Abstract
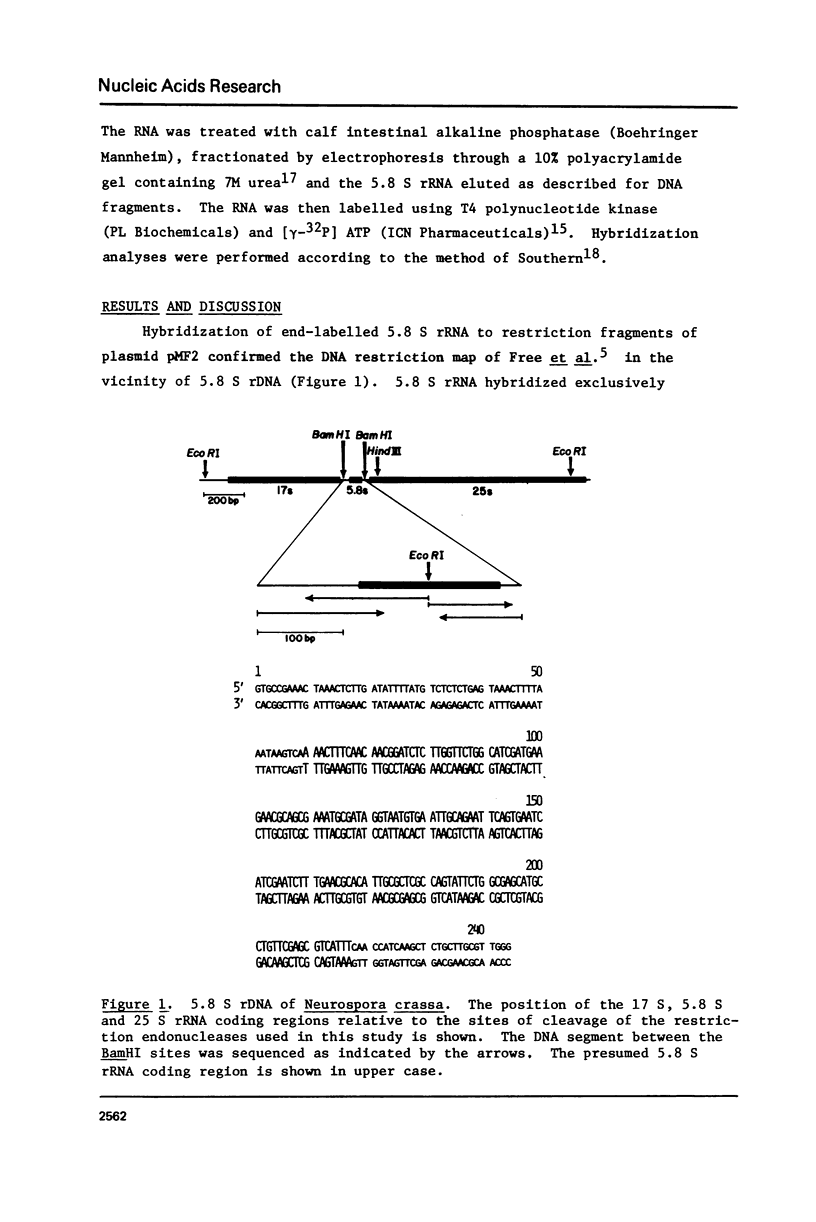
The nucleotide sequence of Neurospora crassa 5.8 S rDNA and adjacent regions has been determined. The deduced 5.8 S rRNA sequence of Neurospora differs from the 5.8 S rRNA sequence of Saccharomyces cerevisiae at 13 of 158 residues. Nine of these differences are clustered in a segment capable of forming a short hairpin secondary structure thought to be involved in the 28 S - 5.8 S rRNA complex. These differences occur in pairs such that the potential secondary structure is preserved.
