Abstract
Multiple properties of plasmonic assemblies are determined by their geometrical organization. While high degree of complexity was achieved for plasmonic superstructures based on nanoparticles (NPs) little is known about the stable and structurally reproducible plasmonic assemblies made up from geometrically diverse plasmonic building blocks. Among other possibilities they open the door for the preparation of regiospecific isomers of nanoscale assemblies significant both from fundamental point of view and optical applications. Here we present a synthetic method for complex assemblies from NPs and nanorods (NRs) based on selective modification of NRs with DNA oligomers. Three types of assemblies denoted as End, Side, and Satellite isomers which display distinct elements of regiospecificity, were prepared with the yield exceeding 85%. Multiple experimental methods independently verify various structural features, uniformity, and stability of the prepared assemblies. The presence of interparticle gaps with finely controlled geometrical parameters and inherently small size comparable with those of cellular organelles fomented their study as intracellular probes. Against initial expectations SERS intensity for End, Side, and Satellite isomers was found to be dependent primarily on the number of the NPs in the superstructures rationalized with the help of electrical field simulations. Incubation of the label-free NP-NR assemblies with HeLa cells indicated sufficient field enhancement to detect structural lipids of mitochondria and potentially small metabolites. This provided the first proof-of-concept data for the possibility of real-time probing of local organelle environment in live cells. Further studies should include structural optimization of the assemblies for multitarget monitoring of metabolic activity and further increase in complexity for applications in transformative optics.
Keywords: nanoparticles, nanorods, plasmons, intracellular, SERS, metabolism, biosensor
INTRODUCTION
Plasmonic nanoparticles (NPs) from noble metals and their assemblies attract a lot of attention as a new tool for biosensing1–5 including such technically complicated and socially important needs as detection of prions6 and rapid assessment of water quality.7–9 The same optical properties also make them promising contrast agents for biomedical imaging.10–12 Versatility of chemical modification of gold surface makes possible the use of Au NPs for difficult tasks of deep tissue imaging,13 drug delivery with release-on-command capabilities,14 chemical15 and photothermal therapy16–18 of cancer. Plasmonic NPs also provide strong competition to other nanoscale materials in the areas of information storage,19 and transformative optics.20,21 Importantly, many of these properties and applications are determined not only by the shapes of individual NPs22 but also by their arrangement in space.23,24,25 This fundamental property combined with the well-developed theory of electromagnetic interactions of metal NPs,26–30 stimulates an overall trend toward constantly increasing three-dimensional (3D) complexity of the plasmonic superstructures. In fact, their sophistication is currently approaching that of molecules, biopolymers, and solid state crystals. A similar trend can also be traced for assemblies of semiconductor NPs.31–33
Consequently, scientists are searching for different approaches to assemble NPs into well-defined predetermined patterns. One of the most powerful and general methods of NP organization is the use of DNA oligomers to connect them into specific geometries.34 Since 199635,36 many different multiparticle constructs were reported using this approach. They are being made primarily from gold NPs and include both discrete24,37–40 and continuous one-dimensional (1D),41,42 two-dimensional (2D),43,44 and 3D superstructures.45–50 Of special interest are the recently made continuous crystalline lattices of plasmonic particles,45,46 due to their dynamic behavior and similarity to atomic/molecular crystals. Among the discrete 3D assemblies of NPs one should also single out assemblies that display chirality,38,51,52 which is a special geometric property typical only to complex purposefully designed atomic structures. Attaining chiral selectivity can be a benchmark for capabilities of organic synthesis methods as well as for NP assembly techniques. Additionally, the superstructures with chiral bands in plasmonic spectral region can be quite useful for negative refractive index materials53 and potentially for other applications. Importantly, PCR techniques developed recently also allow one to produce such superstructures in large quantities when necessary.38,54
Discrete assemblies from NPs and DNA have topological analogies with classical organic molecules and can potentially have similar structural diversity. Two observations can be made in this respect based on the current state-of-the-art of this field. First, stable discrete assemblies from nanocrystals of different shapes being assembled with the help of DNA or other methods have not been investigated or made yet although theoretical foundation for such studies have been laid down already. Besides bringing more variety to the spectrum of NP superstructures, such assemblies could potentially bring about a new level of parallelism between regular chemical molecules and NP superstructures. As such, they allow for the regioselective attachment of spherical NPs to non-spherical components. Similarly to the chiral assemblies, the regioisomers are expected to be different not only by the geometric placement of the different parts but also by their optical, chemical and potentially other properties. (2) Besides the fundamental value of highly complex plasmonic NP systems with DNA interconnects it would be important to demonstrate some functional significance of such complexity. It was previously demonstrated for continuous assemblies, for instance, supercrystals of gold nanorods6 and hybrid metal-semiconductor systems5,55 but not for discrete plasmonic superstructures. In fact, most applications mentioned above are served fairly well by fairly simple assemblies, such as NP dimers and sometimes even completely disorganized agglomerates.2,56 It would be important to have some idea of potential practical significance of sophisticated molecule-like plasmonic assemblies.
In this work, we are trying to address both of these points and report regiospecific assemblies made from NPs and gold nanorods (NRs). They display a new level of sophistication and a degree of control over the placement of NPs, which is conceptually similar to high regioselectivity in organic chemistry. This feature differentiates them from all previous cases of assemblies between NPs and nanowires/nanorods.5,57–59 Considering the great enhancement of electrical fields in the gaps between NPs and in the ends of NRs,1,29,60–62 they can be viewed as a system engineering platform for nanoscale sensors or “drones” in live cells taking advantage of greatly enhanced Raman scattering. They can also be used in some other applications as well utilizing dynamics of the assembly process, flexibility of the constracts, and biological specificity. Adding further the advantages of both the small size, biocompatibility of gold, and biological linkers present in the superstructures, one of the interesting areas of potential applications of such assemblies would be in-situ monitoring of intracellular metabolism inside the live cells, which was impossible before using assemblies with semiconductor NPs.5,55 In this paper we make the first step toward this goal and demonstrate that the produced superstructures provide a distinct read-out of local intracellular components. Besides being the first successful experiment of this kind, we also show that the intensity of Raman scattering differs significantly for different NP-NR regioisomers, which is associated with the varying number of plasmonic gaps in the regiospecific assemblies. These findings also raise some fundamental questions about the limitations of potential in-silico design of such structures and set up targets for future research work in this area.
EXPERIMENTAL
Materials
All oligonucleotides used were synthesized and purified using polyacrylamide gel electrophoresis by Sangon Biotechnology. The DNA sequences are shown in Table S1. Deionized water from a Milli-Q device (18.2 MΩ, Millipore, Molsheim, France) was used throughout this study. Unless stated otherwise, all other chemicals were purchased from Sigma-Aldrich, and were used without further purification. All glassware used was cleaned with freshly prepared aqua regia and rinsed thoroughly in H2O prior to use.
Instruments
Transmission electron microscopy (TEM) images were obtained using a transmission electron microscope JEOL JEM-2100 operating at an acceleration voltage of 200 kV. Before the TEM examination 10 μL of the samples were dry in air dispersed onto a copper grid coated with the carbon film. All UV/Vis results were acquired on a UNICO 2100 PC UV/Vis spectrophotometer and processed with the Origin Lab software. The zeta potential and size distribution were measured using a Zetasizer Nano ZS system (Malvern). The 633 nm laser was used for the DLS characterization. Synchrotron small angle X-ray scattering (S-SAXS) experiments were performed at the BL16B1 beamline at the Shanghai Synchrotron Radiation Facility (SSRF), Shanghai Institute of Applied Physics (SINAP), China, operated at 3.5 GeV with injection currents of 300 mA. The scattering data were collected with a MAR charge coupled device area detector at wavelength λ=1.5498 Å. SERS experiments were conducted with a LabRam-HR800 Micro-Raman spectrometer with Lab-spec 5.0 software attached to a Leica microscope and a liquid cell. The slit and pinhole were set at 100 and 400 mm, respectively, in a confocal configuration with a holographic grating (600 g/mm) and an air-cooled He-Ne laser for 632.8 nm excitation with a power of ca.8 mW.
