Abstract
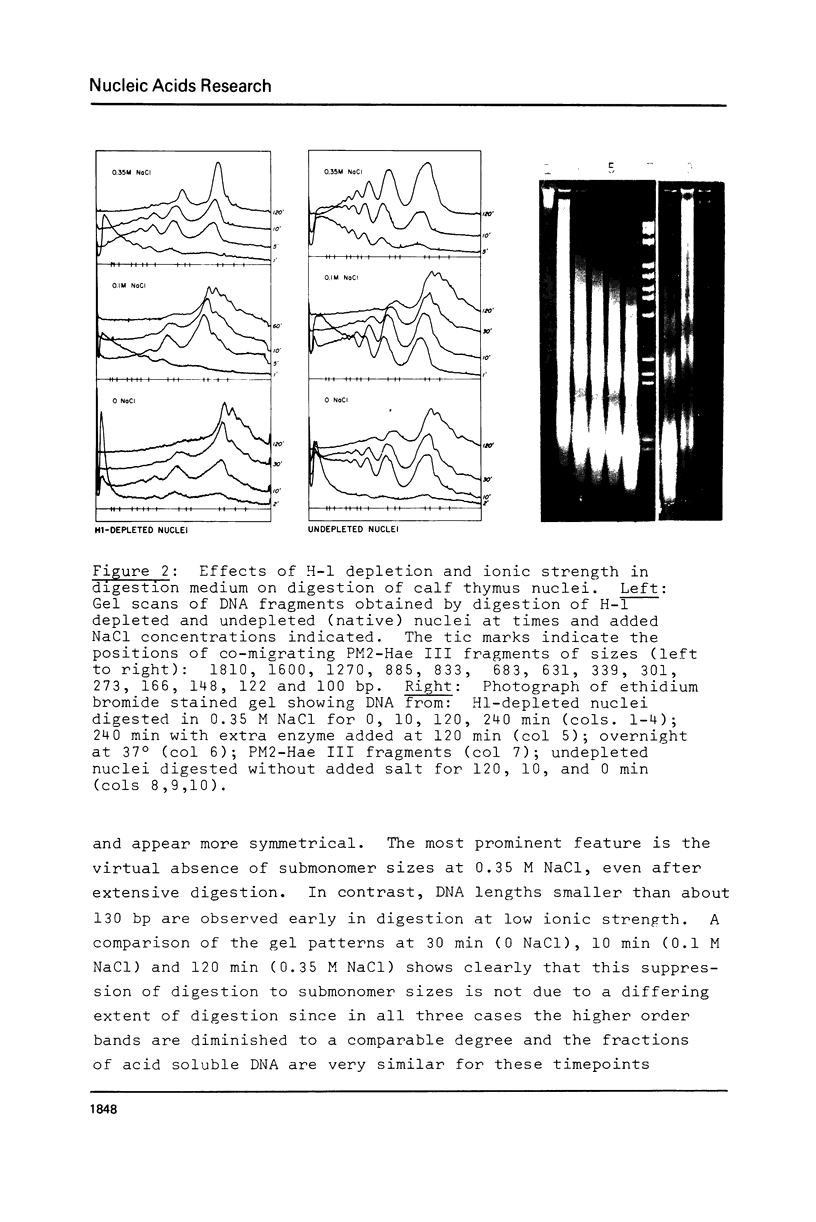
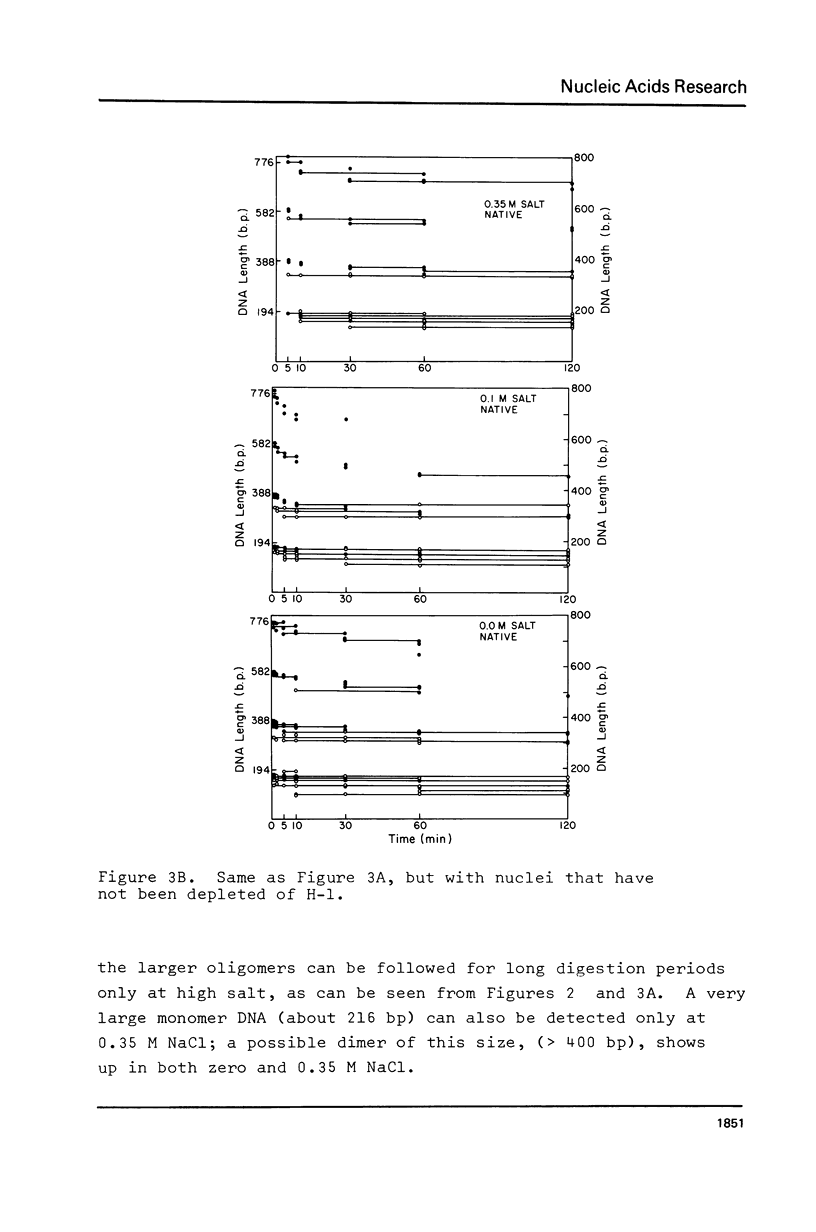
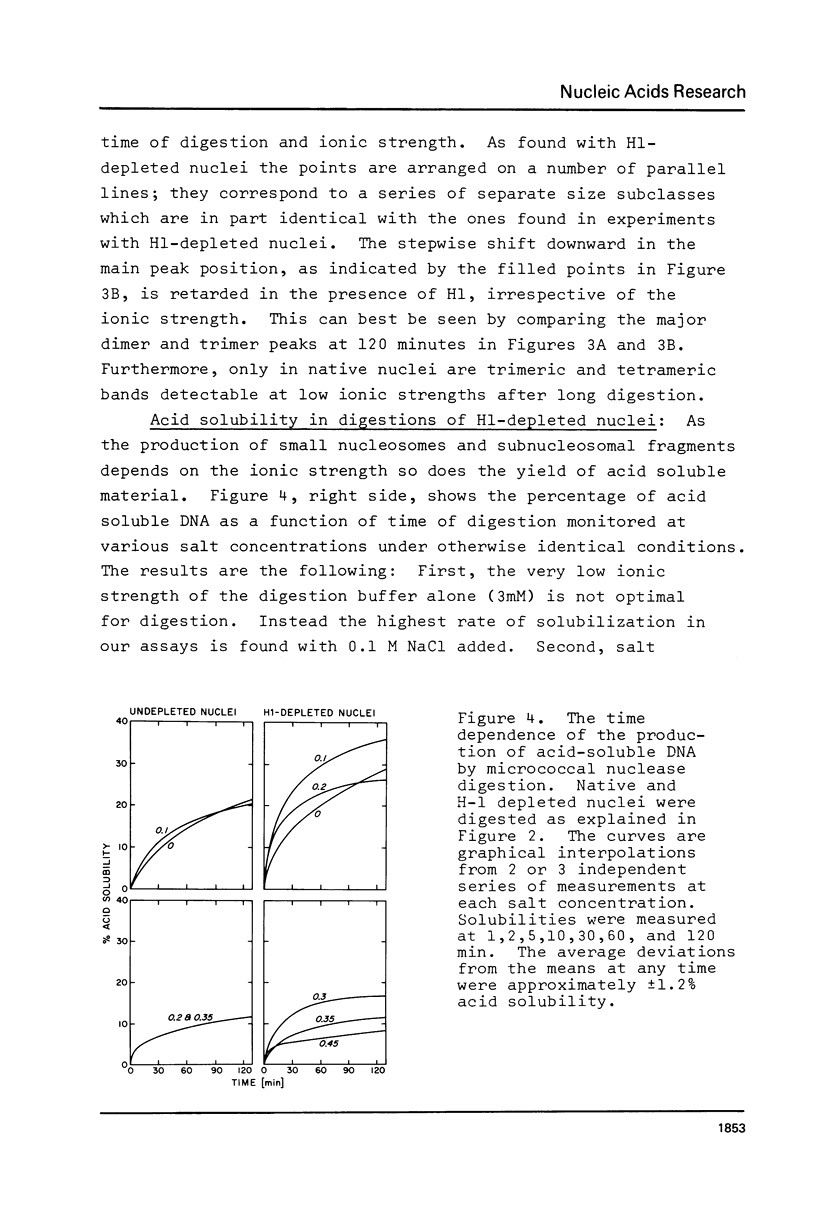
We have removed histone H1 specifically from calf thymus nuclei by low pH treatment, and studied the digestion of such nuclei in comparison with undepleted nuclei. By a number of criteria the nuclei do not appear damaged. The DNA repeat-length in nuclear chromatin is found to be the same (192 +/- 4 bp) in the presence or absence of H1. These experiments demonstrate that the core histone complex of H2A, H2B, H3, and H4 can itself protect DNA sequences as long as 168 bp from nuclease. Our interpretation is that this represents an important structural element in chromatin, carrying two full turns of superhelical DNA. Depending on conditions of digestion this 168 bp fragment may be metastable and is normally rapidly converted by exonucleolytic trimming to the well-known "core-particle" containing 145 bp. Larger stable DNA fragments observed indigestion of H-1 depleted nuclei appear to arise from oligomers assembled from 168 bp cores in close contact exhibiting trimming of 0-20 bp at the ends. Electrophorograms of undepleted nuclear digests reveal oligomer bands in several size classes, each corresponding to one or more combinations of 168 bp particles, H1-protected spacers of about 20 bp length, and particles with ends trimmed to varying degrees.
Full text
PDF















Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Britten R. J., Graham D. E., Neufeld B. R. Analysis of repeating DNA sequences by reassociation. Methods Enzymol. 1974;29:363–418. doi: 10.1016/0076-6879(74)29033-5. [DOI] [PubMed] [Google Scholar]
