Abstract
Wnt signaling is critical for skeletal development and homeostasis. Sclerostin (Sost) has emerged as a potent inhibitor of Wnt signaling and, thereby, bone formation. Thus, strategies to reduce Sclerostin expression may be used to treat osteoporosis or non-union fractures. Transforming growth factor-beta (TGF-β) elicits various effects upon the skeleton both in vitro and in vivo depending on the duration and timing of administration. In vitro and in vivo studies demonstrate that TGF-β increases osteoprogenitor differentiation but decreases matrix mineralization of committed osteoblasts. Because Sclerostin decreases matrix mineralization, this study aimed to examine whether TGF-β achieves such inhibitory effects via transcriptional modulation of Sost. Using the UMR106.01 mature osteoblast cell line, we demonstrated that TGF-β TGF-β1, -β2, -β3 and Activin A increase Sost transcript expression. Pharmacologic inhibition of Alk4/5/7 in vitro and in vivo decreased endogenous Sost expression, and siRNA against Alk4 and Alk5 demonstrated their requirement for endogenous Sost expression. TGF-β1 targeted the Sost bone enhancer ECR5 and did not affect the transcriptional activity of the endogenous Sost promoter. These results indicate that TGF-β1 controls Sost transcription in mature osteoblasts, suggesting that Sclerostin may mediate the inhibitory effect of TGF-β upon osteoblast differentiation.
Keywords: Transforming growth factor-beta, Sost, ECR5, Osteoblast, Bone, Wnt
Introduction
Since the discovery in the past decade that the Wnt glycoprotein co-receptor Lrp5 regulates bone mass [1, 2], tremendous efforts have attempted to elucidate the mechanisms involved. Wnts are ligands for Lrp4, 5, and 6, and a subset of Wnts increase the osteogenic commitment of bone marrow stem cells, enhance matrix formation, and decrease apoptosis of osteoblasts and osteocytes [3, 4]. Regulation of Wnt signaling occurs via secreted decoy receptors (secreted frizzled-related protein [sFRP]) or antagonists (Sclerostin [SOST], Dickkopf [DKK]) that bind to Lrp4 - 6 to prevent Wnt-Lrp interactions, and subsequent signal transduction [5, 6]. As activating mutations in LRP4-6 promote high bone mass (HBM) phenotypes [1, 2, 7, 8], complementary phenotypes emerge from deletion of Lrp4/5/6 antagonists [9–15]: deletion of sFRPs increases trabecular bone [15] and bone mineral density [14], and deletion of DKK isoforms or SOST increases markers of bone formation and bone mass [9, 10].
The influence of SOST on skeletal formation and function is phenotypically observed via loss of Sclerostin protein, which is achieved by two distinct genetic mechanisms. One set of mutations occur within the SOST transcript and comprise either nonsense mutations in exon 2 or aberrant splice sites resulting in null alleles [10]. These mutations cause sclerosteosis in humans (MIM 269500), which is characterized by generalized cortical hyperostosis accompanied by occasional syndactyly of the digits [10]. A highly similar bone mineral density phenotype is observed in van Buchem disease patients (MIM 239100) who also have severe skeletal hyperplasia, but carry no mutations in the SOST gene. Instead, van Buchem results from the deletion of a 52kb non-coding region (also refered to as van Buchem deletion region) that is 35kb downstream of SOST [11]; this van Buchem deletion region functions in cis to enhance SOST transcription in bone. We have previously demonstrated that an evolutionarily-conserved region present within the van Buchem deletion region, termed ECR5, is sufficient to drive reporter assays in bone cells, in vitro and in vivo [13], and confers responsiveness to parathyroid hormone (PTH) [16].
The TGF-β superfamily is composed of more than 40 structurally and functionally related cytokines that regulate a variety of biological processes including morphogenesis, proliferation, stem cell differentiation, apoptosis, and epithelial-to-mesenchymal transition [17]. The superfamily clusters into the subfamilies TGF-β, bone morphogenetic proteins (BMPs), growth and differentiation factors (GDFs), activins and inhibins, and Mullerian inhibiting factor (MIF) [18]. The TGF-β subfamily contains three distinct proteins—TGF-β1, -β2, and -β3— which exert pleiotropic effects upon cells responsible for maintaining or altering skeletal architecture. Indeed, the TGF-β subfamily demonstrates chemotactic effects on osteoprogenitors during endochondral condensation [19], promotes proliferation and differentiation of early osteoprogenitors, yet it also decreases matrix formation in fully-differentiated osteoblasts (reviewed in Janssens et al. [20] and Bonewald et al. [21], amongst others). TGF-β1–3 can interact with osteotropic factors like PTH [22] or prostaglandin E2 [23] to enhance bone formation. Conversely, factors like BMPs [24–26], PTH [16, 27, 28], and prostaglandin E2 [29] regulate Wnt signaling via manipulation of Wnt or Lrp5/6 antagonist expression. BMP signaling through BMPR1A increases Sost expression and decreases Wnt signaling [30], but the influence of other TGF-β superfamily members on Sclerostin expression has not yet been explored. Provided the evidence for a biphasic influence of TGF-β upon osteogenic differentiation, we hypothesized that TGF-β increases Sost transcription in mature osteoblasts, and sought the intracellular mechanisms involved.
