Abstract
Myosin-1d is a monomeric actin-based motor found in a wide range of tissues, but highly expressed in the nervous system. Previous microarray studies suggest that myosin-1d is found in oligodendrocytes where transcripts are upregulated during the maturation of these cells. Myosin-1d was also identified as a component of myelin-containing subcellular fractions in proteomic studies and mutations in MYO1D have been linked to autism. Despite the potential implications of these previous studies, there is little information on the expression and localization of myosin-1d in the developing nervous system. Therefore, we analyzed myosin-1d expression patterns in the peripheral and central nervous systems during postnatal development. In mouse sciatic nerve, myosin-1d is expressed along the axon and in the ensheathing myelin compartment. Analysis of mouse cerebellum prior to myelination at day 3 reveal myosin-1d is present in the Purkinje cell layer, granule cell layer, and region of the cerebellar nuclei. Upon the onset of myelination, myosin-1d enrichment expands along axonal tracts, while still present in the Purkinje and granule cell layers. However, myosin-1d was undetectable in oligodendrocyte progenitor cells at early and late time points. We also show that myosin-1d interacts and is co-expressed with aspartoacylase, an enzyme that plays a key role in fatty acid synthesis throughout the nervous system. Together, these studies provide a foundation for understanding the role of myosin-1d in neurodevelopment and neurological disorders.
Keywords: cytoskeleton, actin, myelin, cerebellum, aspartoacylase
1. Introduction
The myosin superfamily is a diverse collection of actin-binding, ATP-hydrolyzing molecular motors that sort into at least 35 different structural classes (Odronitz and Kollmar, 2007). Class I myosins comprise one of the largest families (eight genes in vertebrates) and are defined by a monomeric heavy chain and the potential to bind directly to acidic phospholipids via a C-terminal basic domain (McConnell and Tyska, 2010). Accordingly, myosins-I have been implicated in a wide range of functions at the actin/membrane interface in a variety tissues (McConnell and Tyska, 2010). Examples include myosin-1a in intestinal epithelia (Collins and Borysenko, 1984), myosin-1c in auditory and vestibular epithelia (Gillespie et al., 1993), Myo1d1 in neurons (Bahler et al., 1994), myosin-1e in kidney (Krendel et al., 2009), and myosins-1f and -1g in a variety of immune cells (Kim et al., 2006; Olety et al., 2010; Patino-Lopez et al., 2010).
Myo1d, previously referred to as myr4 or myosin-Iγ, was first identified in the rat cerebral cortex, spinal cord, brainstem, and cerebellum, in addition to a number of other tissues (Bahler et al., 1994). As a short-tailed class I myosin, Myo1d contains a conserved motor domain, two IQ motifs that bind calmodulin, and a basic C-terminal tail homology-1 (TH1) domain (Bahler et al., 1994). Functional studies suggest that Myo1d plays a role in membrane trafficking (Huber et al., 2000), the control of membrane tension (Nambiar et al., 2009), and the establishment of left-right asymmetry during Drosophila development (Hozumi et al., 2006; Speder et al., 2006). Aside from these initial reports, our understanding of Myo1d function in the context of vertebrate physiology remains largely unexplored.
Three recent lines of evidence suggest that Myo1d plays an important role in nervous system tissues. First, linkage analysis of autistic individuals revealed a potential association with MYO1D (Stone et al., 2007). Second, mass spectrometry studies have identified Myo1d as an enriched component of the myelin proteome (Ishii et al., 2009; Jahn et al., 2009; Yamaguchi et al., 2008). Third, Myo1d is a significantly upregulated transcript during oligodendrocyte maturation, along with other classical myelin-associated components (Cahoy et al., 2008; Nielsen et al., 2006). All of these investigations implicate Myo1d in neurodevelopment and further suggest that this motor plays a role in the process of myelination. However, there is currently no cell biological data to validate or extend the results derived from these broad screening studies.
The goal of this study was to investigate the expression, localization, and function of Myo1d during neurodevelopment. Here, we show that Myo1d is present in both peripheral (PNS) and central nervous systems (CNS). In the CNS, our analysis focused on the cerebellum, where Myo1d expression is limited to neurons, exhibiting a punctate distribution along axons and in cell bodies. This motor was not found in glial cells as expected based on previous studies (Cahoy et al., 2008; Nielsen et al., 2006). We also identified aspartoacylase as a putative binding partner for Myo1d in Purkinje cells. Aspartoacylase functions in fatty acid synthesis and mutations in this protein lead to leukodystrophy (Namboodiri et al., 2006). Together, these findings hold implications for understanding the contribution of Myo1d to neurodevelopment and neurological disorders such as autism or Canavan disease.
2. Results
2.1 Myo1d is present in myelinating and non-myelinating cells of the PNS
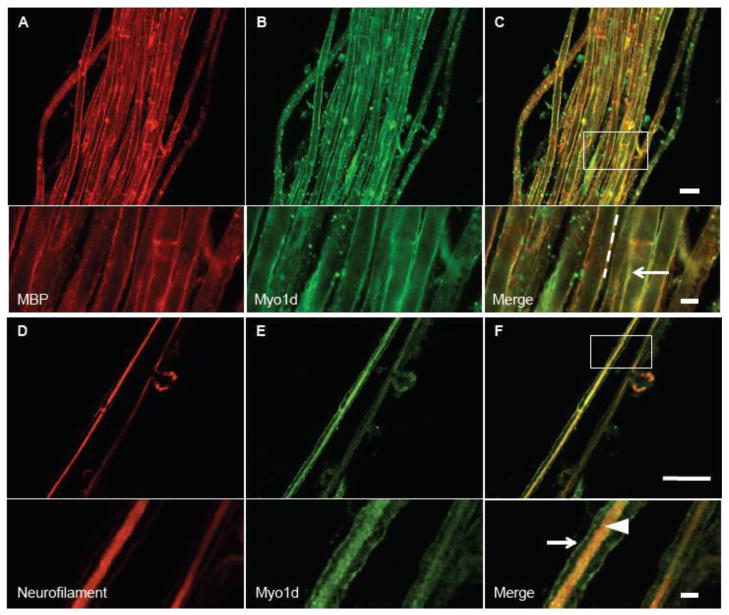
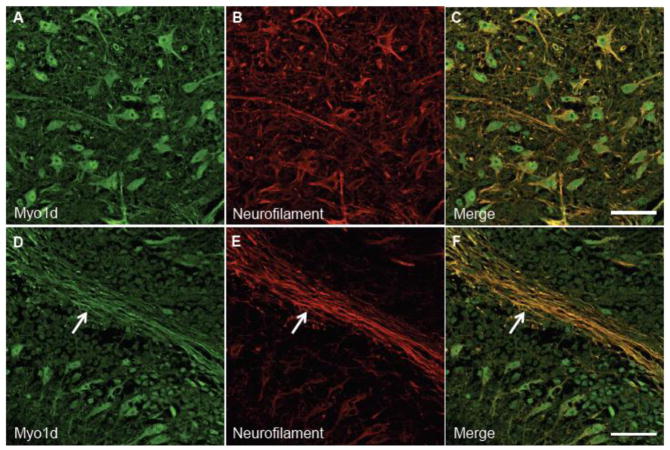
Myo1d was originally identified in the rat cerebrum, spinal cord (Bahler et al., 1994), and sciatic nerve (Lund et al., 2005). Recently, microarray studies demonstrated that Myo1d transcripts are present in oligodendrocytes (Cahoy et al., 2008), and proteomic studies suggest that this motor is also associated with myelin (Ishii et al., 2009; Jahn et al., 2009; Yamaguchi et al., 2008). To further develop our understanding Myo1d function in myelinating cells and neurons, we used high-resolution confocal imaging to characterize the distribution of this motor in the PNS and CNS. To this end, we first dissected mouse sciatic nerve bundles for immuno-fluorescence labeling and confocal imaging. To visualize the distribution of Myo1d in sciatic nerve, the nerve bundle was teased into constituent fibers (a single axon wrapped by Schwann cells), which were then stained with antibodies targeting Myo1d, myelin basic protein (MBP) to label Schwann cells (Mirsky et al., 1980), or neurofilament light chain to label axons (Fabrizi et al., 1997; Sotelo-Silveira et al., 2000). Interestingly, Myo1d exhibited robust co-localization with MBP along the myelin sheath enveloping neurons (Fig. 1A–C). In teased fibers that were exposed to higher levels of Triton X-100 (1%) to increase permeabilization, the motor maintained localization along the myelin sheath, but also co-localized with neurofilament labeling along the length of axons (Fig. 1D–F). These high-resolution confocal images indicate that Myo1d is present in both neurons and myelinating cells in the PNS. These data are also consistent with Western blots of sciatic nerve samples (data not shown), brightfield studies demonstrating Myo1d is in the sciatic nerve (McQuarrie and Lund, 2009), and proteomic studies suggesting that Myo1d is present in myelinating cells (Ishii et al., 2009; Jahn et al., 2009; Yamaguchi et al., 2008).
Figure 1. Myo1d colocalizes with MBP in Schwann cells.
