Abstract
Small-angle x-ray scattering (SAXS) is a powerful method for obtaining quantitative structural information on the size and shape of proteins, and it is increasingly used in kinetic studies of folding and association reactions. In this mini-review, we discuss recent developments in using SAXS to obtain structural information on the unfolded ensemble and early folding intermediates of proteins using continuous-flow mixing devices. Interfacing of these micromachined devices to SAXS beamlines has allowed access to the microsecond time regime. The experimental constraints in implementation of turbulence and laminar flow based mixers with SAXS detection and a comparison of the two approaches are presented. Current improvements and future prospects of microsecond time-resolved SAXS and the synergy with ab initio structure prediction and molecular dynamics simulations are discussed.
Introduction
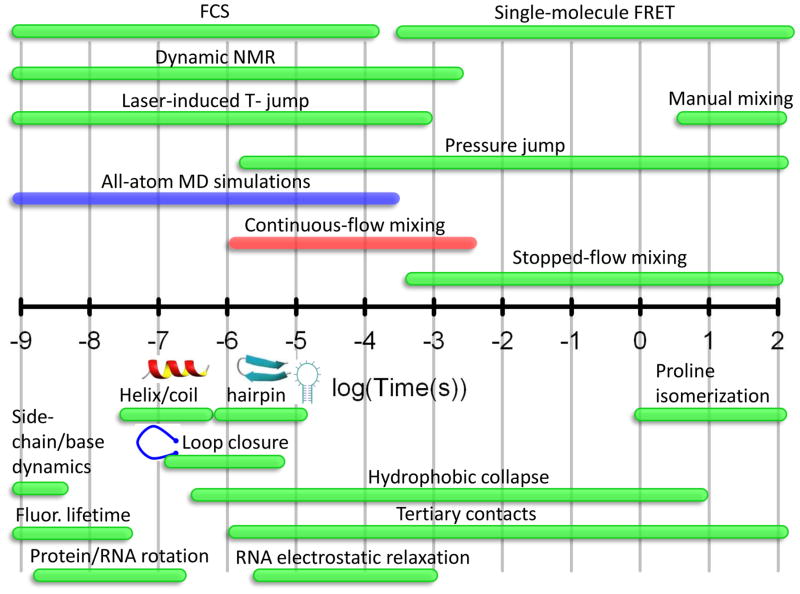
Obtaining information on the structure and dynamics of biological macromolecules is of fundamental importance in understanding their biological function and mechanism. This is especially true for protein folding, a process that involves a large dynamic range in both length and time scales (Figure 1). The kinetics of many fundamental protein building blocks such as α-helices, β–turns and loops, and those of numerous proteins have been resolved with sub-μs and μs time resolution with a wide range of techniques.1–3 However, the structural information often sought in these studies is on the marginally stable states in the upper reaches of the folding energy landscape. This information is generally difficult to access by equilibrium methods, and one approach to populating these states or basins is by monitoring refolding kinetics initiated from an unfolded ensemble. The transient nature of these states precludes many high-resolution solution methods for obtaining quantitative geometric information. Additionally, some of the sub-ensembles are often composed of partially or completely disordered regions, and the language for describing these structures is framed in terms of distributions or structural biases rather than specific structures. Small-angle x-ray scattering (SAXS) is a versatile tool that fulfills many of the requirements for quantitatively probing transiently populated protein structures. In this mini-review, we focus on the use of time-resolved SAXS to obtain geometric insights into the folding reactions of proteins. Recent developments are highlighted in which continuous-flow mixers have extended the time range of SAXS to gain structural insights into early folding events.
Figure 1.
Timescales for folding events and the techniques used to study them. The bottom half of the log-scale timeline depicts the folding processes and the range of timescales over which they occur. The most common experimental techniques and their applicable time ranges are shown in the top half. The time range for all-atom molecular dynamics simulations are shown in red, illustrating the overlap with microfluidic mixing methods.
Why SAXS?
SAXS has seen a tremendous increase in popularity in the last decade as judged by the number of publications (ISI Web of Knowledge [v.4.10], http://apps.isiknowledge.com). Quantitative information about the size, shape and oligomeric state of macromolecular complexes can be obtained from SAXS without the need for extrinsic labeling.4 Synergistic advances in hardware and software have also played an important role in this explosive growth. The quality of data available at 3rd generation synchrotrons (e.g., APS, SSRL, Spring8, ESRF) has allowed unprecedented signal-to-noise ratios which has, in turn, motivated efforts at ab initio modeling of macromolecular structures.5,6 The high brilliance x-ray sources and new detectors have also made possible the time-resolved SAXS experiments reviewed here. These developments are ushering in an era that brings us increasingly closer toward our goal of obtaining structural snapshots of macromolecular processes such as folding, ligand binding and association/oligomerization reactions on timescales from microseconds to hundreds of seconds.
