Abstract
Myogenesis has proven to be a powerful paradigm for understanding cell fate specification and differentiation in many model organisms. This includes the nematode C. elegans for which the genetic, cellular, and molecular tools have allowed an in-depth understanding of muscle development. One tool not yet available in C. elegans is a robust, pure and prolific cell culture system to study myogenesis. As an alternative, this chapter describes a method by which the cell fates of early, uncommitted blastomeres in the embryo are converted to a myogenic lineage. This technique permits the nearly synchronous induction of myogenesis in vivo with the potential to generate a nearly homogeneous population of cells. Coupled with the RNA isolation and cDNA amplification methods that are also described, one can now profile gene expression throughout myogenesis using any platform of choice (e.g. expression arrays, next generation sequencing). Although limited by the artificial nature of this developing mass of muscle inside the eggshell, blastomere conversion and transcriptional profiling is a very powerful tool to investigate changes in gene expression associated with myogenesis in C. elegans that is applicable to many different cell types. When coupled with next generation sequencing, the method has the potential to yield a very high-resolution map of changes in gene expression throughout myogenesis.
Keywords: Myogenesis, C. elegans, Blastomere Conversion, Cell Fate, HLH-1, Transcription
1. Introduction
Muscle is arguably one of the most studied and understood tissues across multiple model systems, including the nematode C. elegans. The two major muscle types in C. elegans are the body-wall and pharyngeal muscles. Of the 558 embryonic cells at hatching, 81 are body-wall muscle (another 14 are added post-embryonically) while 20 are pharyngeal [1]. Both of these muscle groups are striated and, based on structure, function, contractile properties and transcriptional ontogeny, the body-wall muscle is homologous to mammalian somatic muscle whereas the pharyngeal muscle resembles cardiac muscle [2–5]. From the earliest genetic studies in the worm, mutations affecting muscle structure and function were isolated and characterized and the cloning of the mutant genes and studies of these gene products provided extensive mechanistic information on sarcomere assembly and regulated contraction [2]. The relatively large mass of muscle in the worm compared to other tissues also permits biochemistry on this tissue type that nicely complements the genetics of the system. Thus, muscle biology in C. elegans, like many other systems, is well understood at molecular, genetic, and cellular levels.
Despite the many advantages of C. elegans for myogenic studies, one of the areas in which the worm lags behind some other systems is the lack of a pure source of muscle cells due to the inability to isolate in sufficient quantity specific muscles cells from the intact animal and the absence of a prolific culture system for the different cell types. One way to overcome these obstacles is to tag muscle cells (or nuclei using INTACT) and isolate them in large quantities by subsequent dissociation and cell sorting [6–8]. Embryonic blastomeres isolated this way can be analyzed immediately or can be cultured for a limited amount of time. However, cultured cells quickly differentiate and fail to proliferate; no stable cell line for any tissue type in C. elegans has been established. Recent reports suggest that some post-embryonic tissues can also be cultured, which may provide a novel and useful source of material for future studies, although no stable postembryonic cell lines have been established [9]. Of course, concerns about the influences of culture conditions and tissue dissociation on cellular functions must be taken into account when studying myogenesis by these methods.
Another approach to isolate nearly pure embryonic muscle cells from C. elegans is the in vivo myogenic conversion of blastomeres, the subject of this methods article. This technique alters gene expression in early C. elegans embryos such that many, if not all, cells adopt the myogenic fate of choice [10, 11]. Depending on the strength and/or penetrance of altered gene function, embryos that consists of 200 to 300 muscle cells, each synchronously executing the differentiation program, are routinely achieved. Importantly, after terminal differentiation, these masses of muscle remaining inside the eggshell are viable for a day or more without signs of degradation. Of course there are several caveats associated with muscle generated by blastomere conversion. For example, these cells are not fully functional and develop in an atypical environment. Muscles generated by blastomere conversion assemble myofilaments, but they lack organized sarcomeres and do not efficiently contract. This reflects the lack of normal extrinsic signaling and polarity cues due to the absence of hypodermis (and perhaps other tissue types) in these converted embryos [12]. Despite these drawbacks, the ability to isolate a large numbers of nearly homogenous muscle tissue in vivo has many advantages for studying muscle development, particularly in the area of gene expression analysis.
