Abstract
Recent experimental and theoretical work clarifying the physical chemistry of blood-protein adsorption from aqueous-buffer solution to various kinds of surfaces is reviewed and interpreted within the context of biomaterial applications, especially toward development of cardiovascular biomaterials. The importance of this subject in biomaterials surface science is emphasized by reducing the “protein-adsorption problem” to three core questions that require quantitative answer. An overview of the protein-adsorption literature identifies some of the sources of inconsistency among many investigators participating in more than five decades of focused research. A tutorial on the fundamental biophysical chemistry of protein adsorption sets the stage for a detailed discussion of the kinetics and thermodynamics of protein adsorption, including adsorption competition between two proteins for the same adsorbent immersed in a binary-protein mixture. Both kinetics and steady-state adsorption can be rationalized using a single interpretive paradigm asserting that protein molecules partition from solution into a three-dimensional (3D) interphase separating bulk solution from the physical-adsorbent surface. Adsorbed protein collects in one-or-more adsorbed layers, depending on protein size, solution concentration, and adsorbent surface energy (water wettability). The adsorption process begins with the hydration of an adsorbent surface brought into contact with an aqueous-protein solution. Surface hydration reactions instantaneously form a thin, pseudo-2D interface between the adsorbent and protein solution. Protein molecules rapidly diffuse into this newly-formed interface, creating a truly 3D interphase that inflates with arriving proteins and fills to capacity within milliseconds at mg/mL bulk-solution concentrations CB. This inflated interphase subsequently undergoes time-dependent (minutes-to-hours) decrease in volume VI by expulsion of either-or-both interphase water and initially-adsorbed protein. Interphase protein concentration CI increases as VI decreases, resulting in slow reduction in interfacial energetics. Steady-state is governed by a net partition coefficient . In the process of occupying space within the interphase, adsorbing protein molecules must displace an equivalent volume of interphase water. Interphase water is itself associated with surface-bound water through a network of transient hydrogen bonds. Displacement of interphase water thus requires an amount of energy that depends on the adsorbent surface chemistry/energy. This “adsorption-dehydration” step is the significant free-energy cost of adsorption that controls the maximum amount of protein that can be adsorbed at steady state to a unit adsorbent-surface area (the adsorbent capacity). As adsorbent hydrophilicity increases, protein adsorption monotonically decreases because the energetic cost of surface dehydration increases, ultimately leading to no protein adsorption near an adsorbent water wettability (surface energy) characterized by a water contact angle θ → 65°. Consequently, protein does not adsorb (accumulate at interphase concentrations greater than bulk solution) to more hydrophilic adsorbents exhibiting θ < 65° . For adsorbents bearing strong Lewis acid/base chemistry such as ion-exchange resins, protein/surface interactions can be highly favorable, causing protein to adsorb in multilayers in a relatively thick interphase. A straightforward, three-component free energy relationship captures salient features of protein adsorption to all surfaces predicting that the overall free energy of protein adsorption is a relatively small multiple of thermal energy for any surface chemistry (except perhaps for bioengineered surfaces bearing specific ligands for adsorbing protein) because a surface chemistry that interacts chemically with proteins must also interact with water through hydrogen bonding. In this way, water moderates protein adsorption to any surface by competing with adsorbing protein molecules. This Leading Opinion ends by proposing several changes to the protein-adsorption paradigm that might advance answers to the three core questions that frame the “protein-adsorption problem” that is so fundamental to biomaterials surface science.
Keywords: Review, protein adsorption, blood, solution depletion, QCM, tensiometry, contact angles
1.0 Introduction
A working hypothesis that has emerged as a fundamental biomaterials-surface-science tenet from more than five decades of focused research is that protein adsorption is the first step in the acute biological response to artificial materials. Furthermore, it is widely held that adsorbed protein catalyzes, mediates, or moderates subsequent biochemical reactions that ultimately control biocompatibility [1-10]. In light of these seemingly incontrovertible facts, it is apparent that a full-and-quantitative understanding of how proteins arrive at, and adsorb to, biomaterial surfaces from complex biological milieu is essential to prospective biomaterials design for advanced medical devices. If the number and kind of proteins adsorbed to a surface is not clearly known, then evidence-based biochemical mechanisms of the biological response to materials cannot be responsibly proposed. And if mechanisms of the biological response to materials remain obscure, then structure-property relationships cannot be formulated, leaving biomaterials development dependent on design-directed or trial-and-error approaches [11-13]. Thus, the entirety of biomaterials surface science seems critically dependent on a thorough understanding of protein adsorption.
As a consequence of the above need, the biophysical mechanism of protein adsorption has been the focus of intense research. A vast literature has emerged characterized by limited consensus among investigators and vigorous contention of the most basic aspects of how proteins adsorb to materials immersed in protein solutions [14]. This lack of consensus among numerous, highly-qualified investigators emphasizes a need for continued research on the topic; a need made urgent by the pragmatic importance of understanding fundamentals of protein adsorption toward prospective biomaterial design for numerous advanced medical-device applications.
And yet, in spite of continued need, research into mechanisms of protein adsorption is now widely greeted with palpable distain; first from research-funding agencies and second from researchers themselves, each logically funding and researching other topics with higher odds of success. Many within the biomaterials community are well bored with protein adsorption, choosing to ignore and work around the problem. Indeed, many young biomedical scientists seem oblivious to the fact that early, formative years of biomaterials was nearly entirely consumed by this exceedingly contentious scientific issue that has by now faded into a smoldering background. In spite of this unpopularity, there remain a few researchers from various disciplines who do not give up on a problem that refuses to yield. And new comers are rediscovering unanswered questions in protein adsorption. All are applying increasingly sophisticated experimental/theoretical methods in hope that improved resolution or computer models will provide the much-needed breakthrough, proliferating a burgeoning literature that defies consumption in the process. Remarkably, still others declare that protein adsorption is, in fact, fully understood but simply not subject to any useful control (which is not functionally different than ignoring the problem). Whatever the point-of-view on this “protein-adsorption problem” may be, it seems inescapable that three broad questions of great practical significance to human healthcare remain unanswered, giving rise to related unresolved biomedical issues:
(i) What are the quantitative structure-property relationships connecting surface chemistry/energy to the extent and specificity of protein adsorption?
Issue: The biophysical chemistry of protein adsorption remains obscure with only a few general “rules of thumb” available to guide biomaterials design for specific applications.
(ii) How does protein selectively collect at biomaterial surfaces from multi-component protein solutions such as blood?
Issue: We cannot predict composition of protein layer(s) adsorbed from complex biological milieu.
(iii) Exactly how does adsorbed protein catalyze/mediate/moderate the biological response to artificial materials?
Issue: Explanations of the role that adsorbed protein plays in the biological response to materials are anecdotal in nature and are vague, at best, or incorrect altogether when the surface science of the process is examined in detail [6, 15].
Resolution of these questions and issues would enable prospective, rule-based surface engineering of materials with favorable interactions with proteins and permit engineering of biocompatibility through, in the words of Professor Buddy Ratner, “…by exploiting proteins and cells of the body to meet a specific performance goal…” or as stated by Professor David Williams “…by control of interactions with components of living systems…” [7, 16]. The result would be advanced biomaterials with customized biocompatibility for a wide variety of biomedical devices. This objective cannot be achieved without focused research on the above three categories. Unfortunately, efforts of the few research stalwarts choosing to persist in the face of an indifferent audience are further frustrated by lack of help from the funding agencies. Thus, prospects for near-term resolution of the above-stated problems are not good. A motivation of this Leading Opinion is to advocate for more attention to the protein-adsorption problem from both funding agencies and biomedical researchers.
2.0 Scope of this Leading Opinion
Herein I review and broadly interpret approximately 25 years of my research into blood-protein adsorption. It is emphasized that blood proteins are the exclusive focus of this work and extrapolation of these results to other types of proteins dissolved in fluid phases other than high-ionic-strength aqueous-buffer solutions should be made with great caution. As discussed in the following section, what one observes about protein adsorption is highly dependent on analytical methods applied and, as a consequence, results summarized herein do not necessarily agree with those of many other competent researchers working on the protein-adsorption problem; past, present, and little doubt future as well. But then the collected work of these other competent researchers is itself internally inconsistent, so there should be no surprise about diversity in opinion. Future researchers should be aware at the outset that their results will be woven into the warp of a very complex fabric of information and misinformation, interpretation and misinterpretation.
This Leading Opinion occasionally cites supporting work from the literature with the understanding from the following Section 3 that numerous examples of un-cited opposing literature can undoubtedly be found. Failure to cite opposing literature represents no serious omission because inclusion of contrary citations would greatly complicate text and only serve to amplify the main point that there is little consensus in the protein-adsorption literature to be had. Readers are encouraged to consider the work reviewed herein in its totality and draw conclusions regarding veracity and relevance to biomaterials surface science.
2.1 Experimental Strategy
Experimental measurements applied in my work fall into the Group 2 category discussed below in Section 3.1 and were made using either interfacial tensiometry or the solution-depletion method. These methods were applied to study both kinetics and steady-state adsorption. The overarching motivation of using interfacial energetics and solution depletion was to achieve energy-and-mass balance. Interfacial energetics was used to predict partition coefficients (through surface thermodynamics) and mass measurements were used to predict adsorption energetics (through partition coefficients). The hand-shaking obtained between methods engenders confidence that these two very different methods are in substantial agreement and not seriously affected by conspiring experimental factors discussed below in Section 3.2.
A broad selection of blood proteins spanning three decades in molecular weight (MW) were used in these studies, occasionally supplemented with small proteins such as lysozyme or ubiquitin to sample the MW range below the kidney cutoff (approximately 30 kDa). Adsorption was scaled as a function of protein MW, revealing trends in protein size that were not typically objectives of previous work. Adsorption of particular proteins were scaled as a function of adsorbent hydrophilicity (surface energy) revealing trends in surface energy that were not typically objectives of previous work. Taken together, a rather comprehensive understanding of protein adsorption was obtained.
2.2 Experimental Method
Tensiometry (contact angles and wettability methods, Section 6.1-6.2; see ref. [17] for a review specific to biomaterials and citations therein) measure interfacial energetics of adsorption when scaled as a function of solution concentration [17-19]. Either a manual or automated tensiometer was used to measure interfacial tensions and contact angles of protein solutions at varying concentrations [6, 20-26], using distilled de-ionized water as a reference standard that was further verified using Wilhelmy balance tensiometry (see, for examples, [27, 28] for a statistical comparison of tensiometric methods). Tensiometric data was interpreted using Gibbsian surface thermodynamics (Sections 4.1, 6,1; see also ref. [29] for a particularly lucid treatment) and some modifications thereof [18, 19, 30].
Solution depletion is taken to be an absolute method of measuring adsorbed mass, requiring no complex instrumentation other than that used in measuring unknown protein solution concentration such as UV-Vis spectroscopy or electrophoresis (Section 6.3) [31-36]. As applied herein, the solution-depletion method required no separation of the fluid and adsorbent phases and protein was not labeled. Interpretive theory for solution depletion was simply the arithmetic of mass balance [31].
A wide variety of surfaces were used in these studies; the buffer-air (liquid vapor, lv) interface, self-assembled monolayers (SAMs) supported on gold-coated electronic-grade semiconductor surfaces, silanized glass particles, and sepharose-based chromatographic media. These polymeric materials were chosen as model biomaterials to test various hypotheses (relationships among protein size and adsorbent surface chemistry/energy) that were in a form suitable for interfacial tension or solution-depletion measurements. The hand-shaking between various proteins and surfaces engendered confidence that results were not unique to a particular blood protein or polymeric adsorbent type. However these polymeric test materials did not include minerals (such as mica or hydroxyapatite) which may well exhibit very different protein-adsorption properties. Extrapolation of results summarized herein to adsorbents not falling into the general family of polymeric materials should be made with caution.
3.0 A Reflection on the Protein-Adsorption Literature
Mentioned above is a vast ocean of literature on the subject of protein adsorption that has emerged over the last five decades. Open fundamental issues include the reversibility/irreversibility of protein adsorption [31, 32, 34, 37, 38], mechanism of the so-called Vroman effect [1, 21, 39-58], capacity of proteins to adsorb in multilayers [6, 20, 24, 31-35, 53, 59-68], energetics of protein adsorption [6, 20-26, 28, 32, 59], and the applicability of thermodynamic/computational models [34]. Anyone closely examining this literature in its entirety will be struck by a lack of consensus so extreme that nearly no general conclusions can be extracted. There is general agreement that hydrophobic surfaces have a higher adsorbent capacity than equal surface area of hydrophilic surface [1, 69], however hydrophobic and hydrophilic may be specifically defined [70, 71]. But even here one can find exactly opposite statements. Thus it seems that for every factoid one or more anti-factoids can be found; for every yin there seems to be an equally-compelling yang [14].
Critical analysis further finds that it is not generally possible to style any particular researcher’s work as “right” or “wrong”; the best is the same as the rest – different. Consequently, there seems little purpose in a systematic review of this literature jumble until-and-unless it can be discovered how it comes to be that this collection of work, performed by so many highly-qualified researchers over so many years with increasingly powerful analytical tools, has become so internally inconsistent. The following subsections attempt to find reasons underlying the chaos.
3.1 Two Groups of Protein-Adsorption Research
Protein-adsorption research seems to fall into two broad groups employing two different experimental strategies. Some consensus might be found within each group but certainly not between these groups. The primary reason for this is because Group 1 effectively applies a different understanding of the adsorption process than Group 2.
Group 1 chooses to measure only that fraction of adsorbed protein that remains bound to an adsorbent surface after application of an adsorbent-rinsing protocol. Adsorption here is thus operationally defined as a process that causes protein to become irreversibly or strongly bound to the adsorbent surface. The putative loosely-bound fraction removed by application of a (usually arbitrarily-designed) rinsing step is deemed unimportant.
In sharp contrast, Group 2 defines adsorption as a process that leads to the concentration of protein within a surface region separating bulk solution from the physical adsorbent surface (a.k.a. interphase), in general agreement with classical texts on surface physical chemistry. According to Group 2 think, adsorbed protein subsumes all that has been removed from solution by contact with adsorbent and includes both strongly- and loosely-bound fractions. For this reason, Group 2 insists that the interphase must not be perturbed in any way in the measurement of adsorption [14, 17], again in sharp contrast to Group 1. Techniques that might not overtly perturb the interphase include modern in-situ ellipsometry (adsorbed thickness in contact with solution [72]), the solution-depletion method (mass balance before-and-after contact with adsorbate [31]), interfacial tensiometry (contact angle and wetting techniques [6, 14, 17-26, 28, 30]), quartz-crystal microbalance (QCM, [73-78]), and various types of spectroscopy (especially including attenuated total reflection ATR [79-81] and surface plasmon resonance SPR [82-84]). Citations provided here are arbitrarily selected from many available.
3.1.1 Protein Labels
Group 1 frequently uses radioactive [85-93] or dye [94-96] labels as a detection tool. Protein labels can, and do, significantly affect the structure and amphilicity of the protein which, in turn, affect adsorption outcomes (see ref. [85] and citations therein). The extent to which adsorption measurements are actually affected by labels is hotly contested by advocates, some of whom have invested years of research using I125, typically introduced into the protein by the Chloramine T method [97-99]. Recently it has become popular to use fluorescent dyes (e.g. Alexa Fluor by Invitrogen Corporation) with little-or-no consideration of the potential labeling artifacts.
In fact, it is not so easy to compare experimental methods using labels to those not using labels. Unlabelled protein cannot be used in protocols requiring a label and very few studies have been performed using labeled protein in protocols that do not require label (partly because of safety issues in the use of radiolabels; see as an exception ref. [85]). Hence, the effect of protein labels on adsorption measurements remains in the realm of opinion among biomaterials surface scientists rather than demonstrable fact. But it seems only reasonable to suggest that it is the responsibility protein-labeling advocates to prove that labeling does not strongly influence results rather than placing that burden on those who use neither labels nor adsorbent rinsing. After all, this latter group is innocent of any modification of protein.
3.1.2 Adsorbent Rinsing
Adsorbent rinsing is nearly universally applied when the protein is labeled with radioactive tags or dyes for the simple reason that labeled proteins within the bulk solution must be removed in order to clearly resolve the adsorbed fraction. How effective/efficient various rinsing protocols actually are at the interfacial level appears not to have been systematically studied. Presumably a dip rinse is less effective than a spray rinse which is less effective than sonication in water or buffer or detergent solution.
Adoption of a particular rinsing protocol from the many choices available as a “standard method” to be applied for the sake of consistency is an inadequate experimental strategy until-and-unless it is shown that this standard rinsing protocol works with equal efficiency for all different proteins, protein-solution concentrations, and adsorbent surfaces to be studied. But then one needs a standard rinsing protocol to carry out such a study in the first place. So it seems that experimental verification of Group 1 adsorbent-rinsing methods is caught up in a difficult experimental loop – a standard rinsing protocol is required to test against all different proteins, protein-solution concentrations, and adsorbent surfaces to be studied but development of this standard protocol requires testing against all different proteins, protein-solution concentrations, and adsorbent surfaces. Who knows, could get lucky in just a few turns of a very long loop.
Experimental verification aside, use of adsorbent rinsing implicitly assumes that protein adsorption is inherently strong or irreversible so that adsorbed protein will persist after adsorbent rinsing, as already discussed in Section 3.1 as the feature distinguishing Group 1 from Group 2. This assumption is apparently based on a preconceived notion of how adsorption actually works which, like most preconceived notions, involves an element of logical circularity. Needless to say, perhaps, adsorbent rinsing will only confirm assumption of strongly-bound protein, quite independent of the actual protein-adsorption mechanism, because only strongly-bound protein persists after rinsing. This preconceived notion is locked into a second level of circularity with certain theories of adsorption premised on the idea of irreversible adsorption (see Section 4.5); Group 1 experiment shows that protein is strongly surface bound, because that is all that remains after rinsing, which corroborates theoretical expectations, which reinforces veracity of Group 1 methods. These two levels of circularity have propagated a great deal of misinformation about protein adsorption over decades of research that will likely require decades to overcome because of the tenacity by which both experimentalists and theoreticians cling to the idea that protein adsorption is mediated by strong protein/surface interactions (see further Section 4.5). Experimental proof that protein adsorption is not irreversible, or even strong for that matter (Section 6.3.3), is either ignored or rejected out-of-hand because accepting this fact effectively undermines a very large body of Group 1 research.
3.1.3 Which Adsorbed Protein Fraction is Important?
Quite aside from the merits of different analytical methods, it is not at all obvious which fraction of adsorbed protein is actually most important in the biological response to materials. It may well be found that only the strongly-bound fraction measured by Group 1 is primarily involved in triggering biological cascades such as blood coagulation, complement activation, and immune responses. If so, the total adsorbed fraction purportedly measured by Group 2 studies may not be directly relevant to biomaterial problems after all. Studies of the contact activation of blood coagulation, for example, demonstrating a strong effect of adsorption competition among plasma proteins suggests this is not the case [100], but much more work is required to make a definitive statement in this regard. In any event, as a matter of fundamental investigation into the biophysics of protein adsorption, it seems self evident that all of the adsorbed protein must be included in order to achieve complete energy and mass balance that is so critical to formulating adsorption mechanisms. As a consequence, this Leading Opinion will not further consider results obtained by Group 1 but rather will focus attention on lack of consensus within Group 2 studies, searching for factors underlying discord among many competent Group 2 investigators.
3.2 Lack of Consensus within Group 2
A detailed examination of Group 2 literature strongly suggests that lack of consensus therein arises substantially from a failure to interpret available data on an internally-consistent basis. And, as it turns out, formulating an internally-consistent basis of comparison is quite elusive because of a number of conspiring cross-related factors convolving that which is measured with analytical methods employed. In other words, the bulk of Group 2 literature is “right” on an individual study basis, but these studies defy collective interpretation because of a bias imposed by the measurement method itself; all complicated by numerous factors that affect how protein adsorbs to test surfaces. Perhaps the most perplexing issue encountered in comparing Group 2 methods on an internally-consistent basis is the myriad of analytical methods in widespread use.
Table 1 is an organizational tool intended to help unravel this conspiracy between data and analytics. It is important to note that the clarifying act of separating various factors that influence interpretation of protein adsorption data automatically obscures how separated factors conspire in the final outcome. But once individual influences are identified, it is easier to see how various combinations of factors create interpretive difficulties. The following briefly considers some of these factors in row order of Table 1 with each section keyed to the sequential rows of the Experimental Variable column.
Table 1.
Conspiring Factors that Influence Measurement of Protein Adsorption
| Experimental Variable | Factor | Impact | Comment |
|---|---|---|---|
| Protein | Molecular weight | Size matters | Adsorbent capacity is a function of MW [31, 33, 34, 101] and larger proteins can occupy multiple layers in the adsorbed state [31, 101]. |
| Solution concentration |
Kinetics and subsequent protein- surface interactions |
Solution concentration should be compared to that which saturates adsorbent surface which is seldom reported in literature. Adsorption from concentrated solution is rapid compared to dilute solution. Protein denaturation is relatively rapid on sub-saturated solution with decreasing rate of denaturation with increasing solution concentration [38]. |
|
| Source | Molecular shape/volume |
Blood proteins are oblate spheroids in solution [102-107]. Other proteins (such as environmental or food proteins) may not share this commonality. Degree of glycosylation or de-lipidization. |
|
| Number of proteins in solution |
Adsorption competition | Adsorption from binary solution is vastly more complex than adsorption from purified protein solutions [13, 35]. |
|
|
| |||
| Adsorbent | Hydrophilic v. hydrophobic |
Terminology | Little agreement among investigators on terminology [71]. Broad categorization of surfaces can ignore important differences in surface chemistry. |
| Surface characterization |
Chemistry matters | Surface functional groups have different Lewis acid/base strength that affects interaction with water and proteins [110]. Hydrophilic surface functionalities strong Lewis acid/base strength (electric field) can adsorb protein by ion-exchange not available to surface functionalities such as hydroxyl, carboxyl, ether, etc. [34]. |
|
| ADsorption v. ABsorption |
On v. In | Protein entrapped IN the matrix of porous or water-swollen surfaces can appear to be adsorbed. Adsorption and absorption can be difficult to differentiate and can frequently occur in hydrophilic materials [17, 110]. |
|
|
| |||
| Aqueous Phase | Media ionic strength |
Electrostatic screening | Electrostatic interactions of proteins with adsorbents is shielded in high ionic strength media unless surface functional groups exhibit very high Lewis acid/base properties [34]. |
| Surface hydration | Role of water | High vacuum spectroscopies do not account for hydration reactions. | |
|
| |||
| Protocol | Adsorbent rinsing | Perturbation of the interfacial region |
“Dip-rinse-measure” protocol destroys integrity of the interphase [14] and removes loosely-bound protein that interacts with more strongly bound protein [96], underestimating total amount adsorbed [85, 218]. Efficiency of rinsing at interfacial dimensions is unclear and untested. |
| Protein labeling | Experimental artifacts | Radio [85-93] and fluorescent [94-96] protein labels significantly affect protein structure and adsorption properties. |
|
| Adsorption Isotherm |
Scaling: moles v. weight concentration |
Complete characterization of adsorption requires measurement of a full adsorption isotherm. One or a few arbitrarily-selected solution concentrations is usually an inadequate basis for general conclusions. Should adsorption be compared on a molar or mass basis? |
|
| Gravimetry and Spectroscopy |
Surface Sensitivity and Selectivity |
Gravimetric methods measure the same mass? Evanescent wave methods must capture entire interphase depth and resolve bulk solution contribution. |
|
3.2.1 Protein - Molecular Weight
In working through Table 1, it is useful to bear in mind the simple experimental observation that a greater mass of larger blood proteins (higher MW) adsorbs to a particular fixed adsorbent surface area than smaller proteins (lower MW), but more moles of the smaller protein adsorb than the larger protein [101]. Of course, this is a natural outcome of the fact that bigger proteins weigh more than smaller proteins but more smaller molecules can fit on a unit surface area than bigger proteins. A compelling question arising from this observation, seldom addressed in the literature, asks how adsorption of two different proteins to the same adsorbent should be compared. That is, how should protein adsorption be scaled (see further Section 3.2.2)?
Multi-layer adsorption is among the more controversial issues in the protein adsorption literature (see citations in Section 3). This old controversy is due, at least in part, to the fact that Group 1 analytical methods remove all-or-nearly-all protein putatively adsorbed in secondary layers that are not tightly bound to the adsorbent. Hence, Group 1 does not “see” adsorbed multi-layers and therefore typically rejects the reality of the multi-layering phenomenon. Furthermore, if the preconceived notion is that protein adsorption is an inherently tenacious or irreversible process due to strong protein/surface physicochemical interactions (see Section 4.5), then it follows that multi-layering is not possible because second (or higher order) protein layers are not close enough to be bound by these strong surface forces. In this regard, Group 1 methods engage in a self-fulfilling prophesy already mentioned in Section 3.1.2; protein adsorption is presumed to be controlled by strong protein/surface interactions, which justifies use of Group 1 adsorbent-rinsing methods that measure only strongly-bound protein, which only verify that protein adsorption is controlled by strong protein/surface interactions. This circular logic leads directly to the conclusion that multilayer adsorption is not possible.
Exacerbating this multi-layering issue is the fact that both Group 1 and Group 2 protein adsorption literature substantially focuses on smaller proteins < 200 kDa, effectively inferring protein-adsorption behavior of all proteins from a select group of smaller proteins [101]. So, if multi-layer adsorption occurs only in the case of larger proteins, then multi-layering will not be observed by studying behavior of smaller proteins. Furthermore, if solution concentration (molar or weight) is dilute relative to that required to saturate the adsorbent surface (which cannot be known without an adsorption isotherm), then larger proteins may not adsorb in multi-layers, even though multi-layers might form at or near surface saturation.
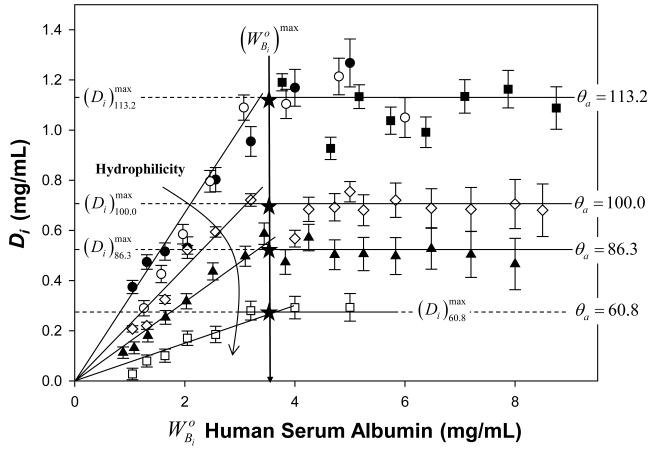
My work comparing adsorbent capacities of proteins with varying MW clearly shows that human blood proteins larger than albumin can occupy multiple layers in the adsorbed state whereas smaller proteins adsorb as a full or partial monolayer at surface saturation. This work confirms that of others reported decades ago, adding that MW plays an important role. Krishnan et al. [20], Noh et al. [31], and Parhi et al. [101] showed that protein layers progressively increased with protein MW nearly 6 fold in going from albumin (66 kDa) to IgM (1000 kDa), suggesting that many protein layers was possible. In fact, these experimental data only show that up to 6 albuminoid layers would be required to constitute the same thickness occupied by IgM at surface-saturation. Kao et al. [36] applied a rational model of how proteins assemble within the interphase of a hydrophobic adsorbent and concluded that, when protein size and different adsorbed-protein packing scenarios are considered, 6 albuminoid layers can be obtained with two authentic layers of large proteins such as fibrinogen (341 kDa), IgG (160 kDa), and IgM (1000 kDa) at surface saturation. Kao’s model accounted for adsorbed mass of proteins measured by both solution depletion and QCM nearly exactly.
