Abstract
Studies of tumors from human familial adenomatous polyposis, sporadic colon cancer, and mouse and rat models of intestinal cancer indicate that the majority of early adenomas develop through loss of normal function of the Adenomatous polyposis coli (APC) gene. In murine models of familial adenomatous polyposis, specifically the multiple intestinal neoplasia mouse (Min) and the polyposis in the rat colon (Pirc) rat, most adenomas have lost their WT copy of the Apc gene through loss of heterozygosity by homologous somatic recombination. We report that large colonic adenomas in the Pirc rat have no detectable copy number losses or gains in genomic material and that most tumors lose heterozygosity only on the short arm of chromosome 18. Examination of early mouse and rat tumors indicates that a substantial subset of tumors shows maintenance of heterozygosity of Apc in genomic DNA, apparently violating Knudson's two-hit hypothesis. Sequencing of the Apc gene in a sampling of rat tumors failed to find secondary mutations in the majority of tumors that maintained heterozygosity of Apc in genomic DNA. Using quantitative allele-specific assays of Apc cDNA, we discovered two neoplastic pathways. One class of tumors maintains heterozygosity of ApcMin/+ or ApcPirc/+ RNA expression and may involve haploinsufficiency for Apc function. Another class of tumors exhibits highly biased monoallelic expression of the mutant Apc allele, providing evidence for a stochastic or random process of monoallelic epigenetic silencing of the tumor suppressor gene Apc.
Keywords: epigenetics, genomic stability, loss of imprinting, X-inactivation
Understanding the earliest genetic, epigenetic, and environmental events in adenoma formation may illuminate the range of mechanisms of tumor initiation and subsequent pathways of progression. In most sporadic cases of colon cancer, the first genetic lesion involves the gatekeeper tumor suppressor gene Adenomatous polyposis coli (APC), leading to growth of the adenoma (1). Studies of tumors and normal epithelium from patients with familial adenomatous polyposis (FAP), heterozygous for an APC mutation, indicate that a single genetic lesion may itself have a biological effect—the one-hit hypothesis (2). Frank tumor formation seems to require a balance of mutant APC alleles—the just-right hypothesis (3). Early colonic adenomas with APC mutations can persist for up to a decade without further advancement (4) and may only progress to invasive metastatic disease by acquiring additional mutations or genomic instability.
Loss of function of the APC gene can occur through multiple alternative genetic mechanisms representing many different pathways of pathogenesis (5–7). The acquisition of some form of genetic instability may be required to accumulate sufficient mutations or imbalances for cancer (8). Reports of such instability include amplifications, deletions, gene fusions, and microsatellite instability caused by germ-line or somatic mutations in mismatch repair genes leading to errors in replication. Advanced colonic cancers in humans often exhibit chromosomal instability with large losses and gains of genetic material (9). Except for microsatellite instability (10), genomic instability is remarkably missing in most mouse models of intestinal cancer (11, 12). Homologous somatic recombination is a more subtle form of genomic instability observed in human FAP and its murine models (13, 14). Conservative somatic recombination between the centromere and the APC locus causes loss of heterozygosity (LOH); the resultant adenoma carries two mutant copies of APC/Apc. Is such conservative somatic recombination elevated over the whole genome in the cancer lineage?
Beyond genetic and genomic changes that trigger the initiation and progression of cancer, it is important to identify epigenetic differences associated with the cancer lineage. Epigenetic changes involve transmissible changes in phenotype that are not directly caused by changes in DNA sequence. On one hand, an epigenetic change can be caused by a developmentally programmed alteration of an upstream effector (i.e., a transcription factor) acting on downstream targets to control the cancer phenotype (14). On the other hand, epimutation—either silencing or activation—can involve a somatically heritable effect on a single allele of a gene that impacts cancer.
The work by Grady et al. (15) reported an example of silencing of the E-cadherin gene in gastric cancer reflected by 100% levels of methylation of key CpG sites within the promoter region. The work also reported that a subset of tumors exhibits 50% methylation in these sites (15). This observation may reflect monoallelic silencing in the neoplastic lineage or an admixture of normal stroma in the tumor. Programmed epigenetic change is expected to affect both alleles at a locus, whereas rare stochastic epigenetic change is expected to affect only a single allele. The heterozygous diploid condition can distinguish between these alternatives.
The involvement of epigenetic instability in cancer is shown by monoallelic changes in expression in the absence of changes in DNA sequence. The silent allele at an imprinted locus is activated rarely in colon cancer (16). This change in state implies the loss of a maintenance function. By contrast, this report involves cases in which the genomically stable multiple intestinal neoplasia (Min) mouse and polyposis in the rat colon (Pirc) rat models of familial intestinal neoplasia generate adenomas through random epigenetic silencing of the WT Apc allele. Our detailed analysis of these exceptional cases shows that the WT Apc allele is silenced and maintained in a stable, monoallelically active state without detectable changes in DNA sequence.
Results
Genomic and Epigenetic Changes at the Apc Locus in the Rb9-Min Mouse.
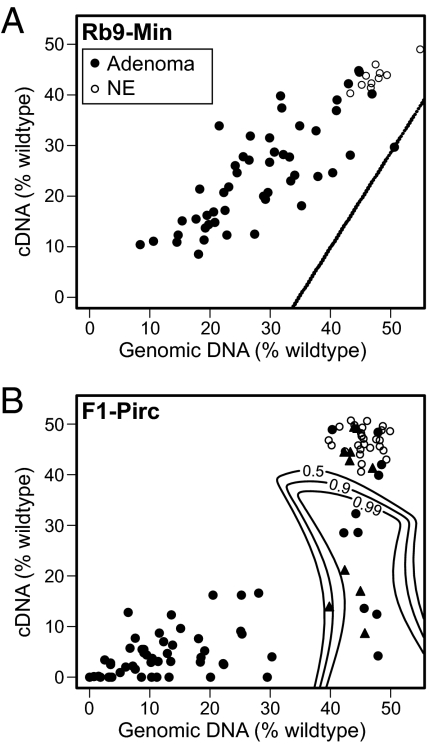
To assess LOH in genomic DNA (gLOH) at the heterozygous Apc locus, we examined the percent contribution of each allele in gDNA from tumors arising in the Rb9-Min mouse (Materials and Methods). A total of 52 tumors and 10 normal adjacent tissue samples were examined for the ratio of their Apc alleles. The ratios of Min to WT center near 1:1 in gDNA from the control normal tissues, signaling maintenance of heterozygosity in gDNA (gMOH). In tumors, by contrast, the contribution of the WT allele varied from 8% to 50% without a clear delineation between the gLOH and gMOH classes of tumors. In an earlier report (17), the distribution of allelic ratios had been bimodal.
