Abstract
ErbB4 is a member of the ErbB family of receptor tyrosine kinases. This family includes ErbB2 (HER2/Neu), a validated therapeutic target in breast cancer. Several studies indicate that ErbB4 functions as a tumor suppressor in breast cancer, whereas others indicate that ErbB4 functions as an oncogene. Here the authors explore the context in which ErbB4 functions as an oncogene. Silencing expression of either ErbB2 or ErbB4 in breast tumor cell lines results in reduced stimulation of anchorage independence and cell motility by the ErbB4 agonist neuregulin 2β. ErbB2 tyrosine kinase activity, but not ErbB4 tyrosine kinase activity, is required for neuregulin 2β to stimulate cell proliferation. Moreover, sites of ErbB4 tyrosine phosphorylation, but not sites of ErbB2 tyrosine phosphorylation, are required for neuregulin 2β to couple to cell proliferation. These data suggest that targeting ErbB2 expression or tyrosine kinase activity may be effective in treating ErbB4-dependent breast tumors, even those tumors that lack ErbB2 overexpression.
Keywords: ErbB2, ErbB4, crosstalk, breast cancer, invasiveness, motility
Introduction
ErbB4 (HER4) is a member of the ErbB family of receptor tyrosine kinases. This family also includes the epidermal growth factor (EGF) receptor (ErbB1/HER1), ErbB2 (Neu/HER2), and ErbB3 (HER3). ErbB4 contains an extracellular ligand-binding domain, a hydrophobic transmembrane domain, an intracellular tyrosine kinase domain, and carboxyl-terminal tyrosine residues that serve as sites of phosphorylation.1 Ligand binding to ErbB4 stimulates ErbB4 homodimerization or heterodimerization of ErbB4 with another ErbB receptor. Dimerization enables ErbB4 transphosphorylation on tyrosine residues and coupling of the phosphorylated tyrosine residues to signaling effectors and biological responses.1-4 ErbB4 signaling is commonly accompanied by cleavage and release of the ErbB4 cytoplasmic domain (4ICD) from the membrane, after which this motif may traffic to the nucleus and mitochondria.5,6
A wealth of evidence indicates that ErbB4 may function as a tumor suppressor. ErbB4 expression is typically absent in aggressive cancers of the prostate, pancreas, and larynx.7-9 ErbB4 expression in breast cancers correlates with sensitivity to endocrine therapies,10,11 and ErbB4 expression in cervical, ovarian, and breast cancers correlates with a more favorable prognosis.8,9,12,13 The ErbB4 Q646C mutant undergoes ligand-independent homodimerization and tyrosine phosphorylation.14-16 Moreover, it inhibits clonogenic colony formation by human breast, prostate, and pancreatic tumor cell lines.14-16 Likewise, the constitutively active ErbB4 I658Q mutant stimulates apoptosis in breast, prostate, and ovarian tumor cell lines,17 and the cytoplasmic domain of ErbB4 (4ICD) exhibits constitutive signaling activity and inhibits cell proliferation in breast carcinoma cell lines.18
However, ErbB4 also appears to possess oncogenic activities. ErbB4 is overexpressed in medulloblastomas and ependymomas.1 Indeed, co-(over)expression of ErbB2, ErbB4, and the ErbB4 ligand NRG1β in medulloblastoma correlates with increased metastasis.19 ErbB4 overexpression in lung carcinomas correlates with increased proliferation.9,20 Co-(over)expression of ErbB4 with EGF receptor (EGFR) and ErbB2 in breast cancers correlates with poor prognosis, whereas overexpression of ErbB4 alone correlates with a more favorable outcome.21 Similarly, silencing endogenous ErbB4 expression in MCF7 and T47D breast tumor cell lines reduces stimulation of anchorage-independent proliferation by an ErbB4 ligand.22 Finally, ErbB4 mutants found in melanomas exhibit greater tyrosine phosphorylation and signaling activity than does wild-type ErbB4.23
ErbB2 (HER2/Neu), another ErbB family receptor tyrosine kinase, may be responsible for the fact that ErbB4 possesses tumor suppressor and oncogenic activities. There are no soluble ligands for ErbB2; however, ErbB4 ligands stimulate ErbB4 crosstalk with ErbB2, particularly ErbB2 tyrosine phosphorylation and coupling to effectors.24,25 Gain-of-function ErbB2 mutants have been found in lung carcinomas.9,26 Moreover, ErbB2 is overexpressed in a variety of cancers, particularly breast carcinomas.9,26 Indeed, ErbB2 is a validated target for the breast cancer chemotherapeutics trastuzumab and lapatinib.26,27
The oncogenic activities of ErbB2 and the fact that ErbB2 can heterodimerize with ErbB4 and signal in response to stimulation with ErbB4 ligands24,25 have led us to hypothesize that ErbB2 mediates the oncogenic activities of ErbB4 and ErbB4 ligands. We have evaluated this hypothesis by assessing the effects of silencing endogenous ErbB2 or ErbB4 in human breast tumor cell lines. We have also assessed the effects of co-expressing ErbB2 and ErbB4 in a heterologous lymphoid (BaF3) model system. Silencing either ErbB2 or ErbB4 results in reduced stimulation of migration and anchorage-independent growth by an ErbB4 ligand. Moreover, ErbB2 tyrosine kinase activity and the carboxyl-terminal sites of ErbB4 tyrosine phosphorylation are both required for an ErbB4 ligand to simulate proliferation in a heterologous lymphoid model system. Thus, our results suggest that ErbB2 expression may define the biological responses to ErbB4 ligands and that crosstalk between ErbB2 and ErbB4 (even in contexts in which ErbB2 is not overexpressed) may be a target for therapeutic intervention.
Results
Silencing expression of ErbB4 or ErbB2 reduces NRG2β stimulation of anchorage-independent proliferation in the T47D human breast tumor cell line
We have previously demonstrated that the constitutively active ErbB4 Q646C mutant possesses tumor suppressor activity in a number of human tumor cell lines, including human breast tumor cell lines.14-16 However, we have also demonstrated that the expression of wild-type ErbB4 in human pancreatic tumor cell lines devoid of endogenous ErbB4 expression potentiates neuregulin stimulation of oncogenic activities, suggesting that ErbB4 also possesses oncogenic activities.16
To address this dichotomy, we first assessed the effect of silencing endogenous ErbB4 transcription in the T47D human breast tumor cell line. We generated a pooled cell line by infecting T47D cells with the pLKO-ErbB4 4915 shRNA recombinant lentivirus, which targets the 3′ UTR of the endogenous ErbB4 transcript in T47D cells. This cell line (ErbB4 shRNA) displays a 65.5% ± 1.9% decrease in ErbB4 expression (Table 1 and Fig. 1a). The ErbB4 ligand neuregulin 2β (NRG2β) stimulates anchorage-independent proliferation of the control T47D GFP shRNA cell line. In contrast, NRG2β fails to stimulate anchorage-independent proliferation of the T47D ErbB4 shRNA cell line (Fig. 1b,c). (The control and various experimental cell lines yielded a similar number of anchorage-independent colonies; the differences observed appear to be restricted to the size of the colonies.) This effect is consistent with the observation that ErbB4 possesses oncogenic activities in some contexts.1,9,19-23
Table 1.