Materials and methods
Cell culture
UMR106.01 cells were kindly provided by Dr. Alexander Robling (Indiana University School of Medicine). Cells were cultured in MEM with Earle’s Salts (Invitrogen), which was supplemented with 10% fetal bovine serum (FBS; Invitrogen) and 1% penicillin-streptomycin (Invitrogen). Cells were maintained in a standard humidified incubator at 37°C/95% air / 5% CO2, and were routinely sub-cultured with 0.05% trypsin when 75–90% confluent. Unless otherwise indicated, cells were seeded for experiments at 5k/cm2, and growth factor supplements were added two days later.
Reagents
TGF-β1, TGF-β2, TGF- β3, and Activin A (R&D Systems) and human parathyroid hormone(1–34) (hPTH(1–34); Bachem) were dissolved in 0.1% BSA in PBS and stored at −20°C. Cycloheximide was purchased from Sigma; SIS3 and SB431542 were from Calbiochem. Purified RNA from adult murine calvariae or femora (Zyagen), used to examine Alk4/5/7 expression were purchased from Zyagen.
Quantitative PCR (qPCR)
At indicated time points, cell culture samples were washed with PBS, collected in RLT buffer with 2-mercaptoethanol (Qiagen), from which RNA was purified using RNeasy Kit (Qiagen). RNA purity was tested by measuring the absorbance at 260 and 280nm. One microgram of RNA was reverse-transcribed with QuantiTect Reverse Transcription kit (Qiagen), which includes genomic DNA elimination. qPCR was performed using primers listed in Table 1 and either QuantiFast Probe PCR Kit (Qiagen) or QuantiFast SYBR Green PCR Kit (Qiagen). Cycling conditions were 95°C for 3 minutes (5 minutes for SYBR), followed by 40 cycles of 95°C for 3 seconds (10 seconds for SYBR) and 30 seconds at 60°C. qPCR results were calculated relative to internal control (Rpl32 or Tbp; 2−ΔCt) with the exception of Figure 3A and 3B, results were further normalized to control, time-matched conditions (2−ΔΔCt) [31].
TABLE 1.
qPCR primers
| Target | Source | Assay no. | Species | Chemistry |
|---|---|---|---|---|
| Sost | Applied Biosystems | Rn0057791_m1 | Rat | TaqMan |
| Rpl32 | Applied Biosystems | Rn00820748_g1 | Rat | TaqMan |
| Alk4 (Acvr1b) | Qiagen | Mm_Acvr1b_1_SG | Mouse | SYBR Green |
| Alk5 (Tgfbr1) | Qiagen | Mm_Tgfbr1_1_SG | Mouse | SYBR Green |
| Alk7 (Acvr1c) | Qiagen | Mm_Acvr1c_2_SG | Mouse | SYBR Green |
| Tbp | Qiagen | Mm_Tbp_1_SG | Mouse | SYBR Green |
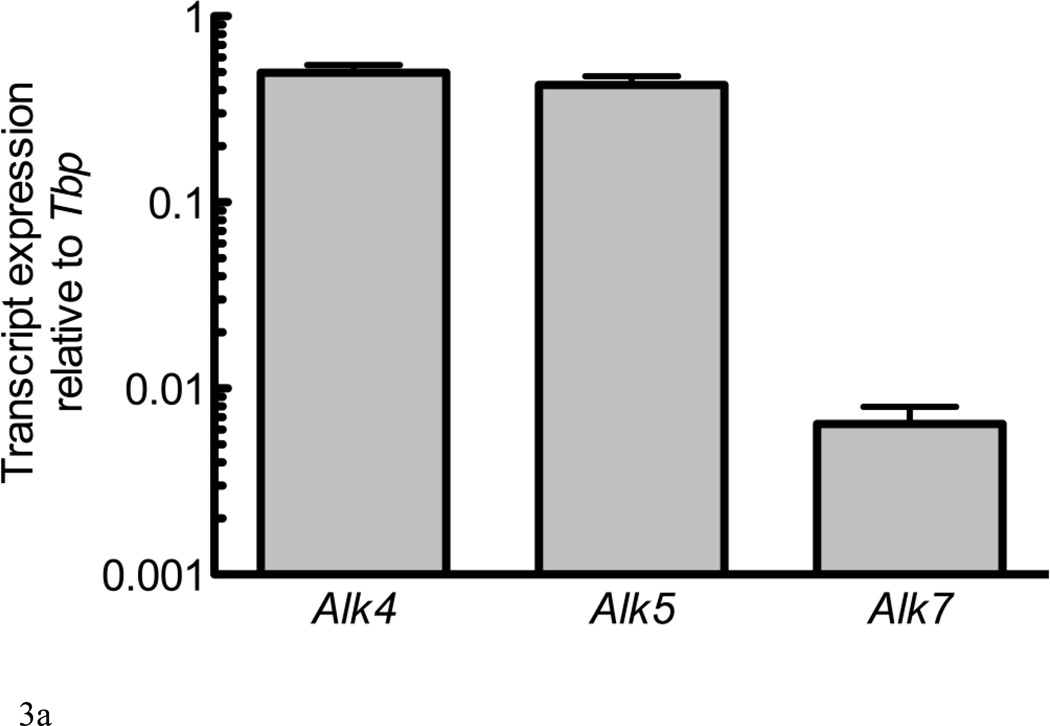
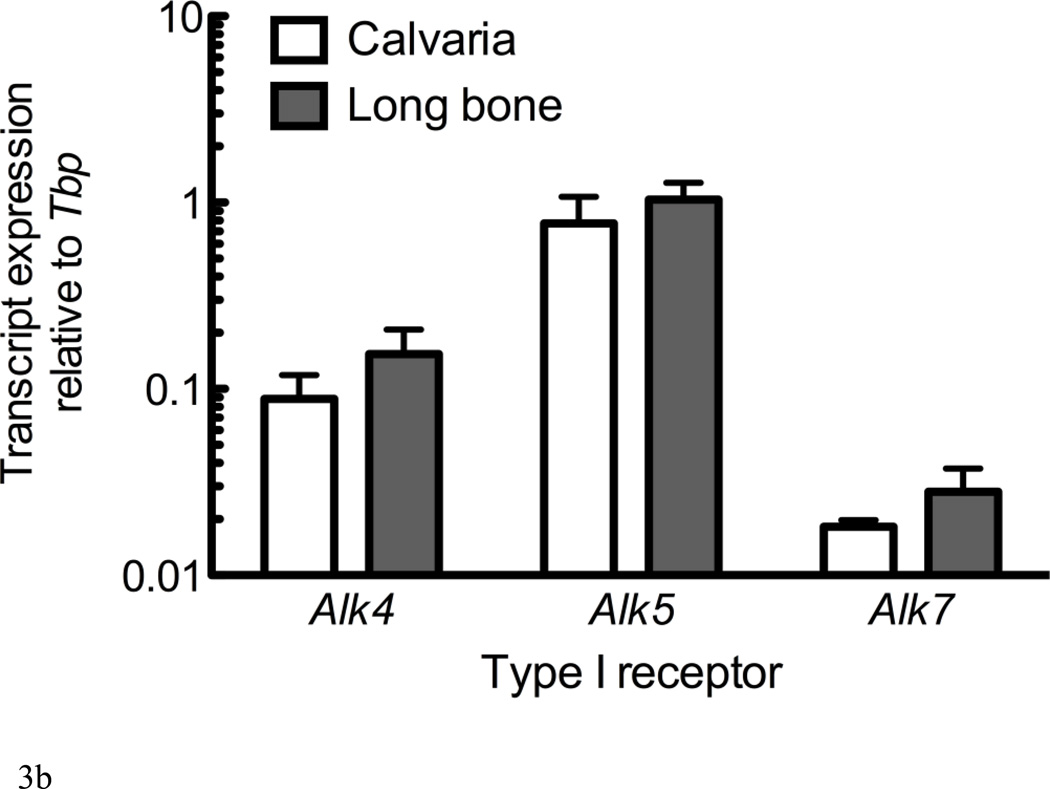
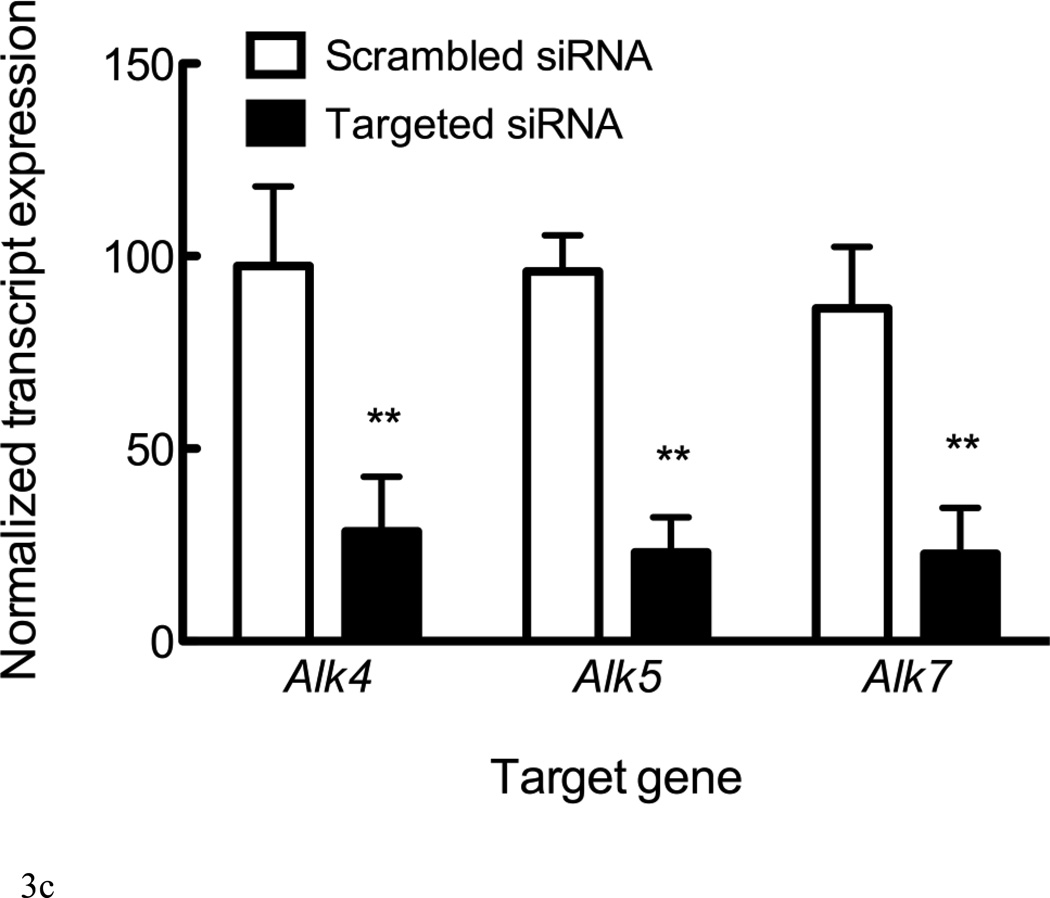
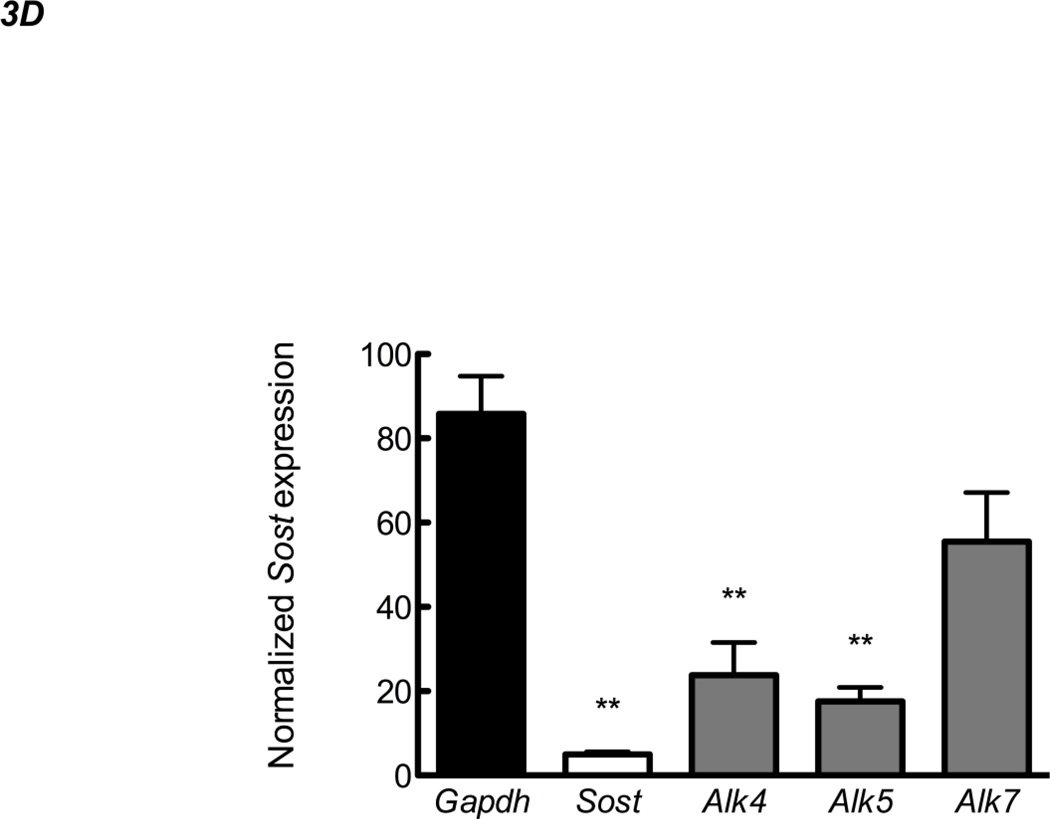
Figure 3. Alk4 and Alk5 regulate Sost expression.
RNA from UMR106.01 cells (A) and (B) murine calvaria and long bone was analyzed by qPCR for expression of TGF-β receptors Alk4, Alk5, and Alk7 relative to Tbp. (C) UMR106.01 cells were transfected with custom siRNA against Alk4, Alk5, or Alk7, or scrambled control. 24 hours later, RNA was isolated for qPCR analysis of Alk4, Alk5, or Alk7 relative to Gapdh. (D) RNA from (C) was analyzed for Sost transcript by qPCR relative to Gapdh. * indicates p < 0.05 compared to Vehicle, ** indicates p < 0.01 compared to Vehicle, *** indicates p < 0.001 compared to Vehicle.
siRNA transfection
siRNA (Qiagen) were designed against murine Sost, Gapdh, Alk4, Alk5, and Alk7; specificity was confirmed with BLAST. Cells were seeded at a density of 6,000 cells per well in 48-well plates. One hour later, 10nM siRNA and Fugene 6 (Roche) were diluted into Opti-MEM (Invitrogen) and were gently added to the culture plate. Samples were collected 48 hours later and were processed for qPCR analysis of target knock-down.
TGF-βrI kinase inhibitor treatment in vivo
Eight-week old male C57BL/6 mice were treated for 24 hours with vehicle (HBSS), or SD-208 (60 mg/kg twice daily, one injection every 12 hours; Tocris Biosciences) by intraperitoneal delivery (IP). No adverse effects of SD-208 on mouse health were detected during the study. At 24 hours after the first injection animals were euthanized humanely. Femoral shafts were scraped of soft tissue and skeletal muscle, flushed with ice-cold HBSS with a 25 gauge needle to remove the bone marrow before placing into RNAlater (Qiagen) and stored at 4°C. For RNA isolation, samples were removed from RNAlater and homogenized in 1 mL Qiazol and purified using Rneasy mini-kit using manufacture’s guidelines (Qiagen). Approvals for work conducted on the mice used in this study were granted by Lawrence Livermore National Laboratory Institutional Animal Care and Use Committee, under application no. 168. Animals were treated humanely; housing and experimental procedures followed the guidelines outlined in the National Institute of Health 'Principles of Laboratory Care'.