Comparison to other structural probes
SAXS is an attractive complement to other probes of geometric structure such as Förster resonance energy transfer (FRET),7,8 double electron-electron resonance (DEER),9 paramagnetic relaxation enhancement (PRE) NMR,10–12 residual dipolar coupling and pulse field gradient (PFG) NMR.13 Ab initio based bead modeling and ensemble optimization has allowed low resolution structures (or more precisely, ensemble of structures) to be determined at a 6–15 Å resolution, making solution SAXS an attractive alternative to cryo-electron microscopy.4–6 This approach has also been implemented in time-resolved experiments14 and forms the motivation for obtaining structural insights into microsecond kinetic folding events using SAXS. Low-resolution structural models are possible from SAXS because the scattering profile contains contributions, weighted by their form factors, from all pairs of electron density centers in the macromolecule. This averaging is distinct from that in dipole-dipole based distance readouts (e.g., FRET, DEER, NOE) in which distances in an ensemble are 1/r6 weighted. A potential drawback of SAXS is that a weighted average is typically obtained over many distances, yielding no specific information for residue pairs. However, this limitation can be circumvented with the use of gold nanocrystals and contrast matching, an approach that shows great promise in equilibrium and time-resolved studies owing to the significantly greater range of distances that can be probed.15,16 It is worth noting that the timescale over which SAXS probes structure is on the electronic timescale (<fs), orders of magnitude faster than molecular motion (~ps).4 In comparison, dipole-dipole based approaches (e.g. FRET) probe over much longer timescales (~ns).17
With the dramatic improvements in computational power, all-atom simulations with explicit solvent have become possible on larger systems, ~60 residues, to times well beyond a microsecond (Figure 1).18,19 Although there is a paucity of experimental structural metrics to compare with simulations, fast timescale SAXS measurements are an excellent tool to fill this need. Calculated observables, such as the radius of gyration (Rg), pair distribution function of the dominant structural ensembles and coarse grained structures obtained from hidden Markov models, can be compared with experimental results. Although the folding landscape is complex,20 an intriguing finding from recent simulations is that a limited number of pathways govern the accessibility of the native state.19 This suggests that intermediates resolved in experiments can be related to those from simulations. The label-free nature of SAXS measurements and the robust global probes obtained from the technique are an advantage in that simulations are generally carried out in the absence of extrinsic labels. Increasing the overlap in timescales between simulation and SAXS is likely to be important in this regard.
Development of fast time-resolved SAXS
Historical developments of sub-ms SAXS
Design principles for microsecond continuous-flow turbulent mixing devices were outlined nearly 25 years ago by Regenfuss et al. 21 and initially applied to folding studies by the Rousseau, Eaton and Roder groups in the mid-1990’s.22–24 The first integration with SAXS detection utilized a slight modification of this design to achieve a 14 ms dead time.25 A substantial improvement in time resolution and the breaking of the millisecond-barrier was achieved by Takahashi and coworkers by using micromachined mixers to achieve complete mixing within 70 μs.26 This design has been subsequently adopted in most continuous-flow SAXS work. The x-ray beam geometry, however, limited the time resolution of the experiment to ~160 μs, and this metric that has not been improved.
Concurrent with application of turbulent mixers to protein folding studies, laminar mixing using hydrodynamic focusing was pioneered by the Austin group.27,28 Hydrodynamic focusing offers the promise of significant savings in sample consumption, and its physics is much more amenable to computational fluid dynamics simulations than turbulent mixing. Pollack and coworkers pioneered the merger of laminar mixing devices with SAXS detection.29 The time resolution of laminar mixing SAXS methods has historically trailed those achieved using turbulent methods (~1 ms vs. ~200 μs), primarily because of the slower flow speeds and beam geometry as discussed below.
Comparison of turbulent and laminar flow SAXS
Turbulence based mixers utilize high Reynolds number flow (Re>103) in a micromachined channel to reduce the size of the largest eddies to ~0.1 μm (Figure 2).2,21 The last and rate-limiting step in mixing is diffusion over this distance, which is determined by the diffusion time, td=λ2/D, where λ=diffusion length and D is the translational diffusion coefficient (typically ~10−5 cm2/s for small molecules and ~10−7 cm2/s for biological macromolecules). Small solvent molecules and additives, such as chemical denaturants and metal ions, diffuse over this length scale within 10 μs. In contrast, laminar mixing is achieved using hydrodynamic focusing of the solution containing the macromolecule of interest (e.g., unfolded protein) down to a sheath that is 0.1 to 1 μm wide. Mixing on the microsecond timescale occurs via the rapid diffusion of solvent across this sheath, within 10 μs. The biological macromolecules, which are considerably larger, diffuse over a much longer timescale. In both methods, distance along the flow channel is converted to the reaction time using the known flow rate and dimensions of the channel.
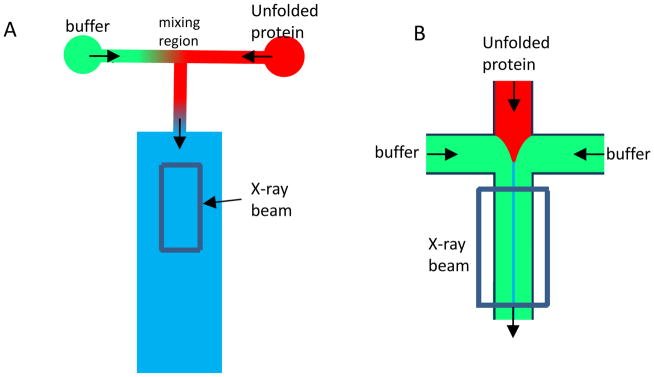
Figure 2.
Schematics of turbulent and laminar flow mixers. In Turbulent mixing, two solutions are brought together at either a T-junction (shown in A) or at 45° angles at high flow rates. The unfolded protein typically flowing at 1–2 ml/min is mixed at the intersection of the two input channels with dilution buffer flowing at 10–20 ml/min. Mixing typically takes place within the first 1 mm of the ~20 mm long central observation channel. Distance along the channel corresponds to folding time at a conversion rate of ~250–300 μs/mm. In laminar mixing (B), a 4-way junction is used and the two side channels containing buffer focus the unfolded protein in the central input channel down to a sheath of ~0.1 to 1 μm in width. The solvent molecules diffuse nearly 2 orders of magnitude faster than the protein. The flow velocity along the channel is typically ~0.1 mm/ms (~10 ms/mm).