Tumors were then examined for allelic biases in expression by assaying cDNAs generated from tumor RNA. Two tumors scored as gMOH yielded no detectable cDNA from the Apc gene, whereas cDNA from the control housekeeping gene ribonuclear protein Rpl10a was present (Fig. S1). Thus, the level of APC RNA in these two tumors lay below the detection limit of the RT-PCR pyrosequencing assay. For the majority of the gMOH Rb9-Min tumors, the percent contribution of WT allele in both gDNA and cDNA varied from 8% to 45%. Interestingly, in 1 of these 52 tumors, the contribution of the WT allele in cDNA was significantly reduced compared with the contribution in gDNA (Fig. 1A). We have designated this exceptional class of tumor as gMOH/cLOH.
Fig. 1.
Percentage of the WT Apc allele compared with the Pirc mutant allele in gDNA (x axis) and cDNA (y axis) from either Rb9-Min (A) or F1-Pirc (B) adenomas (filled symbols) or normal epithelium (NE; open circles). Tumors within the designated significance boundaries maintain heterozygosity of the gDNA but have lost heterozygosity in their cDNA. Rb9-Min significance lines (0.5–0.99) are superimposed on each other. F1-Pirc tumors that were sequenced for Apc are represented as filled triangles.
Genomic and Epigenetic Changes in Colonic Tumors of the F1-Pirc Rat.
Stability of copy number and heterozygosity.
The predominant early somatic genetic event in the formation of early adenomas in the Pirc (ApcPirc/+) rat is also somatic recombination with LOH at the Apc locus (13, 18). We have investigated whether these conservative events at the Apc locus are accompanied by genome-wide changes that affect diploid copy number. Array comparative genomic hybridization analysis was performed on DNA from two large (0.8 and 1.0 cm) colonic adenomas from two F344-Pirc animals (Fig. S2). No significant alterations in copy number were detected.
Relative copy number can be conserved in the face of widespread LOH involving homologous somatic recombination. To determine whether LOH at the Apc locus is accompanied by genome-wide LOH, we examined the incidence of LOH across the genome in F1-Pirc rat tumors using pyrosequencing of allelic ratios at informative SNP sites (Materials and Methods). We chose SNP sites located at the distal ends of chromosomes to capture all proximal somatic recombination events. We previously showed that LOH in tumors in the Pirc rat involves the short (p) arm of chromosome (Chr) 18, resulting in two copies of the mutant allele in the tumor lineage. We tested a mode of 23 tumors from each of three or more independent F1-Pirc animals, examining 13 SNP sites on seven chromosomes (1, 4, 7, 10, 15, 16, and 18q). This survey revealed that nearly all of the SNP sites maintained heterozygosity, showing allelic ratios close to 1:1, including SNPs on the long arm of chromosome 18 (Fig. S3 and Table S1). One exceptional tumor showed loss of the ACI alleles for most of the distal arm of Chr 1 (Table S1). Thus, even very large adenomas in the Pirc rat can develop without detectable widespread copy number instability or LOH at loci other than Apc.
Allelic ratios of gDNA and cDNA in colonic tumors of the F1-Pirc rat.
We analyzed the percent WT contribution in gDNA and cDNA from normal colonic tissue and tumors from F1-Pirc rats. Material came from either spontaneously arising tumors in untreated rats or tumors in animals that were treated with the ulcerogenic inflammatory agent dextran sulfate sodium (DSS) to increase the number of colonic tumors. After harvesting a portion of each tumor for RNA and DNA extraction, the remaining portion of each tumor was fixed and sectioned for histological analysis. One of the gMOH tumors from an untreated rat was determined by histological examination to contain nearly 90% stromal cells, with very little epithelial contribution, and therefore, it was removed from further analysis. DNA from 67 tumors (34 DSS-treated and 33 spontaneous tumors) revealed that the majority (47) lost the WT Apc allele (gLOH), whereas 20 maintained heterozygosity of the Apc locus (gMOH) (Fig. 1B). The distribution between gLOH and gMOH classes was similar between untreated vs. DSS-treated F1-Pirc rats.
Quantitative pyrosequencing of the allelic ratio at the Pirc site was performed on cDNAs paired with each tumor gDNA sample. These assays showed that, regardless of treatment, each tumor that lost heterozygosity in gDNA also showed significant allelic loss in cDNA. Among the gMOH class of F1-Pirc rat tumors, one-half (10/20) were also cMOH, with nearly equal contributions of mutant and WT Apc alleles. The remaining one-half (10/20) were gMOH/cLOH tumors with expression at the Apc locus that was dramatically skewed to the mutant Pirc allele.
RT-PCR quantitation of mRNA levels.
The relative level of transcripts from the Apc gene was measured by quantitative RT-PCR (qRT-PCR) (Table 1). gMOH/cLOH tumors showed a relative reduction in Apc transcripts compared with gMOH/cMOH tumors (Wilcoxon rank sum test, P = 0.003) and gLOH/cLOH tumors (P = 0.05). Interestingly, gMOH/cMOH tumors did not show a significant difference in the relative level of Apc transcripts compared with gLOH/cLOH tumors (P = 0.84). This observation reflects the conservative maintenance of somatic Apc copy number by the homologous somatic recombination process.