Synthesis of gold nanoparticles (NPs)
Au NPs were synthesized by reduction of HAuCl4 using trisodium citrate.63–65 Typically, aqueous trisodium citrate solution (2.4 mL, 1% by weight, freshly prepared) was quickly added to a boiling aqueous solution of HAuCl4 (100 mL, 0.25 mM) under vigorous stirring and reflux. After several minutes, the color of the solution changed from blue to brilliant red. After boiling for 15 min, the heat source was removed to allow the reaction solution to cool to room temperature, and it was subsequently stored at 4 °C.
Synthesis of gold nanorods (NRs)
Au NRs with an aspect ratio of ca. 3 were prepared by a slightly modified seed-mediated growth procedure based on that proposed by Murphy and El-Sayed.66–68 Initially, a 5 mM HAuCl4 solution (0.1 mL) was added to 0.20 M CTAB solution (1 mL) that was kept at a constant temperature of 28.0 °C. Immediately, a deep-orange colored solution was obtained. Then, freshly prepared 0.01 M NaBH4 solution (0.12 mL) was added in one portion and the solutions were mixed by inversion. After stirring rapidly for 2 min, the solution turned pale-brown in color. To prepare the Au NR, 0.005 M HAuCl4 solution (5 mL) was mixed with 0.2 M CTAB solution (5 mL) and 4 mL of water. After that 65 μL of 0.1 M ascorbic acid solution and 0.125 mL of 0.01 M AgNO3 solution were added to the reaction media followed by with mixing for about 2 min, which resulted in a colorless solution. Finally, 0.05 mL of the seed solution was added with gentle mixing for about 20 s. The Au NRs could be used after ageing for 4 h.
Preparation of single-strand DNA-modified Au NPs
Au NPs were functionalized with alkanethiol derivative of DNA according to Ref 69. Briefly, DNA oligomers ASY2 with a sequence of TAG GAA TAG TTA TAA AAA AAA AAA A C(6)SH was added to Au NPs in 0.01 M phosphate buffer with 0.01% of sodium dodecylsulfate (SDS). The solution of ASY2 and Au NP was incubated at room temperature for 20 min. The concentration of NaCl was increased to 0.05 M using 2M NaCl and 0.01 M pH 8.0 phosphate buffer, while maintaining SDS concentration of 0.01%. The solution of DNA and Au NP was then sonicated for ~10 s followed by a 20 min incubation period at room temperature. This process was repeated for one more increment of 0.05 M NaCl and for every 0.1 M NaCl increment thereafter until a concentration of 0.7 M NaCl was reached. The ionic strength adaptation process was followed by incubation overnight at room temperature. To remove the excess of ASY2, Au NPs were centrifuged and the supernatant was removed, leaving a pellet of Au NPs at the bottom. The particles were then re-suspended in 0.01% SDS. This washing process was repeated for five times. 4-Aminothiophenol (4-ATP) modification was carried out in a similar procedures with a ratio of 4-ATP to NPs of about 600 after the NPs modified with DNA.
Fabrication of single-strand DNA-modified Au NRs
Firstly, Au NRs were centrifuged at 7500 rpm for 15 min in order to remove the excess ascorbic acid, AgNO3, and small spherical particles. The Au NRs were re-suspended and concentrated tenfold in the appropriate volume of 0.005 M CTAB solution. The re-dispersed Au NRs were used in all of the following experiments.
CTAB is preferentially bound along the {100} facets on the side of NRs as opposed to {111} facets at the ends of the NRs. Therefore the ends of the NRs are more reactive in respect to thiolated DNA.7 To obtain superstructures denoted as Satellite assemblies (see below) ASY1 oligomer with a sequence of TTA TAA CTA TTC CTA AAA AAA AAA A C(6)SH was added to Au NRs with a ratio of DNA to Au NR of ca 500. The mixture was allowed to react at room temperature for 12 h with gentle shaking. After the reaction, the conjugate formed was collected after three consecutive purification cycles consisting of centrifugation for 10 min at 6000 rpm and re-dispersion in 0.005 M CTAB buffer. The conjugate was stored at 4 °C.
To obtain superstructures denoted as Side assemblies (see below) non-target DNA with a sequence of AAA AAA AAA AAA C(6)SH abbreviated as “Helper” with a ratio of DNA to Au NR of about 100 was used to block the end sites of NR from the beginning of the synthesis. The conjugate was subsequently purified by three consecutive centrifugation-redispersion cycles identical to those used for Satellite assemblies. After that ASY1 was added to NR keeping the ratio of DNA to Au NR equal to 500.
To make superstructures denoted as End assemblies a similar synthetic process was used with the difference that the target DNA was conjugated to the ends of Au NRs. The only difference was that ASY1 was first added to a solution of Au NRs and then the “Helper” DNA was conjugated with the side surfaces of the Au NRs.
Modification with 4-aminothiophenol (4-ATP) was employed for surface enhanced Raman scattering (SERS) experiments and was carried out following the identical procedure with a ratio of 4-ATP to NPs or NRs of 550.
Self-assembly of Au NRs and Au NPs
In order to form the Satellite assemblies, 1 μL of Au NR-ASY1 were mixed with 5 μL of Au NP-ASY2 in 6 μL of 0.001 M Tris-HCl buffer (0.01% SDS; MgCl2 20 mM, pH 8.0). The mixture was incubated for 12 h at room temperature. Side assemblies were prepared according to a similar method as the Satellite assemblies. To obtain End assemblies, the volume ratio of NPs and NRs was about 1. The components were mixed well in the hybridized buffer (containing 0.0005 M Tris-HCl, 0.005% SDS, 10 mM MgCl2) and incubated for 12 h at room temperature.
FDTD Simulations of Plasmonic Properties
Field enhancement properties of NP-NR assemblies were calculated taking advantage of recently developed finite difference time domain (FDTD) method. An FDTD package from CST STUDIO SUITE™ 2010 utilizing the Drude model was used for the calculation in the vicinity of the plasmonic for media with electrical permittivity of ε=1 and magnetic permeability of μe= 1. The End, Side, and Satellite plasmonic assemblies were modeled by 16 nm diameter Au NPs and 16 nm (diameter) x 50 nm (length) NR separated by a gap of 4–8 nm. The electrical permittivity (ε) and plasma frequency (ωp) of gold employed in the calculations were taken to be constant and equal to 1.0 and 1.37 × 1016 rad/s, respectively. The local electromagnetic fields were calculated for 632.8 nm excitation and parallel orientation of rods to the external field.
RESULTS AND DISCUSSION
Preparation of NP-NR Assemblies with Regiospecific Geometries
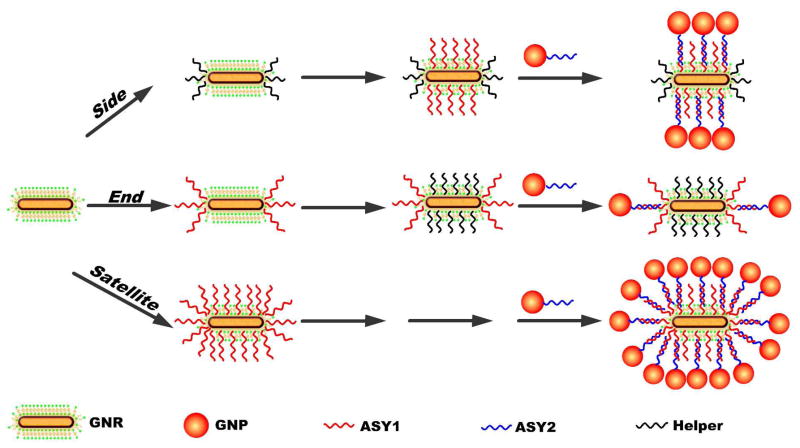
Let us consider different potential geometries for the placement of NPs around a NR. In the first approximation there are three basic options for such assemblies. NPs can be attached only to the ends, only to the sides, and both to the ends and to the sides of NRs. These three types of superstructures will be referred to as End, Side, and Satellite assemblies respectively (Scheme 1). Notably, one can and should consider finer differences between geometrical placements and the number of the NPs attached. They include, for instance, the case of a single NP attached to one end of the NR and a variety of asymmetric placements of NPs on the sides of NR. These superstructures are quite enigmatic and, when possible, we shall address their preparation and properties. At this stage, however, it will be probably more prudent to limit ourselves to simpler cases. We will focus, therefore, on these more basic varieties of the superstructures, methods of their preparations, principle distinctions of optical properties, and related differences in potential applications.