- Camerini-Otero R. D., Sollner-Webb B., Felsenfeld G. The organization of histones and DNA in chromatin: evidence for an arginine-rich histone kernel. Cell. 1976 Jul;8(3):333–347. doi: 10.1016/0092-8674(76)90145-8. [DOI] [PubMed] [Google Scholar]
- Camerini-Otero R. D., Sollner-Webb B., Felsenfeld G. The organization of histones and DNA in chromatin: evidence for an arginine-rich histone kernel. Cell. 1976 Jul;8(3):333–347. doi: 10.1016/0092-8674(76)90145-8. [DOI] [PubMed] [Google Scholar]
- Clark R. J., Felsenfeld G. Chemical probes of chromatin structure. Biochemistry. 1974 Aug 13;13(17):3622–3628. doi: 10.1021/bi00714a034. [DOI] [PubMed] [Google Scholar]
- Clark R. J., Felsenfeld G. Structure of chromatin. Nat New Biol. 1971 Jan 27;229(4):101–106. doi: 10.1038/newbio229101a0. [DOI] [PubMed] [Google Scholar]
- Cole R. D., Lawson G. M., Hsiang M. W. H1 histone and the condensation of chromatin and DNA. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 1):253–263. doi: 10.1101/sqb.1978.042.01.027. [DOI] [PubMed] [Google Scholar]
- Compton J. L., Bellard M., Chambon P. Biochemical evidence of variability in the DNA repeat length in the chromatin of higher eukaryotes. Proc Natl Acad Sci U S A. 1976 Dec;73(12):4382–4386. doi: 10.1073/pnas.73.12.4382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaubatz J. W., Chalkley R. Distribution of H1 histone in chromatin digested by micrococcal nuclease. Nucleic Acids Res. 1977 Oct;4(10):3281–3301. doi: 10.1093/nar/4.10.3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohr D., Corden J., Tatchell K., Kovacic R. T., Van Holde K. E. Comparative subunit structure of HeLa, yeast, and chicken erythrocyte chromatin. Proc Natl Acad Sci U S A. 1977 Jan;74(1):79–83. doi: 10.1073/pnas.74.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noll M., Thomas J. O., Kornberg R. D. Preparation of native chromatin and damage caused by shearing. Science. 1975 Mar 28;187(4182):1203–1206. doi: 10.1126/science.187.4182.1203. [DOI] [PubMed] [Google Scholar]
