figure 1B). In contrast, high CD24 and CD49f expression at E18.5 identifies a basal-epithelial compartment with negligible staining in the fat pad and surrounding mesenchyme, suggesting the utility of these markers for fMaSC enrichment (Figure 1B). Consistent with this, flow cytometry demonstrated that most E13.5 and E15.5 rudiment cells expressed CD24 and CD49f (Figure S1B). At E18.5, however, these markers delineate a distinct subpopulation comprising approximately 5% of the cells following endothelial and hematopoietic lineage exclusion (Figure 1C and S1B). We then compared the CD24 and CD49f staining profiles of admixed and co-processed eGFP− adult and eGFP+ E18.5 fetal mammary cells (Figure 1C). Cells derived from E18.5 rudiments express higher levels of CD24 than the previously reported CD24medCD49fhigh aMaSC-enriched population and higher levels of CD49f than the CD24highCD49flow adult luminal CFC population (Colony Forming Cells) (Figure 1C) (Stingl et al., 2006). This direct comparison with adult mammary cells shows that the fetal population is CD24highCD49fhigh.
The CD24highCD49fhigh subpopulation contained all fMaSC activity by transplantation analyses (Figure 1A; Table 1). The remaining fetal population exhibits lower CD24 levels, is devoid of fMaSC activity, and is enriched in stromal cells as defined by cellular morphology, protein and gene expression analyses (see below). Therefore, we call this population the fetal stroma-enriched population (fSTR). As few as 5-10 fMaSCs reproducibly enabled robust mammary gland repopulation, while up to 3,000 cells from the fSTR consistently failed to generate outgrowths, even with Matrigel addition (Figure 1A; Table 1 and S1). We estimate the stem cell frequency in the fMaSC-enriched population to be 1 in 14 with Matrigel (Table 1 and S1; Figure 1A), which is an ~4-fold enrichment over the aMaSC frequency using immune-compromised hosts in the presence of Matrigel [1/50 (aMaSC) versus 1/14 (fMaSC), p<0.001) (Table 1 and S1)]. Importantly, the aMaSC frequency with Matrigel using allo-transplantation into immune-compromised hosts was similar to that obtained using an immune competent, syngeneic model (C57BL6) in the absence of Matrigel (Table S1 and S3), and host immune competence did not significantly affect fMaSC frequency in the presence of Matrigel (Table S3).
fMaSCs generated morphologically normal, fully arborized ductal structures that produced casein positive alveolar structures upon induction of pregnancy (Figure 1D). Mammary outgrowths exhibited the expected localized expression of luminal and myoepithelial keratins (K8 and K14, respectively) (Figure 1D), the phenotypic cellular heterogeneity of wild-type adult mammary glands (Figure 1E), and contained cells able to self-renew based on serial transplantation analyses (Figure 1F). Thus, the fMaSC-enriched population exhibits the multi-lineage cell differentiation and self-renewal characteristics expected of aMaSCs, but at considerably higher concentration (Shackleton et al., 2006; Stingl et al., 2006).
fMaSCs are multipotent and co-express markers of multiple mammary lineages
We evaluated the ability of individual fMaSCs to generate multiple lineages in vitro (Dontu et al., 2003). While fMaSCs had negligible sphere forming efficiency (SFE) using a conventional non-adherent sphere forming protocol at low seeding density (1,000 cells per cm2) (SFE ≈ 0.1%; Figure 2A and 2B), fSTR formed numerous spherical clusters using identical conditions (SFE=1.4%) (Figure 2A and 2B). However, just as Matrigel profoundly increased transplantation efficiency, even low percentages of Matrigel (2%) enabled the fMaSC population to generate spheres with an SFE of 9.4% when plated at low density (Figure 2A and 2B). The primary fMaSC-derived spheres were morphologically similar to the colonies previously reported for the aMaSC population (Figure 2A) (Stingl et al., 2006). In addition, fMaSC-derived primary spheres expressed markers associated with both myoepithelial (e.g., cytokeratin 14, K14) and luminal (cytokeratin 8, K8) epithelial lineages of the mammary gland (Figure 2C).
Figure 2.
Individual cells from the fMaSC population generate clonal, multi-lineage spheres that can be serially propagated, and co-express markers of multiple lineages. (A) Morphology of structures generated from fMaSC (i) and fSTR (ii) populations grown under non-adherent conditions in vitro, in the presence and absence of Matrigel. (iii) Confocal image of an fSTR polyclonal sphere derived by mixing fSTR cells from WT and actin-eGFP transgenic embryos showing Vimentin immunofluorescence (red), nuclear counterstain DAPI (blue), and actin-eGFP (green). Scale bar, 50μm. (B) Quantification of clonal, primary fMaSC-derived sphere growth, secondary and tertiary sphere growth, and fSTR-derived sphere growth. Error bars, standard deviation. (C) Confocal immunofluorescence analysis of spheres derived from the fMaSC population showing the expression of K8 (red), K14 (green) or both (yellow) with nuclear counterstain DAPI (blue) and tabular summary of sphere types observed. Type 1 spheres consist of cells expressing either K14 (i.e. sphere periphery) or K8 (i.e. middle of the sphere), while type 2 spheres consist mainly of cells co-expressing K8 and K14 (yellow cells). Inset, secondary antibody control. Scale bars, 25μm. Inset, secondary antibody control. Scale bars, 50μm. (D) Summary of the percentage of cells in the fMaSC and fSTR populations expressing K8, K14, and/or Vimentin. Error bars, standard deviation. See also Figure S2.
We used two independent strategies to determine whether primary spheres arise from clonal expansion from a single cell or from cell aggregation. First, we seeded single cells from the fMaSC population into individual wells. Primary spheres formed (SFE=10.7%) in 2% Matrigel, similar to the 9.4% SFE observed when the cells were plated at low density (Figure 2B and S2A). Secondary and tertiary spheres were also formed with similar SFEs (~10%; Figure 2B). The fSTR did not generate spheres in the presence of Matrigel, and instead produced cultures of dispersed cells resembling fibroblasts and neurons (Figure 2Aii and 2B). Second, we mixed single cell suspensions of eGFP+ and eGFP− cells from the fMaSC population, and then grew them at low density (Figure 2B and S2B). As 199 out of 200 spheres were a single color, the vast majority must derive from single cells (Figure S2C). Approximately 60% of fSTR derived clusters were overtly polyclonal when cultured in non-adherent conditions without Matrigel (Figure 2Aiii), indicating they arise by aggregation. Taken together, these data show that ~10% of the fMaSC population exhibits the stem cell properties of multipotent differentiation and self-renewal in vitro (Figure 2B and 2C).
Co-expression of proteins associated with multiple lineages has been proposed to indicate plasticity in the normal mammary gland and in breast cancers (Creighton et al., 2009; Livasy et al., 2006; Petersen and Polyak, 2010; Sun et al., 2010; Thomas et al., 1999). We detected cells that co-express the K14 and K8 from as early as E13.5 in the developing mammary gland (Figure S2D,E). Approximately 30% of the cells within the fMaSC population were K14+K8+, and we frequently detected such double positive cells in fMaSC-derived spheres (Figure 2C and 2D). We also analyzed vimentin expression as it has been associated with the myoepithelial and mesenchymal lineages of the normal mammary gland and with aggressive disease when co-expressed with luminal epithelial markers in breast cancer patients (Creighton et al., 2009; Thomas et al., 1999). Approximately 70% of cells within the K14+K8+ fMaSC population also expressed vimentin (Figure 2D).
