A classic system for studying gene activation in eukaryotes has been the regulation of galactose gene expression in S. cerevisiae, in which galactose activates transcription through regulation of a transcriptional activator, a repressor, and a transducer. Joshua-Tor and colleagues have solved the structure of a key complex between the repressor Gal80p, the transducer Gal3p, the inducer galactose, and ATP. The authors found that the interaction between the transducer and the repressor occurs only when Gal3p is in the closed state, and this is induced by binding of galactose and ATP. These results provide novel insight into the phenotypes of several important Gal80 and Gal3 mutants.
Keywords: transcription regulation, activation, structural biology, transcriptional repression
Abstract
A wealth of genetic information and some biochemical analysis have made the GAL regulon of the yeast Saccharomyces cerevisiae a classic model system for studying transcriptional activation in eukaryotes. Galactose induces this transcriptional switch, which is regulated by three proteins: the transcriptional activator Gal4p, bound to DNA; the repressor Gal80p; and the transducer Gal3p. We showed previously that NADP appears to act as a trigger to kick the repressor off the activator. Sustained activation involves a complex of the transducer Gal3p and Gal80p mediated by galactose and ATP. We solved the crystal structure of the complex of Gal3p–Gal80p with α-D-galactose and ATP to 2.1 Å resolution. The interaction between the proteins occurs only when Gal3p is in a “closed” state induced by ligand binding. The structure of the complex provides a rationale for the phenotypes of several well-known Gal80p and Gal3p mutants as well as the lack of galactokinase activity of Gal3p.
Saccharomyces cerevisiae responds to galactose in its environment by activating the coordinated transcription of a family of genes known as the GAL genes. The proteins responsible for metabolizing galactose include Gal1p, a galactokinase; Gal7p, a galactose-1-phosphate-uridyl transferase; Gal10p, an epimerase; and Gal2p, a galactose permease. Transcription is regulated by the GAL regulatory switch which consists of three key protein components: Gal4p, the transcriptional activator; Gal80p, the repressor; and Gal3p, the transducer. The interplay between Gal3p, Gal80p, and Gal4p determines the on/off status of the switch. Gal4p binds the upstream activating sequences of GAL genes (UASGAL) (Bram et al. 1986) through its N-terminal DNA-binding domain (Keegan et al. 1986; Johnston 1987; Baleja et al. 1992; Marmorstein et al. 1992) and activates transcription through interaction of its C-terminal transcription activation domain (TAD) with the transcription machinery (Keegan et al. 1986; Johnston et al. 1987; Ma and Ptashne 1987). During noninducing conditions, Gal80p binds to the TAD of Gal4p to repress transcription. Repression is relieved upon induction with galactose, mediated by the transducer Gal3p (Sil et al. 1999). Gal3p forms a complex with Gal80p, galactose, and ATP. Biochemical and in vivo experiments suggest that NAD facilitates S. cerevisiae Gal80p binding to Gal4p, while NADP destabilizes this interaction and appears to be the initial trigger for activation (Kumar et al. 2008; Li et al. 2010).
The molecular mechanism by which the induction of the yeast GAL genes occurs has been the subject of debate. It was initially thought that upon binding galactose, Gal3p enters the nucleus and disrupts the Gal80p–Gal4p interaction, thereby releasing the Gal4p TAD to interact with the transcriptional machinery. More recently, Gal3p was shown to reside exclusively in the cytoplasm and is apparently excluded from the nucleus, whereas Gal80p is present in both the cytoplasm and the nucleus (Diep et al. 2008). Indeed, when Gal3p was anchored to the cell plasma membrane, it retained its induction function (Peng and Hopper 2000). In addition, targeting Gal80p to the nucleus impairs GAL gene induction, but impaired induction can be partially suppressed by targeting Gal3p to the nucleus as well. This led to a proposal that Gal80p exhibits nucleocytoplasmic shuttling and is sequestered by Gal3p in the cytoplasm under inducing conditions (Peng and Hopper 2000, 2002). Other reports claim that Gal3p does enter the nucleus in the presence of galactose, and induction occurs through the association of a tripartite complex formed between Gal4p, Gal80p, and Gal3p (Platt and Reece 1998; Wightman et al. 2008; Jiang et al. 2009). Recently, Hopper and colleagues (Egriboz et al. 2011) suggested that there is sufficient Gal3p in the nucleus prior to galactose induction to account for the rapid induction kinetics.
Gal3p is highly related to the S. cerevisiae galactokinase Gal1p. It is 74% identical in sequence but lacks the galactokinase activity of the Gal1p enzyme (Bhat et al. 1990). Both Gal1p and Gal3p are members of the galactokinase, homoserine kinase, mevalonate kinase, and phosphomevalonate kinase (GHMP) superfamily (Bork et al. 1993), and, as the rest of the GHMP superfamily members, Gal3p has a phosphate-binding loop (spanning residues 156–162), which serves as the binding site for the ATP phosphoryl tail. The lack of galactokinase activity has been attributed to the absence of two amino acids in the GLSSSA(A/S)(F/L/I) motif typical of all functional galactokinases, since Gal3p contains the sequence 161GLSSAF in that position. Gal3p can, however, be converted into a galactokinase through the addition of two amino acids, serine and alanine, after Ser 164 (Platt et al. 2000). It should be noted that in Kluyveromyces lactis, a single protein, Gal1p, serves both functions: as the transducer and galactokinase.
Based on a model constructed using the structure of S. cerevisiae Gal1p (Thoden et al. 2005a), it was proposed that, upon binding ATP and galactose, Gal3p adopts a rigid conformation that allows its interaction with Gal80p.
Here, we report on the structure of the complex of Gal3p, Gal80p, ATP, galactose, and Mg2+ as well as the structure of Gal3p in its apo form. These structures allow us to address several open questions regarding the GAL transcription regulatory system.