Observation of multi-layer adsorption is important because it shows that strong protein/surface interactions cannot be the sole driving force for adsorption as contemplated by various protein-adsorption theories (see further Section 4.5). Protein size, solution concentration, and multi-layer adsorption are convolved variables that require a very careful accounting.
3.2.2 Protein - Solution Concentration
Section 3.2.1 raised the question of how adsorption of different proteins with different MW should be scaled: equal molar- or weight-solution concentration basis? This invites a second question…which solution concentration(s) should be used for protein-adsorption studies? Physiologic concentration or some arbitrary multiple thereof? If the knee-jerk answer is physiologic concentration, then the respondent must explain how physiology and biophysical chemistry of protein adsorption are connected. In fact, these two factors are not directly connected. If the response is some arbitrary concentration, then the respondent must rationalize that particular choice from an infinite selection up to the solubility limit. Clearly, this primitive question of scaling requires an answer before different studies can be compared on an internally consistent basis.
Curiously, solution concentration can affect the fate of adsorbed protein. If solution concentration is such that the adsorbent surface is sub-saturated, then room is readily available for proteins to unfold (denature). By contrast, proteins occupying saturated adsorbents must displace adsorbed neighbors to occupy more room, which requires energy, and this slows the denaturation process. Siegel et al. [38] developed a mathematical model of protein denaturation and used this model to extract denaturation rate constants from experimental data. Not surprisingly perhaps, at least in retrospect, these investigators found that denaturation was much more rapid when protein was adsorbed from sub-saturating-solution concentrations compared to surface-saturating solution concentrations. It thus becomes apparent that comparison of adsorption at different solution concentrations (relative to that required to saturate the adsorbent surface) can lead to conclusions that appear to be in conflict.
3.2.3 Protein - Source
Molecular shape is an important variable and care must be exercised when comparing adsorption behavior of proteins from different sources – blood, environmental, food, etc. Blood proteins are a unique class of proteins in that these proteins can be treated as oblate spheroids in solution as a good first approximation [102-107]; a property that may not be shared among different protein sources (see Section 4.3.1). Although adsorption of blood proteins of different mammalian species appear to be similar [22, 25], different degrees of glycosylation can introduce differences in adsorption behavior (e.g. bovine vs. human serum albumin [108]). Frequently one finds that chemically-treated proteins are used as surrogates for natural forms, such as in delipidized fatty-acid free (FAF) albumin compared to “fraction V” (FV) albumin. These proteins are not the same and comparisons among different classes of proteins or chemically-treated must be made with great caution.
3.2.4 Protein - Number of Proteins in Solution
Adsorption competition between different proteins in solution is very much more complicated than adsorption from single solution (for examples, refs. [3, 21, 33, 46, 79, 109]). Experimental results drawn from single-protein solutions and solutions containing two-or-more proteins are very difficult to reconcile [13]. A reason underlying the complexity of protein-adsorption competition appears to be that individual proteins in a mixture diffuse toward a newly-created interface at a rate dependent upon diffusion constants specific to these individual proteins and specific to individual concentration gradients. This is opposed to all proteins at dissimilar solution concentrations diffusing against a collective concentration gradient [13, 35] (see further Section 6.3.4-6.3.5).
Diffusion constants depend on size according to the Stokes-Einstein-Sutherland equation such that, for approximately spherical blood proteins, the diffusion constant ratio for two proteins (see further Section 4.3.1), predicting that albumin (HSA, 66 kDa) diffuses about 1.3X faster than immunoglobulin G (IgG, 160 kDa). Hence, it is quite apparent that both protein size and solution concentration must be taken into account in any comparison of different adsorption-competition experiments or comparison of adsorption from multiple-protein solutions to single-protein solutions. It is not at all clear how such comparisons can be quantitatively made [13]. Understanding protein adsorption from biological milieu is an important issue of the protein-adsorption problem that currently exceeds our collective reach (see Section 1).
3.2.5 Adsorbent - Hydrophilicity and Surface Chemistry
Another conspiring factor that can cause data to be inconsistently compared is adsorbent surface chemistry. Frequently, adsorbents are categorized as either hydrophilic or hydrophobic. This terminology has been a cause of considerable confusion in the literature because there is no generally-accepted standard of comparison that measures these terms [17, 70, 71, 110]. Although hydrophilic/hydrophobic terminology is now so embedded in scientific lexicon that use of these terms seems unavoidable, the extent of water interaction with a surface is simply not an adequate characterization of surface chemistry.
For example, a fully-water-wettable (hydrophilic) adsorbent may bear surface functional groups that span a great range of Lewis acid/base strength. Strong Lewis acid/base functional groups can exhibit ion-exchange properties and adsorb protein through an ion-exchange mechanism unavailable to weaker acids/bases [34]. In this connection, it is worth noting that muscovite mica, widely used in protein-adsorption studies by atomic force microscopy (AFM), exhibits ion-exchange properties [111] related to the dissolution potassium from the mineral surface [112]. Adsorption by ion exchange is very much different than adsorption to surfaces with “ordinary” surface functionalities such as hydroxyl, carboxyl, ether, etc. where charge interactions are effectively screened in high-ionic strength buffered media typically used in protein adsorption studies [34, 110]. Thus, direct comparison of protein adsorption to mica and other polymeric surfaces should be made with all due caution.
3.2.6 Adsorbent - ADsorption v. ABsorption
Adsorption and absorption are two very different phenomena that are not always easily distinguished, especially for hydrophilic adsorbents into which water can penetrate and swell the adsorbent surface. Absorption can cause entrapment of protein into a swollen surface that might not otherwise adsorb protein, leading to erroneous conclusions about adsorption to hydrophilic surfaces. In principle, protein should not adsorb to water-wetted surfaces because of the energetic cost of displacing surface-bound water (see Sections 6). And indeed protein adsorption onto water-wettable surfaces such as clean glass or oxidized polymers is not detected (see, for examples, refs. [19, 30, 32, 101, 113] and citations therein). Nevertheless, protein ad/absorption onto (into?) hydrogels, especially hydrogels used in contact lenses, has been reported over the years by researchers applying both Group 1 and Group 2 type analytical protocols (see, for example, [114-118]). Blurring the picture further is the finding that hydrogel-like surfaces offer variable resistance to protein adsorption depending on numerous physicochemical details (see, for examples, refs. [119-128]). It seems reasonable to suppose that much of the inconsistency regarding protein ad/absorption on (or in) hydrogel surfaces can be traced to analytical problems. But one thing seems certain: hydrogels are a unique class of hydrophilic materials and protein ad/absorption to hydrogels should not be extrapolated to other kinds of hydrophilic materials that are not water swollen.
3.2.7 Aqueous Phase - Ionic Strength and Role of Water
The vast majority of protein-adsorption studies in the biomaterials literature are performed in high-ionic strength buffer, such as phosphate-buffered saline (PBS). However, chemists and physicists sometimes study protein adsorption in water or low-ionic strength media in search of the very charge interactions that are screened in PBS. Adsorption outcomes thus depend strongly on both surface chemistry and media ionic strength and these factors must be carefully weighed when comparing results.
The greatest oversight of the protein-adsorption literature is the almost universal disregard for the role of water in the adsorption process (see Sections 4.4 and 4.7). This oversight not only greatly effects the formulation and interpretation of protein-adsorption mechanisms, but also affects choice of analytical instruments applied. High vacuum spectroscopies such as Electron Spectroscopy for Chemical Analysis (ESCA, a.k.a XPS, x-ray photoelectron spectroscopy) are favorite surface-science tools because of the exquisite surface sensitivity. Use of these tools in biomaterials science frequently falls into the Group 1 category because surface rinsing, and worse, drying in vacuum, is widely applied. The dry state chemistry observed by these tools fails to measure the great effect surface hydration has on interfacial properties [17, 129].
3.2.8 Protocol - Adsorbent Rinsing and Protein Labeling
Group 1 is not necessarily unique in the application of surface rinsing and protein labels, except that application of labeling and rinsing in Group 2 studies is a bit more subtle. For example, a common use of surface plasmon resonance (SPR) is to flow protein solution over the detector-chip surface followed by flow of buffer rinse to remove bulk solution. Not quite the same as Group 1 “dip-rinse-measure” but a perturbation of the interphase region nonetheless with unknown consequences. Protein labels are used as well. For example, labeled protein is frequently used with internal-reflection infrared (FTIR) experiments, with-or-without surface rinsing. Any time a rinse step is applied, there is a distinct possibility of destroying or perturbing the interphase and removing loosely-adsorbed protein, which is part of the adsorbed fraction. Any time proteins are labeled there is the distinct possibility that amphiphilic properties are significantly altered, and that alteration is not necessarily the same for different sized proteins (more-or-less label per molecule or more-or-less impact per protein).
3.2.9 Protocol - Adsorption Isotherms
Group 2 literature is replete with detailed, painstaking studies performed with one or only a few arbitrarily-selected solution concentrations, typically prepared on a w/v basis (e.g. mg/mL). Measurement of adsorption at one or a few concentrations is insufficient expect for the most qualitative of studies. I strongly advocate that comparison of the adsorption of different proteins to particular adsorbents at fixed adsorbent surface area should be made only on the basis of complete adsorption isotherms. Protein adsorption isotherms are all-too-seldom reported in the literature.
Examination of adsorption isotherms reveals a useful and relevant benchmark for comparing adsorption of different proteins - the solution concentration at which available adsorbent surface area becomes saturated with adsorbed protein (the adsorbent capacity for a particular protein). Without this information, which is almost never reported, there is no way to know what fraction of available adsorbent capacity remains available at a particular solution concentration (mass or molar) or what portion of available protein has become adsorbed – two factors essential to formulating mass balance.
3.2.10 Protocol - Gravimetry and Spectroscopy
Seemingly inconsistent results in Group 2 literature can arise from very subtle interpretive issues among analytical methods. As a relevant example, consider two gravimetric methods of measuring protein adsorption; the solution-depletion method and the Quartz Crystal Microbalance (QCM). The basic idea behind the venerable depletion method was mentioned in Section 2; measure the protein-solution concentration before-and-after contact with adsorbent particles (mass balance) while adsorbent remains in contact with solution (see, for example, refs. [13, 31-35, 101] as recent application to measuring protein adsorption; Section 6.3). This method is one of the most unambiguous methods of measuring adsorption and falls within the gravimetric category because it measures adsorbed mass directly from concentration measurements.
The QCM method uses a crystal oscillator to measure changes in the resonant frequency caused by adsorption onto the oscillator surface, which may bear a well-defined surface chemistry prepared by various surface-engineering methods (see refs. [73-78] arbitrarily drawn from many). QCM is typically considered to be a gravimetric method because it calculates adsorbed mass from frequency measurements using the well-known Sauerberry equation or variants thereof (see, for example, ref. [130] and citations therein).
At first glance, solution depletion and QCM should give identical measures of adsorption because both purport to measure adsorbed mass. That is true with the depletion method but not quite true with QCM [36]. The reason is that QCM actually measures the viscoelastic response of the adsorbed layer that critically depends not only on the adsorbed mass of protein itself but also “trapped” [131] or “intra-layer”[132] or “hydrodynamically coupled”[133] water. Hence, outcomes of the depletion method and QCM turn out not to be directly comparable. QCM measures both mass of protein and coupled water. Depletion measures change in adsorbed mass in terms of change in solution concentration induced by adsorption. Direct comparison of QCM to depletion requires some knowledge of the amount of coupled water, which requires a model of how protein packs within the interphase (see Section 4.8).
Evanescent wave spectroscopies, especially including attenuated total reflection (ATR; see, for examples, refs. [79-81]) and surface plasmon resonance (SPR; see, for examples, refs. [82-84]), have come into routine use in measuring protein adsorption. The analysis depth of these methods must be matched to the thickness of the adsorbed layer(s), which can change depending the size of the protein and the solution concentration relative to that required to saturate the sensor surface area (Sections 3.2.2 and 3.2.3). Here too there are many opportunities for comparing results on an inconsistent basis. Already mentioned is that SPR frequently employs adsorbent rinsing using a buffer flow which can perturb the interphase region, possibly rinsing away adsorbate and thereby underestimating the amount of protein adsorbed.
3.3 Summary
In full view of the discussion of this section and conspiring factors listed in Table 1, it becomes evident why little consensus is to be found in the protein-adsorption literature. The very nature of the adsorption process is in dispute and the way protein adsorption is measured substantially depends on the investigator’s understanding of that process. If the preconceived notion is that adsorption is an irreversible process or a process that leads to a strongly-bound monolayer of adsorbed protein, then it follows that adsorbent rinsing is an acceptable analytical protocol (Group 1). But if the more conservative assumption is that protein is not so strongly bound, or that protein adsorption is possibly reversible, or that adsorption is a process that can lead to adsorbed multilayers, then adsorbent rinsing will be anathema (Group 2). Group 1 is caught up in a self-verifying interpretive circularity from which there is no escape without testing the underlying preconceived notion. Group 2 methods generally, but not in every case, avoids this circularity. But Group 2 applies so many different analytical methods that internally-consistent interpretation is a bad dream turned into a nightmare by the subtle factors convolving that which is measured with analytical method employed. Scientific opinions held by Group 1 and Group 2 researchers are firmly entrenched and there seems little chance that a consensus opinion will soon be formulated. The best that can be expected is that investigators agree to disagree in a cordial, professionally-dispassionate manner. But anyone who has worked in the field of biomaterials for very long knows better than to expect much of that.
4.0 Technical Background
In anticipation of technical issues that frequently arise in discussion of the biophysics of protein-adsorption, this section provides some general background deemed essential to understanding the kinetics and thermodynamics involved. This biophysics nominally includes interactions among the major system constituents – adsorbent surface, protein, and water (specifically the aqueous phase including ions). In turn, these obvious interactive constituents implicate protein/surface, protein/protein, protein/water, and water/surface as the primary pair-wise influences on the protein-adsorption process. This section is organized against this scheme, discussing first general characteristics of the system constituents and then interactions among constituents (protein/protein interactions are beyond the intended scope of this Leading Opinion and will not be considered further herein). But first, it is essential to carefully discuss and define the basics of adsorption because, according to Section 3.1, there is lack of agreement on what adsorption means in biomaterials surface science.
4.1 The Adsorption Process
For the purposes of this Leading Opinion, the term adsorption subsumes all physicochemical events leading to an excess accumulation of either solute or solvent at the interface between two mutually insoluble phases, where bulk-solution concentration is the standard of comparison [17, 29]. Examples pertinent to this Leading Opinion are adsorption to the solution-air (liquid-vapor or lv) interface and adsorption to the solid-solution (sl) interface. Descriptors such as binding, charge interactions, directed assembly, ion-exchange, and the like are nothing more than specific ways surface-active solutes such as proteins might adsorb at an interface (depending on protein and adsorbent surface chemistry) [134] and are not different processes than adsorption [14]. Adsorption can be detected and quantified by a great number of different sensitive techniques mentioned in Section 3 but only a few of which are directly comparable.
4.1.1 Adsorption as Partioning into an Interphase
Adsorption is formally defined as the partitioning of a chemical species (protein in the present case) between a bulk phase and an interface [29]. Both underscored terms need careful definition and consideration. It is expedient to discuss the second term first. An interface is a boundary region between any two mutually insoluble phases. The term ‘boundary region’ conveys the fact that the interface has a finite thickness, at least as large as the molecules that occupy it. When solute molecules are large compared to solvent, it is convenient to use the term ‘interphase’ to stress the volumetric aspect of the boundary region.
The two enduring models of the interphase arise from J. Willard Gibbs [135] and E. A. Guggenheim [136]. Gibbs’ approach has great practical utility but is not as intuitive as that of Guggenheim. Both are very useful in biomaterials surface science and both contemplate surface region as a 3D space (interphase). The primary difference between the Gibbs’ and Guggenheim’s approach lies in the way the chemical potential of solvent is dealt with in development of the thermodynamic equations describing the interphase (see [29] for an exceptionally lucid comparison). Gibbs calculates adsorption excess (above or below bulk concentration) within the interphase and reports this amount per-unit-area of a purely hypothetical 2D dividing plane located somewhere within the interphase by a mathematical convention that eliminates consideration of solvent chemical potential. Guggenheim eliminates solvent chemical potential by clever application of the Gibbs’-Duhem relationship. The two treatments give identical results, at least for dilute solutions.
Gibbs’ approach is frequently misinterpreted to mean that adsorbate necessarily forms a pseudo-2D layer on a planar interface. Although this might occur, especially when there is a strong chemical interaction between adsorbate and adsorbent surface, a 2D layer is not an underlying proposition of Gibbsian surface thermodynamics. Importantly, a 2D concept is inappropriate for proteins because these molecules are quite large in comparison to water molecules or even synthetic surfactants, and proteins can occupy multiple layers in the adsorbed state, as mentioned in Section 3.2.1. For this reason, the Guggenheim interphase construction is a particularly appealing for the study of protein adsorption, but both Gibbsian and Guggenheim approaches are very useful in this pursuit.
The other underscored term from above, partitioning, is to be understood in both positive and negative senses. Typically, adsorption is thought of in the positive sense wherein solute concentrates within the interphase relative to bulk solution. But partitioning can be negative, leading to lower interphase concentrations compared to bulk solution; as occurs, for example, in the partitioning of NaCl to the liquid-vapor surface from concentrated (molar) solutions. Importantly, adsorption always requires that solute and solvent molecules partition in opposite directions because objects cannot occupy the same space at the same time. Thus, proteins accumulating at a surface by adsorption from aqueous solution must displace interphase water and this requires energy to accomplish. Given the relative dimensions of proteins (large, see Section 4.3.1) and water (very small), it becomes immediately evident that adsorption of a protein molecule must displace many hundreds-to-thousands of water molecules depending on protein size (MW).
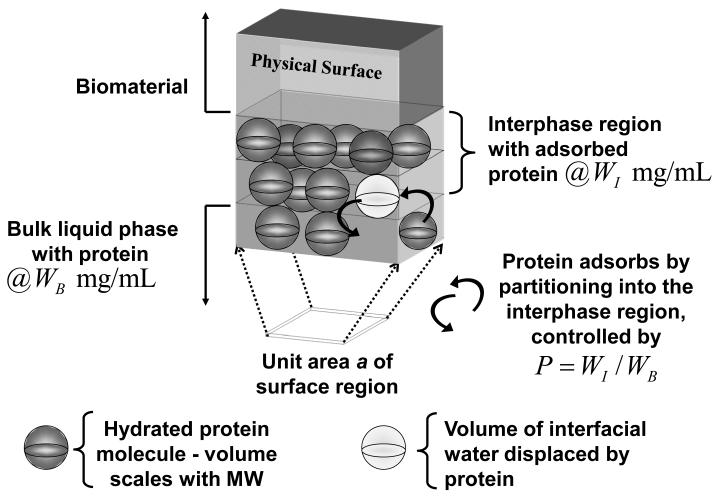
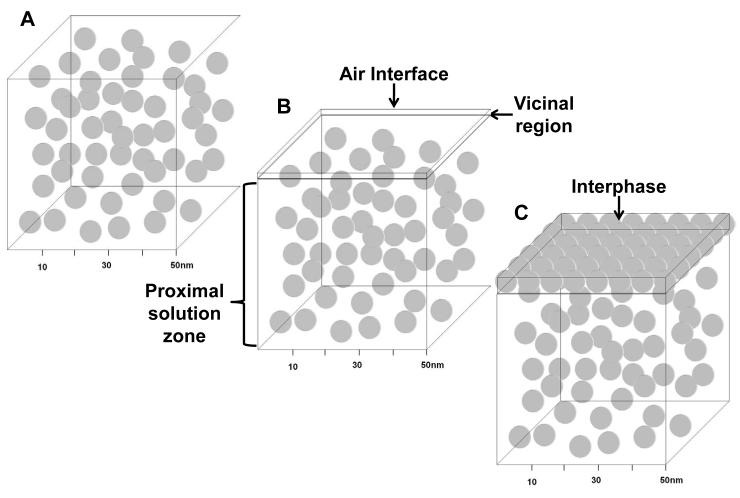
Fig. 1 diagrams partitioning of a protein into an interphase separating bulk solution from the physical surface of a biomaterial, where a protein has been approximated as a sphere (see Section 4.3.1). Arrows indicate that as a protein molecule partitions into (or out of) the interphase, an equivalent volume of interphase water is necessarily displaced. Proteins typically accumulate within the interphase at a concentration WI(mg/mL) that exceeds that of bulk solution at concentration WB. Partitioning is implicit in the thermodynamics of adsorption, as discussed in the following subsection.
Figure 1.
Partitioning of a spherical protein into the interphase separating bulk solution from the physical surface of a biomaterial adsorbent. The thin unit area of surface a in contact with bulk solution is expanded to reveal a three-dimensional interphase containing two hypothetical protein layers occupying an interphase volume VI at weight concentration WI (mg/mL) adsorbed from bulk solution at concentration WB. Curved arrows indicate that a protein partitioning into the interphase from bulk solution must displace a volume of interphase water equivalent to the volume of the hydrated protein because two objects cannot occupy the same space at the same time. The volume of displaced interphase water depends on the size of the protein (MW) and may involve hundreds-to-thousands of water molecules per adsorbed protein molecule. Steady-state is controlled by the partition coefficient .
4.1.2 Thermodynamics of Partitioning
The fundamental equilibrium adsorption equation for a simple two component system (solvent component “1”, solute component “2”; i.e. water and protein, respectively) according to the Guggenheim surface construction [29, 136] reads:
| (1) |
where γ is interfacial energy (ergs/cm2 = mJ/m2 equivalent to interfacial tension, dyne/cm = mN/m) Γ ≡(nI/a) measures the mole number within the interphase nI surrounding a unit area a of adsorbent (moles/cm2), and nB is the mole number within bulk solution. The subscripts B and I track bulk solution and interphase, respectively. Needless to say, perhaps, this equation refers to a circumstance that is highly simplified compared to real biomedical situations in which proteins adsorb to biomaterials. In the real world, the aqueous phase contains a number of different dissolved salts and a biological milieu usually contains more than a single protein, and may contain other different surface-active constituents as well. Laboratory studies of the adsorption of single proteins from purified buffer solutions might approach the simplicity of Eq. (1) but steady state is not thermodynamic equilibrium (see following Section 4.1.3). Nevertheless, it is instructive to further develop Eq. (1) to attain an appreciation for the general biophysics of protein adsorption and to explore the idea of adsorption as a partitioning of solute from the bulk phase to the interphase.
At equilibrium, solute chemical potential μ2 in bulk solution is equal to that within the interphase:
| (2) |
where A is solute activity. Neglecting the term in Eq. (1) in view of the fact that nB,1 >> nB,2 for nearly all practical protein solutions allows the fundamental adsorption equation to be rewritten as:
| (3) |
Eq. (3) thus concludes that or that an incremental change in solution activity leads to an incremental change in interfacial activity. This is the essence of partitioning; a change in bulk-solution concentration induces a change in interphase concentrations. The relevance of this becomes clearer when activity is converted to more intuitive molar concentrations C by using the relationships AI,2≡σI,2XI,2 and AB,2≡σB,2XB,2 with the mole-fraction approximations and appropriate for dilute solute solutions and where σ terms are activity (fugacity) coefficients. Accordingly, and so that:
| (4) |
Eq. (4) defines the dimensionless partition coefficient that measure relative concentrations B within the interphase and bulk-solution phase for solvent and solute. As solute interphase concentration increases due to adsorption (p2 > 1), the solvent concentration must decrease commensurately (p1 < 1, fractional) because two molecules cannot occupy the same space at the same time. In fact, for any particular protein, the amount of water displaced from the interphase upon adsorption will principally depend on the excluded volume of the protein (i.e. molecular size, see Section 4.3.1). Thus, p2 is functionally related to p1 through protein excluded volume. The partition coefficient p2 is identical to the experimental partition coefficient for a single protein adsorbing from purified solution .
Eqs. (3) and (4) emphasize that interfacial energies follow interphase concentrations dictated by partition coefficients for both solute and solvent. The role of partition coefficients in controlling adsorption can be made yet more explicit by writing p1 and p2 in terms of molar compositions and substituting molar partition coefficients into the fundamental equation Eq. (1):
| (5) |
where the volumes of the bulk phase VB and interphase VI have been used to write concentrations explicitly in terms of moles. Eq. (1) then becomes:
| (6) |
Noting that , the interphase thickness, and that (nB,2/VB) = CB,2 leads to simplified versions of Eq. (6):
| (7) |
where the second form of Eq. (7) converts chemical potential to concentrations as detailed in Appendix A. Notice from Eq. (7) that the rate-of-change in interfacial energetics with solution concentration is proportional to interphase thickness ΩI; it takes energy to inflate the interphase region.
Using these same identities, it is evident from Eq. (3) that (Appendix A), and, by substitution into Eq. (7), it is clear that the amount of protein in the interphase depends on how the bulk concentration partitions into the interphase:
| (8) |
When P >> p1, as is the case when protein adsorption is significant, the amount of protein within the interphase is almost a direct proportion of the bulk concentration; Γ2 = PCB,2ΩI. In this case, Eq. (8) predicts a simple Henry isotherm which is, in fact, the general form of isotherms measured by the solution depletion method (see Section 6.3). Thus, experiment vindicates assumptions leading to Eq. (8) and first-principles theory explains why protein adsorption from dilute solutions is controlled by a partition coefficient at steady state.
Failure of a real protein system to follow expectations of partitioning is diagnostic that protein does not adsorb reversibly (i.e. an adsorbed protein does not have a finite probability of desorbing from the surface into solution). Such a circumstance is signaled by P → ∞, meaning that every molecule in solution binds to the surface and CB → 0. Conversely, if a real protein system follows expectations of partitioning, then protein adsorption is most probably reversible. Such a circumstance is signaled by finite partition coefficients (50 < P2 < 500 for different blood proteins [20, 31]). The latter does not necessarily guarantee that every protein in a large collection of proteins adsorbs reversibly from solution, only that the majority of the proteins adsorbed from solution adsorb in a reversible manner.
4.1.3 Applicability of Thermodynamics to Protein Adsorption
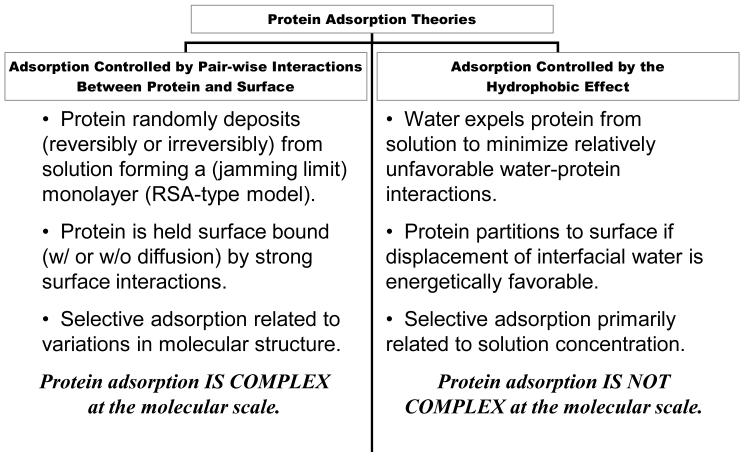
The utility of equilibrium thermodynamics in protein-adsorption research has been a matter of debate for decades within the biomaterials community. Many within the Group 1 research category (see Section 3.1) insist that thermodynamics is useless because protein adsorption is either irreversible or at least does not reach equilibrium. Group 1 researchers thus sometimes claim that analysis of interfacial energies (Sections 6.1 - 6.2), for example, is based on faulty application of thermodynamics and that this is a cause for lack of agreement between those Group 1 and Group 2. But the fault actually lies with Group 1 methods that only measure strongly-or- irreversibly-bound protein (Section 3.2.8). That is to say, the perception that thermodynamics is useless in protein-adsorption research is based on the circularity in reasoning discussed in Section 3.1.2.