Laminar and turbulent mixing based continuous-flow SAXS methods each have distinct advantages. The plug flow in turbulent mixing gives rise to a relatively uniform reaction time in the channel orthogonal to the flow direction, making interfacing to SAXS relatively straightforward. This is further facilitated by the relatively large observation region, typically having dimensions of 200×400 μm (Figure 2). With a well-focused x-ray beam, signal-to-noise ratios comparable to those from static equilibrium experiments can be achieved with a total acquisition time of ~10 s/point. Sample concentrations down to 1 mg/mL have yielded good signal-to-noise ratios (Figure 3).30,31 Another advantage of turbulent mixers is that the linear flow velocity is typically ~4 m/s (or ~250 μs/mm), resulting in a small dead-time (~150–200 μs). The beam exposure times are also over an order of magnitude below the onset of effects from radiation damage and/or heating artifacts (~1–3 ms of beam exposure), essentially eliminating this artifact. The trade-off is that large amounts of sample are required for a successful experiment. Typically, 50–100 mg have been required for the acquisition of a single time point with ~15×1 s exposures. Significant increases in duty cycle are likely to reduce this amount by well over an order of magnitude (see Future directions, below). Because the flow rate requirements are dictated by mixing times and mixing efficiency rather than by the radiation damage threshold, sample usage is far from optimal in current implementations.
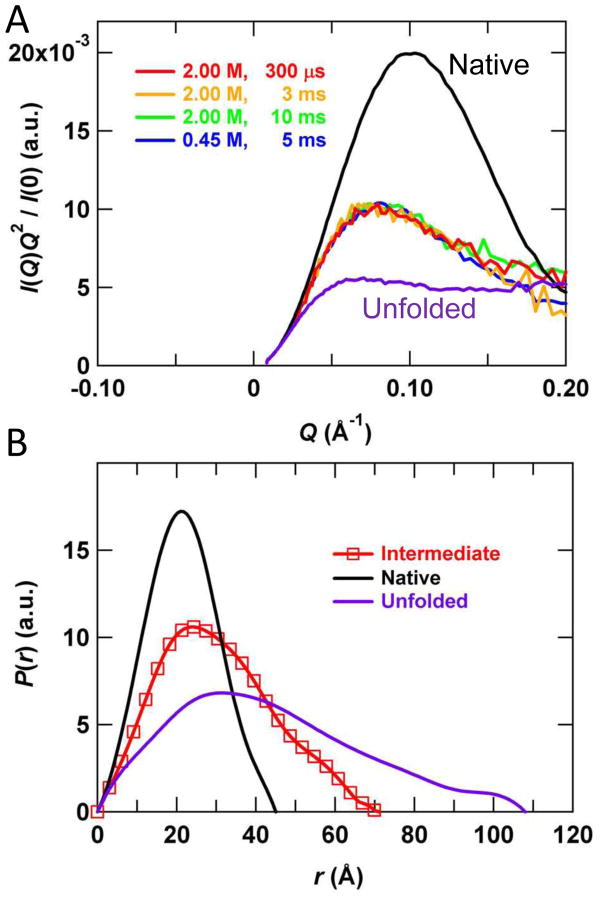
Figure 3.
Representative continuous-flow SAXS data. Refolding of dihydrofolate reductase following a 4.5 M to 0.45 M urea jump is shown at a final protein concentration of ~1 mg/mL. Each scattering curve corresponds to approximately 15×1s images acquired at a final flow rate of 20 ml/min in a 200×350 μm2 flow channel. Although the sample consumption corresponding to this acquisition time is relatively modest (20 ml/min × 1 mg/ml × 15×1 s=5 mg), the duty cycle of the experiment is <10%, requiring >50 mg for each point. Future optimizations are expected to bring the duty cycle to >95%. The figure is reproduced from Reference 30, with permission.
Laminar mixing approaches, by contrast, can achieve nearly perfect sample optimization, consuming significantly less sample. However, Laminar mixing is technically challenging because of the thinness of the sample containing sheath (~1 μm) and the much larger beam dimensions at biological SAXS beamlines (~50–100 μm). Consideration also needs to be given to the parabolic flow in laminar mixers.27 Additionally, the flow rate in laminar mixing systems is typically over an order of magnitude slower than in turbulent mixers (~100 mm/s vs. ~4 m/s) making the size of the x-ray beam a more significant factor in ultimate time resolution of the experiment compared to turbulent mixers.29,32 For example, for a 100 mm/s flow rate and 100 μm beam, the beam focus limits time resolution to approximately 1 ms, which has thus far also been the time resolution in laminar continuous-flow mixing SAXS experiments.29 This hurdle is likely to be overcome in the near future, however, with improved focusing of x-ray beams using microfocus setups or newer synchrotrons such as NSLSII (http://www.bnl.gov/nsls2/sciOps/LifeSci/structDynamics.asp). The excellent time resolution of laminar mixers achieved using optical techniques33 is a goal for SAXS measurements.