Reporter gene assays
pGL3-based reporter plasmids (Promega) containing ECR5 upstream of the human SOST or the SV40 promoter were previously described [13, 16]. A putative SMAD site was predicted within a multiple sequence alignment of human and mouse ECR5 sequences using power weight matrices available from TRANSFAC and utilized by MultiTF (http://multitf.dcode.org/). ECR5 was PCR cloned into the EcoR1 site of pGL3-promoter vector, as well as in a pGL3 vector where the SV40 promoter has been replaced by a 2kb fragment of the human sclerostin promoter. Subsequently the SMAD or MEF2 site was deleted using site-directed mutagenesis according to the manufacturer's instructions (Quickchange Site-directed Mutagenesis kit; Stratagene). UMR106.01 cells were seeded at 20k/well into 48-well plates. On the following day, media was removed, replaced with Opti-MEM, and transfected with Fugene 6, the reporter of interest (250ng/well), and pRL-TK (50ng/well; Promega) as a transfectant control. 24 hour later, media was removed and replaced with TGF-β1. Samples were analyzed 24 hours later using Dual-Luciferase Reporter System (Promega) and TD-20/20 luminometer (Turner Systems).
Statistical analysis
Each experiment was performed a minimum of 3 times, each time in duplicate or triplicate. Unless otherwise noted, data are presented as mean ± standard error of the mean. Statistical significance was assessed by two-tailed Student’s t test or ANOVA for non-repeated measurements followed by a Dunnet post-hoc analysis compared to control (vehicle). p < 0.05 was considered statistically significant.
Results
TGF-β superfamily members modulate Sost expression in mature osteoblastic cells
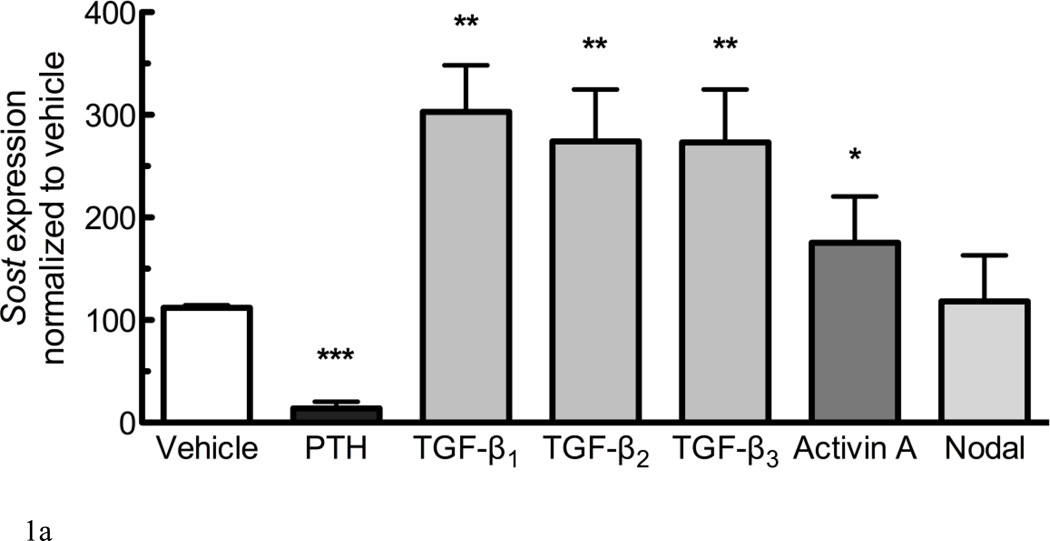
A described mode of modulating Wnt signaling involves altering the expression levels of Lrp5/6 antagonists, as both deletion of Dkk1 and Sost have been shown to promote aggressive bone overgrowth characterized by hyperactive osteoblast activity due to elevated Wnt signaling. We evaluated whether the expression of Sost was influenced by members of the TGF-β superfamily. UMR106.01 cells were cultured in the presence of 10ng/mL of TGF-β1, -β2, -β3, Activin A, or Nodal for 6 hours; these growth factors were chosen because they differentially activate signaling through the Alk4, Alk5, or Alk7 TGF-β type I receptors. Alternately, cells were cultured with 100nM hPTH(1–34), which decreases Sost expression [27–29]. Each TGF-β isoform increased Sost expression nearly 3-fold compared to vehicle control (Figure 1A); Activin A was less effective than TGF-β1–3, and Nodal did not affect Sost expression at the dose examined.
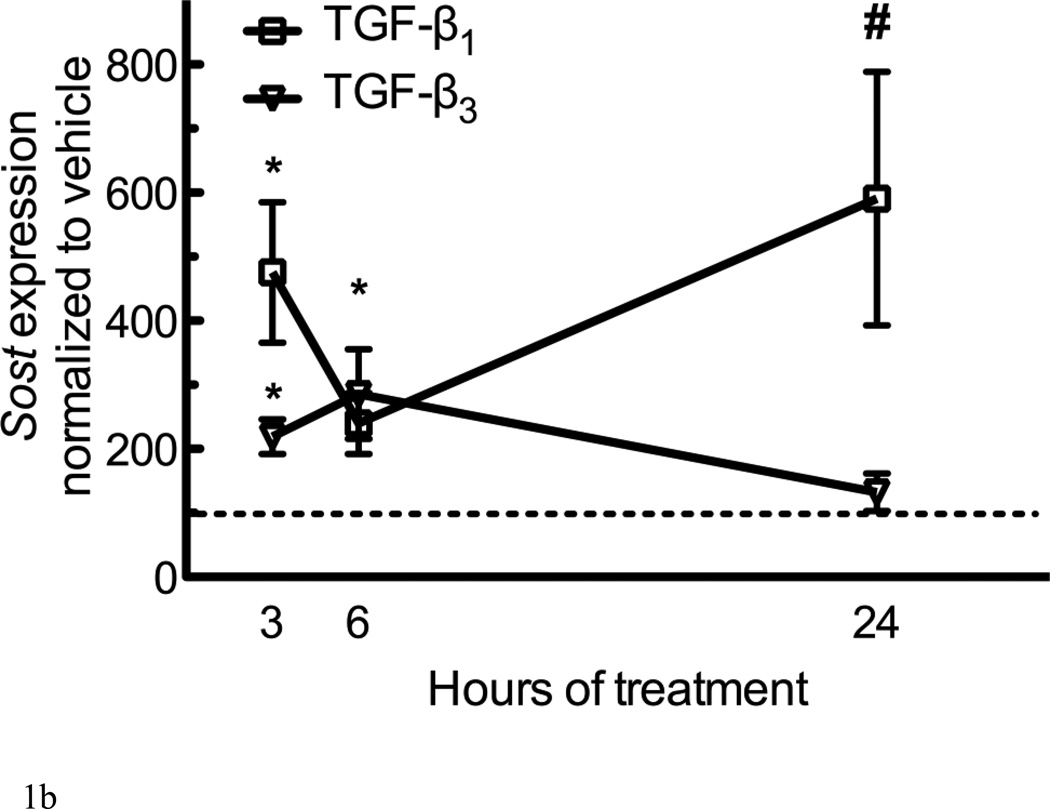
Figure 1. TGF-β superfamily members regulate expression of Sost.
(A) UMR106.01 mature osteoblasts were cultured for 6 hours with vehicle or 10ng/mL of indicated ligand; 100nM hPTH(1–34) was used as a positive control for Sost suppression. RNA was isolated for qPCR analysis of Sost expression relative to Rpl32. (B) UMR 106.01 mature osteoblasts were cultured for 3, 6, or 24h in the presence of 10ng/mL growth factor. For the sake of clarity, only the influence of TGF-β1 and -β3 upon Sost are shown. Data are mean ± SEM of 5 independent experiments. * indicates p < 0.05 compared to Vehicle, ** indicates p < 0.01 compared to Vehicle, *** indicates p < 0.001 compared to Vehicle, and # indicates p < 0.05 compared to TGF-β3.
Dose- and time-dependent effects of TGF-β family members upon Sost expression
To investigate dose-dependence of Sost expression in response to TGF-β1, -β2, -β3, and Activin A, UMR106.01 cells were cultured in the presence of increasing concentrations of growth factor (1–10,000 pg/mL) for 6 hours. TGF-β1, -β2, -β3, effective at Sost induction (257 – 301% increase) at 10ng/mL, whereas Activin A was about half as effective (TABLE 2). There were differences in potency amongst the four proteins: TGF-β1 and -β3 demonstrated a similar EC50 for Sost induction (29 and 34pg/mL, respectively), whereas TGF-β2 (186 pg/mL) and Activin A (1371 pg/mL) were less potent. These data indicate that TGF-β1, -β2, -β3 and Activin A, members of the TGF-β superfamily, are capable of inducing Sost expression.
TABLE 2.
TGF-β isoform effects upon Sost
| Ligand | Sost EC50 (ng/mL) | Maximum induction (percent of vehicle) |
|---|---|---|
| TGF-β1 | 29 | 257 |
| TGF-β2 | 186 | 301 |
| TGF-β3 | 34 | 297 |
| Activin A | 1371 | 175 |
The temporal nature of TGF-β-induced Sost expression was examined by treating cells with 10ng/mL of each growth factor, after which RNA was collected 3, 6, or 24h later. TGF-β1 exerted a rapid increase upon Sost expression after 3 hours of culture (Figure 1B); there was a trend (p<.12) for decreased Sost induction after 6 hours of culture, although expression remained significantly elevated after 6 or 24 hours of culture compared to vehicle control. TGF-β2 exerted a gradual increase in Sost induction over the time course examined (data not shown). The effect of TGF-β3 upon Sost expression was transient, as it was significantly elevated after 3 or 6 hours of treatment, but regressed toward baseline after 24 hours of treatment (Figure 1B). The effect of Activin A mimicked that of TGF-β3 (data not shown).