2. Methodological Overview
The developmental plasticity of early embryonic blastomeres allows them to be re-programmed into a number of different cell types, including pharyngeal or body-wall muscle, although conversion to the latter is much more easily done and more efficient at present. The most effective way to convert early C. elegans embryonic blastomeres to body-wall muscle precursors is by the over expression of the master myogenic regulator, HLH-1 [11]. HLH-1 is the only worm homolog of the vertebrate Myogenic Regulatory Factors (MRFs), including Myf-5, MyoD, MRF-4, and Myogenin, and it operates at the nodal point of blastomere commitment to the body-wall muscle fate in C. elegans [13]. Thus, HLH-1 expression alone is sufficient to trigger potent positive feed forward and feedback loops that induce myogenesis in most blastomeres (Figure 1). By using the heat shock promoter to drive hlh-1 expression from an integrated transgene, high levels of HLH-1 are generated in all blastomeres, with the exception of the early germline precursors, which are refractory to heat shock promoter activation and/or mRNA translation [11]. Early blastomeres exposed to HLH-1 at the appropriate time continue to proliferate mitotically to ~250 cells prior to exiting the cell cycle and synchronously executing body-wall muscle-like differentiation. Thus, all events downstream of the HLH-1-mediated developmental trigger point can be assayed with these embryos using methods that profile the entire development process or analyze the terminally differentiated and arrested muscle mass.
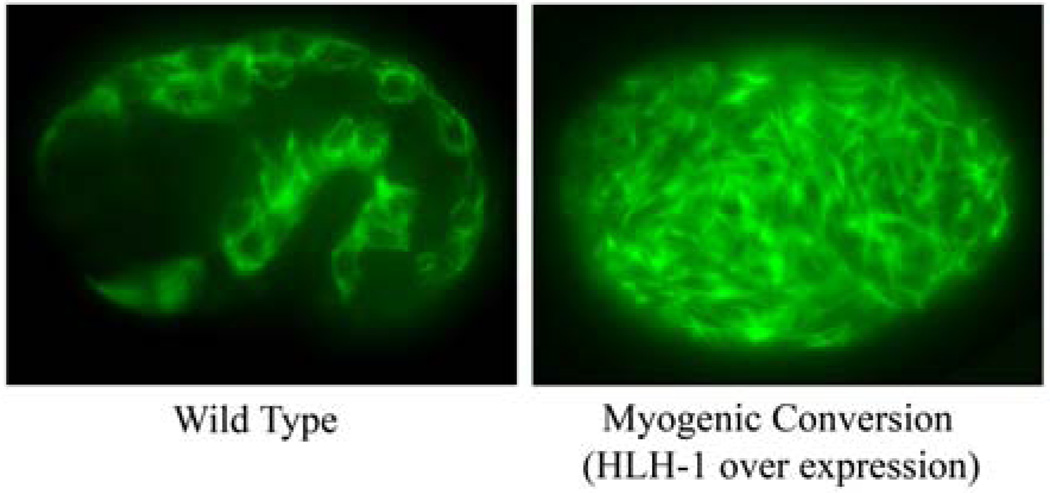
Figure 1.
Comparison of myosin staining in a wild-type and a manipulated embryo that has undergone myogenic conversion. The embryo at left is a left lateral view of a wild-type embryo at the 1.5-fold stage of embryogenesis showing the typical myosin heavy chain antibody staining pattern in the body-wall muscles. At right is a transgenic embryo treated with a pulse of heat shock to induce the expression of the master myogenic regulator HLH-1 resulting in efficient myogenic conversion of most blastomeres. The body-wall muscle-like cells are visible throughout the manipulated embryo with robust myosin levels in disorganized filament-like structures.