Folding studies
Insights into the unfolded ensemble under native conditions from SAXS
Significant effort has been devoted to understanding the unfolded ensemble of proteins and the biases in the energy landscape that direct folding to the native state. Measurements of the radius of gyration, Rg, of unfolded proteins by SAXS have demonstrated that their overall geometry follows statistical scaling relationships expected for a self-avoiding random coil under strongly denaturing conditions (e.g., ≥6 M GdnHCl).34 Despite obeying random coil statistics overall, spectroscopic probes sensitive to local structure have detected persistent biases in the unfolded ensemble.10,35 The dimensions of the unfolded ensemble under native conditions, however, have been the subject of some debate.36 At low denaturant concentrations (e.g., ≤1 M GdnHCl) the native state is thermodynamically strongly favored and the unfolded ensemble comprises a small fraction of the population at a given snapshot in time. Single-molecule FRET experiments are able to selectively probe this small population under equilibrium conditions. Recent single molecule FRET studies of several two-state folding proteins have brought into focus the idea that the unfolded ensemble undergoes compaction under strongly folding conditions.37–41 Rapid dilution kinetic experiments represent an alternative method of probing the unfolded ensemble under native favoring conditions. The generality of the finding from single-molecule FRET studies has been challenged by continuous-flow SAXS results showing that barrierless compaction of the unfolded chain upon transfer to a low-denaturant solvent environment may not be obligatory.42 The earliest observable species at 2.5 ms for both ubiquitin and common-type acyl phosphatase (ctAcP) over a wide range of denaturant concentrations in refolding was shown to have dimensions comparable to the high denaturant unfolded ensemble. Access to low denaturant concentrations in denaturant dilution refolding experiments requires excellent signal-to-noise ratios because of the generally high final protein concentrations, 1 mg/mL or higher, required in the ten-fold dilution. A recent study on barnase also showed that the denatured ensemble is expanded under native conditions (0.7 M GdnHCl).43 A much earlier stopped-flow study on protein L came to a similar conclusion, although insufficiently low denaturant concentrations may have prevented compaction.44
Because parallel SAXS and single-molecule FRET studies have yet to be carried out on a protein with the same extent of extrinsic labeling, it is not clear whether the conflicting results obtained with these techniques is attributable to differences in distance weighting between SAXS and FRET. The Rg metric, for example, because of the r2-averaging, tends to weight longer distances more heavily45 and does not preclude local structure formation.46,47 A study currently underway on monomeric superoxide dismutase48 (C. Kayatekin and O. Bilsel, unpublished results), utilizing both lifetime resolved FRET and SAXS, supports the hypothesis that chain contraction is not obligatory. Furthermore, with one possible exception,30 the unfolded baseline in unfolding denaturation titrations is insensitive to denaturant when Rg is monitored (Figure 4), consistent with an expanded unfolded ensemble at low denaturant concentrations.42–44 With the availability of higher sample throughput via autosamplers at most beamlines today, higher resolution titrations can test these hypotheses with greater accuracy on various point mutants with large differences in stabilities. Recent optical studies have highlighted the role of electrostatics in modulating the compactness of the denatured ensemble in intrinsically disordered proteins.38,49 As noted above, caution should also be exercised in using a single parameter such as Rg, since it can mask local structural changes that are readily apparent in the full scattering curve, particularly at higher scattering angles as shown in Figure 4.
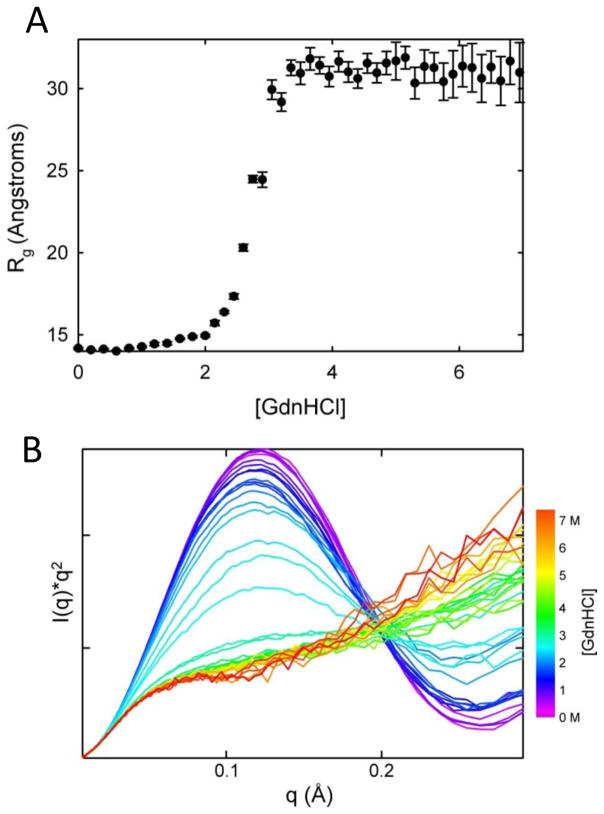
Figure 4.
Equilibrium unfolding titration of horse heart cytochrome c. The equilibrium unfolding titration of cytochrome c illustrates the quality and density of data currently available with autosamplers at the BioCAT beamline. The data in panel (A) illustrates the insensitivity of the radius of gyration to denaturant concentration in the unfolded baseline. However, the Kratky curves in panel (B) hint at local structural changes over the same denaturant concentration range (green to red curves). These data, acquired in the presence of 0.2 M imidazole and at a protein concentration of 4 mg/mL, support the conclusions of Segel et al.66 The Rg in panel (A) was calculated using the Guinier approximation, which uses the low angle region of the scattering curve satisfying the relation Rg*q≤1.3. The Rg can alternatively be calculated from the pair distribution function obtained by transformation of the full scattering curve. However, because the Rg is a root-mean-square average over all pairwise distances the Rg will be more heavily weighted by the longer pairwise distances.
Insights into early collapsed intermediates
A series of continuous-flow SAXS experiments with sub-millisecond time resolution were carried out by the Takahashi group on four proteins of different structural class and chain lengths.26,43,50–52 These studies have demonstrated an initial collapse reaction in the burst-phase of continuous-flow mixers with development of varying degrees of secondary structure and tertiary interactions. Refolding was initiated from either the acid denatured state (cytochrome c26, apomyoglobin51 and heme oxygenase52) or the alkaline denatured state (single chain monellin53) by a rapid pH jump to refolding conditions. Initiation of refolding by pH jump is advantageous because a lower dilution ratio can be used compared to the typically 10-fold dilution needed for urea or GdnHCl jumps. The drawback, however, is that the statistical random-coil unfolded ensemble is not always achieved with pH induced unfolding.26 In all four proteins, large deviations in Rg from the respective unfolded values are observed within the dead time of the experiments (160 μs – 600 μs). Consistent with the observed compaction, the size of the burst-phase species scales with chain length as predicted for a poor solvent by Flory (exponent of 1/3).52 A scaling exponent of 1/3 is expected for a sphere and is also observed for the native states of proteins,34,45 suggesting that the burst phase intermediates are, on average, compact solvent-excluding ensembles. A scaling exponent of 0.588 is expected and observed for statistical random coil unfolded proteins.34 The SAXS studies were complemented with sub-millisecond resolution optical methods such as circular dichroism and infrared spectroscopy to monitor development of secondary structure. Interestingly, the secondary structure content did not correlate with the amount of compaction or with the structural class of proteins. The results are summarized in Table 1 and Figure 5.
