Abstract
Membrane fusion within the eukaryotic endomembrane system depends on the initial recognition of Rab GTPase on transport vesicles by multisubunit tethering complexes and subsequent coupling to SNARE-mediated fusion. The conserved vacuolar/lysosomal homotypic fusion and vacuole protein sorting (HOPS) tethering complex combines both activities. Here we present the overall structure of the fusion-active HOPS complex. Our data reveal a flexible ≈30-nm elongated seahorse-like structure, which can adopt contracted and elongated shapes. Surprisingly, both ends of the HOPS complex contain a Rab-binding subunit: Vps41 and Vps39. The large head contains in addition to Vps41 the SNARE-interacting Vps33, whereas Vps39 is found in the bulky tip of its tail. Vps11 and Vps18 connect head and tail. Our data suggest that HOPS bridges Ypt7-positive membranes and chaperones SNAREs at fusion sites.
Eukaryotic cells rely on an elaborate and dynamic vesicular transport system to direct proteins and lipids to their destinations in the cell. Membrane fusion within the endomembrane system of eukaryotic cells is a multistage reaction, which includes an initial recognition, termed tethering, followed by mixing of the membrane bilayers (1, 2).
Tethering in most cases is controlled by Rab GTPases, which exist in an inactive membrane-bound or soluble GDP form and an active membrane-bound GTP form. Once activated by their respective guanine nucleotide exchange factor, such as the well-characterized transport protein particle (TRAPP) complex that acts on Rab1, GTP-Rabs residing on vesicles or organelles can bind to tethering factors, including long coiled-coil proteins like the early endosome antigen (EEA1), which captures vesicles at a distance and promotes their subsequent fusion (1, 3, 4). Multisubunit tethering complexes (MTCs), which are found on all endomembranes, have additional functions such as SNARE binding (1, 2). Of all known complexes, only the Dsl1 complex at the endoplasmic reticulum (ER) does not depend on a Rab GTPase for its apparent activity but rather is anchored to the ER via SNAREs (5, 6). All other MTCs, such as the exocyst, Golgi-associated retrograde protein (GARP) complex, or COG, bind their respective Rab-GTP and require this interaction for function (1). Some MTCs, like COG, the exocyst, depend on SLY1-20 (Dsl) complex, and homotypic fusion and vacuole protein sorting (HOPS), also bind SNAREs (1, 2) and support their assembly during membrane fusion (7). Structural data on coiled-coil and Rab-independent tethers (1) and the Rab-binding domain of the exocyst subunit Sec15 exist (8). However, molecular insight into the overall structure of a Rab-binding MTC has been lacking to date.
Within the endolysosomal system, two similar heterohexameric MTCs, the endosomal class C core vacuole/endosome tethering (CORVET) complex and the vacuolar HOPS, have been identified (9–11). They share four of their six subunits, namely the class C subunits Vps11, Vps16, Vps18, and the Sec1/Munc18-like Vps33, which interacts with SNAREs (12). The endosomal CORVET contains in addition the two subunits Vps3 and Vps8, which both bind the Rab5 homolog Vps21 in its GTP form (11, 13, 14). HOPS binds the Rab7 GTPase Ypt7 via its subunits Vps41 and Vps39 (15, 16) and promotes fusion of late endosomes, AP-3 vesicles, and autophagosomes with vacuoles, as well as homotypic fusion (2, 17). All HOPS and CORVET subunits—except Vps33—likely consist of an N-terminal β-propeller and a C-terminal α-solenoid region (13, 16). They thus differ structurally from the CATCHR (complex associated with tethering containing helical rods) complexes such as Dsl1, COG, GARP, and exocyst (1). HOPS is also the only complex for which an in vitro fusion assay has been established (7, 18). Moreover, HOPS is limiting for Ypt7-dependent fusion of proteoliposomes carrying vacuolar SNAREs (19, 20). HOPS—and likely CORVET—are conserved across species (21–23). Thus, HOPS is a prime candidate to unravel the mechanism of Rab-mediated tethering and the promotion of SNARE-mediated fusion.
Here we analyzed the HOPS complex by electron microscopy (EM) combined with single-particle analysis and tomography. HOPS forms an elongated particle with two Rab-binding sites formed by Vps39 and Vps41 at opposite ends, connected by a flexible linker between the two terminal lobes. The SNARE-binding site is proximal to one Rab-binding site, suggesting that HOPS coordinates Rab-mediated tethering with SNARE-driven fusion.
Results
HOPS Is an Elongated and Flexible Complex.