Sost expression is inhibited by antagonists of Alk4/5/7 in vitro and in vivo
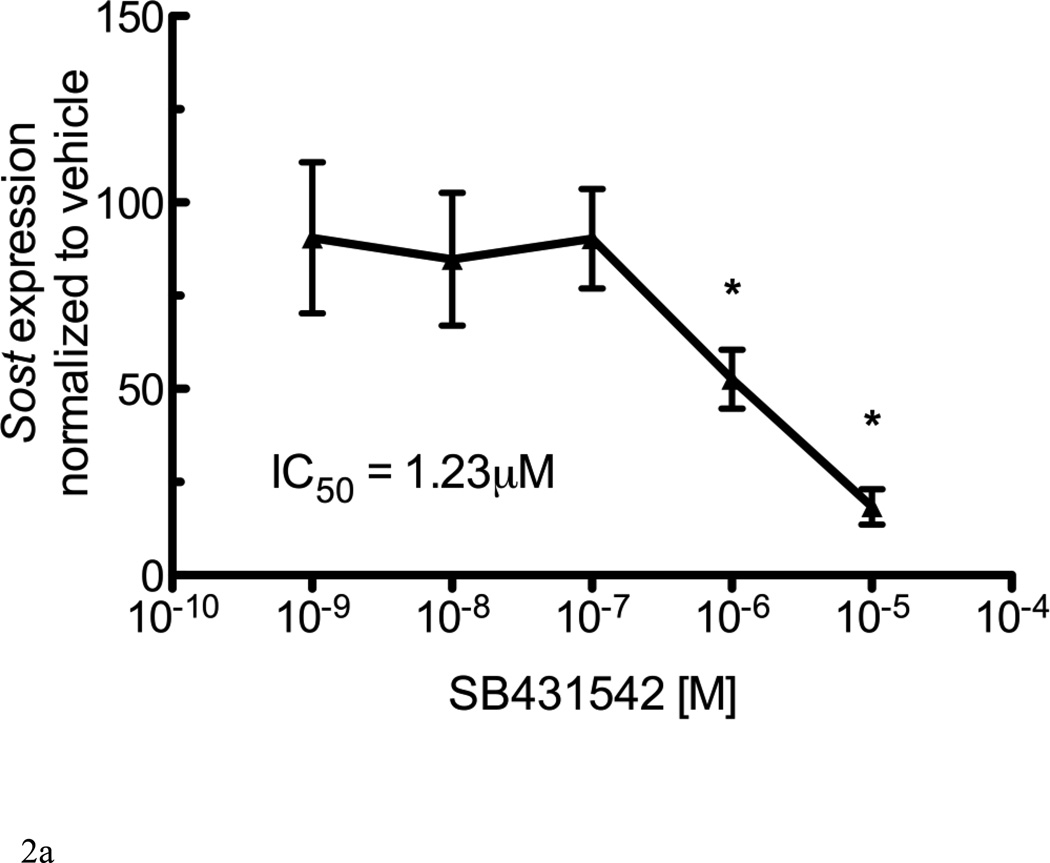
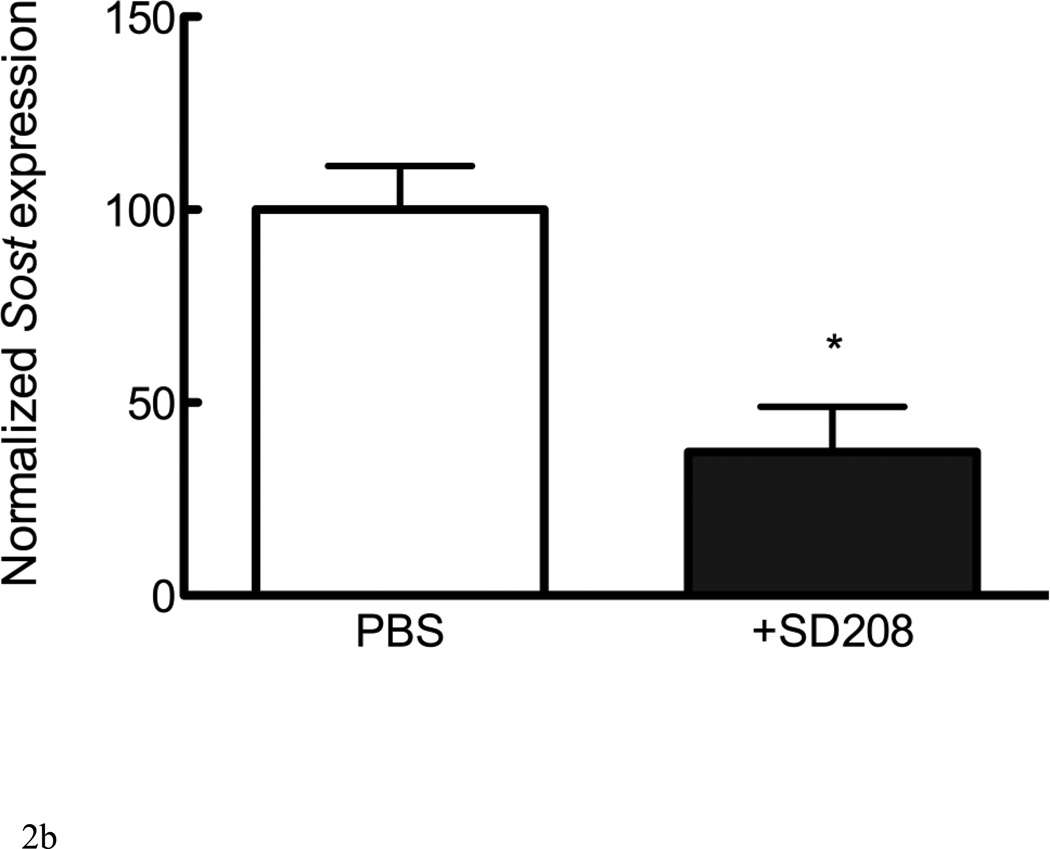
TGF-β and Activin A activate intracellular signaling cascades once bound to type I and type II receptors. TGF-β type I receptors include Alk4, Alk5, and Alk7. The addition of the Alk4/5/7 antagonist SB431542 to UMR 106.01 cells decreased endogenous Sost expression (Figure 2A). We next examined whether inhibition of Alk4/5/7 similarly influenced Sost expression, in vivo. Wild-type mice treated with a related Alk4/5/7 kinase inhibitor, SD-208 (60mg/kg), for 24 hours [32] exhibited a 63% decrease in Sost expression compared to PBS-treated controls (Figure 2B).
Figure 2. Sost expression is inhibited by Alk4/5/7 antagonists.
(A) UMR106.01 cells cultured for 6 hours in the presence of the Alk4/5/7 antagonist SB431542. RNA was isolated for qPCR analysis of Sost expression relative to Rpl32. (B) Mice were injected with either PBS or 20mg/kg SD208. 24 hours later, femorae were extracted and processed by qPCR for Sost expression relative to Rpl32. Data are mean ± SEM of 3 independent experiments. * indicates p < 0.05 compared to Vehicle
Alk4 and Alk5 mediate induction of Sost
Expression of the three TGF-β type I receptors (Alk4, Alk5, and Alk7) was confirmed in vitro in UMR106.01 osteoblasts (Figure 3A) and in vivo from femur and calvariae RNA (Figure 3B). Alk4 (receptor for Activin A) and Alk5 (receptor for TGF-β1, -β2, -β3) were consistently expressed at higher levels compared to Alk7 (receptor for Nodal). siRNA directed against Alk4, Alk5, or Alk7 decreased target gene expression by 72–77% relative to non-silencing RNA controls (Figure 3C). siRNA directed against Alk4 and Alk5 significantly decreased endogenous Sost production by 76 and 81%, respectively, while siRNA directed against Alk7 only modestly influenced Sost levels (Figure 3D). These data indicate that Sost expression is in part regulated by Alk5 and Alk4 receptors, respectively, whereas in these experiments Alk7 does not appear to play a significant role in the regulation of Sost expression.
TGF-β1 induction of Sost involves Smad3 and is antagonized by PTH
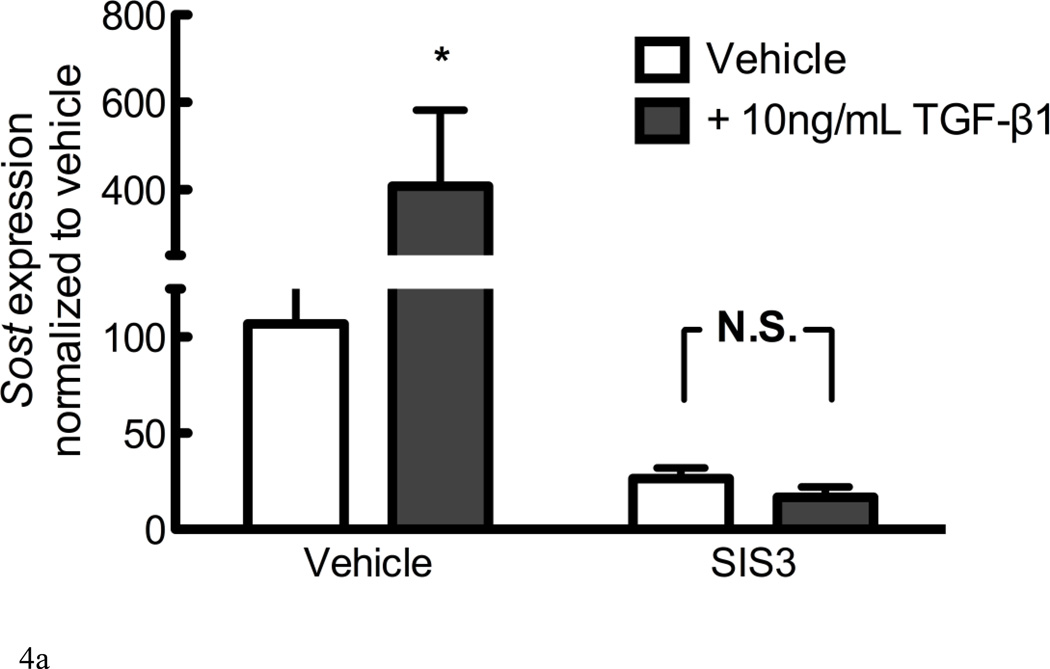
Direct regulation of gene expression by TGF-β involves Smad2 or Smad3 transcription factors. SIS3 is a cell-permeant Smad3 inhibitor that reduces Smad3 phosphorylation and DNA binding, without influencing Smad2 [33]. The addition of 10µM SIS3 to cultures significantly reduced Sost expression in both vehicle and 10ng/mL TGF-β1-treated cells (Figure 4A), indicating that both endogenous and growth factor-induced Sost expression is dependent on Smad3 phosphorylation and DNA binding.