ErbB4 shRNA or ErbB2 shRNA Specifically Reduces Expression of the Targeted Receptor in the T47D Cell Line
| T47D Cell Lines | ErbB2 Expression Relative to Parental, % | n | ErbB4 Expression Relative to Parental, % | n |
|---|---|---|---|---|
| GFP shRNA | 111.4 ± 13.5 | 3 | 93.9 ± 5.8 | 4 |
| ErbB4 shRNA | 91.8 ± 18.2 | 3 | 34.5 ± 1.9a | 4 |
| ErbB2 shRNA | 10.2 ± 1.0a | 3 | 116.5 ± 9.7 | 4 |
GFP = green fluorescent protein.
Marked loss of expression.
Figure 1.
Silencing endogenous ErbB4 or ErbB2 transcription in the T47D human breast tumor cell line reduces stimulation of anchorage-independent proliferation by NRG2β. (a) ErbB2 expression (upper panel) and ErbB4 expression (lower panel) were evaluated in T47D cell lines that express the ErbB4 4915 shRNA (ErbB4 shRNA), the ErbB2 4355 shRNA (ErbB2 shRNA), or a control shRNA specific for green fluorescence protein (GFP shRNA). Samples generated from parental T47D cells were used to construct calibration curves for quantification. (b) The indicated T47D cell lines were seeded in semisolid medium supplemented with either 10 nM NRG2β or phosphate-buffered saline (diluent control). Fifteen days later, anchorage-independent colonies were photographed, counted, and measured. Representative photomicrographs are provided for the samples treated with NRG2β. (c) For each sample, we report the distribution of colonies whose diameter is less than 70 µm, 70 to 140 µm, or greater than 140 µm (upper right panel). The effect of NRG2β stimulation on anchorage independence was determined using a chi-squared test. P values are reported. NS indicates not significant. To highlight differences in anchorage independence, we also present an expanded depiction of the percentage of colonies whose diameter is greater than 140 µm (*) for each of the samples (lower left panel).
Analyses of ErbB4 function in human pancreatic tumor cell lines suggest that crosstalk between ErbB2 and ErbB4 be necessary for NRG2β stimulation of anchorage independence and other malignant phenotypes.16 Here we directly tested that hypothesis by assessing the effect of silencing endogenous ErbB2 transcription in the T47D human breast tumor cell line. We generated a pooled cell line by infecting T47D cells with the pLKO-ErbB2 4355 shRNA recombinant lentivirus, which targets the 3′ UTR of the endogenous ErbB2 transcript in T47D cells. This cell line (ErbB2 shRNA) displays an 89.8% ± 1.0% decrease in ErbB2 expression (Table 1 and Fig. 1a). This decrease in ErbB2 expression is biologically significant. NRG2β fails to stimulate anchorage-independent proliferation of the T47D ErbB2 shRNA cell line (Fig. 1b,c). (The control and various experimental cell lines yielded a similar number of anchorage-independent colonies; the differences observed appear to be restricted to the size of the colonies.)
Silencing expression of ErbB4 or ErbB2 reduces NRG2β stimulation of migration in the MCF7 human breast tumor cell line
Next, we explored whether the effects of silencing ErbB4 or ErbB2 transcription on T47D cells were specific to that cell line. We generated clonal MCF7 human breast tumor cell lines that express the ErbB4 4915 shRNA. Relative to a pooled MCF7 cell line that expresses a corresponding scrambled shRNA sequence, the most promising ErbB4 4915 shRNA cell lines exhibited only a modest reduction in ErbB4 expression. Con- sequently, we reinfected the most promising clonal ErbB4 4915 shRNA cell line with the ErbB4 4915 shRNA lentivirus and cloned cell lines from the resulting pool. Two resulting clonal cell lines (ErbB4 shRNA CL 4 and ErbB4 shRNA CL 12) display a marked decrease (89.3% ± 1.4% and 86.2% ± 4.3%, respectively) in ErbB4 expression (Table 2 and Fig. 2a). Relative to the control MCF7 cell lines, the MCF7 ErbB4 shRNA CL 4 and CL 12 cell lines exhibit a modest but statistically significant (P < 0.05) reduction in motility (wound healing) in the absence of exogenous ErbB4 ligand (Fig. 2b). These results are consistent with the observation that ErbB4 possesses oncogenic activities1,9,19-23 and suggest that MCF7 cells endogenously express an ErbB4 ligand.
Table 2.
ErbB4 shRNA or ErbB2 shRNA Specifically Reduces Expression of the Targeted Receptor in the MCF7 Cell Line
| MCF7 Cell Lines | ErbB2 Expression Relative to Parental, % | n | ErbB4 Expression Relative to Parental, % | n |
|---|---|---|---|---|
| ErbB4sc shRNA | 69.3 ± 2.0 | 3 | 42.1 ± 17.9 | 3 |
| ErbB4 CL 4 shRNA | 96.6 ± 3.6 | 3 | 10.7 ± 1.4a | 5 |
| ErbB4 CL 12 shRNA | 90.4 ± 7.4 | 3 | 13.8 ± 4.3a | 5 |
| ErbB2sc shRNA | 69.9 ± 3.9 | 3 | 59.6 ± 16.6 | 3 |
| ErbB2 shRNA | 13.0 ± 3.8a | 3 | 93.9 ± 6.2 | 3 |
Marked loss of expression.
Figure 2.
Silencing endogenous ErbB4 or ErbB2 transcription in the MCF7 human breast tumor cell line reduces stimulation of motility by NRG2β. (a) ErbB2 expression (upper panel) and ErbB4 expression (lower panel) were evaluated in MCF7 cell lines that express the ErbB4 4915 shRNA (ErbB4 shRNA CL 4 and 12), the ErbB2 4355 shRNA (ErbB2 shRNA), a scrambled ErbB4 4915 shRNA sequence (ErbB4sc shRNA), or a scrambled ErbB2 4355 shRNA sequence (ErbB2sc shRNA). Samples generated from parental MCF7 cells were used to construct calibration curves for quantification. (b) Confluent monolayers of the indicated MCF7 cell lines were subjected to laser ablation to create a roughly circular area devoid of cells. Following treatment with NRG2β or a diluent control, cell migration into the wounded areas (wound healing) was monitored over a 96-hour period by time-lapse microscopy. (c) Wound healing in the presence of NRG2β was quantified by digital image processing and plotted as a function of recovery time. (d) Two-way analysis of variance with a Bonferroni posttest was used to evaluate the effect of NRG2β on wound healing in each cell line at 48 hours. P values are reported. NS indicates not significant.
Silencing ErbB4 expression in MCF7 cells also reduces the effect of an exogenous ErbB4 ligand on motility. NRG2β stimulates motility (wound healing) by the parental MCF7 cell line and the MCF7 cell line (ErbB4sc shRNA) that expresses the scrambled shRNA sequence that corresponds to the ErbB4 4915 shRNA (Fig. 2b-d). In contrast, the effect of NRG2β on motility in the MCF7 ErbB4 shRNA CL 4 and CL 12 cell lines is reduced (Fig. 2c,d) to the point of being statistically insignificant (Fig. 2b). Taken together, these results strongly indicate that ErbB4 is coupled to motility in MCF7 cells.
Next we explored the hypothesis that endogenous ErbB2 expression and ErbB2/ErbB4 crosstalk may be necessary for stimulation of motility by NRG2β in the MCF7 breast tumor cell line. We generated a pooled cell line by infecting MCF7 cells with the pLKO-ErbB2 4355 shRNA recombinant lentivirus, which targets the 3′ UTR of the endogenous ErbB2 transcript in MCF7 cells. This cell line (ErbB2 shRNA) displays an 87.0% ± 3.8% decrease in ErbB2 expression (Table 2 and Fig. 2a). Relative to the control MCF7 cell lines, the ErbB2 shRNA MCF7 cell line exhibits a minor, statistically insignificant reduction (P > 0.05) in motility in the absence of an ErbB4 ligand (Fig. 2b). In contrast, silencing ErbB2 expression mutes NRG2β stimulation of motility (Fig. 2b-d). This latter result strongly indicates that ErbB2 is required for an ErbB4 ligand to stimulate motility in MCF7 cells; thus, our studies indicate that ErbB2 is required for ErbB4 coupling to biological responses in some contexts.