Scheme 1.
Schematics of synthetic method for regiospecific NPs assemblies.
Standard 16 nm (diameter) × 50 nm (length) NRs were synthesized by the well-known seed-mediated growth method.67 After that they were surface modified with single-stranded DNA (ssDNA) in three different ways (see Experimental and Scheme 1). Cetyltrimethylammonium bromide (CTAB) coating is thinner in their ends.7,70 So, when thiolated DNA is present in low concentrations, the covalent attachment of the oligonucleotides occurs exclusively in the “end” positions. Increase of the concentration of ssDNA leads to the surface modification of NRs both in the ends and sides and therefore to the Satellite route in these assemblies (Scheme 1).
However, one can use different oligonucleotides for the modification steps with low and high concentration of ssDNA. As such, we used 25-base thiolated ssDNA denoted ASY1 (red in Scheme 1) with a sequence of TTA TAA CTA TTC CTA AAA AAA AAA A C(6)SH, and a “helper” DNA (black in Scheme 1) with a simple sequence of AAA AAA AAA AAA C(6)SH. ASY1 is used to attach NPs via complementary ASY2 strand with a sequence of TAG GAA TAG TTA TAA AAA AAA AAA A C(6)SH. The helper DNA does not have any matching oligomers in this process; it is used for the purpose of controlling regioselectivity of the assembly process because otherwise, thiolated ssDNA attached to the ends might spontaneously diffuse all over the NR surface scrambling the selectivity of the NP placement later on. Covalently attached coating of the helper DNA presents much stronger barrier to DNA diffusion than a layer of ionically adsorbed CTAB. So, when we initially expose NRs to low concentration of helper DNA, the ends are becoming blocked. Subsequent addition of ASY1 results in the attachment along the sides. The NPs will be eventually attached to the sides, too. In Scheme 1, this synthetic method is denoted as a route to Side regioisomers (Scheme 1).
When the NRs are exposed to low concentrations of ASY1 first, and after that to high concentration of helper DNA, the sides of the NR become blocked while ASY1 oligomers are localized in the ends. Therefore, the NPs attach only to the ends of NRs. The corresponding assemblies will be designated as End isomers. Using the fluorescence-based method69 described in Supporting Information (SI), the numbers of ASY1 units grafted onto the surface of one NR were estimated to be 41±5, 32±3, and 6±1 for the Satellite, Side, and End assemblies, respectively.
Classical citrate-protected 16 nm diameter NPs were functionalized with 25-base thiolated ssDNA of type ASY2 with a sequence of TAG GAA TAG TTA TAA AAA AAA AAA A C(6)SH. The number of ssDNA units on the surface of each NP was estimated to be 64±6.38 The concentrations of NPs and NRs in as-prepared dispersions were calculated to be 2.1 and 1.2 nM, respectively (see SI). These data are used to control the NP/NR ratio to obtain nearly high synthetic yield of designated regioisomers in respect to original NRs.
Structural Characterization
Following a 12 h incubation of NPs and NRs resulting in the hybridization of ASY1 and ASY2 strands, NRs and NPs assemblies in different geometrical arrangements were structurally examined by UV/Vis spectroscopy, dynamic light-scattering (DLS), transmission electron microscopy (TEM) as well as synchrotron small-angle X-ray scattering (S-SAXS). The representative TEM images clearly demonstrate the three types of regiospecific NP-NR assemblies. One can also point out fairly accurate control of the geometries, sizes of assemblies, and interparticle gaps between adjacent NPs. Note that the obtained NP-NR superstructures were stable for at least several weeks in aqueous solutions with appropriate precautions against biological contamination with DNA cleaving enzymes. In control experiments with NRs and NPs bearing non-complementary ssDNAs no assembles were formed (Figure S3). No spontaneous organization of NPs and NRs on the substrate solely due to drying and capillary forces was observed as well.
From TEM images, the average size of the junction between adjacent NRs and NPs can be determined to be 3.8±0.2 nm. The total length of two fully stretched 25mers of DNA connected via complementary 15 base-pairs at each terminal is ca. 10.3 nm. The full extension of DNA linkages is unlikely even in solution. The substantial reduction of the NP-NR gap due to DNA coiling in the dry state characteristic for TEM grids is quite expected.
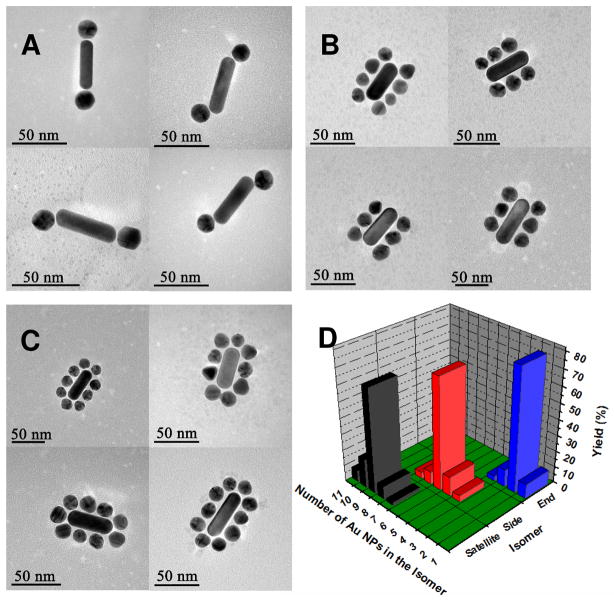
TEM also provides the possibility to assess the dispersity of the assembled structures. As one can see in Figure 1D, there is always the strong dominance of very specific structure with 9, 6, and 2 NPs attached to NRs for Satellite, Side, and End assemblies, with the yields of 56.8%, 60.3%, and 75.7%, respectively (Figure 1D). Note also that despite some variability in the number of NPs attached to one NR especially for Satellite related probably to the statistical distribution of the NP diameters and their packing pattern around the NR. For instance, a total of 6–8 NPs can be accommodated on the sides of NRs in Figure 1B, which represent different regioisomers. More accurate control of the number of NPs could be the next challenge for synthesis of such superstructures. At the same time, there is virtually no overlap in the types of assemblies (aka isomers) made by the specific route indicated in Scheme 1. The yield for isomers classified as Side, End, and Satellite is always greater than 85% and in many cases above 90%. The self-assembly processes of NP and NRs have conceptually similarity to one-step regiospecific reactions in organic chemistry with exception that atoms or chemical groups are replaced with spherical NPs.71–73 In a crude approximation some specific chemical groups such as aromatic rings and their substituent in o-, m-, or p-positions can be viewed as geometrically different atomic groups. Note also, that yield of specific isomers in such reactions rarely exceeds 50% unless it is catalyzed by an enzyme. In the latter case it could be as high as 96.0%,71–73 which is still comparable to the yields obtained in assembly reactions here.
Figure 1.
(A–C): Representative TEM images of (A) End, (B) Side and (C) Satellite assemblies; (D) Statistical analysis of the number of Au NPs assembled following different synthetic routes. Statistical analysis was made on the basis of 150, 145, 160 of assemblies counted for Satellite, Side, and End isomers, respectively.