Table 1.
qRT-PCR results for rat tumor classes
| Type | N | ΔCt (mean ± SD) |
| gLOH/cLOH | 3 | 6.02 ± 0.52 |
| gMOH/cMOH | 8 | 5.97 ± 0.44 |
| gMOH/cLOH | 7 | 7.24 ± 0.65 |
Results are expressed as ΔCt using Gapdh as the control gene for F1-Pirc tumors for each allele-specific class. P = 0.007 for the null hypothesis that all of the samples are taken from the same population (Kruskal–Wallis). Pairwise P values are 0.05 for gLOH/cLOH vs. gMOH/cLOH, 0.003 for gMOH/cLOH vs. gMOH/cMOH, and 0.84 for gLOH/cLOH vs. gMOH/cMOH (Wilcoxon rank sum test).
Search for secondary Apc mutations in gMOH F1-Pirc tumors.
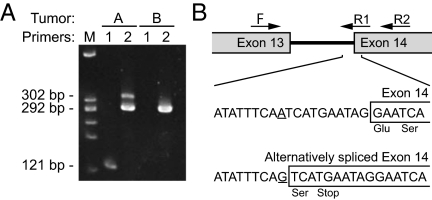
To determine whether a second genetic hit had occurred in the gMOH tumors, we used traditional fluorescent Sanger sequencing to analyze gDNA from 6 of 10 gMOH/cMOH and 4 of 10 gMOH/cLOH F1-Pirc rat tumors. Tumor DNA was amplified using PCR with primers for all 15 exons of the rat Apc gene plus exon–intron boundaries and the minimal promoter (∼1 kb) region. Beyond the Pirc mutation, two of gMOH/cMOH tumors carried mutations in Apc exon 15 that created premature stop codon mutations—one from a nontreated animal and one from a DSS-treated F1-Pirc animal (Table S2). No sequence changes in the Apc gene were found in gDNA from the other four gMOH/cMOH tumors. Of the four gMOH/cLOH tumors, a single mutation in Apc intron 13 was found in one tumor (Table S2), creating a splice acceptor that led to an alternatively spliced transcript that was shown by RT-PCR (Fig. 2). This transcript carries a premature stop codon in exon 14 that is highly likely to be subject to nonsense-mediated decay, unlike the two exon 15 nonsense mutations described above and the Min and Pirc nonsense alleles. No sequence changes in the Apc gene were found in any of the three remaining gMOH/cLOH tumors, providing strong evidence for the existence of an epigenetic monoallelic silencing process.
Fig. 2.
RT-PCR confirmation of alternative splice forms of the Apc transcript generated by a splice acceptor mutation in intron 13. (A) RT-PCR of tumors A and B using primers spanning intron 13. The PCR product from the normal splice form (primer set 2) is shown in tumor B (292 bp); it is also present in tumor A. Primer set 1 is specific to the alternative splice form created by the A–G transition mutation in intron 13. The 121-bp band is present only in tumor A. The differences in band intensity for primer set 2 in tumor A indicate that the alternative splice form is less abundant than the normal splice form. (B) Map and PCR primers spanning intron 13. Sequencing of the alternative splice form identified the sequence and position of the mutation that creates a splice acceptor.
Search for extragenic second mutations in gMOH/cMOH F1-Pirc rat tumors.
The molecular basis for the interesting gMOH/cMOH F1-Pirc rat tumors was investigated. In the absence of Apc mutations, constitutive mutations in the β-catenin gene (Ctnnb1) have been shown to activate the WNT pathway. However, no sequence changes were found in the region of Ctnnb1 associated with such activating mutations. Similarly, no mutations in Kras were found in any of the sequenced tumors. None of the gMOH/cMOH F1-Pirc rat tumors gave evidence for biallelic silencing by qRT-PCR assays of cDNA. Possible molecular bases for these gMOH/cMOH tumors will be discussed.
Histopathology of F1-Pirc and Rb9-Min tumors.
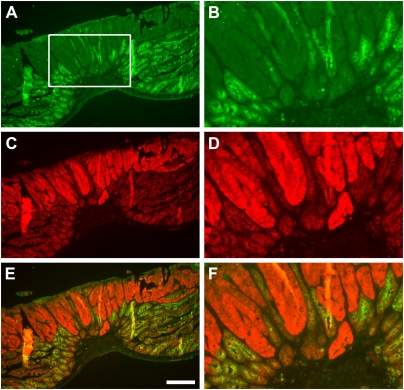
Histological analysis of adenomas has failed to find major differences between these gLOH and gMOH classes of F1-Pirc rat adenomas. Immunohistochemical staining for β-catenin showed similar enhanced accumulation of the antigen in the cytoplasm and nucleus, with coordinate loss of the lateral membrane-bound localization. Additionally, the pattern of staining by H&E showed no pronounced difference between the classes, with similar low levels of normal epithelial cell contribution except in the single case cited above where there was substantial stromal and minimal tumor epithelial contributions. In Rb9-Min tumors, staining for WT APC protein showed that the pervasive loss of normal APC antigen and increase in cytoplasmic and nuclear β-catenin are similar between gMOH/cLOH and gLOH/cLOH cases (Fig. 3). Significantly, APC staining was undetectable in the nonepithelial stromal contribution within the tumor or surrounding normal tissue.
Fig. 3.
Immunohistochemistry was performed with antibodies to (A and B) Apc protein (green) and (C and D) β-catenin (red; merged in E and F). B, D, and F are enlargements of the box in A and equivalent areas in C and E, respectively. Loss of Apc protein and an increase in cytoplasmic and nuclear β-catenin in tumor epithelial cells were observed for the gMOH/cLOH tumor as previously reported for MOH and LOH tumors (13). Apc protein cannot be detected in normal stroma; it is detectable primarily in normal intestinal epithelium. (Scale bar: A, C, and E, 200 μm.)
Discussion
Conservative Genome of Early Rb9-Min and F1-Pirc Neoplasms.
The early stages of colonic neoplasia in the F1-Pirc rat model of familial colon cancer show no evidence of extensive changes in copy number over the genome (Fig. S2). These observations mirror prior studies in the Min mouse, including invasive stages of tumors in the small intestine of long-lived (SWR × B6)F1-Min mice (11). The present study extends these observations of the stable genome to investigate conservative LOH over the genome in F1-Pirc rat tumors.