Teased mouse sciatic nerve bundles were treated with 0.1% Triton X-100 and antibodies targeting MBP (red) and Myo1d (green). A) MBP localizes along the axonal ensheathment and colocalizes with Myo1d (B). C) Both proteins target to ensheathment around the axon, arrow. D-F) Teased sciatic nerve fibers were treated with 1% Triton X-100 and antibodies specific for neurofilament (red) (D), and Myo1d (E). F) Myo1d and neurofilament colocalize along the axon (arrowhead) while Myo1d maintains localization on the axon ensheathment (arrow). A–C) Bar, 20 μm; inset bar is 5 μm. D–F) 50 μm, inset bar is 5 μm.
2.2 Myo1d exhibits a developmentally regulated distribution in the cerebellum
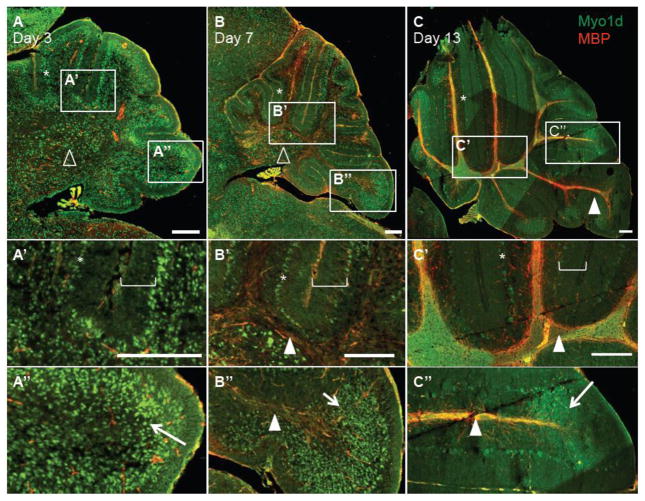
To explore the cellular distribution and sub-cellular localization of Myo1d in the CNS, we applied a similar staining strategy to frozen sections of the developing mouse brain. At postnatal day 7 (P7), Myo1d exhibits broad expression throughout the whole brain, with enrichment in select regions, including the hippocampal formation and cerebellar Purkinje cell layer (Fig. 2). We noticed prominent axonal tract staining in the cerebellum and pontine region in agreement with previous biochemical analyses (Bahler et al., 1994). We chose to focus further studies on the cerebellum as this region undergoes dramatic maturation of both neuronal and myelinating cell populations to form highly organized cell layers during early mouse postnatal development. Beginning with P3 prior to myelination (Skoff et al., 1976), Myo1d expression is present throughout the cerebellum in the Purkinje and granule cell layers, and in the region of the deep nuclei (Fig. 3A–A″). However, expression is largely absent from the molecular cell layer demonstrating that Myo1d expression is not present in all neural cell types (Fig. 3A′). At P7, after the onset of myelination (Skoff et al., 1976), Myo1d expression expands along tracts that co-label with MBP, a marker for myelin (Kornguth and Anderson, 1965)(Fig. 3B–B″). At this time point, Myo1d expression is maintained in the Purkinje cell layer, but in the granule cell layer, expression has shifted to the apex of the cerebellar lobules (Fig. 3B″). Myo1d exhibits a similar expression pattern at P13, with pronounced expression in Purkinje cells and a subset of granule cells (Fig. 3C–C″). These data indicate that Myo1d expression patterns are developmentally regulated, as suggested by previous biochemical studies (Bahler et al., 1994). In particular, Myo1d enrichment along axons increases concomitantly with maturation and expression in granule cells becomes restricted to the apex of cerebellar lobules.
Figure 2. Myo1d is expressed throughout the mouse brain.
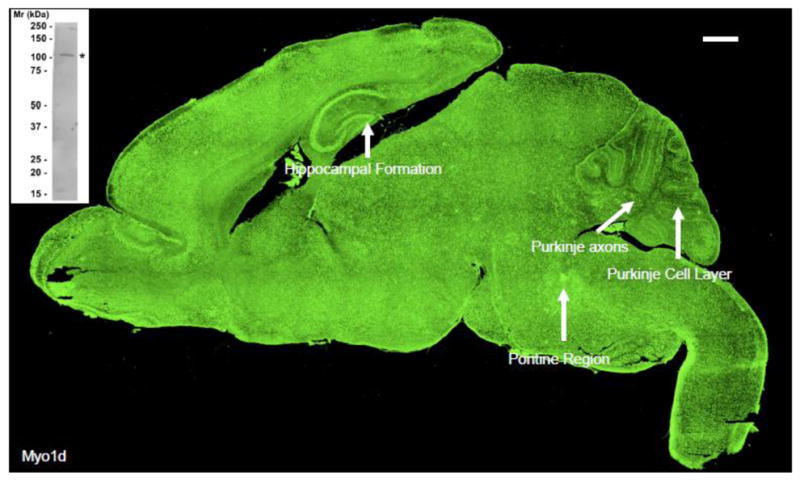
P7 mouse whole brain was stained with antibody targeting Myo1d. Myo1d expression is seen throughout the brain with enrichment in select regions including the hippocampal formation and Purkinje cell layer. Interestingly, axonal tract labeling is evident in the pontine region and cerebellum. Control experiments with secondary only or non-immune primary revealed that the intense choroid plexus staining observed here represents non-specific reactivity. Bar, 500 μm. Inset shows a Western blot of a whole adult mouse brain lysate using the same H60 antibody employed for tissue staining experiments. The H60 probe recognized a single band of the appropriate molecular weight, ~116 kDa.
Figure 3. Myo1d distribution in the cerebellum is developmentally regulated.
Mouse cerebella were stained with Myo1d (green) and MBP (red) at P3, P7, and P13. A) Myo1d is expressed throughout the cerebellum in the Purkinje (asterisk) and, region of deep nuclei (open triangle). A′) The motor is absent from the molecular cell layer (bracket). A″) Myo1d is also present throughout the granule cell layer (arrow). B–B″) Upon the onset of myelination at day 7, Myo1d expression expands along axonal tracts (arrowhead). B′–B″) Myo1d is present in the Purkinje cell layer and has a restricted expression pattern in the granule cell layer. C–C″) At day 13, Myo1d expression is maintained in the Purkinje and granule cell layers, and is distinctly present along axonal tracts. Bar, 200 μm.
2.3 Myo1d is mostly absent from oligodendrocyte precursors and oligodendrocytes
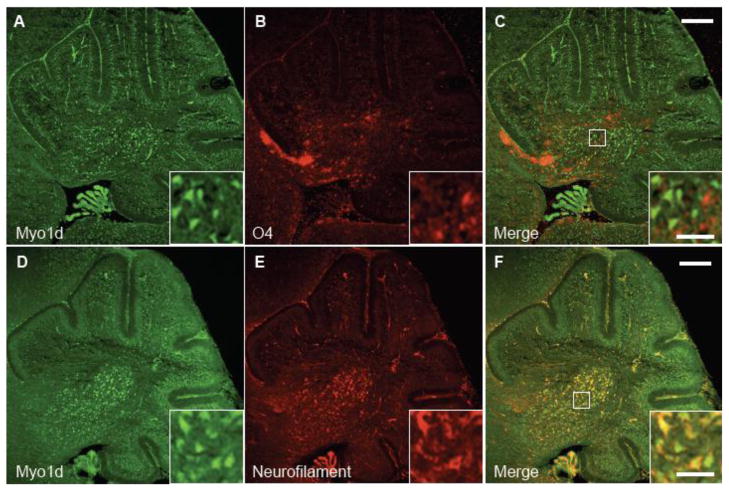
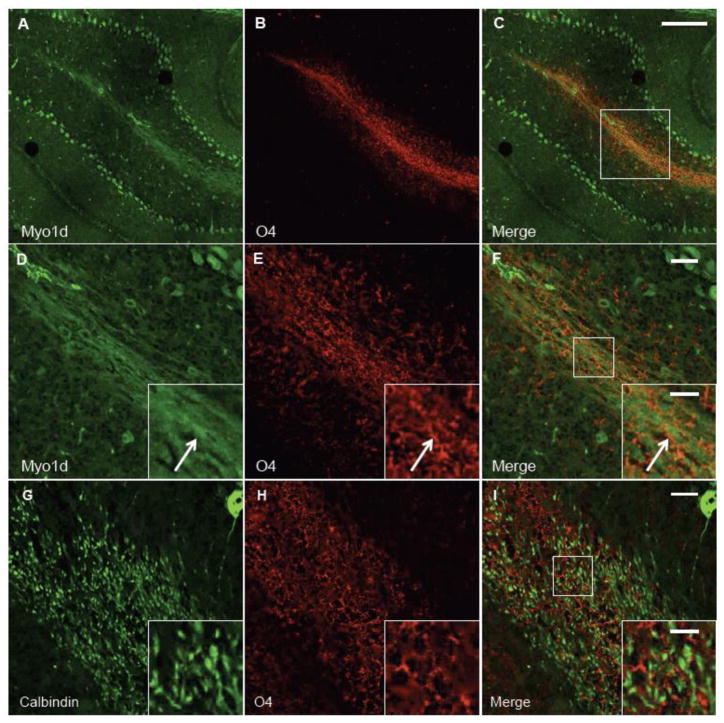
Upon the onset of myelination at P7, Myo1d expression expands along axonal tracts suggesting that this motor is found in Purkinje cell axons or in oligodendrocyte processes that myelinate these axons (or both). To distinguish among these possiblities, we performed high-resolution confocal microscopy on labeled P3 and P14 mouse brains with an antibody targeting O4, the earliest marker known for mature oligodendrocytes (Schachner et al., 1981; Sommer and Schachner, 1981). At P3, O4 appears in an arc at the base of the cerebellum in the region of the deep nuclei and there is minimal expression overlap with Myo1d (Fig. 4A–C). In fact, O4-positive and Myo1d-positive cells appear interspersed in a mutually exclusive pattern. We also analyzed O4 and Myo1d labeling in P14 mice cerebella (Fig. 5). Interestingly, Myo1d was present along axons, whereas O4 labeled cellular processes clearly enveloped axons in these sections. As a control, we stained mouse cerebella with an antibody targeting calbindin, a Purkinje specific marker (Baimbridge and Miller, 1982; Jande et al., 1981; Roth et al., 1981), and O4. This data shows O4-positive processes interwoven among Purkinje axons (Figure 5G–I), in the same manner observed in Myo1d-labeled sections. Taken together, our high-resolution imaging data indicate that at early and intermediate time points in cerebellar development, Myo1d is either not present or is expressed at very low levels in oligodendrocytes.