In fact, it is easy to show that protein adsorption is “not irreversible” using the solution-depletion method of measuring adsorption isotherms (Section 6.3). However, experimental demonstration of “not irreversible” neither proves thermodynamic reversibility nor achievement of thermodynamic equilibrium, and does not necessarily recommend use of thermodynamics in-and-of-itself. Fortunately, it is not necessary to prove adsorption reversibility or attainment of equilibrium to justify application of thermodynamics. The reason is that, using ordinary bench-top methods of measuring protein adsorption, thermodynamic reversibility and equilibrium is not possible achieve. The test tube is an open system subject to both mass and energy loss/gain. Thermodynamic equilibrium is out of the question and along with that the notion of thermodynamic reversibility. Perhaps careful work with a calorimeter might approach thermodynamic reversibility and equilibrium [66], but such experimental systems are almost never applied in protein-adsorption studies, at least not in the biomaterials world. Thermodynamic ideality is ideal only in the sense that it is very difficult or impossible to achieve. The fact that a chemical system does not adhere to ideality does not invalidate use of thermodynamics as a modeling tool [17]. Were this not the case, thermodynamics would be useless in any practical pursuit, including biomaterials, which of course is not at all the case.
The more interesting question is whether protein undergoes adsorption and desorption in a free partition between interphase and bulk solution. Experimental demonstration of “not irreversible” shows that free partitioning is indeed the case for a variety of blood proteins adsorbed to variety adsorbent surfaces. Now then, if protein adsorption is “not irreversible”, or at least not strongly bound, then Group 1 experimental methods employing adsorbent rinsing are to be held suspect - not use of thermodynamics as a modeling tool.
4.2 The Adsorbent Surface
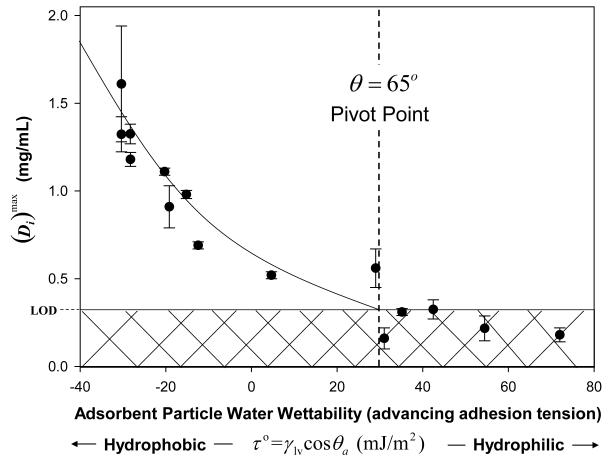
Section 3.2.5 has already introduced essential features of adsorbent surface chemistry which is typically categorized as hydrophilic or hydrophobic, depending on water wetting characteristics. As a matter of practical convenience, this Leading Opinion defines hydrophilic as all solid surfaces that support advancing water (or buffer) contact angles θ < 65° and hydrophobic as solid surfaces supporting θ > 65° [110]. The basis of this definition is the observation that biological responses such as bioadhesion, protein adsorption, and blood plasma coagulation seem to “pivot” from high-to-low or vice-versa within an approximately ±15° range around θ = 65° [11, 14, 69, 137] (note that these citations include reviews of a broad literature).
A notable example mentioned above is contact activation of blood plasma coagulation. It is a common observation in hematology laboratories that coagulation induced by contact with hydrophobic surfaces (as defined above) is so much less efficient than by an equal area of hydrophilic surface that materials effectively fall into one of two groups; efficiently-activating hydrophilic surfaces and inefficiently-activating hydrophobic surfaces [14, 137, 138]. This is perhaps the most profound example of hydrophilic/hydrophobic contrast in the biological response to materials. Pursuant to discussion of Section 3.2.7, hydrogels are purposely excluded from this categorization scheme because of the difficulty in differentiating ABsorption from ADsorption and the inherent ambiguity in rating water-swollen materials on any sensible water-wetting (surface-energy) scale. Another example mentioned above is the adhesion of mammalian cells to surfaces wherein it is generally found that hydrophobic surfaces to not support cell adhesion whereas cells readily attach to, and proliferate, on hydrophilic surfaces [69, 139, 140].
4.2.1 Surface Chemistry
The surface chemistry giving rise to hydrophilicity is sometimes referred to as “anionic” or “cationic” in the literature, presumably referring to a preponderance of negatively- or positively-charged surface-functional groups, respectively. These designations are, by themselves, incomplete specification of Lewis acid/base strength that dominates the interaction of the surface with water and solutes [17, 110].
Extensive studies demonstrate that anionic-hydrophilic surfaces bearing relatively weak Lewis-base functional groups (e.g. oxidized functionalities such as hydroxyl, carbonyl; conjugate bases such as ionized carboxyl, etc.) resist protein adsorption by hydrogen-bonding to water so strongly that protein cannot displace interphase water and enter the adsorbed state (see, for examples, refs. [19, 30, 32, 113] and citations therein; see further Sections 4.7 and 5.2). This finding is in sharp contrast to a common biomaterials anecdote that protein adsorbs to all materials, including hydrophilic materials. This anecdote possibly arises from poorly-executed Group 1 adsorption measurements by which drying of the surface between rinse steps causes denaturation of protein onto to the adsorbent [113] or is the result of protein-labeling artifacts. However this anecdote arises, it is clear that both claims – all and hydrophilic – are too broad to be seriously considered in view of the facts that not “all” have been, or will ever be, fully tested and the notorious lack of a quantitative rating scale for hydrophobic/hydrophilic terminology [71, 110, 141]. Furthermore, there is need for systematic categorization of hydrophilic materials according to class (hydrogel, Lewis acid/base strength, etc.) for a complete comparison of protein adsorption to hydrophilic surfaces. As discussed in Section 3.2.6, for example, extrapolating results of measuring protein adsorption (or absorption) to hydrogels to other non-hydrogel hydrophilic materials is completely unwarranted.
In summary, an adsorbent surface may bear a variable surface density of Lewis acid/base functional groups capable of hydrogen bonding with water, increasing from 0 functional groups/nm2 (a hydrophobic surface such as polyethylene) to a surface packed with functional groups (a hydrophilic surface such as oxidized polyethylene or a perhaps a clean glass surface bearing silanol functionalities). The Lewis acid/base strength of surface functional groups can vary from relatively weak (amine, hydroxyl, carboxyl, etc.) to strong for ion-exchange functionalities such as sulfopropyl (−CH2−SO3−), carboxymethyl (−CH2−COOH), quarternary ammonium (NR4+), and dimethyl aminoethyl ((CH3)2-N-(CH24CH2)−) [34, 142, 143].
4.2.2 Water Wettability
Increasing surface functional group density increases the extent of hydrogen bonding of water to a surface, which manifests in a decreasing contact angle from about 120° (a smooth hydrophobic surface such as polytetrafluoroethylene) to fully wetted 0° (as might be observed on clean glass or oxidized polymers for example). Wettability can be characterized by the Dupre’ work of water adhesion to surfaces W = γlv (1+cosθ), where the interfacial energy of water γlv = 71.97 mJ/m2 at 25 °C and thus 36 ≤ W ≤ 144 mJ/m2 over the 120° contact angle range stipulated above [17]. Use of this Dupre’ work-of-adhesion is limited to surfaces where contact angles are well defined [70, 110], which notably excludes hydrogels that ABsorb water and create a deformable surface. The θ = 65° hydrophobic/hydrophilic dividing line (Section 4.2) corresponds to W = 108 mJ/m2, meaning that approximately 75% of the experimentally-observed W range is deemed hydrophobic and the remaining 25% hydrophilic, again bearing in mind that hydrophilic subsumes a broad range of materials. This closely corresponds to the 70:30 hydrogen-bonding-to-dispersion-force contributions to the self association of water (see further Section 4.4.1).
This categorization of water wetting into two groups implies that surfaces falling on (or near) θ = 65° are neither hydrophobic nor hydrophilic in nature. Water vicinal (in close proximity) to these dividing-line surfaces is neither deficient in hydrogen bonds, as occurs at hydrophobic surfaces that do not hydrogen bond efficiently with water, nor excessively hydrogen bonded to hydrophilic surfaces bearing many Lewis acid/base sites; where efficiency of hydrogen bonding is measured against the hydrogen bonding per-water-molecule that occurs in bulk water [14, 137]. Surfaces near the dividing line thus do not significantly perturb the structure of vicinal water. This vicinal water is thus “water like” with structure and reactivity similar to bulk water, unperturbed by the presence of an imposed surface [14].
The reason that perturbation of water networking at a surface is thought to be influential in the biological response to materials is that solvent properties correlate with the extent of self association by hydrogen bonding [14, 137]. Water is a relatively poor solvent at low temperatures near the density maximum (3.98 °C) because nearly all hydrogen bonds are involved in self association. On the other hand, water steam (100 °C) is quite corrosive because nearly all hydrogen bonds are available to do chemical work. Thus it may be anticipated that changes in hydrogen bonding induced by contact with surfaces at ambient temperatures will have a significant effect on vicinal-water solvent properties [110] which, in turn, will influence the distribution of ions near the water-contacting surface [14, 137, 144], and possibly affect pH within the vicinal-water region. A biological entity such as a protein or a cell entering the vicinal-water region can encounter significantly different chemistry than experienced in bulk solution depending on the extent to which self association of vicinal water has been affected by the presence of the surface. Water vicinal to pivot-point surfaces is chemically similar to bulk water, unperturbed by the presence of the imposed surface [14].
4.3 Proteins - The Blood Plasma Proteome
It is useful to briefly consider the protein composition of blood plasma as a means of putting the problem of protein adsorption from blood plasma in perspective. Hematological research of the late 1800’s, perhaps beginning with Franz Hofmeister’s precipitation of bovine blood proteins with various salts [14, 145] through to that of the early 1900’s, discovered about 30 different proteins in blood. These proteins are now referred to as the “classical plasma proteins” [146]. By the year 2000, about 490 proteins were identified using greatly-improved protein-separation methods. By this decade, more than 1000 blood proteins have been identified using combined chromatography and mass spectroscopy. Individual protein concentrations span more than 10 decades achieving about 8% w/v total plasma concentration. Clearly then, blood plasma is a complex and quite concentrated mixture of proteins. Relevance of the typical laboratory study of single, purified-protein adsorption from buffer solutions at 1/10th to 1/1000th physiological concentrations to the adsorption of proteins from the plasma milieu must be carefully considered before interpreting laboratory studies in terms of biomaterial performance in the clinical setting. Given the insuperable difficulties understanding single-protein adsorption discussed in Section 3, it is clear that biomaterials science has a very long way to go before protein adsorption from blood plasma to biomedical-device surfaces can be fully, or even vaguely, understood.
4.3.1 Blood Proteins as Little Spheres
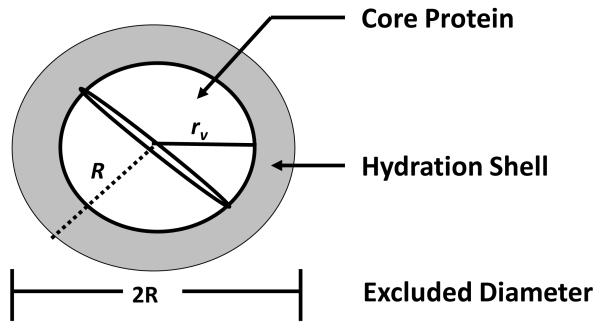
A good first-level of approximation is that blood proteins are oblate spheroids in solution [102-107] with a conserved partial specific volume ν° falling between 0.70 <ν° < 75 cm3/g (protein density falls within a 1.3 < ρ < 1.4 g/cm3 range) [147, 148]. Sphericity is a direct outcome of the fact that the packed polypeptide chain has a spherical radius rν between 1 < rν < 7 nm (rv = 6.72×10−8 MW1/3 for radius in cm and MW in kDa) [102]. In other words, protein volume is directly proportional to MW. Using this radius, it is easy to show that ν° = 0.77 cm3/g would correspond to a perfectly spherical protein [101], which is just outside the experimental ν° range. Proteins are polyelectrolytes with an excluded volume surrounding the protein [149] that forms an osmolaric barrier to the overlapping of hydration shells. This excluded radius is estimated to be about 1/3 larger than the packed radius rν based on electrophoretic mobility and dynamic light scattering (see ref. [20] and citations therein), so that the net radius of a protein sphere R = 1.3rv.
Even if proteins are viewed as rods, or any other shape for that matter, the excluded volume surrounding such an object rotating in free space is spherical. Consequently, when considering early stages of protein adsorption involving diffusion to the surface region and incorporation into the interphase separating bulk solution and the physical surface, it is useful to contemplate proteins as little spheres. For example, as mentioned in Section 3.2.4, the Stokes-Einstein-Sutherland equation predicts that the diffusion-constant ratio for two spherical proteins will follow [13]. Fig. 2 shows that this relationship fits many different proteins (not just blood proteins) spanning a broad range of MW (see inset and refs. [13, 150, 151] for technical details). Clearly, proteins diffuse through solution as anticipated for little spheres. These considerations give rise to a core-shell model of a hydrated protein diagramed in Fig. 3 that is a useful approximation in calculating how protein molecules might pack at an adsorbent surface [36, 101].
Figure 2.
Diffusion coefficient ratio as a function of molecular weight ratio follows the relationship predicted by the Stokes-Einstein-Sutherland equation (line through data) applied to a spherical model of proteins with good fidelity for the proteins listed in the table annotation. Proteins from different sources with different shape exhibit a spherical excluded volume in solution.
Figure 3.
Core-shell model of a hydrated spherical protein with hydrated radius R that is 1.3X larger than the packed amino-acid core with radius rv , creating an excluded diameter 2R. The packed radius scales as MW1/3 so that protein volume scales directly with MW. The hydration shell surrounding a polyelectrolyte protein is an osmolaric barrier to physical contact between two proteins.
Proteins in contact with a surface might become less mobile over time and lose the spherical excluded-volume shape. Change in protein tertiary structure upon adsorption is sometimes referred to as “denaturation” wherein tertiary structure is degraded to a significant extent (see refs. [127, 128, 152, 153] as examples drawn from many). Denaturation can change biological properties and cause the protein to become more-or-less active in promoting a biological response. As mentioned in Section 3.2.2, denaturation occurs at a rate that depends on the extent to which the surface is saturated with protein (room available for the protein to spread). Denaturation has been widely used as an explanation why adsorbent capacity is lower than various theories predict but, careful analysis suggests that this explanation has been over emphasized [101]. Nevertheless, it seems evident that modeling proteins as little spheres is an approximation with a limited timeframe of applicability in the adsorption process. Length of that timeframe is not known and probably is highly dependent on convolved variables identified in Section 3.2.
Thinking of proteins as little spheres is in sharp contrast to a typical literature representation of proteins as irregular-shaped globs, or as tiny crystallographic structures, or something between these views. There are many studies that compute occupied area of adsorbed proteins based specific geometrical shapes and draw conclusions about the distribution of adsorbed conformations – side on, end on, up-or-down configuration (see, for example, ref. [154]). A complete review of this topic is beyond the intended scope of this Leading Opinion, other than perhaps to comment that we have obtained excellent agreement between QCM and solution-depletion measurement of adsorbent capacity of different blood proteins (HSA, 66kDa; IgG, 160 kDa; fibrinogen, 341 kDa; IgM, 1000 kDa) using simple-sphere packing models [36, 101] with no conformational considerations involved. To my knowledge, there has been no systematic comparison of adsorbed-protein-packing models that evaluate the virtues of more complex geometrical considerations over sphere packing. Given the difficulty in comparing different protein-adsorption studies discussed in Section 3.0, it would seem such details are beyond our collective technical reach.
4.3.2 Protein Amphilicity
Blood proteins are comprised of the 20 amino acids of the mammalian proteome, some of which have ionizable (hydrophilic) functionalities. These functionalities are non-uniformly distributed on the solvent-exposed surface which confers a net amphilicity to proteins in solution. That is to say, proteins have certain patches and domains that are well solubilized in water and others that are not-so-well solubilized by water. Those portions of the protein that are relatively poorly-water solubilized tend to be expelled from water solution. Hence, amphilicity is ultimately responsible for the adsorption of proteins from solution to surfaces immersed in a protein solution. This tendency to adsorb from aqueous solution is sometimes referred to as “biosurfactancy” in reference to the similarity in behavior to synthetic surfactants [17]. The experimental observation that proteins adsorb from solution is prima facie evidence that proteins are amphiphiles with biosurfactant properties [6, 18, 20, 22-26, 30, 59]. These biosurfactant properties reduce blood γlv interfacial energies from the ~72 mJ/m2 of pure water (buffer) to about 45 mJ/m2 (see ref. [22] and citations therein). Likewise for serum-containing culture media typically used in the art of cell culture (see, for examples, refs. [69, 155-159]).
4.3.3 A Gedanken Model of Blood-Plasma Proteins
It is useful to generate a mental image of the distribution of proteins in blood plasma. A significant simplification of human plasma, yet out of quantitative experimental reach in modern protein-adsorption studies, is to consider only the five most abundant blood proteins (excluding hemoglobin): serum albumin (HSA, 66.3 kDa, ~45 mg/mL), total various forms of immunoglobulin G (IgG ,160 kDa, ~10 mg/mL), fibrinogen (Fib, 341 kDa, ~3 mg/mL), transferrin (Tr, 77 kDa, ~3 mg/mL), and total immunoglobulin A (IgA, 160 kDa, ~1 mg/mL). It is noteworthy that albumin concentration is close to the solubility limit near 50 mg/mL. Nominal plasma concentrations given above vary significantly [146, 160] depending on age, gender, and mammalian species in consideration. All taken together, it is evident that blood proteins are large biological macromolecules that are surprisingly soluble in water for such high-molecular-weight organic compounds.
Using protein dimensions discussed in Section 4.3.1 together with nominal plasma concentrations, it is possible to render a hypothetical cube of blood plasma that visualizes the five most abundant proteins in plasma solution. Fig. 4 shows the relative sizes of these proteins and computes the number of each protein that would reside in a 50×50×50 nm cube. A much larger cube would be required to capture the 30 classical proteins because plasma concentrations decrease nearly exponentially from that of HSA [146, 160].
Figure 4.
Rendering of a hypothetical 50×50×50 nm cube of blood plasma showing distribution and relative sizes of spheres representing the 5 most abundant blood proteins. A much larger cube would be required to capture all of the 30 classical blood proteins. Numbers of molecules of each protein type that would fall within the cube are listed on the bottom right. Human serum albumin (HSA) greatly out numbers all other blood protein molecules; immunoglobulins type G (IgG), transferrin (Tr), Fibrinogen (Fib), and immunoglobulins type A (IgA).
Using nominal blood concentrations mentioned above, it works out that 10% v/v is a reasonable estimate of the volume fraction occupied by the five classical proteins in blood plasma. At a hematocrit of approximately 45% by volume [161], human blood is then a 45% v/v suspension of formed cellular elements and 10% protein, where water (plus salts) occupies the remaining 45%. Compared to 70% total water content of blood and 60% total whole-body water (including water within cells) [162, 163], it is quite apparent that blood is a highly-concentrated tissue when viewed on a phase-distribution basis. One might conclude by inspection of Fig. 4 that adsorption from plasma would be completely dominated by albumin and that all but IgG and Tr could be ignored. This might be true in some circumstances, but it is not at all obvious how proteins of different size compete in the process of adsorbing from concentrated solution to a particular kind of adsorbent [13, 33] or what rules guide adsorption selectivity as a function of relative protein solution concentration (see further Section 6.3.5). Adsorption of dilute blood enzymes such as blood Factor XII (FXII, Hageman factor) that seems at least partly responsible for sluggish blood coagulation induced by contact with hydrophobic surfaces [164] suggests that adsorption of minor blood constituents in the overwhelming presence of all other blood proteins cannot be ignored.
4.4 Water - The Aqueous Phase
Water is the universal life solvent system in which biology is dissolved or suspended [165]. This statement of obvious truth would be unnecessary were it not for two related facts. The first fact is that the interaction of water with surfaces remains one of the most important and substantially unsolved mysteries of materials science [110, 165]. The second fact is that the role of water in protein adsorption is almost universally ignored. Look in nearly any general textbook on biomaterials (refs. [166] or [167] as examples from many) and one finds various diagrams of protein adsorbing to surfaces with no mention of the aqueous phase whatever. It is as if protein adsorbs from vacuum. Many theories of protein adsorption also do not include any statement about water. It is all too easy to ignore water because water is ubiquitous and this familiarity tends to obscure the importance as a life solvent system [165].
4.4.1 Self Association
The single physicochemical feature that distinguishes water from all other fluids, and property that makes water such a difficult solvent system to fully comprehend, is the extent to which water self associates through hydrogen bonding [14]. Effectively, water is a transiently-connected network of molecules with something like 75% of all molecules in room temperature liquid connected to three-or-four nearest neighbors [165, 168]. At approximately 2 kJ/mole (0.5 kCal/mole) of hydrogen bonds in liquid water [169], there is a strong energetic preference for the self associated state due to hydrogen bonding (which can be treated as a partially electrostatic and partially chemical Lewis acid/base (AB) interaction [14]). Any solute imposed into water interferes with this hydrogen-bonded network to some extent and will tend to be excluded from water solution by what amounts to be the hydrophobic effect [14, 137, 170]. That is, water expels the solute from solution. This applies to ions of the lyotropic (Hofmeister) series as well [144].
Water also self associates through Lifshitz-van der Waals (LW) dispersion-force interactions, as is true for all condensed matter. The extent of water self association due to LW interactions can be estimated from interfacial-tension-component theory and measurement of interfacial tensions between water (phase 1) and saturated hydrocarbons (phase 2) for which no AB interaction across the interface between phase 1 and 2 (12) is possible. Choosing hexane as an example, it is found experimentally that and [171, 172]. Using for water at 20 °C in the traditional geometric-mean equation () [171, 173, 174] calculates that , or about 29% of the total interfacial tension of water. LW interactions are thus hardly negligible in water, in part because water is such a small molecule that packs efficiently into a rather dense liquid phase. We return to this point in following sections considering water interactions with solutes and surfaces.
4.4.2 Role of Water in Protein Adsorption
Inclusion of water in computational models of protein adsorption is, at best, very challenging. Water is such a small molecule (approximately the dimensions of atomic oxygen, only about 0.25 nm in the longest direction) that the mole ratio of water-to-protein is huge. Thinking of a 45 mg/mL HSA solution (such as in blood plasma) as a specific example, the water:protein mole ratio works out to be 8×104 meaning that there are nearly 105 water molecules for every albumin molecule. This perspective invites an important question that asks just how much water must be included in models of protein adsorption: None at all? Just the outermost hydration shell surrounding protein and adsorbent surface? Or perhaps the entire volume of water displaced by a protein adsorbing to a surface from aqueous solution?
The answer to these questions is not clear because details about the structure and reactivity of water vicinal to different kinds of surfaces and solutes remains the subject of active experimental and theoretical research. This “depth of the surface zone of a liquid”, as it was titled by Henniker in 1949 [175], has actually been a matter of intense speculation and research for more than 200 years, harkening back to the beginnings of chemistry as a discipline (see refs. [14, 110, 137] for reviews and especially [14] for biomedical interpretation). The problem is that much more complex for biological fluids where protein adsorption is involved. If the depth of the interphase region is not known and the depth over which vicinal water properties might be different than bulk solution, then it is not at all clear how to compute the energetic impact of displacement of interphase water by an adsorbing protein (see further Section 4.7.2). But it certainly seems safe to conclude that some water must be included in modeling the role of water in protein adsorption. The following logic suggests that much more than just water of hydration (of both protein and surface) must be considered.
Water in direct contact with an adsorbent surface is physicochemically bound to that surface to an extent that depends on the adsorbent surface chemistry/energy (Section 4.2.2) [17, 110]. In turn, this vicinal water is transiently hydrogen bonded to proximal water molecules which are hydrogen bonded to adjacent water molecules and so on, networking out into solution. Hence, adjacent water is bound to proximal water which is bound to vicinal water which is bound to the adsorbent surface. So it follows then that water within the near surface region (interphase) into which protein partitions is indirectly bound to the adsorbent surface to an extent that presumably depends on adsorbent surface energy (see Appendix B of ref. [34] for a slightly more quantitative discussion).
Thus, displacement of water from a surface by an adsorbing protein is not without energetic consequence. Unless there are specific reasons to the contrary, it therefore seems necessary to consider the energetics of displacing a volume of water at least equivalent to the volume of the adsorbing protein in modeling the protein-adsorption process. This amount of displaced water depends critically on protein size (MW, Section 4.3.1), how many protein layers are occupied in the adsorbed state (which depends both on solution concentration and protein MW), and how protein molecules are organized in adsorbed layer(s) [36]. Inclusion of this many water molecules is quite outside the computational power available to most theoreticians (see further Section 4.8.3).
The unique properties of water strongly influence pair-wise interactions mentioned in the introduction of this section and as further elaborated below. As a consequence of these factors, it becomes apparent that water is a (the) significant driver of protein adsorption and can be ignored only at risk of completely misunderstanding a (the most) fundamental aspect of protein adsorption. This Leading Opinion has it that ignoring the role of water in protein adsorption has been a fatal flaw in much of the existing literature, further invalidating Group 1 results along with the many interpretive theories of protein adsorption based on Group 1 data. Because water is so easy to overlook and so difficult to incorporate into conceptual paradigms, ignoring water most probably will contaminate future work as well.
4.5 Protein/Surface Interactions
Protein/surface interactions have been a primary focus of modern theoretical work because the energetics involved have long been presumed to be large compared to thermal energy (see, for example, ref. [176] and citations therein; also theory underlying the popular Random Sequential Adsorption (RSA) model [177, 178]). Quantitatively, the phrase “large compared to thermal energy” depends on the type of theory applied and specifics considered, so considerable care must be exercised in comparing different expressions for interaction energetics used in the literature (e.g. free energies vs. potential energies). A limited survey of the literature suggests that typical protein/surface interactions contemplated by theory are of the order −20RT (free energy, −50 kJ/mol) and −80RT (potential energy, −200 kJ/mol) [179-182]; where R is the gas constant, T is Kelvin temperature taken to be 298.15 °K, and the negative sign denotes attraction between protein and surface.
Interestingly, in this regard, the apparent free energy of protein adsorption is not found to be large compared to thermal energy experimentally. Indeed, ~ −5±1RT (12.5 kJ/mole) for the adsorption of different proteins adsorbing to different hydrophobic surfaces (buffer-air interface, self-assembled monolayers, silanized glass, and octyl sepharose (OS) chromatographic packing; see Section 6 and ref. [32] and citations therein for more discussion). This modest free energy is entirely consistent with the generic, weak “biosurfactancy” observed for different purified proteins and protein mixtures such as blood plasma/serum (see refs [6, 20-25] and citations therein), as well as the commensurately small quantity of protein that adsorbs to various materials (generally falling within the 2-10 mg/m2 range for hydrophobic surfaces depending on protein MW [101]). In some applications, such as the assembly of colloidal units into various kinds of macroscopic structures, −5RT may be considered to be quite large. But −5RT is quite small in the world of biological binding constants. It is also observed that protein adsorption decreases with increasing adsorbent hydrophilicity or, in other words, increases to zero with increasing hydrophilicity (see refs. [26, 32, 101] and citations therein).
Thus it appears that experimentally-measured standard free energies of protein adsorption to hydrophobic surfaces are at least 4X smaller than theoretical protein/surface interaction energies. Clearly, for both experiment and theory to be simultaneously correct, one-or-more of the other pair-wise influences mentioned in the introduction of this main section must involve compensating interaction energetics, leaving a small net-negative residual out of a presumably large attractive protein/surface interaction-energy budget.
Or it may be true that energetics of protein/surface interactions are not, in fact, large compared to thermal energy. The following attempts to decide between these two alternatives, leading to the conclusion that protein/surface interactions are not large compared to thermal energy unless the adsorbent surface is surrounded by a strong electric field, such as occurs in the case of ion exchange resins for example [34], or perhaps if protein adsorption is carried out in pure water where surface charges are not screened, as occurs in high-ionic strength buffer solutions that are most relevant in the biomedical context.