A disadvantage of using HLH-1 to convert blastomeres to body-wall muscle is that all upstream developmental processes are missed. The capture of earlier developmental events controlling myogenesis, requires the over expression of upstream regulators of hlh-1. Unfortunately, not all 81 embryonic body-wall muscle cells use a common transcriptional pathway for hlh-1 activation. In the embryonic body-wall muscle derived from posterior founder blastomeres (C & D), there is only one known transcription factor acting upstream of hlh-1 and that is the Caudal-related factor PAL-1 [14, 15]. Over expression of PAL-1 alone converts about one half of all early blastomeres to body-wall muscle-like precursors, with the other half taking on a hypodermal-like fate. This is because the developmental program triggered by PAL-1 is influenced by the activity of another nuclear factor, POP-1, which is related to mammalian TCF/LEF-1 factors. In the presence of low or absent POP-1 activity, PAL-1 directs the body-wall muscle fate whereas in the presence of high POP-1, PAL-1 directs a hypodermal fate [15]. Thus, for PAL-1 to efficiently convert most blastomeres to bodywall muscle, the POP-1 levels must be down-regulated throughout the embryo. This can be done most effectively via RNAi-mediated knockdown in the maternal germline by injection of pop-1 dsRNA into the parental gonad.
The combined action of pop-1 RNAi and PAL-1 over expression effectively converts most early blastomeres to D-like and posterior C-like fates that give rise to body-wall muscle. Using similar approaches, one can manipulate the early embryo into various other founder blastomere cell fates (see Table 1). For example, combinatorial RNAi can be used to generate an embryo of mostly MS-like blastomeres (Figure 2). In this case, mex-1 RNAi converts the AB lineage to MS [16], lit-1 RNAi will raise POP-1 levels [17, 18], converting E to MS, and elimination of PAL-1 activity by RNAi prevents the posterior blastomeres (C & D) executing any fate decision [19]. Thus, a triple mex-1, lit-1, pal-1 RNAi treated embryo at the 8-cell stage has six MS-like blastomeres in the anterior and several unspecified/differentiated cells, along with the germline precursor, in the posterior. Since pharyngeal muscle is a large fraction of the MS descendents, this approach yields embryos greatly enriched for the pharyngeal muscle developmental program.
Table 1.
Induction conditions for myogenic conversion.
| Muscle Type |
Factor | Strain | Embryo Incubation Time Prior to Induction |
Heat Shock Condition |
Gene(s) to RNAi |
|---|---|---|---|---|---|
| Body-wall | HLH-1 | KM267, KM438 | 1- to 4-cell embryos, 60 min, 22°C | 34°C, 60 min | none |
| Body-wall (C & D) | PAL-1 | JA1179, JA1180 | 2-cell embryos, 20 min, 22°C | 34°C, 30 min | pop-1, skn-1 |
| Body-wall | UNC-120 | KM405, KM406 | 1- to 4-cell embryos, 60 min, 22°C | 34°C, 60 min | pop-1, mex-1 or pop-1, pal-1, skn-1 |
| Body-wall | HND-1 | KM403, KM404 | 2-cell embryos, 25 min, 22°C | 34°C, 25 min | pop-1, mex-1 or pop-1, pal-1, skn-1 |
| Pharyngeal (MS) | None | Wild Type | N/A | N/A | lit-1, mex-1, pal-1 |
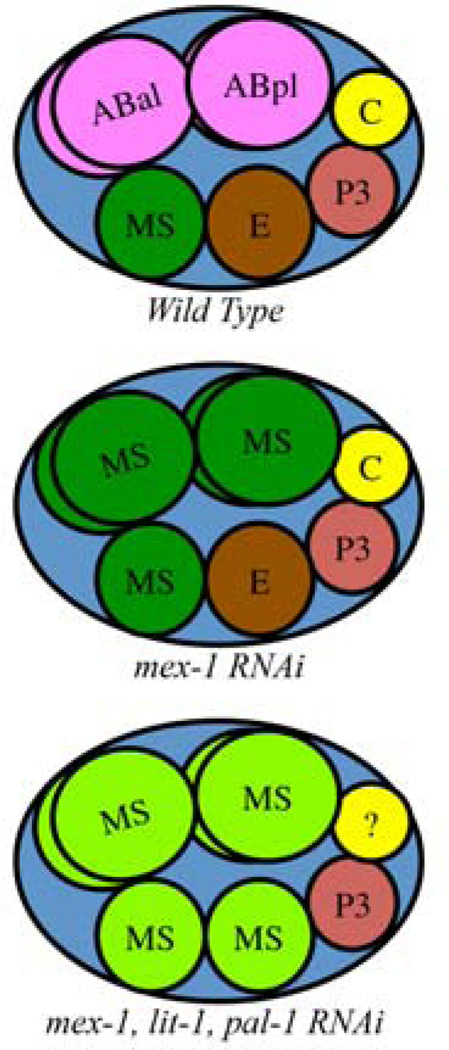
Figure 2.
