Abstract
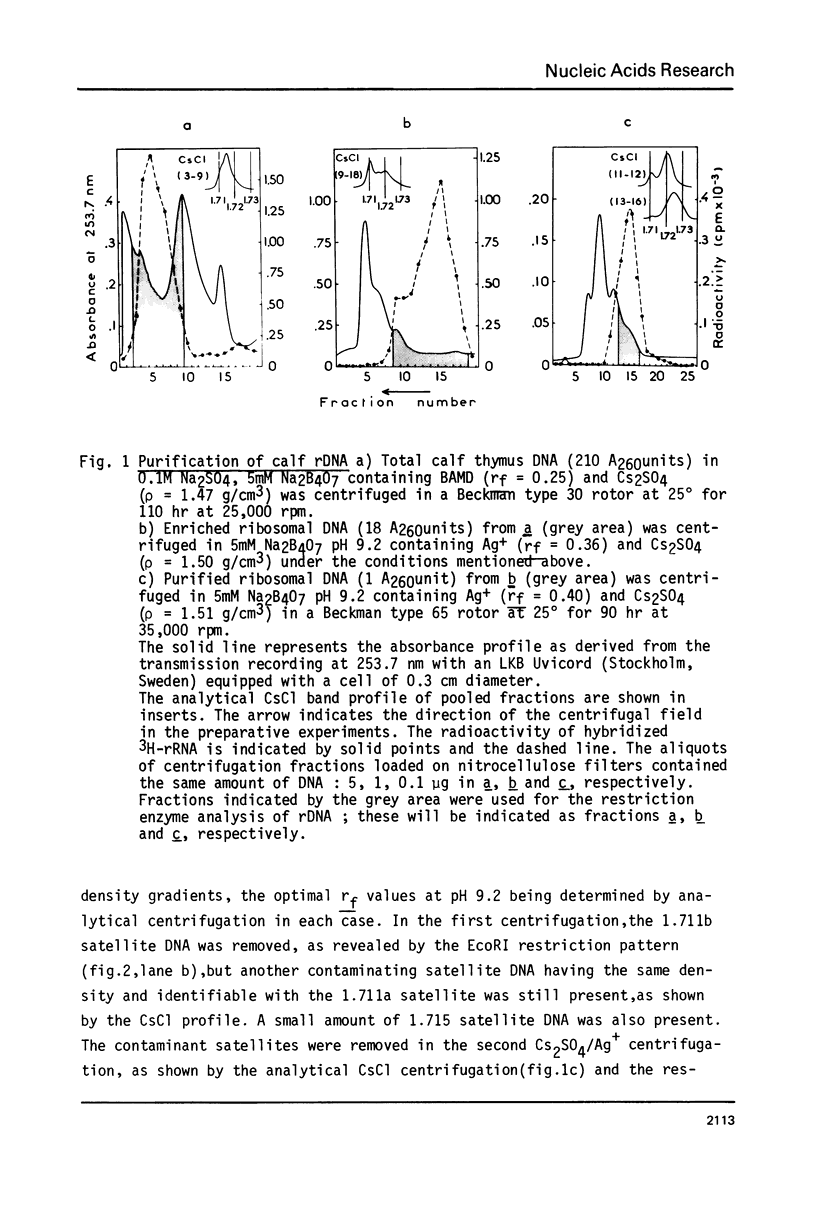
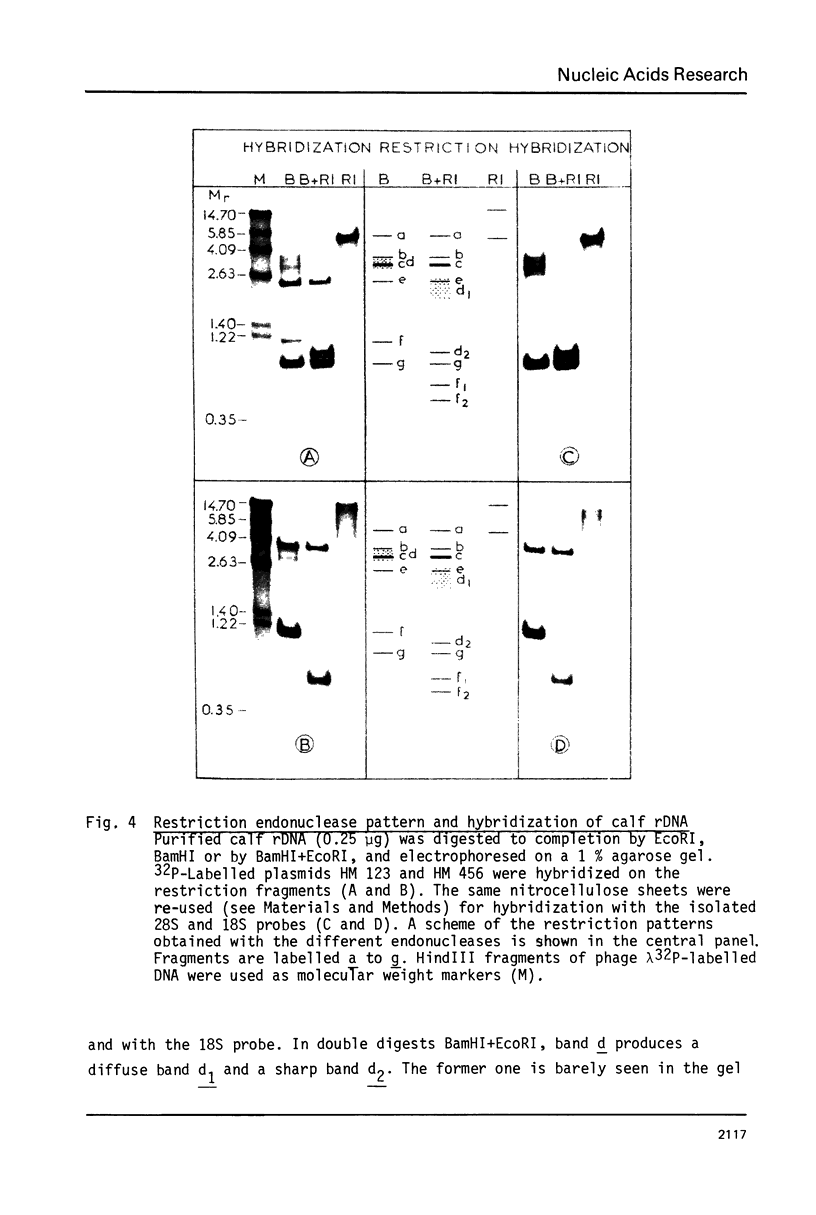
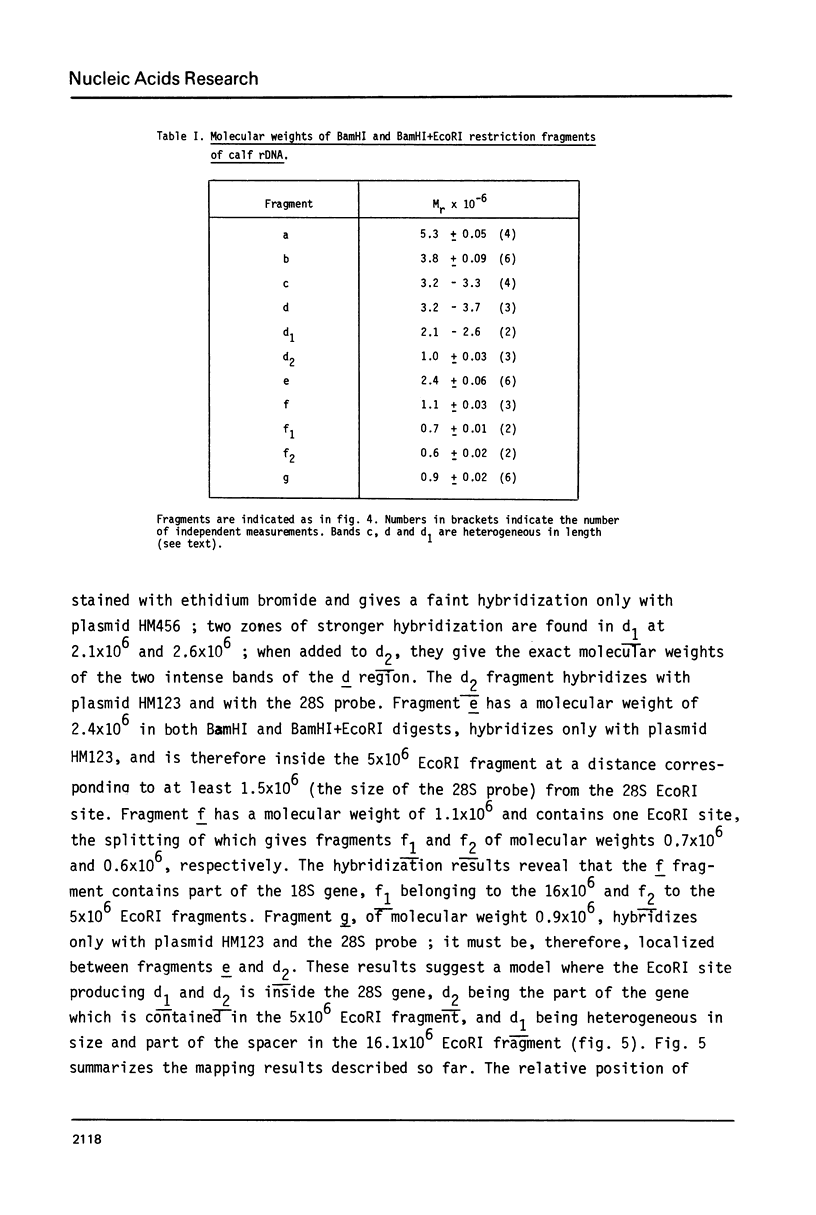
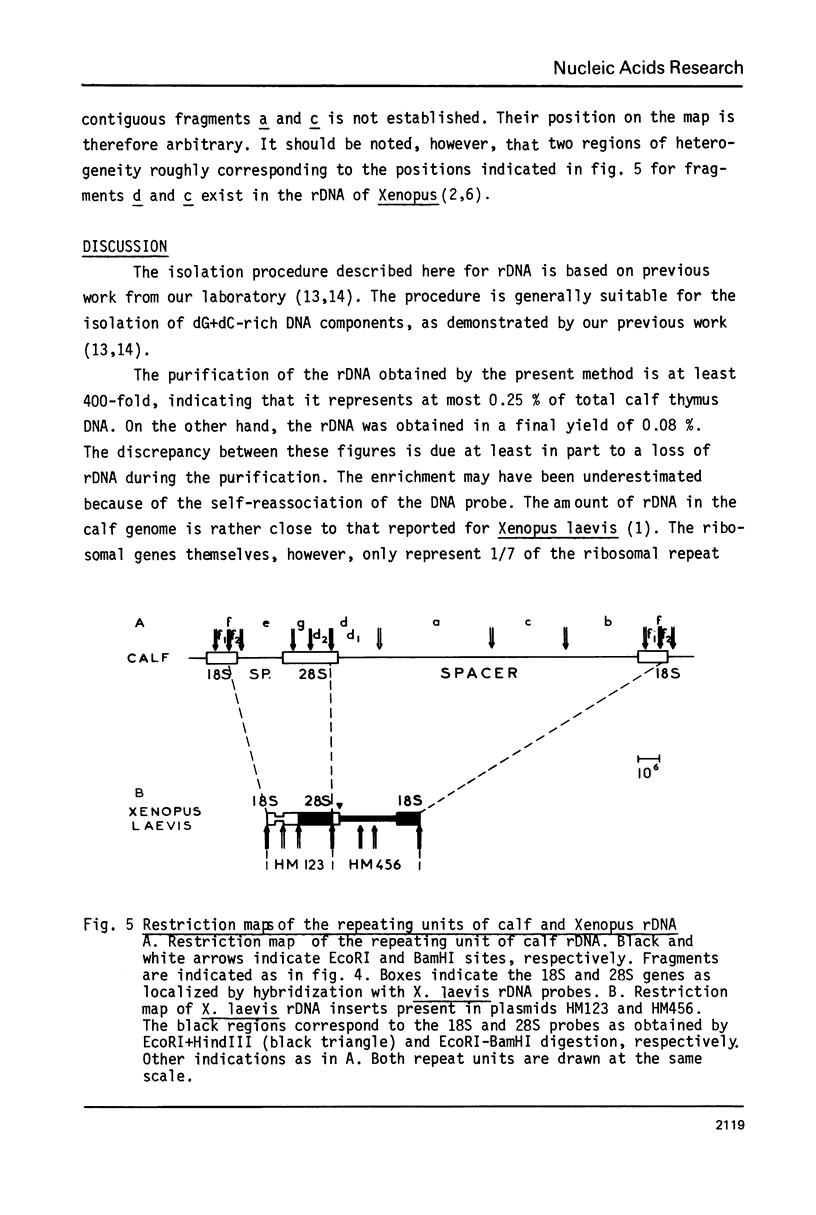
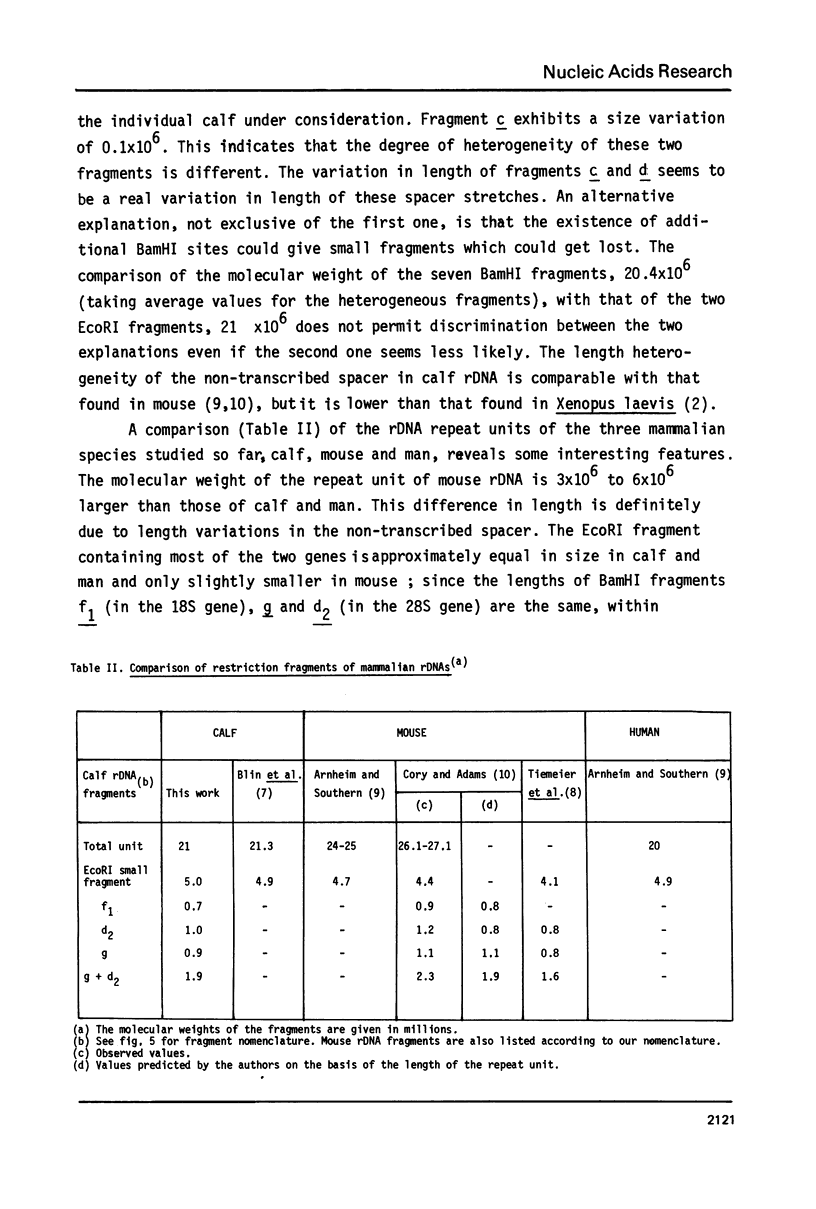
Ribosomal DNA (rDNA) from calf was isolated by three density gradient centrifugations. The first centrifugation in Cs2S04/BAMD was used to obtain partially resolved dG+dC-rich fractions from total DNA. The second and third centrifugations, in Cs2S04/Ag+, led to the isolation of an rDNA fraction characterized by a symmetrical band in CsCl, p = 1.724 g/cm3. This new procedure appears to be generally suitable for the isolation of rDNA and other dG+dC-rich repeated genes. The organization of isolated calf rDNA has been studied by restriction enzyme digestion and by hybridization with cloned rDNA from Xenopus laevis. The repeat unit of calf rDNA has a molecular weight of 21x10(6) and is split by EcoR1 into two fragments, 16x10(6) and 5.0x10(6), and by BamHI into seven fragments. EcoRI and BamHI sites have been mapped. Most of the 18S and 28S RNA genes and the transcribed spacer are contained in the