Results
The structure of the unbound transducer Gal3p
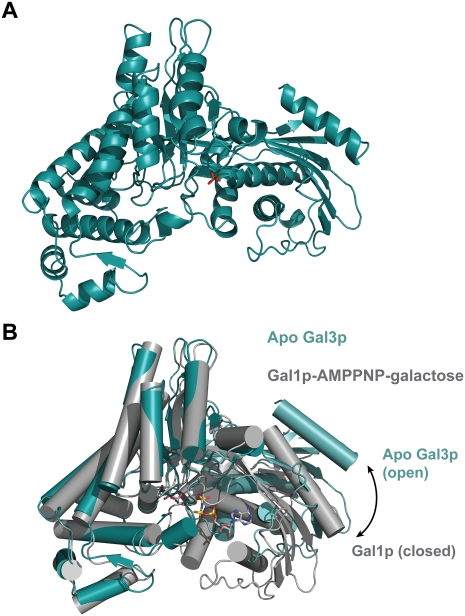
The structure of a 16-residue N-terminal truncation of Gal3p was determined to 2.1 Å resolution (Fig. 1A). There are two molecules in the asymmetric unit, and although they interact, this dimeric form does not appear to be of biological significance, and the protein is monomeric in solution. It is different from the dimeric form that interacts with Gal80p described below. Gal3p is comprised of two domains, with the ligand-binding sites located between the two domains. The N-terminal domain has a central six-stranded mixed β sheet surrounded by α helices. The larger C-terminal domain is largely helical, with a β sheet close to the domain interface. As anticipated, given the high sequence identity, the overall structure of Gal3p is very similar to the structure of Gal1p that was determined in complex with α-D-galactose and Mg2+-AMPPNP (Thoden et al. 2005a), with an important difference. Gal3p displays a more open conformation than Gal1p in the complex (Fig. 1B). The N-terminal domains of the two structures superimpose well, with a root-mean-square deviation (rmsd) of 1.12 Å for 212 structurally equivalent α-carbon atoms, while the rmsd of the C-terminal domains is 1.27 Å for 255 α-carbons. However, there is an ∼30° rotation, which opens the cleft between the two domains in Gal3p compared with Gal1p. Therefore, we captured Gal3p in an open state, while Gal1p in complex with both ligands is in a closed state. This is most likely due to the absence of ligands in the Gal3p structure, rather than any differences between the two proteins. We should note that a sulfate ion occupies the γ-phosphate-binding pocket in the Gal3p structure, but this does not appear to be sufficient to induce the closed state in Gal3p.
Figure 1.
Apo Gal3p adopts an open conformation. (A) Ribbon diagram of the structure of the Apo form of Gal3p. A sulfate ion that occupies the γ-phosphate-binding pocket is shown in stick representation, with the sulfur in orange and oxygen atoms in red. (B) Superposition of the Apo form of Gal3p (teal) and the ligand-bound form of Gal1p (gray). The superposition was carried out using only the C-terminal domain of each protein (residues 241–510 for Gal3p, and 249–519 for Gal1p). Gal1p's ligands (AMPPNP and galactose) are shown in stick representation and are colored by atom. (Gray) Carbons; (blue) nitrogens; (red) oxygens; (yellow) phosphorous.
Overall structure of the Gal3p–Gal80p complex
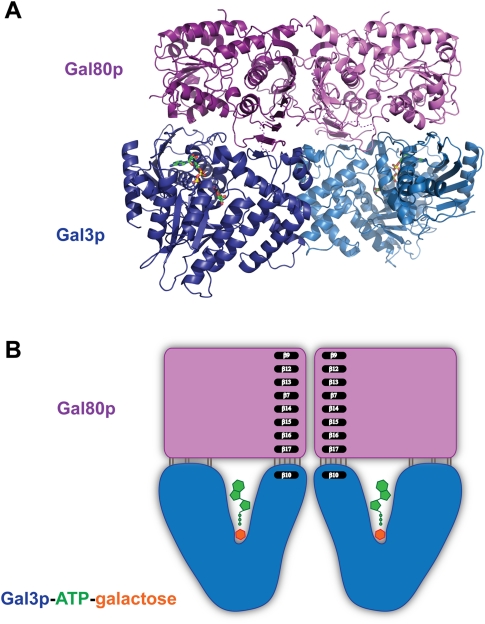
The induced form of this classic transcriptional switch involves a complex between the transducer Gal3p, the repressor Gal80p, galactose, and ATP. In order to understand the nature of this interaction, we determined the structure of this quaternary complex. The Gal3p–Gal80p–α-D-galactose–Mg2+–ATP complex crystallized as a heterotetramer in space group P1 and its structure was determined to 2.1 Å resolution (Fig. 2). The interaction between the two Gal80p monomers is similar to what we observed in the complex with Gal4–AD (Kumar et al. 2008) and occurs largely through the interaction between two large β sheets, forming a very extensive dimerization surface. The interaction between the two Gal3p molecules is not as extensive and occurs largely through water molecules. Two sets of direct protein–protein interactions occur between the Glu 364 side chain and both the backbone and side chain of Ser 361 due to the dimeric twofold symmetry. In addition, the backbone of Asn 315 from one monomer interacts with the side chain of Gln 316 of the second, and the side chain of Arg 299 from the first monomer interacts with the backbone oxygen of Asn 359 of the second. This dimeric interface forms only upon interaction with Gal80p and no doubt contributes to the stabilization of the heterotetrameric complex.
Figure 2.
The structure of the Gal80p–Gal3p–ATP–galactose heterotetramer complex. (A) A ribbon diagram of the complex of a dimer of Gal80p (in two shades of purple) and a dimer of Gal3p (in two shades of blue) is shown with the ATP and galactose ligands. ATP and galactose are shown in stick representations and are colored by atom as in Figure 1B, except for carbons that are shown in green. (B) A schematic representation of the Gal80p–Gal3p–ATP–galactose heterotetramer complex illustrating the mode of binding. Gal3p binds Gal80p in a closed conformation. A β strand (β10) from Gal3p extends the large β sheet that forms the Gal80p dimerization surface.
The two Gal3p monomers (rmsd 0.5 Å) and the two Gal80p monomers (rmsd 0.3 Å) are very similar. The main differences lie in two loop regions of Gal3p, spanning residues 317–320 and 404–407. In order to simplify the discussion, we only refer to the first monomer in the coordinates file.
Gal80p interacts with the closed state of Gal3p
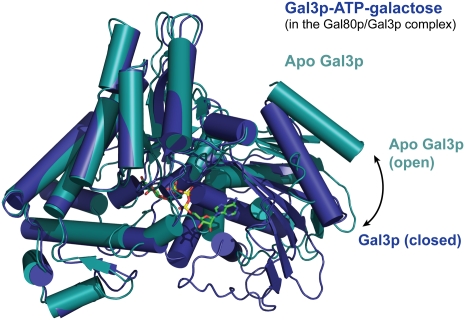
As alluded to earlier, Gal3p can adopt distinct “open” and “closed” states, and these depend on the presence of ligands. When comparing Gal3p in the Gal3p–Gal80p–galactose–MgATP complex with apo-Gal3p, Gal3p in the complex is clearly in a closed state (Fig. 3). The outer regions of the protein, previously termed “lips” (Menezes et al. 2003), move closer together, with residue Lys 241 as the approximate pivot point, as in the ligand-bound Gal1p structure (Supplemental Fig. 1). Indeed, the distance between Lys 81 on the N-terminal lip and Gln 378 on the C-terminal lip is 64.3 Å in unbound Gal3p, and this distance is reduced by >10 Å, to 53.5 Å, in the complex with galactose, ATP, and Gal80p. This is very similar to the equivalent distance of 53.8 Å in ligand-bound Gal1p (between Gal1p Lys 87 and Gln 386). Both ligands are probably necessary to induce this closed state, since binding of a nucleotide alone is probably insufficient, as seen in a nucleotide-bound Gal3p structure (data not shown). Gal80p thus interacts with this closed conformation, as was suggested previously (Thoden et al. 2005a).