4.5.1 Physicochemical Interactions
It is of interest to contemplate the sorts of interactions that are possible between a protein and a surface. A good deal of physical-chemistry rigor is required to probe this subject in depth, but the basics can be discovered by considering just the two general sorts of interactions – Lewis acid/base (AB) interactions and Lifshitz-van der Waals (LW) interactions mentioned in Section 4.4 in reference to water. In this simplified approach, any kind of chemical interactions involving sharing of electron density (polarity) is categorized AB and any kind of electrodynamic interaction (dispersion forces) are categorized as LW. Electrostatics is not relevant in ionic solution and repulsion can be neglected at ambient pressures.
Significant AB interactions between protein and adsorbent requires a superficial density of AB-type functional groups on both protein and adsorbent sufficient to affect over-and-above ubiquitous LW interactions considered separately below. Such adsorbents would fall into the hydrophilic category as defined in Section 4.2 because water would hydrogen bond to such surfaces. In other words, AB interactions between protein and adsorbent will be significantly larger than LW only when the adsorbent is hydrophilic. And, in order for protein to engage in AB reaction with the adsorbent surface, the water between protein and adsorbent must be displaced in order to get the protein close enough to the surface to engage in the AB interaction. This requires that the energetics of protein/surface AB interactions must be greater than water/adsorbent interactions.
These latter conditions are not likely to be met in most practical biomaterials applications. First of all, energetics of displacing water (surface dehydration) is quite high and overall free energy of protein adsorption quite low (see further Section 4.7). Second, the solution phase of interest in biomaterial applications is high-ionic strength fluid such as blood (0.1 M NaCl plus a multitude of other ions), meaning that charged groups on both surface and protein are screened to a Debye length of less than 0.5 nm [183]. Counter charges thus block AB interaction of a protein with charged groups on an adsorbent surface. These factors undoubtedly contribute to the fact that adsorbent capacity decreases as a function of hydrophilicity to immeasurably small amounts near the 65° dividing line defining hydrophilic and hydrophobic (see Section 4.2 and refs. [26, 30, 32, 101]). The only exception to this generality seems to be when the adsorbent surface bears ion-exchange functionalities with extraordinary Lewis AB strength [34].
Setting this exceptional case aside for the moment, it seems apparent that if AB interactions were, in fact, the significant energetic driver of protein adsorption, then (ordinary) hydrophilic surfaces would have higher (not lower) adsorbent capacity than hydrophobic surfaces. The fact that experiment reveals this not to be true is persuasive evidence that there are no strong AB interactions between surfaces and protein, at least as this applies to protein adsorption from buffer solution to adsorbents bearing relatively weak AB functional groups. That is to say, protein/surface interactions are not typically large compared to thermal energy due to AB type chemistry.
Given the electronic composition of matter it is evident that LW interactions between proteins and surfaces must occur to some extent, independent of adsorbent hydrophilicity. As it is observed experimentally that hydrophobic surfaces have much greater adsorbent capacity than hydrophilic surfaces, it seems safe to conclude that LW are responsible for this high-adsorbent capacity because hydrophobic surfaces do not bear surface functional groups that can bond with protein. Protein adsorption to hydrophobic surfaces requires only displacement of relatively weakly-bound vicinal water and there is no electrical double layer to block close approach of a protein and a hydrophobic surface (although there is some evidence that aqueous surfaces are inherently charged, see refs. [184, 185] and citations therein). Even so, energetics of LW-mediated protein/surface interactions forces cannot be large relative to thermal energy because LW forces are of the same order of magnitude as thermal energy at ambient temperatures. That is to say, surfaces exhibiting the highest adsorbent capacities attain this capacity with the weakest of interaction energetics.
These qualitative arguments taken together lead to the conclusion that protein/surface interactions are not generally large compared to thermal energy, except perhaps in the case of surfaces bearing very strong Lewis AB functional groups. It may well be true that protein/surface interactions are significantly larger than thermal energy in the vacuum of a computer simulation leaving out water and ions dissolved therein but, for practical biomaterial applications, quite the opposite seems closer to the truth.
4.6 Protein/Water Interaction
Amphiphilic properties briefly mentioned in Section 4.3.2 are responsible for relatively weak protein biosurfactancy (compared to many synthetic surfactants). Amphiphiles such as proteins (or ions for that matter [144]) are excluded from solution to recover hydrogen bonds among water molecules otherwise separated by solutes in solution by what amounts to be the hydrophobic effect (see further Section 4.4.1). Repetition of the 20 amino acids of the mammalian proteome in construction of different proteins causes blood proteins to have similar amphilicity (see further Section 6.1). Consequently, the free energy change associated with this hydrophobic effect is approximately constant (on a weight basis) for all globular proteins because the partial specific volume v° of proteins falls within the conserved range of 0.70 ≤ v° ≤ 0.75 cm3/g protein mentioned in Section 4.3. In other words, the number of hydrogen bonds recovered by water by expulsion of protein is approximately constant per-gram-protein because protein volume/gram (density) is nearly constant for any particular blood protein or mixtures of blood protein). Thus it happens that the interfacial energetics of protein adsorption are very similar among different purified proteins, binary mixtures, and plasma/serum from different mammalian species when scaled on a w/v concentration basis [6, 18, 20, 22-26, 30, 59].
The fact that water/protein interactions and water/surface interactions dominates protein adsorption greatly simplifies the physical chemistry involved. The primary driving force for adsorption, expulsion from solution, is effectively generic for all proteins when scaled by protein mass. Likewise, surface interactions are generically related to ubiquitous LW interactions, with little-or-no specific chemical AB interactions (Section 4.5). Were this not the case, the protein-adsorption problem would be complex at the molecular scale, requiring detailed microscopic information at the level of protein-surface-water interaction potentials (computation) that would be different for each protein and surface combination under consideration (see further Sections 6.1.4 and 6.2.4, refs. [186-188] and citations therein). Such complexity would paint a rather gloomy forecast for understanding protein adsorption from complex biological milieu such as blood containing a thousand different proteins with widely-varying natural abundances (Section 4.3).
4.7 Water/Surface Interaction
Section 4.4 qualitatively discussed the important role of water in the protein adsorption process in terms of the energetic cost of displacement of vicinal water vicinal by adsorbing protein. This section quantitatively expands this theme by calculating this energetic cost in tangible energetic terms using a thermodynamic theory developed by C. Extrand [189, 190]. Extrand theory permits conversion of wetting measurements in units of energy/unit area (as in mJ/m2 for example) to kJ/mole-of-interactive-sites.
4.7.1 Energetics of Surface Dehydration
Extrand developed the concept of “molar surface area” [190] by envisioning a polymeric surface with smooth area A comprised of a repeat unit with molecular weight Mo. The surface area occupied by a repeat unit Asite is the 2/3 power of site volume (Vsite)2/3 which, in turn, is related to Mo and density ρ by Vsite = (Mo / ρNA), where NA is the Avogadro number. The molar surface area is obtained by multiplying by NA, yielding with units of cm2/mole-of-surface-sites. For SiO2 glass, as an example, with ρ = 2.2 g/cm2 and Mo = 60.1 g/mole monomer unit, it works out that = 7.7×108 cm2/mole-SiO2-sites (1.3 nmole-SiO2-sites/cm2).
Extrand applied this molar-surface concept in deriving a relationship between the free energy ΔGE of wetting a mole of surface sites and the advancing contact angle θa of a wetting liquid on that surface, concluding that in units of kJ/mole of-surface-sites [189]; where the “E” subscript identifies the free energy as that calculated from Extrand theory and RT the product of the gas constant and Kelvin temperature. It is thus evident that ΔGE scales directly with temperature (which is taken to be 298.15 °K in the following, see ref. [11] for further discussion of Extrand theory related to biomaterials). This equation was used to calculate free energy as a function of cosθa shown in Fig. 5. ΔGE was interpreted as the strength of interaction of the wetting fluid with the surface and was equated with liquid-solid adhesion [189], or (negative of) the work required to remove a wetting fluid from the surface (dehydration energy).
Figure 5.
Free energy of dehydrating a surface ΔGE as a function of surface site wettability measured by cosine of the advancing contact angle, cosθa. Wetting energy increases as surface sites become more hydrophilic (left-to-right on the abscissa), crossing a boundary near a θa = 65° nominal contact angle (cosθa = 0.42, dotted vertical line within the gray band) that differentiates hydrophobic from hydrophilic and where biological responses to materials pivot from high-to-low or low-to-high (see Section 4.2). The wetting energy corresponding to this pivot point (dotted horizontal line) defines hydrophilicity on an energetic basis: hydrophilic wetting expends > 1.3 kJ/mole-of-surface-sites whereas hydrophobic wetting expends < 1.3 kJ/mole-of-surface-sites (see vertical arrow annotations, left-hand axis). The dashed line annotation running diagonally along the ΔGE trend emphasizes the linear-like increase in ΔGE through the hydrophobic range of water wettability which increases sharply increases through the hydrophilic range.
In agreement with chemical intuition, the energetic cost of displacing water from a surface (dehydration) rises sharply with hydrophilicity of the surface sites measured by the advancing contact angle cosθa. Wetting energetics rise in a linear-like way (dashed line) with cosθa through the range considered to be hydrophobic (cosθa < 0.42, θa > 65°; see Section 2.3). Wetting energetics rise more steeply within the hydrophilic range (cosθa > 0.42, θa < 65° ). Extrand’s wetting theory thus affords an additional specification on hydrophilicity; hydrophilic wetting expends > 1.3 kJ/mole-of-surface-sites whereas hydrophobic wetting expends < 1.3 kJ/mole-of-surface-sites (see arrow annotations).
It is of interest that the transition from the low slope, linear-like increase in ΔGE (dashed line) to the more sharply rising bend in the curve of Fig. 5) occurs within the pivot-point range where a sharp change in the biological response to materials occurs (gray band, see Section 4.2). In particular, protein adsorption is observed to decrease to immeasurably small amounts within this pivot-point range (see, for examples, refs. [19, 30, 32, 101, 113] and citations therein). Apparently, at the hydrophilic/hydrophobic dividing line, the energetic cost of displacing interphase water nearly equals the net energy gained by partitioning a protein from bulk solution into the interphase and the adsorption process becomes energetically unfavorable, as will be discussed in more detail in Section 5.2.
4.7.2 Displacement of Interphase Water by Adsorbing Proteins
A principal interest in hydration energetics discussed in the preceding sections is calculation of the energy expended in the displacement of interphase water by a protein molecule partitioning into the interphase, briefly discussed in Sections 4.4.1 and 4.4.2 in reference to Fig. 1 (a.k.a. surface dehydration, ΔGdehydration; see Section 5.2 and refs. [31, 32]). Even knowing hydration energetics, however, this calculation is not at all straightforward. The complicating factor is that displaced interphase water almost assuredly involves more than just the first few water layers vicinal to the adsorbent surface that are directly affected by hydration reactions, possibly including the entire excluded volume of the protein (Section 4.4.2).
At a partial specific volume ν° = 0.75 cm3/g protein (an arbitrary choice lying between the low-end experimental range and the high-end calculated for a perfectly spherical protein; see Section 4.3.1), the molar volume (cm3/mole for MW in kDa). By comparison, the molar volume of water is if water density ρHOH = 1 g/cm3. It follows that . Thus, 625 moles of water are displaced per mole of a small protein such as lysozyme (15 kDa) and 41,700 moles of water are displaced by a mole of a large protein such as IgM (1000 kDa). Clearly neither the volume of the interphase nor amount of water within that interphase can be the same for different size molecules [36]. It further seems reasonable to assume that the molar dehydration energetics cannot be the same for different proteins.
The nagging question is how much energy is required to displace both vicinal water and interphase water proximal to these vicinal-water layers? Interphase water is in a different chemical state than bulk water, not only because it is proximal to the physical surface but also because the interphase water concentration can be substantially lower than bulk water concentration (because interphase protein concentration is substantially higher than in the bulk phase). Thus, the chemical potential of interphase water must be different than that of bulk water. How much this contributes to the energetic cost of interphase water displacement and how this depends on protein size is entirely unclear. Hence calculation of the energetics of interphase dehydration remains an open question.
4.8 Summary
Protein adsorption is controlled by a combination of protein/surface, protein/protein, protein/water, and water/surface interactions. Each of these interactions is complex and the biophysical chemistry of each active areas of research. As a consequence, combination of these interactions into an overall biophysical mechanism of protein adsorption remains at the cutting edge of biomaterials surface science. Section 5 attempts to put these pieces of the protein-adsorption puzzle together into a reasonably straightforward kinetic and thermodynamic description appropriate to the adsorption of blood proteins that accommodates experimental measurements discussed in Section 6. But before moving to these specifics, it is instructive to expand the gedanken model of blood-plasma proteins of Section 4.3.3 into a gedanken model of the adsorption of the most abundant plasma protein, human serum albumin (HSA), using the buffer-air surface an example surface.
4.8.1 A Gedanken Model of Albumin Adsorption
Fig. 6A was created by removing all proteins other than HSA from Fig. 4 thereby rendering an image of a 50×50×50 nm cube of HSA solution at physiologic concentration. Fig. 6B contemplates an air interface instantaneously created at the upper surface of the cube but before movement of any protein molecules. Imposition of this hydrophobic air interface is a significant energetic perturbation that would cause structuring of water within the first few molecular layers directly adjacent to the surface, in the manner described in Section 4.7, with commensurate redistribution of ions [5]. This hydration reaction would create a thin vicinal-water region adjacent to a thin pseudo-2D interface that separates the physical (air) surface from the bulk solution. The interface of Fig. 6B is empty because protein molecules have not yet been allowed to move in this gedanken experiment. At this moment, it useful to further define a ‘proximal solution zone’ as that portion of the bulk solution directly adjacent to the interface from which proteins will move as soon as the stop-motion constraint is removed. Depth of this proximal zone within the imaginary HSA-solution cube will be estimated subsequently.
Figure 6.
Sequence of events leading to the adsorption of human serum albumin (HSA, gray spheres) to a liquid-air surface that interprets neutron reflectometry (NR) data (Section 4.8.2). Panel A is derived from Fig. 4 by removing all other proteins from the hypothetical 50X50X50 nm cube of blood plasma. Panel B represents the instant of creating an air interface on top of Panel A but before any movement of HSA occurs. Panel C releases the stop-motion constraint allowing HSA molecules to diffuse into an inflating interphase, ultimately achieving the interphase concentration detected by NR. Calculations suggest that a packed hexagonal array of molecules is required to achieve NR concentration, as depicted in Panel C.
When the stopped-motion constraint is released, protein molecules will move to the final state depicted in Fig. 6C. The interface has inflated into a truly 3D interphase to accept adsorbed HSA molecules. This interphase has a steady-state thickness governed by the size and packing density of adsorbed protein molecules, as well as the number of molecular layers that might be occupied by adsorbed protein (sees Section 3.2.1). Important questions ask how much protein is adsorbed at steady state, how the adsorbed HSA molecules are actually arranged within the interphase, and how close the rendering of Fig. 6C might be to reality.
4.8.2 Thickness of the Interphase
Neutron reflectometry (NR) is specialized technique that provides some answer to the above questions. This method will not be discussed further herein other than to say that NR of HSA adsorption to the (lv) surface resolves 2.1±0.3 mg/m2 (HSA mass per unit area interface) in a single 4.8 nm thick layer [108], which is almost as large as the hydrated molecules shown in Fig. 6 (constructed in accordance with the diagram of Fig. 3). Physiologic concentration was not used in the NR experiments of ref. [108], but the solution concentration was sufficient to saturate the (lv) surface [20]. Adding still more protein to a surface-saturating solution concentration does not lead to additional adsorption because there is no room at the surface for additional adsorbate. Thus, for the purpose of this illustrative example, it can be assumed that the (lv) surface imagined in Fig. 6C would approximate the circumstance investigated by NR…one layer of HSA molecules sufficient to accumulate 2.1 mg/m2.
Forcing this mass of HSA into a 4.8 nm thick pool creates a concentration = 6.7 mM = 442 mg/mL, where the ‘max’ superscript emphasizes that the interphase is saturated with adsorbate. This calculated concentration is nearly 8X the HSA solubility limit! Similar outcomes are deduced from disparate studies of protein adsorption [14], leading to the conclusion that interphase concentrations at surface saturation are generally large compared to solubility limits and that the NR outcome is not at all unusual. Indeed, the proteinaceous interphase is an exotic region by any ordinary measure with a tangible viscoelasticity resulting from the interactions between adsorbed molecules at the (lv) surface [59].
Knowledge of interphase dimensions, concentration, and size of hydrated HSA molecules (Fig. 3 and Section 4.3.1) permits construction of the saturated (lv) interphase shown in Fig. 6C. The 4.8 nm interphase layer at 442 mg/mL would contain 48.2 HSA molecules in the 4.8×50×50 nm box. Assembling these molecules into the interphase requires very close packing of hydrated diameters as shown in Fig. 6C. In fact, hydrated protein occupies 74% of the total interphase volume, displacing 36% of the water within the interphase volume. These considerations lead to the conclusion that HSA adsorbs to the (lv) surface in way that approximates a close-packed hexagonal array. Mathematical modeling HSA adsorption capacity to solid hydrophobic surfaces leads to the same conclusion [36].
This rendering of adsorbed HSA leads to two seemingly divergent views of protein adsorption. On the one hand, 442 mg/mL interphase concentration suggests that protein adsorbs “aggressively”, “tenaciously”, or “avidly” to hydrophobic surfaces and must necessarily deplete bulk solution in the adsorption process. On the other hand, only 2.1 mg/m2 adsorbs to the surface, which is equivalent to about 2×1016 molecules dispersed on a square meter surface area. This can be put into human dimensions by imagining a 10 μL drop of HSA solution at physiologic concentration hanging from a pipette tip. The pendant drop would have an approximate surface area of ~ 0.2 cm2, and a little arithmetic shows that 2.1 mg/m2 corresponds to only 42 ng HSA of the total 450 μg available in the drop. In other words, the solution is depleted by 0.01% to fill the (lv) interphase formed by expelling the drop from the pipette. From this latter perspective, it seems that HSA does not adsorb so aggressively, tenaciously, or avidly to a hydrophobic surface.
The resolution of this apparent dilemma is that a molecularly-thin interphase amplifies ordinary sense and meaning of concentration. Nevertheless, the chemical potential (activity) of adsorbed protein is an explicit function of concentration, not adsorbed mass per unit area [13, 35]. Interphase concentration is thus a highly relevant parameter in consideration of mechanisms of the biological response to materials. With sufficient solution concentration, proteins will pack into a hydrophobic interphase as closely as possible [36] without overlapping hydration spheres surrounding each protein (see also Fig. 3).
4.8.3 Thickness of the Proximal Solution Zone and Rate of Adsorption
Bond and Puls 1937 analysis [191] provides a useful way to estimate the thickness of the proximal solution zone (Fig. 6B) that feeds the HSA protein adsorption process in the gedanken model of HSA adsorption. If Γ is the mass/area adsorbed to a surface and C is the solution concentration, then the depth of solution that must be depleted to fill the interphase (the depth of proximal solution) is (Γ / C) with units of length. At Γ = 2 mg/m2 and physiologic HSA concentration C = 45 mg/mL, then the depth of proximal solution is 44 nm, approximately the thickness of the proximal solution zone sketched in Fig. 6B. In other words, HSA molecules need only move less than 10 molecular diameters to complete the adsorption process. It is also noteworthy that proximal solution depth varies inversely with concentration, sensibly requiring molecules to be swept from greater depth of proximal solution to fill the interphase as solution concentration decreases.
Also according to Bond and Puls, the diffusion half time required to sweep proximal solution (Γ / C) thick is . Using a diffusion constant D ≈ 10−6 generally appropriate for proteins [150], it is apparent that adsorption from concentrated solutions must be over in just a few milliseconds, consistent with moving only a few protein diameters. Thus, the transition from Fig. 6B to Fig. 6C could be quite fast if the HSA molecules can quickly assemble into the close-packed arrangement required by the (lv) adsorption capacity. This latter aspect of protein adsorption will be discussed further in Section 5.
4.8.4 Generalities from the Gedanken Model of Albumin Adsorption
Several generalities can be drawn from the gedanken model that are appropriate for protein adsorption from concentrated solution, where solution concentration is most appropriately measured against that which is required to saturate available adsorbent surface area (see further Section 3.2.1):
Adsorption induces significant movement of both protein and water molecules into-and-out-of the interphase, respectively. Movement of protein into the interphase induces protein movement out of proximal-solution, which in turn induces diffusion into the proximal phase from bulk-solution.
The interphase fills from a proximal solution zone only a few 10’s of nanometers deep.
Mass adsorption is complete within milliseconds. The actual time to steady state will depend on the rate at which adsorbed protein arranges itself into a compact layer or layers.
Adsorbed-protein concentrations can be large compared to solution concentration or even protein solubility limits. At steady state, protein molecules pack into the thinnest possible hydrophobic interphase as closely as possible without overlapping hydration shells. At surface saturation, this packing approximates hexagonal or cubic packing of hydrated molecules.
Adsorption does not significantly deplete bulk solution unless adsorbent surface-area-to-solution-volume is very high because the total capacity of the molecularly-thin interphase is quite low.
These factors in mind, it is apparent that adsorption of proteins to a hydrophobic surface from blood would lead to an interphase filled with albumin and other proteins through a process of adsorption competition that has yet to be clarified. This latter aspect of protein adsorption is among the more important open problems that must be solved before the third of the important questions outlined in Section 1 can be addressed.
5.0 Kinetics and Thermodynamics of Protein Adsorption
With the essentials of Section 4 in mind, it of interest now to expand the gedanken model of Section 4.8.1 in somewhat more detail by discussing the rate at which protein moves from the state represented by Fig. 6B to a final state represented by Fig. 6C and the thermodynamics involved in the process. Furthermore, it is of interest to compare adsorption from a single-protein solution to adsorption from a binary-protein solution so that the impact of protein packing within the interphase can be better assessed.
The following is an interpretation of experimental measurement of interfacial energetics by tensiometry and rates of mass adsorption by solution depletion, discussed in more detail in Section 6. It is emphasized that only the first hour or so of protein/adsorbent contact in stirred or sessile fluid phases is considered. Thus the following subsections do not consider the possibility of long-term structural changes in the adsorbed state (i.e. denaturation) or establishment of “thermodynamic equilibrium”, if such a state is ever attained in ordinary experimental systems (see Section 4.1.3), or adsorption under pulsatile flow, etc.
5.1 Kinetics of Protein Adsorption
Two very significant outcomes were obtained by comparing kinetics of mass adsorption by solution depletion to tensiometric measurement of the rate-of-change in interfacial energetics. The first of these outcomes was that the rate-of-mass-adsorption from single-protein solution to hydrophobic adsorbents was much faster than adsorption from binary-protein solution. In fact, mass adsorption from single-protein solution was faster than minimum time resolution of the depletion method (about 5 min) [35], showing no change in adsorbed mass with time. By contrast, adsorption from binary solution frequently exhibited two pseudo-steady-state adsorption regimes, each lasting 20-30 minutes, separated by a smooth transition period of nearly equal length, depending on the size difference between proteins competing in the adsorption process [13]. The second significant outcome of this work was that the rate-of-change in interfacial energetics of protein adsorption from purified solutions was far slower than the rate-of-mass-adsorption [35]. These results strongly suggest that filling of the interphase by diffusion is indeed prompt, as discussed in Section 4.8, but that organization of adsorbed-protein layer(s) into a final state, like that sketched in Fig. 6C, can require considerable time to achieve.
With these clues in hand, the steps occurring between the moment of adsorbent immersion into protein solution and attainment of steady-state adsorption can be inferred. Fig. 7 supplements Fig. 6 by providing additional details of the process leading from Fig. 6B to Fig. 6C. This process is illustrated for a single-protein solution (upper half figure) and binary-protein mixture (lower half figure) and can be further broken down into six discrete steps that in reality are undoubtedly a much more concerted series of events [13, 35]:
Immersion of an adsorbent surface into a concentrated, multi-component protein solution leads to rapid diffusion of protein molecules from the proximal fluid phase into the interface created by the spontaneous hydration reactions that occur when a material is immersed in an aqueous solution (Fig. 7A, upper and lower figure halves).
Protein molecules diffuse into this interface due to a concentration gradient immediately created by the hydration reactions of the preceding step. Proteins diffusing from the proximal fluid phase create a truly 3D interphase that separates the physical surface from bulk solution. This 3D interphase inflates to accommodate arriving protein (Fig. 7A→B, upper and lower figure halves).
Proteins diffusing into the interphase necessarily displace interphase water (Fig. 7A→B, curved arrows in upper and lower figure halves). Energetics of this “interphase dehydration” controls adsorbent capacity (see Section 5.2). Proteins partitioning into the interphase become effectively trapped by an energetic preference for the adsorbed state.
Simultaneous diffusion of two different-sized proteins into an adsorbing interphase causes a modest size selectivity due to relative rates of diffusion (Fig. 7A→B, lower half figure; see also Fig. 2). Steps 1-4 above require only milliseconds to inflate a relatively disorganized interphase (Fig. 7C, upper and lower figure halves).
The lowest energetic state corresponds to the thinnest possible interphase volume because it requires work to create interphase volume (see Section 4.2.1). This constitutes a driving force for shrinking the initially-swollen interphase (step 4) by packing adsorbed protein into more organized and concentrated interphase layer(s) [36] (Fig. 7C→D, upper and lower figure halves). Shrinkage of the interphase to the steady-state volume occurs by either expulsion of interphase water (Fig. 7C→D, upper half) or both interphase water and initially-adsorbed protein (Fig. 7C→D, lower half).
Reduction of interphase volume concentrates adsorbed protein to a limit controlled by the steady state partition coefficient P (Fig. 7D, upper and lower figure halves; see Sections 4.1.2 and 5.2).
Figure 7.
Graphical illustration of the kinetics of single-protein adsorption (upper half figure) and binary-adsorption competition between i, j proteins for the same hydrophobic adsorbent (lower half figure). Essential steps depicted in both cases are: (A) instantaneous creation of a thin interface between adsorbent (Physical Surface) and protein solution (Bulk Solution); (B) rapid diffusion of proteins from solution into an inflating interphase region with concomitant displacement of interphase water; (C) reorganization and concentration of protein within an interphase that is shrinking by expulsion of either interphase water (upper half figure) or both interphase water and initially-adsorbed protein (lower half figure); and (D) attainment of steady-state interphase protein concentration by entrapment of initially-adsorbed protein in a minimal volume interphase.
5.1.1 Implications of the Six Kinetic Steps
Consideration of the above six kinetic steps precipitates a number of conclusions that are highly relevant to understanding the biological response to materials and the role that proteins play in catalyzing, mediating, or moderating that response:
Individual proteins diffuse into an inflating interphase against individual (not collective) bulk-solution concentrations. The interphase expands to accept incoming protein until no chemical potential difference exists between interphase and bulk solution for each protein in solution. The circumstance of equal chemical potential becomes increasing complex as the interphase fills.
Incoming proteins crowd into the interphase, undergo repulsion (sometimes referred to as “reflection” in the diffusion literature, see refs. [13, 35] and citations therein) and the process begins to deviate sharply from unrestricted Fickian diffusion. That is to say, as proteins approach the interphase, relative rates of diffusion are not well predicted by the SES equation discussed in Section 4.3.1 [13].
The rush individual proteins into the interphase can temporarily create an overinflated (thicker) interphase compared to that of steady state. The total amount of protein adsorbed from binary (or multi-component solutions such as blood) can be higher than otherwise estimated from single-protein studies [13]. Under conditions of flow (mixing in lab experiments or possibly flow in vivo), this over-inflated state persists for many hours, perhaps indefinitely [13]. Mixing prevents or suppresses establishment of the most efficiently-packed 3D interphase rather than facilitating mass transport to-and-from the interphase, as might otherwise seem consistent with ordinary physical intuition [13]. Evidently, adsorption measurements from single-protein solutions or static mixtures are not necessarily good indicators of adsorption from flowing fluids (although relevant to understanding the physical chemistry of adsorption).