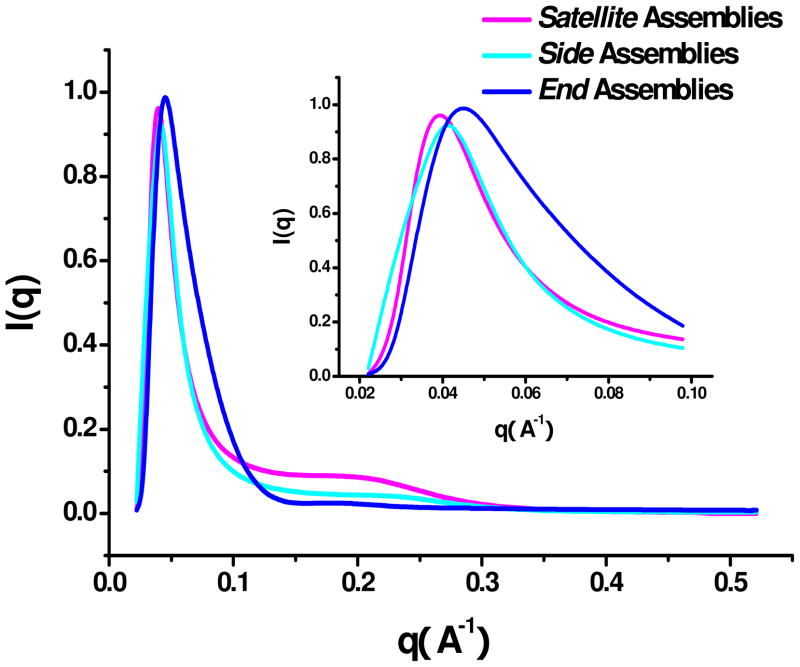
TEM provided indications of remarkable in structural uniformity for all the three different assembly types. To gather more information about the geometry of the NP-NR assemblies in solution and to confirm their organization by an independent experimental technique, we decide to carry out their structural characterization by synchrotron small angle X-ray scattering S-SAXS. The intensity of the standard SAXS equipment was not sufficiently high to provide adequate signal-to-noise ratio. In the small scattering vector q region (q < 0.05 Å−1), distinct scattering patterns were observed for all three types of assemblies. The first peak was observed at q = 0.038, 0.041, and 0.044 Å−1 for Satellite, Side, and End assemblies, respectively (Figure 2). The S-SAXS diameters of the dispersed species associated with these peaks were calculated to be 113 ± 1.5, 72 ± 1.4, and 109 ± 1.2 nm. Note that these data correspond very well with the largest measure of the assemblies seen in the TEM which provides clear confirmation of the uniformity of the produced structures across the entire ensemble of the NP-NR assemblies present in solution. Otherwise the first S-SAXS peak would be very broad or not observed at all. SAXS data also make possible accurate evaluation of the gap between the NP and NR as the second repetitive structural parameter in the system. The positions of the secondary peaks in S-SAXS at q = 0.21, 0.25, and 0.19 Å−1 lead to the corresponding gap distances of 7.8±0.3, 8.1±0.2, 7.7±0.2 nm for Satellite, Side and End assemblies, respectively. These values match very well with the expected distance between NP and NRs controlled by the length of hybridized ASY1-ASY2 DNA. Expectedly, this is longer that the “dry gap” in TEM images, but shorter than the fully stretched DNA linker (see above).
Figure 2.
Synchrotron small angle X-ray scattering (S-SAXS) results for different NR-NPs regiospecific assemblies. Insert: S-SAXS scattering curve in the range of 0.021 ≤ = q ≤ = 0.1.
The ensemble-averaged diameter of the assemblies can also be characterized by dynamic light scattering (DLS) although lower accuracy in size estimates could be expected. The Satellite, Side, and End assemblies showed DLS diameters of 108±12, 79±8, and 103±15 nm, respectively. They are remarkably similar to those determined by S-SAXS, which again must be attributed to remarkable uniformity of the superstructures in dissolved state. Also important, DLS is very sensitive to agglomerated components of the dispersion and the nice correlation between TEM, S-SAXS, and DLS data gives us confidence that the actual species present in solution are indeed those depicted in Scheme 1 and Figure 1.
Similarly to S-SAXS, DLS is the most sensitive to the largest measure of the scattering particles. Taking into account the TEM diameter of NRs and NPs modified with DNA as well as their DLS diameters equal to 68±6 nm and 18±3 nm, respectively, the gap between the NP and NRs can be estimated to be 8 nm. This value is comparable with that determined from S-SAXS data, being most likely an upper limit of the potential gap length.
Optical Properties
UV absorption spectroscopy provides a simple experimental tool for the assessment of surface plasmon coupling between neighboring NPs.24 In turn, such assessment is necessary for better understanding of potential applications of these fairly sophisticated superstructures. Considering the geometry of the synthesized superstructure and demonstrated regioselectivity (Fig. 2), one can consider their utilization as probes in surface-enhanced spectroscopies dependent on the electric field in the gap between the particles,29,74 and nonlinear optical effects.75,20
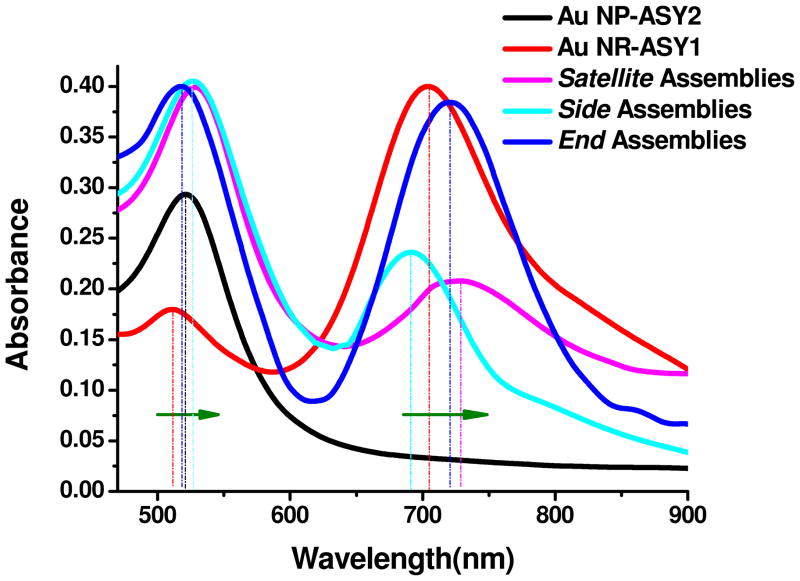
All samples of NP, NRs and constructs from them exhibited two well-resolved plasmon peaks in 500 nm and 700 nm spectral regions. Since the diameter of NPs and NRs are identical, the peak at 520 nm in the assemblies is actually a superposition of the plasmon band of NPs and the transverse band of the NRs. The peak at 701 nm can be easily identified as the longitudinal peak of the NRs.
The behavior of the two bands is markedly different for all the three types of assemblies depending on the coupling mode of the plasmons. The UV spectrum of the End assemblies (Figure 3) displayed a 26 nm red shift of the longitudinal NR band from 701 nm to 727 nm. The superposition band does not change appreciably. Such spectral behavior is consistent with an effective increase of the aspect ratio of the new “composite nanorods” with predominantly longitudinal coupling between the NPs and NRs.7
Figure 3.
UV/Vis spectra of NP, NRs as well as Satellite, Side, and End assemblies.
Side assemblies (Figure 3) display an opposite trend in the plasmon bands. The longitudinal plasmon band of NRs undergoes a blue shift to 690 nm and is accompanied by a clear red shift in the superposition band from 520nm to 526nm. These changes in optical properties can be understood as “fattening” of the original NRs due to attachment of NPs on their sides.
In the case of the Satellite assemblies (Figure 3) the trend is different, too. The longitudinal plasmon band of NRs is red-shifted from 701 to 728 nm, with a concomitant red shift of the superposition band from 520 to 524 nm. Such shift is typical for multiple coupling modes resulting in splitting of the energy level of plasmon oscillations.76
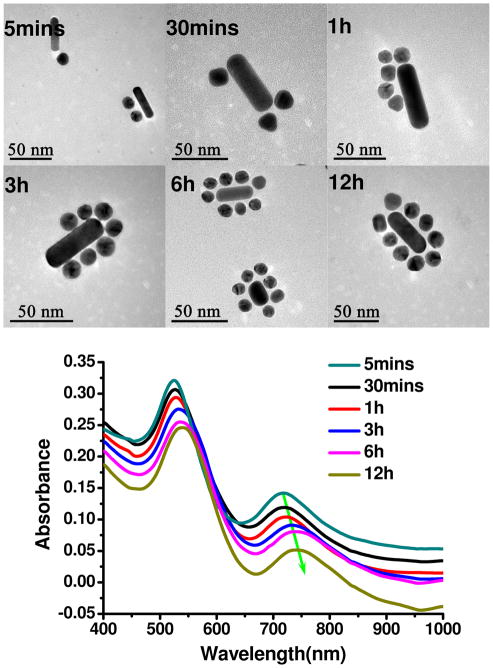
It is also interesting to compare the UV-vis spectra with concurrently acquired TEM images for different time points after the beginning of the DNA hybridization process. It was carried out for the Satellite assemblies because they involve the largest number of “building blocks” and can potentially have the most complex assembly process. Within 5 min after mixing the nanoscale precursors, one-two NPs attached to the surface of NRs. No specific preference in the location of the initially attached NPs was observed (Figure 4). Over a subsequent period of few more hours, NPs gradually assembled around the NR and evolved into asymmetric Satellite assemblies. Satellite assemblies with 1–2 missing NPs were formed after 6 h of hybridization process. Complete superstructures formed in 12 h after the onset of the reaction. Changes in the UV-vis spectra associated with the assembly of NR and NP reflected the stepwise growth of the assemblies: both the superposition peak and the longitudinal plasmon peak gradually red-shift in agreement with the TEM data and expected trend for the plasmonic superstructure.