Figure 4. TGF-β induction of Sost involves Smad3 and is abrogated by PTH.
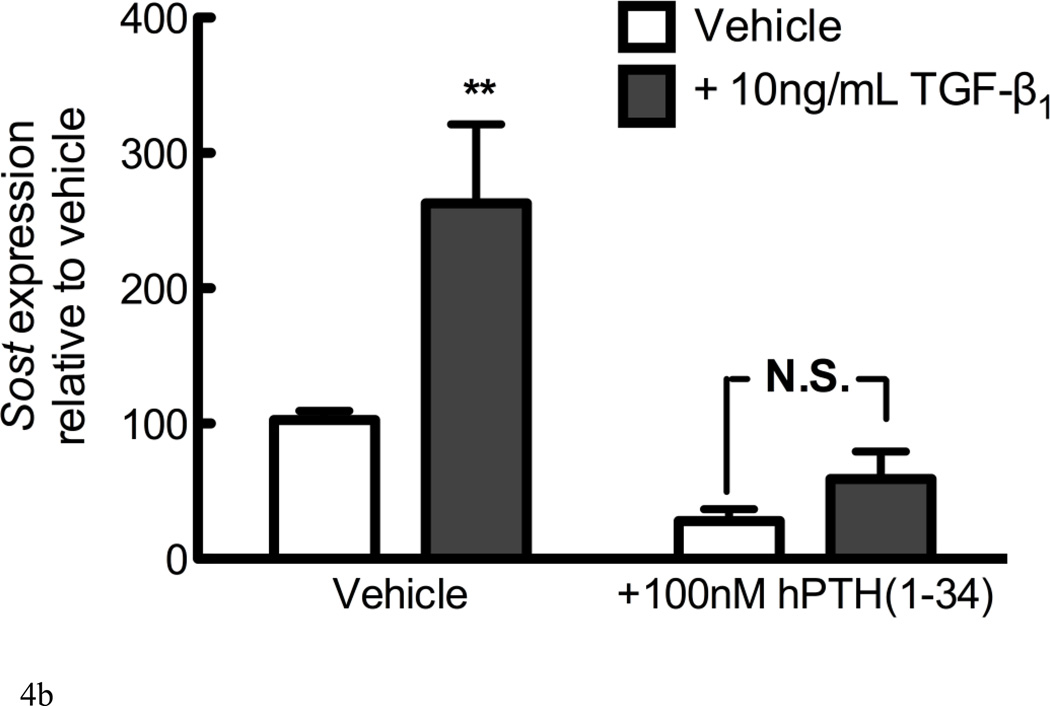
(A) UMR 106.01 cells were pretreated for 1 hour in the absence or presence of the cell-permeant Smad3 inhibitor, SIS3 (10µM), after which 10ng/mL TGF-β1 or vehicle control was added for 6 hours. RNA was isolated for qPCR analysis of Sost expression relative to Rpl32. (B) UMR 106.01 cells were cultured for 6 hours in the absence or presence of 100nM hPTH(1–34) with or without 10ng/mL TGF-β1.
We have previously demonstrated that PTH directly inhibits Sost expression without the need for de novo protein synthesis [28]; thus, we examined the capacity for PTH to interfere with TGF-β1-induced Sost expression. Cells treated with both 100nM hPTH (1–34) and 10ng/mL TGF-β1 for 6 hours revealed no significant increase in Sost expression relative to vehicle controls, suggesting that they regulate Sost expression independently (Figure 4B), and blunted each other’s effect on Sost transcription.
TGF-β1 targets the ECR5 region of the distal Sost enhancer
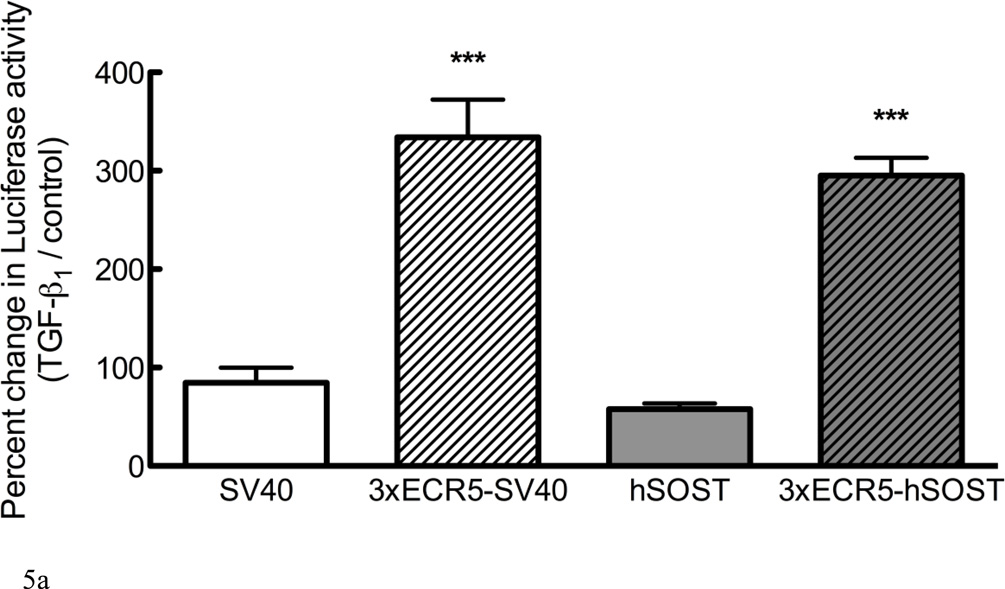
Regulation of Sost by TGF-β1 could involve activation of the proximal promoter, distal enhancer, or a synergistic effect of both. We examined the responsiveness of Luciferase reporter constructs containing a minimal SV40 promoter, the human SOST promoter (hSOST), each with or without the ECR5 distal enhancer [13, 16]. Constructs containing either the minimal SV40 promoter or the hSOST promoter demonstrated no change in Luciferase expression when treated with 10ng/mL TGF-β1, whereas constructs containing the ECR5 region increased the Luciferase activity by approximately 3-fold (Figure 5A), independent of promoter choice. There was no additive effect in plasmids containing ECR5 and hSOST, indicating that these transcriptional regulatory components do not synergize, and that the TGF-β1 response is dependent on the distal enhancer, ECR5.
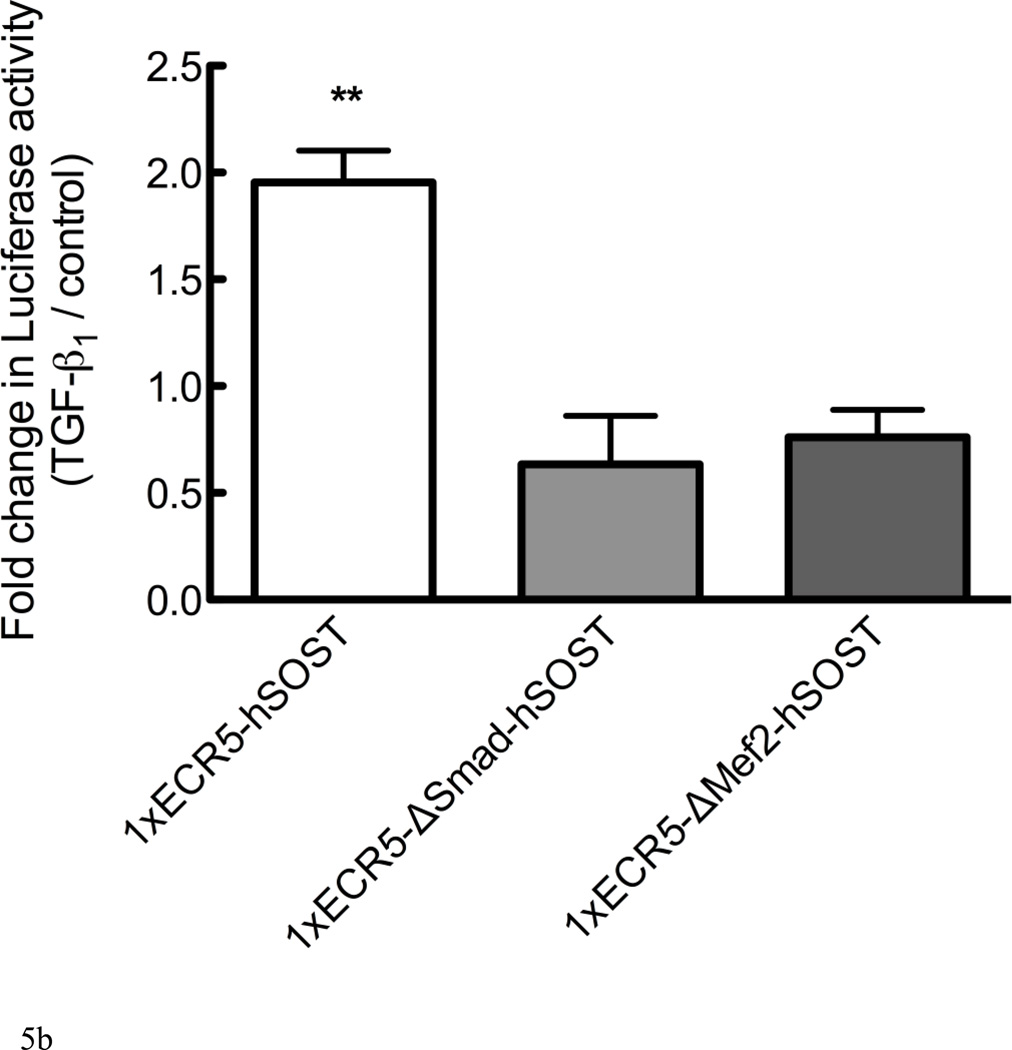
Figure 5. TGF-β1 induces Sost via the ECR5 transcriptional regulatory element through Smad and Mef2.
(A) UMR 106.01 cells were transfected with Luciferase constructs containing minimal SV40 promoter, the human SOST promoter, each with or without the ECR5 distal enhancer and treated with vehicle or 10ng/mL TGF-β1 for 24 hours, after which Luciferase activity was measured. (B) UMR 106.01 cells were transfected with a wild-type 1xECR5-hSOST Luciferase construct or construct with deletions of the Smad (ΔSmad) or Mef2 (ΔMef2) consensus binding site and treated with vehicle or 10ng/mL TGF-β1 for 24 hours, after which Luciferase activity was measured. *** indicates < 0.001 compared to plasmids lacking 3xECR5, ** indicates p < 0.01 compared to plasmids with deletions.