There is little reason to suspect that proximal solution is significantly depleted by adsorption [6], as already discussed in Section 4.8 and amplified by the experimental facts that: (i) surface capacity for protein is actually quite small (typically in the range of 2-10 mg/m2 or μmoles/m2 for kDa-size proteins [101] ), (ii) the total blood-protein concentration is large (50-60 mg/mL, [146, 160]), and (iii) proteins exhibit surprisingly little difference in adsorption energetics across a broad range of blood protein types (3 decades in MW, see Section 6 and refs. [20-26, 31-33, 59] for more discussion). The proximal solution might be depleted for adsorption from dilute protein solutions in contact with high surface area adsorbent, as is sometimes reported in the literature. But for mg/mL solutions, significant reduction of the proximal solution concentration by adsorption is unlikely to occur.
Simultaneous diffusion of any two proteins into an adsorbing interphase causes a modest size selectivity due to relative diffusion rates of bigger-and-smaller proteins (see Section 3.2.4). Relative diffusion rates among proteins are not very large (see Section 4.3.1 and Fig. 2). Hence, relative adsorbed masses of proteins engaged in adsorption competition strongly resembles that of the proximal solution from which adsorption occurs so that “…in mixtures such as blood, the proteins would be adsorbed simply in proportion to their surface collision frequency or concentrations…”; as argued by Brash and Lyman in the early 1960’s [192], with the caveat that the “simple proportion” envisaged by Brash and Lyman occurs as an outcome of a rather complicated diffusion-controlled process (see appendix of ref. [35] for more complete discussion). Diffusion selectivity is not well predicted by the SES equation for the reason discussed in item (2) above but the deviation appears to be systematic, at least for binary-protein solutions [13].
Relative diffusion rates would lead to a number discrimination against larger proteins such that a greater number of smaller proteins adsorb from a mixture of larger-and-smaller proteins [13, 33]. But the popular qualitative explanation for the Vroman Effect based on the idea that lower-MW proteins arriving first at a surface are displaced by higher-MW proteins arriving later [1, 21, 39-58] seems untenable, at least for binary-protein competition. Diffusion from concentrated protein solutions is too rapid [35], only modestly selective (items 2, 5 above), and proteins traverse only short distances from concentrated solutions to arrive at a rapidly-filling surface region (item 1 and ref. [35]). Hence, there seems simply too little time-and-space for mass transfer to exert a significant adsorption-discrimination effect from concentrated protein solution.
Proteins partitioning into the interphase necessarily displace interphase water. Energetics of this “interphase dehydration” controls maximum adsorbent capacity. Maximum adsorbent capacity is consequently observed to be related to adsorbent water wettability [14, 18, 19, 23, 26, 30-32] (with the notable exception of surfaces bearing ion-exchange functionalities [34]). Maximum adsorbent capacity scales with w/v (not molar) solution concentration [20, 31] and is a material property substantially unrelated to the individual identity of protein or proteins adsorbing from purified or mixed solution, respectively.
5.2 A Straightforward Thermodynamic Model of Protein Adsorption
In light of the discussion of the Section 4, I propose that there are three basic components to the overall free energy of protein adsorption [31]:
The free-energy (gain) of the hydrophobic effect t operating on proteins, .
The free-energy (cost) of vicinal water displacement (surface dehydration), .
The free-energy (gain) of protein-protein and protein-surface interactions, .
The first component (1) is the hydrophobic effect that expels protein from solution to recover hydrogen bonds among water molecules otherwise separated by proteins in solution. The hydrophobic effect is approximately constant (on a weight basis) for all globular proteins because, as discussed in Section 4.3.1, the partial specific volume v° of blood proteins falls within a conserved range of 0.70 ≤ v° ≤ 0.75 cm3/g protein. In other words, the number of hydrogen bonds recovered by water by expulsion of protein is approximately constant per-gram-protein because protein volume/gram is nearly constant. Hence it happens that is approximately constant per-gram-protein, for purified proteins and mixtures alike [23, 25], explaining why interfacial energetics of protein adsorption are so similar among various blood proteins and mixtures thereof, when scaled on a w/v concentration basis (see further Section 6).
The second component (2) is the energetic cost of displacing interphase water by adsorbing protein, which increases with increasing adsorbent-surface water wettability (). For a particular surface with a given water wettability, however, is approximately constant per-gram-protein for the same reasons is approximately constant per-gram-protein. And again this helps understand why interfacial energetics of protein adsorption to different adsorbents are so similar among various blood proteins and mixtures thereof, including serum and plasma.
Likewise, the third component (3) must depend in some way on the chemistry of the adsorbent surface. For hydrophobic surfaces, (where “||” denotes absolute value) because protein/surface interactions involve only relatively weak dispersion forces (see Section 4.5). This conclusion is entirely consistent with interfacial rheology which shows that blood proteins adsorbed to the liquid-vapor interface are shear sensitive and exhibit low viscoelasticity (see ref. [193] and citations therein). Low viscoelasticity indicates that intermolecular interactions among adsorbed-protein molecules and with the hydrophobic (lv) surface are relatively weak. And since protein adsorbent capacity decreases with adsorbent surface hydrophilicity, it must be further concluded that holds here too, except perhaps for proteins adsorbed to surfaces bearing ion-exchange functionalities (or possibly minerals) where adsorption by an ion-exchange mechanism is possible [34].
All of the above taken together suggests that the physical chemistry of protein adsorption from purified aqueous solution follows the basic rule , where the approximation is presumably more accurate for relatively hydrophilic adsorbents than hydrophobic adsorbents for the reasons mentioned just above (see also Section 4.5). This proposition is a slightly more formal articulation of the water-displacement idea discussed in Section 4.7: the hydrophobic effect expels protein from solution and expelled protein will partition into the interphase if is not too large (i.e. if ). In fact, the adsorbent surface energy at which occurs near an adsorbent at the pivot-point surface energy characterized by θ = 65° where protein adsorption is observed to decrease to immeasurably low quantities (see Section 4.2). Apparently, at the pivot point, the energetic cost of interphase dehydration just equals the energy gain of expelling protein from solution at this unique adsorbent water wettability.
5.3 Summary
Kinetics and thermodynamics of protein adsorption can be explained with a single interpretive paradigm that has proteins partitioning into a three-dimensional interphase that may contain multiple layers of protein depending on protein MW and solution concentration. Diffusion fills the interphase region which inflates to accept incoming protein. The volume of the interphase can be initially higher than energetically feasible at steady state, leading to time-dependent shrinkage of the interphase by expulsion of interphase water and/or initially-adsorbed protein. Steady state can be described by a straightforward three-component free energy expression in which expulsion of protein from solution by the hydrophobic effect and interaction with the surface and/or among adsorbed proteins are the energetic gains. Energy gains must exceed the energetic cost of moving water out of the interphase by adsorbing protein for adsorption to occur.
6.0 Energy and Mass Balance in Protein Adsorption
This section provides an overview of experimental data supporting the preceding sections. As mentioned in Section 2, the overarching motivation of my experimental program has been to obtain energy and mass balance for the adsorption of blood proteins to polymeric adsorbents (Section 2) incrementally sampling the full range of observable surface energy (water wettability). Adsorption energetics were measured using time-and-concentration-dependent tensiometry (contact angle and wetting methods); interfacial tensions for the buffer-air (liquid-vapor, lv) interface (Sections 6.1) and contact angles for the solid-buffer (solid-liquid, sl) interface (Section 6.2). Adsorbed mass was measured using the solution-depletion method (Section 6.3). In both cases, adsorption isotherms were constructed from experimental measurements. Results obtained were complimentary for a wide range of adsorbent types, adsorbent surface chemistries, and different blood proteins spanning three decades in MW.
6.1 Interfacial Energetics of Protein Adsorption- The Buffer-Air Interface
The buffer-air interface is a molecularly smooth and maximally hydrophobic surface where interfacial energetics can be unambiguously measured by tensiometry. Unlike solid surfaces where tensions at a three-phase (lv, sl, sv) line must be simultaneously considered, the tension at the (lv) surface γlv can be directly measured as a function of solution concentration, temperature, and time [17, 29, 110, 194, 195]. The amount of protein adsorbed to the (lv) surface can be deduced using Gibbsian surface thermodynamics to analyze concentration-dependent γlv (see further Section 6.1.3) [17-19]. The fluid (lv) surface achieves mechanical equilibrium quickly relative to (sl) surfaces. Solid surfaces, by contrast, are in a state of permanent mechanical disequilibrium that results from a meniscus pulling on an (ideally) non-deformable body. Importantly, thermodynamic interpretation of γlv does not depend on the reliability of the Young equation, which is of dubious merit except perhaps at smooth, non-deformable hydrophobic surfaces [70, 110].
We surveyed time-and-concentration dependent γlv of a wide variety of purified blood proteins and protein mixtures, including serum and plasma from a variety of animals [6, 20-22, 24, 59]. Publication and public presentation of the results of this survey encountered a resistant prejudice within the biomaterials community that the (lv) surface was somehow special and not representative of hydrophobic solid surfaces. Furthermore, there was a prevailing opinion that the (lv) surface was irrelevant to biomaterials. However, accumulated experimental results showed that this prejudice and opinion were both unwarranted. As it turns out, the general biophysics of adsorption to hydrophobic solid-liquid (sl) interfaces is not fundamentally different than adsorption to the (lv) surface [6, 18-26, 30, 59]. Viewed in retrospect, this is no particular surprise given that physical interaction between hydrophobic surfaces and proteins is limited to dispersion (Lifshitz-van der Waals) forces which must be substantially similar at hydrophobic (sl) and (lv) interfaces when density of solid surfaces is taken into account (see further Section 4.5). The relevance to biomaterials is that the (lv) surface is the most hydrophobic boundary of a continuum of surface energies that can be explored with the aim of resolving how proteins adsorb to different surfaces. According the discussion of Section 1.1, this is the most fundamentally important problem in biomaterials surface science.
6.1.1 Measurement of Time-and-Concentration-Dependent Liquid-Vapor Interfacial Energies
The three methods most often used for measuring γlv are the Wilhelmy plate, Du Nouy ring, and pendant-drop methods (see ref. [196] for a dated but thorough review and ref. [17] for a review directly relevant to biomaterials). The plate method is complicated by the fact that a solid surface is used to measure tensions at a meniscus where solute deposition and fluid evaporation at the three-phase line greatly affects interpretation of results [17]. Du Nouy was among the first, if not the first, to measure (lv) tensions of biological fluids such as blood serum [71, 197, 198] using a thin ring rather than a plate. Use of a ring overcomes some of the aforementioned plate-method issues. But analysis of the meniscus at a ring is hardly straightforward and shares some technical problems with the plate method, including the fact the interface region is stretched during analysis. Stretching the surface creates new surface area which induces adsorption and mass transport in the manner outlined in Sections 4.8 and 5.0.
The pendant-drop method deduces interfacial energies from the shape of a drop held pendant from a needle or pipette, or possibly supported/suspended on a planar surface. Pendant drop has many advantages for measurement of γlv of protein solutions because it is not complicated by the presence of a plate or ring surface and requires as little as 10 μL of fluid (which is important in studying tensions of expensive or difficult-to-obtain purified proteins). Although Bashford and Adams provided numerical solutions to equations relating shape of pendant drops to interfacial tensions in the late 1800’s [199] and provided look-up tables that permitted interfacial tension to be deduced from a few drop-shape measurements, expansive use of the pendant-drop method awaited development of computerized image acquisition and analysis algorithms [200-202] that reduced labor intensity to a manageable experimental level. Robotics that automate fluid handling and thermostated/humidified sample chambers to control drop evaporation now make the pendant drop a method of choice for studying time-and-concentration dependent interfacial tensions of proteins and surfactants.
6.1.2 Quantifying Protein Adsorption to the Buffer-Air Surface
An issue that arises in the quantitative interpretation of concentration-dependent γlv is that activity coefficients, even of simple hydrocarbon solutes, are not typically unitary as is frequently assumed in ordinary application of Gibbs’ adsorption isotherm. The adsorbed amount calculated from this isotherm is reported as Gibbs’ surface excess Γlv given by:
| (9) |
in units of moles/cm2 and where μ is protein chemical potential (μ = μ° + RTln A, where μ° is chemical potential at standard state and A is protein activity; see also Section 4.1.2). Gibbs’ surface excess is the excess (positive or negative) amount of solute (protein) adsorbed within the interphase, over-and-above the contribution due to bulk solution, calculated as a ratio to adsorbent surface area (formally, area of Gibbs’ dividing plane; see refs. [17, 20, 23] for discussion directly related to biomaterials and refs. [29, 194, 195] as example text books).
Eq. (9) is appropriate for a single, isomerically-pure non-ionizing solute or a polyelectrolyte in swamping salt concentrations of buffer salts [20, 134]. Purified-protein solutions in buffer probably approach these requirements and modified methods are available for chemically-undefined mixtures [30]. But typically, protein activity coefficients σ that translate solution activity to measurable concentration are not available (i.e. A=σX , where X is mole fraction). The ideal-dilute approximation is thus imposed so that solution concentration (mole fraction) can be used as a surrogate for A by insisting on unitary activity coefficients. Therein lies a problem [20, 203, 204].
Proteins are complex polyelectrolyte solutes that are not ideal [205, 206] for which assumption of unit activity coefficients is no doubt quantitatively in error and leads to considerable difference between real surface excess given by Eq. (9) and apparent surface excess surface excess given by:
| (10) |
where CB is dimensionless bulk-solution concentration and the “app” superscript differentiates apparent surface excess from real surface excess. It turns out that is approximately constant for proteins with widely varying MW [20, 23, 26]. Constant strongly suggests that any activity-related discrepancy between and Γlv is also constant among blood proteins studied by tensiometry. Comparison of to Γlv measured by neutron reflectometry revealed that [20]. Using this one-point calibration, tensiometrically measured can be approximately converted to actual adsorbed mass-per-unit-area. In this way, mass deduced from interfacial energetics can be compared to gravimetric measurements obtained by the solution-depletion method to similar surfaces.
6.1.3 Scaling Energetics of Protein Adsorption to the Buffer-Air Interface
Armed with an early version of automated pendant-drop tensiometer, we carried out an extensive survey of time-and-concentration-dependent γlv of purified blood proteins spanning three decades of MW. We compared purified-protein adsorption energetics to surfactant standards and protein mixtures, the latter including blood plasma and serum [6, 20-22]. Data was assembled into 3D graphical representations like that shown in Fig. 8A. These “water fall” diagrams completely characterized time and solution concentration dependence of (lv) interfacial tensions, clearly identified steady state, and provided a comprehensive data set that could be subjected to statistical and thermodynamic interpretation. Time slices from Fig. 8A, C are collected in Fig. 8B, D in a more conventional 2D rendering.
Figure 8.
Time-and-concentration interfacial tensions γlv (Panel A,B) and advancing contact angles θa (Panel C,D) for human serum albumin (fatty-acid free HSA) and prothrombin (blood factor FII), respectively. Panels A,B plot γlv as a function of logarithmic (natural) solution concentration CB scaled as picomoles/L [20]. Panels C,D plot θa of FII solutions on a methyl-terminated SAM surface [23]. Panels A,C display results in 3D coordinates plotting γlv or θa as a function of concentration and analysis time (drop age). Panels B,D show only selected time slices taken from Panels A,C (Panel B: filled circle = 0.25 sec, open circle = 900 sec, filled triangles = 1800 sec, open triangles = 3594 sec. Panel D: filled circle = 0.25 sec, open circle = 900 sec, open triangles = 1800 sec, and open squares = 3594 sec). Notice that kinetics dominates early protein adsorption, requiring 30 min to 1 hr to reach steady state and that surface hydration (Panel D) slowly increases SAM surface wettability (vertical arrow annotation, Panel D).
One of the significant surprises of this survey was that adsorption energetics of diverse proteins were quite similar when interfacial tensions were scaled on w/v solution concentration basis, especially in consideration of the wide range of proteins studied [6] (see further Section 5.2). The significance of this observation is that it shows that the structural variability that confers profoundly different bioactivity does not greatly affect interaction energetics with water that drive adsorption to the hydrophobic (lv) interface (see Section 5.2) [6, 20, 22-25] . At the time of these studies, it was widely anticipated that protein adsorption was complex at the molecular scale, critically dependent on structural differences among proteins. That is to say, most in the biomedical community anticipated that different proteins adsorb to the same surface differently because of differences in molecular structure. Detailed examination of interfacial energetics did indeed reveal differences among various proteins, but these differences seemed much smaller than might otherwise have been expected, again in consideration of the diversity in proteins studied [6]. Many in the biomedical community discounted these observations as being particular to the (lv) surface, but subsequent studies using hydrophobic solid surfaces reviewed in Section 6.2 reproduced these basic findings, controverting the suggestion that the (lv) surface was somehow special.
However, when interfacial energetics were scaled on a molar solution concentration basis, significant differences among proteins were observed [6, 20]. In particular, the molar-solution concentration required to achieve any particular γlv decreased in regular order of increasing protein MW (protein size). This “homologous series in size” was recognized as a recapitulation of the venerable “Traube Rule” of ordinary surfactants. Given that the Traube rule is a signature of the hydrophobic effect (see ref. [20] and citations therein), we postulated that the Traube-rule-like behavior noted in protein adsorption to the (lv) surface was evidence that protein adsorption to the hydrophobic (lv) surface was substantially controlled by water, not protein itself. The rational is that larger proteins displace larger volumes of interphase water than smaller proteins but all blood proteins displace approximately the same volume of water on a per-gram basis because of the conserved partial molar volume (Sections 4.3.1, 5.2).
Hence it happens that concentration-dependent γlv appear very similar (but not identical) among proteins when scaled on a w/v solution concentration basis but these same energetics appear as a homologous series in MW when scaled on a molar basis. These scaling principles, together with the regular spheroidal shape of blood proteins (Section 4.3.1), were exploited in development of a straightforward mathematical model of concentration-dependent γlv that accommodated experimental data with good fidelity [20]. Agreement between relatively simple models of protein adsorption and experimental data strongly suggested that the general biophysics of protein adsorption to hydrophobic surfaces had been successfully captured.
An alternative method of scaling protein adsorption of relevance to biomaterials applications plots interfacial tensions as a function of the dimensionless ratio of solution-concentration-to-physiological-concentration [6, 24] The value of this approach is that it emphasizes the concentration dependence of protein adsorption in the physiologic context. Important proteins known to be directly involved in the biological response to materials (blood factor FXII for example) turn out to be weakly surface active at physiological concentration, suggesting that adsorption to surfaces cannot be solely responsible for activation, especially as adsorbed from a complex milieu of proteins such as plasma ([100, 207], see also ref. [69] for relevance to cell adhesion).
6.1.4 A Provocative Interpretation
It is important to stress that the idea that water controls protein adsorption, a central theme of this Leading Opinion that originated in the above-reviewed studies, remains a radical departure from the conventional interpretation of how proteins adsorb to surfaces from aqueous solution. Two seemingly irreconcilable categories of thought on the matter emerge, as provocatively captured in Fig. 9. Conventional understanding is represented on the left-hand panel asserting that protein-surface interactions are mediated by short-range, pair-wise interactions between protein molecules and adsorbent surfaces. Many theories postulate that these interaction energies are large compared to thermal energy (see Section 4.5) and protein is held in the adsorbed state by large interaction energetics. Accordingly, selective protein adsorption is construed to be due to variations in molecular structure that cause certain proteins to be bound more avidly than others. In its broadest context, this “solute-oriented” paradigm implicitly argues that the protein adsorption process is complex at the molecular scale, requiring detailed microscopic information at the level of statistical mechanics (computation) for complete description of protein/surface interactions (see, for example, refs. [186-188] and citations therein). As pointed out in Section 4.6, complexity at the molecular scale presents itself as a great barrier to understanding protein adsorption from complex biological milieu such as blood containing about 1000 different proteins with widely-varying natural abundances [146].
Figure 9.
A provocative representation of protein-adsorption theories positing that all such theories fall into one of two categories. The leftmost category collects all theories premised on the idea that adsorption is controlled by strong protein/surface interactions whereas the rightmost category collects all theories premised on the idea that the hydrophobic effect is the significant driving force underlying protein adsorption. Subscribers to the former category greatly outnumber the current subscription to the latter. Importantly, the left category has it that protein adsorption is complex at the molecular scale, requiring computations at the statistical-mechanics level to understand the essential biophysics of protein adsorption. By strong contrast, the right category has it that water (solvent) controls protein adsorption and, as a consequence, adsorption of blood proteins can be understood on a much more generic basis. Distinguishing between the two categories is important because most practical biomaterials problems are intractable at the molecular level. Group 1 type analytical methods (Section 3.1) reinforce adherence to the left category through a circular self-fulfilling prediction that protein adsorption is mediated by strong protein/surface interactions (see Section 4.5).
The departure from conventional wisdom represented by the right-hand panel of Fig. 9 contends that protein adsorption is substantially controlled by water through the hydrophobic effect (see Section 5.2). According to this thinking, water expels protein from solution into the interphase separating bulk solution and the physical adsorbent surface (Fig. 1). Protein will enter the adsorbed state if displacement of interphase water is not energetically prohibitive. Selective adsorption is primarily related to solution concentration and differences in protein diffusion coefficients (Section 5.0). This “solvent oriented” perspective explicitly argues that protein adsorption is not complex at the molecular scale and, in fact, has little to do with structural differences among proteins but rather has nearly everything to do with water [14, 17, 110, 137, 170]. Practical biomaterials problems certainly appear much less imposing from this perspective compared to the solute-oriented viewpoint.
Needless to say perhaps, conventional wisdom (left panel) vigorously disputes scientific feasibility of the alternative view (right panel), arguing that protein adsorption is a nanoscopically-localized event between protein and adsorbent surface [127]. These protein/surface interactions are proposed to culminate in a monolayer of adsorbed protein achieving a ‘jamming limit’ of about 55% coverage [178, 208, 209] beyond which adsorption from solution ceases. The alternative view has it that protein adsorption occurs within a 3D interphase separating bulk solution from the adsorbent surface and that displacement of interphase water is the significant energetic cost that caps adsorbent capacity (see further Section 7.3). Adsorption of proteins in multi-layers [20, 31-33] that cannot be explained by protein-surface interactions at close proximity (Section 3.2) is readily understandable from the alternative viewpoint.
The debate between conventional wisdom and new think is exacerbated by the Group 1 vs. Group 2 analytical issues outlined in Section 1.2 that erroneously lead investigators to believe that protein adsorption is mediated by strong protein/surface interactions (Section 4.5). There seems little reason to believe that these two different perspectives on the same process will be resolved any time soon by formulation of a consensus interpretation or mechanism of protein adsorption. But the good news is that there is now a competing theory of protein adsorption that might contribute to solutions to the questions of Section 1.1 that conventional wisdom has failed to answer over the last five decades of effort.
6.2 Interfacial Energetics of Protein Adsorption - The Solid-Liquid Interface
With a firm grasp on the interfacial energetics of protein adsorption to the liquid-vapor surface in hand (Section 6.1), we embarked on a survey of adsorption to solid-surfaces incrementally sampling the full range of observable water wettability. Conventional (by this writing) methods of surface engineering were employed using silane derivatization of glass and self-assembled thiols on gold (SAMs) deposited on electronic-grade semiconductor wafers. Automated tensiometry was once again deployed to measure time-and-concentration interfacial tensions, this time as advancing and receding contact angles θa and θr, respectively. The general observation was that θr was a less reliable measure of wetting than θa and the latter was used in nearly all data interpretation.
6.2.1 Measurement of Time-and-Concentration-Dependent Solid-Liquid Interfacial Tensions
Unlike at the buffer-air interface, interpretation of concentration-dependent contact angles is somewhat ambiguous because tensions at a three-phase (lv, sl, sv) line must be simultaneously considered. Solid tensions are not typically accessible to tensiometry (at least not without additional theory [14, 210] and the inevitable assumptions/simplifications embodied therein). Also, interpretation of solid interfacial tensions almost always depends on the reliability of the Young equation, which is of dubious merit for reasons mentioned in Section 6.1. These issues are especially problematic at hydrophilic surfaces (as defined in Section 4.2) that can swell with water at the interfacial level, causing significant departures from expectations of the Young equation. But, as it turns out, many of these issues are not an issue in measuring protein adsorption at solid surfaces because proteins do not adsorb to hydrophilic surfaces (see Sections 4.2, 4.4, 5.2).
We measured time-and-concentration dependent θa for a wide variety of purified-blood proteins and protein mixtures, including serum and plasma from a variety of animals [23-26] and collected data in water-fall diagrams similar to those described in Section 6.1.3 (see Fig. 8C, D). Publication and public presentation of the results of this survey encountered a strong preconceived notion within the biomaterials community that different proteins adsorb differently to the same solid surfaces. However, as observed in the case of the liquid-vapor surface, the variation observed among diverse blood proteins spanning three decades in MW was much smaller than anticipated, especially in consideration of large differences in protein structure. And, as already mentioned in Section 6.1.1, adsorption of these different proteins to the most hydrophobic surfaces was not significantly different than at the hydrophobic (lv) surface [26].
6.2.2 Quantifying Protein Adsorption to the Solid-Liquid Surface
Practical use of concentration-dependent contact angles as a measure of adsorption to the (sl) interface has been discussed at length elsewhere (see, for examples, refs. [17-19] and citations therein). Briefly, the amount of solute adsorbed to (sv) and (sl) interfaces is measured by the Gibbs’ surface excess quantities Γsv and Γsl, respectively, in units of moles/area (the subscript “a” specifying advancing contact angles is not carried in Γ symbology for the sake of notational compactness). The Gibbs’ adsorption parameter [Γsl − Γsv] (but not separate excess parameters) can be computed from data comprising contact-angle isotherms using Eq.(11):
| (11) |
where (dθa / d ln CB) is the slope of a contact-angle isotherm and is evaluated from Eq. (10). As discussed in Section 6.1.3. The discrepancy between real and apparent can be approximately corrected for by applying as further experimentally confirmed in ref. [23] (see further Section 6.1.2).
For relatively hydrophobic surfaces exhibiting θa > 65° and under experimental conditions that avoid inadvertent mechanical deposition of solute at the (sv) interface, as through contact-angle drop movement on the surface or drop evaporation, for examples, it has been shown that Γsv ~ 0 and [Γsl − Γsv] → Γsl [17-19]. Under the additional restrictions that (i) solute activities at hydrophobic (sl) and (lv) interfaces are approximately equal and (ii) , it can be expected that . Experimental results obtained using automated tensiometry described in refs. [23, 26] confirm that these stringent physical conditions can be attained, leading to the further conclusion that for surfaces exhibiting θ > 65°. As a consequence of all of these factors, concentration-dependent contact angles can be used to measure the approximate amount of protein adsorbed to solid surfaces.
6.2.3 Scaling Energetics of Protein Adsorption to the Solid-Liquid Interface
Use of time-and-concentration dependent tensiometry confirmed that adsorption to hydrophobic surfaces was not different than that observed at the (lv) interface (Section 6.1.4). The “Traube-rule” ordering observed at the (lv) surface was recapitulated at the hydrophobic (sl) interface [23], strongly suggesting that water controlled protein adsorption to (sl) interfaces, as concluded for the (lv) interface and as discussed in Section 4.7 and 5.2.