Figure 4.
Representative TEM images and UV/Vis spectra of Satellite assemblies for different hybridization times.
Differentiation of SERS properties between isomers
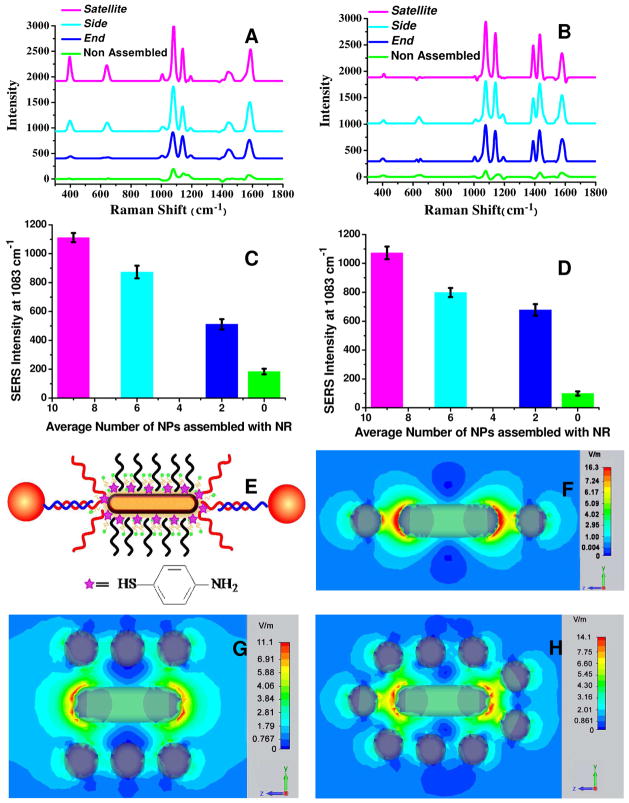
Clear evidence of geometry-dependent plasmonic coupling in the in Side, End, and Satellite regioisomers (Fig. 3, 4), prompted us to consider potential optical functionalities of such structures. Raman scattering is strongly enhanced in the gaps of plasmonic nanostructures due to the concentration of high electrical field22,60,62,77 giving rise to surface enhanced Raman spectroscopy (SERS). The SERS signals obtained from individual NP are rather weak and limited to target molecules possessing large Raman cross-sections.78 This fact highlights the importance of the assembled structures. Therefore, the prepared assemblies can potentially serve as local SERS probes relaying to the observer the information about the chemical composition in the probe surroundings. The potential spatial resolution of such probes should be comparable to their average size in solution, i.e. about 100 nm which is substantially smaller than the diffraction limit for visible light. Additionally, the plasmonic assemblies with highly controllable geometries could be an important path for investigating and optimizing SERS. However, in most studies except probably the most recent ones,6,22 superstructures have been assembled by random aggregation of NPs, making it difficult to correlate the SERS signal with a specific geometry of superstructure and in-silico data on local field enhancement. Developing synthetic approaches to prepare highly uniform multi-NP assemblies in a controlled manner with highly reproducible SERS-active sites is an essential avenue to explore for practical exploitation of SERS. This point can be exemplified by the study of dimers and trimers of NPs and the finding that the relative SERS intensity did not scale with the number of the NP in assemblies and were dependent on geometry.79 In order to ensure that the NP-NR assemblies prepared here are suitable as local SERS probes, a standard Raman reporter molecule, namely 4-aminothiophenol (4-ATP) was attached to NP or NRs. The modified “building blocks” bearing 4-ATP were used to assemble the superstructures (Figure 5A,B,E). SERS spectra of the different superstructures were compared to the intensity of 4-ATP on the surface of NPs in the liquid phase. 4-ATP displayed markedly different intensities for the different NP-NR superstructures (Figure 5A, B). The Raman intensity at the typical 4-ATP frequency of 1083 nm was found to be the highest for the Satellite assemblies. It is also seem to be nearly proportional to the number of NPs involved in the assembly (Figure 5C,D). To understand this dependence better, the electrical field in the prepared superstructures upon illumination was simulated by the comprehensive finite difference time domain (FDTD) method based on Maxwell’s curl equations. As shown in Figure 5F-H, the electric field for different specific superstructures was surprisingly similar. The maximum of field in the hot spots was found to be 16.3, 14.1 and 11.1 V/m for End, Satellite, and Side isomers respectively. Although End assemblies display nominally the highest field but the number of the plasmonic gaps in such isomer is smallest. Statistical probability of the reporter molecule to be in the gap between NP and NR is the highest for Satellite assembly. Given the fairly small difference in the actual value of the electrical field (ca. 13%, Figure 5F–H), this geometrical probability becomes more significant structural parameter for this test. Consequently, this fact brings about the nearly linear dependence of SERS intensity on the number of NPs in the isomer (Figure 5C–D), which would be difficult to predict from the onset of this project.
Figure 5.
SERS of NP-NRs assemblies with SERS reporter molecule made from (A) NPs-ASY2 modified with 4-ATP; (B) NR-ASY1 modified with 4-ATP. Non-assembled samples were made from NP modified with non-complementary “Helper” ssDNA and NR carrying ASY1 with the molar ratio of 11. (C, D) Dependence of SERS intensity on the average number of NPs assembled with NRs for plots in A and B, respectively. (E) Schematic representation of the End assemblies with NRs modified with 4-ATP; (F–H) Finite-difference time-domain (FDTD) simulation of plasmonic properties for (F) End, (G), Side and (H) Satellite assemblies; excitation wavelength is 632.8 nm; the gap between NRs and NP is 8 nm.
In-situ SERS measurements in live cells
Known case studies in the nexus of SERS spectroscopy and cellular biology include NPs probes conjugated with Raman reporter labels similar to 4-ATP and specific tumor-targeting receptors. Such plasmonic tags demonstrated potential for cancer cell detection, which is certainly quite inspiring.82–84 However, the NP probes in these cases were localized at the outer surface of external cell membrane where the majority of common cancer markers are present. Also important that the SERS signals displayed only the SERS signature from the Raman labels extraneously added to the probe and not from the local environment of the membrane. This fact indicates probably fairly weak field enhancement that can be related to sub-optimal choice of probe structure.