Table 1.
Comparisons of Rg from continuous-flow SAXS.a
| Proteinb | SAXS Rgc | Dead time | ||
|---|---|---|---|---|
| Unfolded (Å) | Continuous-flow burst phase (Å) | Native (Å) | ||
| Ub42 | 26 | 25.93 ± 0.03 | 13.9 | 2.5 ms |
| ctAcP42 | 31 | 28.41 ± 0.23 | 14.6 | 2.5 ms |
| DHFR30 | 30.7 | 23.2 ± 0.3 | 16.6 ± 0.1 | 300 μs |
| αTS31 | 43 | 34 | 18.1 | 150 μs |
| cyt c26 | 24 | 20.5 ± 1 | 13.9 | 160 μs |
| apoMb51 | 29.7 ± 1.7 | 23.7 ± 0.9 | 18.2 ± 0.2 | 300 μs |
| SMN53 | 25.5 ± 0.5 | 18.2 ± 0.4 | 15.8 ± 0.2 | 300 μs |
| HO52 | 37.8 ± 1.2 | 26.1 ± 1.1 | 23 ± 1.2 | 600 μs |
Radii of gyration of the burst-phases of different proteins observed by continuous-flow refolding kinetics. The sizes of the native and unfolded states are also given for reference. The dead-time varies between 150 μs and 2.5 ms and is given for each protein.
Ub, ubiquitin; ctAcP, common type acyl-phosphatase; DHFR, E.coli dihydrofolate reductase; αTS, α subunit of tryptophan synthase; cyt c, horse heart cytochrome c; apoMb, apomyoglobin; SMN, single-chain monellin; HO, heme-oxygenase.
When not explicitly given, estimates of the errors for unfolded and continuous-flow burst phase Rg are approximately ±1 Å and those of the native state are approximately ±0.2 Å.
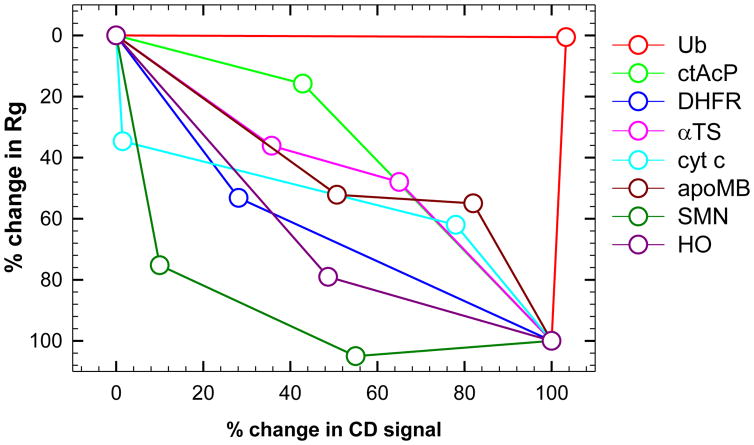
Figure 5.
Secondary structure and Rg reaction coordinate. The reaction coordinate obtained by comparing the relative change in ellipticity from CD measurements to the Rg from SAXS is shown for globular proteins that have been studied by continuous-flow SAXS; ubiquitin (red), common type acyl-phosphatase (green), E.coli dihydrofolate reductase (blue), α subunit of tryptophan synthase (magenta), cytochrome c (cyan), apo-myoglobin (dark red), single-chain monellin (dark green) and heme-oxygenase (dark pink). The values, shown in Table 1, are normalized between unfolded (0%) and Native (100%) states. The values for subsequent intermediates are also shown where available. The profile for ubiquitin has been attributed to the high polyproline-II content of the native state42. The Rg value for the second intermediate of single-chain monellin intermediate is considered within error of the value for the native state (15.4 vs. 15.8 Å) and results in the percentage change slightly exceeding 100%.
Refolding studies with ~250 μs time resolution initiated from the chemically denatured state were carried out on E. coli dihydrofolate reductase (DHFR) and the α-subunit of tryptophan synthase (αTS).30,31 For both proteins, significant collapse is observed within the continuous-flow SAXS dead time. Complementary lifetime-resolved FRET studies suggest that collapse occurs within the ~30 μs dead time of the fluorescence based instrument. In both proteins some secondary structure development is seen in the collapsed intermediates, as suggested by continuous-flow CD, time-resolved anisotropy and ms-timescale pulse labeling hydrogen exchange and mutagenesis analyses.54–57 In both proteins, the compact ensemble is heterogeneous with one tightly packed region and another that is more loosely structured, possibly extended. These regions correspond to the adenosine binding domain (38–106) and disordered loop domain (residues 1–37, 107–159), respectively, in DHFR and an N-terminal region (β1α1β2α2β3α3β4) and a C-terminal region (β6α6β7α7β8α8), respectively, in αTS.31,58 The more locally connected hydrophobic cluster of branched aliphatic side-chains, in both cases, forms first, suggesting a prominent role for hydrophobic “interactions” in early folding events. An intriguing observation in DHFR is that the size (i.e., Rg) of the ~300 μs burst phase species does not appreciably change with final denaturant concentration between 0.45 and 2 M urea as the secondary structure melts away. Resolving the kinetics of these processes, especially by SAXS, will provide insights into the role of the hydrophobic effect and secondary structure in guiding collapse and folding.
Future directions
Enhancements to current approaches
The presence of a burst phase collapse for a number of proteins (e.g., cytochrome c, DHFR, apomyoglobin, αTS; Table 1 and Figure 5) has provided a scientific motivation to access faster timescales (≤30 μs) using SAXS detection. An important experimental parameter affecting time resolution in both turbulent and laminar mixing methods is the beam focus size. In previous experiments at BioCAT, a beam size of ~50 μm full-width at half maximum (FWHM) perpendicular to the flow channel and 150 μm along the flow channel was used.30,31 These dimensions placed constraints on the width of the flow channel, resulting in larger dimensions (200 μm channel width) and greater dead times (150–300 μs) compared to fluorescence studies utilizing narrower channels (<50 μm beam size, 75 μm channel width and ~30 μs dead time). However, recent continuous-flow measurements using a microfocus setup at BioCAT demonstrate that beam dimensions of ~10 μm can significantly reduce the dead time of turbulent mixing experiments to an extent where time resolution is limited by mixing time rather than beam dimensions.