Study of protein adsorption to solid surfaces affords the opportunity to measure how protein-adsorption energetics change as a function of adsorbent wettability. Fig. 10A,B condenses a systematic study of a wide variety of proteins to surfaces incrementally sampling the observable water-wettability range [26]. The data and error bars represent mean and standard deviation, respectively, of ten different blood proteins spanning 3 decades in MW adsorbing to adsorbents incrementally sampling the full range of observable water wettability. The uncorrected Gibbs’ excess parameter [Γsl − Γsv]app is collected in Fig. 10A whereas the combined term (a ratio free of correction for protein activity that uses the (lv) interface as a standard of reference) is plotted in Fig. 10B (see Section 6.2.2 for meaning of excess parameters). It is evident that protein adsorption decreases as function of adsorbent hydrophilicity, passing through zero at the pivot-point surface energy near τ° = 30mJ/m2 (θ 65°, see Section 4.2). Negative [Γsl − Γsv]app and values correspond to surfaces to which protein deposits at the (sv) interface from the evaporating edge of low-contact-angle protein-solution droplets, causing Γsv > Γsl where Γsl → 0 (see refs. [18, 19] for a discussion of the relative magnitudes of surface-excess parameters).
Figure 10.
Apparent Gibbs’ surface excess scaled as a function of adsorbent surface water wettability (surface energy) as measured by the advancing contact angle θa of phosphate buffered saline (PBS) solution, expressed as water (buffer) adhesion tension τ° = γlv cosθa for incrementally sampling the full range of observable water wettability (where buffer interfacial tension γlv = 71.97 mJ/m2; SiOx = oxidized silicon semi-conductor wafer, APTES = aminopropyltriethoxysilane silanized SiOx, PS = polystyrene spun-coated onto SiOx, SAM = 1-hexadecanethiol self-assembled monolayer on gold-coated SiOx). Symbols and error bars represent mean and standard deviation of ten different proteins spanning three orders of MW (ubiquitin, 10.7 kDa; thrombin (FIIa), 35.6 kDa; FV HSA, 66.3 kDa; Hageman factor (FXII), 78 kDa; fibrinogen, 340 kDa; IgG, 160 kDa; C1q, 400 kDa; IgM, 1000 kDa). See ref. [26] for details. Panel A shows that Gibbs’ surface-excess parameter [Γsl − Γsv] decreases monotonically with increasing adsorbent-surface hydrophilicity, projecting [Γsl − Γsv] = 0 near the τ° = 30 mJ/m2 pivot point (θ = 65°, see Section 4.2 and Fig. 5). Likewise, the ratio decreases from +1 to −1 (Panel B) as [Γsl − Γsv] decreases from a maximum [Γsl − Γsv] = −Γlv at the liquid-vapor (lv) interface and hydrophobic SAM surface (τ° = −15 mJ/m2) to a minimum [Γsl − Γsv] = −Γlv at the water-wetted (τ° → 73 mJ/m2 surfaces. Smoothed curves drawn through the data are guides to the eye.
6.2.4 A Provocative Interpretation
Measurement of protein adsorption to solid surfaces over the full range of hydrophilicity strongly suggests that the energetics of surface dehydration (Section 5.2) not only controls the adsorbent capacity but also prevents protein from adsorbing to surfaces more wettable than the θ = 65° pivot point (see Section 4.7.1 and Fig. 10). These findings, which were corroborated by direct measurement of adsorbed protein mass (Section 6.3), have a considerable impact on the interpretation of the biological response to materials.
As mentioned in Section 4.2, biological responses to materials such as blood plasma coagulation [138, 211, 212], blood factor XII activation [15, 207, 213], cell adhesion [69], and protein adsorption [26, 30, 101] are observed to pivot from high-to-low or low-to-high near a water wetting characterized by θ = 65° (note that refs. [69, 100] and [14, 137] are review articles summarizing and comparing work from a broad literature). For examples, mammalian cell adhesion is unfavorable at hydrophobic surfaces with a high-adsorbent capacity but favorable at hydrophilic surfaces that adsorb little-or-no protein (see ref. [69] and citations therein). Contact activation of blood plasma coagulation is low at protein-adsorbent hydrophobic surfaces but high at hydrophilic surfaces that resist protein adsorption (see ref. [100] and citations therein). This means that not all biological responses to materials are catalyzed, mediated, or moderated by adsorbed protein but rather that certain biological responses are catalyzed, mediated, or moderated by lack of adsorbed protein.
Important among this latter class of biological responses are cell adhesion to tissue-culture-grade polystyrene (TCPS) substrata and contact activation of blood coagulation. It is widely believed that cell adhesion is mediated by adhesins adsorbed from serum-containing media. However, TCPS is hydrophilic according to the definition used herein (Section 4.2) and does not adsorb blood proteins. Apparently then, the conventional wisdom that adsorbed proteins mediate cell adhesion cannot be entirely correct [69]. It has been long and widely believed that plasma coagulation is catalyzed by adsorption (sometimes stated as assembly) of blood factor FXII and other proteins of the activation complex onto anionic hydrophilic surfaces. Although both FXII and plasma coagulation are indeed activated in contact with hydrophilic surfaces [100], it is found that FXII does not adsorb to hydrophilic surfaces [26]. Quite apparently, protein adsorption is not necessary for activation to occur. FXII activation also occurs at hydrophobic surfaces, even though FXII is only modestly surface active at physiologic concentrations [6, 24]. It is thus unclear to what extent adsorption to hydrophobic surfaces precedes FXII activation. It is clear, however, that adsorption of plasma proteins competes with FXII/surface contacts and effectively suppresses plasma coagulation by contact with hydrophobic surfaces [207].
In view of the above, it is apparent that the basic tenet underlying biomaterials science articulated in the opening of Section 1, that protein adsorption is the first step in the acute biological response to artificial materials, should be modified in a way that does not directly link protein adsorption to the biological response in a one-for-one way. Perhaps something along the lines surface hydration is the first step in the interaction of a biomaterial with a biological milieu that controls protein adsorption and all other subsequent steps in the biological response to materials. There is yet little doubt that proteins are involved in the biological response to materials. It just happens that physical adsorption to surfaces does not necessarily precede that involvement (see Section 4.2.2).
6.3 Protein Mass Adsorption to Solid Surfaces
Virtues of the solution-depletion method for measuring protein adsorption [31, 32, 34, 101] were briefly mentioned in Section 2; simplicity, no complex instrumentation or interpretive theory required, and applicability to purified protein solutions, mixtures of purified proteins, or chemically-undefined mixtures [69] to reiterate a few of these attributes. A considerable interpretative advantage is that it is not necessary to know adsorbent surface area [31] because results can be calculated in terms of interphase volumes (although it is sometimes useful to know adsorbent surface area for comparison to other methods of measuring adsorption [101] or for estimating protein packing within the interphase [36]). When implemented using electrophoresis as a multiplexing, separation-and-quantification tool, measurement of adsorption competition among of multiple proteins from solution becomes possible [13, 33]. However implemented, solution depletion permits kinetics of mass adsorption to be directly monitored [13, 35]. Implementation of modern “nanodrop” spectrometers opens the method up to spectroscopy without increasing mass of proteins required in the analysis.
Nevertheless, as with any analytical method, there are certain drawbacks to the solution-depletion method. One of the more important of these is that the depletion method is practically limited to particulate adsorbents providing sufficient adsorbent surface area such that a statistically-significant adsorbed mass can be calculated from initial (not in contact with adsorbent) and final (in contact with adsorbent) solution concentrations. Ideally, adsorbent particles should not require separation from solution by centrifugation because these processing steps can introduce experimental artifacts. This latter restriction requires adsorbents larger than colloid particles and excludes particles that float. Solution-handling steps involved with the depletion method practically limits measurement rate to a few minutes, greatly restricting time resolution for kinetic experiments.
In spite of these analytical drawbacks, we found the solution-depletion method could be effectively used with silanized-glass-particle adsorbents and sepharose-based chromatographic materials. Adsorption isotherms of a wide variety of proteins spanning three decades in MW adsorbing to adsorbents incrementally sampling the full range of observable water wettability from purified-protein solution, binary mixtures, and blood serum [13, 31-35, 69, 85, 101]. The sampling-rate issue turned out not to be a problem in comparing mass-adsorption rates to rates-of-change in interfacial energetics because the latter are much slower than mass-adsorption rates (see Section 5.1). The outcome was unambiguous measurement of adsorbed mass obtained without the need to apply Group 1 adsorbent-rinsing steps or protein labeling. The solution-depletion method is thus effectively a gold standard against which all other methods can be benchmarked (see ref. [36] for a revealing comparison to QCM).
6.3.1 Adsorption Isotherms and Adsorbent Capacity
Fig. 11 collects human serum albumin (HSA, 66kDa) adsorption isotherms for five different silanized glass-particle adsorbents exhibiting 113°-60° buffer contact angles obtained using the solution-depletion method [32]. Solution depletion plotted on the ordinate (symbology not to be confused with diffusion coefficients) measures the difference between initial solution concentration before contact with adsorbent particles and final concentration WBi after contact with adsorbent. The subscript i tracks protein identity (HSA in this case) and was used to differentiate from a second protein j used in binary-protein-adsorption studies [13, 33, 34]. Solution concentration and depletion have the same units of mass/volume-bulk-solution VB (mg/mL in Fig.11). The absolute mass adsorbed mi = DiVB and interphase concentration where VIi is the interphase volume occupied by protein i. Thus, Di can be interpreted as either absolute adsorbed mass or concentration adsorbed to a fixed adsorbent surface area a.
Figure 11.
Adsorption isotherms for human serum albumin (HSA, 66kDa) plotting the solution depletion Di against initial solution concentration (mg/mL) for five different silanized glass-particle adsorbents exhibiting 113°-60° advancing buffer contact angles θa obtained using the solution-depletion method (see ref. [32] for details). The adsorbent capacity (Di)max and initial solution concentration at which the adsorbent was saturated by HSA (see star annotations) decreased monotonically with adsorbent hydrophilicity at fixed total surface area (see Fig 12). Lines through the data are guides to the eye.
The data collected in “depletion curves” such as Fig. 11 approximate Henry isotherms for adsorbents with increasing hydrophilicity (see also Section 4.1.2). The adsorbent capacity (Di)max that occurs at a solution concentration decreases monotonically to zero with hydrophilicity through detection limits at the pivot-point wettability near θ = 65° [31] as shown in Fig. 12 (see also Fig. 10 and Section 6.2.3). We have found that higher-molecular-weight proteins exhibit progressively higher (Di)max for the same hydrophobic adsorbent at fixed surface area. In fact, adsorbent capacity for IgM (1000 kDa) has been measured to be ~6X that of HSA, recapitulating result obtained by tensiometry [20], and showing that protein size is an important variable in protein adsorption [31, 101] (see also Section 3.2). Six-fold larger adsorbent capacity means that 6 albuminoid layers would be required to constitute the interphase volume occupied by IgM [101]. A simple analysis [101] shows that adsorbent capacity is capped by a maximum weight (not molar) concentration for all proteins, again corroborating conclusions drawn from tensiometric studies [20]. Furthermore, this analysis shows that interphase thickness ΩIi scales with adsorbent capacity, meaning that the interphase volume increases with protein size or MW (VIi = ΩIia, see also Section 4.3.1). Careful consideration of protein size (Section 4.3.1) and possible packing scenarios reveals that at least two layers of proteins larger than HSA assemble in a saturated hydrophobic interphase [36].
Figure 12.
Adsorbent capacity (Di)max of different silanized glass particles for HSA decreases monotonically with adsorbent hydrophilicity to detection limits (LOD) near the τ° = 30 mJ/m2 (θ = 65°) pivot point (see Section 4.2 and Figs. 5, 10). Lines through the data are guides to the eye.
6.3.2 Adsorption Energetics
The slope of depletion curves Si for various proteins and adsorbent surface energies are typically less than max unity, up to . Si < 1 demonstrates that protein adsorption is neither irreversible, nor especially avid for that matter, confirming tensiometric measurement of modest protein adsorption energetics [6, 20, 22-26]. Irreversible or high-avidity adsorption is characterized by Si →1 because all-or-nearly-all proteins in solution adsorb, creating the circumstance . In fact, we have observed Si →1 only for adsorbents bearing strong Lewis acid/base groups that exhibit ion-exchange properties [34]. Si < 1 does not prove that every adsorbed protein freely partitions between interphase and solution, but quite apparently the majority of molecules enjoy that possibility over a broad range of adsorbent surface energies.
Adsorption avidity can be quantified from solution-depletion isotherms like Fig. 11 in the form of partition coefficients Pi ≡ (WIi /WBi) that measure the ratio of interfacial and bulk-solution-phase w/v (mg/mL) concentrations WI and WB, respectively, at steady-state [31]. For a broad variety of proteins adsorbing to a hydrophobic adsorbents, it was found 45 < P < 520 , meaning that the interphase is concentrated 45-520 fold over bulk concentrations. The perspective arising from these measurements is that of a very concentrated proteinaceous interphase with very different chemical properties than bulk solution (see Fig.7 and discussion thereof in Section 4.8) [14, 17, 137]. Although Pi varies significantly with protein MW, the mean value free energy of adsorption to a hydrophobic adsorbent computed for 6 proteins within 15 < MW < 350 kDa range was −(5±1)RT or about −12.5 kJ/mole protein (see Section 4.6), corroborating tensiometric estimates [20]. Thus, it is evident that protein adsorption to a hydrophobic adsorbent does not really qualify for the term “tenacious” or “avid” as it is sometimes referred to in the literature (see Section 4.8.2), again expect perhaps for surfaces bearing ion-exchange functionalities [34].
6.3.3 Contrasting Solution Depletion to Group 1 Analytical Methods and RSA Theory
The experimental results discussed in the preceding sections show that Group 1 analytical methods (Section 3.1) are inappropriate for measuring protein adsorption. Reversible (or perhaps more accurately “not irreversible”) protein adsorption clearly demonstrated by solution depletion means that some or all adsorbed protein can be rinsed from adsorbent. Thus, Group 1 methods underestimate the amount of protein adsorbed [85]. Multilayer adsorption means that protein molecules adsorbed in a second layer within the interphase are too distant from an adsorbent surface to engage in strong protein/surface interactions (if such strong interactions actually occur, see Section 4.5). Therefore, Group 1 analytical methods not only underestimate the amount of protein adsorbed but also introduce a significant bias against larger proteins.
Many variants of the Random Sequential Adsorption (RSA) theory, especially early versions, are premised on the idea that protein adsorption is mediated by strong protein/surface interactions. Solution depletion shows this is not the case and strong protein/surface interactions cannot be responsible for observed multi-layer protein adsorption (see also Section 4.5). RSA predicts that adsorbent capacity is capped by a “jamming limit” which, when expressed as moles/cm2 , scales as MW−2/3 (see ref. [101] and citations therein). In sharp contrast, experiment for a hydrophobic adsorbent shows that adsorbent capacity follows a roughly linear relationship with MW. It happens that RSA fortuitously predicts adsorption capacity for low-MW proteins with reasonable fidelity but the discrepancy between RSA and experiment increases sharply for MW > 200 kDa. Experimental evidence thus forces the conclusion that Group 1 analytical methods are in serious error and that standard RSA does not capture the essential physics of protein adsorption.
All taken together, it appears that Group 1 analytical methods and RSA have been locked in self-fulfilling circularity for years that has falsely engendered confidence in both. RSA agrees with Group 1 measurements because Group 1 methods wash away loosely-bound protein within the interphase, leaving only strongly-bound protein, which experimentally vindicates use of an RSA model (see Section 3.2.2). Unfortunately, this circularity has reinforced a misunderstanding of the basics of protein adsorption that water controls adsorbent capacity.
6.3.4 Kinetics of Mass Adsorption from Single-Protein Solutions
One of the surprises emerging from recent studies of protein-adsorption kinetics is that mass adsorption is faster than the change in interfacial energetics [13, 35, 214]. The traditional interpretation has been that that the slow change in interfacial tensions is due to slow adsorption into the surface region (interphase). Yet diffusion rates are too fast by orders-of-magnitude to accommodate slow rates of interfacial tension change. So the idea was invented that an (unseen) energy barrier slows the rate at which protein actually at an interfacial plane (see ref. [35] for a brief historical review). There are a number of conceptual difficulties with this idea, especially with regard the construction of the interface region and its relationship to proximal solution (see Section 4.8 and ref. [35]). But imposition of an energy barrier seemed to explain the great discrepancy between measured kinetics (order of hours, see Fig. 8) and rapid Fickian diffusion (order of milliseconds, see Section 4.8.3) for protein adsorption.
Use of the solution-depletion method to measure kinetics of protein mass adsorption clearly shows that partitioning of protein into the interphase is much quicker than change in interfacial tensions for comparable solution concentrations [35], corroborating similar observations using a novel resonant elastomeric surface tension (REST) sensor [214]. As it turns out then, interfacial energetics do not incrementally change due to incremental increase in adsorbed protein mass as protein molecules pass through the energy barrier.
Inspection of Eq. (1) reveals that change in interfacial tension γ is due to change in the chemical potential μ of solute upon partitioning into the interphase. In other words, decrease in interfacial tension follows an increase in concentration of protein adsorbed within the interphase. There are three ways interphase concentration can increase: (i) total adsorbed mass within a fixed interphase volume can increase, (ii) the interphase volume containing a fixed total adsorbed mass can decrease, or (iii) both (i) and (ii) can simultaneously occur. Option (i) is consistent with the traditional theory that protein adsorption is slow due to an energy barrier. But experiment shows that mass adsorption is much quicker than change in interfacial tensions, especially at mg/mL concentrations relevant to biomaterials. Option (i) alone thus cannot explain observed kinetics. Rather, option (ii) must be responsible for slow change in interfacial energetics because option (iii) cannot occur if option (i) is not possible. This leads immediately to the proposition that interphase volume slowly shrinks by expelling interphase water. The scenario outlined in Fig. 7 and six kinetic steps discussed in Section 5.1 thus explains that protein rapidly diffuse into the interphase within milliseconds (Section 4.8.3) rapidly inflating the interphase. This inflated interphase subsequently shrinks in volume, capturing and concentrating the bolus of initially-adsorbed protein and causing decrease in interfacial energetics.
6.3.5 Kinetics of Mass Adsorption from Protein Mixtures
Adsorption of two-or-more proteins in solution to the same adsorbent surface follows the same general outline of events described above for each of non-identical protein. Each diffuses at a characteristic diffusion rate (Sections 3.2.4, 4.3.1) against individual (not collective) concentration gradients. Remarkably, the interphase can, under certain solution conditions, initially fill with far more protein than the steady-state adsorbent capacity for each protein adsorbing individually to the same adsorbent surface [13]. This over-filled interphase condition can persist for long periods, depending on the mixing conditions applied. Mixing adsorbent and protein solution actually appears to prolong the over-filled interphase condition. Under stagnant conditions, the over-filled condition slowly decays to a final state by expulsion of initially-adsorbed protein. To my knowledge, these events have not been monitored for more than two proteins in solution. But extrapolation of results from binary-protein experiments to mixtures such as blood serum would suggest that the interphase in contact with such proteinaceous milieu is very thick and possibly a very diffuse region, as opposed to the organized layers depicted in Figs. 8A,B for single- and binary-protein adsorption cases.
6.4 Summary
Energy-and-mass-balance for blood protein adsorption to surfaces with varying hydrophilicity has been measured by tensiometry and solution depletion. Results from these very different techniques are complimentary, leading to the following conclusions:
Protein adsorption at steady state follows general trends anticipated by equilibrium thermodynamics.
Structural variability that confers profoundly different bioactivity does not greatly affect adsorption energetics (interaction energetics with water).
for hydrophobic surfaces, consistent with weak protein surfactancy - protein does not adsorb “tenaciously” or “avidly” to hydrophobic surfaces.
Adsorbent capacity for protein decreases with increasing hydrophilicity (). Water bound to surfaces exhibiting θa < 65° cannot be displaced by adsorbing protein. Proteins do not adsorb to hydrophilic surfaces as defined herein.
Water orchestrates a systematic pattern in energetics of protein adsorption to surfaces revealing a Traube-rule-like ‘homology in molecular size (MW)’.
Proteins can absorb in multiple layers depending on solution concentration and MW. Adsorbed protein can form a close-packed array at surface saturation.
Interphase dehydration by protein displacement is the key phenomenon controlling protein adsorption to surfaces.
7.0 Revising the Research Paradigm
Evidentially we are collectively missing something. We are not on the right path. Five decades of focused research into the “protein-adsorption problem” defined in Section 1 has made little progress toward resolving open technical issues. The literature is a mess. Rational engineering of biomaterials remains a dream. Crafting a pedagogical approach to teaching biomaterials is elusive because the cornerstone of our understanding – that protein adsorption (or lack thereof) catalyzes, mediates, or moderates the biological response to materials – can be explained only in the vaguest qualitative terms. There is not one literature demonstration of this cornerstone understanding that quantitatively links adsorbed-protein composition to a measured biological response to a materials surface.
Quite apparently there are both experimental and conceptual barriers that impede our progress toward the goal of a comprehensive understanding of protein adsorption in a way that is relevant to biomedical applications. These barriers need to be identified and overcome. And this almost certainly means that certain long-cherished lines of thought and action must be abandoned because these have not been helpful, perhaps even destructive. No single investigator is likely to remedy all of our problems in this regard, but a few suggestions arising from this Leading Opinion might help reset the research compass bearing. The following subsections discuss four changes in course that I feel are critical to moving forward.
7.1 Abandon Group 1 Analytical Methods
Experimental results discussed in Section 6 show that Group 1 analytical methods employing adsorbent rinsing steps are inappropriate for measuring protein adsorption (see Section 3.1). It is easy to show that protein is not strongly bound to polymeric adsorbents defined in Section 2. That means Group 1 methods underestimate the amount of protein adsorbed in an unpredictable, non-systematic way. Experiment further verifies that proteins can adsorb in multi-layers, depending on protein size and solution concentration. Multi-layer adsorption means that protein molecules adsorbed in a second-or-higher layer are too distant from an adsorbent surface to engage in strong protein/surface interactions (if such strong interactions actually occur, see Section 4.5) and are thus easily rinsed from adsorbent surfaces. Therefore, Group 1 analytical methods not only underestimate the amount of protein adsorbed but also introduce a significant bias against larger proteins (MW > 70 kDa) that require at least two layers to saturate hydrophobic adsorbent surfaces. As already discussed in Section 3.1.2, Group 1 analytical methods have propagated a great deal of misinformation about protein adsorption. It is long past time to abandon these methods as a general tool for studying protein adsorption.
7.1.1 Cautiously Replace Group 1 Analytical Methods with Certified Group 2 Methods
It is evident from Section 3.2 that use of analytical methods falling by definition into Group 2 hardly guarantees an internally-consistent data base because of the subtle interpretive factors outlined in Table 1 and those that yet remain to be discovered. No single Group 2 method will service all research needs and this means a variety of analytical methods will be required. These different methods should be “certified” by measurement of full adsorption isotherms for surfaces under study for different proteins spanning a broad MW range and showing that these results compare favorably to those measured by a standard reference method. Attributes of the solution-depletion method mentioned in Sections 2.2 and 6.3 qualifies this method as one such absolute standard of comparison. We can anticipate interpretable uniformity in the literature only when-and-if such certified analytical methods come into widespread use. On the other hand, we can anticipate only more confusion in the literature if different methods with subtle interpretive differences continue to be applied.
7.2 Abandon the 2D Interface Concept
There are at least two related ways adsorption can be theoretically interpreted – occurring on a plane (2D) or in a volume (3D). The 2D model is very well established in the literature, arising from the earliest considerations of diffusion and mass transport, extended and applied by luminaries such as Milner and Langmuir (see ref. [35] and citations therein for a brief historical perspective). The basic idea is that adsorbate binds to a surface through strong adsorbent/adsorbate interactions leading to a pseudo-2D adsorbed layer such that, in the words of Irving Langmuir, “…the adsorbed film should not exceed one molecule in thickness…” [215]. But this is not necessarily how protein adsorption must occur and evidence reviewed in this Leading Opinion strongly suggests that protein adsorption does not occur in this manner. Instead, protein is effectively pushed onto an adsorbent surface and collects within a layer, or layers, that are more flexibly described as a 3D interphase. Adherence to the 2D interface model is an invitation to overly focus on solute at the expense of solvent in a way that predisposes one to ignore other interactive constituents discussed in Section 4. It is long past time to broaden our purview of the protein-adsorption process and escape the conceptual restrictions imposed by the 2D paradigm.
7.2.1 Cultivate the Interphase Concept
The other way to think about adsorption has adsorbate molecules collecting in a near-surface region referred to herein as an interphase discussed in relation to Fig. 1. The interphase is a discrete volume that separates the bulk-solution phase from the physical adsorbent surface phase (see refs. [31-35] and citations therein). Here, the thickness dimension is explicitly considered and adsorption is (or can be) measured in concentration units of per-unit-interphase-volume. In the limit of small-molecule adsorbates such as gas molecules or simple surfactants, 2D and 3D models are substantially equivalent. But as adsorbate dimensions increase to the size of proteins (see Section 4.3.1), the distinction between the models becomes more important because the interfacial energy required to create a thick interphase becomes an increasingly significant component of the overall free energy of adsorption (Section 4.1.2).
The 3D interphase paradigm is hardly new to surface science but this paradigm has not been widely applied in the study of protein adsorption. Significant advantages of the interphase paradigm are retention of the concept of chemical activities (concentrations) essential for a complete understanding of adsorption energetics and consistency with standard surface thermodynamics [35]. Importantly, the interphase model easily accommodates multilayer-protein adsorption and it is possible to explicitly include water. A minor disadvantage of the 3D model is that it requires a slightly different way of thinking about adsorption than usually applied and a commensurately different theoretical treatment.
7.3 Abandon RSA Type Theories
The 2-D interface paradigm has spawned a number of adsorption theories including the venerable Langmuir adsorption-kinetics theory and the Langmuir isotherm derived therefrom. One of the most influential derivatives of Langmuir theory applied to protein adsorption has been the Random Sequential Adsorption (RSA) model [177, 178, 214, 216]. RSA has undergone significant modification and embellishment over the years to find agreement with experiment [214] and to account for various factors such as reversible adsorption, lateral diffusion of adsorbed molecules, protein denaturation, and so on (see, for examples, refs. [38, 178, 214, 217] and citations therein). According to the basic RSA outline, net adsorption is mediated by opposing adsorption/desorption rates. Activated-rate theory has it that these rates are controlled by activation energy barriers, with faster rates corresponding to lower barriers to adsorption/desorption (see further Section 7.4). Thus most, if not all, RSA and computational models of protein adsorption are predicated on the idea that protein/surface interaction energies are large compared to thermal energy [176-178]. But protein adsorption is not mediated by strong protein/surface interactions as a general rule (Section 4.5). Thus, RSA cannot be a general model of protein adsorption.
If strong protein/surface interactions actually were the driving force behind protein adsorption, then one would anticipate increasing adsorbent capacity with increasing surface density of functional groups that can participate in Lewis acid/base interactions between surface and protein. In other words, adsorbent capacity should increase with adsorbent hydrophilicity. But just the opposite is experimentally observed. Thus, RSA type models not only fail to correctly predict molecular-weight dependence of adsorbent capacity (Section 6.3.3) but also incorrectly predicts trends with adsorbent hydrophilicity. The reason is that RSA does not explicitly include the important role of water – the solvent – and misses an essential aspect of the process. It is long past time to move on from RSA type theories of protein adsorption.
7.3.1 Develop Kinetic Theories of the Interphase
Theories that explicitly incorporate the notion of an interphase into which proteins diffuse from bulk solution and displace interphase water can and should be developed to test the ideas of Section 5. These theories will be critical to understanding adsorption competition among proteins adsorbing from mixtures that will help predict composition of the interphase that is linked to participation in the biological response to materials (Section 1). In particular, modified diffusion theories that explain/predict how size influences molecular crowding within the interphase seem essential to formulating a predictive model that can be compared to experiment [13].