Derivation of fMaSC and fStromal specific gene expression signatures
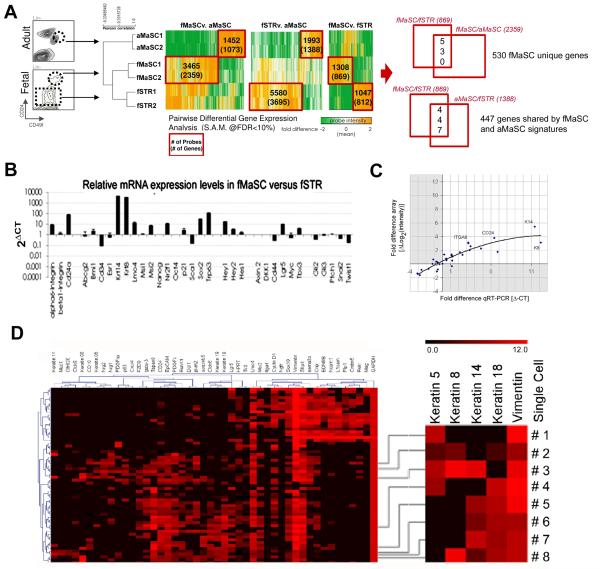
We performed microarray expression analyses on the fMaSC, fSTR, and aMaSC populations to ascertain molecular pathways with potential relevance to fetal mammary development, fMaSC biology, and breast cancer. We obtained reproducible expression profiles from independent biological pools representing each population and identified differentially expressed genes comprising fMaSC, fSTR and aMaSC signatures (Figure 3A; Table S4; Significance Analysis of Microarrays; FDR<10%; (Tusher et al., 2001)). We identified 869 unique genes more highly expressed in the fMaSC population (the fMaSC signature) than in the fSTR, and 812 unique genes more highly expressed in the fSTR population (the fSTR signature) than in the fMaSC. Among the fMaSC signature genes, ~34% were common to both the fMaSC and aMaSC populations when compared to fSTR, but ~40% were significantly overexpressed in the fMaSC relative to the aMaSC (Figure 3A).
Figure 3.
Differential gene expression profiling of fMaSC, fSTR and aMaSC populations. (A) Illustration of sorted populations, Pearson correlation among biological replicates for each cell type and heat maps illustrating the identification of differentially expressed genes (SAM; FDR<10%). (B) qRT-PCR analysis of select stem cell and developmental genes in the fMaSC population relative to fSTR. (C) Expression levels of a representative selection of genes determined by microarray and by qRT-PCR. The expression level in the fMaSC relative to the fSTR is plotted as the fold difference in expression. Fold differences in gene expression were calculated for RT-PCR assuming ideal amplification (fold change = 2ΔCt) and for Nimblegen array data using the normalized probe intensities (fold change = ΔLog2(intensity)). Data were normalized to HPRT. Despite differences in the dynamic range of the two techniques, the pattern of differential expression between the fMaSC and fSTR determined by array was consistent with the pattern determined by qRT-PCR. (D) Microfluidics-based, single-cell, qRT-PCR analyses of cells from the fMaSC population. Right panel, examples of single cells co-expressing various keratins and the mesenchymal marker, vimentin. See also Table S4.
We confirmed the differential gene expression patterns between the fMaSC and fSTR populations on a panel of genes selected from putative stem cell, developmental and cancer relevant pathways (Figure 3B and 3C). Furthermore, high throughput single-cell qRT-PCR analyses confirmed expression of a partially overlapping selection of 46 genes in individual cells of the fMaSC population (Figure 3D). This approach also verified that individual fetal cells co-express luminal keratins, myoepithelial keratins and vimentin (Figure 3D).
Unique expression features of fMaSCs and fSTR
Many well-studied genes were found to be expressed in a manner consistent with the cell types analyzed, indicating the validity of the microarray data (Table S4). However, the fMaSC and fSTR signatures revealed unique gene expression patterns when compared to adult mammary populations or to those isolated earlier in development that we showed to be lower in stem cell content than the fMaSC population (Figures 1 and 4; Table 1, S4 and S5). For example, qRT-PCR analysis showed significant differences in expression of specific stem cell and development related genes between the fMaSC and the E15.5 rudiment (Figure 4A). The gene content in the E18.5 fMaSC and fSTR signatures were also significantly different from those reported for either mouse or human adult mammary, or from E12.5 mouse mammary epithelia (ME) and mammary mesenchyme (MM) (Figure 4B-D, Table S4 and S5) (Kendrick et al., 2008; Lim et al., 2009; Lim et al., 2010; Pece et al., 2010; Stingl et al., 2006; Wansbury et al., 2011). Although the similarities between the fMaSC and previously reported primary mammary epithelial signatures are statistically significant, the majority of genes in the fMaSC signature are not represented in aMaSC signatures (Figure 4B). In contrast, the fSTR signature is similar to the adult mammary stromal signature, and is surprisingly similar to published aMaSC signatures (Figure 4B).
Figure 4.
Unique gene content in the E18.5 fetal mammary. (A) qRT-PCR analysis of select stem cell and developmental genes in fMaSC relative to E15.5 mammary rudiment. (B) Overlap of fMaSC signature genes and their orthologues with previously reported normal adult mammary signatures. The upper panel shows mouse signatures (Lim et al., 2009; Pece et al., 2010). The lower panel shows hMaSC and hStromal signatures (Lim et al., 2010), and a signature from cultured hMaSCs (Lim et al., 2009; Pece et al., 2010); p-values represent the hypergeometric probability based on all 20,309 probes in the mouse array and 19,828 probes in the human arrays. (C) Identification of genes unique to fMaSC and fSTR populations (Venn diagrams) and clustering of expression array data for these genes for fMaSC (f), fSTR (s), aMaSC (a), E15.5 mammary rudiments (b, “buds”), and Lineage depleted adult mammary epithelium (e) (heat maps). (D) Gene ontology enrichment analysis of genes unique to the fMaSC and fSTR signatures. Each globe represents an ontological category and the size of the globe represents the number of genes in the category. Significantly enriched categories are color coded in red for fMaSC and blue for fSTR (Benjamini-Hochberg adjusted FDR=5%). The organic layout algorithm used (Cytoscape), allows visualization of dense ontological data and the observation that many categories are enriched for each signature type. The most highly enriched categories are color coded in orange and the categories with the lowest p-values including, gene names contributing to the most enriched ‘biological process’ for each population, is listed to the right. See also Figure S4 and Table S4,S5.
We delineated genes specific to the fMaSC and fSTR populations by comparing their signatures to composite human/mouse adult MaSC or stromal gene lists and to E12.5 ME and MM signatures (Figure 4C). The sets of genes specific to the fMaSC and fSTR signatures are rich in biological content as indicated by their significant correlation with numerous gene ontology (GO) categories (Figure 4D and S4; Table S4 and S6). Among these, cell signaling and genes associated with the plasma membrane figure prominently (Figure 4D). A selection of the most highly enriched clusters are detailed in Figure 4D, including several genes previously implicated in mammary stem cell function and breast cancer, such as ErbB2 and ErbB4 (Korkaya and Wicha, 2009). Several genes reciprocally expressed in the fSTR and fMaSC populations are suggestive of paracrine signaling and may contribute to stem cell behavior in normal or neoplastic growth. For instance, the fSTR specific signature includes Nodal and Wnt5a. Nodal is a TGF-β family morphogen that can promote oncogenic phenotypes in mammary cells and has been implicated in breast and other cancers (Strizzi et al., 2009). The Wnt5a protein is a non-canonical Wnt implicated in polarity, migration and stem cell maintenance (Kikuchi et al., 2011). Additional processes found in both the fMaSC and fSTR populations are likely to contribute to the unique properties of mammary cells at this stage. For example, changes in chromatin regulation, augmented synthetic metabolism and cell cycle, and the production of distinctive extracellular matrices may contribute to the robust fMaSC function we observe (Figure S4, Table S4).