small EcoRI fragment, while the non-transcribed spacer is localized in the large EcoRI fragment. This spacer showed length heterogeneity within a single individual; such heterogeneity is limited to two regions of the spacer.
Full text
PDF









Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arnheim N., Southern E. M. Heterogeneity of the ribosomal genes in mice and men. Cell. 1977 Jun;11(2):363–370. doi: 10.1016/0092-8674(77)90053-8. [DOI] [PubMed] [Google Scholar]
- Bachmann K. Genome size in mammals. Chromosoma. 1972;37(1):85–93. doi: 10.1007/BF00329560. [DOI] [PubMed] [Google Scholar]
- Birnstiel M. L., Chipchase M., Speirs J. The ribosomal RNA cistrons. Prog Nucleic Acid Res Mol Biol. 1971;11:351–389. doi: 10.1016/s0079-6603(08)60332-3. [DOI] [PubMed] [Google Scholar]
- Blin N., Stephenson E. C., Stafford D. W. Isolation and some properties of a mammalian ribosomal DNA. Chromosoma. 1976 Oct 12;58(1):41–50. doi: 10.1007/BF00293439. [DOI] [PubMed] [Google Scholar]
- Botchan P., Reeder R. H., Dawid I. B. Restriction analysis of the nontranscribed spacers of Xenopus laevis ribosomal DNA. Cell. 1977 Jul;11(3):599–607. doi: 10.1016/0092-8674(77)90077-0. [DOI] [PubMed] [Google Scholar]