Figure 4. Myo1d is predominantly expressed in neurons at P3.
Mouse cerebella were labeled with antibodies specific for Myo1d (A) and oligodendrocyte progenitor marker, O4 (B). C) Myo1d expression does not overlap with O4. However, when cerebella were stained with antibodies for Myo1d (D) or neurofilament (E), there was greater overlap between both proteins (F). Bar, 200 μm; inset, 50 μm.
Figure 5. Myo1d is mostly absent from O4-labeled myelin tracts.
To determine if Myo1d is present in myelin tracts, P14 mouse cerebellum was costained with antibodies for Myo1d (A & D) (green) and O4 (B&E) (red). Confocal image magnified along a white matter tract. Myo1d appears to be specific for axonal tracts (arrow) and Purkinje cells, while O4 is in a separate cell population that wraps around the axons (F). G) Antibodies targeting Calbindin (green) clearly target Purkinje cells, dendrites and axons. H) O4 myelin positive processes form a web-like pattern. I) O4 processes wrap around Calbindin-labeled axons. A–C) Bar, 150 μm; D–I) Bar, 25 μm; inset, 10 μm.
2.4 Myo1d localizes to neuronal cell bodies, processes, and axons
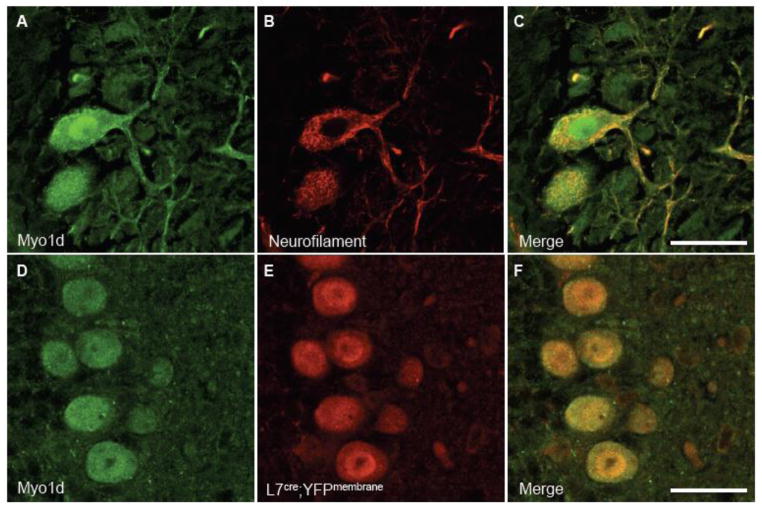
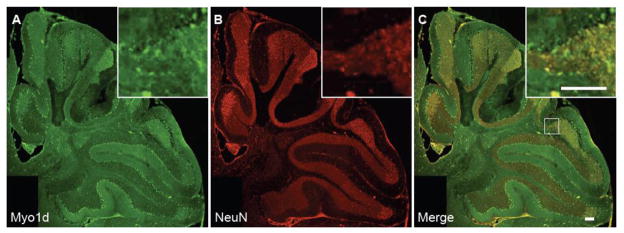
To validate that Myo1d is expressed predominantly in neurons, mouse cerebella at P3 and P7 were costained for this motor and heavy neurofilament. At P3, Myo1d exhibits clear colocalization with neurofilament in the cerebellar nuclei and granule cell layer (Fig. 4D–F). However, at P7, Myo1d and neurofilament exhibit overlapping expression in the Purkijne cell layer, Purkinje axonal tracts, and in the cerebellar nuclei (Fig. 6A–C). High magnification images in the region of the cerebellar nuclei reveal that Myo1d is cytosolic in cell bodies, localizes along processes (Fig. 6D–F), and along axon tracts (Fig. 6G–I). Staining with heavy neurofilament antibodies also revealed Myo1d localization along Purkinje dendrites (Fig. 7A–C). In Purkinje cell bodies, we observed cytosolic Myo1d with punctate labeling distributed throughout the soma as well as regions of enrichment around the cell cortex. We confirmed Myo1d expression in Purkinje cells and axons using a Purkinje specific L7cre;YFPmembrane reporter mouse (Zhang et al., 2001) (Fig. 7D–E). To validate Myo1d expression in the granule cell layer, we stained the mouse cerebella for NeuN, a neuronal specific nuclear protein that is an established marker for this cell population. While the NeuN staining pattern was evident throughout the entire granule cell layer, Myo1d expression is only evident in a subpopulation of granule cells at the apex of the cerebellar lobules (Fig. 8). Together, these results lead us to conclude that, in the context of the cerebellum, Myo1d is expressed in neurons (Bahler et al., 1994), but is unlikely to be present in myelinating cells (Ishii et al., 2009; Jahn et al., 2009; Yamaguchi et al., 2008).
Figure 6. Myo1d exhibits localization along neuronal processes and axons.
P7 cerebella were labeled with antibodies targeting Myo1d (A) (green) and neurofilament (B) (red). Myo1d and neurofilament colocalize along axonal tracts and in the region of the deep nuclei (C). Myo1d has a cytosolic subcellular distribution and is present along neuronal processes. D–F) Myo1d is also expressed along axonal tracts (arrow). A–C) Bar, 50 μm.
Figure 7. Myo1d exhibits cytosolic and dendritic subcellular localization in Purkinje cells.
We colabeled sagittal cross sections of P14 tissue with antibodies for Myo1d (A) and neurofilament (B). A) Myo1d has punctate cytosolic localization pattern and appears along dendrites. The motor appears more densely along the cell cortex. (D–F) Antibodies targeting Myo1d and GFP were applied to coronal cross sections of L7cre;YFPmembrane mouse cerebella. (D) Myo1d is present in Purkinje cells with L7, a specific Purkinje cell marker. Bar, 25 μm.
Figure 8. Myo1d is expressed in a distinct subpopulation of granule cells.
To confirm Myo1d is in the granule cell layer, we stained day 14 cerebella with Myo1d (A) and NeuN (B) specific antibodies. C) Myo1d expression in the granule cell layer is restricted to the apex of the lobules in the granule cell layer. All bars, 100 μm.
2.5 Myo1d interacts and is co-expressed with aspartoacylase, an enzyme critical for fatty acid metabolism in the central nervous system
Performing a yeast-2-hybrid screen with the Myo1d TH1 (i.e. tail) domain as bait, we identified aspartoacylase (Fig. 9), a 313 amino acid protein that catabolizes NAA1 and is expressed in kidney, small intestine, and brain (Kaul et al., 1993; Surendran et al., 2006). To confirm that neither bait nor prey caused auto-activation, we expressed the Myo1d TH1 or aspartoacylase alone in AH109 yeast. Neither construct was capable of initiating auto-activation. The interaction between these two proteins was confirmed with a biochemical pull-down approach; we over-expressed EGFP or EGFP-Myo1d in pig kidney epithelial cells and lysates were incubated with His-tagged aspartoacylase that was purified from BL21 E. coli and captured on a Ni-NTA resin. An interaction between aspartoacylase and Myo1d was detected in pull-down samples (Fig. 9D). EGFP-Myo1d did not bind Ni-NTA resin pre-treated with a native BL21 lysate (Fig. 9E), further suggesting that the interaction between aspartoacylase and Myo1d was specific.
Figure 9. Myo1d interacts with aspartoacylase.