Figure 3.
Ligand-bound Gal3p adopts a closed conformation. Superposition of the ligand-bound form of Gal3p (blue) and the Apo form of Gal3p (teal). The superposition was carried out using only the C-terminal domain of each protein (residues 241–510). Gal3p's ligands (ATP and galactose) are shown in stick representation and are colored as in Figure 2.
Gal80p displays a small yet notable difference when bound to Gal3p. The edge of the β sheet that is also part of the Gal80p dimerization interface reorients slightly compared with its conformation in Gal80S2 and a bit more so compared with Gal80S0 in the complex with Gal4AD and NAD, so as to attach to a β strand from Gal3p (β10) and extend the sheet. A nearby loop spanning residues Gly 307–Leu 317 of Gal80p also becomes well ordered, with an average B factor of 36.4 Å2. This loop has a high B factor (average of 95.5 Å2) in the Gal80pS2 structure and is disordered in the Gal80pS0–Gal4AD–NAD complex. However, the corresponding loop in K. lactis Gal80p does become ordered upon Gal4p–TAD binding (Thoden et al. 2008).
The Gal3p/Gal80p interaction occurs through a cooperative network of hydrogen bonds
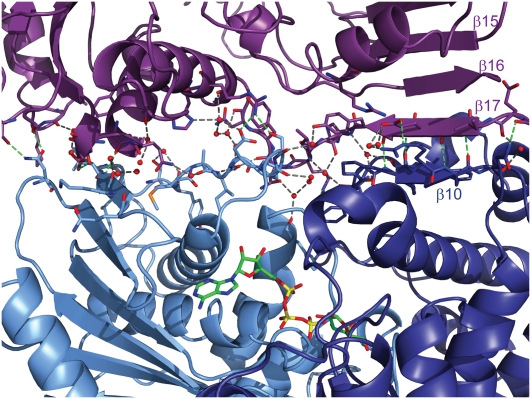
The Gal3p/Gal80p interaction occurs through a complex network of hydrogen bonds. Upon binding, a 2543 Å2 surface area gets buried when a dimer of Gal80p binds the two monomers of Gal3p, and an additional 647 Å2 is buried due to Gal80p-induced Gal3p dimer formation. Water molecules remain in the space between the two proteins and participate in the hydrogen bond network in addition to the direct contacts between the two proteins (Fig. 4). The two Gal3p lip regions described above form the two main regions of the Gal3p/Gal80p interaction. The Gal3p C-terminal lip forms a short β strand (β10; Pro 374–Phe 377) that extends the large β sheet of Gal80p that forms the extensive Gal80p homodimerization interface. Most of the contacts formed through the C-terminal lip are through this β-strand interaction. Four of the six direct hydrogen bonds are backbone–backbone interactions spanning four residues (375–378) (Fig. 2B; Supplemental Fig. 2). The C-terminal lip appears to be anchored better in the structure compared with the N-terminal lip, as it is stabilized by both the interaction with Gal80p and the Gal3p dimerization interaction. This is also reflected in the higher B factor of the N-terminal domain of Gal3p. In addition, with a low concentration of ligands in the crystallization drop, the N-terminal domain dissociates from Gal80p, while the C-terminal domain is still bound (data not shown), and the relative orientation of the two domains with respect to each other is in between the open and closed states. This implies that the two proteins could still bind at high protein concentrations, albeit weakly, without the presence of ligands.
Figure 4.
The Gal80p/Gal3p interaction occurs through a network of hydrogen bonds. Close-up view of the interface between one monomer of Gal80p (purple) and one monomer of Gal3p. (Light blue) N-terminal; (dark blue) C-terminal. Residues that form either direct or water-mediated contacts are shown in stick representation. Water molecules are shown as red spheres. Direct protein–protein interactions are shown as green dashes, and water-mediated interactions are shown as gray dashes. Ligands are shown in stick representation and are colored as in Figure 2.
Gal3p binds to a different surface of Gal80p than the surface for the Gal4p TAD interaction
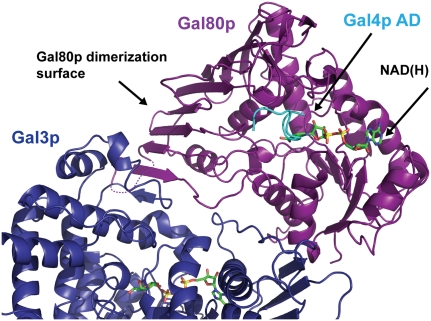
Comparison of the Gal3p–Gal80p–galactose–ATP structure presented here with the Gal80p–Gal4pAD structures from S. cerevisiae that we determined previously (Kumar et al. 2008) and with the Gal80p–Gal4pAD structure from K. lactis (Thoden et al. 2008) shows that Gal3p binds to a different surface on Gal80p than Gal4p–AD (Fig. 5). Although only a small portion of Gal4p is present in these structures, it does not appear that Gal3p and Gal4p binding to Gal80p would be mutually exclusive. Modeling the 9-mer Gal4p peptide as well as the NAD dinucleotide from the S. cervisiae structure resulted in no clashes. In addition, modeling the 14-amino-acid peptide from the K. lactis Gal80p–Gal4pAD structure onto the Gal3p-bound S. cerevisiae Gal80p structure resulted in only minor clashes that could be easily relieved with small adjustments.
Figure 5.
Gal3p binds to a different surface of Gal80p than the surface for the Gal4p TAD interaction. Gal80p from the Gal80p–Gal4p–AD–NAD complex was superimposed on Gal80p in the Gal80p–Gal3p–ATP–galactose complex, placing Gal4p–AD (shown in cyan) and NAD(H) (shown in stick representation) on the complex reported here. ATP and galactose are also shown in stick representation.
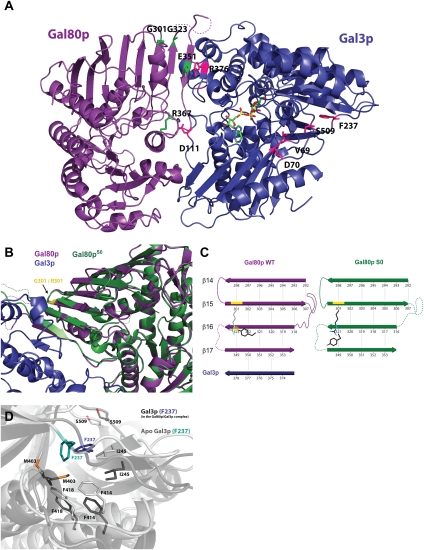
The structural basis for Gal80p and Gal3p mutants
Dominant superrepressor Gal80pS mutations Gal80pS0 (G301R), Gal80pS1 (G323R), and Gal80pS2 (E351K) were shown to produce noninducible phenotypes. This phenotype can be a result of either binding more tightly to the activator Gal4p or reduced interaction with the transducer Gal3p (Douglas and Hawthorne 1972; Nogi et al. 1977; Yano and Fukasawa 1997). In order to understand the basis for these phenotypes, we examined the positions of the residues in question (Fig. 6A) and compared our wild-type Gal80p structure in the Gal3p–Gal80p complex with the structures of Gal80pS0 or Gal80pS2 that we determined previously (Kumar et al. 2008). The side chain of Gal80p Glu 351 (β17) that is mutated to a lysine in the S2 mutant forms a salt bridge with the side chain of Gal3p Arg 376 (on β10 strand). The orientation of the Glu 351 side chain is also stabilized through interactions with Gal80p Arg 160. A mutation of the glutamate to a lysine would not only eliminate the salt bridge interaction, but will likely cause repulsion between the two positively charged side chains, thus reducing the interaction with the transducer, resulting in the superrepressor phenotype (Supplemental Fig. 3).