Cellular interaction is a predicted hallmark of fMaSC function
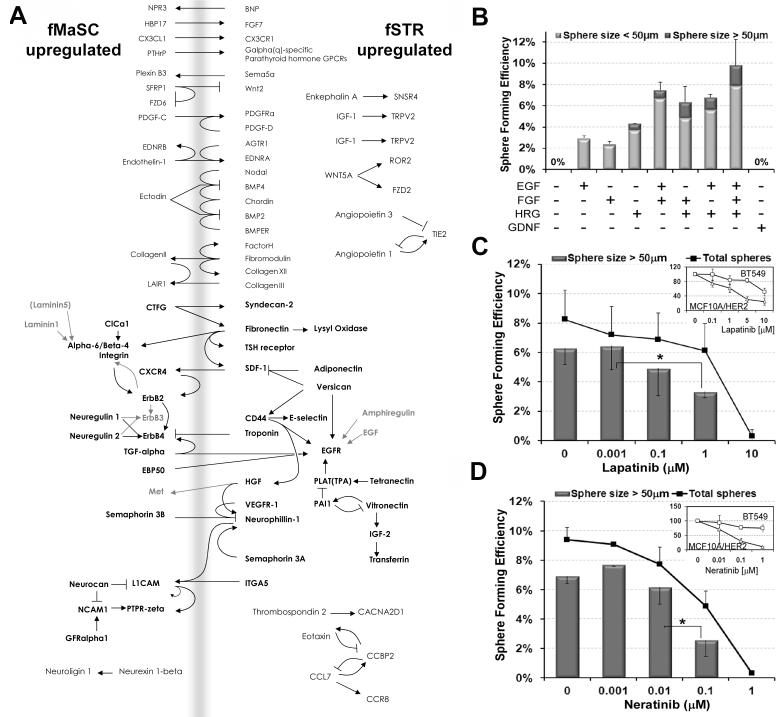
GO enrichment analysis of fetal signatures suggested a prominent role for cell-cell and cell-niche interactions, including cell surface receptor signaling in the fMaSC population (Figure 4; Table S4). We used curated interaction networks in the GeneGo pathway analysis platform to organize the genes comprising the fMaSC and fSTR signatures into potential receptor-ligand interactions. This enabled construction of a hypothetical interaction map based on reported receptor-ligand interactions (Figure 5A).
Figure 5.
Prediction and validation of non-autonomous signaling in fMaSC function. (A) A model constructed from fetal gene signatures filtered for receptors and ligands using the GeneGo pathway analysis platform. The model illustrates candidate protein-protein interactions including receptor-ligand pairs expressed reciprocally in the fMaSC (left) and fSTR (right) populations. Additional gene products of interest predicted to interact with the network are also indicated (gray). The map suggests that ErbB signaling, among other pathways, may play a prominent role in fMaSC function. (B) Quantification of fMaSC-derived spheres in the absence and presence of growth factors suggested by the model in A. (C) Quantification of fMaSC-derived sphere growth upon inhibition of ErbB1/2 signaling by either Lapatinib or inhibition of ErbB1/2/4 signaling by Neratinib. *p < 0.05, Student’s t test. (D) Dose response curves to Lapatinib and Neratinib in resistant human BT549 and sensitive MCF10A/HER2 cell lines (Wang et al., 2006; Weigelt et al., 2010). All error bars, standard deviation. See also Figure S5.
We determined whether predicted pathways are relevant for fMaSC function in vitro (Figure 5B-D and S5). We analyzed ErbB and FGF receptors and their ligands given their cancer relevance, and that growth of adult mammary epithelial cells in vitro requires either EGF or FGF (Dontu et al., 2003). qRT-PCR validated the differential expression of all four ErbB family members, and the hormone receptors ER and PR (Figure S5A, and data not shown). ErbB4 was expressed at a low level but exclusively in the fMaSC population (data not shown). ErbB2 and ErbB3 were expressed more highly in the fMaSC population than either the fSTR or aMaSC populations (Figure S5A), and ErbB2 protein was detected in situ in CD24+ cells in E18.5 mammary rudiments (Figure S5B).
We examined the requirement for ErbB and FGF signaling by growing fMaSC derived spheres in 2% Matrigel culture containing or lacking specific ErbB ligands and FGF. Cultures lacking both EGF and FGF produce no spheres, while either EGF or FGF dramatically stimulated fMaSC-derived sphere formation (Figure 5B). Interestingly, Heregulin 1 (Hrg1, Neuregulin, Neu Differentiation Factor), an ErbB ligand with preference for ErbB3 or ErbB4, stimulated sphere growth in the absence of EGF and FGF (Figure 5B) (Britsch, 2007). The effects of these ligands were additive (SFE ~10% for EGF/FGF/HRG), with Hrg1 showing the most dramatic effect on large sphere production (Figure 5B). By contrast, GDNF, which is not represented in the hypothesized interaction network, did not stimulate sphere formation (Figure 5B).
Consistent with the above results, ErbB and FGFR kinase antagonists inhibited fMaSC-derived sphere growth (Figure 5C,D and S5). Lapatinib is a reversible and highly specific ErbB1/2 dual tyrosine kinase inhibitor (Rusnak et al., 2001), while Neratinib is an irreversible pan-ErbB kinase inhibitor (Rabindran et al., 2004). Lapatinib and Neratinib inhibited sphere growth with a similar dose-dependence as observed for Her2 over-expressing human mammary cells with documented sensitivity to these agents (Figure 5C,D) (Wang et al., 2006). As these drugs have non-overlapping potential off-target effects (Karaman et al., 2008; Rabindran et al., 2004; Rusnak et al., 2001), it is most likely that their effects on sphere growth derive from ErbB pathway antagonism. However, it remains to be determined whether inhibition of one specific ErbB receptor accounts for the observed effects on sphere growth, or whether redundancy in this family necessitates inhibition of multiple receptors for effective fMaSC growth antagonism in vitro. Altogether, these findings substantiate the importance of ErbB and FGF signaling in fMaSC-derived sphere growth in vitro, and they indicate the presence of functionally relevant gene content in the microarray derived fetal mammary signatures.
Molecular links between fetal mammogenesis and breast cancer
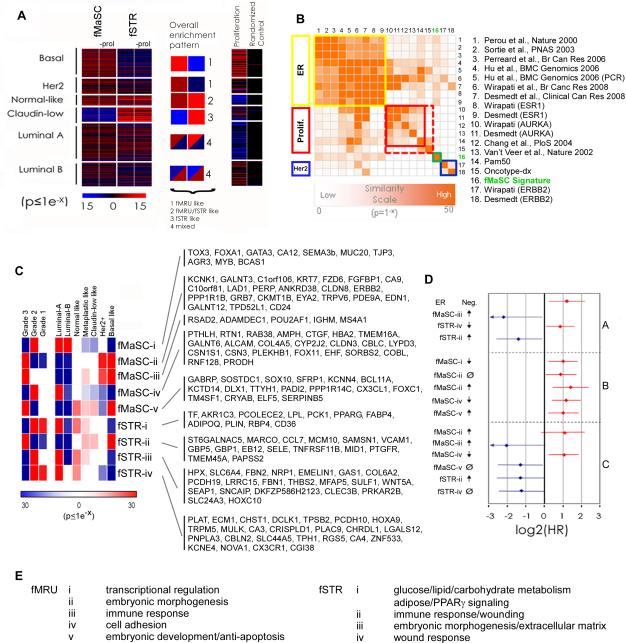
Cancer associated genes (ErbB2, Met, CXCR4, etc.) were prominent among the fetal signatures, pathway analyses, and in unsupervised gene set enrichment analyses (Figure 4,5 and S5; Table S4). Therefore, we determined whether fetal gene expression signatures were enriched in particular human breast cancer intrinsic subtypes using archival tumor microarray data from two independent compendia and human orthologues of the fMaSC and fSTR signature genes (Figure 6 and S6; Table S6) (Ben-Porath et al., 2008; Prat et al., 2010). Enrichment for the fMaSC signature was concentrated among tumors designated as basal-like, which tend to be poorly differentiated and stem-like (Ben-Porath et al., 2008; Mizuno et al., 2010). In addition, many Her2+ tumors showed significant enrichment for the fMaSC signature (Figure 6A and S6A). Enrichment for the fSTR signature often correlated with tumor subtypes characterized by low proliferation and favorable prognoses (Figure 6A and S6A) (Sorlie et al., 2001). However, claudin-low and metaplastic-like tumors, which have also been suggested to be stem-like (Hennessy et al., 2009; Perou et al., 2010; Prat et al., 2010) were generally enriched for the fSTR signature and depleted for the fMaSC signature (Figure 6A and S6A). Breast cancers showing enrichment for fSTR signatures showed nearly identical enrichment patterns for aMaSC signatures (Figure S6B), consistent with the significant gene overlap of aMaSC and fSTR signatures noted above (Figure 4B).