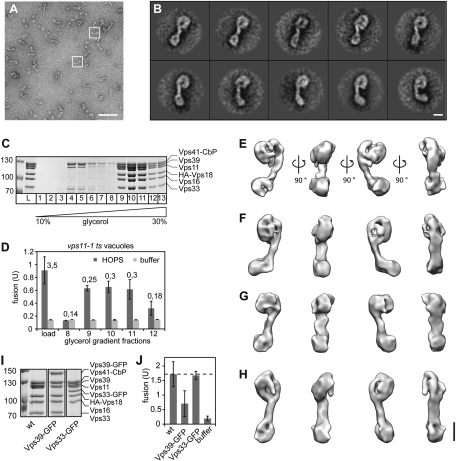
HOPS activity in in vitro fusion assays has been extensively documented (7, 16, 18), although its precise function has remained unclear. We thus reasoned that a functional understanding of tethering requires the elucidation of its architecture. To select its structure, we analyzed the HOPS complex by single-particle EM of negatively stained particles (Fig. 1 A and B). Overproduced HOPS was purified via a tandem affinity purification (TAP) tag on Vps41 from yeast and further resolved on glycerol gradients (Fig. 1C). Using a modified in vitro vacuole fusion assay that depends on the addition of purified HOPS to obtain fusion (18), we confirmed robust fusion activity of the complex in fractions 9–11 of the gradient (Fig. 1D; details in Experimental Procedures). When the complex was analyzed by EM, it partially disintegrated (Fig. S1 A and B). To obtain a sufficient number of intact HOPS particles, we further stabilized HOPS by applying the GraFix method, in which the complexes are centrifuged over a glycerol gradient into an increasing concentration of glutardialdehyde (24). Importantly, mild intramolecular cross-linking occurs slowly while HOPS migrates into the gradient, and intermolecular cross-linking is prevented. As expected, HOPS stability was increased and its integrity conserved (Fig. S1 C and D). In comparison with the non–cross-linked HOPS, we did not notice any overall structural changes (Fig. S1 A–D). We therefore believe that the observed HOPS conformations (see below) reflect its overall flexibility and not cross-linking artifacts. In addition to that, we tested HOPS with GFP tags at different subunits (Fig. 1 A and B and Fig. S1 E and F) and found that HOPS with Vps39-GFP (Fig. 1I) was more stable than wild-type HOPS (Fig. S1 C and D) and HOPS with Vps33-GFP (Fig. 1I and Fig. S1 E and F). Interestingly, it was also only half as active as wild-type HOPS (Fig. 1J). This is not due to GFP tagging per se as HOPS with Vps33-GFP showed robust activity (Fig. 1 I and J). Because of its reproducible remaining activity and higher stability, all subsequent structural analyses were performed with HOPS (Vps39-GFP). The class averages (Fig. 1A and Fig. S2) revealed that HOPS complexes had a seahorse-like shape, with a deeper cavity in its head region and a bulky, highly flexible lobe at the end of its tail (Fig. 1B and Movie S1).
Fig. 1.
Isolation and structure of the HOPS complex. (A) Typical micrograph area of the negatively stained HOPS (Vps39-GFP) tethering complex. Representative particles are boxed in white. (Scale bar, 100 nm.) (B) Representative class averages, each containing between 12 and 27 particles. Fig. S2 shows the complete set of class averages. (Scale bar, 10 nm.) (C) Purification of HOPS via a glycerol gradient. Overproduced HOPS with TAP-tagged Vps41 (16) was purified from yeast via IgG Sepharose as described in Experimental Procedures, then separated on 10–30% glycerol gradients. (D) Fusion activity of HOPS. Indicated fractions of C were applied to reporter vacuoles from vps11-1 strains. Six microliters of each fraction were analyzed per fusion reaction. Numbers indicate the protein concentration in the sample. (E–H) 3D reconstructions of the yeast HOPS complex using the random conical tilt approach and 3D multireference alignment. Four different conformations (E to H) were chosen. Shown are four different side views, after horizontal rotation of 90° around their longest axis. Table S1 shows the volumes of the structures. (Scale bar, 10 nm.) (I) Purification of wild-type and GFP-tagged HOPS complexes. Coomassie-stained SDS/PAGE gels of HOPS, HOPS (Vps33-GFP), and HOPS (Vps39-GFP) are shown. (J) Fusion activity. The different GFP variants were analyzed in fusion as in D. Fusion was compared to wild-type HOPS (wt) was set to 100%. SDs are given, n = 3.
Because of the flexible nature of the complex, we were not able to use cryo-electron microscopy to determine the 3D structure of HOPS. Therefore, we recorded image pairs of negatively stained complexes at tilt angles of 50° and 0° to obtain different views needed to calculate four different 3D density maps of HOPS with the random conical tilt approach (25) (Fig. 1 E–H). Tomography of cryo-negatively stained particles revealed the same overall structures as our reconstructions (Fig. S3). This confirmed that HOPS had not been flattened because of dehydration in our negative stain studies.