ErbB2 kinase activity and sites of ErbB4 tyrosine phosphorylation are required for ligand-induced heterotypic ErbB4 signaling
These data presented here indicate that heterotypic ErbB4 signaling (ErbB2/ErbB4 crosstalk) is required for stimulation of malignant phenotypes by the ErbB4 ligand NRG2β. Co-expression of ErbB4 and ErbB2 enables NRG2β to stimulate IL3-independent proliferation of the BaF3 mouse lymphoid cell line; expression of ErbB4 alone does not permit NRG2β to stimulate IL3 independence.28 Thus, the BaF3 model system is ideal for elucidating the mechanism by which heterotypic ErbB4 signaling as a result of ErbB2/ErbB4 crosstalk couples to proliferation.
Stimulation of IL3 independence by NRG2β is only somewhat diminished (31% of control) in BaF3 cells that stably express wild-type ErbB2 and an ErbB4 mutant (K751M; Fig. 3a)14,29 that lacks tyrosine kinase activity (Table 3). In contrast, there is minimal stimulation of IL3 independence by NRG2β (13% of control) in BaF3 cells that stably express wild-type ErbB2 and an ErbB4 mutant that lacks the 9 putative sites of tyrosine phosphorylation (9Y to F; Fig. 3a)14,29 within the carboxyl terminus (Table 3). Thus, ErbB4 phosphorylation sites are required for coupling heterotypic ErbB4 signaling to proliferation in BaF3 cells, but ErbB4 kinase activity is not.
Figure 3.
In BaF3 ErbB2/ErbB4 cells, anti-ErbB2 and anti-ErbB4 antibodies do not cross-react with the noncognate ErbB receptor. BaF3 ErbB2/ErbB4 cells were starved and stimulated with NRG2β. ErbB2 or ErbB4 was immunoprecipitated using ErbB2- and ErbB4-specific antibodies. (a) BaF3 cells that express various combinations of ErbB2 and ErbB4 constructs were generated by lentiviral and retroviral infections. (b) Immunoblotting was used to assess tyrosine phosphorylation (upper panel), ErbB4 expression (lower panel), and ErbB4 co-precipitation with ErbB2 receptor/antibody complex (lower panel). (c) Immunoblotting was used to assess tyrosine phosphorylation (upper panel), ErbB2 expression (lower panel), and ErbB2 co-precipitation with ErbB4 receptor/antibody complex (lower panel).
Table 3.
NRG2β Activity Requires ErbB2 Kinase Activity and Sites of ErbB4 Tyrosine Phosphorylation but Not ErbB4 Kinase Activity or Sites of ErbB2 Tyrosine Phosphorylation
| ErbB4 Wild-Type, % | ErbB4 K751M, % | ErbB4 9Y to F, % | ||||
|---|---|---|---|---|---|---|
| 0.3 nM NRG2β | − | + | − | + | − | + |
| ErbB2 wild-type | 1.8 ± 0.4 | 100b | 3.7 ± 0.4 | 31 ± 7a | 6.3 ± 2.1 | 13 ± 2 |
| ErbB2 K753A | 3.1 ± 0.5 | 3.3 ± 0.5 | 3.6 ± 0.7 | 3.1 ± 0.2 | 7.3 ± 2.0 | 6.9 ± 2.0 |
| ErbB2 Del1001 | 4.4 ± 1.1 | 40 ± 5a | 3.2 ± 1.0 | 34 ± 6a | 5.3 ± 1.1 | 8.3 ± 2.6 |
Partial activity.
Full activity.
Stimulation of IL3 independence by NRG2β is only somewhat diminished (40% of control) in BaF3 cells that express wild-type ErbB4 and an ErbB2 truncation mutant (Del1001) that lacks all carboxy-terminal sites of tyrosine phosphorylation (Table 3). Moreover, there is minimal stimulation of IL3 independence by NRG2β (3% of control) in BaF3 cells that express wild-type ErbB4 and an ErbB2 mutant (K753A) devoid of tyrosine kinase activity. Thus, ErbB2 kinase activity is required for coupling heterotypic ErbB4 signaling to proliferation in BaF3 cells, but ErbB2 phosphorylation sites are not.
These data led us to predict that ErbB2 phosphorylates ErbB4 in the absence of ErbB4 kinase activity and in the absence of ErbB2 sites of tyrosine phosphorylation. We first confirmed that ErbB2 and ErbB4 antibodies do not recognize the noncognate receptor (Fig. 3b,c). However, an ErbB4 antibody can precipitate ErbB2 following stimulation with NRG2β, suggesting that the ErbB4 antibody precipitates ErbB2/ErbB4 heterodimers that arise as a consequence of stimulation with an ErbB4 agonist (Fig. 3c).
Consistent with our prediction, NRG2β stimulates ErbB4 phosphorylation in the BaF3 ErbB2/ErbB4 K751M and the BaF3 ErbB2 Del1001/ErbB4 K751M cell lines but not in the BaF3 ErbB2 K753A/ErbB4 K751M cell line (Fig. 4). Moreover, NRG2β fails to stimulate detectable ErbB4 phosphorylation in the BaF3 ErbB2/ErbB4 9Y to F cell line (Fig. 4), suggesting that the carboxyl-terminal ErbB4 tyrosine residues are the sites of phosphorylation by ErbB2. Finally, NRG2β fails to stimulate detectable ErbB2 phosphorylation in the BaF3 ErbB2 K753A/ErbB4 cell line. Thus, ErbB2 appears to be capable of phosphorylating ErbB4, but ErbB4 does not appear to be capable of phosphorylating ErbB2.
Figure 4.
In BaF3 ErbB2/ErbB4 cells, ErbB4 tyrosine phosphorylation is abrogated by mutating 9 ErbB4 cytoplasmic tyrosine residues but not by disrupting ErbB4 kinase activity. BaF3 ErbB2/ErbB4 cell lines were starved and stimulated with NRG2β. Immunoprecipitation and immunoblotting were used to assess ErbB4 tyrosine phosphorylation (first panel), ErbB4 expression (second panel), ErbB2 tyrosine phosphorylation (third panel), and ErbB2 expression (fourth panel). Data shown are representative of at least 3 independent experiments.
These data do not rule out the possibility that the inability of the ErbB2 K753A and ErbB4 9Y to F mutants to couple to IL3 independence is due to inadequate expression of these mutants. The expression of the ErbB2 K753A mutant is approximately one-fourth of that displayed by wild-type ErbB2 in the various BaF3 cell lines (Suppl. Fig. S1 and Suppl. Table S1). Similarly, the expression of the ErbB4 9Y to F mutant is approximately one-fifth of that displayed by wild-type ErbB4 in the various BaF3 cell lines (Suppl. Fig. S1 and Suppl. Table S1). The reduction of expression displayed by the ErbB2 K753A and ErbB4 9Y to F mutants is comparable to that displayed by the ErbB2 Del1001 mutant. Because the ErbB2 Del1001 mutant retains coupling to IL3 independence, the reduction of expression displayed by the ErbB2 K753A and ErbB4 9Y to F mutants does not appear to be relevant to the loss of coupling displayed by these mutants. Thus, these biochemical and functional data strongly indicate that phosphorylation of ErbB4 tyrosine residues by ErbB2 is required for ligand-induced heterotypic ErbB4 signaling and coupling to biological responses.