Full text
PDF





Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bell G. I., DeGennaro L. J., Gelfand D. H., Bishop R. J., Valenzuela P., Rutter W. J. Ribosomal RNA genes of Saccharomyces cerevisiae. I. Physical map of the repeating unit and location of the regions coding for 5 S, 5.8 S, 18 S, and 25 S ribosomal RNAs. J Biol Chem. 1977 Nov 25;252(22):8118–8125. [PubMed] [Google Scholar]
- Bennett G. N., Schweingruber M. E., Brown K. D., Squires C., Yanofsky C. Nucleotide sequence of the promoter--operator region of the tryptophan operon of Escherichia coli. J Mol Biol. 1978 May 15;121(2):113–137. doi: 10.1016/s0022-2836(78)80001-1. [DOI] [PubMed] [Google Scholar]
- Boseley P. G., Tuyns A., Birnstiel M. L. Mapping of the Xenopus laevis 5.8S rDNA by restriction and DNA sequencing. Nucleic Acids Res. 1978 Apr;5(4):1121–1137. doi: 10.1093/nar/5.4.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattopadhyay S. K., Kohne D. E., Dutta S. K. Ribosomal RNA genes of Neurospora: isolation and characterization. Proc Natl Acad Sci U S A. 1972 Nov;69(11):3256–3259. doi: 10.1073/pnas.69.11.3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlberg A. E., Dahlberg J. E., Lund E., Tokimatsu H., Rabson A. B., Calvert P. C., Reynolds F., Zahalak M. Processing of the 5' end of Escherichia coli 16S ribosomal RNA. Proc Natl Acad Sci U S A. 1978 Aug;75(8):3598–3602. doi: 10.1073/pnas.75.8.3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdmann V. A. Collection of published 5S and 5.8S RNA sequences and their precursors. Nucleic Acids Res. 1979 Jan;6(1):r29–r44. doi: 10.1093/nar/6.1.419-c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford P. J., Mathieson T. The nucleotide sequences of 5.8-S ribosomal RNA from Xenopus laevis and Xenopus borealis. Eur J Biochem. 1978 Jun 1;87(1):199–214. doi: 10.1111/j.1432-1033.1978.tb12367.x. [DOI] [PubMed] [Google Scholar]
- Free S. J., Rice P. W., Metzenberg R. L. Arrangement of the genes coding for ribosomal ribonucleic acids in Neurospora crassa. J Bacteriol. 1979 Mar;137(3):1219–1226. doi: 10.1128/jb.137.3.1219-1226.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan M. S., Maden B. E. Nucleotide sequence relationships between vertebrate 5.8 S ribosomal RNAs. Nucleic Acids Res. 1977 Jul;4(7):2495–2505. doi: 10.1093/nar/4.7.2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lightfoot D. Thermodynamics of a stable yeast 5.8S rRNA hairpin helix. Nucleic Acids Res. 1978 Oct;5(10):3565–3577. doi: 10.1093/nar/5.10.3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luoma G. A., Marshall A. G. Laser Raman evidence for new cloverleaf secondary structures for eukaryotic 5.8S RNA and prokaryotic 5S RNA. Proc Natl Acad Sci U S A. 1978 Oct;75(10):4901–4905. doi: 10.1073/pnas.75.10.4901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T., Jeffrey A., van deSande H. Chain length determination of small double- and single-stranded DNA molecules by polyacrylamide gel electrophoresis. Biochemistry. 1975 Aug 26;14(17):3787–3794. doi: 10.1021/bi00688a010. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazar R. N., Roy K. L. Nucleotide sequence of rainbow trout (Salmo gairdneri) ribosomal 5.8 S ribonucleic acid. J Biol Chem. 1978 Jan 25;253(2):395–399. [PubMed] [Google Scholar]
- Nazar R. N., Roy K. L. The nucleotide sequence of turtle 5.8 s rRNA. FEBS Lett. 1976 Dec 15;72(1):111–116. doi: 10.1016/0014-5793(76)80824-1. [DOI] [PubMed] [Google Scholar]
- Nazar R. N., Sitz T. O., Busch H. Structural analyses of mammalian ribosomal ribonucleic acid and its precursors. Nucleotide sequence of ribosomal 5.8 S ribonucleic acid. J Biol Chem. 1975 Nov 25;250(22):8591–8597. [PubMed] [Google Scholar]
- Pace N. R., Walker T. A., Schroeder E. Structure of the 5.8S RNA component of the 5.8S-28S ribosomal RNA junction complex. Biochemistry. 1977 Nov 29;16(24):5321–5328. doi: 10.1021/bi00643a025. [DOI] [PubMed] [Google Scholar]
- Pene J. J., Knight E., Jr, Darnell J. E., Jr Characterization of a new low molecular weight RNA in HeLa cell ribosomes. J Mol Biol. 1968 May 14;33(3):609–623. doi: 10.1016/0022-2836(68)90309-4. [DOI] [PubMed] [Google Scholar]
- Perry R. P. Processing of RNA. Annu Rev Biochem. 1976;45:605–629. doi: 10.1146/annurev.bi.45.070176.003133. [DOI] [PubMed] [Google Scholar]
- Rubin G. M. Preparation of RNA and ribosomes from yeast. Methods Cell Biol. 1975;12:45–64. doi: 10.1016/s0091-679x(08)60951-6. [DOI] [PubMed] [Google Scholar]
- Rubin G. M. The nucleotide sequence of Saccharomyces cerevisiae 5.8 S ribosomal ribonucleic acid. J Biol Chem. 1973 Jun 10;248(11):3860–3875. [PubMed] [Google Scholar]
- Russell P. J., Hammett J. R., Selker E. U. Neurospora crassa cytoplasmic ribosomes; ribosomal ribonucleic acid synthesis in the wild type. J Bacteriol. 1976 Aug;127(2):785–793. doi: 10.1128/jb.127.2.785-793.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweizer E., MacKechnie C., Halvorson H. O. The redundancy of ribosomal and transfer RNA genes in Saccharomyces cerevisiae. J Mol Biol. 1969 Mar 14;40(2):261–277. doi: 10.1016/0022-2836(69)90474-4. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Tinoco I., Jr, Borer P. N., Dengler B., Levin M. D., Uhlenbeck O. C., Crothers D. M., Bralla J. Improved estimation of secondary structure in ribonucleic acids. Nat New Biol. 1973 Nov 14;246(150):40–41. doi: 10.1038/newbio246040a0. [DOI] [PubMed] [Google Scholar]
- Young R. A., Steitz J. A. Complementary sequences 1700 nucleotides apart form a ribonuclease III cleavage site in Escherichia coli ribosomal precursor RNA. Proc Natl Acad Sci U S A. 1978 Aug;75(8):3593–3597. doi: 10.1073/pnas.75.8.3593. [DOI] [PMC free article] [PubMed] [Google Scholar]