Figure 6.
Fetal mammary gene expression patterns provide molecular links to human breast cancers. (A) Significant correlation between fMaSC and fSTR gene signatures and human breast cancers (n=337) (Prat et al., 2010) are indicated by horizontal bars, each representing the gene expression profile from an individual tumor sample. Red bars indicate tumors enriched in fetal signature expression, blue bars indicate signature repression. Black bars indicate no significant correlation. Larger colored squares illustrate the trend for each intrinsic subtype. For comparison, a randomized signature of equivalent size and a proliferation signature (Ben-Porath et al., 2008) are shown. (B) A comparison of several signatures and clinical metrics by significance of gene overlap. Most signatures are closely related and are significantly associated with ER (yellow box) or Proliferation (AURKA; red box) related signatures. Because of its size, the small OncotypeDX signature shows modest significance values for the proliferation group, although it includes several proliferation ER and Her2 related genes. The fMaSC signature (green box and arrows) is relatively unique showing no significant overlap with proliferation or ErbB2/Her2 related signatures (blue) and relatively low association with ER related signatures. (C) Significance of enrichment for sub-signatures among diverse breast cancers in a large microarray compendium (n=1211) (Ben-Porath et al., 2008). Enrichments according to subtype and grade are indicated by colored squares that represent probabilities for the percentage of tumors enriched or repressed in each annotation group. Genes comprising each sub-signature are listed. (D) Sub-signatures showing significance in multivariate analysis (p<0.1) are graphed for models including the following categorical clinical variables: A: ER status, Grade, Lymph-node status, Tumor size; B: Grade, Lymph-node status and Tumor size; C: ER status, Lymph-node status and size (NKI295). A positive (or red) value indicates a poorer prognosis, while a negative (or blue) value indicates a better prognosis. The associated hazard for ER-negative tumors is shown in model A for comparison. ↑=sub-signature enrichment, ↓=sub-signature repression, Ø=no significant signature enrichment and/or depletion. (E) Biological functions associated gene constituents of the sub-signatures (gene set enrichment p<0.05). See also Figure S6, S7 and Table S6-8.
Previous studies have used signatures derived either directly from breast cancer array data or from specific biological contexts, such as serum stimulation of fibroblasts (simulating wound healing), to classify breast cancers into different tumor types with distinct clinical features (Fan et al., 2006). The fMaSC signature exhibits relatively little overlap with these signatures (6.5% of fMaSC genes shared, Figure 6B; Table S7). However, the signatures compared in Figure 6B have significant representation of ER and/or proliferation associated genes (Fan et al., 2006; Wirapati et al., 2008). In contrast, the fMaSC and fSTR signatures have little representation of proliferation genes (Figure 6B) since this is a characteristic they share, leading to exclusion from their comparative profiles. Furthermore, removal of the few residual proliferation-related genes from the fMaSC and fSTR signatures did not markedly alter the observed tumor enrichments (Figure 6A and S6A). We cannot rule out the possibility that fetal-like molecular programs are also invoked by other proliferative states in the mammary gland, for instance at puberty, pregnancy or during outgrowth of transplanted material. Regardless, the fMaSC and fSTR signatures clearly identify a novel group of genes associating fetal mammary gland biology and fMaSCs with specific molecularly defined breast cancer subtypes (Figure 6B).
Fetal gene subsets identify patients with diverse prognoses in archival tumor samples
The fMaSC and fSTR signatures can be sub-divided into gene expression modules showing coordinated expression across multiple tumors using hierarchical clustering analysis (Figure S6C,D). We then correlated these sub-signatures with breast cancer grade, progression, subtype or prognosis (Figure 6C, S6 and S7). We subdivided the fMaSC and fSTR signatures into five and four sub-signatures, respectively using the 96 genes with the greatest variance in the compendium. The genes comprising these enriched subsets represent diverse biological processes previously implicated in cancer, including immune response (fMaSC-iii, fSTR-ii), cell survival (fMaSC-v), and wounding (fSTR-ii, fSTR-iv) (Figure 6C,6E and Table S7) (Chang et al., 2004; Perou et al., 2000; Rody et al., 2011).). Other process such as embryonic morphogenesis (fMaSC-ii, fMaSC-v, fSTR-iii) and adhesion (fMaSC-iv, fSTR-iii), which have not been extensively investigated in cancer, were also represented (Figure 6E and Table S7).
These fetal sub-signatures exhibit prognostic relevance in archival breast cancer array data (Figure 6C,D and S7). For instance, enrichment for signature fMaSC-ii or repression of signature fSTR-iv correlated with Her2+ and basal-like tumors, high grade, and reduced probability of patient survival (Figure 6C and S7). This observation is consistent with the predicted outcome of these intrinsic subtypes (Sorlie et al., 2001). In addition, multivariate survival analyses based on enrichment for the fetal sub-signatures showed prognostic value beyond commonly used clinical metrics such as ER status, tumor size, grade and lymph node status (Figure 6D). For example, although enrichment for sub-signature fMaSC-iii correlates with high grade, Her2+ and basal-like tumors, it also correlates with patients having a higher probability of survival in the compendium analyzed (Figure 6C,D). The biological and prognostic relevance of the signatures described here is a function of their biological origin, as randomized signatures are not enriched in a sufficient number of tumors to enable tumor classification and subsequent survival analysis (Figure 6A and S7). However, it may be possible to derive alternative fetal gene sub-signature groupings exhibiting enhanced prognostic value, predictive value, or additional functional biological insight by using alternative statistical approaches. As approximately 60% of the genes comprising these fetal sub-signatures are specifically upregulated relative to the aMaSC population (Table S4, S5 and S7), these signatures provide novel candidates for therapeutic and prognostic strategies that would likely be missed by deriving signatures from the resting adult gland.
Discussion
The existence of fMaSCs has been inferred from studies demonstrating that intact mammary epithelium obtained from as early as E13.5 can fully reconstitute the mammary gland (Sakakura et al., 1979). However, these studies did not quantify or purify mammary stem cells. This left a substantial gap in our understanding of mammary biology and precluded elucidation of the long predicted molecular and genetic links between fetal mammary development, stem cells and breast cancer (Howard and Ashworth, 2006). Here, we provide the first quantitative assessment of mammary stem cell activity during fetal mammogenesis, obtain fetal mammary gene expression profiles and evaluate their relationship to breast cancer.