EGFR kinase activity is required for ligand-induced heterotypic ErbB4 signaling
Co-expression of EGFR and ErbB4 has been noted in breast and meningioma tumor samples.21,30 Moreover, co-expression of ErbB4 and EGFR enables NRG2β to stimulate IL3-independent proliferation of the BaF3 mouse lymphoid cell line; expression of ErbB4 alone does not permit NRG2β to stimulate IL3 independence.28 Thus, the BaF3 model system can be used to characterize heterotypic ErbB4 signaling as a result of EGFR/ErbB4 crosstalk.
The patterns of IL3 independence displayed by BaF3 cell lines that express various combinations of EGFR31 and ErbB4 constructs following stimulation with NRG2β are identical to the patterns of IL3 independence displayed by the BaF3 cells lines that express the corresponding combinations of ErbB2 and ErbB4 constructs. Stimulation of IL3 independence by NRG2β is only somewhat diminished (31% of control) in BaF3 cells that express wild-type EGFR and an ErbB4 mutant (K751M; Fig. 5a) that lacks tyrosine kinase activity (Table 4). In contrast, there is minimal stimulation of IL3 independence by NRG2β (6% of control) in BaF3 cells that express wild-type EGFR and an ErbB4 mutant that lacks the 9 putative sites of tyrosine phosphorylation (9Y to F; Fig. 5a) within the carboxyl terminus (Table 4). Finally, there is minimal stimulation of IL3 independence by NRG2β (5% of control) in BaF3 cells that express wild-type ErbB4 and an EGFR mutant (K721A; Fig. 5a) devoid of tyrosine kinase activity (Table 4). Taken together, these data strongly suggest that phosphorylation of ErbB4 tyrosine residues by EGFR is required for ligand-induced coupling of heterotypic ErbB4 signaling to proliferation in BaF3 cells.
Figure 5.
In BaF3 epidermal growth factor receptor (EGFR)/ErbB4 cells, anti-EGFR and anti-ErbB4 antibodies fail to precipitate the noncognate ErbB receptor. BaF3 EGFR/ErbB4 cells were starved and stimulated with NRG2β. EGFR or ErbB4 was immunoprecipitated using EGFR- and ErbB4-specific antibodies. (a) BaF3 cells that express various combinations of EGFR and ErbB4 constructs were generated by lentiviral and retroviral infections. (b) Immunoblotting was used to assess tyrosine phosphorylation (upper panel), ErbB4 expression (lower panel), and ErbB4 co-precipitation with EGFR receptor/antibody complex (lower panel). (c) Immunoblotting was used to assess tyrosine phosphorylation (upper panel), EGFR expression (lower panel), and EGFR co-precipitation with ErbB4 receptor/antibody complex (lower panel).
Table 4.
NRG2β Activity Requires EGFR Kinase Activity and Sites of ErbB4 Tyrosine Phosphorylation but Not ErbB4 Kinase Activity
| ErbB4 Wild-Type, % | ErbB4 K751M, % | ErbB4 9Y to F, % | ||||
|---|---|---|---|---|---|---|
| 0.3 nM NRG2β | − | + | − | + | − | + |
| EGFR wild-type | 3.5 ± 1.3 | 100b | 5.4 ± 1.3 | 30.5 ± 3.3a | 3.6 ± 0.8 | 5.6 ± 0.01 |
| EGFR K721A | 2.2 ± 0.6 | 5.0 ± 1.8 | — | — | 2.3 ± 0.5 | 1.9 ± 0.8 |
| ErbB2 wild-type | 2.8 ± 0.7 | 130 ± 25b | — | — | — | — |
EGFR = epidermal growth factor receptor; — = not tested.
Partial activity.
Full activity.
These data led us to predict that EGFR phosphorylates ErbB4 in the absence of ErbB4 kinase activity. We first confirmed that EGFR and ErbB4 antibodies do not recognize the noncognate receptor (Fig. 5b,c). Consistent with our prediction, NRG2β stimulates ErbB4 phosphorylation in the BaF3 EGFR/ErbB4 K751M but not in the BaF3 EGFR K721A/ErbB4 K751M cell line (Fig. 6). NRG2β stimulates minimal ErbB4 phosphorylation in the BaF3 EGFR/ErbB4 9Y to F cell line, suggesting that the carboxyl-terminal ErbB4 tyrosine residues are the primary sites of phosphorylation by EGFR (Fig. 6). NRG2β stimulates EGFR phosphorylation in the BaF3 EGFR/ErbB4 9Y to F cell line (Fig. 6). However, NRG2β fails to stimulate IL3 independence in this cell line (Table 4). Thus, these data suggest that EGFR phosphorylation of ErbB4 carboxyl terminal residues is required for ErbB4 coupling to IL3 independence, that ErbB4 kinase activity is not required for ErbB4 coupling to IL3 independence, and that EGFR phosphorylation sites are not sufficient to couple to IL3 independence.
Figure 6.
In BaF3 epidermal growth factor receptor (EGFR)/ErbB4 cells, ErbB4 tyrosine phosphorylation is abrogated by mutating 9 ErbB4 cytoplasmic tyrosine residues but not by disrupting ErbB4 kinase activity. BaF3 EGFR/ErbB4 cell lines were starved and stimulated with NRG2β. Immunoprecipitation and immunoblotting were used to assess ErbB4 tyrosine phosphorylation (first panel), ErbB4 expression (second panel), EGFR tyrosine phosphorylation (third panel), and EGFR expression (fourth panel). Data shown are representative of at least 3 independent experiments.
Nonetheless, these data do not rule out the possibility that the inability of the EGFR K721A and ErbB4 9Y to F mutants to couple to IL3 independence is due to inadequate expression of these mutants in the BaF3 EGFR/ErbB4 cell lines. Indeed, the expression of the EGFR K721A mutant in the various BaF3 EGFR/ErbB4 cell lines is markedly less than the expression of wild-type EGFR in the comparable cell lines (Suppl. Fig. S2 and Suppl. Table S2). Likewise, the expression of the ErbB4 9Y to F mutant in the various BaF3 EGFR/ErbB4 cell lines is markedly less than the expression of wild-type ErbB4 in the comparable cell lines (Suppl. Fig. S2 and Suppl. Table S2). It is interesting to note that the amount of ErbB4 phosphorylation observed in the BaF3 EGFR K721A/ErbB4 cell line is much greater.
Discussion
The work presented here grew out of our observation that homotypic ErbB4 signaling from the constitutively dimerized and active ErbB4 Q646C mutant is coupled to tumor suppressor activities in a number of different human tumor cell lines,14-16 whereas ligand-induced ErbB4 signaling is coupled to proliferation and anchorage independence.16,22,32 Here we present data that may help to resolve this apparent dichotomy. Ligand-induced ErbB4 coupling to anchorage independence, motility, and proliferation appears to be dependent on heterotypic ErbB4 signaling that features ErbB4 crosstalk with either ErbB2 or EGFR. This heterotypic ErbB4 signaling apparently requires ErbB4 tyrosine phosphorylation sites and either EGFR or ErbB2 kinase activity, suggesting that phosphorylation of ErbB4 by EGFR or ErbB2 is critical for ligand-induced ErbB4 coupling to oncogenic activities.