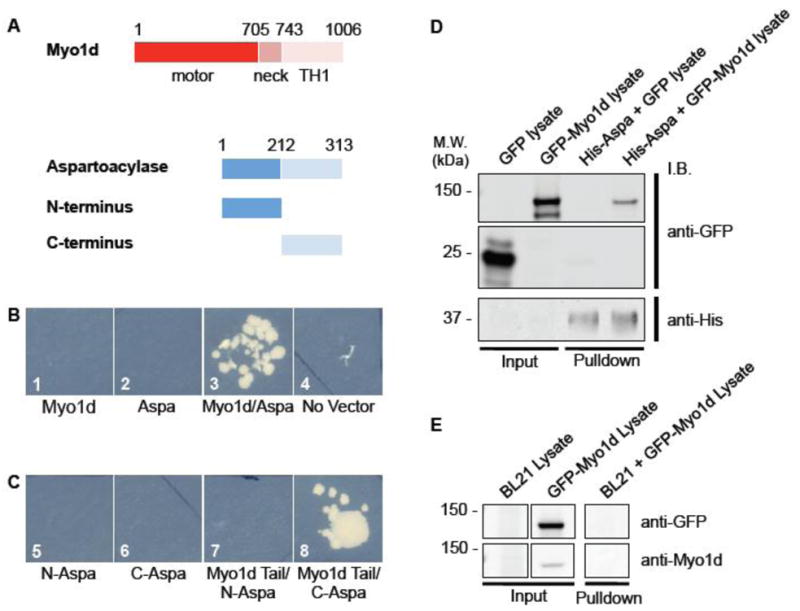
To identify potential binding partners for Myo1d, we performed a yeast 2-hybrid screen. A) The tail domain (amino acids 743–1006) was used as bait in a screen of a human cDNA kidney library. A) Aspartoacylase has two domains: the N-terminus (amino acids 1–212) and C-terminus (213–313). B) Yeast were streaked out onto triple nutrient dropout plates. 1: AH109 yeast expressing Myo1d tail alone, 2: AH109 yeast expressing aspartoacylase alone, 3: Myo1d tail interacts with aspartoacylase, 4: AH109 without any vector. C) Using the Myo1d tail (amino acids 743–1006) as bait, we determined that Myo1d interacts with the C-terminus of aspartoacylase. C) 5: AH109 yeast expressing aspartoacylase N-terminus (N-Aspa), 6: AH109 yeast expressing aspartoacylase C-terminus (C-Aspa), 7: AH109 yeast with both Myo1d tail and aspartoacylase N-terminus, 8: Myo1d and aspartoacylase C-terminus together. D) Myo1d interacts with aspartoacylase in an in vitro pull-down. His-tagged aspartoacylase was expressed in BL21 bacteria, and EGFP or EGFP-Myo1d containing lysates were collected for an in vitro pull-down experiment. Ni-NTA bound His-aspartoacylase interacts with EGFP-Myo1d, but not EGFP alone. E) EGFP-Myo1d does not bind to Ni-NTA resin pre-incubated with lysate from BL21 cells lacking His-aspartoacylase.
Based on structural studies, aspartoacylase has two domains, an N-terminal 212 a.a. domain and a C-terminal 100 a.a. domain (Bitto et al., 2007), which could potentially interact with the Myo1d TH1. Using the same yeast-2-hybrid approach, we determined that Myo1d TH1 interacts with the C-terminus of aspartoacylase (Fig. 9C). Based on the solved aspartoacylase structure and other carboxypeptidase family members the carboxyl-terminus is hypothesized to sterically block catalytic activity (Bitto et al., 2007). Given these results, one possibility is that Myo1d binding may modulate aspartoacylase activity.
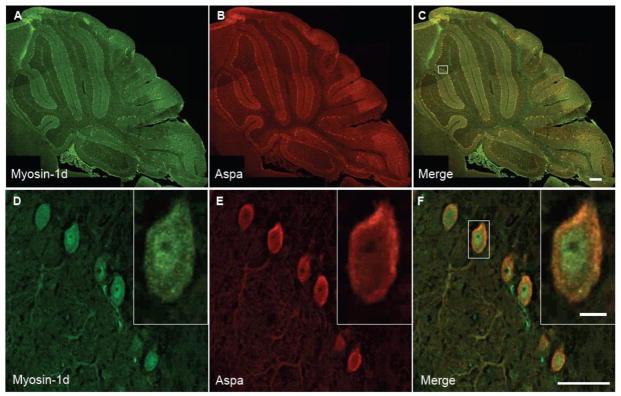
We also performed immuno-fluorescence labeling and confocal imaging of mouse cerebellum to determine if Myo1d and aspartoacylase colocalize in the CNS, which would provide evidence in support an in vivo interaction. Staining mouse frozen sections with antibodies targeting Myo1d and aspartoacylase revealed that these proteins share expression in Purkinje cells and a subpopulation of granule cells (Fig. 10A–C). Aspartoacylase staining was not observed in oligodendrocytes, but the Purkinje cell labeling described here is similar to that reported by the Human Protein Atlas (http://www.proteinatlas.org). Magnification of the Purkinje cell layer revealed that aspartoacylase staining was cytosolic similar to previous work (Madhavarao et al., 2004) (Fig. 10D–F). Moreover, Myo1d and aspartoacylase are present in cell bodies and along dendrites, which suggests a subpopulation of these two proteins are positioned to physically interact in vivo. Taken together, these in vitro and in vivo data provide evidence in support of an interaction between Myo1d and aspartoacylase in neurons of the cerebellum.
Figure 10. Myo1d and aspartoacylase are co-espressed in Purkinje cells.
P14 mouse cerebella were colabeled with antibodies for Myo1d (A & D) (green) and aspartoacylase (B & E) (red). A–C) Myo1d and aspartoayclase expression overlaps in the Purkinje and granule cell layer. D–F) Magnification of the Purkinje cell layer reveals Myo1d and aspartoacylase exhibit a diffuse cytosolic labeling in the cell body and along dendrites. A–C) Bar, 200 μm; D–E) Bar, 10 μm; inset 5μm.
3. Discussion
These studies extend prior immuno-fluorescence analyses that were limited to the sciatic nerve and cerebrum (Bahler et al., 1994; Lund et al., 2005), and more recent microarray and proteomic studies that suggest Myo1d is expressed in myelinating cells (Cahoy et al., 2008; Ishii et al., 2009; Jahn et al., 2009; Yamaguchi et al., 2008). Our high-resolution confocal data indicate that Myo1d is expressed in both myelinating cells and neurons. However, expression in myelinating cells was only detectable in the PNS. In the context of the CNS, our studies are the first to describe the developmental expression and localization of Myo1d in the mouse cerebellum. Here, Myo1d is most highly expressed in neurons, where it localizes throughout cell bodies, axons and other processes. Importantly, Myo1d expression appears as early as P3 in the internal granule layer, becoming detectable at P7 in Purkinje cell axons, and expanding along axon tracts throughout later stages of neurodevelopment. We also report Myo1d binds to aspartoacylase, a protein linked to fatty acid synthesis and Canvan disease (Moffett et al., 2007).
3.1 Myo1d is expressed in myelinating cells of the PNS, but not the cerebellum
Recent proteomic investigations identified Myo1d as a component of mouse and human myelin, suggesting that this motor is present in myelinating cells (Ishii et al., 2009; Jahn et al., 2009; Yamaguchi et al., 2008). Indeed, we observed that Schwann cells clearly express Myo1d, which co-localizes with MBP. However, our studies of the cerebellum demonstrate that Myo1d is not detectable in precursor or mature oligodendrocytes, at least at the level of resolution afforded by confocal microscopy. This difference may reflect a distinct functional requirement for the motor in Schwann cells that may not exist in oligodendrocytes. In fact, studies with myosin-2 have highlighted disparate roles for this motor in oligodendrocyte and Schwann cell myelination (Wang et al., 2008). In oligodendrocytes, decreasing myosin-2 levels facilitates myelination, whereas in Schwann cells myosin-2 deficiency leads to perturbations in cytoskeletal polarity and reduced myelinating activity (Wang et al., 2008). Myo1d may also have disparate roles in oligodendrocytes and Schwann cells, which would be supported by the data described here.
3.2 Roles for Myo1d in myelinating cells of the PNS
What is the role of Myo1d in Schwann cells? Dominant negative studies with another unconventional myosin, myosin-5a, implicate this motor in myelination possibly through a role in transporting VAMP2 along oligodendrocyte processes (Sloane and Vartanian, 2007). Because Myo1d is monomeric, the motor is unlikely to perform processive cargo transport along actin filaments. However, Myo1d does target to distinct membrane compartments (Benesh et al., 2010; Huber et al., 2000) and is able to contribute to membrane-cytoskeleton adhesion (Nambiar et al., 2009). Thus, an alternative possibility is that this motor may facilitate the deformation of membrane relative to F-actin during the extension of myelin-rich processes in Schwann cells. Actin polymerization drives the filopodial and lamellopodial extension of myelin-rich processes in search of axonal targets (Bauer et al., 2009) and Myo1d may help remodel membrane protrusions during these events.