Figure 6.
Structural basis for Gal80p and Gal3p mutants. (A) Location of Gal80p and Gal3p mutation sites. One monomer of Gal3p is shown in blue, and one monomer of Gal80p is shown in purple; ATP and galactose are shown in stick representation and are colored by atom. Highlighted residues are shown in stick representation (pink and green, respectively). Gal80p R367 and Gal3p R376 residues are also highlighted and are discussed in the text. (B) Superposition of Gal80pS0 (dark green) (Protein Data Bank [PDB] code: 3BTV) and Gal80p from the Gal80p–Gal3p complex. (Purple) Gal80p; (blue) Gal3p. The mutation site at position 301 is highlighted in yellow. (C) Schematic representation of the shift in the Gal80p β sheet that occurs upon the G301R mutation. (D) Gal3p F237 moves from its hydrophobic environment upon adopting the closed conformation. Superposition of Apo Gal3p (light gray) and ligand-bound Gal3p from the complex (dark gray) showing F237 (blue for Gal3p from the Gal80p/Gal3p complex, and teal for Apo Gal3p) and the residues surrounding it.
Gly 301, which is mutated to an arginine in the Gal80pS0 superrepressor mutant, is not directly at the interface with Gal3p. It is located three β strands away from that interface (on β15 of Gal80p). However, upon close examination of the differences between wild type and S0, it is clear that an arginine cannot be simply accommodated at this site. In fact, introducing the large arginine side chain causes a significant rearrangement that is transmitted all the way to the interface with Gal3p (Fig. 6B,C; Supplemental Fig. 4). An arginine in place of Gly 301 would clash with Tyr 321 on the neighboring strand (β16; amino acids 318–324). As a result, starting from loop 309–316 (disordered in the Gal80S0 structure), a two-amino-acid shift takes place such that the strand on which Tyr 321 resides now runs from Ser 315 to Tyr 321 in the Gal80S0 structure (this strand runs from Leu 317 to Gly 323 in the wild-type structure), thus moving the otherwise clashing tyrosine out of the way. This is not a simple out-of-register issue due to mistracing of the polypeptide chain in the electron density maps (these were examined again in light of this difference by simulated annealing omit maps), but a real change in structure. Importantly, this β strand moves out of the original β-sheet plane to form a more pronounced twist in the sheet to avoid additional clashes between the Arg 301 in the superrepressor and backbone atoms of the shifted strand. This causes the next strand over to extend the more twisted conformation where it can no longer readily interact with the Gal3p β strand at the interface, as it does with the wild type Gal80p. Unfortunately, we do not have a structure for Gal80pS1; however, a glycine-to-arginine mutation on strand β16 may also result in a similar type of clash and possibly a scenario similar to that of the S0 mutant.
The Gal3p D111C mutation results in a noninducible phenotype (Diep et al. 2008). Gal3p Asp 111 forms a salt bridge with Gal80p Arg 367 (Fig. 6A), and a change to a cysteine would disrupt this interaction, leading to an inability to sustain a complex with Gal80p.
Gal3p constitutive mutants that have a reduced requirement for galactose and ATP have been identified: V69E/D70V, F237Y, D368V, and S509P or S509L (Blank et al. 1997). V69, D70, F237, and S509 are all in the vicinity of the pivot region (Fig. 6A). It was previously speculated (Thoden et al. 2005a) that these mutations would favor the closed state of Gal3p that could interact with Gal80p, thus circumventing the need for the ligands to induce this state. Since we have structures of Gal3p in both the open (apo form) and closed (in complex with Gal80p) conformations, we can compare the two states with respect to these residues. In the open conformation, V69 from the N-terminal domain makes a van der Waals contact with I507 of the C-terminal domain (4.16 Å) across the hinge away from the Gal80p interaction surface. In the closed conformation, this contact is disrupted, with the hinge opening on this side, while the two domains come together on the opposite side to interact with Gal80p. A change to a charged glutamate would disrupt this interaction without the need for the ligands. In the open conformation, F237 sits in a very nice hydrophobic pocket made by F247, M403, L65, F414, F418, and I245 (Fig. 6D). This phenylalanine pops out of that pocket in the closed form to a more hydrophilic environment. A change to a tyrosine (i.e., adding a hydroxyl group) would also remove that side chain out of that pocket and might draw the peptide on which S509 is located toward it, as is the case in the closed form that interacts with Gal80p, again without necessitating ligand binding. All of these changes would achieve the same effect as would binding of ligands.
The ATP-binding site
Gal3p binds galactose and Mg2+-ATP, which induce the closed conformation of the protein and, as described above, enable Gal80p binding. ATP is bound in a canonical fashion similar to other galactokinase family structures (Thoden et al. 2005b). An aromatic cage consisting of Phe 94, Trp 117, and Phe 166 encircle the adenine base of the nucleotide. The base is also anchored to the protein via two hydrogen bonds. The ATP sugar ring adopts a C3′ endo conformation and is anchored through water-mediated hydrogen bonds to the protein. The phosphoryl tail of the nucleotide makes several direct interactions with the protein as well as some water-mediated ones (Supplemental Fig. 5). Comparison with the human galactokinase structure reveals that the phosphoryl tail conformation and its location with respect to the sugar are essentially the same (Thoden et al. 2005b). However, the hydrogen-bonding network that anchors it to the protein is different due to the deletion of conserved serine and alanine residues in the Gal3p phosphate-binding loop. The phosphoryl tail conformation is different, on the other hand, from that found for the ATP analog AMPPNP in the highly related S. cerevisiae Gal1p protein (Thoden et al. 2005a). In that structure, the AMPPNP points ∼6 Å away from the sugar hydroxyl, and the distance is incompatible for catalysis. In contrast, in the current Gal3p–Gal80p–galactose–ATP structure, the Mg2+ ion is octahedrally coordinated by the α-phosphoryl, β-phosphoryl, and Υ-phosphoryl oxygens of the ATP as well as by the carbonyl oxygen of Ser 163 and two water molecules.
Why is Gal3p not a galactokinase?