Previous studies showed that LOH at the APC locus by somatic recombination was often found in early human FAP adenomas (19). Similarly, we have reported in Rb9-Min mouse and F1-Pirc rat tumors a similarly high proportion of LOH events at the Apc locus involving homologous somatic recombination (13, 18). In this report, we tested whether early adenomas that lose heterozygosity of Apc involve genome-wide LOH as in meiosis—a class of somatic instability. Examination of polymorphic SNP loci on multiple chromosomes by quantitative allele-specific pyrosequencing in F1 hybrids, including Chr 18, showed that LOH was largely restricted to the short arm of Chr 18 harboring the Apc locus. This finding indicates that somatic recombination over the whole genome is not enhanced in the tumor lineage but that the LOH events that occur on the short arm of Chr 18 are strongly selected in early adenomagenesis.
Interestingly, in one F1-Pirc tumor, we detected LOH on the long arm of Chr 1 (Table S1). This event may be a passenger event functionally independent of the LOH event at the Apc locus on Chr 18. Alternatively, this region of the genome may contain one or more modifier loci. We note that rat Chr 1 contains many genes with effects on cancer (Igf2, Igf2R, Dkk1, Rab38, and Hras). We note also that many regions on rat Chr 1 are syntenic to imprinted regions in the human genome, including human Chrs 6 (IGF2R), 11 (IGF2/H19), 15 (SNRPN and UBE3A), and 19 (PEG3). Many of these genes regulate the growth of cancers. In this single case of LOH across the q arm of rat Chr 1, the maternal (ACI) allele was lost, plausibly duplicating the paternal expression of many of the imprinted growth-enhancing genes (Igf2 and Igf2r). Indeed, cases of loss of imprinting may instead involve LOH through somatic recombination (20).
Tumors That Maintain Heterozygosity at the Apc Locus (gMOH).
The most interesting groups of tumors in the Rb9-Min and F1-Pirc rat are those groups that maintain heterozygosity of Apc in their gDNA (gMOH). Under Knudson's two-hit hypothesis, one genetic hit alone would be insufficient to cause tumor formation. Interestingly, the gLOH vs. gMOH ratios are more evenly distributed in the set of Rb9-Min tumors (Fig. 1A) than in the F1-Pirc rat tumors (Fig. 1B). This finding may reflect the increased level of normal epithelial and stromal admixture in the nonpedunculate mouse tumors that are dissected from the small intestine. Alternatively, this broad distribution of allelic ratios may reflect the known polyclonality of Min tumors (21) or differences between tumors of the small intestine and the colon. By contrast, the high concordance between the majority of gLOH vs. cLOH allelic ratios in F1-Pirc tumors and between the gMOH vs. cMOH allelic ratios in normal epithelium indicates that these pedunculate colonic rat tumors are dissected with less admixture of normal epithelial and stromal tissue, resulting in more highly defined LOH and MOH classes.
Analysis of Admixture in Rb9-Min and F1-Pirc Tumors.
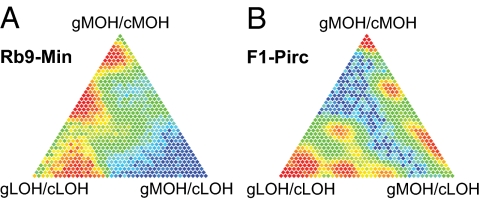
To make rigorous distinctions between MOH and LOH classes, we investigated statistically the role of admixture in the analysis of these tumor classes for both the Rb9-Min small intestinal tumors and the F1-Pirc colonic tumors. The probability that a tumor is composed of any particular mixture of cells from the three classes was estimated by likelihood methods (Fig. 4). This analysis indicates that the Rb9-Min tumors contain a significant admixture between gMOH/cMOH and gLOH/cLOH components (Fig. 4A). It is estimated that 33% of the Rb9-Min tumors have a majority of cells from the gMOH/cMOH class, and 45% have a majority contribution from the gLOH/cLOH class. (The corresponding probabilities for plurality contributions are 45% and 51%, respectively.) These estimates may also reflect a high level of normal epithelial cell contribution to dissected Rb9-Min tumors. The estimated contributions to F1-Pirc tumors are more distinct (Fig. 4B and Table S3). These tumors show a greater gMOH/cLOH contribution admixed with both gMOH/cMOH and gLOH/cLOH classes. Thus, admixture between LOH and MOH components is observed in adenomas of both the Rb9-Min mouse and F1-Pirc rat and may reflect the polyclonal nature of early intestinal cancer (21, 22). A full statistical analysis of admixture is presented in SI Materials and Methods and Fig. S4; custom R code is contained in Dataset S1.
Fig. 4.
Admixture models for Rb9-Min (A) and F1-Pirc (B). Likelihood methods were used to estimate the distribution of tumor composition from cells of three distinct types (corners of the triangles). Plotted here in barycentric coordinates are estimated probabilities that a tumor has a particular fraction of its cells from each cell type, with low probabilities in blue to highest probabilities in red. The estimated distribution also indicates the chance that a tumor is comprised of a majority or plurality of each cell type.
Absence of Detectable Mutations in F1-Pirc Tumors.
The analysis of genomic Apc sequence in gMOH/cMOH F1-Pirc rat tumors revealed only two tumors with secondary mutations in Apc (Table S2). No new mutations in the gene for β-catenin were found, suggesting that there are alternative pathways to early adenoma formation not involving the WNT, Apc, β-catenin axis but requiring either the ApcMin (mouse) or ApcPirc (rat) predisposing mutations. Thus, we consider the interesting possibility that the gMOH/cMOH class of adenomas commonly develops through haploinsufficiency of Apc. The population of Apc RNAs quantified from these tumors (Table 1) is composed of a 1:1 ratio of mutant to WT molecules. The Min phenotype is enhanced by heterozygosity for a KO allele of p27 (23). For example, tumor phenotypes are observed in p53/+ heterozygotes without loss of the WT p53 allele (24). The genomically stable processes of neoplasia in both the Min mouse and Pirc rat models for familial intestinal cancer in the human have opened an opportunity to discover noncomplementing genetic and epigenetic changes that would enhance any haploinsufficiency created by heterozygosity for a mutation in Apc (2).