Our density maps showed that HOPS is an ≈30-nm-long complex that can adopt several conformations without changing its overall composition (Fig. 1 E–H, Table S1, and Movies S2 and S3). Whereas it was obvious from the 2D projections (Fig. 1B) that the end of the tail is very flexible, the 3D reconstructions showed that the head is also intrinsically flexible. The backbone proximal to the large head appears rather stiff and extends into the head. This results in contracted (Fig. 1E) and elongated (Fig. 1H) as well as intermediate shapes (Fig. 1 F and G), in which the distance between the head and tail is almost doubled (from 7 nm in the contracted to 14 nm in the elongated state).
Positioning of the Subunits Within the HOPS Complex.
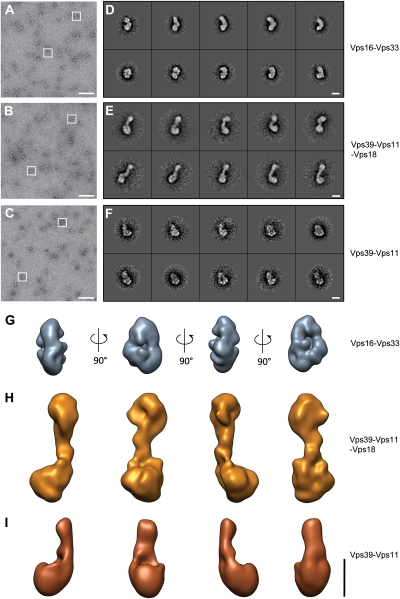
To determine the orientation of subunits within HOPS, we initially turned to subcomplexes composed of Vps16-Vps33, Vps39-Vps11-Vps18, and Vps39-Vps11, each with an apparent equimolar stoichiometry (Fig. 2 and Fig. S4) (16). As for the entire HOPS complex, we stabilized the subcomplexes by mild cross-linking using glutardialdehyde on a glycerol gradient (GraFix). We recorded tilt pairs of negatively stained particles (Fig. 2 A–C) and aligned and classified them (Fig. 2 D–F). Similar to HOPS, its subcomplexes were quite flexible, suggesting hinge regions in the involved proteins (Movies S4 and S5). We then calculated 3D reconstructions of the most prominent class averages (Fig. 2 G–I). The Vps16-Vps33 complex forms a tight complex with a smooth and a rough side, whereas the Vps39-Vps11-Vps18 trimer appears like a smaller HOPS without head. In the absence of Vps18, the remaining dimer looks like the bulky part of the HOPS tail with a part of its backbone.
Fig. 2.
Subcomplexes of HOPS. (A–F) Typical micrograph areas (A–C) and representative class averages (D–F) of negatively stained HOPS subcomplexes Vps16-Vps33 (A and D), Vps39-Vps11-Vps18 (B and F), and Vps39-Vps11 (C and G). Representative particles are boxed in white. Class averages contain approximately 50 particles each. (G–I) 3D reconstructions of HOPS subcomplexes (G) Vps16-33, (H) Vps39-11-18, and (I) Vps39-11. (Scale bars, 100 nm in A–C and 10 nm in D–I.)
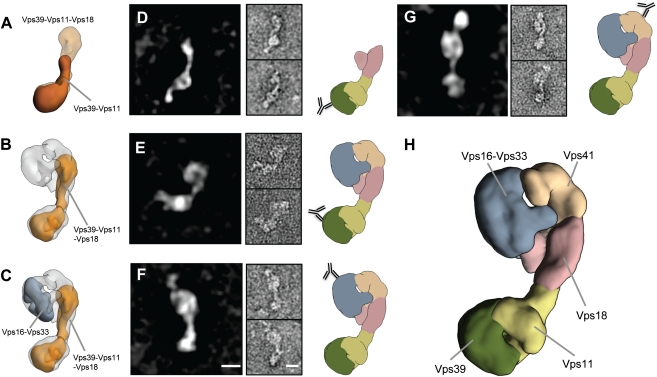
We then used the subcomplex structures and merged them with the HOPS 3D structure (Fig. 3). A comparison of the Vps39-Vps11 dimer with the Vps39-Vps11-Vps18 trimer revealed that Vps18 is oriented toward the large head in the backbone of HOPS (Fig. 3 A and B), whereas Vps39-Vps11 occupies the bulky tail. We confirmed this orientation by antibody staining against the calmodulin-binding peptide (CbP) attached to Vps39 using the Vps39-Vps11-Vps18 trimer (Fig. 3D), and against the GFP-tag for the full HOPS complex (Fig. 3E). Vps16-Vps33 could be nicely mapped to a substructure at the side of the large head (Fig. 3C). This orientation was verified by labeling functional HOPS carrying Vps33-GFP (Fig. 1J and Fig. S5 A–D) with the respective GFP antibody (Fig. 3F). As already apparent (Fig. 3C), Vps41 was localized to the remaining density within HOPS, which was confirmed by antibody labeling of the C-terminal CbP of Vps41 (Fig. 3G). Previously, we and others postulated that Vps41 and Vps39 would cooperate or even interact with each other at one end of the complex to support Rab binding (13, 16). It was thus unexpected that Vps41 and Vps39 are present on opposite ends of the elongated HOPS complex (Fig. 3H and Movie S6).