However, details concerning the mechanism by which heterotypic ErbB4 signaling couples to oncogenic activities remain to be elucidated. One prediction is that heterotypic ErbB4 signaling features direct phosphorylation of ErbB4 by EGFR or ErbB2 (Fig. 7). Biochemical assessment of ErbB4/EGFR and ErbB4/ErbB2 heterodimerization will be needed to test this prediction. Likewise, functional analyses using mutants that affect the regions of the intracellular juxtamembrane and kinase domains that are responsible for ErbB dimerization33 will evaluate whether ErbB heterodimers are indeed responsible for coupling heterotypic ErbB4 signaling to oncogenic activities. Another prediction of our model is that heterotypic ErbB4 signaling features phosphorylation of a set of ErbB4 tyrosine residues that is not identical to the set that is phosphorylated as a result of homotypic ErbB4 signaling. Indirect evidence for such a difference in sites of ErbB4 tyrosine phosphorylation may be gleaned by comparing the patterns of effector signaling as a result of homotypic and heterotypic ErbB4 signaling. Additional indirect evidence may be obtained by using ErbB4 phosphorylation site mutants to identify the sites of tyrosine phosphorylation required for coupling homotypic ErbB4 signaling to tumor suppressor activities and the sites of tyrosine phosphorylation required for coupling heterotypic ErbB4 signaling to oncogenic activities. However, definitive analyses must include biochemical mapping of the sites of ErbB4 tyrosine phosphorylation resulting from homotypic and heterotypic ErbB4 signaling.
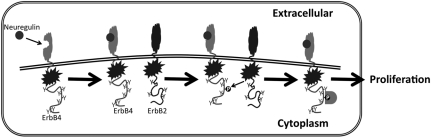
Figure 7.
Crosstalk between ErbB4 and ErbB2 may account for ErbB4 coupling to cell proliferation. We propose that ErbB4/ErbB2 crosstalk occurs through ErbB4 phosphorylation by ErbB2.
Other models have emerged that may account for differences in ErbB4 coupling to biological responses. The ErbB4 transcript contains two sites of alternative splicing. One affects the sequence encoding the cytoplasmic juxtamembrane region; following agonist binding, the canonical ErbB4 JM-a isoform undergoes cleavage, and the soluble cytoplasmic domain of ErbB4 (commonly known as s80 or 4ICD) translocates to the nucleus and the mitochondria.1 In contrast, the ErbB4 JM-b isoform is resistant to cleavage, and the cytoplasmic domain of the JM-b isoform is not released from the plasma membrane and does not translocate to the nucleus or the mitochondria.1,32 This difference in cytoplasmic domain trafficking appears to be functionally significant, as the s80/4ICD fragment of ErbB4 possesses tumor suppressor activities.11,34,35
The other site of alternative splicing of the ErbB4 transcript affects the sequence encoding the portion of the cytoplasmic domain distal to the tyrosine kinase domain (Fig. 3a). The Ct-b/CYT-2 isoform lacks a 16–amino acid sequence present in the canonical CT-a/CYT-1 isoform. This sequence contains a putative site of tyrosine phosphorylation (Tyr1056) that may enable ErbB4 binding to the p85 subunit of PI3 kinase and enable ErbB4 coupling to the PI3 kinase/Akt signaling pathway.14,29 Moreover, the same tyrosine residue absent in the CT-b/CYT-2 isoform may regulate ErbB4 binding to the transcription factor WWOX via the WW domain found in WWOX.5,6,36 ErbB4 coupling to the PI3 kinase/Akt pathway and WWOX may be critical for ErbB4 tumor suppressor activity, as the tumor suppressor activity of the constitutively active ErbB4 Q646C mutant is absent in the context of the Y1056F mutation or the Ct-b/CYT-2 isoform.29 Moreover, the Y1056F mutation alters the trafficking of the cytoplasmic domain of the constitutively active ErbB4 Q646C mutant. Thus, it is tempting to hypothesize that phosphorylation of ErbB4 Tyr1056 is a feature of homotypic ErbB4 signaling but not of heterotypic ErbB4 signaling, thereby accounting for the functional difference between homotypic and heterotypic ErbB4 signaling. This hypothesis is supported by the observation that the ErbB4 Q646C mutant exhibits phosphorylation of Tyr1056.14,29
Regardless of these mechanistic details, these studies shed an additional interesting insight on the roles that ErbB4 and other ErbB receptors may be playing in human tumorigenesis. The failure of EGFR and ErbB2 mutants that lack tyrosine kinase activity to support heterotypic ErbB4 coupling to cell proliferation indicates that blocking EGFR and/or ErbB2 tyrosine kinase activity (using small molecule tyrosine kinase inhibitors, for example) may be an effective strategy for targeting ErbB4-dependent tumors. Moreover, because MCF7 and T47D cells do not overexpress EGFR or ErbB2, our data suggest that overexpression of these receptors may not be necessary for the dependence of these cells on these receptors. MCF7 and T47D cells are not considered to be sensitive to trastuzumab, lapatinib, or erlotinib; nonetheless, therapeutics that target EGFR and ErbB2 may be effective against tumors other than those that overexpress these receptors.37,38 For example, pertuzumab is effective against tumor cells with low and high amounts of ErbB2 expression.39,40 Given that pertuzumab blocks ErbB2 dimerization, including ErbB2 heterodimerization with other ErbB receptors, we postulate that tumors dependent on ErbB4 signaling via ErbB2/ErbB4 crosstalk may comprise part of the population of tumors that respond to pertuzumab. This hypothesis is consistent with the observation that dual-specificity EGFR/ErbB2 inhibitors, which may be targeting the oncogenic EGFR/ErbB4 and ErbB2/ErbB4 heterodimers, appear to be effective in contexts in which targeting EGFR or ErbB2 alone is ineffective.37
Materials and Methods
DNA constructs
The recombinant retroviral expression constructs pLXSN-ErbB2 and pLXSN-EGFR have been described previously.28 We used pLXSN-ErbB2 and pLXSN-EGFR for site-directed mutagenesis using the QuikChange kit (Stratagene, La Jolla, CA). We created an ErbB2 mutant that lacks tyrosine kinase activity (K753A) by changing Lys753 in the putative adenosine triphosphate (ATP) binding pocket of the kinase domain to alanine. We created an ErbB2 mutant that lacks the carboxyl-terminal 255 amino acids (Del1001) by changing the Asp1001 codon to a stop codon. We created an EGFR mutant that lacks tyrosine kinase activity (K721A) by changing Lys721 in the putative ATP binding pocket of the kinase domain to alanine.
The pLKO.1 recombinant lentiviral vector is a generous gift of William Hahn (Dana-Farber Cancer Institute) and features an internal U6 snRNP promoter. The U6 promoter enables us to use this plasmid for the expression of shRNAs. The pLKO-GFP shRNA plasmid, which encodes an shRNA that targets the green fluorescence protein (GFP), has been described previously.41 The sequence that encodes the ErbB4 4915 shRNA (5′-CCG GCA GTT CTC TGT GGT TCA GGA ACT CGA GTT CCT GAA CCA CAG AGA ACT GTT TTT G-3′) and the sequence that encodes the ErbB2 4355 shRNA (5′-CCG GTG TCA GTA TCC AGG CTT TGT ACT CGA GTA CAA AGC CTG GAT ACT GAC ATT TTT G-3′) were subcloned using standard techniques into pLKO. These shRNAs are specific to the 3′ untranslated region of the respective endogenous transcripts and do not target transcripts encoded by the ErbB2 or ErbB4 cDNAs used in experiments described here. The sequence that encodes the ErbB4 4915 shRNA was scrambled using siRNA Wizard (InvivoGen, San Diego, CA), and this scrambled sequence was subcloned into pLKO.1 via standard molecular biology techniques. A sequence generated by scrambling the sequence that encodes the ErbB2 4355 shRNA was subcloned into pLKO.1 using an analogous strategy.