- Buongiorno-Nardelli M., Amaldi F., Beccari E., Junakovic N. Size of ribosomal DNA repeating units in Xenopus laevis: limited individual heterogeneity and extensive population polymorphism. J Mol Biol. 1977 Feb 15;110(1):105–117. doi: 10.1016/s0022-2836(77)80101-0. [DOI] [PubMed] [Google Scholar]
- Cortadas J., Macaya G., Bernardi G. An analysis of the bovine genome by density gradient centrifugation: fractionation in Cs2SO4/3,6-bis(acetatomercurimethyl)dioxane density gradient. Eur J Biochem. 1977 Jun 1;76(1):13–19. doi: 10.1111/j.1432-1033.1977.tb11565.x. [DOI] [PubMed] [Google Scholar]
- Cory S., Adams J. M. A very large repeating unit of mouse DNA containing the 18S, 28S and 5.8S rRNA genes. Cell. 1977 Aug;11(4):795–805. doi: 10.1016/0092-8674(77)90292-6. [DOI] [PubMed] [Google Scholar]
- Denhardt D. T. A membrane-filter technique for the detection of complementary DNA. Biochem Biophys Res Commun. 1966 Jun 13;23(5):641–646. doi: 10.1016/0006-291x(66)90447-5. [DOI] [PubMed] [Google Scholar]