Fig. 3.
Organization of the HOPS complex. (A and B) Electron density of Vps39-Vps11 subcomplex (brown) was fitted into the volume of the Vps39-Vps11-Vps18 subomplex (transparent orange), which was then put into the density of HOPS (transparent gray). (C) The same was done with the 3D density map of the subcomplex Vps16-Vps33 (gray). (D–G) Labeling with antibodies against Vps39-CbP of the subcomplex Vps39-Vps11-Vps18 (D), the HOPS complex with antibody against Vps39-GFP (E), and HOPS labeled with antibody against Vps33-GFP (F) and Vps41-CbP (G). Left: Representative class averages, each containing approximately 10 particles. Center: Raw particle images of the corresponding classes. Right: Localization of each antibody graphically mapped to the model of HOPS derived from one of its 3D densities shown in Fig. 1 (Experimental Procedures). (H) Segmented model of the HOPS tethering complex. The head domain consists of Vps16-Vps33 (gray) and Vps41 (sandy brown), connected to the Vps18 stem (pink). The bulky tail is composed of Vps11 (light olive green) and Vps39 (dark olive green). (Scale bars, 10 nm.)
HOPS Has Two Rab7-Binding Sites at Opposite Ends.
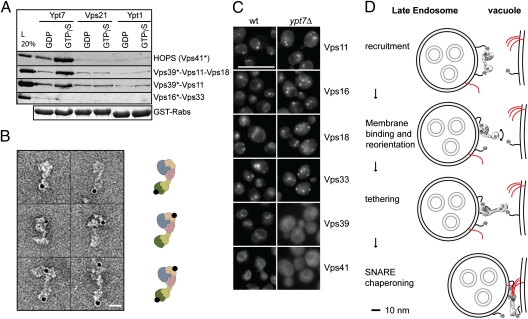
Both Vps41 and Vps39 can bind Ypt7 (13, 15, 16), although our previous data suggested that Vps39 was inactive within HOPS as an Ypt7 interactor (16). We therefore readdressed the Rab-binding ability of Vps39 by GST pull-down, using the available subcomplexes and optimized protocols, and observed efficient binding of subcomplexes containing Vps39 to Ypt7-GTP (Fig. 4A). Importantly, HOPS is at least twice as efficient in Rab binding in comparison with the Vps39 subcomplexes (Fig. 4A). Our data thus agree with two separated Rab-binding sites on two ends of an elongated tethering complex. To further confirm this finding, we labeled cross-linked GTP-Ypt7-HOPS complexes with nano-gold directed against the His-tag on Ypt7, which localized to the identified position of the subunits Vps39 and Vps41 at opposite ends of the complex (Fig. 4B). We then followed all HOPS subunits as GFP fusions in vivo. In agreement with our in vitro observations, Vps39 and Vps41 were the only HOPS subunits that were displaced from membranes in ypt7Δ cells (Fig. 4C). We thus conclude that the HOPS complex contains two binding sites for the GTP-loaded Rab7 GTPase Ypt7 that reside at opposite ends of an elongated and flexible particle.
Fig. 4.
Two Rab-binding sites on a single HOPS complex. (A) Interaction of Ypt7 with Vps39. Rab pull-down with GTPγS- or GDP-loaded Rabs Ypt7, Vps21, and Ypt7 was done with purified HOPS or the HOPS subcomplexes. Bound proteins were eluted and analyzed on SDS/PAGE, followed by Western blotting against the CbP-tag (indicated with *). A representative Coomassie gel shows GST-Rabs present in the pull-down. (B) HOPS and Ypt7-His6 were incubated, cross-linked, and separated on a glycerol gradient. The complexes were then labeled with Ni-NTA-nano-gold and imaged in the electron microscope. Left: Raw particle images. Right: Localization of the gold-label graphically mapped to the model of HOPS as shown in Fig 3. (Scale bar, 10 nm.) (C) Localization of GFP-tagged HOPS subunits. All HOPS subunits were genomically tagged at their C terminus with GFP and localized by fluorescence microscopy in wild-type (wt) and ypt7Δ cells. (Scale bar, 10 μm.) (D) Model of HOPS function in fusion. SNAREs are in red. Details in text.