A limiting factor in turbulent mixing approaches has also been the low duty cycle of the experiments imposed by detector constraints. The latest generation of photon counting x-ray detectors, however, overcome these limitations and will allow for high duty cycle scanning measurements to be recorded using SAXS, analogous to continuous-flow fluorescence methods with uninterrupted time-correlated-single-photon counting.31,59 The increased data density will provide more robust kinetic analysis, determination of species spectra using singular value decomposition data reduction methods60 and subsequent low resolution ab initio bead modeling to obtain a coarse grained structure.14 Significant reductions in sample consumption and simultaneous SAXS/WAXS detection, yielding increases in resolution for structural models, are also expected to accompany these improvements.
Promising advances in microfabrication and microdroplets also present opportunities for reducing sample volume and mixing times. A proof of principle study illustrating the use of ~80 μm diameter microdroplets (268 pL) with stroboscopic SAXS detection using 3×3 μm2 x-ray beam has been applied to cytochrome c.61 Mixing times are still in the ms range, but microsecond mixing times are anticipated as the technology matures. Novel microfabrication methods also provide strategies for significantly improving mixing efficiencies for both turbulent and laminar flow mixing approaches. 62,63
Temperature-jump pump probe methods
Access to the shortest timescales will necessitate approaches to initiation of the folding reaction other than by turbulent or laminar mixing. Temperature-jump initiated pump-probe experiments have provided access to these faster timescales using fluorescence and infrared spectroscopy and their integration with SAXS has recently been published.64 An excellent overlap in timescale between temperature-jump methods and all-atom simulations would provide valuable validation for all-atom simulations.
Site-specific information from time-resolved SAXS
A limitation of SAXS studies has been the lack of residue specificity in the structural information. For this reason, lifetime resolved FRET has been used in conjunction with SAXS to obtain this information.31 However, comparisons between SAXS and FRET have not been straightforward because of the different weighting in the two techniques. For example, a recent study on a TIM barrel using tryptophan-AEDANS as a FRET pair noted that FRET was sensitive only to relatively compact conformations with end-to-end distances <40 Å.31 However, these limitations can potentially be overcome with the recent development of nanocrystalline gold particles for site-specific labeling at exposed cysteine residues.15,16 This will allow pairwise and triangulated distance measurements, with a single type of extrinsic label that is comparable in size to an organic fluorophore. Pairwise distance measurements using SAXS are sensitive to a significantly broader range of distances (limited by the q-range of the detection setup) without dynamical averaging, and the application of this technique to time-resolved SAXS is a very promising quantitative tool.
Outlook
The combination of continuous-flow time-resolved SAXS with other spectroscopic techniques to probe early folding events has revealed that a strong bias is present in the unfolded ensemble for many proteins. This bias rapidly directs the formation of specific, in some cases native-like, contacts in heterogeneous compact conformations that appear within 30 μs into the folding reaction. One of the unresolved aspects of the folding problem is why this bias is more pronounced in some proteins than in others. The lack of collapse, for example, in ubiquitin and superoxide dismutase, which maintain unfolded-like dimensions for milliseconds and longer into the folding reaction, is not clear. The entropic and enthalpic solvation penalty for formation of sufficiently large hydrophobic clusters offers a partial explanation for these findings, as reflected in the strong correlations with long range contact order within each class of structure.65 Quantitative structural data are likely to play a key role in our understanding of the guiding principle of these early events, and ultrafast time-resolved SAXS is likely to figure prominently in these efforts.
Acknowledgments
We are indebted to Munehito Arai, Elena Kondrashkina and Can Kayatekin for past collaborations and development of continuous-flow SAXS at BioCAT. We thank Tobin Sosnick, Lois Pollack, Lin Yang, Per Harbury, Jill Zitzewitz and Can Kayatekin for helpful discussions and sharing of unpublished results. This work was supported by grants GM23303 (NIH), GM54836 (NIH) and MCB0721312 (NSF). Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Science, under contract No. W-31-109-ENG-38. BioCAT is a National Institutes of Health-supported Research Center RR-08630.