- Filipski J., Thiery J. P., Bernardi G. An analysis of the bovine genome by Cs2SO4-Ag density gradient centrifugation. J Mol Biol. 1973 Oct 15;80(1):177–197. doi: 10.1016/0022-2836(73)90240-4. [DOI] [PubMed] [Google Scholar]
- Gillespie D., Spiegelman S. A quantitative assay for DNA-RNA hybrids with DNA immobilized on a membrane. J Mol Biol. 1965 Jul;12(3):829–842. doi: 10.1016/s0022-2836(65)80331-x. [DOI] [PubMed] [Google Scholar]
- Lebowitz P., Siegel W., Sklar J. Hemophilus aegyptius restriction edonuclease cleavage map of the simian virus 40 genome and its colinear relation with the hemophilus influenzae cleavage map of SV40. J Mol Biol. 1974 Sep 5;88(1):105–123. doi: 10.1016/0022-2836(74)90297-6. [DOI] [PubMed] [Google Scholar]
- Macaya G., Cortadas J., Bernardi G. An analysis of the bovine genome by density-gradient centrifugation. Preparation of the dG+dC-rich DNA components. Eur J Biochem. 1978 Mar;84(1):179–188. doi: 10.1111/j.1432-1033.1978.tb12155.x. [DOI] [PubMed] [Google Scholar]
- Melli M. Clustering of the DNA sequences complementary to repetitive nuclear RNA of HeLa cells. J Mol Biol. 1975 Mar 25;93(1):23–38. doi: 10.1016/0022-2836(75)90357-5. [DOI] [PubMed] [Google Scholar]