The ECR5 region contains a binding site for the myocyte enhancer family 2 (Mef2) family of transcription factors [34]. This sequence also contains a putative binding site for Smad2/3 [35], transcription factors activated by TGF-β1. To test the role of the Mef2 and Smad binding sites in TGF-β-driven Luciferase activity, reporter constructs were generated that contained mutated binding sites for Mef2 or Smads. Deletion of the Smad binding site prevented Luciferase expression in response to TGF-β1 (Figure 5B); unexpectedly, deletion of the Mef2 binding site also prevented TGF-β1-driven Luciferase activity.
Discussion
Genetic analyses of the rare sclerosing bone dysplasias sclerosteosis and van Buchem disease that either target the SOST transcript, or its distal cis-regulatory elements have positioned SOST as a robust regulator of skeletal homeostasis. Its role on mediating bone accrual and deposition was further confirmed through the generation of Sost [36] or van Buchem region [13] transgenic mice, which are osteopenic compared to wild-type control mice. Sclerostin was initially characterized as a BMP antagonist [36], but was later shown to function through Lrp4-6 [8, 37]. Functionally, a sclerostin-inhibiting antibody enhances bone mineral density and strength [38, 39]. Mechanistically, Sclerostin exerts anti-anabolic effects by decreasing osteoprogenitor proliferation and differentiation, and promoting osteoblast apoptosis [40]. Provided the increasing direct medical costs for osteoporosis-related or other traumatic fractures, it is critical to understand the mechanisms involved in Sost expression because of its function in vivo as an inhibitor of bone formation.
The TGF-β family represents an important superfamily involved in regulating bone metabolism since a significant number of its members are produced locally within the skeleton [41] where they accumulate within the matrix [42] and are released by osteoclasts during remodeling [43]. Germline deletion of individual TGF-β genes yields multiple, non-overlapping developmental defects [44]. Of the three TGF-β proteins, TGF-β1 is the isoform most abundantly expressed in bone [45]. It has opposing effects on osteoprogenitors compared to osteoblasts, inducing the migration, proliferation and differentiation of the former, but inhibiting matrix accumulation of the latter [20]. Little is known as to how TGF-β elicits these differential effects, although Alliston et al. have demonstrated that TGF-β promotes the degradation of the transcription factor essential for osteoblast differentiation, Runx2 [46]. Later, Balooch et al. used a series of transgenic mouse models with alterations in TGF-β signaling to show elevated levels of TGF-β in bone decreased mechanical properties, mineral concentration, and fracture toughness [47]; these trends were reversed in mice expressing a dominant-negative type II TGF-β receptor. Further, Mohammed et al. showed that inhibition of Alk4/5/7 with SD-208 in vivo had catabolic and anabolic effects on the mouse skeleton [48]. These effects included: increased increased bone mineral density and trabecular bone volume independent of cortical bone changes; increased osteoprogenitor differentiation; and decreased osteoclast number. Functionally, Alk4/5/7 inhibition inceased both bone mechanical and material properties, suggesting that Alk4/5/7 inhibition could be used for conditions of skeletal fragility like osteoporosis. These findings combined with the data presented in this manuscript provide compelling evidence that signaling the the Alk4/5/7 axis positively influences Sost expression. We can further speculate that Sclerostin potentially plays an important role in the processes described by Balooch et al. and Mohammed et al. [48].
TGF-β interacts with a variety of signaling pathways implicated in skeletal homeostasis. For example, PTH increases TGF-β1 synthesis and secretion and Smad3 phosphorylation to decrease osteoblast apoptosis [22]. TGF-β generally decreases the capacity for BMPs to induce osteogenic differentiation of osteoprogenitors, but de Gorter et al. recently demonstrated that this inhibitory effect is very much context dependent with respect to such details as the composition of the culture medium used and the duration of growth factor co-culture [49]. Similarly, TGF-β can regulate expression and activity of Wnt signaling. In mesenchymal stem cells, TGF-β1 induces proliferation via Smad3-dependent β-catenin nuclear translocation [50] and increases β-catenin expression [51]. Within, we demonstrate that TGF-β isoforms increase Sost expression in mature osteoblasts. Provided the inhibitory effect of Sclerostin upon matrix mineralization and differentiation [52], this may provide a mechanism to explain the similarly inhibitory effect of TGF-β1 upon differentiation of mature osteoblasts, although further studies are required to fully confirm this.
Nonsense mutations in Sost cause sclerosteosis [9], and a 52kb deletion 35kb downstream of Sost is responsible for van Buchem disease [11]. Using cross-species sequence comparison and enhancer assays, we have previously identified a 255-bp evolutionarily conserved sequence within the van Buchem deletion, termed ECR5, that confers bone-specific expression of SOST [13]. Reporter assays containing the ECR5 enhancer, but not the proximal promoter, are responsive to PTH via Mef2 transcription factors [16]. Within this work, we demonstrate that, similar to PTH, TGF-β1 targets the ECR5 enhancer but not the SOST promoter (Figure 5). Within the ECR5 enhancer, we identified consensus binding sites for Smad2/3, in addition to the previously identified Mef2 sites; surprisingly, both Smad2/3 and Mef2 sites contribute to TGF-β1 transcriptional activation of Sost. We have not observed any changes in expression of Mef2c or Mef2d transcription in response to TGF-βs (data not shown), indicating that the regulation is likely post-transcriptional in nature. One candidate mechanism is direct physical interaction between Mef2 and Smad2. Indeed, Quinn et al. previously demonstrated direct interaction of Smad2 with Mef2 in vivo to enhance Mef2 transcriptional activity [53], as did Ishikawa et al. [54]. How TGF-β1 regulates Mef2 transcriptional activity in osteoblastic cells is currently under investigation.
Similar to TGF-βs, we observed that Activin A, which signals through the Alk4 type I receptor, increases Sost expression, although with less potency and efficacy than TGF-β1,-β2, -β3. Despite the decreased potency, siRNA directed against Alk4 decreased endogenous Sost expression to a similar degree as did siRNA against Alk5. Similar to TGF-β, the role of Activin A in skeletal homeostasis is conflicting, as various reports demonstrate its capacity to increase or decrease osteoblast- and osteoclastogenesis [55, 56]. Pathologically, Activin A and TGF-β are implicated in the progression of osteolytic metastases. Serum Activin A is elevated in patients with multiple myeloma [57], and a soluble Activin receptor IIA fusion protein decreases bone metastasis and resorption [58]. Interestingly, Dkk1, a Wnt antagonist like Sclerostin, is highly expressed in osteolytic cancers such as multiple myeloma [59]; whether Wnt signaling in osteolytic cancers is dependent upon TGF-β is a possibility worthy of investigation.
In conclusion, we demonstrate that members of the TGF-β superfamily beyond BMPs are also capable of inducing Sost expression, as TGF-βs and Activin A demonstrated graded effects on Sost transcription. This is not a general effect of the TGF-β superfamily, as Nodal (Alk7 agonist) had no effect on Sost expression at the concentration examined. Inhibition of Alk4/5/7 with two different antagonists decreased Sost expression in vitro and in vivo, indicating that use of a clonal mature osteoblastic cell line was not a confounding factor in regulation of Sost by TGF-βs. siRNA directed against Alk4 and Alk5 decreased Sost expression. The specific Smad3 inhitor SIS3 attenuated endogenous and TGF-β-stimulated Sost expression, suggesting that traditional TGF-β/Smad, rather than TGF-β/MAPK, signaling is involved in transcription of Sost. Luciferase reporter assays indicated that TGF-β1 targets the ECR5 bone enhancer, and not the proximal promoter, through a mechanism involving both Smad2/3 and Mef2 transcription factors. These results lay the foundation for future work designed to examine the interplay between Alk4 and Alk5 function and Wnt signaling in vivo, and identification of protein intermediates required for TGF-β—dependent induction of Sost transcription.
Highlights.
TGF-β isoforms and Activin A increase Sost expression via Smad3
Inhibition of Alk4 and Alk5 both in vitro and in vivo reduces Sost expression
TGF-β targets ECR5 to enhance Sost
Acknowledgements
This work was supported by Award Number R03AR057547 from the National Institute of Arthritis and Musculoskeletal and Skin Diseases (DCG) and DK075730 from the National Institute of Diabetes and Digestive and Kidney Diseases (DCG, DM, NMC, GGL). This work performed under the auspices of the U. S. Department of Energy by Lawrence Livermore National Laboratory under Contract DE-AC52-07NA27344.