- Olins A. L., Carlson R. D., Wright E. B., Olins D. E. Chromatin nu bodies: isolation, subfractionation and physical characterization. Nucleic Acids Res. 1976 Dec;3(12):3271–3291. doi: 10.1093/nar/3.12.3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renz M., Day L. A. Transition from noncooperative to cooperative and selective binding of histone H1 to DNA. Biochemistry. 1976 Jul 27;15(15):3220–3228. doi: 10.1021/bi00660a010. [DOI] [PubMed] [Google Scholar]
- Renz M., Nehls P., Hozier J. Involvement of histone H1 in the organization of the chromosome fiber. Proc Natl Acad Sci U S A. 1977 May;74(5):1879–1883. doi: 10.1073/pnas.74.5.1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley D., Weintraub H. Nucleosomal DNA is digested to repeats of 10 bases by exonuclease III. Cell. 1978 Feb;13(2):281–293. doi: 10.1016/0092-8674(78)90197-6. [DOI] [PubMed] [Google Scholar]
- Shaw B. R., Herman T. M., Kovacic R. T., Beaudreau G. S., Van Holde K. E. Analysis of subunit organization in chicken erythrocyte chromatin. Proc Natl Acad Sci U S A. 1976 Feb;73(2):505–509. doi: 10.1073/pnas.73.2.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson R. T. Structure of the chromatosome, a chromatin particle containing 160 base pairs of DNA and all the histones. Biochemistry. 1978 Dec 12;17(25):5524–5531. doi: 10.1021/bi00618a030. [DOI] [PubMed] [Google Scholar]