Our studies reveal the surprising finding that mammary rudiments from E15.5 and earlier contain few if any functional fMaSCs. We observed a 200-fold increase in fMaSC activity during the course of fetal mammogenesis that parallels the change in cellular context as the proliferating mammary epithelium begins to invade through the adjacent mesenchyme and interacts with the fat pad microenvironment (Veltmaat et al., 2003). While proliferation during mammogenesis likely contributes to fMaSC abundance, the nine-fold increase in fMaSC frequency between E15.5-E16.5 is difficult to explain solely by cell division. Instead, we propose that stromal interactions during this interval generate signals that act on precursor cells to engender the stem cell competence we assay by transplantation. It is noteworthy that a recent in vivo lineage tracing study also demonstrated the existence of bipotent mammary stem cells in late embryogenesis, and suggested a restriction to unipotent stem/progenitor activity occurring shortly after birth (Van Keymeulen et al., 2011).
The hypothesis that context and extrinsic cues underlie fMaSC functional identity is consistent with studies showing the importance of cell-cell and cell-matrix interactions, and locally produced soluble factors for stem cell function (Jones and Wagers, 2008; Spradling et al., 2001). Direct niche interactions also maintain the stem cell state within various tissues and organisms, such as Drosophila testes and mammalian hair follicle, bone marrow, testes, and intestine (Spradling et al., 2001). Stem cell niches also produce soluble factors, including Wnt, FGF, TGFβ, and EGF ligands, which promote or maintain the stem cell state (Spradling et al., 2001; Zeng and Nusse, 2010). Importantly, the involvement of multiple ErbB receptors and their ligands in mammary morphogenesis (Jackson-Fisher et al., 2008; Jackson-Fisher et al., 2004; Tidcombe et al., 2003; Wansbury et al., 2011) is consistent with our observations implicating this family in fMaSC function in vitro. The ErbB kinase inhibitor studies reported here and gene knockout studies showing that mammary gland development is impaired to differing degrees in various ErbB knockout mouse strains (Jackson-Fisher et al., 2008; Jackson-Fisher et al., 2004; Tidcombe et al., 2003) suggest that interactions with relevant stromal components and growth factor gradients may be important for inducing stem cell activity during development.
The fMaSC population includes cells that co-express luminal and myoepithelial markers with vimentin. The expression of vimentin within epithelial cells of the human adult mammary gland is normally restricted to the myoepithelial lineage and has not been reported to occur in concert with luminal keratin expression (Anbazhagan et al., 1998; Mork et al., 1990). Interestingly, forced co-expression of luminal keratins 8 and 18 with vimentin in human breast cancer cells in vitro increases motility, invasiveness, and proliferation (Hendrix et al., 1997). Similarly, basal-like breast cancers frequently exhibit an undifferentiated phenotype and co-express myoepithelial and luminal epithelial keratins and vimentin (Livasy et al., 2006). Our data suggest that the co-expression of myoepithelial and luminal keratins and vimentin may typify an uncommitted, embryonic, fMaSC-like state. We suggest that the partial epithelial to mesenchymal transition (EMT) commonly observed during aggressive tumorigenesis may represent a reversion to an embryonic-like state resembling the fMaSC and/or fStromal compartments (Hanahan and Weinberg, 2011). EMT has long been recognized as an essential embryonic process required for development beyond the blastula stage (Hay and Zuk, 1995) and may also promote a stem cell-like state in breast cells (Mani et al., 2008; Thiery and Sleeman, 2006).
fMaSC signatures are derived from cells with a defined biological role and have not been analyzed previously for their relationship to cancer. Other signatures representing biological processes such as wound healing and immune response have proven useful for gauging the risk of recurrence in some breast cancer subtypes (Chang et al., 2004; Rody et al., 2011). Thus, we anticipated that our analyses would uncover new genes and pathways related to fetal mammary development, fMaSC function, and breast cancer. Our results suggest that this resource contains new gene sets with prognostic value that may also be useful for predicting which patients will respond to certain treatment strategies. For example, patients receiving ErbB (Her) targeted therapies, such as Herceptin (Trastuzumab) and Lapatinib, are selected based on ErbB2 gene amplification and high level ErbB2 expression within their tumors (Jacobs et al., 1999). However, in the NSABP-31 clinical trial, some patients confirmed by clinical standards to be ErbB2 negative responded to the ErbB targeted treatment regimen (Paik et al., 2008). Our results show that fMaSCs, which would likely also be designated as ErbB2 negative using accepted clinical guidelines, are sensitive to ErbB pathway inhibitors. We speculate that tumors acquiring an fMaSC-like state will rely on ErbB pathway signaling and, therefore, be sensitive to ErbB antagonists despite being clinically designated as ErbB2-negative. The disproportionate representation of proliferation, ER, and Her2 related signaling in many existing prognostic signatures may mask less prominent yet critical signaling pathways that can be uncovered by studying normal developmental paradigms such as the fMaSC and fStromal states that are perturbed in cancer.
Links between embryogenesis and tumorigenesis were first proposed in 1838 by Müller as the stem cell origin of cancer and then extended by Durante and Conheim’s hypothesis that elements remaining in an undifferentiated embryonic state or that reacquire characteristics of this state generate malignancies (Brewer et al., 2009; Sell, 2010). Subsequent descriptions of onco-fetal proteins, identification of embryonic stem cell genes, splice isoforms, microRNAs, and embryonic metabolism in cancer add credence to this concept (Brewer et al., 2009; Christofk et al., 2008; Powers and Mu, 2008; Sell, 2010). Our identification of a novel population of fMaSCs and associated stroma with gene expression signatures enriched in different types of breast cancer further support the importance of understanding both components and their interaction during cancer progression.
We suggest that cells resurrecting the programs that govern fetal tissue stem cells and fetal stroma may subsequently fuel tumor progression in the adult. This raises the question of how cells eliciting such programs arise during tumor progression. In some breast cancers, oncogenic lesions, such as loss of p53, may impart developmental plasticity, either directly or through reprogramming of tumor cells to more primitive states, including those resembling fMaSC or fStroma (Mizuno et al., 2010; Spike and Wahl, 2011). In this regard, the gene expression network we report involving both the fMaSCs and their associated stroma will provide a resource for generating new molecular hypotheses linking development and cancer, developing new diagnostic and prognostic metrics, and identifying candidate therapeutic targets.
Experimental Procedures
Mice and embryos
CD-1 and CB17-SCID were purchased from Charles River. Actin-eGFP mice were maintained on a CD-1 mixed background or pure C57BL/6J (Jackson Labs).
Cell preparation
Adult mammary glands were dissociated according to the Stem Cell Technologies (SCT) protocol. For Fetal mammary glands, Collagenase/Hyaluronidase digestion time was reduced to 90 minutes and the Trypsin treatment was omitted.
Flow cytometry
Single cell suspensions were incubated with DAPI and the following antibodies: Fc receptor (2.4G), biotinylated CD31/CD45/TER119 cocktail, CD24-PE (M1/69), CD49f-FITC all from SCT and streptavidin-PerCPcy5.5 (BD Biosciences).
Mammary transplantation
Mammary transplantation (Deome et al., 1959) was carried out using pulled glass capillaries and mouth pipetting. Transplanted glands were evaluated 6-12 weeks post-surgery.
Immunofluorescence
Whole mounts, paraffin sections, OCT sections, or cytospins were stained with antibodies to: Keratin 14 (AF-64, Covance, 1:1000), Keratin 8 (Troma-1, DSHB, 1:100), CD24 (M1/69, BD Biosciences, 1:1000), CD49f (GoH3, BD Biosciences, 1:1000), Casein (a gift from G.Smith and D. Medina, 1:25), Vimentin (AB5733, Chemicon, 1:1000) and ErbB2 (29D8, Cell Signaling, 1:500).