Gal3p, the transducer of this classic transcriptional switch, is 74% identical in sequence to the galactokinase Gal1p. In fact, in K. lactis, both activities reside in the same protein. Although overexpression of Gal1p can suppress a gal3 mutant (Bhat and Hopper 1992), Gal3p cannot substitute for Gal1p, since it lacks galactokinase activity. As mentioned earlier, this was attributed to a two-amino-acid deletion near the ligand-binding site, and insertion of these two residues converts the protein to a functional enzyme (Platt et al. 2000). However, the ATP and galactose are still in close proximity to one another in Gal3p in the quaternary structure. A direct comparison with the Gal1p structure is difficult because in that structure, the Mg2+-AMPPNP is in a distorted conformation (Thoden et al. 2005a).
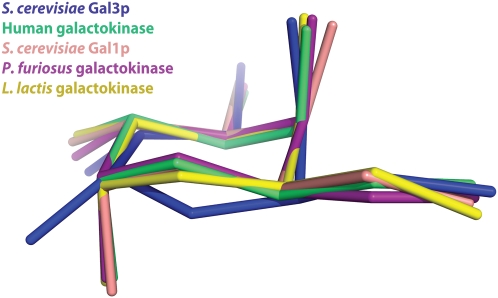
A close examination of the sugar in the Gal3p–Gal80p–galactose–ATP structure shows that the galactose ring is intermediate between an ideal chair and ideal boat conformation. This conformation is clear in the electron density maps and is observed in both molecules of the asymmetric unit. In all four available active galactokinase structures, the conformation of the galactose ring is in a clear chair conformation (Thoden and Holden 2003; Hartley et al. 2004; Thoden et al. 2005a,b). Thus, the conformation of the galactose ring is clearly unique, as seen from an overlay of the sugar of Gal3p on the sugars of other galactokinases (Fig. 7).
Figure 7.
The galactose molecule bound to Gal3p in the complex adopts a unique conformation. Superposition of the galactose molecules from five different galactokinase structures. Gal3p from the Gal80p–Gal3p complex structure is shown in blue, human galactokinase (PDB code: 1WUU) is shown in green, S. cerevisiae galactokinase Gal1p (PDB code: 2AJ4) is shown in light pink, Pyrococcus furious galactokinase (PDB code: 1S4E) is shown in dark pink, and Lactococcus lactis galactokinase (PDB code: 1PIE) is shown in yellow.
As a consequence of the distortion from the chair conformation, the C1 hydroxyl group in Gal3p points in a different direction within the binding site from the direction observed in all of the available galactokinase structures (Supplemental Fig. 6). Two quite different catalytic mechanisms have been proposed for members of the GHMP galactokinase superfamily. According to one mechanism, a conserved aspartate residue (equivalent to Asp 209 in Gal3p) serves to abstract a hydroxyl hydrogen from the galactose substrate, whereas a basic residue such as lysine or arginine (equivalent to Arg 47 in Gal3p) functions to lower the pKa of the hydroxyl group. The activated oxygen of the substrate hydroxyl group can then serve as a nucleophile for attacking the γ-phosphate group of ATP. Mammalian mevalonate kinase (Fu et al. 2002) is an example of a GHMP member protein that probably follows this mechanism. The second mechanism proceeds via a direct attack of the substrate hydroxyl group onto the γ-phosphate group. This mechanism was suggested mainly for proteins that are non-GHMP members (Hasemann et al. 1996). However, homoserine kinase is an example of a GHMP protein that proceeds through this mechanism (Krishna et al. 2001). The basis for assigning the direct mechanism for homoserine kinase is the apparent absence of a catalytic base in the region near the substrate hydroxyl group that is ultimately phosphorylayed during the reaction.
In Gal3p, the galactose C1 hydroxyl group is in direct contact with a Lys 258 side chain nitrogen that could serve in lowering the pKa of the hydroxyl group. However, the distance between Asp 209 and the C1 hydroxyl group oxygen is 4 Å—too long for hydrogen abstraction. This aspartate is stabilized by a nearby arginine (Arg 47), as in active galactokinases; however, in the Gal3p case, it is positioned very stably by bidentate hydrogen bonds to that arginine and through a water-mediated interaction to the Mg2+. That water is in turn well positioned by the Gal3p Ser 164 side chain that is replaced by an alanine in the active galactokinases due to the two-residue deletion in Gal3p. Therefore, the aspartate in Gal3p could not move much to form the three hydrogen bonds to the sugar hydroxyls and to be in close enough proximity to the C1 hydroxyl group for hydrogen abstraction. Instead, it forms only one hydrogen bond to hydroxyl O3 (bound to C2), but this hydroxyl group is too far from the ATP γ-phosphate and is not the hydroxyl that would be phosphorylated in these reactions. In addition, the distance between the C1 hydroxyl group oxygen and the γ-phosphate is 3.57 Å—too long for a direct nucleophilic attack, as in the second mechanism discussed above.
Discussion
The GAL system in the yeast S. cerevisiae has been a model system for transcriptional regulation in eukaryotes for >30 years. However, a clear molecular understanding of how galactose controls this switch has eluded us. Our previous structural and biochemical studies have revealed the unexpected involvement of a small metabolite, NADP, in triggering the system for activation. In this study, we focused on how this initial event is extended to produce sustained activation in the presence of galactose by looking at the complex of the transducer and the repressor mediated by the galactose inducer and ATP: the Gal3p–Gal80p–MgATP–galactose complex.
The structure of the complex shows that a stable interaction between Gal3p and Gal80p must occur while Gal3p is in its closed state. Comparison with the free Gal3p structure, presented here, and a previously determined structure of a galactose- and ATP-bound form of the very similar active galactokinase Gal1p (Thoden et al. 2005a) clearly shows that ligand binding induces the closed state of Gal3p. Furthermore, examination of the sites of Gal3p constitutive mutations shows that these would favor a closed state for Gal3p without necessitating ligand binding. On the other hand, Gal80p superrepressor mutants appear to result from their inability to form a stable complex with Gal3p, rather than tighter association with the activator Gal4p.
Gal3p binds to a different surface of Gal80p than the surface bound by the Gal4p–TAD and is located on an opposite side of the Gal80p extensive β sheet. Gal3p binding appears to cause some conformational changes in Gal80p; namely, some reorientation of the β sheet to optimize interaction with a β strand of Gal3p and extend the sheet and ordering of some loops. However, this would not appear to change the conformation of Gal80p enough to prevent interaction with the Gal4p–TAD. That said, we only have a very small portion of Gal4p in the two crystal structures available (Kumar et al. 2008; Thoden et al. 2008). A tripartite complex between the three proteins can be formed in vitro (Platt and Reece 1998), albeit with a constitutive mutant of Gal3p and a much-truncated form of Gal4p. It is still unclear, however, whether the three proteins would colocalize—whether Gal3p ever enters the nucleus, as discussed above. A recent study suggested that a tripartite complex might form very transiently in the nucleus (Egriboz et al. 2011).