7.4 Abandon the Idea of Energy Barriers to Adsorption
One of the surprises emerging from recent studies of protein-adsorption kinetics is that mass adsorption is much faster than the change in interfacial energetics [13, 35, 214]. This finding is in sharp contrast to the expectations of traditional adsorption-kinetic theories that effectively link mass adsorption to change in interfacial tensions (see ref. [35] for a brief historical review). The core idea behind these kinetic theories is that an energy barrier retards adsorption to the interface from a subsurface zone (Section 6.3.4), rationalizing how it happens that rates-of-change of interfacial tensions are much slower (order of hours for proteins, see Fig. 8) than diffusion rates (order of milliseconds, see Section 4.8.3). Phenomenological rate equations based on this energy-barrier idea seemed to accommodate experimental kinetics data and so the energy-barrier idea was generally accepted, even though this construction of the surface region invites a number of conceptual questions. Among these, why is it that a subsurface zone and an interfacial plane do not together constitute an interphase similar to that employed by Gibbs [135] and Guggenheim [136] in development of surface thermodynamics? And how is it that solute molecules collected in the subsurface zone do not qualify as adsorbed from solution and do not contribute to decay in interfacial tensions?
Now we find that mass adsorption is temporally decoupled from change in interfacial tensions [13, 35, 214]. That is to say, molecules arrive quickly and interfacial energetics change slowly (Section 5.1). Apparently, there is no energy barrier to adsorption that limits the rate-of-change of interfacial tensions due to adsorption. Experimental evidence thus shows that time has come to find an alternative theory that explains fast mass adsorption and slow change of interfacial tension.
7.4.1 Cultivate the Interphase Concept
The dubious assembly of an interfacial plane and subsurface zone can be replaced by the single notion of a 3D interphase that quickly inflates to accommodate protein molecules diffusing from solution which subsequently slowly shrinks by expelling interphase water, as diagrammed in Fig. 7. Change in interfacial tensions is driven by the concentration of protein within the shrinking interphase [35]. This same inflating interphase concept provides an explanation for the unusual kinetics of adsorption competition between two proteins as well (Fig. 7, lower half figure) [13]. Development of the kinetic theories mentioned in Section 7.3.1 would supplant both RSA and energy-barrier theories and provide a route to testing theory against measured interfacial energetics of protein adsorption (Section 6.0).
7.5 Summary
The erroneous idea that protein adsorption is mediated by strong protein/surface interactions remains a key conceptual barrier that impedes our basic understanding of the process. This idea rationalizes use of Group 1 analytical methods that have created widespread misinformation about protein adsorption. This notion has been promulgated into a highly-restrictive 2D interface models of protein adsorption. Adsorption-kinetics theories built upon this 2D interface concept do not accommodate experimental data. Notable among these theories is RSA that incorrectly predicts molecular-weight dependence of adsorbent capacity, does not explain decreasing adsorbent capacity with increasing hydrophilicity, and provides no explanation for multi-layer adsorption. Furthermore, these kinetics theories do not explain the observation that mass-adsorption rates are temporally decoupled from rates-of-change in interfacial tensions. It is easy to show using reliable analytical methods that protein is not strongly bound to adsorbent surfaces, except under special conditions of select surface chemistries and/or fluid phases.
A new research strategy employing certified analytical methods that provide comparable measures of adsorption interpreted in terms of adsorption within a 3D interphase, rather than a pseudo-2D interface, will advance our understanding of protein adsorption toward the goal of predicting the composition of proteins adsorbed from complex biological milieu such as blood.
8.0 Conclusions
Blood-protein adsorption is a fast process by which molecules diffuse from bulk solution into an inflating three-dimensional interphase that separates this bulk solution from the physical-adsorbent surface. Adsorbed protein occupies one-or-more layers within this interphase and necessarily displaces an equivalent volume of interphase water (interphase dehydration) because two objects, water and protein, cannot occupy the same space at the same time. At mg/mL solution concentrations, mass transport into the interphase is over within milliseconds. Thereafter, the interphase slowly shrinks in volume by expelling interphase water and/or initially-adsorbed protein, causing adsorbed protein to concentrate into closely-packed arrangements. Increasing interphase concentrations is directly responsible for the concomitant decrease of interfacial tensions over tens-of-minutes-to-hours, depending on the experimental arrangement. The maximum interphase concentration, and hence adsorbent capacity, is controlled by the energetics of interphase dehydration. This maximum interphase concentration is approximately the same on a w/v (not molar) basis for all blood proteins or mixtures thereof, including serum and plasma, because the partial-specific volume (cm3/g) of blood proteins falls within a conserved range. The net free energy of blood-protein adsorption is favorable by only a few thermal units for hydrophobic adsorbents and decreases with increasing adsorbent hydrophilicity because of the increasing energetic cost of dehydrating the interphase. This energetic cost exceeds the energy gain of partitioning protein into the interphase at an adsorbent surface energy characterized by an advancing contact angle θ = 65° whereupon adsorbent capacity decreases to below detection limits. Protein/surface interactions are not strong compared to thermal energy, as widely believed by many researchers, except perhaps if the surface bears strong Lewis acid/base functional groups or ligands that specifically bind a particular kind of protein. This misconception has proliferated use of analytical methods of measuring protein adsorption that utilize an adsorbent-rinsing step which removes some-or-all of the loosely-adsorbed protein. These “dip-rinse-measure” methods account only for the strongly-bound adsorbed-protein fraction in a circular confirmation of initial misconceptions. This circular reasoning has obscured the important, controlling role of water in the protein adsorption process and has been an impediment to understanding the biophysical mechanisms of biocompatibility.
Acknowledgments
This work was supported by National Institute of Health grant PHS 2R01HL069965. The author is grateful to a long list of collaborators and students with whom he has had the pleasure of exploring the many facets of protein adsorption over many years. Special among these collaborators are Dr. Anandi Krishnan and (now) Professors Hyeran Noh, Naris Barnthip, and Purnendu Parhi. Author thanks Ms. Qiaoling Huang for a careful read of the manuscript. Author appreciates support from the Departments of Bioengineering and Materials Science and Engineering, The Pennsylvania State University.
Appendix A
Conversion of Eq. (7) of Section 4.1.2,
into concentration units uses Eq. (2), , with the identities AB,2≡σB,2XB,2 and ; where σB,2 is an activity (fugacity) coefficient and XB,2 is mole fraction solute in bulk solution. The approximation is appropriate to dilute solute solutions. Differentiating AB,2 leads to the conclusion that because the change in bulk solvent composition in the bulk phase due to protein adsorption can be expected to be very small (dCB,1 → 0, see also Section 4.8.2). Thus it is found that and substitution of this identity into the first form of Eq. (7) leads directly to the second form. Similarly, Eq. (3) can be rewritten as which, when equated with the second form of Eq. (7), leads to Eq. (8) stating that Γ2 = CB,2ΩI[P−p1].
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
A Contribution from the Hematology at Biomaterial Interfaces Research Group
Citations
- [1].Horbett T. Protein adsorption on biomaterials. In: Cooper SL, Peppas NA, Hoffman AS, Ratner BD, editors. Biomaterials: Interfacial Phenomena and Applications. Am. Chem. Soc.; Washington D. C.: 1982. pp. 234–43. [Google Scholar]
- [2].Horbett TA. Principles underlying the role of adsorbed plasma proteins in blood interactions with foreign materials. Cardiovac Pathol. 1993;2:137S–48S. [Google Scholar]
- [3].Horbett TA. The role of adsorbed proteins in tissue response to biomaterials. In: Ratner B, Hoffman A, editors. Biomaterials Science: An Introduction to Materials in Medicine. 2 ed Elsevier Academic Press; San Diego: 2004. pp. 237–46. [Google Scholar]
- [4].Horbett TA. Proteins: structure, properties, and adsorption to surface. In: Ratner BD, Hoffman A, editors. Biomaterials Science: an introduction to materials in medicine. Academic Press; San Diego: 1996. pp. 133–41. [Google Scholar]
- [5].Andrade JD, Hlady V. Protein adsorption and materials biocompatibility: a tutorial review and suggested mechanisms. Adv Polym Sci. 1986;79:3–63. [Google Scholar]
- [6].Krishnan A, Sturgeon J, Siedlecki CA, Vogler EA. Scaled interfacial activity of proteins at the liquid-vapor interface. J Biomed Mat Res. 2004;68A:544–57. doi: 10.1002/jbm.a.20104. [DOI] [PubMed] [Google Scholar]
- [7].Williams DF. On the mechanisms of biocompatibility. Biomaterials. 2008;29:2941–53. doi: 10.1016/j.biomaterials.2008.04.023. [DOI] [PubMed] [Google Scholar]
- [8].Williams DF. General concepts of biocompatibility. In: Black J, Hastings G, editors. Handbook of Biomaterial Properties. Chapman and Hall; London: 1998. pp. 481–8. [Google Scholar]
- [9].Williams DF. The blood-device interface. Medical Device Technol. 1993 September;:8–12. [Google Scholar]
- [10].Williams DF. Review: tissue-biomaterial interactions. J Mater Sci. 1987;22:3421–45. [Google Scholar]
- [11].Vogler EA. The goldilocks surface. Biomaterials. 2011;32:6670–5. doi: 10.1016/j.biomaterials.2011.05.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Brien CP, Stuart SJ, Bruce DA, Latour RA. Modeling of peptide adsorption interactions with a poly(lactic acid) surface. Langmuir. 2008:14115–24. doi: 10.1021/la802588n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Barnthip N, Parhi P, Golas A, Vogler EA. Volumetric interpretation of protein adsorption: kinetics of protein-adsorption competition from binary solution. Biomaterials. 2009;30:6495–513. doi: 10.1016/j.biomaterials.2009.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Vogler EA. Structure and reactivity of water at biomaterial surfaces. Adv Colloid Interfac. 1998;74:69–117. doi: 10.1016/s0001-8686(97)00040-7. [DOI] [PubMed] [Google Scholar]
- [15].Zhuo R, Siedlecki CA, Vogler EA. Autoactivation of blood factor XII at hydrophilic and hydrophobic surfaces. Biomaterials. 2006;27:4325–32. doi: 10.1016/j.biomaterials.2006.04.001. [DOI] [PubMed] [Google Scholar]
- [16].Ratner BD. New ideas in biomaterials science - a path to engineered biomaterials. J Biomed Mat Res. 1993;27:837–50. doi: 10.1002/jbm.820270702. [DOI] [PubMed] [Google Scholar]
- [17].Vogler EA. Interfacial chemistry in biomaterials science. In: Berg J, editor. Wettability. Marcel Dekker; New York: 1993. pp. 184–250. [Google Scholar]
- [18].Vogler EA. Practical use of concentration-dependent contact angles as a measure of solid-liquid adsorption I: theoretical aspects. Langmuir. 1992;8:2005–12. [Google Scholar]
- [19].Vogler EA. Practical use of concentration-dependent contact angles as a measure of solid-liquid adsorption II: experimental aspects. Langmuir. 1992;8:2013–20. [Google Scholar]
- [20].Krishnan A, Siedlecki C, Vogler EA. Traube-rule interpretation of protein adsorption to the liquid-vapor interface. Langmuir. 2003;19:10342–52. [Google Scholar]
- [21].Krishnan A, Siedlecki CA, Vogler EA. Mixology of protein solutions and the vroman effect. Langmuir. 2004;20:5071–8. doi: 10.1021/la036218r. [DOI] [PubMed] [Google Scholar]
- [22].Krishnan A, Wilson A, Sturgeon J, Siedlecki CA, Vogler EA. Liquid-vapor interfacial tension of blood plasma, serum and purified protein constituents thereof. Biomaterials. 2005;26:3445–53. doi: 10.1016/j.biomaterials.2004.09.016. [DOI] [PubMed] [Google Scholar]
- [23].Krishnan A, Liu Y-H, Cha P, Allara DL, Vogler EA. Interfacial Energetics of globular-blood protein adsorption to a hydrophobic surface from aqueous-buffer solution. J R Soc Interface. 2006;3:283–301. doi: 10.1098/rsif.2005.0087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Krishnan A, Liu Y-H, Cha P, Allara DL, Vogler EA. Scaled interfacial activity of proteins at a hydrophobic solid/aqueous-buffer interface. J Biomed Mater Res. 2005;75A:445–57. doi: 10.1002/jbm.a.30444. [DOI] [PubMed] [Google Scholar]
- [25].Krishnan A, Liu Y-H, Cha P, Allara DL, Vogler EA. Interfacial energetics of blood plasma and serum adsorption to a hydrophobic self-assembled monolayer surface. Biomaterials. 2006;27:3187–94. doi: 10.1016/j.biomaterials.2005.12.032. [DOI] [PubMed] [Google Scholar]
- [26].Cha P, Krishnan A, Fiore VF, Vogler EA. Interfacial energetics of protein adsorption from aqueous buffer to surfaces with varying hydrophilicity. Langmuir. 2008;24:2553–63. doi: 10.1021/la703310k. [DOI] [PubMed] [Google Scholar]
- [27].Lander LM, Siewierski LM, Brittain WJ, Vogler EA. A systematic comparison of contact angle methods. Langmuir. 1993;9:2237–9. [Google Scholar]
- [28].Krishnan A, Liu Y-H, Cha P, Allara DL, Vogler EA. An evaluation of goniometric methods. J Colloid Interf Sci. 2005;43:95–8. doi: 10.1016/j.colsurfb.2005.04.003. [DOI] [PubMed] [Google Scholar]
- [29].Aveyard R, Haydon DA. An introduction to the principles of surface chemistry. Cambridge University Press; London: 1973. [Google Scholar]
- [30].Vogler EA, Martin DA, Montgomery DB, Graper JC, Sugg HW. A graphical method for predicting protein and surfactant adsorption properties. Langmuir. 1993;9:497–507. [Google Scholar]
- [31].Noh H, Vogler EA. Volumetric interpretation of protein adsorption: partition coefficients, interphase volumes, and free energies of adsorption to hydrophobic surfaces. Biomaterials. 2006;27:5780–93. doi: 10.1016/j.biomaterials.2006.07.038. [DOI] [PubMed] [Google Scholar]
- [32].Noh H, Vogler EA. Volumetric Interpretation of Protein Adsorption: Mass and energy balance for albumin adsorption to particulate adsorbents with incrementally-increasing hydrophilicity. Biomaterials. 2006;27:5801–12. doi: 10.1016/j.biomaterials.2006.08.005. [DOI] [PubMed] [Google Scholar]
- [33].Noh H, Vogler EA. Volumetric interpretation of protein adsorption: competition from mixtures and the vroman effect. Biomaterials. 2007;28:405–22. doi: 10.1016/j.biomaterials.2006.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Noh H, Vogler EA. Volumetric interpretation of protein adsorption: ion-exchange adsorbent capacity, protein pi, and interaction energetics. Biomaterials. 2008;29:2033–48. doi: 10.1016/j.biomaterials.2008.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Barnthip N, Noh H, Leibner E, Vogler EA. Volumetric interpretation of protein adsorption: kinetic consequences of a slowly-concentrating interphase. Biomaterials. 2008;29:3062–74. doi: 10.1016/j.biomaterials.2008.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Kao P, Parhi P, Krishnan A, Noh H, Haider W, Tadigadapa S, et al. Volumetric interpretation of protein adsorption: interfacial packing of protein adsorbed to hydrophobic surfaces from surface-saturating solution concentrations. Biomaterials. 2010;32:969–78. doi: 10.1016/j.biomaterials.2010.09.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Norde W. Adsorption of proteins from solution at the solid-liquid interface. Adv Colloid Interfac S. 1986;25:267–340. doi: 10.1016/0001-8686(86)80012-4. [DOI] [PubMed] [Google Scholar]
- [38].Seigel RR, Harder P, Dahint R, Grunze M, Josse F, Mrksich M, et al. On-line detection of nonspecific protein adsorption at artificial surfaces. Anal Chem. 1997;69:3321–8. doi: 10.1021/ac970047b. [DOI] [PubMed] [Google Scholar]
- [39].Brash J, Lyman D. Adsorption of proteins and lipids to nonbiological surfaces. In: brash jl., editor. the chemistry of biosurfaces. Marcel Dekker; new York: 1971. pp. 177–222. [Google Scholar]
- [40].Vroman L. What factors determine thrombogenicity? Bull N Y Acad Med. 1972;48:302–10. [PMC free article] [PubMed] [Google Scholar]
- [41].Vroman L, Adams AL, Klings M, Fischer G. Fibrinogen, globulins, albumins, and plasma at interfaces. applied chemistry at protein interfaces. A Symposium at the 166th Meeting of the American Chemical Society; Washington, DC: American Chemical Society; 1975. pp. 255–89. [Google Scholar]
- [42].Cooper SL, Peppas NA, Hoffman AS, Ratner BD. Biomaterials: Interfacial Phenomena and Applications. Am. Chem. Soc.; Washington D. C.: 1982. Protein adsorption on biomaterials; pp. 234–43. [Google Scholar]
- [43].Brash J, Hove Pt. Effect of plasma dilution on adsorption of fibrinogen to solid surfaces. Thromb Haemostas. 1984;51:326–30. [PubMed] [Google Scholar]
- [44].Vroman L, Adams A. Adsorption of proteins out of plasma and solutions in narrow spaces. J Colloid Interf Sci. 1986;111:391–402. [Google Scholar]
- [45].Wojciechowski P, Hove PT, Brash JL. Phenomenology and mechanism of the transient adsorption of fibrinogen from plasma (vroman effect) J Colloid and Interf Sci. 1986;111:455–65. [Google Scholar]
- [46].Elwing H, Askendal A, Lundstrom I. Competition between adsorbed fibrinogen and high-molecular-weight kininogen on solid surfaces incubated in human plasma (the vroman effect): influence of solid surface wettability. J Biomed Mat Res. 1987;21:1023–8. doi: 10.1002/jbm.820210808. [DOI] [PubMed] [Google Scholar]
- [47].Shirahama H, Lyklema J, Norde W. Comparative protein adsorption in model systems. J Colloid Interf Sci. 1990;139:177–87. [Google Scholar]
- [48].Leonard EF, Vroman L. Is the vroman effect of importance in the interaction of blood with artificial materials. J Biomat Sci-Polym E. 1991;3:95–107. doi: 10.1163/156856292x00105. [DOI] [PubMed] [Google Scholar]
- [49].Wahlgren M, Arnebrant T. Protein adsorption to solid surfaces. Trends biotechnol. 1991;9:201–8. doi: 10.1016/0167-7799(91)90064-o. [DOI] [PubMed] [Google Scholar]
- [50].Wojciechowski P, Brash JL. The vroman effect in tube geometry: the influence of flow on protein adsorption measurements. J Bio Sci-Polym E. 1991;2:203–16. doi: 10.1080/09205063.1991.9756660. [DOI] [PubMed] [Google Scholar]
- [51].Brash JL, Hove PT. Protein adsorption studies on "standard" polymeric materials. J Biomat Sci-Polym E. 1993;4:591–9. doi: 10.1163/156856293x00230. [DOI] [PubMed] [Google Scholar]
- [52].Vroman L. Letter to the editors. J Biomat Sci-Polym E. 1994;6:223. [PubMed] [Google Scholar]
- [53].Claesson PM, Blomberg E, Froberg JC, Nylander T, Arnebrant T. Protein interactions at solid surfaces. Adv Colloid Interfac. 1995;57:161–227. [Google Scholar]
- [54].Lin JC, Cooper SL. In vitro fibrinogen adsorption from various dilutions of human blood plasma on glow discharge modified polyethylene. J Colloid Interf Sci. 1996;182:315–25. [Google Scholar]
- [55].Lee JH, Lee HB. Platelet adhesion onto wettability gradient surfaces in the absence and presence of plasma proteins. J Biomed Mat Res. 1998;41:304–11. doi: 10.1002/(sici)1097-4636(199808)41:2<304::aid-jbm16>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- [56].Derand H, Malmsten M. Protein interfacial behavior in microfabricated analysis systems and microarrays. In: Malmsten M, editor. Biopolymers at Interfaces. Marcel Dekker; New York: 1998. pp. 393–413. [Google Scholar]
- [57].Malmsten M. Biopolymers at Interfaces. In: Malmsten M, editor. Surfactant science series v 75. Marcel Dekker; New York: 1998. p. 656. [Google Scholar]
- [58].Jung S-Y, Lim S-M, Albertorio F, Kim G, Gurau MC, Yang RD, et al. The vroman effect: a molecular level description of fibrinogen displacement. J Am Chem Soc. 2003;125:12782–6. doi: 10.1021/ja037263o. [DOI] [PubMed] [Google Scholar]
- [59].Ariola F, Krishnan A, Vogler EA. Interfacial rheology of blood proteins adsorbed to the aqueous-buffer/air interface. Biomaterials. 2006;27:3404–12. doi: 10.1016/j.biomaterials.2006.02.005. [DOI] [PubMed] [Google Scholar]
- [60].Lee S, Ruckenstein E. Adsorption of proteins onto polymeric surfaces of different hydrophilicities - a case study with bovine serum albumin. J Colloid and Interf Sci. 1988;125:365–79. [Google Scholar]
- [61].Jeon J, Superline R, Raghavan S. Quantitative analysis of adsorbed serum albumin on segmented polyurethane using FT-IR/ATR spectroscopy. Appl Spectrosc. 1992;46:1644–8. [Google Scholar]
- [62].Graham DE, Phillips MC. Proteins at liquid interfaces. J Colloid Interf Sci. 1979;70:415–26. [Google Scholar]
- [63].Feijter JAD, Benhamins J, Veer FA. Ellipsometry as a tool to study the adsorption behavior of synthetic and biopolymers at the air-water interface. Biopolymers. 1978;17:1759–72. [Google Scholar]
- [64].Brynda E, Cepalova N, Stol M. Equilibrium adsorption of human serum albumin and human fibrinogen on hydrophobic and hydrophilic surfaces. J Biomed Mat Sci. 1984;18:685–93. doi: 10.1002/jbm.820180609. [DOI] [PubMed] [Google Scholar]
- [65].Lassen B, Malmsten M. Structure of protein layers during competitive adsorption. J Colloid Interf Sci. 1996;180:339–49. [Google Scholar]
- [66].Chen H-MH Wen-Yih, Chien-Chen Lin, Fu-Yung Lin, Yu-Chia Chan. Effect of temperature on hydrophobic interaction between proteins and hydrophobic adsorbents: studies by isothermal titration calorimetry and the van’t hoff equation. Langmuir. 2003;19:9395–403. [Google Scholar]
- [67].Zhou C, Friedt J-M, Angelova A, Choi K-H, Laureyn W, Frederix F, et al. Human immunoglobulin adsorption investigated by means of quartz crystal microbalance dissipation, atomic force microscopy, surface acoustic wave, and surface plasmon resonance techniques. Langmuir. 2004;20:5870–8. doi: 10.1021/la036251d. [DOI] [PubMed] [Google Scholar]
- [68].Beverung C, Radke C, Blanch H. Protein adsorption at the oil/water interface: characterization of adsorption kinetics by dynamic interfacial tension measurements. Biophys Chem. 1999;81:59–80. doi: 10.1016/s0301-4622(99)00082-4. [DOI] [PubMed] [Google Scholar]
- [69].Parhi P, Golas A, Vogler EA. Role of water and proteins in the attachment of mammalian cells to surfaces: a review. J Adhes Sci Technol. 2010;24:853–88. [Google Scholar]
- [70].Gao L, McCarthy TJ. Wetting 101. Langmuir. 2009;25:14105–15. doi: 10.1021/la902206c. [DOI] [PubMed] [Google Scholar]
- [71].Vogler EA. On the origins of water wetting terminology. In: Morra M, editor. Water in Biomaterials Surface Science. John Wiley and Sons; New York: 2001. pp. 150–82. [Google Scholar]
- [72].Elwing H. Protein absorption and ellipsometry in biomaterial research. Biomaterials. 1998;19:397–406. doi: 10.1016/s0142-9612(97)00112-9. [DOI] [PubMed] [Google Scholar]
- [73].Ballantine DS. Acoustic wave sensors-theory, design, and physico-chemical applications. Academic Press; San Diego: 1997. [Google Scholar]
- [74].Lu C, Czanderna AW. Applications of piezoelectric quartz crystal microbalances. Elsevier; New York: [Google Scholar]
- [75].Henry C. Measuring the masses: quartz crystal microbalances. Anal Chem. 1996:625A. [Google Scholar]
- [76].Handley J. Quartz crystal microbalances. Anal Chem. 2001:225A. doi: 10.1021/ac012432d. [DOI] [PubMed] [Google Scholar]
- [77].Sullivan CKO, Guilbault GG. Commercial quartz crystal microbalances-theory and applications. Biosens Bioelectron. 1999;14:663–70. [Google Scholar]
- [78].Kanazawa K, Gordon J. The oscillation frequency of a quartz resonator in contact with liquid. Anal Chim Acta. 1985;175:99–105. [Google Scholar]
- [79].Vaidya SS, Ofol RY. Adsorption and interaction of fibronectin and human serum albumin at the liquid-liquid interface. Langmuir. 2005;21:5852–58. doi: 10.1021/la046766k. [DOI] [PubMed] [Google Scholar]
- [80].Lassen B, Malmsten M. Competitive protein adsorption at plasma polymer surfaces. J Colloid Interf Sci. 1997;186:9–16. doi: 10.1006/jcis.1996.4529. [DOI] [PubMed] [Google Scholar]
- [81].Van Wagenen RA, Rockhold S, Andrade JD. Probing protein adsorption: total internal reflection intrinsic fluorescence. biomaterials: interfacial phenomena and applications. Am Chem Soc; Washington, D. C.: 1982. pp. 351–70. [Google Scholar]
- [82].Mrksich M, Sigal GB, Whitesides GM. Surface plasmon resonance permits in situ measurement of protein adsorption on self-assembled monolayers of alkanethiolates on gold. Langmuir. 1995;11:4383–5. [Google Scholar]
- [83].Stenberg E, Persson B, Roos H, Urbaniczky C. Quantitative determination of surface concentration of protein with surface plasmon resonance using radiolabeled proteins. J Colloid Interf Sci. 1991;143:513–26. [Google Scholar]
- [84].Green RJ, Davies J, Davies MC, Roberts CJ, Tendler SJB. Surface plasmon resonance for real time in situ analysis of protein adsorption to polymer surfaces. Biomaterials. 1997;18:405–13. doi: 10.1016/s0142-9612(96)00141-x. [DOI] [PubMed] [Google Scholar]