Discussion
The structural and functional analysis of the yeast HOPS complex provides a major advance in understanding Rab-dependent homotypic tethering at the molecular level. Our data reveal that HOPS is a flexible, 28- to 35-nm-long seahorse-like structure with its two Rab-binding sites on opposite ends. Flexibility is likely required for functionality. We observed that HOPS carrying GFP-tagged Vps39 binds Ypt7 and SNAREs efficiently but is less fusogenic than wild-type HOPS (Fig. 1 I and J). In our EM analysis, this complex was also more stable than the wild-type variant. It is possible that additional conformations, including contacts between the head and the tail, exist, which would explain some of the previously reported interaction (13).
Head and tail in HOPS are likely reserved for different functions. The large head contains both the SM-like Vps33 protein, which binds SNAREs (26, 27), its interactor Vps16, and Vps41. Overall, our structure largely confirms the previously mapped interactions (10, 12, 13, 16). Surprisingly, interactions of Vps41 with Vps16 or Vps33 have not been reported but could be reproduced by pull-downs using a C-terminal 400-residue fragment of Vps41 (Fig. S5E). In the structure, Vps41 is also proximal to Vps18, in agreement with earlier findings (16), and Vps18 forms the backbone of HOPS, bridging its head and tail. In fact, HOPS falls apart into the Vps16-Vps33 and Vps39-Vps11 dimer in vps41Δ cells (16). How does HOPS then function in fusion? Our model is summarized in Fig. 4D. HOPS is recruited to late endosomes by binding Ypt7 via Vps39 and Vps41 (Fig. 4C) (28). At the endosome, HOPS may be reoriented at the membrane, as Vps41 in addition to Ypt7-GTP also binds to lipids via its ALPS (amphipathic lipid packaging) motif (27, 28), and the entire HOPS has a preference for phosphoinositides (18). This membrane binding seems tight enough to prevent efficient Rab inactivation (29, 30). Such a reorientation of HOPS would require that Vps39 let go of endosomal Ypt7, to then be free for binding Ypt7 on the vacuole surface. In agreement with this model, HOPS requires Ypt7 to tether proteoliposomes in vitro (31) and promote their Ypt7-dependent fusion (19, 20). At present we cannot explain the observed interaction between Vps41 and Vps39 (13), although it is possible that HOPS undergoes a closed conformation whereby the two subunits get in touch. Tethering of the membranes may be facilitated by the ability of HOPS to stretch outward (Fig. 1 E–H and Movies S4 and S5). Elongated structures between endosomes and lysosomes have indeed been observed in ultrastructural studies, which could represent HOPS (32, 33). Closer apposition of the membranes would then allow Vps33 to facilitate SNARE assembly before membrane mixing. It is likely that the flexibility of HOPS also supports this process.
In addition to homotypic fusion, HOPS also acts in the fusion of AP-3 vesicles with vacuoles by binding the δ-ear domain of the AP-3 subunit Apl5 (28, 34). For this interaction, the HOPS subunit Vps41 has to be phosphorylated by the vacuolar kinase Yck3. Phosphorylation at its membrane-interacting ALPS motif releases Vps41 from its close membrane apposition and thus exposes the Apl5 binding site (28). On the basis of our structural insights, we can now start to unravel the precise function of Vps41/HOPS in this process.
How does HOPS compare with known EM structures of tethering complexes? To date, EM structures are available of the Dsl1 complex, which operates without Rab binding, and the Golgi-localized COG complex. Both belong to the CATCHR family of helical tethering complexes. The Dsl1 complex also forms an elongated particle, which requires ER-localized SNAREs for its membrane localization (5), and may capture COPI vesicles via coat binding, whereas COG forms a three-legged structure that is reminiscent of the clathrin legs (35).
HOPS shows some similarity to the Dsl complex but also differs strongly. The Dsl complex seems to be stiffer, owing to its rod-like subunits, whereas HOPS has six, likely interconnected subunits. Five of the six HOPS subunits are related to known vesicle coat components and nuclear pore complexes (36–39), which form polymeric structures unlike HOPS. It is thus likely that additional hinge regions exist between these subunits, which would explain its flexibility. COG with its three flexible legs seems more like a coat but could also span membranes with its similar length of some 40 nm (35). It will thus be important to determine in the future how each complex behaves on membranes. To understand HOPS in detail, high-resolution structures of the subunits and mapping of their interfaces will be critical to determine its precise organization and test the proposed tethering model.
Experimental Procedures
Details are provided in SI Experimental Procedures.
Biochemical Procedures.