Abbreviations
- TGF-β
Transforming Growth Factor-beta
- Lrp5
Low-density lipoprotein-related receptor 5
- Wnt
Wingless int
- ECR5
Evolutionarily Conserved Region 5
- PTH
Parathyroid Hormone
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Gong Y, Slee RB, Fukai N, Rawadi G, Roman-Roman S, Reginato AM, Wang H, Cundy T, Glorieux FH, Lev D, Zacharin M, Oexle K, Marcelino J, Suwairi W, Heeger S, Sabatakos G, Apte S, Adkins WN, Allgrove J, Arslan-Kirchner M, Batch JA, Beighton P, Black GC, Boles RG, Boon LM, Borrone C, Brunner HG, Carle GF, Dallapiccola B, De Paepe A, Floege B, Halfhide ML, Hall B, Hennekam RC, Hirose T, Jans A, Juppner H, Kim CA, Keppler-Noreuil K, Kohlschuetter A, LaCombe D, Lambert M, Lemyre E, Letteboer T, Peltonen L, Ramesar RS, Romanengo M, Somer H, Steichen-Gersdorf E, Steinmann B, Sullivan B, Superti-Furga A, Swoboda W, van den Boogaard MJ, Van Hul W, Vikkula M, Votruba M, Zabel B, Garcia T, Baron R, Olsen BR, Warman ML. LDL receptor-related protein 5 (LRP5) affects bone accrual and eye development. Cell. 2001;l107:513–523. doi: 10.1016/s0092-8674(01)00571-2. [DOI] [PubMed] [Google Scholar]
- 2.Little RD, Carulli JP, Del Mastro RG, Dupuis J, Osborne M, Folz C, Manning SP, Swain PM, Zhao SC, Eustace B, Lappe MM, Spitzer L, Zweier S, Braunschweiger K, Benchekroun Y, Hu X, Adair R, Chee L, FitzGerald MG, Tulig C, Caruso A, Tzellas N, Bawa A, Franklin B, McGuire S, Nogues X, Gong G, Allen KM, Anisowicz A, Morales AJ, Lomedico PT, Recker SM, Van Eerdewegh P, Recker RR, Johnson ML. A mutation in the LDL receptor-related protein-gene results in the autosomal dominant high-bone-mass trait. Am. J. Hum. Genet. 2002;l70:11–19. doi: 10.1086/338450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Almeida M, Han L, Bellido T, Manolagas SC, Kousteni S. Wnt proteins prevent apoptosis of both uncommitted osteoblast progenitors and differentiated osteoblasts by beta-catenin- dependent and -independent signaling cascades involving Src/ERK and phosphatidylinositol 3-kinase/AKT. J. Biol. Chem. 2005;l280:41342–41351. doi: 10.1074/jbc.M502168200. [DOI] [PubMed] [Google Scholar]
- 4.Bodine PV, Billiard J, Moran RA, Ponce-de-Leon H, McLarney S, Mangine A, Scrimo MJ, Bhat RA, Stauffer B, Green J, Stein GS, Lian JB, Komm BS. The Wnt antagonist secreted frizzled-related protein-1 controls osteoblast and osteocyte apoptosis. J. Cell. Biochem. 2005;l96:1212–1230. doi: 10.1002/jcb.20599. [DOI] [PubMed] [Google Scholar]
- 5.Williams BO, Insogna KL. Where Wnts went: the exploding field of Lrp5 and Lrp6 signaling in bone. J. Bone Miner. Res. 2009;l24:171–178. doi: 10.1359/jbmr.081235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Choi HY, Dieckmann M, Herz J, Niemeier A. Lrp4, a novel receptor for Dickkopf 1 and sclerostin, is expressed by osteoblasts and regulates bone growth and turnover in vivo. PLoS ONE. 2009;l4:e7930. doi: 10.1371/journal.pone.0007930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kato M, Patel MS, Levasseur R, Lobov I, Chang BH, Glass DA, 2nd, Hartmann C, Li L, Hwang TH, Brayton CF, Lang RA, Karsenty G, Chan L. Cbfa1-independent decrease in osteoblast proliferation, osteopenia, and persistent embryonic eye vascularization in mice deficient in Lrp5, a Wnt coreceptor. J. Cell Biol. 2002;l157:303–314. doi: 10.1083/jcb.200201089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leupin O, Piters E, Halleux C, Hu S, Kramer I, Morvan F, Bouwmeester T, Schirle M, Bueno-Lozano M, Fuentes FJ, Itin PH, Boudin E, de Freitas F, Jennes K, Brannetti B, Charara N, Ebersbach H, Geisse S, Lu CX, Bauer A, Van Hul W, Kneissel M. Bone overgrowth-associated mutations in the LRP4 gene impair sclerostin facilitator function. J. Biol. Chem. 2011;l286:19489–19500. doi: 10.1074/jbc.M110.190330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Balemans W, Ebeling M, Patel N, Van Hul E, Olson P, Dioszegi M, Lacza C, Wuyts W, Van Den Ende J, Willems P, Paes-Alves AF, Hill S, Bueno M, Ramos FJ, Tacconi P, Dikkers FG, Stratakis C, Lindpaintner K, Vickery B, Foernzler D, Van Hul W. Increased bone density in sclerosteosis is due to the deficiency of a novel secreted protein (SOST) Hum. Mol. Genet. 2001;l10:537–543. doi: 10.1093/hmg/10.5.537. [DOI] [PubMed] [Google Scholar]
- 10.Brunkow ME, Gardner JC, Van Ness J, Paeper BW, Kovacevich BR, Proll S, Skonier JE, Zhao L, Sabo PJ, Fu Y, Alisch RS, Gillett L, Colbert T, Tacconi P, Galas D, Hamersma H, Beighton P, Mulligan J. Bone dysplasia sclerosteosis results from loss of the SOST gene product, a novel cystine knot-containing protein. Am. J. Hum. Genet. 2001;l68:577–589. doi: 10.1086/318811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Balemans W, Patel N, Ebeling M, Van Hul E, Wuyts W, Lacza C, Dioszegi M, Dikkers FG, Hildering P, Willems PJ, Verheij JB, Lindpaintner K, Vickery B, Foernzler D, Van Hul W. Identification of a 52 kb deletion downstream of the SOST gene in patients with van Buchem disease. J. Med. Genet. 2002;l39:91–97. doi: 10.1136/jmg.39.2.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kusu N, Laurikkala J, Imanishi M, Usui H, Konishi M, Miyake A, Thesleff I, Itoh N. Sclerostin is a novel secreted osteoclast-derived bone morphogenetic protein antagonist with unique ligand specificity. J. Biol. Chem. 2003;l278:24113–24117. doi: 10.1074/jbc.M301716200. [DOI] [PubMed] [Google Scholar]
- 13.Loots GG, Kneissel M, Keller H, Baptist M, Chang J, Collette NM, Ovcharenko D, Plajzer-Frick I, Rubin EM. Genomic deletion of a long-range bone enhancer misregulates sclerostin in Van Buchem disease. Genome Res. 2005;l15:928–935. doi: 10.1101/gr.3437105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nakanishi R, Shimizu M, Mori M, Akiyama H, Okudaira S, Otsuki B, Hashimoto M, Higuchi K, Hosokawa M, Tsuboyama T, Nakamura T. Secreted frizzled-related protein 4 is a negative regulator of peak BMD in SAMP6 mice. J. Bone Miner. Res. 2006;l21:1713–1721. doi: 10.1359/jbmr.060719. [DOI] [PubMed] [Google Scholar]
- 15.Bodine PV, Zhao W, Kharode YP, Bex FJ, Lambert AJ, Goad MB, Gaur T, Stein GS, Lian JB, Komm BS. The Wnt antagonist secreted frizzled-related protein-1 is a negative regulator of trabecular bone formation in adult mice. Mol. Endocrinol. 2004;l18:1222–1237. doi: 10.1210/me.2003-0498. [DOI] [PubMed] [Google Scholar]
- 16.Leupin O, Kramer I, Collette NM, Loots GG, Natt F, Kneissel M, Keller H. Control of the SOST bone enhancer by PTH using MEF2 transcription factors. J. Bone Miner. Res. 2007;l22:1957–1967. doi: 10.1359/jbmr.070804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kanaan RA, Kanaan LA. Transforming growth factor beta1, bone connection. Med Sci Monit. 2006;l12:RA164–RA169. [PubMed] [Google Scholar]
- 18.Santibanez JF, Quintanilla M, Bernabeu C. TGF-beta/TGF-beta receptor system and its role in physiological and pathological conditions. Clin. Sci. 2011;l121:233–251. doi: 10.1042/CS20110086. [DOI] [PubMed] [Google Scholar]
- 19.Origuchi N, Ishidou Y, Nagamine T, Onishi T, Matsunaga S, Yoshida H, Sakou T. The spatial and temporal immunolocalization of TGF-beta 1 and bone morphogenetic protein-2/-4 in phallic bone formation in inbred Sprague Dawley male rats. In Vivo. 1998;l12:473–480. [PubMed] [Google Scholar]
- 20.Janssens K, ten Dijke P, Janssens S, Van Hul W. Transforming growth factor-beta1 to the bone. Endocr. Rev. 2005;l26:743–774. doi: 10.1210/er.2004-0001. [DOI] [PubMed] [Google Scholar]
- 21.Bonewald LF, Dallas SL. Role of active and latent transforming growth factor beta in bone formation. J. Cell. Biochem. 1994;l55:350–357. doi: 10.1002/jcb.240550312. [DOI] [PubMed] [Google Scholar]
- 22.Sowa H, Kaji H, Iu MF, Tsukamoto T, Sugimoto T, Chihara K. Parathyroid hormone-Smad3 axis exerts anti-apoptotic action and augments anabolic action of transforming growth factor beta in osteoblasts. J. Biol. Chem. 2003;l278:52240–52252. doi: 10.1074/jbc.M302566200. [DOI] [PubMed] [Google Scholar]
- 23.Pilbeam CC, Kawaguchi H, Voznesensky OS, Alander CB, Raisz LG. Regulation of inducible prostaglandin G/H synthase by interleukin-1, transforming growth factors-alpha and - beta, and prostaglandins in bone cells. Adv. Exp. Med. Biol. 1997;l400B:617–623. [PubMed] [Google Scholar]
- 24.Sutherland MK, Geoghegan JC, Yu C, Winkler DG, Latham JA. Unique regulation of SOST, the sclerosteosis gene, by BMPs and steroid hormones in human osteoblasts. Bone. 2004;l35:448–454. doi: 10.1016/j.bone.2004.04.019. [DOI] [PubMed] [Google Scholar]
- 25.Kamiya N, Ye L, Kobayashi T, Mochida Y, Yamauchi M, Kronenberg HM, Feng JQ, Mishina Y. BMP signaling negatively regulates bone mass through sclerostin by inhibiting the canonical Wnt pathway. Development. 2008;l135:3801–3811. doi: 10.1242/dev.025825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Papanicolaou SE, Phipps RJ, Fyhrie DP, Genetos DC. Modulation of sclerostin expression by mechanical loading and bone morphogenetic proteins in osteogenic cells. Biorheology. 2009;l46:389–399. doi: 10.3233/BIR-2009-0550. [DOI] [PubMed] [Google Scholar]
- 27.Kulkarni NH, Halladay DL, Miles RR, Gilbert LM, Frolik CA, Galvin RJ, Martin TJ, Gillespie MT, Onyia JE. Effects of parathyroid hormone on Wnt signaling pathway in bone. J. Cell. Biochem. 2005;l95:1178–1190. doi: 10.1002/jcb.20506. [DOI] [PubMed] [Google Scholar]