Standard molecular biology techniques and the shuttle vector pENTR1A (Invitrogen, Carlsbad, CA) were used to subclone wild-type and mutant ErbB4 cDNAs into the recombinant lentiviral expression vector pLenti6/V5/DEST (Invitrogen). Note that each of these cDNA sequences includes a stop codon that terminates protein translation prior to the V5 epitope tag sequence present in the expression vector. Wild-type ErbB4 was subcloned from pCH4M2.42 An ErbB4 mutant that lacks tyrosine kinase activity (K751M) was subcloned from pLXSN-ErbB4 K751M.14 An ErbB4 mutant in which the 9 putative sites of tyrosine phosphorylation in the carboxyl terminus (Tyr1022, Tyr1150, Tyr1056, Tyr1162, Tyr1188, Tyr1202, Tyr1242, Tyr1258, and Tyr1284) were mutated to phenylalanine (YChg9F) was subcloned from pLXSN-ErbB4 YChg9F.14
Cell lines and cell culture
Mouse ψ2 and PA317 recombinant retrovirus packaging cell lines and the BaF3 mouse pro-B-lymphocyte cell line are generous gifts of Daniel DiMaio (Yale University). These cells were cultured essentially as described previously.28,43,44 MCF7 and T47D human breast tumor cell lines were obtained from American Type Culture Collection (Manassas, VA) and were cultured as recommended. HEK293FT cells were obtained from Invitrogen and cultured as recommended. Cell culture media and supplements were obtained from Invitrogen, Sigma Scientific (St. Louis, MO), and Mediatech (Herndon, VA). Fetal bovine serum, G418, and puromycin were obtained from Gemini Bioproducts (Woodland, CA). Blasticidin was obtained from Sigma Scientific. Recombinant NRG2β was expressed and purified as previously described.45 Other biochemicals were obtained from Sigma Scientific.
ErbB4 lentiviruses
The pLenti-ErbB4,16 pLenti-ErbB4 K751M,16 and pLenti-ErbB4 YChg9F constructs and the pLenti6/V5/DEST vector control were packaged into recombinant lentiviral particles by transient co-transfection with the packaging vectors pLP1, pLP2, and pLP/VSVG into the HEK293FT lentiviral packaging cell line (Invitrogen). We transfected the cells and recovered the recombinant lentiviruses as recommended.
A 24-well plate was seeded with 4 × 105 BaF3 cells in 500 µL complete medium supplemented with 6 µg/mL polybrene. A pLenti lentiviral stock (500 µL) was added to each well, and the cells were incubated overnight at 37°C. The cells were recovered by gentle centrifugation and were resuspended in complete medium supplemented with 12 µg/mL blasticidin to select for stably infected, recombinant BaF3/pLenti cell lines.
Recombinant retroviruses
Briefly, the recombinant amphotropic retroviruses LXSN-ErbB2 (ErbB2 WT), LXSN-ErbB2 K753A, LXSN-ErbB2 Del1001, LXSN-EGFR (EGFR WT), and LXSN-EGFR K721A were packaged using the ψ2 ecotropic retrovirus packaging cell line46 and the PA317 amphotropic retrovirus packaging cell line47 essentially as previously described.43 BaF3/pLenti cell lines were infected with these recombinant retroviruses using procedures essentially identical to those used to generate the BaF3/pLenti cell lines. We used 6 µg/mL G418 to select for stably infected, BaF3/pLenti/LXSN recombinant cell lines.
shRNA lentiviruses
The pLKO.1, pLKO-GFP shRNA, pLKO-ErbB4 4915 shRNA, pLKO-ErbB2 4355 shRNA, and scrambled shRNA recombinant lentiviral constructs were packaged by transient co-transfection with the packaging vectors pCMV-dR8.91 and pMD2G-VSVG into the HEK293FT lentiviral packaging cell line (Invitrogen). Recombinant pLKO lentiviruses were recovered from the conditioned medium of these transfected HEK293FT cells. Following infection with the recombinant pLKO lentiviruses, MCF7 and T47D human breast tumor cells were incubated with 1 µg/mL puromycin to select for stably infected, recombinant pLKO cell lines. Thereafter, MCF7 clones that express the pLKO-ErbB4 4915 shRNA were generated via limited dilution cloning, and ErbB4 expression was analyzed in the resulting clonal cell lines. The clone displaying the least ErbB4 expression was reinfected with the pLKO-ErbB4 4915 shRNA recombinant lentivirus, and clones of superinfected cells were selected for further analysis. Clones 4 and 12 exhibit the least ErbB4 expression.
Analysis of ErbB2 and ErbB4 expression and tyrosine phosphorylation by immunoblotting and immunprecipitation
In experiments that featured ligand stimulation, ErbB receptor tyrosine phosphorylation was assayed using cells starved in basal medium for 16 hours. Cells were stimulated for 7 minutes on ice with 10 nM NRG2β or phosphate-buffered saline (PBS; diluent control) and lysed. In experiments that did not feature ligand stimulation, cells were grown to saturation density in complete medium and were lysed.
ErbB receptor expression and tyrosine phosphorylation were assayed by immunoprecipitation and immunoblotting essentially as described.28,48,49 Briefly, ErbB2 was precipitated using an anti-ErbB2 mouse monoclonal antibody (OP-39; EMD Chemicals, Inc., Gibbstown, NJ); ErbB4 was precipitated using an anti-ErbB4 mouse monoclonal antibody (SC-8050; Santa Cruz Biotechnology, Santa Cruz, CA). Immunoblotting antibodies include the SC-283 rabbit anti-ErbB2 polyclonal antibody (Santa Cruz Biotechnology; used to assess endogenous ErbB2 expression in MCF7 and T47D cells), the AF1129 goat anti-ErbB2 polyclonal antibody (R&D Systems, Minneapolis, MN; used to assess ectopic ErbB2 expression in BaF3 cells), the SC-283 rabbit anti-ErbB4 polyclonal antibody (Santa Cruz Biotechnology; used to assess endogenous and ectopic ErbB4 expression), and the 4G10 mouse antiphosphotyrosine monoclonal antibody (Millipore, Billerica, MA). The resulting chemilumigrams were digitized, and National Institutes of Health Image J software was used to quantify receptor expression. Statistical analyses were performed using Microsoft Excel. The data are averages of at least 3 independent experiments. The standard error of the mean was determined for the expression of ErbB2 and ErbB4 of each cell line.
Anchorage independence assay
The 2 × 104 cells were seeded in semisolid medium that consisted of Dulbecco’s modified Eagle’s medium (DMEM) and 0.3% LMP agarose supplemented with either 10 nM NRG2β or PBS (diluent control). Fifteen days later, 6 microscopic fields were selected at random and photographed. Using these photomicrographs, we measured the colonies and recorded the number of colonies whose diameter was less than 70 µm, 70 to 140 µm, and greater than 140 µm. For each experimental condition, approximately 225 to 350 colonies were measured. The data from 4 independent experiments were pooled, and the effect of NRG2β stimulation on anchorage-independent proliferation (distribution of colony size) was determined by chi-squared analysis.