- Morrow J. F., Cohen S. N., Chang A. C., Boyer H. W., Goodman H. M., Helling R. B. Replication and transcription of eukaryotic DNA in Escherichia coli. Proc Natl Acad Sci U S A. 1974 May;71(5):1743–1747. doi: 10.1073/pnas.71.5.1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polisky B., Greene P., Garfin D. E., McCarthy B. J., Goodman H. M., Boyer H. W. Specificity of substrate recognition by the EcoRI restriction endonuclease. Proc Natl Acad Sci U S A. 1975 Sep;72(9):3310–3314. doi: 10.1073/pnas.72.9.3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prunell A., Kopecka H., Strauss F., Bernardi G. The mitochondrial genome of wild-type yeast cells. V. Genome evolution. J Mol Biol. 1977 Feb 15;110(1):17–47. doi: 10.1016/s0022-2836(77)80096-x. [DOI] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Sinclair J. H., Brown D. D. Retention of common nucleotide sequences in the ribosomal deoxyribonucleic acid of eukaryotes and some of their physical characteristics. Biochemistry. 1971 Jul 6;10(14):2761–2769. doi: 10.1021/bi00790a017. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Long range periodicities in mouse satellite DNA. J Mol Biol. 1975 May 5;94(1):51–69. doi: 10.1016/0022-2836(75)90404-0. [DOI] [PubMed] [Google Scholar]
- Thomas M., Davis R. W. Studies on the cleavage of bacteriophage lambda DNA with EcoRI Restriction endonuclease. J Mol Biol. 1975 Jan 25;91(3):315–328. doi: 10.1016/0022-2836(75)90383-6. [DOI] [PubMed] [Google Scholar]
- Tiemeier D. C., Tilghman S. M., Leder P. Purification and cloning of a mouse ribosomal gene fragment in coliphage lambda. Gene. 1977;2(3-4):173–191. doi: 10.1016/0378-1119(77)90016-6. [DOI] [PubMed] [Google Scholar]
- Wellauer P. K., Dawid I. B., Brown D. D., Reeder R. H. The molecular basis for length heterogeneity in ribosomal DNA from Xenopus laevis. J Mol Biol. 1976 Aug 25;105(4):461–486. doi: 10.1016/0022-2836(76)90229-1. [DOI] [PubMed] [Google Scholar]
- Wellauer P. K., Reeder R. H. A comparison of the structural organization of amplified ribosomal DNA from Xenopus mulleri and Xenopus laevis. J Mol Biol. 1975 May 15;94(2):151–161. doi: 10.1016/0022-2836(75)90074-1. [DOI] [PubMed] [Google Scholar]