Abstract
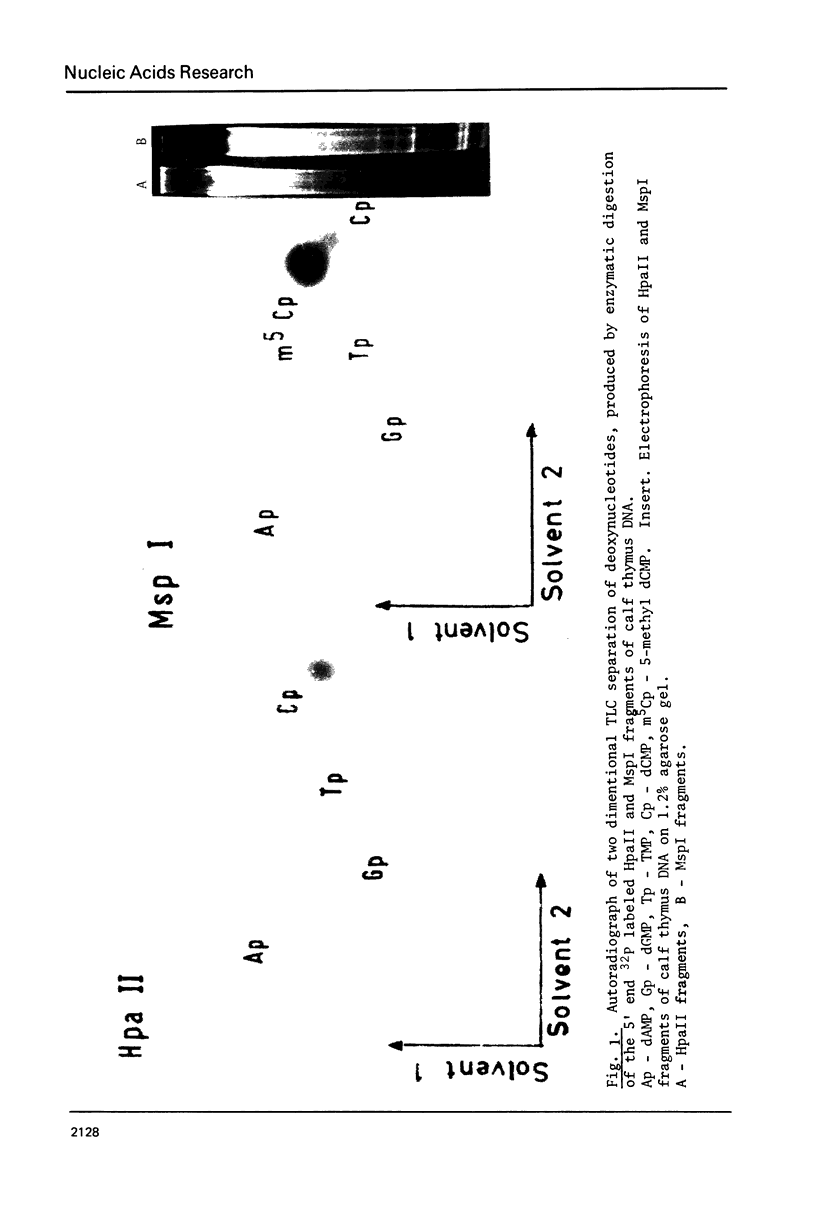
The extent of methylation of the internal C in the sequence CCGG in DNA from various eukaryotic sources has been determined using the restriction enzyme MspI known to be specific for this sequence. The methylation of the CCGG sequence is reflected in the restriction pattern obtained by DNA treated with MspI and its isoschizomer HpaII and analyzed by gel electrophoresis. A direct method for detection 5-methylcytosine in the sequence CCGG has been deviced. DNA fragments obtained with MspI were radioactively labeled at their 5' ends and subsequently degraded to the corresponding 5'-deoxyribonucleoside monophosphates. 5 methylcytidylic acid has been found in most of the 5' ends of MspI fragments of calf thymus DNA (about 90%) indicating heavy methylation of the sequence CCGG in calf thymus DNA. The results also reveal a symmetric methylation of both strands at this sequence in calf thymus DNA. In contrast, the CCGG sequence in other eukaryotic DNAs from organisms like Neurospora, Drosophila and Herpes virus proved to be undermethylated at this sequence.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bird A. P. Use of restriction enzymes to study eukaryotic DNA methylation: II. The symmetry of methylated sites supports semi-conservative copying of the methylation pattern. J Mol Biol. 1978 Jan 5;118(1):49–60. doi: 10.1016/0022-2836(78)90243-7. [DOI] [PubMed] [Google Scholar]
- DOSKOCIL J., SORM F. Distribution of 5-methylcytosine in pyrimidine sequences of deoxyribonucleic acids. Biochim Biophys Acta. 1962 Jun 11;55:953–959. doi: 10.1016/0006-3002(62)90909-5. [DOI] [PubMed] [Google Scholar]
- Gautier F., Bünemann H., Grotjahn L. Analysis of calf-thymus satellite DNA: evidence for specific methylation of cytosine in C-G sequences. Eur J Biochem. 1977 Oct 17;80(1):175–183. doi: 10.1111/j.1432-1033.1977.tb11869.x. [DOI] [PubMed] [Google Scholar]
- Grippo P., Iaccarino M., Parisi E., Scarano E. Methylation of DNA in developing sea urchin embryos. J Mol Biol. 1968 Sep 14;36(2):195–208. doi: 10.1016/0022-2836(68)90375-6. [DOI] [PubMed] [Google Scholar]
- Kelly T. J., Jr, Smith H. O. A restriction enzyme from Hemophilus influenzae. II. J Mol Biol. 1970 Jul 28;51(2):393–409. doi: 10.1016/0022-2836(70)90150-6. [DOI] [PubMed] [Google Scholar]
- Mann M. B., Smith H. O. Specificity of Hpa II and Hae III DNA methylases. Nucleic Acids Res. 1977 Dec;4(12):4211–4221. doi: 10.1093/nar/4.12.4211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SINSHEIMER R. L. The action of pancreatic deoxyribonuclease. II. Isomeric dinucleotides. J Biol Chem. 1955 Aug;215(2):579–583. [PubMed] [Google Scholar]
- Waalwijk C., Flavell R. A. DNA methylation at a CCGG sequence in the large intron of the rabbit beta-globin gene: tissue-specific variations. Nucleic Acids Res. 1978 Dec;5(12):4631–4634. doi: 10.1093/nar/5.12.4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waalwijk C., Flavell R. A. MspI, an isoschizomer of hpaII which cleaves both unmethylated and methylated hpaII sites. Nucleic Acids Res. 1978 Sep;5(9):3231–3236. doi: 10.1093/nar/5.9.3231. [DOI] [PMC free article] [PubMed] [Google Scholar]