HOPS was purified from 2 L of YP medium with 2% (vol/vol) d-galactose that were grown for 48 h. Purification of HOPS and HOPS subcomplexes was done via the TAP protocol, as previously described (16). For preparation of HOPS for EM analyses, the buffer was as follows: 1 M NaCl, 50 mM Hepes/NaOH (pH 7.5), and 10% (vol/vol) glycerol. Glycerol gradient centrifugation with simultaneous cross-linking (GraFix) with glutardialdehyde was performed according to Kastner et al. (24). Vacuole fusion was done as described with vacuoles carrying the vps11-1 allele (18).
EM and Image Processing.
Samples were either directly prepared for EM as previously described (40) or first cross-linked on a glycerol gradient as described above. All images were taken with a JEOL JEM-1400 electron microscope equipped with a LaB6 filament at an operation voltage of 120 kV. Particles were manually selected, aligned, and classified using reference-free alignment and K-means classification procedures implemented in SPARX and EMAN2 (41).
For the 3D reconstruction of HOPS (Vps39-GFP), image pairs were collected at tilt angles of 50° and 0°, and 4,800 particle pairs were selected from 200 image pairs using WEB, the display program associated with the SPIDER software package (42), which was used for further image processing steps. Random conical tilt reconstructions of the tilted particles were calculated from the seven best class averages by back-projection, followed by back-projection refinement. The resulting reconstructions were then submitted to several rounds of 3D multireference projection matching against the dataset of 24,900 single particles using SPARX. Four reconstructions remained stable during the refinement process with a resolution of ≈29 Å according to the 0.5 criterion (Fig. S6).
For visualization, analysis, and preparation of the figures, we used Chimera (43). Initial segmentation of the HOPS complex was performed automatically using the module Segment Map of the Chimera software package (44).
To locate subunits in the HOPS complex, we labeled HOPS (Vps33-GFP), HOPS (Vps39-GFP), HOPS-Vps41-CbP (wild type), and HOPS subcomplex Vps39-CbP-Vps11-Vps18 with anti-GFP and anti-CbP antibodies, respectively. The data sets were submitted to reference-free alignment and k-means classification (40–50 particles per class) using SPARX. To determine the position of HOPS Ypt7 binding, TAP-purified HOPS complexes and 150 μg purified His6-tagged Ypt7, preloaded with GTPγS according to Ostrowicz et al. (16), was incubated at 1:1 ratio for 1 h at 4 °C. After subsequent cross-linking by glutardialdehyde and separation on a glycerol gradient, the sample was labeled with Ni-NTA-nano-gold (Nanoprobes) for 1 min.
Supplementary Material
Acknowledgments
We thank Jacob Piehler (University of Osnabrück) for suggestions and discussions, Oliver Hofnagel for assistance at the electron microscope, Tatjana ter Braak and Clemens Ostrowicz for initial contributions, and Fulvio Reggiori and Francis Barr for useful comments on the manuscript. S.R. thanks Roger S. Goody for continuous support. This work was supported by Deutsche Forschungsgemeinschaft (DFG) Grant UN 111/5-2, Sonderforschungsbereich 944, the Hans-Mühlenhoff Foundation (C.U.), DFG Grant RA 1781/1-1, and the Max Planck Society (S.R.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1117797109/-/DCSupplemental.
References
- 1.Yu IM, Hughson FM. Tethering factors as organizers of intracellular vesicular traffic. Annu Rev Cell Dev Biol. 2010;26:137–156. doi: 10.1146/annurev.cellbio.042308.113327. [DOI] [PubMed] [Google Scholar]
- 2.Bröcker C, Engelbrecht-Vandré S, Ungermann C. Multisubunit tethering complexes and their role in membrane fusion. Curr Biol. 2010;20:R943–R952. doi: 10.1016/j.cub.2010.09.015. [DOI] [PubMed] [Google Scholar]
- 3.Barr F, Lambright DG. Rab GEFs and GAPs. Curr Opin Cell Biol. 2010;22:461–470. doi: 10.1016/j.ceb.2010.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim YG, et al. The architecture of the multisubunit TRAPP I complex suggests a model for vesicle tethering. Cell. 2006;127:817–830. doi: 10.1016/j.cell.2006.09.029. [DOI] [PubMed] [Google Scholar]