Silencing in the Absence of Detectable Mutation in Apc.
We investigated the possibility that adenoma formation in the gMOH tumors was owing to loss of function of Apc through epigenetic silencing. The two Rb9-Min tumors that lacked sufficient cDNA from Apc to amplify in the RT-PCR assay may have silenced both alleles through a programmed epigenetic mechanism. However, the limited amount of material obtained from Min tumors limited additional resequencing. F1-Pirc rat gMOH/cMOH tumors did not show a significant reduction of Apc transcript levels as judged by qRT-PCR (Table 1). This finding excludes the possibility that these F1-Pirc tumors arose by biallelic silencing of Apc.
The remaining 10 MOH/cLOH F1-Pirc rat tumors provide the strongest evidence for monoallelic epigenetic silencing. This class did show a twofold reduction in the expression of Apc by qRT-PCR (Table 1). Allele-specific analysis of cDNAs by pyrosequencing indicated that, as expected, the mutant Pirc allele was preferentially expressed. Sequence analysis of 4 of 10 tumors found that only 1 of these gMOH/cLOH tumors carried a new mutation in the Apc locus: an intronic mutation that leads to an alternative splice form. This mutant transcript likely involves the WT allele and would be subjected to nonsense-mediated decay, resulting in two prematurely truncated alleles. APC mutations resulting in alternatively spliced transcripts have been observed in FAP patients, with some of these mutations subject to nonsense-mediated decay (25). Although we sequenced the exons and exon/intron boundaries of Apc in only four gMOH/cLOH tumors, it seems unlikely that the remaining six F1-Pirc gMOH/cLOH tumors all have intronic mutations that lead to nonsense-mediated decay. The major result is finding three independent cases of monoallelic epigenetic silencing.
Possible Mechanisms of Monoallelic Epigenetic Silencing.
Do the cases of Apc silencing represent programmed events or the result of a stochastically initiated silencing event? Global changes in gene expression through a developmental programming may explain the two Rb9-Min gMOH tumors that failed to express Apc mRNA. By contrast, a stochastic epigenetic event would occur at random, be fixed early in tumorigenesis, and act in cis at the allele level. We interpret the adenomas that exhibit monoallelic expression of Apc to be a product of such a stochastic silencing event that was then fixed and propagated with sufficient stability to support selection for the neoplastic process.
Monoallelic expression of a locus can reflect one of several conditions: constitutional polymorphism for a regulatory element controlling the level of the transcript from the locus, an evolved regulatory mechanism that generates allelic exclusion, or an altered structure of one allele that can be somatically propagated. The fact that the Apc locus is expressed from both alleles in normal epithelial tissue from the intestine of the Rb9-Min mouse and F1-Pirc rat rules out a functional constitutional polymorphism. An evolved mechanism of allelic exclusion (in the tumor lineage) is unlikely, because many of the tumors maintain heterozygous expression of APC transcripts. Finally, the failure to detect a new mutation in the WT Apc allele in three independent colonic tumors of the F1-Pirc rat forces us to consider a cis-acting epigenetic mechanism that can be propagated in tumors.
X inactivation in eutherian mammals is the paradigm for random, monoallelic epigenetic silencing that can be stably propagated in the soma (26). The molecular events associated with X inactivation involve expression from the inactive X chromosome, Xi, of the long, noncoding RNA X-inactive specific transcript (XIST) that coats the inactive X (27). Interesting recent studies of reprogramming in mouse epiblast stem cells through nuclear transfer show that, when XIST is removed from the inactive X, genes on the inactive X are not reactivated. Instead, the Xi state may be maintained through the histone variant macroH2A that also coats the inactive X but is not removed during reprogramming (28). Finally, cytosine residues in CpG dinucleotide sequences are often methylated at key regulatory sequences and act as a cis-limited mark of a silent allele. Cytosine methylation of the APC gene has been reported in microsatellite-unstable tumors in humans (29). However, examination of the mouse Apc promoter provides no evidence of methylation changes in tumors or during aging (30, 31). It remains to be determined if a differential methylation mark is present at the rat Apc locus. Importantly, de novo CpG methylation is not required for the initiation or propagation of X inactivation in mammals (32). Alternatively, long-range protein-based silencing mechanisms (for example, with macroH2A) invoke cooperative binding. A paradigm for this behavior is the clustered binding of phage T4's gp32 protein to its substrate (33). Altogether, a range of molecular mechanisms can be investigated to explain the cis-acting monoallelic silencing observed in the gMOH/cLOH tumors. This report illustrates that the range of possible monoallelic and biallelic silencing mechanisms can be uncovered first through the analysis of cDNA from F1 genotypes.
We have developed informative models for familial colon cancer in both the mouse and the rat and have used quantitative assays for LOH in gDNA and cDNA of these tumors. Beyond the previously documented pathway of tumorigenesis involving LOH by homologous somatic recombination, we provide evidence for tumorigenesis by monoallelic and biallelic silencing of the Apc tumor suppressor locus in the absence of genomic instability. The F1-Pirc rat provides a large proportion of colon tumors that are highly enriched for epithelial tumor material. The residual level of admixture between LOH and MOH classes may be owing to the nonmedullary nature of all intestinal tumors, or it may reflect polyclonality as documented in the mouse and human (21, 22, 34).
The discovery of monoallelic silencing at the Apc locus indicates that this platform can be extended to investigate the occurrence and phenotypic consequences of allele-specific genetic and epigenetic events over the entire genome. Informative F1 genotypes enhance the dynamic range by which to quantify allele-specific genetic or epigenetic events. It will be interesting to expand the narrow window of the Apc locus in this study to a genome-wide epigenetic analysis. Identifying the full range of neoplastic pathways and stages that involve monoallelic silencing can power studies of the genetic and environmental influences on this apparently random epigenetic process. Continued investigation by whole-genome and transcriptome sequencing of the gMOH tumor classes discovered in this study is also called for to determine the spectrum of mutations that would combine synthetically with the proposed haploinsufficient state of Apc.