3.3 Possible functions for Myo1d in neurons
In the context of neurons, myosin superfamily members have proposed roles ranging from organelle transport to orchestrating actin rearrangements with consequences for migration and synaptic function (Hirokawa et al., 2010). Myo1d was identified in pyramidal neurons of the cerebral cortex and thalamus (Bahler et al., 1994), and was also shown to be upregulated at lesions in sciatic nerve (Lund et al., 2005). However, the expression pattern of Myo1d in the cerebellum has not been described. Our studies demonstrate Myo1d is found in cerebellar neurons including Purkinje and granule cells. Intriguingly, Myo1d is only expressed in a subpopulation of granule cells at the apex of the cerebellar lobules, which receive inputs from the pontocerebellar fibers (Voogd and Glickstein, 1998).
Previous studies revealed that Myo1d exhibits cytosolic and punctate sub-cellular localization in cell bodies, and also localizes along axons and dendrites in the cortex (Bahler et al., 1994; Lund et al., 2005). We report similar sub-cellular localization in cerebellar Purkinje neurons. While the motor is expressed in neurons of the cortex and cerebellum, the function(s) for Myo1d in these cells remains uncharacterized. Observed Myo1d puncta might represent vesicles or other small membranous organelles, as described for other class I myosins (Bose et al., 2002). Myo1d does exhibit punctate staining in the C6 glial cell line (Bahler et al., 1994) and MDCK cells, where it may facilitate early endosomal trafficking (Huber et al., 2000). However, punctate staining has also been linked to functions other than vesicle transport in neurons. For example, myosins-1b exhibits a punctate distribution in growth cones where it may control retrograde flow (Lewis and Bridgman, 1996). It has also been proposed that Myo1d might associate with larger organelles such as the smooth endoplasmic reticulum, to enable the ‘ratcheting’ of F-actin through sciatic nerve (McQuarrie and Lund, 2009). Additional studies will be needed to fully understand the nature and functional implications of the punctate staining observed in the current work.
Our data indicate that Myo1d becomes enriched along axons at the onset of myelination, which may reflect a developmental stage-dependent function for this motor. Indeed, myelination coincides with neuronal maturation, and includes clustering of lipids and ion channels to axonal subdomains, which facilitates conductance (Barres and Raff, 1999; Simons and Trajkovic, 2006). Since class I myosins interact with specific lipid species (Hokanson et al., 2006) and retain transmembrane proteins within lipid rafts (Tyska and Mooseker, 2004), one possibility is that Myo1d may help orchestrate lipid or protein clustering along axons. Interestingly, studies in Drosophila reveal that Myo31DF (a MYO1D homolog) co-localizes with β-catenin at adherens junctions of enterocytes (Speder et al., 2006). This interaction is proposed to regulate cell-cell contacts because mutations in Myo31DF give rise to left-right asymmetry defects during development (Hozumi et al., 2006; Speder et al., 2006). Neurons also depend on the β-catenin/cadherin complex for cell adhesion and synaptic plasticity (Murase et al., 2002; Togashi et al., 2002), and therefore might rely on Myo1d in a similar manner, to facilitate these processes during neuronal maturation.
3.4 Myo1d interacts with aspartoacylase in vitro
Our data suggests that Myo1d interacts with aspartoacylase, which catabolizes N-acetyl-L-aspartate (NAA). Myo1d binds to the C-terminus of aspartoacylase (a.a. 212–313), which may sterically hinder the active site as suggested by structural studies of aspartoacylase function (Bitto et al., 2007). Our data also demonstrate that Myo1d and aspartoacylase are co-expressed in Purkinje neurons. While aspartoacylase NAA catalytic activity is found predominantly in oligodendrocytes (Baslow et al., 1999), aspartoacylase protein is also expressed in large neurons (Madhavarao et al., 2004; Moffett et al., 2011). Although published studies have not observed Purkinje cell aspartoacylase expression (Madhavarao et al., 2004), staining catalogued in the Human Protein Atlas corroborates our findings.
In addition to contributing acetic acid for fatty acid synthesis (Chakraborty et al., 2001; Madhavarao et al., 2005), NAA is also hypothesized to have roles important in maintaining viable neuronal populations. Many neuropathies such as schizophrenia, multiple sclerosis, and epilepsy are associated with altered CNS NAA levels (Moffett et al., 2007). Proposed functions for neuronal NAA include regulating osmolarity (Baslow and Yamada, 1997; Baslow, 1998), neuron-glia interactions (Baslow, 2000), and energy metabolism (Miller et al., 1996). Most notably, mutations in aspartoacylase can lead to higher NAA levels and the lethal leukodystrophy, Canavan disease (Namboodiri et al., 2006). Given the number of neuropathies associated with altered NAA levels, maintaining the proper concentration of NAA in neurons and oligodendrocytes appears to be essential for normal CNS function.
Most studies of aspartoacylase activity have focused on oligodendrocytes or myelin, and therefore a neuronal role for the protein has not been reported to date. Since several studies including our own demonstrate aspartoacylase expression in neurons (Madhavarao et al., 2004; Moffett et al., 2011), it is possible that the neuronal aspartoacylase may provide a mechanism for buffering NAA concentrations, which would contribute to replenishing acetate and aspartate levels. In addition, we propose that Myo1d modulates aspartoacylase activity, either through sequestering the protein to discrete locations or directly interfering with activity at the aspartoacylase catalytic site. Indeed, our immuno-fluorescence data demonstrates that Myo1d exhibits a punctate cytosolic distribution and localizes with aspartoacylase around Purkinje cell soma.
Recent genetic studies of autistic individuals revealed multiple SNP variants with strong association to MYO1D (Stone et al., 2007). Other work has shown that autistic individuals have decreased NAA levels throughout the brain (Friedman et al., 2003; Levitt et al., 2003). While autism etiology is unknown, the disease is considered a developmental neurological disorder influenced by multiple genetic and environmental factors (DSM-IV, 2000). Given that Myo1d exhibits expression throughout the brain (cortex, brain stem, and cerebellum), which coincides with axonal maturation, this motor is well-suited to make wide-ranging contributions to CNS development and function.
In conclusion, Myo1d is present in myelinating cells of the PNS, but not CNS suggesting distinct roles for this motor in these different tissues. However, Myo1d expression is present in both sciatic and cerebellar neurons. We also demonstrate that the cerebellar distribution of Myo1d is developmentally regulated in both Purkinje and granule cells. Specifically, during early postnatal development Myo1d expression becomes restricted to cerebellar lobule apexes. Moreover, Myo1d subcellular enrichment increases along Purkinje cell axons during early postnatal maturation. Finally, we identified aspartoacylase, an enzyme critical for maintaining proper levels of NAA in the brain, as a potential Myo1d binding partner. The data presented here provide the foundation for functional assays, which will be required for fully understanding the role of Myo1d during neurodevelopment, and its potential link to neurological disorders such as autism and Canavan disease.
4. Experimental Procedures
4.1 Sciatic nerve tissue preparation
Following a published protocol (Spiegel et al., 2007), sciatic nerve was dissected from adult mouse, and then fixed in 4% paraformaldehyde/PBS for 30 min at 4° C. The nerve was washed in 1 M sucrose/Tris buffered saline (TBS) and then placed in glycerol until the tissue was teased apart under a dissecting microscope. After individual fibers were separated, isolated material was placed on Superfrost®/Plus microscope slides (Fisher Scientific) and washed 3 times with PBS to remove residual glycerol.
4.2 Brain preparation
129 WT Mice 14 days old and younger were sacrificed according to Vanderbilt IACUC guidelines. Briefly, whole brains were removed and placed in 4% paraformaldehyde/Phosphate buffered saline (PBS; 50 mM EGTA, 137 mM NaCl, 7 mM Na2HPO4, and 3 mM NaH2PO4, pH 7.2) and allowed to rotate at 4° C for 4, 6, or 8 hours for mice that were 3, 7, and 13/14 days old, respectively. Next, tissue was cryoprotected overnight in 1 M sucrose/TBS (50 mM Tris, 150 mM NaCl) at 4° C. The following day, sucrose was washed out with OCT (Sakura Finetek) and then frozen in OCT. Samples were sectioned at 15 μm thickness with a Leica CM 1900 cryostat and applied to Superfrost®/Plus microscope slides for further analysis. For generation of the L7cre;YFPmembrane reporter mouse, Rosa-YFPmembrane mice (Jackson Laboratories,) were crossed with L7cre (Jackson Laboratories), and genotyped. Tissue was processed similarly as wildtype.
4.3 Immunofluorescence
Sciatic nerves were permeabilized with 0.1% or 1% Triton® X-100 (Sigma-Aldrich) for 30 min, and rinsed three times in PBS. Next, fibers were blocked with 5% bovine serum albumin, fraction V (BSA; Research Products International Corp.) in PBS. Antibodies targeting Myo1d (H60, polyclonal, 1:50, Santa Cruz Biotechnology, Inc.), myelin basic protein (SMI-94, monoclonal, 1:100, Covance), and light-neurofilament (DA2, monoclonal, 1:100, Cell Signaling) were applied to the samples overnight at 4° C. We have previously shown that the H60 antibody specifically recognizes Myo1d (Benesh et al., 2010), and consistently provides the best signal to noise results for immunofluoresence and Western blots. The next day, unbound primary antibodies were removed with three 5-min washes of PBS. Secondary antibody was added to each sample for 45 min (Alexa Fluor® 488 or 568 goat anti-mouse [IgG] or goat anti-rabbit, Invitrogen Molecular Probes). Samples were washed with PBS three times and adhered to glass slides with Prolong® Gold antifade (Invitrogen).