References
- 1.Kubelka J, Hofrichter J, Eaton WA. Curr Opin Struct Biol. 2004;14:76–88. doi: 10.1016/j.sbi.2004.01.013. [DOI] [PubMed] [Google Scholar]
- 2.Roder H, Maki K, Cheng H. Chemical Reviews. 2006;106:1836–1861. doi: 10.1021/cr040430y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eaton WA, Munoz V, Hagen SJ, Jas GS, Lapidus LJ, Henry ER, Hofrichter J. Annual Review of Biophysics and Biomolecular Structure. 2000;29:327–359. doi: 10.1146/annurev.biophys.29.1.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Svergun DI, Koch MHJ. Reports on Progress in Physics. 2003;66:1735–1782. [Google Scholar]
- 5.Bernado P, Mylonas E, Petoukhov MV, Blackledge M, Svergun DI. Journal of the American Chemical Society. 2007;129:5656–5664. doi: 10.1021/ja069124n. [DOI] [PubMed] [Google Scholar]
- 6.Takahashi Y, Nishikawa Y, Fujisawa T. J Appl Crystallography. 2003;36:549–552. [Google Scholar]
- 7.Haas E. Chemphyschem. 2005;6:858–870. doi: 10.1002/cphc.200400617. [DOI] [PubMed] [Google Scholar]
- 8.Schuler B, Eaton WA. Current Opinion in Structural Biology. 2008;18:16–26. doi: 10.1016/j.sbi.2007.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sale K, Song LK, Liu YS, Perozo E, Fajer P. Journal of the American Chemical Society. 2005;127:9334–9335. doi: 10.1021/ja051652w. [DOI] [PubMed] [Google Scholar]
- 10.Shan B, Eliezer D, Raleigh DP. Biochemistry. 2009;48:4707–4719. doi: 10.1021/bi802299j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iwahara J, Schwieters CD, Clore GM. Journal of the American Chemical Society. 2004;126:5879–5896. doi: 10.1021/ja031580d. [DOI] [PubMed] [Google Scholar]
- 12.Wu KP, Baum J. Journal of the American Chemical Society. 2010;132:5546. doi: 10.1021/ja9105495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gabel F, Simon B, Nilges M, Petoukhov M, Svergun D, Sattler M. Journal of Biomolecular Nmr. 2008;41:199–208. doi: 10.1007/s10858-008-9258-y. [DOI] [PubMed] [Google Scholar]
- 14.Vestergaard B, Groenning M, Roessle M, Kastrup JS, van de Weert M, Flink JM, Frokjaer S, Gajhede M, Svergun DI. PLoS Biol. 2007;5:e134. doi: 10.1371/journal.pbio.0050134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mathew-Fenn RS, Das R, Harbury PA. Science. 2008;322:446–449. doi: 10.1126/science.1158881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mathew-Fenn RS, Das R, Silverman JA, Walker PA, Harbury PA. PLoS One. 2008;3:e3229. doi: 10.1371/journal.pone.0003229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lakowicz JR. Principles of fluorescence spectroscopy. Springer; New York: 2006. [Google Scholar]
- 18.Voelz VA, Bowman GR, Beauchamp K, Pande VS. J Am Chem Soc. 2010;132:1526–1528. doi: 10.1021/ja9090353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shaw DE, Maragakis P, Lindorff-Larsen K, Piana S, Dror RO, Eastwood MP, Bank JA, Jumper JM, Salmon JK, Shan Y, Wriggers W. Science. 2010;330:341–346. doi: 10.1126/science.1187409. [DOI] [PubMed] [Google Scholar]
- 20.Noe F, Schutte C, Vanden-Eijnden E, Reich L, Weikl TR. Proc Natl Acad Sci U S A. 2009;106:19011–19016. doi: 10.1073/pnas.0905466106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Regenfuss P, Clegg RM, Fulwyler MJ, Barrantes FJ, Jovin TM. Review of Scientific Instruments. 1985;56:283–290. [Google Scholar]
- 22.Chan CK, Hu Y, Takahashi S, Rousseau DL, Eaton WA, Hofrichter J. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:1779–1784. doi: 10.1073/pnas.94.5.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Takahashi S, Yeh SR, Das TK, Chan CK, Gottfried DS, Rousseau DL. Nat Struct Biol. 1997;4:44–50. doi: 10.1038/nsb0197-44. [DOI] [PubMed] [Google Scholar]
- 24.Shastry MC, Luck SD, Roder H. Biophysical Journal. 1998;74:2714–2721. doi: 10.1016/S0006-3495(98)77977-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Segel DJ, Bachmann A, Hofrichter J, Hodgson KO, Doniach S, Kiefhaber T. Journal of Molecular Biology. 1999;288:489–499. doi: 10.1006/jmbi.1999.2703. [DOI] [PubMed] [Google Scholar]
- 26.Akiyama S, Takahashi S, Kimura T, Ishimori K, Morishima I, Nishikawa Y, Fujisawa T. Proc Natl Acad Sci USA. 2002;99:1329–1334. doi: 10.1073/pnas.012458999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brody JP, Yager P, Goldstein RE, Austin RH. Biophysical Journal. 1996;71:3430–3441. doi: 10.1016/S0006-3495(96)79538-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Knight JB, Vishwanath A, Brody JP, Austin RH. Physical Review Letters. 1998;80:3863–3866. [Google Scholar]
- 29.Pollack L, Tate MW, Darnton NC, Knight JB, Gruner SM, Eaton WA, Austin RH. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:10115–10117. doi: 10.1073/pnas.96.18.10115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Arai M, Kondrashkina E, Kayatekin C, Matthews CR, Iwakura M, Bilsel O. J Mol Biol. 2007;368:219–229. doi: 10.1016/j.jmb.2007.01.085. [DOI] [PubMed] [Google Scholar]
- 31.Wu Y, Kondrashkina E, Kayatekin C, Matthews CR, Bilsel O. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:13367–13372. doi: 10.1073/pnas.0802788105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Park HY, Qiu XY, Rhoades E, Korlach J, Kwok LW, Zipfel WR, Webb WW, Pollack L. Analytical Chemistry. 2006;78:4465–4473. doi: 10.1021/ac060572n. [DOI] [PubMed] [Google Scholar]
- 33.Yao S, Bakajin O. Analytical Chemistry. 2007;79:5753–5759. doi: 10.1021/ac070528n. [DOI] [PubMed] [Google Scholar]