- 5.Ren Y, et al. A structure-based mechanism for vesicle capture by the multisubunit tethering complex Dsl1. Cell. 2009;139:1119–1129. doi: 10.1016/j.cell.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meiringer CT, et al. The Dsl1 protein tethering complex is a resident endoplasmic reticulum complex, which interacts with five soluble NSF (N-ethylmaleimide-sensitive factor) attachment protein receptors (SNAREs): Implications for fusion and fusion regulation. J Biol Chem. 2011;286:25039–25046. doi: 10.1074/jbc.M110.215327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mima J, Hickey CM, Xu H, Jun Y, Wickner W. Reconstituted membrane fusion requires regulatory lipids, SNAREs and synergistic SNARE chaperones. EMBO J. 2008;27:2031–2042. doi: 10.1038/emboj.2008.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu S, Mehta SQ, Pichaud F, Bellen HJ, Quiocho FA. Sec15 interacts with Rab11 via a novel domain and affects Rab11 localization in vivo. Nat Struct Mol Biol. 2005;12:879–885. doi: 10.1038/nsmb987. [DOI] [PubMed] [Google Scholar]
- 9.Seals DF, Eitzen G, Margolis N, Wickner WT, Price A. A Ypt/Rab effector complex containing the Sec1 homolog Vps33p is required for homotypic vacuole fusion. Proc Natl Acad Sci USA. 2000;97:9402–9407. doi: 10.1073/pnas.97.17.9402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wurmser AE, Sato TK, Emr SD. New component of the vacuolar class C-Vps complex couples nucleotide exchange on the Ypt7 GTPase to SNARE-dependent docking and fusion. J Cell Biol. 2000;151:551–562. doi: 10.1083/jcb.151.3.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peplowska K, Markgraf DF, Ostrowicz CW, Bange G, Ungermann C. The CORVET tethering complex interacts with the yeast Rab5 homolog Vps21 and is involved in endo-lysosomal biogenesis. Dev Cell. 2007;12:739–750. doi: 10.1016/j.devcel.2007.03.006. [DOI] [PubMed] [Google Scholar]
- 12.Rieder SE, Emr SD. A novel RING finger protein complex essential for a late step in protein transport to the yeast vacuole. Mol Biol Cell. 1997;8:2307–2327. doi: 10.1091/mbc.8.11.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Plemel RL, et al. Subunit organization and Rab interactions of Vps-C protein complexes that control endolysosomal membrane traffic. Mol Biol Cell. 2011;22:1353–1363. doi: 10.1091/mbc.E10-03-0260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Markgraf DF, et al. The CORVET subunit Vps8 cooperates with the Rab5 homolog Vps21 to induce clustering of late endosomal compartments. Mol Biol Cell. 2009;20:5276–5289. doi: 10.1091/mbc.E09-06-0521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brett CL, et al. Efficient termination of vacuolar Rab GTPase signaling requires coordinated action by a GAP and a protein kinase. J Cell Biol. 2008;182:1141–1151. doi: 10.1083/jcb.200801001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ostrowicz CW, et al. Defined subunit arrangement and rab interactions are required for functionality of the HOPS tethering complex. Traffic. 2010;11:1334–1346. doi: 10.1111/j.1600-0854.2010.01097.x. [DOI] [PubMed] [Google Scholar]
- 17.Nickerson DP, Brett CL, Merz AJ. Vps-C complexes: Gatekeepers of endolysosomal traffic. Curr Opin Cell Biol. 2009;21:543–551. doi: 10.1016/j.ceb.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stroupe C, Collins KM, Fratti RA, Wickner W. Purification of active HOPS complex reveals its affinities for phosphoinositides and the SNARE Vam7p. EMBO J. 2006;25:1579–1589. doi: 10.1038/sj.emboj.7601051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hickey CM, Stroupe C, Wickner W. The major role of the Rab Ypt7p in vacuole fusion is supporting HOPS membrane association. J Biol Chem. 2009;284:16118–16125. doi: 10.1074/jbc.M109.000737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stroupe C, Hickey CM, Mima J, Burfeind AS, Wickner W. Minimal membrane docking requirements revealed by reconstitution of Rab GTPase-dependent membrane fusion from purified components. Proc Natl Acad Sci USA. 2009;106:17626–17633. doi: 10.1073/pnas.0903801106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huizing M, et al. Molecular cloning and characterization of human VPS18, VPS 11, VPS16, and VPS33. Gene. 2001;264:241–247. doi: 10.1016/s0378-1119(01)00333-x. [DOI] [PubMed] [Google Scholar]
- 22.Schonthaler HB, et al. The zebrafish mutant lbk/vam6 resembles human multisystemic disorders caused by aberrant trafficking of endosomal vesicles. Development. 2008;135:387–399. doi: 10.1242/dev.006098. [DOI] [PubMed] [Google Scholar]
- 23.Zlatic SA, Tornieri K, L'Hernault SW, Faundez V. Clathrin-dependent mechanisms modulate the subcellular distribution of class C Vps/HOPS tether subunits in polarized and nonpolarized cells. Mol Biol Cell. 2011;22:1699–1715. doi: 10.1091/mbc.E10-10-0799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kastner B, et al. GraFix: Sample preparation for single-particle electron cryomicroscopy. Nat Methods. 2008;5:53–55. doi: 10.1038/nmeth1139. [DOI] [PubMed] [Google Scholar]