Wound-healing (migration) assay
Derivatives of the MCF7 human breast cancer cell line were seeded into a plastic, flat-bottom, 96-well culture plate (C-lect Stem Cell Manager; Cyntellect, Inc., San Diego, CA) and allowed to grow to confluence. They were then starved in basal media for 48 hours. The plate was loaded into a LEAP instrument (Cyntellect, Inc.), and 25% of the cell monolayer in each well was cleared by laser ablation. The LEAP instrument fired a series of 7-µJ laser pulses that had a 25-µm grid spacing to form the “wound” area. These pulses killed the cells, resulting in wounds in which 25% of the monolayer in each well was cleared. The wells were washed to remove the dead cells, and fresh basal medium was added that contained 10 nM NRG2β or PBS (diluent control). The cells were incubated at 37°C, and the LEAP instrument imaged each well by bright-field microscopy every 24 hours for a 96-hour period.50
A custom MATLAB script and these images were used to quantify the recovery (healing) of the wounded areas. The results are expressed as a percent of wound healing relative to the 0-hour time point for each well. The data shown represent 3 independent trials, each of which represents 5 independent wells per condition. The mean and standard error of the mean (SEM) were calculated for each condition and time point, and differences in wound healing were assessed using a two-way analysis of variance with the Bonferroni posttest.
IL3 independence assay
These experiments were performed essentially as previously described.51 Briefly, the BaF3 cells were seeded in a 24-well plate at a density of 1 × 105 cells/mL in medium lacking IL3 but supplemented with either 0.3 nM NRG2β or PBS (diluent control). Cells were incubated for 96 hours, after which viable cell density was determined by counting using a hemacytometer, and the percent stimulation was calculated relative to mock-stimulated cells (Prism; GraphPad Software, San Diego, CA). The data shown are the mean and SEM for 3 independent experiments.
Supplementary Material
Acknowledgments
The authors thank Fred Koller at Cyntellect and Ianthe Bryant for essential reagents and supplies.
Footnotes
The author(s) declared the following potential conflicts of interest with respect to the research, authorship, and/or publication of this article: Jeffrey Settleman is an employee of Genentech. However, Genentech did not otherwise sponsor this work, Genentech holds no interest in this work, and no Genentech products were used, discussed, or alluded to in this work.
This work was supported by the US Army Medical Research and Materiel Command Breast Cancer Research Program [DAMD17-00-1-0415, DAMD17-00-1-0416 to D.J.R.], the US Army Medical Research and Materiel Command Prostate Cancer Research Program [DAMD17-02-1-0130 to D.J.R.], the National Institutes of Health [R21CA080770 and R01CA114209 to DJR; S10RR023651 to J.F.L.; K08DE020139 to S.M.R.], the American Cancer Society [IRG 58-006-40], the Showalter Trust, the Purdue University Center for Cancer Research, the Indiana Elks Foundation, the Purdue School of Pharmacy, and the Carroll County (Indiana) Cancer Association.
Supplementary material for this article is available on the Genes & Cancer website at http://ganc.sagepub.com/supplemental.
References
- 1. Carpenter G. ErbB-4: mechanism of action and biology. Exp Cell Res. 2003;284:66-77 [DOI] [PubMed] [Google Scholar]
- 2. Jones RB, Gordus A, Krall JA, MacBeath G. A quantitative protein interaction network for the ErbB receptors using protein microarrays. Nature. 2006;439:168-74 [DOI] [PubMed] [Google Scholar]
- 3. Schulze WX, Deng L, Mann M. Phosphotyrosine interactome of the ErbB-receptor kinase family. Mol Syst Biol. 2005;1:20050008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. van der Geer P, Hunter T, Lindberg RA. Receptor protein-tyrosine kinases and their signal transduction pathways. Annu Rev Cell Biol. 1994;10:251-337 [DOI] [PubMed] [Google Scholar]
- 5. Jones FE. HER4 intracellular domain (4ICD) activity in the developing mammary gland and breast cancer. J Mammary Gland Biol Neoplasia. 2008;13:247-58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sundvall M, Iljin K, Kilpinen S, et al. Role of ErbB4 in breast cancer. J Mammary Gland Biol Neoplasia. 2008;13:259-68 [DOI] [PubMed] [Google Scholar]
- 7. Saglam O, Shah V, Worsham MJ. Molecular differentiation of early and late stage laryngeal squamous cell carcinoma: an exploratory analysis. Diagn Mol Pathol. 2007;16:218-21 [DOI] [PubMed] [Google Scholar]
- 8. Thybusch-Bernhardt A, Beckmann S, Juhl H. Comparative analysis of the EGF-receptor family in pancreatic cancer: expression of HER-4 correlates with a favourable tumor stage. Int J Surg Investig. 2001;2:393-400 [PubMed] [Google Scholar]
- 9. Uberall I, Kolar Z, Trojanec R, Berkovcova J, Hajduch M. The status and role of ErbB receptors in human cancer. Exp Mol Pathol. 2008;84:79-89 [DOI] [PubMed] [Google Scholar]
- 10. Ghayad SE, Vendrell JA, Larbi SB, et al. Endocrine resistance associated with activated ErbB system in breast cancer cells is reversed by inhibiting MAPK or PI3K/Akt signaling pathways. Int J Cancer. 2010;126:545-62 [DOI] [PubMed] [Google Scholar]
- 11. Naresh A, Long W, Vidal GA, et al. The ERBB4/HER4 intracellular domain 4ICD is a BH3-only protein promoting apoptosis of breast cancer cells. Cancer Res. 2006;66:6412-20 [DOI] [PubMed] [Google Scholar]
- 12. Gilmour LM, Macleod KG, McCaig A, et al. Expression of erbB-4/HER-4 growth factor receptor isoforms in ovarian cancer. Cancer Res. 2001;61:2169-76 [PubMed] [Google Scholar]
- 13. Lee CM, Shrieve DC, Zempolich KA, et al. Correlation between human epidermal growth factor receptor family (EGFR, HER2, HER3, HER4), phosphorylated Akt (P-Akt), and clinical outcomes after radiation therapy in carcinoma of the cervix. Gynecol Oncol. 2005;99:415-21 [DOI] [PubMed] [Google Scholar]
- 14. Pitfield SE, Bryant I, Penington DJ, Park G, Riese DJ., II Phosphorylation of ErbB4 on tyrosine 1056 is critical for ErbB4 coupling to inhibition of colony formation by human mammary cell lines. Oncol Res. 2006;16:179-93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Williams EE, Trout LJ, Gallo RM, et al. A constitutively active ErbB4 mutant inhibits drug-resistant colony formation by the DU-145 and PC-3 human prostate tumor cell lines. Cancer Lett. 2003;192:67-74 [DOI] [PubMed] [Google Scholar]
- 16. Mill CP, Gettinger KL, Riese DJ., II Ligand stimulation of ErbB4 and A constitutively-active ErbB4 mutant result in different biological responses in human pancreatic tumor cell lines. Exp Cell Res. 2011;317:392-404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Vidal GA, Clark DE, Marrero L, Jones FE. A constitutively active ERBB4/HER4 allele with enhanced transcriptional coactivation and cell-killing activities. Oncogene. 2007;26:462-6 [DOI] [PubMed] [Google Scholar]
- 18. Linggi B, Cheng QC, Rao AR, Carpenter G. The ErbB-4 s80 intracellular domain is a constitutively active tyrosine kinase. Oncogene. 2006;25:160-3 [DOI] [PubMed] [Google Scholar]