Brain slices were thawed to room temperature and a Super HT Pap Pen (Research Products International Corp.) was used to draw a hydrophobic boundary around tissue sections. Samples were washed briefly in PBS to remove residual OCT, and then permeabilized with cold methanol for 30 min at 4° C; in the case of O4 staining, samples were permeabilized with 0.1% Triton® X-100 (Sigma-Aldrich) for 30 min at RT. After three 5-min washes with PBS, samples were blocked with 5% BSA/PBS for 30 min at RT. Primary antibodies used in study: anti-Myo1d, anti-myelin basic protein, anti-neurofilament-L, anti-neurofilament-H (RMdO 20, monoclonal, 1:200, Cell Signaling), anti-calbindin (C26D12, polyclonal, 1:200, Cell Signaling), anti-O4 (MAB1326, monoclonal, 1:200, Research and Development Systems), anti-neun (MAB377, monoclonal, 1:200, Millipore), anti-aspartoacylase (sc-109208, goat, 1:50, Santa Cruz). Samples were incubated overnight at 4° C. Tissue was then washed in three 5-min washes with PBS before adding the appropriate secondary antibody species (Alexa Fluor® 488 or 568 goat anti-mouse (IgG or IgM) or goat anti-rabbit, Invitrogen Molecular Probes) for 45 min at RT in darkness. The secondary antibody was washed out in three 5-min washes before sealing a coverslip over the sample with Prolong® Gold antifade. Brain slices were imaged on a Leica TCS-SP5 confocal microscope (Leica Microsystems) with 10x and 63x objectives. Brain slices seen in Figures 2 and 3 were imaged on an Ariol® SL-50 platform (Genetix) with the assistance of Joseph Roland in the Epithelial Biology Center, Vanderbilt University Medical Center. All images were pseudo-colored, contrast enhanced, and cropped in ImageJ 1.44j (National Institutes of Health, http:/imagej.hig.gov/ij). 10x images were stitched together in Photoshop CS5 using the ‘Automerge’ feature.
4.4 Yeast 2-hybrid assay
A human kidney Matchmaker cDNA library was screened according to the manufacturer’s instructions (Clontech). Briefly, Myo1d tail was subcloned (nucleotides 2230–3021) into the pGBKT-7 vector with a forward primer containing an EcoR1 site (AGCAGAATTCAAAGCCAGGCGATTCCACGGGGTC) and a reverse primer containing a BamH1 site (ATCGGGATCCATTCCCGGGCACACTGAGGAT). Myo1d tail pGBKT-7 transformed AH109 yeast tested negative for leaky HIS3 expression and auto-transcriptional activation. The bait-containing AH109 strain was mated with the Y187 yeast pre-transformed library in liquid culture overnight. Mating mixtures were streaked onto synthetic dropout plates (without Histidine, Leucine, and Tryptophan) and incubated for a week at 30° C. Colonies were replica plated on quadruple synthetic dropout plates (without Histidine, Leucine, Tryptophan, Adenine). DNA was extracted from clones picked from quadruple synthetic dropout plates, amplified by PCR, and sequenced according to manufacturer’s instructions.
4.5 In vitro pull-down
BL21-Gold(DE3)pLysS (Stratagene, #230134) bacterial cells were transformed with pQE-32 vector (Qiagen, #32915) containing His-tagged Hs aspartoacylase cDNA, and streaked on plates to enable single colony selection. Colonies were picked and grown in 5 ml LB broth (FisherScientific, BP1426-2) overnight to test expression levels. 50 μl of a high expressing clone was used to inoculate a 50 ml starter culture that was grown overnight. As a negative control, we cultured in parallel BL21-Gold(DE3)pLysS lacking the His-tagged aspartoacylase expression vector. The following day, 25 ml of starter culture was added to 500 ml LB broth and grown until the OD600 reached 0.6. To induce over-expression, isopropyl β-D-1-thiogalactopyranoside (IPTG) (500 μm, Sigma-Aldrich, 15502) was added to cultures, which were then allowed to grow for an additional 3–4 hours. Bacteria were then pelleted in a Beckman X-15R at 5,000 × g for 20 minutes, 4° C. The pellet was snap-frozen in liquid N2 and stored at −80° C. To begin lysis, pellets were thawed and resuspended in 20 ml of Lysis buffer (100 mM KCl, 10% glycerol, 20 mM Tris-HCl, 10 mM Imidazole, fresh 10 mM β-mercaptoethanol, 0.2 ml fresh chicken lysozyme/gram of pellet, 1 mM Pefabloc, pH 8.5). Resuspended bacteria were agitated for 15 min at room temperature, and then sonicated with a Branson Sonicator 250 at 300W, 50% duty cycle five times for 10 sec. After bacterial lysis, the supernatant was clarified by centrifugation at 20,000 × g (Sorvall, SS-34 rotor). To bind the 6x-His tag aspartoacylase constructs to Ni-NTA resin (Qiagen), 2.5 ml of resin was equilibrated with Lysis buffer; the bacterial lysate was then added to resin and rotated for 1 hr at 4° C. The supernatant flow-through was removed by centrifugation (5 min at 500 × g). Resin was washed three times in Wash Buffer (300 mM KCl, 20 mM Tris-HCl, 20 mM Imidazole, pH 8.5). His-tagged aspartoacylase was then incubated with LLC-PK1-CL4 (CL4) cell lysates containing either EGFP or EGFP-Myo1d. CL4 homogenates were prepared as previously described (Tyska and Mooseker, 2004). Briefly, confluent CL4 cells in a T-75 flask were washed three times with 37° C TBS, scraped, and then pelleted (500 × g, 10 min). Next, CL4 cells were resuspended in nine pellet volumes of Homogenization Buffer (2 mM EGTA, 150 mM KCl, 5 mM MgCl2, 1 mM Dtt, 1 mM Pefabloc, 40 mM imidazole, 1 mM ATP, 1% Triton X-100, pH 7.2) and homogenized in a dounce. The homogenate was then centrifuged at 15,000 × g for 20 min at 4° C. The CL4 supernatant was incubated with the His-tagged aspartoacylase bound Ni-NTA resin overnight at 4° C. The following day, the resin was washed three times with Wash Buffer and then treated with boiling Laemmli sample buffer to prepare associated proteins for SDS-PAGE.
4.6 Immunoblotting
Protein samples were separated using SDS-PAGE. After electrophoresis, gels were transferred to a nitrocellulose membrane overnight (35V, 4° C). Membranes were rinsed three times with deionized water, blocked with nonfat dry milk in PBS, and then incubated with primary antibodies anti-GFP (1:500, Molecular Probes, A11122) or anti-His (1:1,000, Cell Signaling, 27E8) for 1 hr at RT. Blots were then washed with PBS-Tween 0.1% three times before adding secondary antibody (1:1000,IRDye® 680 goat anti-mouse, 827–11080, IRDye® 800 goat anti-rabbit, 827–08365, LI-COR) for 30 min. After washing three more times with PBS-Tween 0.1%, blots were imaged with an Odyssey LI-COR Imaging System.
Highlights.
Myo1d expression was observed in Schwann cells but not oligodendrocytes.
Myo1d distribution in cerebellar neurons is developmentally regulated.
Myo1d interacts with aspartoacylase in vitro.
Myo1d is co-expressed with aspartoacylase in Purkinje and granule cells.
Acknowledgments
The authors would like to thank all members of the Tyska laboratory for their advice and support, and the VUMC Cell Imaging Shared Resource. We also thank the Wente and Coffey laboratories for their technical advice. This work was supported by grants from the National Institutes of Health (R01-DK075555, MJT) and the American Heart Association (pre-doctoral fellowship, AEB; 09GRNT2310188, MJT).
Footnotes
Abbreviations: Myosin-1d (Myo1d), tail homology-1 (TH1), N-acetyl-l-aspartate (NAA)
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Andrew E. Benesh, Email: andrew.e.benesh@vanderbilt.edu.
Jonathan T. Fleming, Email: jonathan.t.fleming@vanderbilt.edu.
Chin Chiang, Email: chin.chiang@vanderbilt.edu.
Bruce D. Carter, Email: bruce.carter@vanderbilt.edu.
Matthew J. Tyska, Email: matthew.tyska@vanderbilt.edu.