- 25.Radermacher M, Wagenknecht T, Verschoor A, Frank J. Three-dimensional reconstruction from a single-exposure, random conical tilt series applied to the 50S ribosomal subunit of Escherichia coli. J Microsc. 1987;146:113–136. doi: 10.1111/j.1365-2818.1987.tb01333.x. [DOI] [PubMed] [Google Scholar]
- 26.Pieren M, Schmidt A, Mayer A. The SM protein Vps33 and the t-SNARE H(abc) domain promote fusion pore opening. Nat Struct Mol Biol. 2010;17:710–717. doi: 10.1038/nsmb.1809. [DOI] [PubMed] [Google Scholar]
- 27.Krämer L, Ungermann C. HOPS drives vacuole fusion by binding the vacuolar SNARE complex and the Vam7 PX domain via two distinct sites. Mol Biol Cell. 2011;22:2601–2611. doi: 10.1091/mbc.E11-02-0104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cabrera M, et al. Phosphorylation of a membrane curvature-sensing motif switches function of the HOPS subunit Vps41 in membrane tethering. J Cell Biol. 2010;191:845–859. doi: 10.1083/jcb.201004092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.LaGrassa TJ, Ungermann C. The vacuolar kinase Yck3 maintains organelle fragmentation by regulating the HOPS tethering complex. J Cell Biol. 2005;168:401–414. doi: 10.1083/jcb.200407141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cabrera M, et al. Vps41 phosphorylation and the Rab Ypt7 control the targeting of the HOPS complex to endosome-vacuole fusion sites. Mol Biol Cell. 2009;20:1937–1948. doi: 10.1091/mbc.E08-09-0943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hickey CM, Wickner W. HOPS initiates vacuole docking by tethering membranes before trans-SNARE complex assembly. Mol Biol Cell. 2010;21:2297–2305. doi: 10.1091/mbc.E10-01-0044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Futter CE, Pearse A, Hewlett LJ, Hopkins CR. Multivesicular endosomes containing internalized EGF-EGF receptor complexes mature and then fuse directly with lysosomes. J Cell Biol. 1996;132:1011–1023. doi: 10.1083/jcb.132.6.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Row PE, Prior IA, McCullough J, Clague MJ, Urbé S. The ubiquitin isopeptidase UBPY regulates endosomal ubiquitin dynamics and is essential for receptor down-regulation. J Biol Chem. 2006;281:12618–12624. doi: 10.1074/jbc.M512615200. [DOI] [PubMed] [Google Scholar]
- 34.Angers CG, Merz AJ. HOPS interacts with Apl5 at the vacuole membrane and is required for consumption of AP-3 transport vesicles. Mol Biol Cell. 2009;20:4563–4574. doi: 10.1091/mbc.E09-04-0272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lees JA, Yip CK, Walz T, Hughson FM. Molecular organization of the COG vesicle tethering complex. Nat Struct Mol Biol. 2010;17:1292–1297. doi: 10.1038/nsmb.1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brohawn SG, Leksa NC, Spear ED, Rajashankar KR, Schwartz TU. Structural evidence for common ancestry of the nuclear pore complex and vesicle coats. Science. 2008;322:1369–1373. doi: 10.1126/science.1165886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Devos D, et al. Components of coated vesicles and nuclear pore complexes share a common molecular architecture. PLoS Biol. 2004;2:e380. doi: 10.1371/journal.pbio.0020380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Harrison SC, Kirchhausen T. Structural biology: Conservation in vesicle coats. Nature. 2010;466:1048–1049. doi: 10.1038/4661048a. [DOI] [PubMed] [Google Scholar]
- 39.Lee C, Goldberg J. Structure of coatomer cage proteins and the relationship among COPI, COPII, and clathrin vesicle coats. Cell. 2010;142:123–132. doi: 10.1016/j.cell.2010.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ohi M, Li Y, Cheng Y, Walz T. Negative staining and image classification—powerful tools in modern electron microscopy. Biol Proced Online. 2004;6:23–34. doi: 10.1251/bpo70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hohn M, et al. SPARX, a new environment for Cryo-EM image processing. J Struct Biol. 2007;157:47–55. doi: 10.1016/j.jsb.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 42.Frank J, et al. SPIDER and WEB: Processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- 43.Pettersen EF, et al. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 44.Pintilie GD, Zhang J, Goddard TD, Chiu W, Gossard DC. Quantitative analysis of cryo-EM density map segmentation by watershed and scale-space filtering, and fitting of structures by alignment to regions. J Struct Biol. 2010;170:427–438. doi: 10.1016/j.jsb.2010.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.