Wild-type and blastomere transformed embryos at the 8-cell stage of embryogenesis. Top: A wild-type embryo at the 8-cell stage showing the founder blastomeres (MS, E, C, P3) and four AB descendents. Middle: Treatment of the parental hermaphrodite with mex-1 RNAi results in conversion of the four AB descendents into MS-like founder blastomeres that execute an MS-like fate [16]. Bottom: Treatment the parental hermaphrodite with mex-1, lit-1, pal-1 triple RNAi results in an embryo comprised mostly of MS-like blastomeres. The fate of the C founder cannot be specified in the absence of PAL-1 [14].
3. Blastomere Conversion Method
When inducing a myogenic factor to drive myogenesis, developmental timing of induction is critical for efficient conversion of early embryonic blastomeres to muscle. For this reason, it is advisable to stage the embryos carefully by collecting early embryos to chilled buffer, slowing down development and providing sufficient time to pick the needed number of embryos for the experiment. Once the temperature is returned to near 22°C, the embryos resume normal development with the synchronicity dependent on how selective you are in staging the selected embryos. By selecting only 1- or 2-cell stage embryos, all embryos develop nearly synchronously with the timing controlled by the point at which they were returned to higher temperature to resume normal development.
3.1 RNAi treatment
Many manipulations of early embryonic blastomere fates require the knockdown of maternal gene products by RNAi. The most efficient RNAi-mediated knockdown in our hands is by injection of double stranded RNA (dsRNA) into the body cavity of young adults [20]. Feeding RNAi [21] also works, but it is less efficient and results in lower rates of blastomere cell fate conversion. One reason to consider feeding over injection RNAi is that a much larger population of animals can be processed, yielding a much larger population of affected embryonic progeny. Using RNAi injection, we have successfully knocked down up to three genes simultaneously for blastomere conversion assays; we have not tried more than three. The concentration of injected dsRNA is as follows: 1 ug/µl for a single gene, 500 ng/µl each for two genes, and 333 ng/µl each for three genes. A fraction of RNAi-treated embryos should be assayed visually in each experiment for expected phenotypic consequences and/or by hatching rate to confirm the effectiveness of the gene knock downs.
3.2 Isolation of embryos for transcriptional analysis
Wild-type, transgenic, or RNAi treated gravid adult worms are transferred into 30 µl of phosphate buffered saline (PBS) solution in one of two wells of a hanging drop glass slide (e.g. Fisher Scientific, cat# 12-565B). A typical target number of embryos to collect is 80 at the 1- to 4-cell stage, thus 60–100 gravid adults are picked for dissection. The glass slide is kept on an aluminum block that is kept chilled on ice when it is not under microscopic observation. Under a dissecting microscope, cut the animals in half with a surgical blade to release the embryos and return the slide to the chilled aluminum block. Add an additional 30 µl of PBS followed by 70 µl of diluted (1:10) commercial bleach and mix well by stirring with a pipette tip. After incubation for 3 min on the chilled aluminum block, add 80 µl of a 5% bovine serum albumin (BSA) in PBS to neutralize the bleach. Mix the solution well by gentle stirring and incubate on ice for a minute to let the embryos settle to the bottom of solution. Using a drawn out capillary tube and mouth pipetting, collect the appropriately staged embryos and transfer them to 250 µl of PBS in the other well of the hanging drop slide. This entire process should take about 30 min to ensure tight synchrony among the embryos.