Gene regulation is ultimately and always dictated by molecular interactions between regulatory molecules. A full understanding of these transcriptional switches requires an intimate knowledge of these contacts and the consequences of the conformational changes they induce. Here, we presented another glimpse into the machinations of a classic regulatory switch.
Materials and methods
Protein expression and purification
Gal3p (17–520)
The gene encoding full-length Gal3p was amplified by PCR from S. cerevisiae genomic DNA. The amplification products were identified, isolated, and digested with both NdeI and SalI. The fragments were ligated into the pET28a vector (Novagen), from which the DNA encoding Gal3p (17–520 residues) was amplified by PCR and cloned into the pSMT3 vector (a generous gift from Dr. Christopher Lima) with BamHI and SalI restriction endonucleases. The vector was transformed into Escherichia coli DH5α cells. The plasmid with the insert was isolated and verified by restriction enzyme analysis and then transformed into E. coli BL21(DE3) RIPL cells for expression. Truncated Gal3p was expressed as a Smt3 fusion protein with an N-terminal hexahistidine (His)6 tag. Starter culture from a single colony was grown overnight at 37°C in LB medium supplemented with kanamycin (34 μg/mL). Subsequently, ∼8 mL of the mixture was transferred to 1 L of LB medium supplemented with kanamycin (34 μg/mL) and grown at 30°C to ∼1 OD at 600 nm and then induced with 50 μM IPTG and incubated overnight at 17°C. The cells were harvested by centrifugation and frozen at −70°C. Frozen cells were thawed in lysis buffer (20 mM Tris-Cl at pH 8.0, 10 mM imidozole, 0.5 M NaCl, 10% glycerol, 10 mM β-mercaptoethanol [BME],1 mM phenylmethanesulphonylfluoride [PMSF], 1 mg/mL lysozyme) and sonicated. Cell debris was removed by centrifugation, and the clarified supernatant was collected. Ammonium sulfate fractionation of the supernatant was carried out at 70% saturation. The pellet was resuspended in a buffer (20 mM Tris-Cl at pH 8.0, 10 mM imidozole, 0.5 M NaCl, 10% glycerol, 10 mM BME, 1 mM PMSF) and applied to an Ni-NTA agarose (Qiagen) column that had been equilibrated previously with lysis buffer. The column was then washed with wash buffer (20 mM Tris-Cl at pH 8.0, 10 mM imidozole, 0.5 M NaCl, 10% glycerol, 10 mM BME) until the absorbance at 280 nm reached background level. The Smt3-truncated Gal3p fusion protein was eluted with 250 mM imidazole in the wash buffer, and Smt3 was removed using Ulp1 protease (a generous gift from Dr. Christopher Lima) by dialysis against 50 mM Tris-Cl (pH 8.0), 300 mM NaCl, and 10 mM DTT overnight at 4°C. The truncated Gal3p was further purified by heparin column chromatography. The protein was eluted at 25 mM Tris-HCl (pH 8.0) with a linear gradient of NaCl. The protein was further purified by gel filtration using a Superdex75 column with 10 mM Tris-Cl (pH 8.0), 200 mM NaCl, and 10 mM dithiotreitol (DTT) as the elution buffer.
Gal3p (2–520) and Gal80p (1–435)
Gal80p (1–435) was generated from S. cerevisiae genomic DNA using standard PCR protocols. Gal80p was cloned into pET28a, yielding a construct that expresses Gal80p with an N-terminal (His)6 tag that can be cleaved by thrombin protease. Gal3p (2–520) was cloned into a modified pET28a vector containing a (His)6-Smt3 tag, which can then be cleaved by Ulp1 protease. Each protein was expressed separately in BL21(DE3)-RIPL E. coli cells grown in LB medium to an OD600 of 1 at 32°C and induced with 0.25 mM IPTG. Cells were then grown for an additional 16 h at 18°C. The cells were harvested by centrifugation and were frozen at −80°C. The cells were thawed and suspended in buffer A (20 mM Tris-Cl at pH 8.0, 10 mM imidazole, 0.1 M NaCl, 5% glycerol, 5 mM galactose) supplemented with protease inhibitor (Complete, Roche), benzonase, 10 mM MgCl2, and lysosyme. Following cell lysis by a cell disruputer (Avestin), the cell debris was removed by centrifugation. Cleared lysates were passed through a Ni-NTA agarose column (Qiagen) equilibrated with buffer A. After extensive washing with buffer A, it was replaced by a large excess of buffer B (20 mM Tris-Cl at pH 8.0, 0.1 M NaCl, 5% glycerol, 5 mM galactose) to facilitate thrombin cleavage of the His6 tag (for Gal80p) or Ulp1 cleavage of the His6-Smt3 tag (for Gal3p) and subsequent purification. On-bead digestions with thrombin (Gal80p purification) or Ulp1 (Gal3p purification) were carried out for 20 h at room temperature, and the tag-free proteins were eluted with buffer B. The proteins were then purified using two in-tandem 5-mL prepacked heparin columns coupled to an ÄKTA Explorer purification system (GE Healthcare). The proteins were subjected to a gradient of 0–2 M NaCl in the presence of 20 mM Tris-Cl (pH 8.0), 2.5 mM DTT, 5% glycerol, and 5 mM galactose. Gal80p elutes between 0.5 and 0.7 M NaCl. Gal3p elutes between 0.3 and 0.5 M NaCl. Fractions containing Gal80p or Gal3p were concentrated and stored at −80°C.
Gal3p–Gal80p–galacotose–Mg2+ATP complex
The complex was formed by incubating equimolar quantities of Gal3p and Gal80p in the presence of 20 mM Tris-Cl (pH 8.0), 0.1 M NaCl, 5 mM galactose, 5 mM ATP, 10 mM MgCl2, and 2.5 mM DTT on ice. The complex was purified by gel filtration on a Superdex 200 (10/300) column and eluted at a volume that corresponds to a heterotetramer of Gal3p/Gal80p (confirmed by MALS Wyatt). Fresh purified protein complex was used for crystallization.
Crystallization
Gal3p (17–520) crystals were obtained by the hanging-drop vapor diffusion method by mixing 1 μL of protein at a concentration of 28 mg/mL (with or without 5mM ATP or AMPPNP) with 1 μL of reservoir solution containing 10 mM MgCl2, 50 mM MES (pH 5.8–6.0), and 1.8–1.9 M Li2SO4. Crystals grew to 0.9 × 0.2 × 0.1 mm in size and were removed from the mother liquor with a fiber loop and immediately frozen in liquid nitrogen.
Gal3p–Gal80p–galactose–Mg2+ATP complex crystals were obtained by the same method by mixing 1 μL of protein at 5.9 mg/mL and 1 μL of reservoir solution containing 10% PEG-3350 and 0.1 M disodium succinate (pH 7.6). Crystals were obtained after 5 d, grew to 130 × 60 × 30 μm at 17°C, were scooped and placed in a cryoprotectant solution composed of 20% glycerol in the reservoir supplemented with 2.5 mM ATP and 2.5 mM galactose, and were immediately plunged into liquid nitrogen.