Materials and Methods
Mouse and Rat Maintenance.
We bred mice carrying the Min allele in cis to the Rb9 (7:18) translocation on the C57BL/J background (Rb9-Min) to increase the proportion of adenomas that maintain heterozygosity at the Apc locus (13).
Rats carrying the Pirc allele were bred on an F1 hybrid background that permits examination of allele-specific genetic and epigenetic events at all informative loci in the genome. Inbred F344N/Tac (Taconic) male rats coisogenic for the ApcPirc mutation (F344-Pirc) were bred to ACI/Hsd (ACI; Harlan) females to generate (ACI × F344)F1 ApcPirc/+ (F1-Pirc) hybrids. The Pirc allele was genotyped as previously described (18).
Rats and mice were fed 5020 chow (Purina) with access to an automatic supply of acidified water. One-half of the rats were treated with DSS in the drinking water. These animals were given two 7-d cycles of 4% DSS (500,000 kDa molecular mass) starting at 40 d of age, with a 7-d break between treatments. Animals were maintained in standard cages under a university-approved animal protocol in a facility approved by the Association for Assessment and Accreditation of Laboratory Animal Care.
Tumor Harvesting.
We designed the protocol for tumor dissection to optimize both the molecular and histological analysis of each tumor. Speed of dissection, quantity of material, and minimization of nontumor tissue are each important. Tumors were harvested at necropsy immediately after CO2 asphyxiation. Colons in the rat and both colons and small intestines in the mouse were washed with Dulbecco's PBS. One-half of each small-intestinal mouse tumor (not including the muscularis) and a one-quarter wedge of tumor from the upper one-half of each rat colonic tumor were put into RLTplus buffer (Qiagen) and homogenized. This method reduced the extent of nontumor tissue in the resulting RNA and DNA extractions. Intestines with the remaining tumors were washed with 70% ethanol, fixed in 10% buffered formalin for ∼16 h (mouse) or 24–40 h (rat), and then stored in 70% ethanol. When a portion of a tumor had been taken into RLTplus buffer, the residual tissue was excised and fixed for histological analysis. Purity of the dissected material was assessed from sections of the fixed tissue either stained with H&E or analyzed by immunochemistry for enhanced β-catenin expression.
Extraction of DNA and RNA from Tumors.
DNA and RNA were extracted from each RLTplus sample using the standard Qiagen RNA/DNA extraction kit with the manufacturer's protocol. DNA contamination of RNA samples was reduced to background levels by DNase treatment of the RNA during the extraction using the recommended on the column DNase treatment (Qiagen). cDNA was then generated from the resulting RNA using the Invitrogen SuperScript III reverse transcriptase kit (Invitrogen).
Identification and Confirmation of Polymorphisms and Mutations in the Rat Genome.
Standard fluorescent Sanger sequencing was used to confirm SNPs in the rat genome and identify new mutations in particular genes of interest in colonic tumors of the F1-Pirc rat. We identified SNPs using the Rat Genome Database SNPlotyper and chose those SNPs near the telomeric ends of chromosomes to capture any proximal somatic recombination events. To identify new mutations in tumors, all 15 exons, exon–intron boundaries, and 1 kb of the upstream promoter and alternative promoter of the rat Apc locus were sequenced as well as the hotspot regions of K-Ras (the sequence around the codon for amino acid G12) and β-catenin (Ctnnb1; exons 2, 3, and 4). All primers were designed using Primer 3 v0.4.0 (Table S4) (35). PCR protocols were optimized for Mg2+ concentration and annealing temperature. Products were confirmed by agarose gel electrophoresis and treated with exonuclease I (0.5 U) and shrimp alkaline phosphatase (0.25 U) at 37 °C for 30 min followed by heat inactivation at 80 °C for 15 min. A volume of 1–2 μL was used to sequence in both forward and reverse directions with Applied Biosystems Big Dye v3.1, and the products were purified on Sephadex G50 columns. All products were run by the University of Wisconsin Biotechnology Core Sequencing Facility.
Determination of Allelic Ratios.
To follow genetic and epigenetic changes at the Apc locus, we used pyrosequencing to quantify the ratios of the Min and Pirc single-base Apc mutations vs. their WT alleles. Confirmed rat SNPs covering the genome were used to detect any changes of allelic ratios in F1-Pirc tumor samples. For pyrosequencing, gDNA and cDNA samples were used in a PCR mix with the following final concentrations: 1× GoTaq clear buffer, 1.2 mM MgCl2, 0.2 mM dNTPs, 264 pM each primer, 0.6 U GoTaq Flexi (Promega), 8 μL DNA, and ddH2O to 50 μL (18, 36). The PCR cycling profile was 94 °C for 3 min followed by 50 cycles of 94 °C for 15 s, 57 °C for 1.5 min, and 72 °C for 2 min, with a final elongation step at 72 °C for 10 min. Pyrosequencing was performed according to the manufacturer's protocols using Pyro Gold Reagents with a PSQ96 or Qiagen Pyromark MD machine and PSQ 96 v2.1 software (Biotage) (Table S4); 40 μL PCR product were used per well. For samples run in the PSQ96, only sequence reads with single-base peak heights of over 120 units were included. All pyrosequencing assays were run in duplicate. If results differed by more than 5%, samples were rerun.
Statistical Analysis of Allelic Ratios.
Two statistical methods—cluster and admixture analyses—were developed and applied to pyrosequencing allelic ratio data from gDNA and cDNA. In the cluster analysis, tumors were separated into three classes—gMOH/cMOH, gLOH/cLOH, and gMOH/cLOH—using computational and mathematical tools from the theory of Gaussian mixture models (37, 38). Computations were performed in the R system (39). Specifically, the gDNA, cDNA data pair from each tumor was considered to be a random draw from a mixture of three bivariate normal components, one for each of the named classes (SI Materials and Methods). From the fitted probability model, various predictions were computed, including contours indicating the posterior probability that a tumor arose from the gMOH/cLOH class given the gDNA/cDNA data. This mixture of bivariate normal distributions extends a simpler univariate normal mixture used previously for MOH/LOH analysis to available data (17).