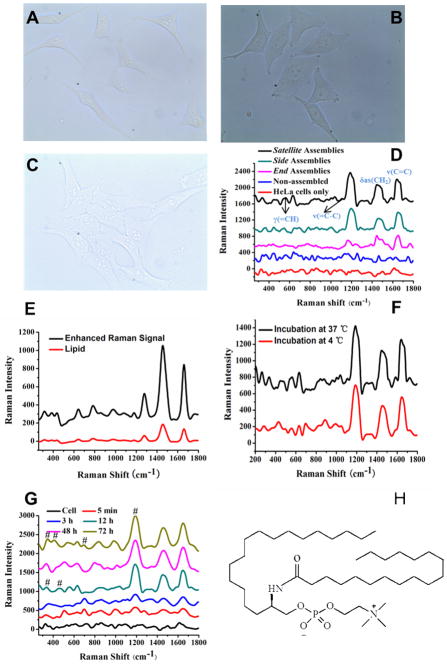
It would be both challenging and important for many researchers in cell biology or drug discovery to be able to monitor cellular processes with single cell resolution and, better yet, at the level of cellular substructures without any traditional fluorescent labels or antibodies which are known to interfere with intracellular processes. Additionally many small molecules essential for metabolism do not have any antibodies or are impossible to produce for biological reasons (toxicity). SERS probes based on the discrete plasmonic superstructures with the size comparable with cellular organelles can potentially provide such possibility. With this idea in mind, the three kinds of assemblies (0.416 nM of each type of assembly, referring to the concentration of NRs) were mixed with 100 μL of a cell culture medium for HeLa cells and incubated at 37 °C under 5% CO2 for 12 h in a 96-well cell culture plate. Over 95% of the HeLa cells remained viable in their presence (Figure 6A–C) after incubation for 12 h. The shapes characteristic for strong adhesion of the cells to substrate and the live-dead assay indicated that these three kinds of assemblies were not toxic to the HeLa cells; this fact was not entirely unexpected but needed to be tested regardless of large amount of previous data. After incubation with the NR-NP superstructures (Figure 6D), strong SERS signals from the cell culture can be seen at 1186 cm−1, 1462 cm−1 and 1655 cm−1. They correspond to HC= in-plane deformations, CH2 deformations, and cis-unsaturated C=C stretching vibrations, respectively. They are likely to originate from intracellular unsaturated lipids located in the mitochondria.80,81 Incubation of the assemblies with the neat lipid (Sigma) revealed identical Raman spectra. These positively charged lipids are known to bind to negatively charged DNA in the gaps between the NPs and NR through electrostatic attraction.39 The intensities of which decreased in the order of the Satellite, Side, and End assemblies which is identical to the order observed for 4-ATP. This observation both confirms localization of the lipids in the NP-NR gaps and indicates consistency in the plasmonic behavior of the assemblies. The latter is inherently related to the stability of NP-NR assemblies in the cellular environment, which far from being obvious observation due to the abundance of DNA-cleaving enzymes inside the cells. The structural uniformity of the produced isomers also plays a role here. Similarly to 4-ATP, the total number of the NP-NR gaps appears to be the dominating structural parameter responsible for the overall SERS intensity, which opens the door for structural tuning and optimization of SERS probes in ex-vivo experimental setting.
Figure 6.
The optical images of HeLa cell with different binary ensembles incubated after 12 hours. (A) Satellite, (B) Side, and(C) End assemblies; (D) Raman spectra from the Hela cells incubated for 12 h with three types of assemblies (no extraneous Raman labels); Non-aseembled: the mixture of NPs carrying with ASY1 and the entire surface of NR modified with the noncomplementary Helper ssDNA with the molar ratio of 11. (E–G) Raman spectra from(E) lipid and the mixture of Satellite assemblies and lipid, (F) Hela cells incubation at 37 °C and 4 °C, (G) Hela cells incubation on differentive time, respectively. The bands marked # belong to other cellular components or metabolites than the lipid in H found in mitochondria. (H) The chemical structure of N-Stearoyl-D-sphingomyelin.
Note that steric hindrance due to the presence of the DNA chains does not appear to be the problem for the large lipid molecules (Figure 6H), which display clear Raman signal with the assemblies. The same conclusion can be made from TEM images and the DLS data indicating that inclusion of lipid does not change very much the geometrical dimensions of the assemblies (Figure S8). These probes can be particularly useful for imaging of small metabolites for which no antibodies can be made. Such metabolites can be exemplified by compounds as NO, glucose, NADPH/NADPH+, polyamines, phosphate and others. The concentration of positively charged analytes, can be even enhanced in the NP-NR gaps.
To understand better the transport of assemblies into the cells, the Satellite assemblies were incubated with HeLa cells at 4 °C when the endocytosis is blocked (5% CO2 for 12 h, 96-well cell culture plate). This did not change the intensity or the position of main bands in Raman spectrum compared to incubation at 37 °C (Figure 6F). This clearly indicate that Satellite assemblies can penetrate through the cellular membrane entering directly the cytoplasm (Figure 6G). This is important finding that open the road for further applications of these structures as intracellular probes.
After traversing the outer membrane of the cell the assemblies the probes can be transported to different parts of the cells and encounter cell heterogeneity. In part we see the reflection this process in the variance of the weaker bands of the Raman spectra obtained in the low frequency region. Some of these weaker bands and nuances of the two spectra can be the signature(s) of small metabolites mentioned above, such as NO adducts. Their low intensity and their low signal-to-noise ratio do not allow at the moment their attribution (Figure 6E).
In summary, we have demonstrated successful synthesis of new plasmonic nanoscale superstructures which combine building blocks of different shapes connected by DNA oligomers. They were denoted as Satellite, Side, and End NP-NR assemblies and display distinct elements of regiospecificity and high synthetic yield. Multiple experimental methods including TEM, S-SAXS, UV/Vis, and DLS cross-validate their organization and structural uniformity. Detailed SERS study combined with simulation of electrical fields inside the assemblies demonstrated the unexpected structural trends for SERS intensity. In-vivo data indicate a realistic possibility of their utilization as label-free intracellular probes to acquire real-time information about local metabolic processes especially for low molecular weight metabolites. The degree of structural control available for these assemblies makes possible the development of cellular “drones” for detection of metabolic products essential for different aspects of cellular activity. Multiple simple molecules for which antibodies are not possible to produce, for instance, NO, NADPH+, glucose, phosphates, ATP, amino acids etc, can be potentially detected simultaneously.
Further development of the method is also possible and may include dynamic multi-NP multi-NR superstructures with increased topological complexity and regiospecificity. One potential research target could be NP-NR assemblies in the shape of spirals53 that can lead to fascinating optical properties and negative refractive index.
Supplementary Material
Acknowledgments
This work is financially supported by the National Natural Science Foundation of China (21071066, 20835006, 91027038, 21101079, 21175034), the Key Programs from MOST (2010AA06Z302, 2010DFB3047, 2008BAK41B03, 2009BAK61B04, 2010GB2C100167), and grants from Natural Science Foundation of Jiangsu Province, MOF and MOE (BK2010001, BK2010141, JUSRP10921, JUSRP11019, JUDCF10026).
This work is based upon work partially supported by the Center for Solar and Thermal Energy Conversion, an Energy Frontier Research Center funded by the U.S. Department of Energy, Office of Science, and Office of Basic Energy Sciences under Award Number #DE-SC0000957 (NAK). We acknowledge support from NSF under grant ECS-0601345; EFRI-BSBA 0938019; CBET 0933384; CBET 0932823; and CBET 1036672. (NAK) The work is also partially supported by AFOSR MURI 444286-P061716 and NIH 1R21CA121841-01A2.