Diffraction data collection and processing
Data sets for all structures were collected at beam lines X26C (Gal3p) and X25 (complex) at the National Synchrotron Light Source (NSLS) at Brookhaven National Laboratory (BNL). Data collection was carried out under cryogenic conditions (100K). Data were integrated and scaled with the HKL-2000 package (see Supplemental Table 1 for data collection statistics; Otwinowski and Minor 1997).
Structure determination and refinement
Gal3p structures were solved by molecular replacement with the program PHASER (McCoy et al. 2007) using the coordinates of S. cerevisiae Gal1p (Protein Data Bank [PDB] code: 2AJ4) as a search model. Solvent flattening and NCS averaging were performed with RESOLVE (Terwilliger 1999) and improved map quality. The models were initially built automatically with the program ARP/wARP (Perrakis et al. 1999). Additional manual model building was carried out using the program O (Jones and Kjeldgaard 1997). The models were initially refined using CNS (Brünger et al. 1998) with iterative model building using O. This was followed by refinement using REFMAC (Murshudov et al. 1997) and then phenix.refine (Adams et al. 2010), with iterative model building using the program Coot (Emsley and Cowtan 2004). TLS refinement was introduced with groups defined as follows: group 1: chain A (20–240 and 510–520); group 2: chain A (241–509); group 3: chain B (20–240 and 510–520); group 4: chain B (241–509). The final model is missing the first two residues (18–19) of each protein chain and residues of internal loop 259–261 in chain A and 257–261 in chain B. Coordinates and structure factors have been deposited in the PDB under accession code 3V5R.
The Gal3p–Gal80p–galactose–Mg2+ATP structure was solved by molecular replacement with the program PHASER (McCoy et al. 2007) using the coordinates of full-length S. cerevisiae Gal80pS0 (PDB code: 3BTS) and the C-terminal domain of the apo form structure of Gal3p (residues 259–520) as search models. Two Gal80p molecules and two copies of the C-terminal domain of Gal3p were placed in the asymmetric unit. Rigid body refinement, followed by individual atom refinement, was carried out using phenix.refine (Adams et al. 2010) accompanied by iterative model building using Coot (Emsley and Cowtan 2004). After building 92% of the protein and refining individual atoms and grouped isotropic ADPs, the ATP and galactose ligands were incorporated into the model using LigandFit (Supplemental Fig. 7; Terwilliger et al. 2006). A restraint file for the ligands was created using eLBOW (Moriarty et al. 2009) and was used for the refinement. Mg2+ ions were modeled into high positive peaks (>6σ, Fo − Fc) in the vicinity of the ATP and galactose. Individual isotropic ADP refinement was introduced, followed by anisotropic ADP refinement of the ligands and the Mg2+ ions. Further building was done until 97% of the protein was modeled. Water molecules were modeled using the automatic “ordered solvent” utility in phenix.refine and were manually examined. TLS refinement was introduced with groups defined as follows: Gal80p groups: group 1: chain A (1–148); group 2: chain A (149–183, 206–234, and 260–435); group 3: chain A (184–205 and 235–259); group 4: chain B (1–148); group 5: chain B (149–183, 206–234, and 260–435); group 6: chain B (184–205 and 235–259); and Gal3p groups: group 7: chain C (1–240 and 510–520); group 8: chain C (241–509); group 9: chain D (1–240 and 510–520); group 10: chain D (241–509). Validation of water molecules was done by Coot (Emsley and Cowtan 2004). Two glycerol molecules were modeled at the interface between Gal80p and Gal3p. The final model is missing the following residues: chain A (1–14 and 327–338), chain B (1–14 and 326–344), chain C (1 and 12–14), and chain D (1 and 11–15). Coordinates and structure factors have been deposited in the PDB under accession code 3V2U.
Figures were produced using the PyMOL (Schrödinger, LLC) molecular graphics system.
Acknowledgments
We thank David Wah, Eric Enemark, Alexander Gann, Amy Caudy, and members of the Joshua-Tor laboratory for discussions and advice, and A. Héroux for help at the National Synchrotron Light Source at Brookhaven National Laboratory, which is supported by the Department of Energy, Office of Basic Energy Sciences. This work was supported by NIH grant GM074075 and the Louis Morin Charitable Trust (to L.J.). L.J. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.182691.111.
References
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. 2010. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66: 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baleja JD, Marmorstein R, Harrison SC, Wagner G 1992. Solution structure of the DNA-binding domain of Cd2-GAL4 from S. cerevisiae. Nature 356: 450–453 [DOI] [PubMed] [Google Scholar]
- Bhat PJ, Hopper JE 1992. Overproduction of the GAL1 or GAL3 protein causes galactose-independent activation of the GAL4 protein: Evidence for a new model of induction for the yeast GAL/MEL regulon. Mol Cell Biol 12: 2701–2707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat PJ, Oh D, Hopper JE 1990. Analysis of the GAL3 signal transduction pathway activating GAL4 protein-dependent transcription in Saccharomyces cerevisiae. Genetics 125: 281–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blank TE, Woods MP, Lebo CM, Xin P, Hopper JE 1997. Novel Gal3 proteins showing altered Gal80p binding cause constitutive transcription of Gal4p-activated genes in Saccharomyces cerevisiae. Mol Cell Biol 17: 2566–2575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bork P, Sander C, Valencia A 1993. Convergent evolution of similar enzymatic function on different protein folds: The hexokinase, ribokinase, and galactokinase families of sugar kinases. Protein Sci 2: 31–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bram RJ, Lue NF, Kornberg RD 1986. A GAL family of upstream activating sequences in yeast: Roles in both induction and repression of transcription. EMBO J 5: 603–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. 1998. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr 54: 905–921 [DOI] [PubMed] [Google Scholar]
- Diep CQ, Tao X, Pilauri V, Losiewicz M, Blank TE, Hopper JE 2008. Genetic evidence for sites of interaction between the Gal3 and Gal80 proteins of the Saccharomyces cerevisiae GAL gene switch. Genetics 178: 725–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas HC, Hawthorne CD 1972. Uninducible mutants in the gal i locus of Saccharomyces cerevisiae. J Bacteriol 109: 1139–1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egriboz O, Jiang F, Hopper JE 2011. The rapid GAL gene switch of Saccharomyces cerevisiae depends on nuclear Gal3, not nucleo-cytoplasmic trafficking of Gal3 and Gal80. Genetics 189: 825–836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K 2004. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60: 2126–2132 [DOI] [PubMed] [Google Scholar]