In the admixture data analysis, we recognized that each tumor sample could be an unequal mixture of three pure cell types—the same three types (classes) as in the cluster analysis, except that now these classes were viewed as properties of constituent cells rather than whole tumors. Furthermore, we reasoned that measurement error in the gDNA and cDNA channels should to be uncorrelated and that apparent within-class correlation in the first analysis would be attributable to admixture of different cell types within each tumor. Unknown parameters in the admixture model include class-specific mean values for the three bivariate pure cell classes constrained as in the cluster analysis, channel-specific measurement error variance, and tumor-specific mixing proportions over the three classes. To facilitate inference, we treated each tumor's vector of three mixing proportions as a draw from an unknown distribution over proportion vectors (a distribution that characterizes the tumor population in question [Rb9-Min or F1-Pirc]). Markov chain Monte Carlo computations were then developed to estimate the unknown parameters of the admixture model (SI Materials and Methods).
qRT-PCR.
We performed qRT-PCR to determine whether overall APC RNA expression was diminished in MOH tumors. We used predesigned assays for APC RNA (TaqMan:Rn00560714 FAM-dye-labeled MGB probe; Applied Biosystems) and control GAPDH RNA (4352338E VIC dye-labeled MGB probe). The two assays were combined and run together for each sample. We used the ABI Taqman Gene Expression Master Mix at 1× concentration in 20-μL reactions with cDNA generated as described above. The samples were run in duplicate and analyzed on an ABI 7900HT machine using the recommended cycling conditions of 40 cycles with a 15-s 95 °C denaturing step and a 1-min 60 °C annealing/extension. Data were analyzed using the ABI program DataAssist.
Supplementary Material
Acknowledgments
The University of Wisconsin Comprehensive Cancer Center's Histology Shared Service has provided talented support, both technical (Jane Weeks and Harlene Edwards) and conceptual (Ruth Sullivan). We thank Norman Drinkwater and James Shull for critical comments and Alexandra Shedlovsky and Jennifer Pleiman for clarification of this report. We thank two reviewers for critiques that have significantly enhanced this report. Finally, we pay tribute to the long-term impact of Mary Lyon on mammalian genetics and epigenetics. This research was supported by American Cancer Society Postdoctoral Fellowship PF-08-077-01 (to J.M.A.-L.), a training grant from the National Institute on Environmental Health Sciences (to A.A.I.), National Cancer Institute Grants R01 CA63677 and R01 CA125591 (to W.F.D.), and National Institutes of Health/National Cancer Institute Grant P30 CA014520 (University of Wisconsin Comprehensive Cancer Center Support). This is publication no. 3647 from the Laboratory of Genetics.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1120753109/-/DCSupplemental.
References
- 1.Fearnhead NS, Britton MP, Bodmer WF. The ABC of APC. Hum Mol Genet. 2001;10:721–733. doi: 10.1093/hmg/10.7.721. [DOI] [PubMed] [Google Scholar]
- 2.Yeung AT, et al. One-hit effects in cancer: Altered proteome of morphologically normal colon crypts in familial adenomatous polyposis. Cancer Res. 2008;68:7579–7586. doi: 10.1158/0008-5472.CAN-08-0856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lewis A, et al. Severe polyposis in Apc(1322T) mice is associated with submaximal Wnt signalling and increased expression of the stem cell marker Lgr5. Gut. 2010;59:1680–1686. doi: 10.1136/gut.2009.193680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jones S, et al. Comparative lesion sequencing provides insights into tumor evolution. Proc Natl Acad Sci USA. 2008;105:4283–4288. doi: 10.1073/pnas.0712345105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheng YW, et al. CpG island methylator phenotype associates with low-degree chromosomal abnormalities in colorectal cancer. Clin Cancer Res. 2008;14:6005–6013. doi: 10.1158/1078-0432.CCR-08-0216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Georgiades IB, Curtis LJ, Morris RM, Bird CC, Wyllie AH. Heterogeneity studies identify a subset of sporadic colorectal cancers without evidence for chromosomal or microsatellite instability. Oncogene. 1999;18:7933–7940. doi: 10.1038/sj.onc.1203368. [DOI] [PubMed] [Google Scholar]
- 7.Toyota M, et al. CpG island methylator phenotype in colorectal cancer. Proc Natl Acad Sci USA. 1999;96:8681–8686. doi: 10.1073/pnas.96.15.8681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Loeb LA, Loeb KR, Anderson JP. Multiple mutations and cancer. Proc Natl Acad Sci USA. 2003;100:776–781. doi: 10.1073/pnas.0334858100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lengauer C, Kinzler KW, Vogelstein B. Genetic instabilities in human cancers. Nature. 1998;396:643–649. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- 10.Edelmann W, et al. Tumorigenesis in Mlh1 and Mlh1/Apc1638N mutant mice. Cancer Res. 1999;59:1301–1307. [PubMed] [Google Scholar]
- 11.Halberg RB, et al. Long-lived Min mice develop advanced intestinal cancers through a genetically conservative pathway. Cancer Res. 2009;69:5768–5775. doi: 10.1158/0008-5472.CAN-09-0446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fodde R, et al. Mutations in the APC tumour suppressor gene cause chromosomal instability. Nat Cell Biol. 2001;3:433–438. doi: 10.1038/35070129. [DOI] [PubMed] [Google Scholar]
- 13.Haigis KM, Dove WF. A Robertsonian translocation suppresses a somatic recombination pathway to loss of heterozygosity. Nat Genet. 2003;33:33–39. doi: 10.1038/ng1055. [DOI] [PubMed] [Google Scholar]