References
- Bahler M, Kroschewski R, Stoffler HE, Behrmann T. Rat myr 4 defines a novel subclass of myosin I: identification, distribution, localization, and mapping of calmodulin-binding sites with differential calcium sensitivity. J Cell Biol. 1994;126:375–89. doi: 10.1083/jcb.126.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baimbridge KG, Miller JJ. Immunohistochemical localization of calcium-binding protein in the cerebellum, hippocampal formation and olfactory bulb of the rat. Brain Res. 1982;245:223–9. doi: 10.1016/0006-8993(82)90804-6. [DOI] [PubMed] [Google Scholar]
- Barres BA, Raff MC. Axonal control of oligodendrocyte development. The Journal of cell biology. 1999;147:1123–8. doi: 10.1083/jcb.147.6.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baslow MH, Yamada S. Identification of N-acetylaspartate in the lens of the vertebrate eye: a new model for the investigation of the function of N-acetylated amino acids in vertebrates. Exp Eye Res. 1997;64:283–6. doi: 10.1006/exer.1996.0205. [DOI] [PubMed] [Google Scholar]
- Baslow MH. Function of the N-acetyl-L-histidine system in the vertebrate eye. Evidence in support of a role as a molecular water pump. J Mol Neurosci. 1998;10:193–208. doi: 10.1007/BF02761774. [DOI] [PubMed] [Google Scholar]
- Baslow MH, Suckow RF, Sapirstein V, Hungund BL. Expression of aspartoacylase activity in cultured rat macroglial cells is limited to oligodendrocytes. J Mol Neurosci. 1999;13:47–53. doi: 10.1385/JMN:13:1-2:47. [DOI] [PubMed] [Google Scholar]
- Baslow MH. Functions of N-acetyl-L-aspartate and N-acetyl-L-aspartylglutamate in the vertebrate brain: role in glial cell-specific signaling. J Neurochem. 2000;75:453–9. doi: 10.1046/j.1471-4159.2000.0750453.x. [DOI] [PubMed] [Google Scholar]
- Bauer NG, Richter-Landsberg C, Ffrench-Constant C. Role of the oligodendroglial cytoskeleton in differentiation and myelination. Glia. 2009;57:1691–705. doi: 10.1002/glia.20885. [DOI] [PubMed] [Google Scholar]
- Benesh AE, Nambiar R, McConnell RE, Mao S, Tabb DL, Tyska MJ. Differential Localization and Dynamics of Class I Myosins in the Enterocyte Microvillus. Mol Biol Cell. 2010 doi: 10.1091/mbc.E09-07-0638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitto E, Bingman CA, Wesenberg GE, McCoy JG, Phillips GN., Jr Structure of aspartoacylase, the brain enzyme impaired in Canavan disease. Proc Natl Acad Sci U S A. 2007;104:456–61. doi: 10.1073/pnas.0607817104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bose A, Guilherme A, Robida SI, Nicoloro SM, Zhou QL, Jiang ZY, Pomerleau DP, Czech MP. Glucose transporter recycling in response to insulin is facilitated by myosin Myo1c. Nature. 2002;420:821–4. doi: 10.1038/nature01246. [DOI] [PubMed] [Google Scholar]
- Cahoy JD, Emery B, Kaushal A, Foo LC, Zamanian JL, Christopherson KS, Xing Y, Lubischer JL, Krieg PA, Krupenko SA, Thompson WJ, Barres BA. A Transcriptome Database for Astrocytes, Neurons, and Oligodendrocytes: A New Resource for Understanding Brain Development and Function. J Neurosci. 2008;28:264–278. doi: 10.1523/JNEUROSCI.4178-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty G, Mekala P, Yahya D, Wu G, Ledeen RW. Intraneuronal N-acetylaspartate supplies acetyl groups for myelin lipid synthesis: evidence for myelin-associated aspartoacylase. J Neurochem. 2001;78:736–45. doi: 10.1046/j.1471-4159.2001.00456.x. [DOI] [PubMed] [Google Scholar]
- Collins JH, Borysenko CW. The 110,000-dalton actin- and calmodulin-binding protein from intestinal brush border is a myosin-like ATPase. J Biol Chem. 1984;259:14128–35. [PubMed] [Google Scholar]
- DSM-IV APATFo; DSM-IV-TR. Diagnostic and statistical manual of mental disorders. American Psychiatric Association; Washington, DC: 2000. [Google Scholar]
- Fabrizi C, Kelly BM, Gillespie CS, Schlaepfer WW, Scherer SS, Brophy PJ. Transient expression of the neurofilament proteins NF-L and NF-M by Schwann cells is regulated by axonal contact. J Neurosci Res. 1997;50:291–9. doi: 10.1002/(SICI)1097-4547(19971015)50:2<291::AID-JNR17>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- Friedman SD, Shaw DW, Artru AA, Richards TL, Gardner J, Dawson G, Posse S, Dager SR. Regional brain chemical alterations in young children with autism spectrum disorder. Neurology. 2003;60:100–7. doi: 10.1212/wnl.60.1.100. [DOI] [PubMed] [Google Scholar]
- Gillespie PG, Wagner MC, Hudspeth AJ. Identification of a 120 kd hair-bundle myosin located near stereociliary tips. Neuron. 1993;11:581–94. doi: 10.1016/0896-6273(93)90071-x. [DOI] [PubMed] [Google Scholar]
- Hirokawa N, Niwa S, Tanaka Y. Molecular motors in neurons: transport mechanisms and roles in brain function, development, and disease. Neuron. 2010;68:610–38. doi: 10.1016/j.neuron.2010.09.039. [DOI] [PubMed] [Google Scholar]
- Hokanson DE, Laakso JM, Lin T, Sept D, Ostap EM. Myo1c binds phosphoinositides through a putative pleckstrin homology domain. Molecular biology of the cell. 2006;17:4856–65. doi: 10.1091/mbc.E06-05-0449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hozumi S, Maeda R, Taniguchi K, Kanai M, Shirakabe S, Sasamura T, Speder P, Noselli S, Aigaki T, Murakami R, Matsuno K. An unconventional myosin in Drosophila reverses the default handedness in visceral organs. Nature. 2006;440:798–802. doi: 10.1038/nature04625. [DOI] [PubMed] [Google Scholar]
- Huber LA, Fialka I, Paiha K, Hunziker W, Sacks DB, Bahler M, Way M, Gagescu R, Gruenberg J. Both calmodulin and the unconventional myosin Myr4 regulate membrane trafficking along the recycling pathway of MDCK cells. Traffic. 2000;1:494–503. doi: 10.1034/j.1600-0854.2000.010607.x. [DOI] [PubMed] [Google Scholar]
- Ishii A, Dutta R, Wark GM, Hwang SI, Han DK, Trapp BD, Pfeiffer SE, Bansal R. Human myelin proteome and comparative analysis with mouse myelin. Proc Natl Acad Sci U S A. 2009;106:14605–10. doi: 10.1073/pnas.0905936106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahn O, Tenzer S, Werner HB. Myelin proteomics: molecular anatomy of an insulating sheath. Mol Neurobiol. 2009;40:55–72. doi: 10.1007/s12035-009-8071-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jande SS, Tolnai S, Lawson DE. Immunohistochemical localization of vitamin D-dependent calcium-binding protein in duodenum, kidney, uterus and cerebellum of chickens. Histochemistry. 1981;71:99–116. doi: 10.1007/BF00592574. [DOI] [PubMed] [Google Scholar]
- Kaul R, Gao GP, Balamurugan K, Matalon R. Cloning of the human aspartoacylase cDNA and a common missense mutation in Canavan disease. Nat Genet. 1993;5:118–23. doi: 10.1038/ng1093-118. [DOI] [PubMed] [Google Scholar]
- Kim SV, Mehal WZ, Dong X, Heinrich V, Pypaert M, Mellman I, Dembo M, Mooseker MS, Wu D, Flavell RA. Modulation of cell adhesion and motility in the immune system by Myo1f. Science. 2006;314:136–9. doi: 10.1126/science.1131920. [DOI] [PubMed] [Google Scholar]
- Kornguth SE, Anderson JW. Localization of a basic protein in the myelin of various species with the aid of fluorescence and electron microscopy. J Cell Biol. 1965;26:157–66. doi: 10.1083/jcb.26.1.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krendel M, Kim SV, Willinger T, Wang T, Kashgarian M, Flavell RA, Mooseker MS. Disruption of Myosin 1e promotes podocyte injury. J Am Soc Nephrol. 2009;20:86–94. doi: 10.1681/ASN.2007111172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitt JG, O’Neill J, Blanton RE, Smalley S, Fadale D, McCracken JT, Guthrie D, Toga AW, Alger JR. Proton magnetic resonance spectroscopic imaging of the brain in childhood autism. Biological psychiatry. 2003;54:1355–66. doi: 10.1016/s0006-3223(03)00688-7. [DOI] [PubMed] [Google Scholar]
- Lewis AK, Bridgman PC. Mammalian myosin I alpha is concentrated near the plasma membrane in nerve growth cones. Cell Motil Cytoskeleton. 1996;33:130–50. doi: 10.1002/(SICI)1097-0169(1996)33:2<130::AID-CM5>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- Lund LM, Machado VM, McQuarrie IG. Axonal isoforms of myosin-I. Biochem Biophys Res Commun. 2005;330:857–64. doi: 10.1016/j.bbrc.2005.02.187. [DOI] [PubMed] [Google Scholar]