- 19. Gilbertson RJ, Clifford SC, MacMeekin W, et al. Expression of the ErbB-neuregulin signaling network during human cerebellar development: implications for the biology of medulloblastoma. Cancer Res. 1998;58:3932-41 [PubMed] [Google Scholar]
- 20. Burgess AW. EGFR family: structure physiology signalling and therapeutic targets. Growth Factors. 2008;26:263-74 [DOI] [PubMed] [Google Scholar]
- 21. Abd El-Rehim DM, Pinder SE, Paish CE, et al. Expression and co-expression of the members of the epidermal growth factor receptor (EGFR) family in invasive breast carcinoma. Br J Cancer. 2004;91:1532-42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tang CK, Concepcion XZ, Milan M, et al. Ribozyme-mediated down-regulation of ErbB-4 in estrogen receptor–positive breast cancer cells inhibits proliferation both in vitro and in vivo. Cancer Res. 1999;59:5315-22 [PubMed] [Google Scholar]
- 23. Prickett TD, Agrawal NS, Wei X, et al. Analysis of the tyrosine kinome in melanoma reveals recurrent mutations in ERBB4. Nat Genet. 2009;41:1127-32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Graus-Porta D, Beerli RR, Daly JM, Hynes NE. ErbB-2, the preferred heterodimerization partner of all ErbB receptors, is a mediator of lateral signaling. EMBO J. 1997;16:1647-55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wang LM, Kuo A, Alimandi M, et al. ErbB2 expression increases the spectrum and potency of ligand-mediated signal transduction through ErbB4. Proc Natl Acad Sci U S A. 1998;95:6809-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kruser TJ, Wheeler DL. Mechanisms of resistance to HER family targeting antibodies. Exp Cell Res. 2010;316:1083-100 [DOI] [PubMed] [Google Scholar]
- 27. Jones KL, Buzdar AU. Evolving novel anti-HER2 strategies. Lancet Oncol. 2009;10:1179-87 [DOI] [PubMed] [Google Scholar]
- 28. Riese DJ, II, van Raaij TM, Plowman GD, Andrews GC, Stern DF. The cellular response to neuregulins is governed by complex interactions of the erbB receptor family. Mol Cell Biol. 1995;15:5770-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gallo RM, Bryant I, Fry R, Williams EE, Riese DJ., II Phosphorylation of ErbB4 on Tyr1056 is critical for inhibition of colony formation by prostate tumor cell lines. Biochem Biophys Res Commun. 2006;349:372-82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Andersson U, Guo D, Malmer B, et al. Epidermal growth factor receptor family (EGFR, ErbB2-4) in gliomas and meningiomas. Acta Neuropathol. 2004;108:135-42 [DOI] [PubMed] [Google Scholar]
- 31. Honegger AM, Dull TJ, Felder S, et al. Point mutation at the ATP binding site of EGF receptor abolishes protein-tyrosine kinase activity and alters cellular routing. Cell. 1987;51:199-209 [DOI] [PubMed] [Google Scholar]
- 32. Junttila TT, Sundvall M, Lundin M, et al. Cleavable ErbB4 isoform in estrogen receptor–regulated growth of breast cancer cells. Cancer Res. 2005;65:1384-93 [DOI] [PubMed] [Google Scholar]
- 33. Red Brewer M, Choi SH, Alvarado D, et al. The juxtamembrane region of the EGF receptor functions as an activation domain. Mol Cell. 2009;34:641-51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Feng SM, Sartor CI, Hunter D, et al. The HER4 cytoplasmic domain, but not its C terminus, inhibits mammary cell proliferation. Mol Endocrinol. 2007;21:1861-76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Muraoka-Cook RS, Sandahl M, Husted C, et al. The intracellular domain of ErbB4 induces differentiation of mammary epithelial cells. Mol Biol Cell. 2006;17:4118-29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Aqeilan RI, Donati V, Gaudio E, et al. Association of Wwox with ErbB4 in breast cancer. Cancer Res. 2007;67:9330-6 [DOI] [PubMed] [Google Scholar]
- 37. Karamouzis MV, Badra FA, Papavassiliou AG. Breast cancer: the upgraded role of HER-3 and HER-4. Int J Biochem Cell Biol. 2007;39:851-6 [DOI] [PubMed] [Google Scholar]
- 38. Tiseo M, Loprevite M, Ardizzoni A. Epidermal growth factor receptor inhibitors: a new prospective in the treatment of lung cancer. Curr Med Chem Anticancer Agents. 2004;4:139-48 [DOI] [PubMed] [Google Scholar]
- 39. Albanell J, Codony J, Rovira A, Mellado B, Gascon P. Mechanism of action of anti-HER2 monoclonal antibodies: scientific update on trastuzumab and 2C4. Adv Exp Med Biol. 2003;532:253-68 [DOI] [PubMed] [Google Scholar]
- 40. Johnson BE, Janne PA. Rationale for a phase II trial of pertuzumab, a HER-2 dimerization inhibitor, in patients with non–small cell lung cancer. Clin Cancer Res. 2006;12:4436s-40s. [DOI] [PubMed] [Google Scholar]
- 41. Rothenberg SM, Engelman JA, Le S, et al. Modeling oncogene addiction using RNA interference. Proc Natl Acad Sci U S A. 2008;105:12480-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Plowman GD, Culouscou JM, Whitney GS, et al. Ligand-specific activation of HER4/p180erbB4, a fourth member of the epidermal growth factor receptor family. Proc Natl Acad Sci U S A. 1993; 90:1746-50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Leptak C, Ramon y, Cajal S, Kulke R, et al. Tumorigenic transformation of murine keratinocytes by the E5 genes of bovine papillomavirus type 1 and human papillomavirus type 16. J Virol. 1991;65:7078-83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Riese DJ, 2nd , DiMaio D. An intact PDGF signaling pathway is required for efficient growth transformation of mouse C127 cells by the bovine papillomavirus E5 protein. Oncogene. 1995;10:1431-9 [PubMed] [Google Scholar]
- 45. Wilson KJ, Mill CP, Cameron EM, et al. Inter-conversion of neuregulin2 full and partial agonists for ErbB4. Biochem Biophys Res Commun. 2007;364:351-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Mann R, Mulligan RC, Baltimore D. Construction of a retrovirus packaging mutant and its use to produce helper-free defective retrovirus. Cell. 1983;33:153-9 [DOI] [PubMed] [Google Scholar]
- 47. Miller AD, Buttimore C. Redesign of retrovirus packaging cell lines to avoid recombination leading to helper virus production. Mol Cell Biol. 1986;6:2895-902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Penington DJ, Bryant I, Riese DJ., II Constitutively active ErbB4 and ErbB2 mutants exhibit distinct biological activities. Cell Growth Differ. 2002;13:247-56 [PubMed] [Google Scholar]
- 49. Riese DJ, II, Komurasaki T, Plowman GD, Stern DF. Activation of ErbB4 by the bifunctional epidermal growth factor family hormone epiregulin is regulated by ErbB2. J Biol Chem. 1998;273:11288-94 [DOI] [PubMed] [Google Scholar]
- 50. Zordan MD, Mill CP, Riese DJ, II, Leary JF. A high throughput, interactive imaging, bright-field wound healing assay. Cytometry A. 2011;79A:227-32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hobbs SS, Cameron EM, Hammer RP, et al. Five carboxyl-terminal residues of neuregulin2 are critical for stimulation of signaling by the ErbB4 receptor tyrosine kinase. Oncogene. 2004;23:883-93 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.