- 14.Sieber OM, et al. Homologous somatic recombination is a more subtle form of genomic instability observed in human FAP and in its murine models. Proc Natl Acad Sci USA. 2002;99:16910–16915. [Google Scholar]
- 15.Grady WM, et al. Methylation of the CDH1 promoter as the second genetic hit in hereditary diffuse gastric cancer. Nat Genet. 2000;26:16–17. doi: 10.1038/79120. [DOI] [PubMed] [Google Scholar]
- 16.Cui H, et al. Loss of IGF2 imprinting: A potential marker of colorectal cancer risk. Science. 2003;299:1753–1755. doi: 10.1126/science.1080902. [DOI] [PubMed] [Google Scholar]
- 17.Shoemaker AR, et al. A resistant genetic background leading to incomplete penetrance of intestinal neoplasia and reduced loss of heterozygosity in ApcMin/+ mice. Proc Natl Acad Sci USA. 1998;95:10826–10831. doi: 10.1073/pnas.95.18.10826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Amos-Landgraf JM, et al. A target-selected Apc-mutant rat kindred enhances the modeling of familial human colon cancer. Proc Natl Acad Sci USA. 2007;104:4036–4041. doi: 10.1073/pnas.0611690104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Crabtree M, et al. Refining the relation between ‘first hits’ and ‘second hits’ at the APC locus: The ‘loose fit’ model and evidence for differences in somatic mutation spectra among patients. Oncogene. 2003;22:4257–4265. doi: 10.1038/sj.onc.1206471. [DOI] [PubMed] [Google Scholar]
- 20.Baba Y, et al. Hypomethylation of the IGF2 DMR in colorectal tumors, detected by bisulfite pyrosequencing, is associated with poor prognosis. Gastroenterology. 2010;139:1855–1864. doi: 10.1053/j.gastro.2010.07.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thliveris AT, et al. Polyclonality of familial murine adenomas: Analyses of mouse chimeras with low tumor multiplicity suggest short-range interactions. Proc Natl Acad Sci USA. 2005;102:6960–6965. doi: 10.1073/pnas.0502662102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thirlwell C, et al. Clonality assessment and clonal ordering of individual neoplastic crypts shows polyclonality of colorectal adenomas. Gastroenterology. 2010;138:1441–1454. doi: 10.1053/j.gastro.2010.01.033. [DOI] [PubMed] [Google Scholar]
- 23.Payne SR, Kemp CJ. Tumor suppressor genetics. Carcinogenesis. 2005;26:2031–2045. doi: 10.1093/carcin/bgi223. [DOI] [PubMed] [Google Scholar]
- 24.Hagstrom SA, Dryja TP. Mitotic recombination map of 13cen-13q14 derived from an investigation of loss of heterozygosity in retinoblastomas. Proc Natl Acad Sci USA. 1999;96:2952–2957. doi: 10.1073/pnas.96.6.2952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.De Rosa M, et al. Alternative splicing and nonsense-mediated mRNA decay in the regulation of a new adenomatous polyposis coli transcript. Gene. 2007;395:8–14. doi: 10.1016/j.gene.2006.10.027. [DOI] [PubMed] [Google Scholar]
- 26.Lyon MF. Gene action in the X-chromosome of the mouse (Mus musculus L.) Nature. 1961;190:372–373. doi: 10.1038/190372a0. [DOI] [PubMed] [Google Scholar]
- 27.Brown CJ, et al. A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature. 1991;349:38–44. doi: 10.1038/349038a0. [DOI] [PubMed] [Google Scholar]
- 28.Pasque V, Gillich A, Garrett N, Gurdon JB. Histone variant macroH2A confers resistance to nuclear reprogramming. EMBO J. 2011;30:2373–2387. doi: 10.1038/emboj.2011.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Derks S, et al. Integrated analysis of chromosomal, microsatellite and epigenetic instability in colorectal cancer identifies specific associations between promoter methylation of pivotal tumour suppressor and DNA repair genes and specific chromosomal alterations. Carcinogenesis. 2008;29:434–439. doi: 10.1093/carcin/bgm270. [DOI] [PubMed] [Google Scholar]
- 30.Linhart HG, et al. Dnmt3b promotes tumorigenesis in vivo by gene-specific de novo methylation and transcriptional silencing. Genes Dev. 2007;21:3110–3122. doi: 10.1101/gad.1594007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maegawa S, et al. Widespread and tissue specific age-related DNA methylation changes in mice. Genome Res. 2010;20:332–340. doi: 10.1101/gr.096826.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sado T, Okano M, Li E, Sasaki H. De novo DNA methylation is dispensable for the initiation and propagation of X chromosome inactivation. Development. 2004;131:975–982. doi: 10.1242/dev.00995. [DOI] [PubMed] [Google Scholar]
- 33.Delius H, Mantell NJ, Alberts B. Characterization by electron microscopy of the complex formed between T4 bacteriophage gene 32-protein and DNA. J Mol Biol. 1972;67:341–350. doi: 10.1016/0022-2836(72)90454-8. [DOI] [PubMed] [Google Scholar]
- 34.Novelli MR, et al. Polyclonal origin of colonic adenomas in an XO/XY patient with FAP. Science. 1996;272:1187–1190. doi: 10.1126/science.272.5265.1187. [DOI] [PubMed] [Google Scholar]
- 35.Rozen S, Skaletsky HJ. In: Bioinformatics Methods and Protocols: Methods in Molecular Biology. Krawetz S, Misener S, editors. Totowa, NJ: Humana Press; 2000. pp. 365–386. [Google Scholar]
- 36.Kwong LN, et al. Identification of Mom7, a novel modifier of Apc(Min/+) on mouse chromosome 18. Genetics. 2007;176:1237–1244. doi: 10.1534/genetics.107.071217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McLachlan G, Peel D. Finite Mixture Models. New York: Wiley; 2000. [Google Scholar]
- 38.Fraley C, Raftery AE. Model-based clustering, discriminant analysis, and density estimation. J Am Stat Assoc. 2002;97:611–631. [Google Scholar]
- 39.R Development Core Team . R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing; 2011. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.