- Madhavarao CN, Moffett JR, Moore RA, Viola RE, Namboodiri MA, Jacobowitz DM. Immunohistochemical localization of aspartoacylase in the rat central nervous system. J Comp Neurol. 2004;472:318–29. doi: 10.1002/cne.20080. [DOI] [PubMed] [Google Scholar]
- Madhavarao CN, Arun P, Moffett JR, Szucs S, Surendran S, Matalon R, Garbern J, Hristova D, Johnson A, Jiang W, Namboodiri MA. Defective N-acetylaspartate catabolism reduces brain acetate levels and myelin lipid synthesis in Canavan’s disease. Proc Natl Acad Sci U S A. 2005;102:5221–6. doi: 10.1073/pnas.0409184102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell RE, Tyska MJ. Leveraging the membrane - cytoskeleton interface with myosin-1. Trends in cell biology. 2010;20:418–26. doi: 10.1016/j.tcb.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQuarrie IG, Lund LM. Intra-axonal myosin and actin in nerve regeneration. Neurosurgery. 2009;65:A93–6. doi: 10.1227/01.NEU.0000338593.76635.32. [DOI] [PubMed] [Google Scholar]
- Miller SL, Daikhin Y, Yudkoff M. Metabolism of N-acetyl-L-aspartate in rat brain. Neurochem Res. 1996;21:615–8. doi: 10.1007/BF02527761. [DOI] [PubMed] [Google Scholar]
- Mirsky R, Winter J, Abney ER, Pruss RM, Gavrilovic J, Raff MC. Myelin-specific proteins and glycolipids in rat Schwann cells and oligodendrocytes in culture. J Cell Biol. 1980;84:483–94. doi: 10.1083/jcb.84.3.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffett JR, Ross B, Arun P, Madhavarao CN, Namboodiri AM. N-Acetylaspartate in the CNS: from neurodiagnostics to neurobiology. Prog Neurobiol. 2007;81:89–131. doi: 10.1016/j.pneurobio.2006.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffett JR, Arun P, Ariyannur PS, Garbern JY, Jacobowitz DM, Namboodiri AM. Extensive aspartoacylase expression in the rat central nervous system. Glia. 2011 doi: 10.1002/glia.21186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murase S, Mosser E, Schuman EM. Depolarization drives beta-Catenin into neuronal spines promoting changes in synaptic structure and function. Neuron. 2002;35:91–105. doi: 10.1016/s0896-6273(02)00764-x. [DOI] [PubMed] [Google Scholar]
- Nambiar R, McConnell RE, Tyska MJ. Control of cell membrane tension by myosin-I. Proc Natl Acad Sci U S A. 2009;106:11972–7. doi: 10.1073/pnas.0901641106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namboodiri AM, Peethambaran A, Mathew R, Sambhu PA, Hershfield J, Moffett JR, Madhavarao CN. Canavan disease and the role of N-acetylaspartate in myelin synthesis. Mol Cell Endocrinol. 2006;252:216–23. doi: 10.1016/j.mce.2006.03.016. [DOI] [PubMed] [Google Scholar]
- Nielsen JA, Maric D, Lau P, Barker JL, Hudson LD. Identification of a novel oligodendrocyte cell adhesion protein using gene expression profiling. J Neurosci. 2006;26:9881–91. doi: 10.1523/JNEUROSCI.2246-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odronitz F, Kollmar M. Drawing the tree of eukaryotic life based on the analysis of 2,269 manually annotated myosins from 328 species. Genome biology. 2007;8:R196. doi: 10.1186/gb-2007-8-9-r196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olety B, Walte M, Honnert U, Schillers H, Bahler M. Myosin 1G (Myo1G) is a haematopoietic specific myosin that localises to the plasma membrane and regulates cell elasticity. FEBS Lett. 2010;584:493–9. doi: 10.1016/j.febslet.2009.11.096. [DOI] [PubMed] [Google Scholar]
- Patino-Lopez G, Aravind L, Dong X, Kruhlak MJ, Ostap EM, Shaw S. Myosin 1G is an abundant class I myosin in lymphocytes whose localization at the plasma membrane depends on its ancient divergent pleckstrin homology (PH) domain (Myo1PH) J Biol Chem. 2010;285:8675–86. doi: 10.1074/jbc.M109.086959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth J, Baetens D, Norman AW, Garcia-Segura LM. Specific neurons in chick central nervous system stain with an antibody against chick intestinal vitamin D-dependent calcium-binding protein. Brain Res. 1981;222:452–7. doi: 10.1016/0006-8993(81)91054-4. [DOI] [PubMed] [Google Scholar]
- Schachner M, Kim SK, Zehnle R. Developmental expression in central and peripheral nervous system of oligodendrocyte cell surface antigens (O antigens) recognized by monoclonal antibodies. Dev Biol. 1981;83:328–38. doi: 10.1016/0012-1606(81)90478-4. [DOI] [PubMed] [Google Scholar]
- Simons M, Trajkovic K. Neuron-glia communication in the control of oligodendrocyte function and myelin biogenesis. Journal of cell science. 2006;119:4381–9. doi: 10.1242/jcs.03242. [DOI] [PubMed] [Google Scholar]
- Skoff RP, Price DL, Stocks A. Electron microscopic autoradiographic studies of gliogenesis in rat optic nerve. II. Time of origin. J Comp Neurol. 1976;169:313–34. doi: 10.1002/cne.901690304. [DOI] [PubMed] [Google Scholar]
- Sloane JA, Vartanian TK. Myosin Va controls oligodendrocyte morphogenesis and myelination. J Neurosci. 2007;27:11366–75. doi: 10.1523/JNEUROSCI.2326-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer I, Schachner M. Monoclonal antibodies (O1 to O4) to oligodendrocyte cell surfaces: an immunocytological study in the central nervous system. Dev Biol. 1981;83:311–27. doi: 10.1016/0012-1606(81)90477-2. [DOI] [PubMed] [Google Scholar]
- Sotelo-Silveira JR, Calliari A, Kun A, Benech JC, Sanguinetti C, Chalar C, Sotelo JR. Neurofilament mRNAs are present and translated in the normal and severed sciatic nerve. J Neurosci Res. 2000;62:65–74. doi: 10.1002/1097-4547(20001001)62:1<65::AID-JNR7>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- Speder P, Adam G, Noselli S. Type ID unconventional myosin controls left-right asymmetry in Drosophila. Nature. 2006;440:803–7. doi: 10.1038/nature04623. [DOI] [PubMed] [Google Scholar]
- Spiegel I, Adamsky K, Eshed Y, Milo R, Sabanay H, Sarig-Nadir O, Horresh I, Scherer SS, Rasband MN, Peles E. A central role for Necl4 (SynCAM4) in Schwann cell-axon interaction and myelination. Nature neuroscience. 2007;10:861–9. doi: 10.1038/nn1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone JL, Merriman B, Cantor RM, Geschwind DH, Nelson SF. High density SNP association study of a major autism linkage region on chromosome 17. Hum Mol Genet. 2007;16:704–715. doi: 10.1093/hmg/ddm015. [DOI] [PubMed] [Google Scholar]
- Surendran S, Matalon R, Tyring SK. Upregulation of aspartoacylase activity in the duodenum of obesity induced diabetes mouse: implications on diabetic neuropathy. Biochemical and biophysical research communications. 2006;345:973–5. doi: 10.1016/j.bbrc.2006.04.179. [DOI] [PubMed] [Google Scholar]
- Togashi H, Abe K, Mizoguchi A, Takaoka K, Chisaka O, Takeichi M. Cadherin regulates dendritic spine morphogenesis. Neuron. 2002;35:77–89. doi: 10.1016/s0896-6273(02)00748-1. [DOI] [PubMed] [Google Scholar]
- Tyska MJ, Mooseker MS. A role for myosin-1A in the localization of a brush border disaccharidase. J Cell Biol. 2004;165:395–405. doi: 10.1083/jcb.200310031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voogd J, Glickstein M. The anatomy of the cerebellum. Trends Neurosci. 1998;21:370–5. doi: 10.1016/s0166-2236(98)01318-6. [DOI] [PubMed] [Google Scholar]
- Wang H, Tewari A, Einheber S, Salzer JL, Melendez-Vasquez CV. Myosin II has distinct functions in PNS and CNS myelin sheath formation. J Cell Biol. 2008;182:1171–84. doi: 10.1083/jcb.200802091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y, Miyagi Y, Baba H. Two-dimensional electrophoresis with cationic detergents: a powerful tool for the proteomic analysis of myelin proteins. Part 2: analytical aspects. J Neurosci Res. 2008;86:766–75. doi: 10.1002/jnr.21549. [DOI] [PubMed] [Google Scholar]
- Zhang X, Baader SL, Bian F, Muller W, Oberdick J. High level Purkinje cell specific expression of green fluorescent protein in transgenic mice. Histochemistry and cell biology. 2001;115:455–64. doi: 10.1007/s004180100283. [DOI] [PubMed] [Google Scholar]