- 28.Keller H, Kneissel M. SOST is a target gene for PTH in bone. Bone. 2005;l37:148–158. doi: 10.1016/j.bone.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 29.Genetos DC, Yellowley CE, Loots GG. Prostaglandin E(2) Signals Through PTGER2 to Regulate Sclerostin Expression. PLoS ONE. 2011;l6:e17772. doi: 10.1371/journal.pone.0017772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kamiya N, Ye L, Kobayashi T, Lucas DJ, Mochida Y, Yamauchi M, Kronenberg HM, Feng JQ, Mishina Y. Disruption of BMP signaling in osteoblasts through type IA receptor (BMPRIA) increases bone mass. J. Bone Miner. Res. 2008;l23:2007–2017. doi: 10.1359/JBMR.080809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;l25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 32.Yingling JM, Blanchard KL, Sawyer JS. Development of TGF-beta signalling inhibitors for cancer therapy. Nat. Rev. Drug Discov. 2004;l3:1011–1022. doi: 10.1038/nrd1580. [DOI] [PubMed] [Google Scholar]
- 33.Jinnin M, Ihn H, Tamaki K. Characterization of SIS3, a novel specific inhibitor of Smad3, and its effect on transforming growth factor-beta1-induced extracellular matrix expression. Mol. Pharmacol. 2006;l69:597–607. doi: 10.1124/mol.105.017483. [DOI] [PubMed] [Google Scholar]
- 34.Fickett JW. Quantitative discrimination of MEF2 sites. Mol. Cell. Biol. 1996;l16:437–441. doi: 10.1128/mcb.16.1.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dennler S, Itoh S, Vivien D, ten Dijke P, Huet S, Gauthier JM. Direct binding of Smad3 and Smad4 to critical TGF beta-inducible elements in the promoter of human plasminogen activator inhibitor-type 1 gene. EMBO J. 1998;l17:3091–3100. doi: 10.1093/emboj/17.11.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Winkler DG, Sutherland MK, Geoghegan JC, Yu C, Hayes T, Skonier JE, Shpektor D, Jonas M, Kovacevich BR, Staehling-Hampton K, Appleby M, Brunkow ME, Latham JA. Osteocyte control of bone formation via sclerostin, a novel BMP antagonist. EMBO J. 2003;l22:6267–6276. doi: 10.1093/emboj/cdg599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Semenov M, Tamai K, He X. SOST is a ligand for LRP5/LRP6 and a Wnt signaling inhibitor. J. Biol. Chem. 2005;l280:26770–26775. doi: 10.1074/jbc.M504308200. [DOI] [PubMed] [Google Scholar]
- 38.Li X, Ominsky MS, Warmington KS, Morony S, Gong J, Cao J, Gao Y, Shalhoub V, Tipton B, Haldankar R, Chen Q, Winters A, Boone T, Geng Z, Niu QT, Ke HZ, Kostenuik PJ, Simonet WS, Lacey DL, Paszty C. Sclerostin antibody treatment increases bone formation, bone mass, and bone strength in a rat model of postmenopausal osteoporosis. J. Bone Miner. Res. 2009;l24:578–588. doi: 10.1359/jbmr.081206. [DOI] [PubMed] [Google Scholar]
- 39.Ominsky MS, Vlasseros F, Jolette J, Smith SY, Stouch B, Doellgast G, Gong J, Gao Y, Cao J, Graham K, Tipton B, Cai J, Deshpande R, Zhou L, Hale MD, Lightwood DJ, Henry AJ, Popplewell AG, Moore AR, Robinson MK, Lacey DL, Simonet WS, Paszty C. Two doses of sclerostin antibody in cynomolgus monkeys increases bone formation, bone mineral density, and bone strength. J. Bone Miner. Res. 2010;l25:948–959. doi: 10.1002/jbmr.14. [DOI] [PubMed] [Google Scholar]
- 40.van Bezooijen RL, ten Dijke P, Papapoulos SE, Lowik CW. SOST/sclerostin, an osteocyte-derived negative regulator of bone formation. Cytokine Growth Factor Rev. 2005;l16:319–327. doi: 10.1016/j.cytogfr.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 41.Robey PG, Young MF, Flanders KC, Roche NS, Kondaiah P, Reddi AH, Termine JD, Sporn MB, Roberts AB. Osteoblasts synthesize and respond to transforming growth factor-type beta (TGF-beta) in vitro. J. Cell Biol. 1987;l105:457–463. doi: 10.1083/jcb.105.1.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Seyedin SM, Thompson AY, Bentz H, Rosen DM, McPherson JM, Conti A, Siegel NR, Galluppi GR, Piez KA. Cartilage-inducing factor-A. Apparent identity to transforming growth factor-beta. J. Biol. Chem. 1986;l261:5693–5695. [PubMed] [Google Scholar]
- 43.Mundy GR, Bonewald LF. Role of TGF beta in bone remodeling. Ann. N. Y. Acad. Sci. 1990;l593:91–97. doi: 10.1111/j.1749-6632.1990.tb16102.x. [DOI] [PubMed] [Google Scholar]
- 44.Sanford LP, Ormsby I, Gittenberger-de Groot AC, Sariola H, Friedman R, Boivin GP, Cardell EL, Doetschman T. TGFbeta2 knockout mice have multiple developmental defects that are non-overlapping with other TGFbeta knockout phenotypes. Development. 1997;l124:2659–2670. doi: 10.1242/dev.124.13.2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hering S, Isken E, Knabbe C, Janott J, Jost C, Pommer A, Muhr G, Schatz H, Pfeiffer AF. TGFbeta1 and TGFbeta2 mRNA and protein expression in human bone samples. Exp. Clin. Endocrinol. Diabetes. 2001;l109:217–226. doi: 10.1055/s-2001-15109. [DOI] [PubMed] [Google Scholar]
- 46.Alliston T, Choy L, Ducy P, Karsenty G, Derynck R. TGF-beta-induced repression of CBFA1 by Smad3 decreases cbfa1 and osteocalcin expression and inhibits osteoblast differentiation. EMBO J. 2001;l20:2254–2272. doi: 10.1093/emboj/20.9.2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Balooch G, Balooch M, Nalla RK, Schilling S, Filvaroff EH, Marshall GW, Marshall SJ, Ritchie RO, Derynck R, Alliston T. TGF-beta regulates the mechanical properties and composition of bone matrix. Proc. Natl. Acad. Sci. U. S. A. 2005;l102:18813–18818. doi: 10.1073/pnas.0507417102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mohammad KS, Chen CG, Balooch G, Stebbins E, McKenna CR, Davis H, Niewolna M, Peng XH, Nguyen DH, Ionova-Martin SS, Bracey JW, Hogue WR, Wong DH, Ritchie RO, Suva LJ, Derynck R, Guise TA, Alliston T. Pharmacologic inhibition of the TGF-beta type I receptor kinase has anabolic and anti-catabolic effects on bone. PLoS ONE. 2009;l4:e5275. doi: 10.1371/journal.pone.0005275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.de Gorter DJ, van Dinther M, Korchynskyi O, ten Dijke P. Biphasic effects of transforming growth factor beta on bone morphogenetic protein-induced osteoblast differentiation. J. Bone Miner. Res. 2011;l26:1178–1187. doi: 10.1002/jbmr.313. [DOI] [PubMed] [Google Scholar]
- 50.Jian H, Shen X, Liu I, Semenov M, He X, Wang XF. Smad3-dependent nuclear translocation of beta-catenin is required for TGF-beta1-induced proliferation of bone marrow-derived adult human mesenchymal stem cells. Genes Dev. 2006;l20:666–674. doi: 10.1101/gad.1388806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Maeda S, Hayashi M, Komiya S, Imamura T, Miyazono K. Endogenous TGF-beta signaling suppresses maturation of osteoblastic mesenchymal cells. EMBO J. 2004;l23:552–563. doi: 10.1038/sj.emboj.7600067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Atkins GJ, Rowe PS, Lim HP, Welldon KJ, Ormsby R, Wijenayaka AR, Zelenchuk L, Evdokiou A, Findlay DM. Sclerostin is a locally acting regulator of late-osteoblast/preosteocyte differentiation and regulates mineralization through a MEPE-ASARM-dependent mechanism. J. Bone Miner. Res. 2011;l26:1425–1436. doi: 10.1002/jbmr.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Quinn ZA, Yang CC, Wrana JL, McDermott JC. Smad proteins function as comodulators for MEF2 transcriptional regulatory proteins. Nucleic Acids Res. 2001;l29:732–742. doi: 10.1093/nar/29.3.732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ishikawa F, Miyoshi H, Nose K, Shibanuma M. Transcriptional induction of MMP-10 by TGF-beta, mediated by activation of MEF2A and downregulation of class IIa HDACs. Oncogene. 2010;l29:909–919. doi: 10.1038/onc.2009.387. [DOI] [PubMed] [Google Scholar]
- 55.Centrella M, McCarthy TL, Canalis E. Activin-A binding and biochemical effects in osteoblast-enriched cultures from fetal-rat parietal bone. Mol. Cell. Biol. 1991;l11:250–258. doi: 10.1128/mcb.11.1.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kawabata N, Kamiya N, Suzuki N, Matsumoto M, Takagi M. Changes in extracellular activin A:follistatin ratio during differentiation of a mesenchymal progenitor cell line, ROB-C26 into osteoblasts and adipocytes. Life Sci. 2007;l81:8–18. doi: 10.1016/j.lfs.2007.04.011. [DOI] [PubMed] [Google Scholar]
- 57.Vallet S, Mukherjee S, Vaghela N, Hideshima T, Fulciniti M, Pozzi S, Santo L, Cirstea D, Patel K, Sohani AR, Guimaraes A, Xie W, Chauhan D, Schoonmaker JA, Attar E, Churchill M, Weller E, Munshi N, Seehra JS, Weissleder R, Anderson KC, Scadden DT, Raje N. Activin A promotes multiple myeloma-induced osteolysis and is a promising target for myeloma bone disease. Proc. Natl. Acad. Sci. U. S. A. 2010;l107:5124–5129. doi: 10.1073/pnas.0911929107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chantry AD, Heath D, Mulivor AW, Pearsall S, Baud'huin M, Coulton L, Evans H, Abdul N, Werner ED, Bouxsein ML, Key ML, Seehra J, Arnett TR, Vanderkerken K, Croucher P. Inhibiting activin-A signaling stimulates bone formation and prevents cancer-induced bone destruction in vivo. J. Bone Miner. Res. 2010;l25:2633–2646. doi: 10.1002/jbmr.142. [DOI] [PubMed] [Google Scholar]
- 59.Hall CL, Daignault SD, Shah RB, Pienta KJ, Keller ET. Dickkopf-1 expression increases early in prostate cancer development and decreases during progression from primary tumor to metastasis. Prostate. 2008;l68:1396–1404. doi: 10.1002/pros.20805. [DOI] [PMC free article] [PubMed] [Google Scholar]