- [85].Leibner ES, Barnthip N, Chen W, Baumrucker CR, Badding JV, Pishko M, et al. Human serum albumin adsorption to expanded polytetrafluoroethylene. Acta Biomater. 2009;5:1389–98. doi: 10.1016/j.actbio.2008.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Desbuquo B, Aurbach GD. Effects of iodination on distribution of peptide hormones in aqueous 2-phase polymer systems. Biochem J. 1974;143:83–91. doi: 10.1042/bj1430083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Holmberg M, Stibiusa KB, Ndonia S, Larsena NB, Kingshottb P, Houc XL. Protein aggregation and degradation during iodine labeling and its consequences for protein adsorption to biomaterials. Anal Biochem. 2007;361:120–5. doi: 10.1016/j.ab.2006.11.016. [DOI] [PubMed] [Google Scholar]
- [88].Anraku M, Kragh-Hansen U, Kawai K, Maruyama T, Yamasaki Y, Takakura Y, et al. Validation of the chloramine-T induced oxidation of human serum albumin as a model for oxidative damage in vivo. Pharmaceut Res. 2003;20:684–92. doi: 10.1023/a:1023219420935. [DOI] [PubMed] [Google Scholar]
- [89].Efimova YM, Wierczinski B, Haemers S, van Well AA. Changes in the secondary structure of proteins labeled with I-125: CD spectroscopy and enzymatic activity studies. J Radioanal Nucl Chem. 2005;264:91–6. [Google Scholar]
- [90].Osborne JC, Schaefer EJ, Powell GM, Lee NS, Zech LA. Molecular-properties of radioiodinated apolipoprotein-a-i. J Biol Chem. 1984;259:347–53. [PubMed] [Google Scholar]
- [91].Sheardown H, Cornelius RM, Brash JL. Measurement of protein adsorption to metals using radioiodination methods: a caveat. Colloid Surface B. 1997;10:29–33. [Google Scholar]
- [92].Hunter JR, Kilpatrick PK, Carbonell RG. Lysozyme adsorption at the air/water interface. J Colloid and Interf Sci. 1989;137:462–82. [Google Scholar]
- [93].Scheer ATVD, Feijen J, Elhorst JK, Dagneaux PGLCK, Smolders CA. The Feasibility of radiolabeling for human serum albumin adsorption studies. J Colloid and Interf Sci. 1978;66:136–45. [Google Scholar]
- [94].Teske CA, Schroeder M, Simon R, Hubbuch Jr. Protein-labeling effects in confocal laser scanning microscopy. J Phys Chem B. 2005;109:13811–7. doi: 10.1021/jp050713+. [DOI] [PubMed] [Google Scholar]
- [95].Steiner RF, Edelhoch H. Fluorescent protein conjugates. Chem Rev. 1962;62:457–83. [Google Scholar]
- [96].Clark DC, Coke M, Mackie AR, Pinder AC, Wilson DR. Molecular diffusion and thickness measurements of protein-stabilized thin liquid films. J Colloid Interf Sci. 1990;138:207–19. [Google Scholar]
- [97].Hussain AA, Awad R, Crooks PA, Dittert LW. Chloramine-t in radiolabeling techniques .1. kinetics and mechanism of the reaction between chloramine-t and amino-acids. Anal Biochem. 1993;214:495–9. doi: 10.1006/abio.1993.1528. [DOI] [PubMed] [Google Scholar]
- [98].Hussain AA, Jona JA, Yamada A, Dittert LW. chloramine-t in radiolabeling techniques .2. a nondestructive method for radiolabeling biomolecules by halogenation. Anal Biochem. 1995;224:221–6. doi: 10.1006/abio.1995.1033. [DOI] [PubMed] [Google Scholar]
- [99].Walker JM. The protein protocols handbook. 2nd ed Humana Press; Totowa, N.J.: 2002. [Google Scholar]
- [100].Vogler EA, Siedlecki CA. Contact activation of blood plasma coagulation. Biomaterials. 2009;30:1857–69. doi: 10.1016/j.biomaterials.2008.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Parhi P, Golas A, Barnthip N, Vogler EA. Volumetric interpretation of protein adsorption: capacity scaling with adsorbate molecular weight and adsorbent surface energy. Biomaterials. 2009;30:6814–24. doi: 10.1016/j.biomaterials.2009.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Richards FM. Areas, volumes, packing and protein structure. Ann Rev Biophys Bioeng. 1977;6:151–76. doi: 10.1146/annurev.bb.06.060177.001055. [DOI] [PubMed] [Google Scholar]
- [103].Chothia C. Structural invariants in protein folding. Nature. 1975;254:304–8. doi: 10.1038/254304a0. [DOI] [PubMed] [Google Scholar]
- [104].Miller S, Lesk A, Janins J, Chothia C. The accessible surface area and stability of oligomeric proteins. Nature. 1987;328:834–6. doi: 10.1038/328834a0. [DOI] [PubMed] [Google Scholar]
- [105].Miller S, Janin J, Lesk A, Chothia C. Interior and surface of monomeric proteins. J Mol Biol. 1987;196:641–56. doi: 10.1016/0022-2836(87)90038-6. [DOI] [PubMed] [Google Scholar]
- [106].Tsai J, Taylor R, Chothia C, Gerstin M. The packing density in proteins: standard radii and volumes. J Mol Bio. 1999;290:253–66. doi: 10.1006/jmbi.1999.2829. [DOI] [PubMed] [Google Scholar]
- [107].Gerstein M, Chothia C. Packing at the protein-water interface. Proc Natl Acad Sc. 1996;93:10167–72. doi: 10.1073/pnas.93.19.10167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Lu JR, Su TJ, Penfold J. Adsorption of serum albumins at the air/water interface. Langmuir. 1999;15:6975–83. [Google Scholar]
- [109].Pugnaloni LA, Ettelaie R, Dickinson E. Do mixtures of proteins phase separate at interfaces? Langmuir. 2003;19:1923–6. [Google Scholar]
- [110].Vogler EA. How water wets biomaterials. In: Morra M, editor. Water in Biomaterials Surface Science. John Wiley and Sons; New York: 2001. pp. 269–90. [Google Scholar]
- [111].Gaines GL. The Ion-exchange properties of muscovite mica. J Phys Chem. 1957;61:1408–13. [Google Scholar]
- [112].Terashima H. A direct measurement of the ion-exchange capacity of muscovite mica using a mettler ultramicrobalance. J Colloid Interf Sci. 2002;245:81–5. doi: 10.1006/jcis.2001.7984. [DOI] [PubMed] [Google Scholar]
- [113].Vogler EA, Graper JC, Sugg HW, Lander LM, Brittain WJ. Contact activation of the plasma coagulation cascade.2. protein adsorption on procoagulant surfaces. J Biomed Mat Res. 1995;29:1017–28. doi: 10.1002/jbm.820290814. [DOI] [PubMed] [Google Scholar]
- [114].Castillo E, Koenig J, Anderson J. Characterization of protein adsorption on soft contact lenses i. conformational changes of adsorbed human serum albumin. Biomaterials. 1984;5:319–25. doi: 10.1016/0142-9612(84)90029-2. [DOI] [PubMed] [Google Scholar]
- [115].Castillo E, Koenig J, Anderson J. Protein adsorption on hydrogels ii. reversible and irreversible interactions between lysozyme and soft contact lens surfaces. Biomaterials. 1985;6:338–45. doi: 10.1016/0142-9612(85)90089-4. [DOI] [PubMed] [Google Scholar]
- [116].Castillo E, Koenig J, Anderson J, Jentoft N. Protein adsorption on soft contact lenses. iii. mucin. Biomaterials. 1986;7:9–16. doi: 10.1016/0142-9612(86)90081-5. [DOI] [PubMed] [Google Scholar]
- [117].Castillo EJ, Koenig JL, Anderson JM. Characterization of protein adsorption on soft contact lenses iv. comparison of in vivo spoilage with the in vitro adsorption of tear proteins. Biomaterials. 1986;7:89–96. doi: 10.1016/0142-9612(86)90062-1. [DOI] [PubMed] [Google Scholar]
- [118].Horbett TA, Hoffman AS. Bovine plasma protein adsorption onto radiation-grafted hydrogels based on hydroxyethyl methacrylate and n-vinylpyrrolidone. Applied Chemistry at Protein Interfaces: Am Chem Soc. 1975:230–54. [Google Scholar]
- [119].Feldman K, Hahneer G, Spencer ND, Harder P, Grunze M. Probing resistance to protein adsorption of oligo(ethylene glycol)-terminated self-assembled monolayers by scanning force microscopy. J Am Chem Soc. 1999;12:10134–41. [Google Scholar]
- [120].Chapman RG, Ostuni E, Takayama S, Homin RE, Yan L, Whitesides GM. Surveying for surfaces that resist the adsorption of proteins. J Am Chem Soc. 2000;122:8303–4. [Google Scholar]
- [121].Ktano H, Ichikawa K, Ide M, Fukuda M, Mizuno W. Fourier transform infrared study on the state of water sorbed to polyethylene glycol films. Langmuir. 2001;17:1889–95. [Google Scholar]
- [122].Harder P, Grunze M, Dahint R, Whitesides GM, Laibinis PE. Molecular conformation in oligo(ethylene glycol)-terminated self-assembled monolayers on gold and silver surfaces determines their ability to resist protein adsorption. J Phys Chem B. 1998;102:426–36. [Google Scholar]
- [123].Pertsin AJ, Grunze M. Computer simulation of water near the surface of oligo(ethylene glycol)-terminated alkanethiol self-assembled monolayers. Langmuir. 2000;16:8829–41. [Google Scholar]
- [124].Prime KL, Whitesides GM. Adsorption of proteins onto surfaces containing end-attached oligo ethylene oxide: a model system using self-assembled monolayers. J Am Chem Soc. 1993;115:10714–21. [Google Scholar]
- [125].Chapman RG, Ostuni E, Liang MN, Meluleni G, Kim E, Yan L, et al. Polymeric thin films that resist the adsorption of proteins and the adhesion of bacteria. Langmuir. 2001;17:1225–33. [Google Scholar]
- [126].Kane RS, Deschatelets P, Whitesides GM. Kosmotropes form the basis of protein-resistant surfaces. Langmuir. 2003;19:2388–91. [Google Scholar]
- [127].Ostuni E, Grzybowski BA, Mrksich M, Roberts CS, Whitesides GM. Adsorption of proteins to hydrophobic sites on mixed self-assembled monolayers. Langmuir. 2003;19:1861–72. [Google Scholar]
- [128].Sethuraman A, Han M, Kane RS, Belfort G. Effect of surface wettability on the adhesion of proteins. Langmuir. 2004;20:7779–88. doi: 10.1021/la049454q. [DOI] [PubMed] [Google Scholar]
- [129].Vogler EA. On the biomedical relevance of surface spectroscopy. J Electron Spectroscopy and Related Phenomenon. 1996;81:237–47. [Google Scholar]
- [130].Kao P, Allara DL, Tadigadapa S. Fabrication and performance characteristics of high-frequency micromachined bulk acoustic wave quartz resonator arrays. Meas Sci Technol. 2009;20:124007–15. [Google Scholar]
- [131].Hook F, Kasemo B. Variations in coupled water, viscoelastic properties, and film thickness of a mefp-1 protein film during adsorption and cross-linking: a quartz crystal microbalance with dissipation monitoring, ellipsometry, and surface plasmon resonance study. Anal Chem. 2001;73:5796–804. doi: 10.1021/ac0106501. [DOI] [PubMed] [Google Scholar]
- [132].Hook F, Rodahl M, Brzezinski P, Kasemo B. Energy Dissipation kinetics for protein and antibody-antigen adsorption under shear oscillation on a quartz crystal microbalance. Langmuir. 1998;14:729–34. [Google Scholar]
- [133].Hook F, Voros J, Rodahl M, Kurrat R, Boni P, Ramsden JJ, et al. A comparative study of protein adsorption on titanium oxide surfaces using in situ ellipsometry, optical waveguide lightmode spectroscopy, and quartz crystal microbalance/dissipation. Colloid Surface B. 2002;24:155–70. [Google Scholar]
- [134].Rosen MJ. Surfactants and interfacial phenomena. Wiley; New York: 1978. [Google Scholar]
- [135].Gibbs JW. The scientific papers of J. Willard Gibbs. Dover Publications; New York: 1961. [Google Scholar]
- [136].Guggenheim EA. Thermodynamics: an advanced treatment for chemists and physicists. 5 ed Wiley; New York: 1967. [Google Scholar]
- [137].Vogler EA. Water and the acute biological response to surfaces. J Biomat Sci Polym Edn. 1999;10:1015–45. doi: 10.1163/156856299x00667. [DOI] [PubMed] [Google Scholar]
- [138].Vogler EA, Graper JC, Harper GR, Lander LM, Brittain WJ. Contact activation of the plasma coagulation cascade.1. procoagulant surface energy and chemistry. J Biomed Mat Res. 1995;29:1005–16. doi: 10.1002/jbm.820290813. [DOI] [PubMed] [Google Scholar]
- [139].Liu X, Lim JY, Donahue HJ, Dhurjhati R, Andrea MM, Vogler EA. Influence of substratum surface hydrophilicity/hydrophobicity on osteoblast adhesion, morphology, and focal adhesion assembly. Biomaterials. 2007;28:4535–50. [Google Scholar]
- [140].Lim JY, Liu X, Vogler EA, Donahue HJ. Systematic variation in osteoblast adhesion and phenotype with substratum surface characteristics. J Biomed Mat Res. 2004;68A:504–12. doi: 10.1002/jbm.a.20087. [DOI] [PubMed] [Google Scholar]
- [141].Gao L, McCarthy TJ. Teflon is Hydrophilic. comments on definitions of hydrophobic, shear versus tensile hydrophobicity, and wettability characterization. Langmuir. 2008;24:9183–8. doi: 10.1021/la8014578. [DOI] [PubMed] [Google Scholar]
- [142].Xu X-HN, Yeung ES. Long-range electrostatic trapping of single-protein molecules at a liquid-solid interface. Science. 1998;281:1650–3. doi: 10.1126/science.281.5383.1650. [DOI] [PubMed] [Google Scholar]
- [143].Xu X, Lenhoff AM. A predictive approach to correlating protein adsorption isotherms on ion-exchange media. J Phys Chem B. 2008 doi: 10.1021/jp0754233. [DOI] [PubMed] [Google Scholar]
- [144].Collins KD. Sticky ions in biological systems. Proc Natl Acad Sci USA. 1995;92:5553–7. doi: 10.1073/pnas.92.12.5553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [145].Hofmeister F. zur lehre von der wirkung der salze (to the theory of the effect of salts) Archiv fur Experimentelle Pathologie und Pharmakologie (Arch Exp Path Pharm) 1888;24:247–69. [Google Scholar]
- [146].Anderson NL, Anderson NG. The human plasma proteome: history, character, and diagnostic prospects. Mol Cell Proteomics. 2002;1:845–67. doi: 10.1074/mcp.r200007-mcp200. [DOI] [PubMed] [Google Scholar]
- [147].Chalikian TV, Totrov M, Abagyan R, Breslauer KJ. The hydration of globular proteins as derived from volume and compressibility measurements: cross correlating thermodynamic and structural data. J Mol Biol. 1996;260:588–603. doi: 10.1006/jmbi.1996.0423. [DOI] [PubMed] [Google Scholar]
- [148].Chalikian TV, Breslauer KJ. On volume changes accompanying conformational transitions of biopolymers. Biopolymers. 1996;39:619–26. doi: 10.1002/(sici)1097-0282(199611)39:5<619::aid-bip1>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- [149].Durchschlag H, Zipper P. Comparative investigations of biopolymer hydration by physicochemical and modeling techniques. Biophys Chem. 2001;93:141–57. doi: 10.1016/s0301-4622(01)00217-4. [DOI] [PubMed] [Google Scholar]
- [150].Young ME, Carroad PA. Estimation of diffusion coefficients of proteins. Biotechnol Bioeng. 1980;22:947–55. [Google Scholar]
- [151].Walters RR, Graham JF, Moore RM, Anderson DJ. Protein diffusion coefficient measurements by laminar flow analysis: method and applications. Anal Biochem. 1984;140:190–5. doi: 10.1016/0003-2697(84)90152-0. [DOI] [PubMed] [Google Scholar]
- [152].Fernsler J. A quasi-equilibrium theory of protein adsorption. Langmuir. 2011;27:148–57. doi: 10.1021/la103316j. [DOI] [PubMed] [Google Scholar]
- [153].Xu L-C, Siedlecki CA. Effects of surface wettability and contact time on protein adhesion to biomaterial surfaces. Biomaterials. 2007;28:3273–83. doi: 10.1016/j.biomaterials.2007.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Adamczyk Z, Barbasz J, Ciesìla M. Kinetics of fibrinogen adsorption on hydrophilic substrates. Langmuir. 2011;26:11,934–11,945. doi: 10.1021/la101261f. [DOI] [PubMed] [Google Scholar]
- [155].Horbett TA, Klumb LA. Cell culturing: surface aspects and considerations. In: Brash JL, Wojciechowski PW, editors. Interfacial Phenomena and Bioproducts. Marcel Dekker; New York: 1996. pp. 351–445. [Google Scholar]
- [156].Barngrover D. Substrata for anchorage-dependent cells. In: Thilly WG, editor. Mammalian Cell Technology. Butterworths; Boston: 1986. pp. 131–49. [Google Scholar]
- [157].Kruse PF, Patterson MK. Tissue culture : methods and applications. Academic Press; New York: 1973. [Google Scholar]
- [158].Paul J. Cell and tissue culture. Livingstone; Edinburgh: 1965. [Google Scholar]
- [159].Freshney RI. Culture of animal cells: a manual of basic technique. Alan R. Liss, Inc.; New York: 1983. [Google Scholar]
- [160].Putnam FW. Alpha, beta, gamma, omega - the roster of the plasma proteins. In: Putnam FW, editor. The Plasma Proteins: Structure, Function, and Genetic Control. Academic Press; New York: 1975. pp. 58–131. [Google Scholar]
- [161].Purves WK. Life, the science of biology. 7th ed W.H. Freeman and Co.; Sunderland, Mass.: 2004. [Google Scholar]
- [162].Armstrong KF, Jackson SM. Anatomy & physiology for nurses. 8th ed Tindall; London: 1972. [Google Scholar]
- [163].Guyton AC, Hall JE. Textbook of medical physiology. 11th ed Elsevier Saunders; Philadelphia: 2006. [Google Scholar]
- [164].Vogler EA, Graper JC, Harper GR, Sugg HW, Lander LM, Brittain WJ. Contact activation of the plasma coagulation cascade I. procoagulant surface chemistry and energy. J Biomed Mat Res. 1995;29:1005–16. doi: 10.1002/jbm.820290813. [DOI] [PubMed] [Google Scholar]
- [165].Vogler EA. Biological properties of water. In: Morra M, editor. Water in Biomaterials Surface Science. John Wiley and Sons; New York: 2001. pp. 4–24. [Google Scholar]
- [166].Ratner BD, Hoffman AS, Schoen FJ, Lemmons JE. Biomaterials science: an introduction to materials in medicine. 2 ed Elsevier Academic Press; New York: 2004. [Google Scholar]
- [167].Park JB. Biomaterials science and engineering. Plenum Press; New York: 1984. [Google Scholar]
- [168].Robinson GW, Zhu S-B, Singh S, Evans MW. Water in biology, chemistry, and physics. World Scientific; London: 1996. [Google Scholar]
- [169].Vinogradov SN, Linnell RH. Hydrogen bonding. Van Nostrand Reinhold Co.; New York: 1971. [Google Scholar]
- [170].Yaminsky VV, Vogler EA. Hydrophobic hydration. Curr opin Colloid In. 2001;6:342–9. [Google Scholar]
- [171].Zettlemoyer AC. Hydrophobic surfaces. In: Fowkes FM, editor. Hydrophobic Surfaces. Academic Press; New York: 1969. pp. 1–27. [Google Scholar]
- [172].Harkins WD, Brown FE, Davies ECH. The structure of the surfaces of liquids, and solubility as related to the work done by the attraction of two liquid surfaces as they approach each other. Surface Tension. V. J Am Chem Soc. 1917;39:354–64. [Google Scholar]
- [173].Kamusewitz H, Possart W, Paul D. Measurement of Thermodynamic quantities by means of solid/water vapor and solid/water/hexadecane systems. In: Mittal KL, Lee KW, editors. Polymer Surfaces and Interfaces: Characterization, Modification, and Application. VSP; 1997. pp. 125–43. [Google Scholar]
- [174].Fowkes FM. Role of acid-base interfacial bonding in adhesion. J Adhes Sci Tech. 1987;1:7–27. [Google Scholar]
- [175].Henniker JC. The depth of the surface zone of a liquid. Rev Mod Phys. 1949;21:322. [Google Scholar]
- [176].Fang F, Szleifer I. Competitive adsorption in model charged protein mixtures: Equilibrium isotherms and kinetics behavior. J Chem Phys. 2003;119:1053–65. [Google Scholar]
- [177].Sagvolden G, Glaever I, Feder J. Characteristic protein adhesion forces on glass and polystyrene substrates by atomic force microscopy. Langmuir. 1998;14:5984–7. [Google Scholar]
- [178].Slomkowski S, Sosnowski S, Przerwa E. Reversible Adsorption of spherical particles from binary mixtures: long-time behavior. C R Chimie. 2003;6:1393–401. [Google Scholar]
- [179].Szleifer I. Protein adsorption on surfaces with grafted polymers: a theoretical approach. Biophys J. 1997;72:595–612. doi: 10.1016/s0006-3495(97)78698-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [180].Satulovsky J, Carignano MA, I. S. Kinetic and thermodynamic control of protein adsorption. Proc Natl Acad Sci. 2000;97:9037–41. doi: 10.1073/pnas.150236197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [181].Gong P, Szleifer I. Competitive adsorption of model charged proteins: the effect of total charge and charge distribution. J Colloid Interf Sci. 2004;278:81–90. doi: 10.1016/j.jcis.2004.05.023. [DOI] [PubMed] [Google Scholar]
- [182].Latour RA. Thermodynamic perspectives on the molecular mechanisms providing protein adsorption resistance that include protein-surface interactions. J Biomed Mater Res. 2006;78A:843–54. doi: 10.1002/jbm.a.30818. [DOI] [PubMed] [Google Scholar]
- [183].Hartvig RA, Weert Mvd, Østergaard J, Jorgensen L, Jensen H. Protein adsorption at charged surfaces: the role of electrostatic interactions and interfacial charge regulation. Langmuir. 2011;27:2634–43. doi: 10.1021/la104720n. [DOI] [PubMed] [Google Scholar]
- [184].Yaminsky VV, Ohnishi S, Vogler EA, Horn RG. Stability of aqueous films between bubbles. part 1. the effect of speed on bubble coalescence in purified water and simple electrolyte solutions. Langmuir. 2010;26:8061–74. doi: 10.1021/la904481d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [185].Yaminsky VV, Ohnishi S, Vogler EA, Horn RG. Stability of aqueous films between bubbles. part 2. effects of trace impurities and evaporation. Langmuir. 2010;26:8075–80. doi: 10.1021/la904482n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [186].Vasquez M, Nemethyt G, Scheraga HA. Conformational energy calculations on polypeptides and proteins. Chem Rev. 1994;94:2183–239. [Google Scholar]
- [187].Head-Gordon T, Hura G. Water structure from scattering experiments and simulation. Chem Rev. 2002;102:2651–70. doi: 10.1021/cr0006831. [DOI] [PubMed] [Google Scholar]
- [188].Cramer CJ, Truhlar DG. Implicit solvation models: equilibria, structure, spectra, and dynamics. Chem Rev. 1999;99:2161–220. doi: 10.1021/cr960149m. [DOI] [PubMed] [Google Scholar]
- [189].Extrand CW. A Thermodynamic model for wetting free energies from contact angles. Langmuir. 2003;19:646–9. [Google Scholar]
- [190].Extrand CW. A Thermodynamic model for contact angle hysteresis. J Colloid and Interf Sci. 1998;207:11–9. doi: 10.1006/jcis.1998.5743. [DOI] [PubMed] [Google Scholar]
- [191].Bond WN, Puls HO. The change of surface tension with time. Phil Mag. 1937;24:864–88. [Google Scholar]
- [192].Brash JL, Lyman DJ. Adsorption of plasma proteins in solution to uncharged, hydrophobic polymer surfaces. J Biomed Mat Res. 1969;3:175–89. doi: 10.1002/jbm.820030114. [DOI] [PubMed] [Google Scholar]
- [193].Benjamins J, Lyklema J, Lucassen-Reynders EH. Compression/expansion rheology of oil/water interfaces with adsorbed proteins. comparison with the air/water surface. Langmuir. 2006;22:6181–8. doi: 10.1021/la060441h. [DOI] [PubMed] [Google Scholar]
- [194].Hiemenz PC. Principles of colloid and surface chemistry. M. Dekker; New York: 1977. [Google Scholar]
- [195].Birdi KS. Handbook of surface and colloid chemistry. CRC Press; Boca Raton, FL: 1997. p. 763. [Google Scholar]
- [196].Neumann AW, Good RJ. Techniques of Measuring contact angles. Surf and Colloid Sci. 1979;11:31–91. [Google Scholar]
- [197].duNouy PL. Surface equilibria of biological and organic colloids. The Chemical Catalog Co.; New York: 1926. [Google Scholar]
- [198].duNouy PL. Surface Tension of Serum. XIII. On certain physicochemical changes in serum as a result of immunization. J Exp Med. 1925;41:779–93. doi: 10.1084/jem.41.6.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [199].Bashforth F, Adams JC. An attempt to test the theories of capillary action of drops of fluid with an explanation of the method of integration employed in constructing the tables which give the theoretical forms of such drops. Cambridge University Press; Cambridge: 1883. [Google Scholar]
- [200].Girault HH, Schiffrin DJ, Smith BDV. The measurement of interfacial tension of pendant drops using a video image profile digitizer. J Colloid Interf Sci. 1984;101:257–66. [Google Scholar]
- [201].Lin SY, McKeigue K, Maldrelli C. Diffusion-controlled surfactant adsorption studied by pendant drop digitization. AIChE J. 1990;36:1785–95. [Google Scholar]
- [202].Hansen FK, Rodsrud G. Surface tension by pendant drop. J Colloid Interf Sci. 1991;141:1–9. [Google Scholar]
- [203].Frommer MA, Miller IR. Adsorption of dna at the air-water interface. J Phys Chem. 1968;72:2862–6. doi: 10.1021/j100854a029. [DOI] [PubMed] [Google Scholar]
- [204].Strey R, Vilsanen Y, Aratono M, Kratohvil JP, Yin Q, Friberg SE. On the necessity of using activities in the gibbs equation. J Phys Chem B. 1999;103:9112–6. [Google Scholar]
- [205].Wills PR, Comper WD, Winzor DJ. Thermodynamic non-ideality in macromolecular solutions: interpretation of viral coefficients. Archiv Biochem Biophys. 1993;300:206–12. doi: 10.1006/abbi.1993.1029. [DOI] [PubMed] [Google Scholar]
- [206].Knezic D. Thermodynamic properties of supersaturated protein solutions [Ph. D.] Polytechnic University; New York: 2002. [Google Scholar]
- [207].Golas A, Parhi P, Dimachkie ZO, Siedlecki CA, Vogler EA. Surface-energy dependent contact activation of blood factor XII. Biomaterials. 2010;31:1068–79. doi: 10.1016/j.biomaterials.2009.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [208].Feder J. Random sequential adsorption. J Theor Biol. 1980;87:237–54. [Google Scholar]
- [209].Swendsen RH. Dynamics of random sequential adsorption. Phys Rev A. 1981;24:504–8. [Google Scholar]
- [210].Ghasemi H, Ward CA. Sessile-water-droplet contact angle dependence on adsorption at the solid-liquid interface. J Phys Chem C. 2010;114:5088–100. [Google Scholar]
- [211].Zhuo R, Colombo P, Pentane C, Vogler EA. Silicon oxycarbide glasses for blood-contact applications. Acta Biomater. 2005;1:583–9. doi: 10.1016/j.actbio.2005.05.005. [DOI] [PubMed] [Google Scholar]
- [212].Zhuo R, Miller R, Buzzard KM, Siedlecki CA, Vogler EA. Procoagulant stimulus processing by the intrinsic pathway of blood plasma coagulation. Biomaterials. 2005;26:2965–73. doi: 10.1016/j.biomaterials.2004.08.008. [DOI] [PubMed] [Google Scholar]
- [213].Zhuo R, Siedlecki CA, Vogler EA. Competitive-protein adsorption in contact activation of blood factor XII. Biomaterials. 2007;28:4355–69. doi: 10.1016/j.biomaterials.2007.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [214].Clark AJ, Colicky A, Haynes CA, Whitehead LA. A new model of protein adsorption kinetics derived from simultaneous measurement of mass loading and changes in surface energy. Langmuir. 2007;23:5591–600. doi: 10.1021/la0635350. [DOI] [PubMed] [Google Scholar]
- [215].Langmuir I. The adsorption of gases on plane surfaces of glass, mica, and platinum. 1918. pp. 1361–403.
- [216].Feeder J, Giaever I. Adsorption of ferritin. J Colloid Interf Sci. 1980;78:144–54. [Google Scholar]
- [217].Robeson JL, Tilton RD. Spontaneous reconfiguration of adsorbed lysozyme layers observed by total internal reflection fluorescence with a ph-sensitive fluorophore. Langmuir. 1996;12:6104–13. [Google Scholar]
- [218].Marcel S, Vogler EA, Ferment L, Watt T, Hayne S, Saga DY. Peptide, Protein, and cellular interactions with self-assembled monolayer model surfaces. J Biomed Mat Res. 1993;27:1463–76. doi: 10.1002/jbm.820271202. [DOI] [PubMed] [Google Scholar]