Three-dimensional, in vitro culture
Suspension mammosphere culture: freshly sorted cells were plated on ultra-low adherence plates (Corning) at 1000 cells/cm2 in Epicult-B mouse media containing B-supplement, rhEGF, rhbFGF, Heparin and Penicillin/Streptomycin. 2% Matrigel culture: cells were plated on ultra-low adherence plates (Corning) at 1000cells/cm2 in mammosphere media supplemented with 2% Matrigel (growth factor reduced, BD Biosciences). 100% Matrigel culture: freshly sorted cells were seeded on top of a 30ul bed of Matrigel (growth factor reduced) in 2% Matrigel media. Vehicle (DMSO), Lapatinib (LC Laboratories), Neratinib (HKI-272, Pfizer) or FGFR inhibitor (PD173074, gift from the Verma lab, Salk Institute) were added at the indicated doses. Clonal sphere culture. Single cells were sorted into 96 well, low adherence plates at a single cell per well density in 2% Matrigel media. eGFP+ and eGFP− mixing experiments: The eGFP+ and eGFP− fMaSC and fSTR populations were mixed in a 1:1 or 1:4 ratio. fMaSCs were seeded at low density on top of Matrigel (growth factor reduced) in 2% Matrigel media and fSTR cells were seeded at both low and high densities in mammosphere media.
Microarray and bioinformatic analysis
RNA was linearly T7-amplified and gene expression was measured using a Nimblegen Array (12x135k MM9; Roche Nimblegen). The data is available at the gene expression omnibus at http://www.ncbi.nlm.nih.gov/under accession GSE27027. Data was RMA normalized and processed using Excel, TIGR-MeV, Genomica, Cytoscape and MedCalc softwares, and the DAVID website. Differential expression was determined using SAM at FDR<10% (Tusher et al., 2001). Detailed experimental and statistical methods accompany this manuscript as supplemental information.
Supplementary Material
Acknowledgements
We dedicate this work to Evelyn Lauder, visionary founder and inspirational leader of the BCRF, and Madame Corinne Mentzelopoulos for generously supporting the Salk Institute. We thank J.C. Izpisua Belmonte, T. Bonnefoix, G. Cunha, C. Eaves, P. Eirew, L. Ellies, P. Gray, J. Green, S. Kinkel, A. Legler, D. Medina, C.M. Perou, A. Prat, A. Rodrigues, B. Vonderhaar, and Wahl Lab members for invaluable advice, Pfizer for Neratinib, and BCRF, DOD-BCRP, Susan G. Komen for the Cure, the G. Harold and Leila Y. Mathers Charitable Foundation, T32 training grants CA009523 (BTS), GM07240 (DDE), CA009370 (JCL), California Breast Cancer Research Program 15GB-0015 (DDE) for funding, and NCI 5P30CA014195 for core resource support.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anbazhagan R, Osin PP, Bartkova J, Nathan B, Lane EB, Gusterson BA. The development of epithelial phenotypes in the human fetal and infant breast. J Pathol. 1998;184:197–206. doi: 10.1002/(SICI)1096-9896(199802)184:2<197::AID-PATH992>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- Ben-Porath I, Thomson MW, Carey VJ, Ge R, Bell GW, Regev A, Weinberg RA. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet. 2008;40:499–507. doi: 10.1038/ng.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnefoix T, Bonnefoix P, Verdiel P, Sotto JJ. Fitting limiting dilution experiments with generalized linear models results in a test of the single-hit Poisson assumption. J Immunol Methods. 1996;194:113–119. doi: 10.1016/0022-1759(96)00077-4. [DOI] [PubMed] [Google Scholar]
- Brewer BG, Mitchell RA, Harandi A, Eaton JW. Embryonic vaccines against cancer: an early history. Exp Mol Pathol. 2009;86:192–197. doi: 10.1016/j.yexmp.2008.12.002. [DOI] [PubMed] [Google Scholar]
- Britsch S. The neuregulin-I/ErbB signaling system in development and disease. Adv Anat Embryol Cell Biol. 2007;190:1–65. [PubMed] [Google Scholar]
- Chang HY, Sneddon JB, Alizadeh AA, Sood R, West RB, Montgomery K, Chi JT, van de Rijn M, Botstein D, Brown PO. Gene expression signature of fibroblast serum response predicts human cancer progression: similarities between tumors and wounds. PLoS Biol. 2004;2:E7. doi: 10.1371/journal.pbio.0020007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christofk HR, Vander Heiden MG, Harris MH, Ramanathan A, Gerszten RE, Wei R, Fleming MD, Schreiber SL, Cantley LC. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature. 2008;452:230–233. doi: 10.1038/nature06734. [DOI] [PubMed] [Google Scholar]
- Creighton CJ, Li X, Landis M, Dixon JM, Neumeister VM, Sjolund A, Rimm DL, Wong H, Rodriguez A, Herschkowitz JI, et al. Residual breast cancers after conventional therapy display mesenchymal as well as tumor-initiating features. Proceedings of the National Academy of Sciences. 2009;106:13820–13825. doi: 10.1073/pnas.0905718106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunha GR, Hom YK. Role of mesenchymal-epithelial interactions in mammary gland development. J Mammary Gland Biol Neoplasia. 1996;1:21–35. doi: 10.1007/BF02096300. [DOI] [PubMed] [Google Scholar]
- Deome KB, Faulkin LJ, Jr., Bern HA, Blair PB. Development of mammary tumors from hyperplastic alveolar nodules transplanted into gland-free mammary fat pads of female C3H mice. Cancer Res. 1959;19:515–520. [PubMed] [Google Scholar]
- Desmedt C, Haibe-Kains B, Wirapati P, Buyse M, Larsimont D, Bontempi G, Delorenzi M, Piccart M, Sotiriou C. Biological processes associated with breast cancer clinical outcome depend on the molecular subtypes. Clin Cancer Res. 2008;14:5158–5165. doi: 10.1158/1078-0432.CCR-07-4756. [DOI] [PubMed] [Google Scholar]
- Dontu G, Al-Hajj M, Abdallah WM, Clarke MF, Wicha MS. Stem cells in normal breast development and breast cancer. Cell Prolif. 2003;36(Suppl 1):59–72. doi: 10.1046/j.1365-2184.36.s.1.6.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan C, Oh DS, Wessels L, Weigelt B, Nuyten DS, Nobel AB, van’t Veer LJ, Perou CM. Concordance among gene-expression-based predictors for breast cancer. N Engl J Med. 2006;355:560–569. doi: 10.1056/NEJMoa052933. [DOI] [PubMed] [Google Scholar]
- Fan C, Prat A, Parker JS, Liu Y, Carey LA, Troester MA, Perou CM. Building prognostic models for breast cancer patients using clinical variables and hundreds of gene expression signatures. BMC Med Genomics. 2011;4:3. doi: 10.1186/1755-8794-4-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haibe-Kains B, Desmedt C, Piette F, Buyse M, Cardoso F, Van’t Veer L, Piccart M, Bontempi G, Sotiriou C. Comparison of prognostic gene expression signatures for breast cancer. BMC Genomics. 2008;9:394. doi: 10.1186/1471-2164-9-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Hay ED, Zuk A. Transformations between epithelium and mesenchyme: normal, pathological, and experimentally induced. Am J Kidney Dis. 1995;26:678–690. doi: 10.1016/0272-6386(95)90610-x. [DOI] [PubMed] [Google Scholar]