- Fu Z, Wang M, Potter D, Miziorko HM, Kim JJ 2002. The structure of a binary complex between a mammalian mevalonate kinase and ATP: Insights into the reaction mechanism and human inherited disease. J Biol Chem 277: 18134–18142 [DOI] [PubMed] [Google Scholar]
- Hartley A, Glynn SE, Barynin V, Baker PJ, Sedelnikova SE, Verhees C, de Geus D, van der Oost J, Timson DJ, Reece RJ, et al. 2004. Substrate specificity and mechanism from the structure of Pyrococcus furiosus galactokinase. J Mol Biol 337: 387–398 [DOI] [PubMed] [Google Scholar]
- Hasemann CA, Istvan ES, Uyeda K, Deisenhofer J 1996. The crystal structure of the bifunctional enzyme 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase reveals distinct domain homologies. Structure 4: 1017–1029 [DOI] [PubMed] [Google Scholar]
- Jiang F, Frey BR, Evans ML, Friel JC, Hopper JE 2009. Gene activation by dissociation of an inhibitor from a transcriptional activation domain. Mol Cell Biol 29: 5604–5610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston M 1987. Genetic evidence that zinc is an essential co-factor in the DNA binding domain of GAL4 protein. Nature 328: 353–355 [DOI] [PubMed] [Google Scholar]
- Johnston SA, Salmeron JM Jr, Dincher SS 1987. Interaction of positive and negative regulatory proteins in the galactose regulon of yeast. Cell 50: 143–146 [DOI] [PubMed] [Google Scholar]
- Jones TA, Kjeldgaard M 1997. Electron-density map interpretation. Methods Enzymol 277: 173–208 [DOI] [PubMed] [Google Scholar]
- Keegan L, Gill G, Ptashne M 1986. Separation of DNA binding from the transcription-activating function of a eukaryotic regulatory protein. Science 231: 699–704 [DOI] [PubMed] [Google Scholar]
- Krishna SS, Zhou T, Daugherty M, Osterman A, Zhang H 2001. Structural basis for the catalysis and substrate specificity of homoserine kinase. Biochemistry 40: 10810–10818 [DOI] [PubMed] [Google Scholar]
- Kumar PR, Yu Y, Sternglanz R, Johnston SA, Joshua-Tor L 2008. NADP regulates the yeast GAL induction system. Science 319: 1090–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Chen G, Liu W 2010. Multiple metabolic signals influence GAL gene activation by modulating the interaction of Gal80p with the transcriptional activator Gal4p. Mol Microbiol 78: 414–428 [DOI] [PubMed] [Google Scholar]
- Ma J, Ptashne M 1987. The carboxy-terminal 30 amino acids of GAL4 are recognized by GAL80. Cell 50: 137–142 [DOI] [PubMed] [Google Scholar]
- Marmorstein R, Carey M, Ptashne M, Harrison SC 1992. DNA recognition by GAL4: Structure of a protein-DNA complex. Nature 356: 408–414 [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ 2007. Phaser crystallographic software. J Appl Crystallogr 40: 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menezes RA, Amuel C, Engels R, Gengenbacher U, Labahn J, Hollenberg CP 2003. Sites for interaction between Gal80p and Gal1p in Kluyveromyces lactis: Structural model of galactokinase based on homology to the GHMP protein family. J Mol Biol 333: 479–492 [DOI] [PubMed] [Google Scholar]
- Moriarty NW, Grosse-Kunstleve RW, Adams PD 2009. electronic Ligand Builder and Optimization Workbench (eLBOW): A tool for ligand coordinate and restraint generation. Acta Crystallogr D Biol Crystallogr 65: 1074–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr 53: 240–255 [DOI] [PubMed] [Google Scholar]
- Nogi Y, Matsumoto K, Toh-e A, Oshima Y 1977. Interaction of super-repressible and dominant constitutive mutations for the synthesis of galactose pathway enzymes in Saccharomyces cerevisiae. Mol Gen Genet 152: 137–144 [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 276: 307–326 [DOI] [PubMed] [Google Scholar]
- Peng G, Hopper JE 2000. Evidence for Gal3p's cytoplasmic location and Gal80p's dual cytoplasmic-nuclear location implicates new mechanisms for controlling Gal4p activity in Saccharomyces cerevisiae. Mol Cell Biol 20: 5140–5148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng G, Hopper JE 2002. Gene activation by interaction of an inhibitor with a cytoplasmic signaling protein. Proc Natl Acad Sci 99: 8548–8553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrakis A, Morris R, Lamzin VS 1999. Automated protein model building combined with iterative structure refinement. Nat Struct Biol 6: 458–463 [DOI] [PubMed] [Google Scholar]
- Platt A, Reece RJ 1998. The yeast galactose genetic switch is mediated by the formation of a Gal4p–Gal80p–Gal3p complex. EMBO J 17: 4086–4091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt A, Ross HC, Hankin S, Reece RJ 2000. The insertion of two amino acids into a transcriptional inducer converts it into a galactokinase. Proc Natl Acad Sci 97: 3154–3159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sil AK, Alam S, Xin P, Ma L, Morgan M, Lebo CM, Woods MP, Hopper JE 1999. The Gal3p–Gal80p–Gal4p transcription switch of yeast: Gal3p destabilizes the Gal80p–Gal4p complex in response to galactose and ATP. Mol Cell Biol 19: 7828–7840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger TC 1999. Reciprocal-space solvent flattening. Acta Crystallogr D Biol Crystallogr 55: 1863–1871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger TC, Klei H, Adams PD, Moriarty NW, Cohn JD 2006. Automated ligand fitting by core-fragment fitting and extension into density. Acta Crystallogr D Biol Crystallogr 62: 915–922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoden JB, Holden HM 2003. Molecular structure of galactokinase. J Biol Chem 278: 33305–33311 [DOI] [PubMed] [Google Scholar]
- Thoden JB, Sellick CA, Timson DJ, Reece RJ, Holden HM 2005a. Molecular structure of Saccharomyces cerevisiae Gal1p, a bifunctional galactokinase and transcriptional inducer. J Biol Chem 280: 36905–36911 [DOI] [PubMed] [Google Scholar]
- Thoden JB, Timson DJ, Reece RJ, Holden HM 2005b. Molecular structure of human galactokinase: Implications for type II galactosemia. J Biol Chem 280: 9662–9670 [DOI] [PubMed] [Google Scholar]
- Thoden JB, Ryan LA, Reece RJ, Holden HM 2008. The interaction between an acidic transcriptional activator and its inhibitor. The molecular basis of Gal4p recognition by Gal80p. J Biol Chem 283: 30266–30272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wightman R, Bell R, Reece RJ 2008. Localization and interaction of the proteins constituting the GAL genetic switch in Saccharomyces cerevisiae. Eukaryot Cell 7: 2061–2068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano K, Fukasawa T 1997. Galactose-dependent reversible interaction of Gal3p with Gal80p in the induction pathway of Gal4p-activated genes of Saccharomyces cerevisiae. Proc Natl Acad Sci 94: 1721–1726 [DOI] [PMC free article] [PubMed] [Google Scholar]