3.3 Transcription factor induction
For blastomere cell fate conversions driven by over expression of a transcription factor, it is best to use an integrated transgenic strain to avoid mosaic activation of the transgene among blastomeres (see Table 1). Extrachromosomal arrays can be used, if they are mitotically very stable, but this is not recommended because blastomeres lacking the array are not converted. Prior to transcription factor induction, isolate staged embryos in 250 µl of PBS and incubate at room temperature to allow development to proceed to the optimal stage for the particular factor to effect blastomere conversion (see Table 1). To induce transgene expression, incubate the solution containing embryos at 34°C; a dry heat hybridization oven works well for this. The optimal length of time before heat shock treatment should be empirically determined, although it is typically between 25 min and 60 min. Post-induction, put the embryos at 22°C to develop to the desired stage for analysis. Most embryos left to develop overnight remain as a ball of cells and appear healthy by low power microscopic observation and muscle factor antibody staining.
4. Transcriptional Profiling of Manipulated Embryos
Embryos undergoing nearly synchronous and uniform myogenesis following blastomere cell fate conversion are used to profile transcription by a variety of methods, including expression arrays or next generation sequencing. Although there are examples of expression profiles being derived from only a few embryos [14, 22], we prefer using between 60 and 100 embryos per sample so that variations in blastomere conversion efficiency are averaged in each sample. As with any transcriptional study, confidence in the resulting data is increases by observing a similar profile from multiple, independent biological replicates. A minimum of three biological replicates for each strain and treatment ensures statistical power during subsequent data analysis. In the case of a time course experiment following transcription factor induction, samples of embryos prior to heat shock treatment serve as a useful reference of maternal contributions and for changes in gene expression.
4.1 RNA Isolation
Isolate total RNA from collected embryos (60–100) at the appropriate stage of development. This procedure was developed for C. elegans embryos by Baugh and Hunter [23]. Although only slightly modified below, additional and extensive details related to their protocol are found at <http://www.mcb.harvard.edu/Hunter/ProtData/>. As they suggest, a negative control processed in parallel to the experimental samples is essential when preparing these samples. In our experience, mock preparations (lacking biological material) carried through to hybridization on whole genome chips yielded signals that pass Affymetrix quality control thresholds. Thus, negative controls are essential to discriminate biologically relevant signals from background.
Collect the embryos into 30 µl or less of aqueous buffer (e.g. phosphate buffered saline (PBS)) in a 1.5 ml microfuge tube and immediately add 300 µl TRIzol reagent (Invitrogen, cat#15596-026). Mix thoroughly by shaking and/or pipetting the solution up and down several times.
Add 5 µg of linear polyacrylamide (GenElute LPA, Sigma-Aldrich cat#56575) and mix by vortexing on the highest setting. The LPA serves as a carrier for the nucleic acid through subsequent steps and is preferred over the traditional nucleic acid carriers because of the cDNA PCR amplification steps usually needed for subsequent analysis. Note: If your mouth pipetting skills are good, add the picked embryos with a drawn out capillary tube directly into 30 µl of aqueous buffer containing LPA in the cap of a 1.5 ml microfuge tube that also contains 300 µl TRIzol. After all embryos have been added to the cap, close the cap and immediately vortex and spin to ensure the embryos are in the solution at the bottom of the tube.
Add 60 µl chloroform and vortex 30 sec on the highest setting.
Spin at full speed in a microfuge (~14,000 rpm) for 5 min.
Transfer the aqueous phase to a clean, RNAse-free, 1.5 ml microfuge tube.
Add 0.8 volumes of isopropanol and mix well by repeatedly vortexing on the highest setting repeatedly. Place at −20°C overnight to precipitate nucleic acids.