- Hendrix MJ, Seftor EA, Seftor RE, Trevor KT. Experimental co-expression of vimentin and keratin intermediate filaments in human breast cancer cells results in phenotypic interconversion and increased invasive behavior. Am J Pathol. 1997;150:483–495. [PMC free article] [PubMed] [Google Scholar]
- Hennessy BT, Gonzalez-Angulo AM, Stemke-Hale K, Gilcrease MZ, Krishnamurthy S, Lee JS, Fridlyand J, Sahin A, Agarwal R, Joy C, et al. Characterization of a naturally occurring breast cancer subset enriched in epithelial-to-mesenchymal transition and stem cell characteristics. Cancer Res. 2009;69:4116–4124. doi: 10.1158/0008-5472.CAN-08-3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herschkowitz J, Simin K, Weigman V, Mikaelian I, Usary J, Hu Z, Rasmussen K, Jones L, Assefnia S, Chandrasekharan S, et al. Identification of conserved gene expression features between murine mammary carcinoma models and human breast tumors. Genome Biology. 2007;8:R76. doi: 10.1186/gb-2007-8-5-r76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard B, Ashworth A. Signalling pathways implicated in early mammary gland morphogenesis and breast cancer. PLoS Genet. 2006;2:e112. doi: 10.1371/journal.pgen.0020112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Smyth GK. ELDA: extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J Immunol Methods. 2009;347:70–78. doi: 10.1016/j.jim.2009.06.008. [DOI] [PubMed] [Google Scholar]
- Jackson-Fisher AJ, Bellinger G, Breindel JL, Tavassoli FA, Booth CJ, Duong JK, Stern DF. ErbB3 is required for ductal morphogenesis in the mouse mammary gland. Breast Cancer Res. 2008;10:R96. doi: 10.1186/bcr2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson-Fisher AJ, Bellinger G, Ramabhadran R, Morris JK, Lee KF, Stern DF. ErbB2 is required for ductal morphogenesis of the mammary gland. Proc Natl Acad Sci U S A. 2004;101:17138–17143. doi: 10.1073/pnas.0407057101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs TW, Gown AM, Yaziji H, Barnes MJ, Schnitt SJ. Specificity of HercepTest in determining HER-2/neu status of breast cancers using the United States Food and Drug Administration-approved scoring system. J Clin Oncol. 1999;17:1983–1987. doi: 10.1200/JCO.1999.17.7.1983. [DOI] [PubMed] [Google Scholar]
- Jones DL, Wagers AJ. No place like home: anatomy and function of the stem cell niche. Nat Rev Mol Cell Biol. 2008;9:11–21. doi: 10.1038/nrm2319. [DOI] [PubMed] [Google Scholar]
- Karaman MW, Herrgard S, Treiber DK, Gallant P, Atteridge CE, Campbell BT, Chan KW, Ciceri P, Davis MI, Edeen PT, et al. A quantitative analysis of kinase inhibitor selectivity. Nat Biotechnol. 2008;26:127–132. doi: 10.1038/nbt1358. [DOI] [PubMed] [Google Scholar]
- Kendrick H, Regan JL, Magnay FA, Grigoriadis A, Mitsopoulos C, Zvelebil M, Smalley MJ. Transcriptome analysis of mammary epithelial subpopulations identifies novel determinants of lineage commitment and cell fate. BMC Genomics. 2008;9:591. doi: 10.1186/1471-2164-9-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi A, Yamamoto H, Sato A, Matsumoto S. Wnt5a: its signalling, functions and implication in diseases. Acta Physiol (Oxf) 2011 doi: 10.1111/j.1748-1716.2011.02294.x. [DOI] [PubMed] [Google Scholar]
- Korkaya H, Wicha MS. HER-2, notch, and breast cancer stem cells: targeting an axis of evil. Clin Cancer Res. 2009;15:1845–1847. doi: 10.1158/1078-0432.CCR-08-3087. [DOI] [PubMed] [Google Scholar]
- Lim E, Vaillant F, Wu D, Forrest NC, Pal B, Hart AH, Asselin-Labat ML, Gyorki DE, Ward T, Partanen A, et al. Aberrant luminal progenitors as the candidate target population for basal tumor development in BRCA1 mutation carriers. Nat Med. 2009;15:907–913. doi: 10.1038/nm.2000. [DOI] [PubMed] [Google Scholar]
- Lim E, Wu D, Pal B, Bouras T, Asselin-Labat M-L, Vaillant F, Yagita H, Lindeman G, Smyth G, Visvader J. Transcriptome analyses of mouse and human mammary cell subpopulations reveal multiple conserved genes and pathways. Breast Cancer Research. 2010;12:R21. doi: 10.1186/bcr2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livasy CA, Reading FC, Moore DT, Boggess JF, Lininger RA. EGFR expression and HER2/neu overexpression/amplification in endometrial carcinosarcoma. Gynecol Oncol. 2006;100:101–106. doi: 10.1016/j.ygyno.2005.07.124. [DOI] [PubMed] [Google Scholar]
- Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno H, Spike BT, Wahl GM, Levine AJ. Inactivation of p53 in breast cancers correlates with stem cell transcriptional signatures. Proc Natl Acad Sci U S A. 2010;107:22745–22750. doi: 10.1073/pnas.1017001108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mork C, van Deurs B, Petersen OW. Regulation of vimentin expression in cultured human mammary epithelial cells. Differentiation. 1990;43:146–156. doi: 10.1111/j.1432-0436.1990.tb00441.x. [DOI] [PubMed] [Google Scholar]
- Paik S, Kim C, Wolmark N. HER2 status and benefit from adjuvant trastuzumab in breast cancer. N Engl J Med. 2008;358:1409–1411. doi: 10.1056/NEJMc0801440. [DOI] [PubMed] [Google Scholar]
- Paik S, Tang G, Shak S, Kim C, Baker J, Kim W, Cronin M, Baehner F, Watson D, Bryant J. Gene expression and benefit of chemotherapy in women with node-negative, estrogen receptor-positive breast cancer. J Clin Oncol. 2006;24:3726–3734. doi: 10.1200/JCO.2005.04.7985. [DOI] [PubMed] [Google Scholar]
- Pal SK, Childs BH, Pegram M. Triple negative breast cancer: unmet medical needs. Breast Cancer Res Treat. 2011;125:627–636. doi: 10.1007/s10549-010-1293-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pece S, Tosoni D, Confalonieri S, Mazzarol G, Vecchi M, Ronzoni S, Bernard L, Viale G, Pelicci PG, Di Fiore PP. Biological and Molecular Heterogeneity of Breast Cancers Correlates with Their Cancer Stem Cell Content. Cell. 2010;140:62–73. doi: 10.1016/j.cell.2009.12.007. [DOI] [PubMed] [Google Scholar]
- Perou CM, Parker JS, Prat A, Ellis MJ, Bernard PS. Clinical implementation of the intrinsic subtypes of breast cancer. Lancet Oncol. 2010;11:718–719. doi: 10.1016/S1470-2045(10)70176-5. author reply 720-711. [DOI] [PubMed] [Google Scholar]
- Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- Petersen OW, Polyak K. Stem cells in the human breast. Cold Spring Harb Perspect Biol. 2010;2:a003160. doi: 10.1101/cshperspect.a003160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers S, Mu D. Genetic similarities between organogenesis and tumorigenesis of the lung. Cell Cycle. 2008;7:200–204. doi: 10.4161/cc.7.2.5284. [DOI] [PubMed] [Google Scholar]