Footnotes
Supporting Information Available: The results, characterization of the DNA coverage of binary Au NPs conjugation, and three regiospecific assemblies. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Cao YWC, Jin RC, Mirkin CA. Science. 2002;297:1536. doi: 10.1126/science.297.5586.1536. [DOI] [PubMed] [Google Scholar]
- 2.Elghanian R, Storhoff JJ, Mucic RC, Letsinger RL, Mirkin CA. Science. 1997;277:1078. doi: 10.1126/science.277.5329.1078. [DOI] [PubMed] [Google Scholar]
- 3.Storhoff JJ, Lucas AD, Garimella V, Bao YP, Muller UR. Nat Biotechnol. 2004;22:883. doi: 10.1038/nbt977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sepulveda B, Angelome PC, Lechuga LM, Liz-Marzan LM. Nano Today. 2009;4:244. [Google Scholar]
- 5.Lee J, Hernandez P, Lee J, Govorov AO, Kotov NA. Nat Mater. 2007;6:291. doi: 10.1038/nmat1869. [DOI] [PubMed] [Google Scholar]
- 6.Alvarez-Puebla RA, Agarwal A, Manna P, Khanal BP, Aldeanueva-Potel P, Carbo-Argibay E, Pazos-Perez N, Vigderman L, Zubarev ER, Kotov NA, Liz-Marzan LM. P Natl Acad Sci USA. 2011;108:8157. doi: 10.1073/pnas.1016530108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang LB, Zhu YY, Xu LG, Chen W, Kuang H, Liu LQ, Agarwal A, Xu CL, Kotov NA. Angew Chem Int Edit. 2010;49:5472. doi: 10.1002/anie.200907357. [DOI] [PubMed] [Google Scholar]
- 8.Walker DA, Browne KP, Kowalczyk B, Grzybowski BA. Angew Chem Int Edit. 2010;49:6760. doi: 10.1002/anie.201002558. [DOI] [PubMed] [Google Scholar]
- 9.Klajn R, Olson MA, Wesson PJ, Fang L, Coskun A, Trabolsi A, Soh S, Stoddart JF, Grzybowski BA. Nat Chem. 2009;1:733. doi: 10.1038/nchem.432. [DOI] [PubMed] [Google Scholar]
- 10.Jain PK, Huang XH, El-Sayed IH, El-Sayed MA. Accounts Chem Res. 2008;41:1578. doi: 10.1021/ar7002804. [DOI] [PubMed] [Google Scholar]
- 11.Wang LV. Nat Photonics. 2009;3:503. doi: 10.1038/nphoton.2009.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mallidi S, Larson T, Tam J, Joshi PP, Karpiouk A, Sokolov K, Emelianov S. Nano Lett. 2009;9:2825. doi: 10.1021/nl802929u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang XD, Shao X, Agarwal A, Rajian JR, Kotov NA. Nanotechnology. 2011;22:135102. doi: 10.1088/0957-4484/22/13/135102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen W, Xu NF, Xu LG, Wang LB, Li ZK, Ma W, Zhu YY, Xu CL, Kotov NA. Macromol Rapid Comm. 2010;31:228. doi: 10.1002/marc.200900793. [DOI] [PubMed] [Google Scholar]
- 15.Podsiadlo P, Sinani VA, Bahng JH, Kam NWS, Lee J, Kotov NA. Langmuir. 2007;24:568. doi: 10.1021/la702782k. [DOI] [PubMed] [Google Scholar]
- 16.Huang X, El-Sayed IH, Qian W, El-Sayed MA. J Am Chem Soc. 2006;128:2115. doi: 10.1021/ja057254a. [DOI] [PubMed] [Google Scholar]
- 17.Giljohann DA, Seferos DS, Daniel WL, Massich MD, Patel PC, Mirkin CA. Angew Chem Int Edit. 2010;49:3280. doi: 10.1002/anie.200904359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bardhan R, Lal S, Joshi A, Halas NJ. Accounts Chem Res. 2011;44:936. doi: 10.1021/ar200023x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zijlstra P, Chon JWM, Gu M. Nature. 2009;459:410. doi: 10.1038/nature08053. [DOI] [PubMed] [Google Scholar]
- 20.Klein MW, Enkrich C, Wegener M, Linden S. Science. 2006;313:502. doi: 10.1126/science.1129198. [DOI] [PubMed] [Google Scholar]
- 21.Agarwal A, Lilly GD, Govorov AO, Kotov NA. J Phys Chem C. 2008;112:18314. [Google Scholar]
- 22.Zhu Z, Meng H, Liu W, Liu X, Gong J, Qiu X, Jiang L, Wang D, Tang Z. Angew Chem Int Edit. 2011;123:1631. doi: 10.1002/anie.201005493. [DOI] [PubMed] [Google Scholar]
- 23.Pramod P, Thomas KG. Adv Mater. 2008;20:4300. [Google Scholar]
- 24.Maye MM, Nykypanchuk D, Cuisinier M, van der Lelie D, Gang O. Nat Mater. 2009;8:388. doi: 10.1038/nmat2421. [DOI] [PubMed] [Google Scholar]
- 25.Sonnichsen C, Reinhard BM, Liphardt J, Alivisatos AP. Nat Biotechnol. 2005;23:741. doi: 10.1038/nbt1100. [DOI] [PubMed] [Google Scholar]
- 26.Schmucker AL, Harris N, Banholzer MJ, Blaber MG, Osberg KD, Schatz GC, Mirkin CA. Acs Nano. 2010;4:5453. doi: 10.1021/nn101493t. [DOI] [PubMed] [Google Scholar]
- 27.Kleinman SL, Ringe E, Valley N, Wustholz KL, Phillips E, Scheidt KA, Schatz GC, Van Duyne RP. J Am Chem Soc. 2011;133:4115. doi: 10.1021/ja110964d. [DOI] [PubMed] [Google Scholar]
- 28.McMahon JM, Gray SK, Schatz GC. Phys Rev B. 2011;83:115428. [Google Scholar]
- 29.Lee A, Andrade GFS, Ahmed A, Souza ML, Coombs N, Tumarkin E, Liu K, Gordon R, Brolo AG, Kumacheva E. J Am Chem Soc. 2011;133:7653. doi: 10.1021/ja2015179. [DOI] [PubMed] [Google Scholar]
- 30.Wang ZB, Luk’yanchuk BS, Guo W, Edwardson SP, Whitehead DJ, Li L, Liu Z, Watkins KG. J Chem Phys. 2008;128:094705. doi: 10.1063/1.2835598. [DOI] [PubMed] [Google Scholar]
- 31.Srivastava S, Santos A, Critchley K, Kim KS, Podsiadlo P, Sun K, Lee J, Xu CL, Lilly GD, Glotzer SC, Kotov NA. Science. 2010;327:1355. doi: 10.1126/science.1177218. [DOI] [PubMed] [Google Scholar]
- 32.Tang ZY, Kotov NA, Giersig M. Science. 2002;297:237. doi: 10.1126/science.1072086. [DOI] [PubMed] [Google Scholar]
- 33.Tang ZY, Zhang ZL, Wang Y, Glotzer SC, Kotov NA. Science. 2006;314:274. doi: 10.1126/science.1128045. [DOI] [PubMed] [Google Scholar]
- 34.Aldaye FA, Palmer AL, Sleiman HF. Science. 2008;321:1795. doi: 10.1126/science.1154533. [DOI] [PubMed] [Google Scholar]
- 35.Alivisatos AP, Johnsson KP, Peng XG, Wilson TE, Loweth CJ, Bruchez MP, Schultz PG. Nature. 1996;382:609. doi: 10.1038/382609a0. [DOI] [PubMed] [Google Scholar]
- 36.Mirkin CA, Letsinger RL, Mucic RC, Storhoff JJ. Nature. 1996;382:607. doi: 10.1038/382607a0. [DOI] [PubMed] [Google Scholar]
- 37.Maye MM, Kumara MT, Nykypanchuk D, Sherman WB, Gang O. Nat Nanotechnol. 2010;5:116. doi: 10.1038/nnano.2009.378. [DOI] [PubMed] [Google Scholar]
- 38.Chen W, Bian A, Agarwal A, Liu LQ, Shen HB, Wang LB, Xu CL, Kotov NA. Nano Lett. 2009;9:2153. doi: 10.1021/nl900726s. [DOI] [PubMed] [Google Scholar]
- 39.Fu AH, Micheel CM, Cha J, Chang H, Yang H, Alivisatos AP. J Am Chem Soc. 2004;126:10832. doi: 10.1021/ja046747x. [DOI] [PubMed] [Google Scholar]