Spin full speed in a microfuge for 30 min, preferably at 4°C.
Carefully remove the supernatant with a pipet without touching the pellet.
Wash the pellet once with 500 µl 75% ethanol. The ethanol will make the pellet appear opaque and easier to see.
Spin full speed in a microfuge for 10 min and carefully remove the supernatant with a pipet. Briefly spin to top speed and remove the residual ethanol without touching the pellet.
Allow the pellet to air dry for 1–2 min prior to dissolving the pellet in 14 µl of RNAse-free water. Tap the side of the tube and vortex briefly to help dissolve the pellet. Briefly spin to collect the solution at the bottom of the tube and let stand 5 min at which time it is ready to process or freeze for later use.
4.2 cDNA Synthesis and Amplification
Because the amount of total RNA from 50–100 embryos is relatively small, the entire nucleic acid preparation is used in the cDNA synthesis reaction. The method below is a modification of the protocols originally described in the Super SMART PCR cDNA Synthesis Kit (BD Clontech; marketed currently as SMARTer™ PCR cDNA Synthesis Kit) and the published work of Gustincich and colleagues [24].
-
To 14 µl of total RNA in a 200 µl PCR tube, add 3 µl of 12 µM Smart7T27 primer (modified from the original 3’ SMART CDS Primer II from BD Clontech) and 3 µl of 12 µM of SmartIIA primer (BD Clontech) and mix well.
SmartIIA: 5’- AAGCAGTGGTATCAACGCAGAGTACGCGGG
Smart7T27:5’- TGAAGCAGTGGTAACAACGCAGAGTAATACGACTCACTATAGGGAGAAGC(T)27VN (IUPAC single letter code : V(=A or C or G) and N(=A or C or G or T))
Incubate the 20 µl mixture in a thermal cycler machine at 65°C for 2 min and then reduce the temperature to 42°C.
-
Add the following to the reaction:
- 8 µl of 5xFirst-Strand cDNA Buffer (BD Clontech)
- 4 µl of 20mM DDT (BD Clontech)
- 4 µl of 10 mM dNTP (Invitrogen)
- 2 µl of RNase Inhibitor (40U/µl) (e.g. RNase OUT, Invitrogen)
- 2 µl of SMARTScribe Reverse Transcriptase (BD Clontech)
Mix gently by pipetting up and down and incubate at 42°C for 90 min.
Add 2 µl of 0.5M EDTA to stop the reaction.
Purify the cDNA from unincorporated nucleotides and very small (<100 bp) cDNA fragments by phenol-chloroform extraction and standard ethanol precipitation (0.3M NaOAc, pH5.2, and ethanol), overnight at −20°C. Alternatively, one can use a commercial column for purification following manufacturers instructions (e.g. NucleoSpin Extraction II Kit, Clontech), although the efficiency of recovery may be lower than simple precipitation.
Recover the cDNA by spinning at top speed in a microfuge for 30 min (or eluting from column purification) and resuspending the pellet in 64 µl water in preparation for cDNA amplification.
- For many applications, the cDNA products require amplification prior to use. Any amplification has the possibility of introducing a bias in the distribution of products; limiting the number of amplification cycles minimizes this potential bias. For PCR amplification, add the following to your 64 µl cDNA in a 200 µl PCR tube:
- 10 µl 10X GC-Melt solution (BD Clontech)
- 20 µl Advantage® 2 PCR buffer (BD Clontech)
- 2 µl 50X dNTP (10mM) (Invitrogen)
- 2 µl 5’ PCR primer IIA (12 µM) (BD Clontech)
- 2 µl Advantage® 2 PCR Polymerase (BD Clontech)
- 5’ PCR primer IIA: 5’- AAGCAGTGGTAACAACGCAGAGT
- Use the following PCR conditions that include 20 cycles of amplification with increasing extension times (5 sec) per cycle:
95°C 1 min 20 cycles at 95°C 10 sec 65°C 5 sec 68°C 6 min (add 5 sec/cycle) 65°C 5 min Clean the PCR reaction prior to use. If performing analysis by whole-genome expression arrays, purify the reaction products using the Affymetrix double stranded cDNA protocol. For library construction and subsequent next generation sequencing, ethanol precipitate the amplified cDNA products.