- Prat A, Parker JS, Karginova O, Fan C, Livasy C, Herschkowitz JI, He X, Perou CM. Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast Cancer Res. 2010;12:R68. doi: 10.1186/bcr2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prat A, Perou CM. Deconstructing the molecular portraits of breast cancer. Mol Oncol. 2011;5:5–23. doi: 10.1016/j.molonc.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabindran SK, Discafani CM, Rosfjord EC, Baxter M, Floyd MB, Golas J, Hallett WA, Johnson BD, Nilakantan R, Overbeek E, et al. Antitumor activity of HKI-272, an orally active, irreversible inhibitor of the HER-2 tyrosine kinase. Cancer Res. 2004;64:3958–3965. doi: 10.1158/0008-5472.CAN-03-2868. [DOI] [PubMed] [Google Scholar]
- Raouf A, Zhao Y, To K, Stingl J, Delaney A, Barbara M, Iscove N, Jones S, McKinney S, Emerman J, et al. Transcriptome analysis of the normal human mammary cell commitment and differentiation process. Cell Stem Cell. 2008;3:109–118. doi: 10.1016/j.stem.2008.05.018. [DOI] [PubMed] [Google Scholar]
- Rody A, Karn T, Liedtke C, Pusztai L, Ruckhaeberle E, Hanker L, Gaetje R, Solbach C, Ahr A, Metzler D, et al. A clinically relevant gene signature in triple negative and basal-like breast cancer. Breast Cancer Res. 2011;13:R97. doi: 10.1186/bcr3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusnak DW, Lackey K, Affleck K, Wood ER, Alligood KJ, Rhodes N, Keith BR, Murray DM, Knight WB, Mullin RJ, et al. The effects of the novel, reversible epidermal growth factor receptor/ErbB-2 tyrosine kinase inhibitor, GW2016, on the growth of human normal and tumor-derived cell lines in vitro and in vivo. Mol Cancer Ther. 2001;1:85–94. [PubMed] [Google Scholar]
- Sakakura T, Nishizuka Y, Dawe CJ. Capacity of mammary fat pads of adult C3H/HeMs mice to interact morphogenetically with fetal mammary epithelium. J Natl Cancer Inst. 1979;63:733–736. doi: 10.1093/jnci/63.3.733. [DOI] [PubMed] [Google Scholar]
- Sell S. On the stem cell origin of cancer. Am J Pathol. 2010;176:2584–2494. doi: 10.2353/ajpath.2010.091064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shackleton M, Vaillant F, Simpson KJ, Stingl J, Smyth GK, Asselin-Labat ML, Wu L, Lindeman GJ, Visvader JE. Generation of a functional mammary gland from a single stem cell. Nature. 2006;439:84–88. doi: 10.1038/nature04372. [DOI] [PubMed] [Google Scholar]
- Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey SS, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotiriou C, Piccart MJ. Taking gene-expression profiling to the clinic: when will molecular signatures become relevant to patient care? Nat Rev Cancer. 2007;7:545–553. doi: 10.1038/nrc2173. [DOI] [PubMed] [Google Scholar]
- Spike BT, Wahl GM. p53, Stem Cells, and Reprogramming: Tumor Suppression beyond Guarding the Genome. Genes Cancer. 2011;2:404–419. doi: 10.1177/1947601911410224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spradling A, Drummond-Barbosa D, Kai T. Stem cells find their niche. Nature. 2001;414:98–104. doi: 10.1038/35102160. [DOI] [PubMed] [Google Scholar]
- Stingl J, Eirew P, Ricketson I, Shackleton M, Vaillant F, Choi D, Li HI, Eaves CJ. Purification and unique properties of mammary epithelial stem cells. Nature. 2006;439:993–997. doi: 10.1038/nature04496. [DOI] [PubMed] [Google Scholar]
- Strizzi L, Hardy KM, Seftor EA, Costa FF, Kirschmann DA, Seftor RE, Postovit LM, Hendrix MJ. Development and cancer: at the crossroads of Nodal and Notch signaling. Cancer Res. 2009;69:7131–7134. doi: 10.1158/0008-5472.CAN-09-1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun P, Yuan Y, Li A, Li B, Dai X. Cytokeratin expression during mouse embryonic and early postnatal mammary gland development. Histochem Cell Biol. 2010;133:213–221. doi: 10.1007/s00418-009-0662-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiery JP, Sleeman JP. Complex networks orchestrate epithelial-mesenchymal transitions. Nat Rev Mol Cell Biol. 2006;7:131–142. doi: 10.1038/nrm1835. [DOI] [PubMed] [Google Scholar]
- Thomas PA, Kirschmann DA, Cerhan JR, Folberg R, Seftor EA, Sellers TA, Hendrix MJ. Association between keratin and vimentin expression, malignant phenotype, and survival in postmenopausal breast cancer patients. Clin Cancer Res. 1999;5:2698–2703. [PubMed] [Google Scholar]
- Tidcombe H, Jackson-Fisher A, Mathers K, Stern DF, Gassmann M, Golding JP. Neural and mammary gland defects in ErbB4 knockout mice genetically rescued from embryonic lethality. Proc Natl Acad Sci U S A. 2003;100:8281–8286. doi: 10.1073/pnas.1436402100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van ’t Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, Peterse HL, van der Kooy K, Marton MJ, Witteveen AT, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- van de Vijver MJ, He YD, van’t Veer LJ, Dai H, Hart AA, Voskuil DW, Schreiber GJ, Peterse JL, Roberts C, Marton MJ, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- Van Keymeulen A, Rocha AS, Ousset M, Beck B, Bouvencourt G, Rock J, Sharma N, Dekoninck S, Blanpain C. Distinct stem cells contribute to mammary gland development and maintenance. Nature. 2011;479:189–193. doi: 10.1038/nature10573. [DOI] [PubMed] [Google Scholar]
- Veltmaat JM, Mailleux AA, Thiery JP, Bellusci S. Mouse embryonic mammogenesis as a model for the molecular regulation of pattern formation. Differentiation. 2003;71:1–17. doi: 10.1046/j.1432-0436.2003.700601.x. [DOI] [PubMed] [Google Scholar]
- Wang SE, Narasanna A, Perez-Torres M, Xiang B, Wu FY, Yang S, Carpenter G, Gazdar AF, Muthuswamy SK, Arteaga CL. HER2 kinase domain mutation results in constitutive phosphorylation and activation of HER2 and EGFR and resistance to EGFR tyrosine kinase inhibitors. Cancer Cell. 2006;10:25–38. doi: 10.1016/j.ccr.2006.05.023. [DOI] [PubMed] [Google Scholar]
- Wansbury O, Mackay A, Kogata N, Mitsopoulos C, Kendrick H, Davidson K, Ruhrberg C, Reis-Filho JS, Smalley MJ, Zvelebil M, et al. Transcriptome analysis of embryonic mammary cells reveals insights into mammary lineage establishment. Breast Cancer Res. 2011;13:R79. doi: 10.1186/bcr2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigelt B, Lo AT, Park CC, Gray JW, Bissell MJ. HER2 signaling pathway activation and response of breast cancer cells to HER2-targeting agents is dependent strongly on the 3D microenvironment. Breast Cancer Res Treat. 2010;122:35–43. doi: 10.1007/s10549-009-0502-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirapati P, Sotiriou C, Kunkel S, Farmer P, Pradervand S, Haibe-Kains B, Desmedt C, Ignatiadis M, Sengstag T, Schutz F, et al. Meta-analysis of gene expression profiles in breast cancer: toward a unified understanding of breast cancer subtyping and prognosis signatures. Breast Cancer Res. 2008;10:R65. doi: 10.1186/bcr2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng YA, Nusse R. Wnt Proteins Are Self-Renewal Factors for Mammary Stem Cells and Promote Their Long-Term Expansion in Culture. Cell Stem Cell. 2010;6:568–577. doi: 10.1016/j.stem.2010.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.