- Simpson R. T., Whitlock J. P., Jr Chemical evidence that chromatin DNA exists as 160 base pair beads interspersed with 40 base pair bridges. Nucleic Acids Res. 1976 Jan;3(1):117–127. doi: 10.1093/nar/3.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sollner-Webb B., Felsenfeld G. A comparison of the digestion of nuclei and chromatin by staphylococcal nuclease. Biochemistry. 1975 Jul;14(13):2915–2920. doi: 10.1021/bi00684a019. [DOI] [PubMed] [Google Scholar]
- Steinmetz M., Streeck R. E., Zachau H. G. Closely spaced nucleosome cores in reconstituted histone.DNA complexes and histone-H1-depleted chromatin. Eur J Biochem. 1978 Feb;83(2):615–628. doi: 10.1111/j.1432-1033.1978.tb12131.x. [DOI] [PubMed] [Google Scholar]
- Tatchell K., Van Holde K. E. Compact oligomers and nucleosome phasing. Proc Natl Acad Sci U S A. 1978 Aug;75(8):3583–3587. doi: 10.1073/pnas.75.8.3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd R. D., Garrard W. T. Two-dimensional electrophoretic analysis of polynucleosomes. J Biol Chem. 1977 Jul 10;252(13):4729–4738. [PubMed] [Google Scholar]
- Whitlock J. P., Jr, Simpson R. T. Removal of histone H1 exposes a fifty base pair DNA segment between nucleosomes. Biochemistry. 1976 Jul 27;15(15):3307–3314. doi: 10.1021/bi00660a022. [DOI] [PubMed] [Google Scholar]
- Whitlock J. P., Jr Staphylococcal nuclease and pancreatic DNase cleave the DNA within the chromatin core particle at different sites. J Biol Chem. 1977 Nov 10;252(21):7635–7639. [PubMed] [Google Scholar]
- Wittig B., Wittig S. Nucleosome mono, di, tri-, and tetramers from chicken embryo chromatin. Nucleic Acids Res. 1977 Nov;4(11):3901–3917. doi: 10.1093/nar/4.11.3901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray W., Stubblefield E. A new method for the rapid isolation of chromosomes, mitotic apparatus, or nuclei from mammalian fibroblasts at near neutral pH. Exp Cell Res. 1970 Mar;59(3):469–478. doi: 10.1016/0014-4827(70)90656-7. [DOI] [PubMed] [Google Scholar]