5. Concluding Remarks
The techniques in this chapter provide a convenient method to study transcriptional changes associated with myogenesis (and other developmental programs) in C. elegans. The advantages include the ability to profile expression in proliferating myoblasts, differentiated muscle cells, and all stages in between. Moreover, one can control the synchronicity of myogenic induction with the potential to generate nearly homogeneous cell populations in vivo. The disadvantage of all blastomere conversion-based approaches is that development occurs in the absence of other tissue types that would normally provide cues during differentiation. Therefore, some aspects of myogenesis are undoubtedly absent in this system while novel patterns of gene expression may be present. The current description and past application of the method relies on relatively small numbers of embryos. However, next generation sequencing and improved efficiencies in isolating and amplifying RNA from single cells means this technique can be pushed further, likely allowing profiles from single embryos of choice at any stage of myogenesis. Continued refinements in the methodologies will open new windows into myogenic transcriptional profiling and new insights into how muscle cell development is regulated.
Acknowledgements
This work was supported by the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) of the Intramural Research Program of the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Sulston JE, Schierenberg E, White JG, Thomson JN. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev. Biol. 1983;100:64–119. doi: 10.1016/0012-1606(83)90201-4. [DOI] [PubMed] [Google Scholar]
- 2.Waterston RH. Muscle. In: Wood WB, editor. The nematode Ceanorhabditis elegans. Vol. 17. Cold Spring Harbor: Cold Spring Harbor Laboratory; 1988. pp. 281–336. [Google Scholar]
- 3.Okkema PG, Fire A. The Caenorhabditis elegans NK-2 class homeoprotein CEH-22 is involved in combinatorial activation of gene expression in pharyngeal muscle. Development. 1994;120:2175–2186. doi: 10.1242/dev.120.8.2175. [DOI] [PubMed] [Google Scholar]
- 4.Fukushige T, Brodigan TM, Schriefer LA, Waterston RH, Krause M. Defining the transcriptional redundancy of early bodywall muscle development in C. elegans: evidence for a unified theory of animal muscle development. Genes Dev. 2006;20:3395–3406. doi: 10.1101/gad.1481706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Haun C, Alexander J, Stainier DY, Okkema PG. Rescue of Caenorhabditis elegans pharyngeal development by a vertebrate heart specification gene. Proc. Natl. Acad. Sci. U. S. A. 1998;95:5072–5075. doi: 10.1073/pnas.95.9.5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roy PJ, Stuart JM, Lund J, Kim SK. Chromosomal clustering of muscle-expressed genes in Caenorhabditis elegans. Nature. 2002;418:975–979. doi: 10.1038/nature01012. [DOI] [PubMed] [Google Scholar]
- 7.Fox RM, Watson JD, Von Stetina SE, McDermott J, Brodigan TM, Fukushige T, Krause M, Miller DM., 3rd The embryonic muscle transcriptome of Caenorhabditis elegans. Genome Biol. 2007;8:R188. doi: 10.1186/gb-2007-8-9-r188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deal RB, Henikoff S. The INTACT method for cell type-specific gene expression and chromatin profiling in Arabidopsis thaliana. Nat. Protoc. 2011;6:56–68. doi: 10.1038/nprot.2010.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang S, Banerjee D, Kuhn JR. Isolation and culture of larval cells from C. elegans. PLoS One. 2011;6:e19505. doi: 10.1371/journal.pone.0019505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhu J, Fukushige T, McGhee JD, Rothman JH. Reprogramming of early embryonic blastomeres into endodermal progenitors by a Caenorhabditis elegans GATA factor. Genes Dev. 1998;12:3809–3814. doi: 10.1101/gad.12.24.3809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fukushige T, Krause M. The myogenic potency of HLH-1 reveals wide-spread developmental plasticity in early C. elegans embryos. Development. 2005;132:1795–1805. doi: 10.1242/dev.01774. [DOI] [PubMed] [Google Scholar]