- 34.Kohn JE, Millett IS, Jacob J, Zagrovic B, Dillon TM, Cingel N, Dothager RS, Seifert S, Thiyagarajan P, Sosnick TR, Hasan MZ, Pande VS, Ruczinski I, Doniach S, Plaxco KW. Proc Natl Acad Sci USA. 2004;101:12491–12496. doi: 10.1073/pnas.0403643101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Klein-Seetharaman J, Oikawa M, Grimshaw SB, Wirmer J, Duchardt E, Ueda T, Imoto T, Smith LJ, Dobson CM, Schwalbe H. Science. 2002;295:1719–1722. doi: 10.1126/science.1067680. [DOI] [PubMed] [Google Scholar]
- 36.Sosnick TR, Barrick D. Curr Opin Struct Biol. doi: 10.1016/j.sbi.2010.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hoffmann A, Kane A, Nettels D, Hertzog DE, Baumgartel P, Lengefeld J, Reichardt G, Horsley DA, Seckler R, Bakajin O, Schuler B. Proc Natl Acad Sci U S A. 2007;104:105–110. doi: 10.1073/pnas.0604353104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Muller-Spath S, Soranno A, Hirschfeld V, Hofmann H, Ruegger S, Reymond L, Nettels D, Schuler B. Proc Natl Acad Sci U S A. 107:14609–14614. doi: 10.1073/pnas.1001743107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nettels D, Gopich IV, Hoffmann A, Schuler B. Proc Natl Acad Sci U S A. 2007;104:2655–2660. doi: 10.1073/pnas.0611093104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.O’Brien EP, Ziv G, Haran G, Brooks BR, Thirumalai D. Proc Natl Acad Sci U S A. 2008;105:13403–13408. doi: 10.1073/pnas.0802113105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ziv G, Thirumalai D, Haran G. Phys Chem Chem Phys. 2009;11:83–93. doi: 10.1039/b813961j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jacob J, Krantz B, Dothager RS, Thiyagarajan P, Sosnick TR. J Mol Biol. 2004;338:369–382. doi: 10.1016/j.jmb.2004.02.065. [DOI] [PubMed] [Google Scholar]
- 43.Konuma T, Kimura T, Matsumoto S, Goto Y, Fujisawa T, Fersht AR, Takahashi S. J Mol Biol. 2011;405:1284–1294. doi: 10.1016/j.jmb.2010.11.052. [DOI] [PubMed] [Google Scholar]
- 44.Plaxco KW, Millett IS, Segel DJ, Doniach S, Baker D. Nat Struct Biol. 1999;6:554–556. doi: 10.1038/9329. [DOI] [PubMed] [Google Scholar]
- 45.Goldenberg DP. J Mol Biol. 2003;326:1615–1633. doi: 10.1016/s0022-2836(03)00033-0. [DOI] [PubMed] [Google Scholar]
- 46.Fitzkee NC, Rose GD. Proc Natl Acad Sci USA. 2004;101:12497–12502. doi: 10.1073/pnas.0404236101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jha AK, Colubri A, Freed KF, Sosnick TR. Proc Natl Acad Sci U S A. 2005;102:13099–13104. doi: 10.1073/pnas.0506078102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Svensson AKE, Bilsel O, Kondrashkina E, Zitzewitz JA, Matthews CR. Journal of Molecular Biology. 2006;364:1084–1102. doi: 10.1016/j.jmb.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 49.Mao AH, Crick SL, Vitalis A, Chicoine CL, Pappu RV. Proc Natl Acad Sci U S A. 107:8183–8188. doi: 10.1073/pnas.0911107107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kimura T, Akiyama S, Uzawa T, Ishimori K, Morishima I, Fujisawa T, Takahashi S. J Mol Biol. 2005;350:349–362. doi: 10.1016/j.jmb.2005.04.074. [DOI] [PubMed] [Google Scholar]
- 51.Uzawa T, Akiyama S, Kimura T, Takahashi S, Ishimori K, Morishima I, Fujisawa T. Proc Natl Acad Sci U S A. 2004;101:1171–1176. doi: 10.1073/pnas.0305376101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Uzawa T, Kimura T, Ishimori K, Morishima I, Matsui T, Ikeda-Saito M, Takahashi S, Akiyama S, Fujisawa T. J Mol Biol. 2006;357:997–1008. doi: 10.1016/j.jmb.2005.12.089. [DOI] [PubMed] [Google Scholar]
- 53.Kimura T, Uzawa T, Ishimori K, Morishima I, Takahashi S, Konno T, Akiyama S, Fujisawa T. Proc Natl Acad Sci USA. 2005;102:2748–2753. doi: 10.1073/pnas.0407982102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu Y, Vadrevu R, Kathuria S, Yang X, Matthews CR. J Mol Biol. 2007;366:1624–1638. doi: 10.1016/j.jmb.2006.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wu Y, Vadrevu R, Yang X, Matthews CR. J Mol Biol. 2005;351:445–452. doi: 10.1016/j.jmb.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 56.Jones BE, Matthews CR. Protein Sci. 1995;4:167–177. doi: 10.1002/pro.5560040204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.O’Neill JC, Jr, Robert Matthews C. J Mol Biol. 2000;295:737–744. doi: 10.1006/jmbi.1999.3403. [DOI] [PubMed] [Google Scholar]
- 58.Vadrevu R, Wu Y, Matthews CR. J Mol Biol. 2008;377:294–306. doi: 10.1016/j.jmb.2007.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bilsel O, Kayatekin C, Wallace LA, Matthews CR. Review of Scientific Instruments. 2005;76 [Google Scholar]
- 60.Henry ER, Hofrichter J. Methods in Enzymology. 1992;210:129–192. [Google Scholar]
- 61.Graceffa R, Burghammer M, Davies RJ, Ponchut C, Riekel C. Applied Physics Letters. 2009;94 [Google Scholar]
- 62.Dootz R, Evans H, Koster S, Pfohl T. Small. 2007;3:96–100. doi: 10.1002/smll.200600288. [DOI] [PubMed] [Google Scholar]
- 63.Palacios E, Ocola LE, Joshi-Imre A, Bauerdick S, Berse M, Peto L. J Vac Sci Technol B. 2010;28 [Google Scholar]
- 64.Cho HS, Dashdorj N, Schotte F, Graber T, Henning R, Anfinrud P. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:7281–7286. doi: 10.1073/pnas.1002951107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Istomin AY, Jacobs DJ, Livesay DR. Protein Sci. 2007;16:2564–2569. doi: 10.1110/ps.073124507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Segel DJ, Fink AL, Hodgson KO, Doniach S. Biochemistry. 1998;37:12443–12451. doi: 10.1021/bi980535t. [DOI] [PubMed] [Google Scholar]