- 40.Xu XY, Rosi NL, Wang YH, Huo FW, Mirkin CA. J Am Chem Soc. 2006;128:9286. doi: 10.1021/ja061980b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Deng Z, Tian Y, Lee S-H, Ribbe AE, Mao C. Angewandte Chemie. 2005;117:3648. doi: 10.1002/anie.200463096. [DOI] [PubMed] [Google Scholar]
- 42.Lo PK, Karam P, Aldaye FA, McLaughlin CK, Hamblin GD, Cosa G, Sleiman HF. Nat Chem. 2010;2:319. doi: 10.1038/nchem.575. [DOI] [PubMed] [Google Scholar]
- 43.Cheng WL, Campolongo MJ, Cha JJ, Tan SJ, Umbach CC, Muller DA, Luo D. Nat Mater. 2009;8:519. doi: 10.1038/nmat2440. [DOI] [PubMed] [Google Scholar]
- 44.Cheng W, Park N, Walter MT, Hartman MR, Luo D. Nat Nano. 2008;3:682. doi: 10.1038/nnano.2008.279. [DOI] [PubMed] [Google Scholar]
- 45.Nykypanchuk D, Maye MM, van der Lelie D, Gang O. Nature. 2008;451:549. doi: 10.1038/nature06560. [DOI] [PubMed] [Google Scholar]
- 46.Park SY, Lytton-Jean AKR, Lee B, Weigand S, Schatz GC, Mirkin CA. Nature. 2008;451:553. doi: 10.1038/nature06508. [DOI] [PubMed] [Google Scholar]
- 47.Jones MR, Macfarlane RJ, Lee B, Zhang J, Young KL, Senesi AJ, Mirkin CA. Nat Mater. 2010;9:913. doi: 10.1038/nmat2870. [DOI] [PubMed] [Google Scholar]
- 48.Sharma J, Chhabra R, Cheng A, Brownell J, Liu Y, Yan H. Science. 2009;323:112. doi: 10.1126/science.1165831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen CL, Zhang PJ, Rosi NL. J Am Chem Soc. 2008;130:13555. doi: 10.1021/ja805683r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rosi NL, Chen CL. Angew Chem Int Edit. 2010;49:1924. doi: 10.1002/anie.200903572. [DOI] [PubMed] [Google Scholar]
- 51.Mastroianni AJ, Claridge SA, Alivisatos AP. J Am Chem Soc. 2009;131:8455. doi: 10.1021/ja808570g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Guerrero-Martínez A, Auguié B, Alonso-Gómez JL, Džoliæ Z, Gómez-Graña S, Žiniæ M, Cid MM, Liz-Marzán LM. Angew Chem Int Edit. 2011;50:5499. doi: 10.1002/anie.201007536. [DOI] [PubMed] [Google Scholar]
- 53.Lilly GD, Agarwal A, Srivastava S, Kotov NA. Small. 2011;7:2004. doi: 10.1002/smll.201100536. [DOI] [PubMed] [Google Scholar]
- 54.Kuang H, Zhao S, Chen W, Ma W, Yong Q, Xu L, Wang L, Xu C. Biosen Bioelectron. 2011;26:2495. doi: 10.1016/j.bios.2010.10.043. [DOI] [PubMed] [Google Scholar]
- 55.Lee J, Govorov AO, Kotov NA. Angew Chem Int Edit. 2005;44:7439. doi: 10.1002/anie.200501264. [DOI] [PubMed] [Google Scholar]
- 56.Chen JIL, Chen Y, Ginger DS. J Am Chem Soc. 2010;132:9600. doi: 10.1021/ja103240g. [DOI] [PubMed] [Google Scholar]
- 57.Lee J, Govorov AO, Dulka J, Kotov NA. Nano Lett. 2004;4:2323. [Google Scholar]
- 58.Lee J, Govorov AO, Kotov NA. Nano Lett. 2005;5:2063. doi: 10.1021/nl051042u. [DOI] [PubMed] [Google Scholar]
- 59.Pramod P, Joseph STS, Thomas KG. J Am Chem Soc. 2007;129:6712. doi: 10.1021/ja071536o. [DOI] [PubMed] [Google Scholar]
- 60.Nie S, Emory SR. Science. 1997;275:1102. doi: 10.1126/science.275.5303.1102. [DOI] [PubMed] [Google Scholar]
- 61.Wustholz KL, Henry AI, McMahon JM, Freeman RG, Valley N, Piotti ME, Natan MJ, Schatz GC, Van Duyne RP. J Am Chem Soc. 2010;132:10903. doi: 10.1021/ja104174m. [DOI] [PubMed] [Google Scholar]
- 62.Li JF, Huang YF, Ding Y, Yang ZL, Li SB, Zhou XS, Fan FR, Zhang W, Zhou ZY, Wu DY, Ren B, Wang ZL, Tian ZQ. Nature. 2010;464:392. doi: 10.1038/nature08907. [DOI] [PubMed] [Google Scholar]
- 63.Turkevich J, SPC, Hillier J. Discuss Faraday Soc. 1951;11:55. [Google Scholar]
- 64.Frens G. Nat Phys Sci. 1973;241:20. [Google Scholar]
- 65.Xu L, Zhu Y, Ma W, Chen W, Liu L, Kuang H, Wang L, Xu C. J Phys Chem C. 2011;115:3243. [Google Scholar]
- 66.Nikoobakht B, El-Sayed MA. Chem Mater. 2003;15:1957. [Google Scholar]
- 67.Gole A, Murphy CJ. Chem Mater. 2004;16:3633. [Google Scholar]
- 68.Murphy CJ, San TK, Gole AM, Orendorff CJ, Gao JX, Gou L, Hunyadi SE, Li T. J Phys Chem B. 2005;109:13857. doi: 10.1021/jp0516846. [DOI] [PubMed] [Google Scholar]
- 69.Hurst SJ, Lytton-Jean AKR, Mirkin CA. Anal Chem. 2006;78:8313. doi: 10.1021/ac0613582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nie ZH, Fava D, Rubinstein M, Kumacheva E. J Am Chem Soc. 2008;130:3683. doi: 10.1021/ja711150k. [DOI] [PubMed] [Google Scholar]
- 71.Pikus JD, Studts JM, McClay K, Steffan RJ, Fox BG. Biochemistry. 1997;36:9283. doi: 10.1021/bi971049t. [DOI] [PubMed] [Google Scholar]
- 72.Chen H, Schlecht S, Semple TC, Hartwig JF. Science. 2000;287:1995. doi: 10.1126/science.287.5460.1995. [DOI] [PubMed] [Google Scholar]
- 73.Stork G, Danheiser RL. The Journal of Organic Chemistry. 1973;38:1775. [Google Scholar]
- 74.Chen T, Wang H, Chen G, Wang Y, Feng Y, Teo WS, Wu T, Chen H. Acs Nano. 2010;4:3087. doi: 10.1021/nn100269v. [DOI] [PubMed] [Google Scholar]
- 75.Xia Y, Song L, Zhu C. Anal Chem. 2011;83:1401. doi: 10.1021/ac1028825. [DOI] [PubMed] [Google Scholar]
- 76.Fan JA, Wu CH, Bao K, Bao JM, Bardhan R, Halas NJ, Manoharan VN, Nordlander P, Shvets G, Capasso F. Science. 2010;328:1135. doi: 10.1126/science.1187949. [DOI] [PubMed] [Google Scholar]
- 77.Fang Y, Seong N-H, Dlott DD. Science. 2008;321:388. doi: 10.1126/science.1159499. [DOI] [PubMed] [Google Scholar]
- 78.Bell SEJ, Sirimuthu NMS. Chem Soc Rev. 2008;37:1012. doi: 10.1039/b705965p. [DOI] [PubMed] [Google Scholar]
- 79.Chen G, Wang Y, Yang MX, Xu J, Goh SJ, Pan M, Chen HY. J Am Chem Soc. 2010;132:3644. doi: 10.1021/ja9090885. [DOI] [PubMed] [Google Scholar]
- 80.Hamaguchi H, Onogi C, Motoyama M. J Raman Spectrosc. 2008;39:555. [Google Scholar]
- 81.Nantes IL, Rodrigues T, de Franca LP, Kawai C, de Faria PA, Mugnol KCU, Braga FM, Tersariol ILS, Smaili SS. J Biol Chem. 2007;282:25577. doi: 10.1074/jbc.M700009200. [DOI] [PubMed] [Google Scholar]
- 82.Qian X, Peng X-H, Ansari DO, Yin-Goen Q, Chen GZ, Shin DM, Yang L, Young AN, Wang MD, Nie S. Nat Biotech. 2008;26:83. doi: 10.1038/nbt1377. [DOI] [PubMed] [Google Scholar]
- 83.Samanta A, Maiti KK, Soh K-S, Liao X, Vendrell M, Dinish US, Yun S-W, Bhuvaneswari R, Kim H, Rautela S, Chung J, Olivo M, Chang Y-T. Angew Chem Int Edit. 2011;50:6089. doi: 10.1002/anie.201007841. [DOI] [PubMed] [Google Scholar]
- 84.El-Sayed IH, Huang XH, El-Sayed MA. Nano Lett. 2005;5:829. doi: 10.1021/nl050074e. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.