- 12.Moerman DG, Fire A. Muscle: Structure, Function, and Development. In: Riddle DL, Blumenthal T, Meyer BJ, Priess JR, editors. C. elegans II. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 1997. Chapter 16. [PubMed] [Google Scholar]
- 13.Krause M. MyoD and myogenesis in C. elegans. Bioessays. 1995;17:219–228. doi: 10.1002/bies.950170308. [DOI] [PubMed] [Google Scholar]
- 14.Baugh LR, Hill AA, Claggett JM, Hill-Harfe K, Wen JC, Slonim DK, Brown EL, Hunter CP. The homeodomain protein PAL-1 specifies a lineage-specific regulatory network in the C. elegans embryo. Development. 2005;132:1843–1854. doi: 10.1242/dev.01782. [DOI] [PubMed] [Google Scholar]
- 15.Lei H, Liu J, Fukushige T, Fire A, Krause M. Caudal-like PAL-1 directly activates the bodywall muscle module regulator hlh-1 in C. elegans to initiate the embryonic muscle gene regulatory network. Development. 2009;136:1241–1249. doi: 10.1242/dev.030668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mello CC, Draper BW, Krause M, Weintraub H, Priess JR. The pie-1 and mex-1 genes and maternal control of blastomere identity in early C. elegans embryos. Cell. 1992;70:163–176. doi: 10.1016/0092-8674(92)90542-k. [DOI] [PubMed] [Google Scholar]
- 17.Rocheleau CE, Yasuda J, Shin TH, Lin R, Sawa H, Okano H, Priess JR, Davis RJ, Mello CC. WRM-1 activates the LIT-1 protein kinase to transducer anterior/posterior polarity signals in C. elegans. Cell. 1999;97:717–726. doi: 10.1016/s0092-8674(00)80784-9. [DOI] [PubMed] [Google Scholar]
- 18.Shin TH, Yasuda J, Rocheleau CE, Lin R, Soto M, Bei Y, Davis RJ, Mello CC. MOM-4, a MAP kinase kinase kinase-related protein, activates WRM-1/LIT-1 kinase to transduce anterior/posterior polarity signals in C. elegans. Mol. Cell. 1999;4:275–280. doi: 10.1016/s1097-2765(00)80375-5. [DOI] [PubMed] [Google Scholar]
- 19.Hunter CP, Kenyon C. Spatial and temporal controls target pal-1 blastomere-specification activity to a single blastomere lineage in C. elegans embryos. Cell. 1996;87:217–226. doi: 10.1016/s0092-8674(00)81340-9. [DOI] [PubMed] [Google Scholar]
- 20.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 21.Timmons L, Court DL, Fire A. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene. 2001;263:103–112. doi: 10.1016/s0378-1119(00)00579-5. [DOI] [PubMed] [Google Scholar]
- 22.Baugh LR, Hill AA, Slonim DK, Brown EL, Hunter CP. Composition and dynamics of the Caenorhabditis elegans early embryonic transcriptome. Development. 2003;130:889–900. doi: 10.1242/dev.00302. [DOI] [PubMed] [Google Scholar]
- 23.Baugh LR, Hill AA, Brown EL, Hunter CP. Quantitative analysis of mRNA amplification by in vitro transcription. Nucleic Acids Res. 2001;29:E29. doi: 10.1093/nar/29.5.e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gustincich S, Contini M, Gariboldi M, Puopolo M, Kadota K, Bono H, LeMieux J, Walsh P, Carninci P, Hayashizaki Y, Okazaki Y, Raviola E. Gene discovery in genetically labeled single dopaminergic neurons of the retina. Proc. Natl. Acad. Sci. U. S. A. 2004;101:5069–5074. doi: 10.1073/pnas.0400913101. [DOI] [PMC free article] [PubMed] [Google Scholar]