Abstract
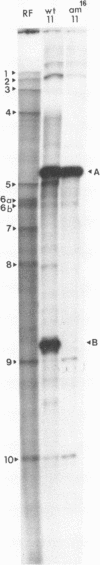
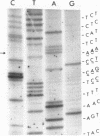
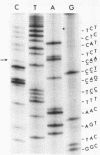
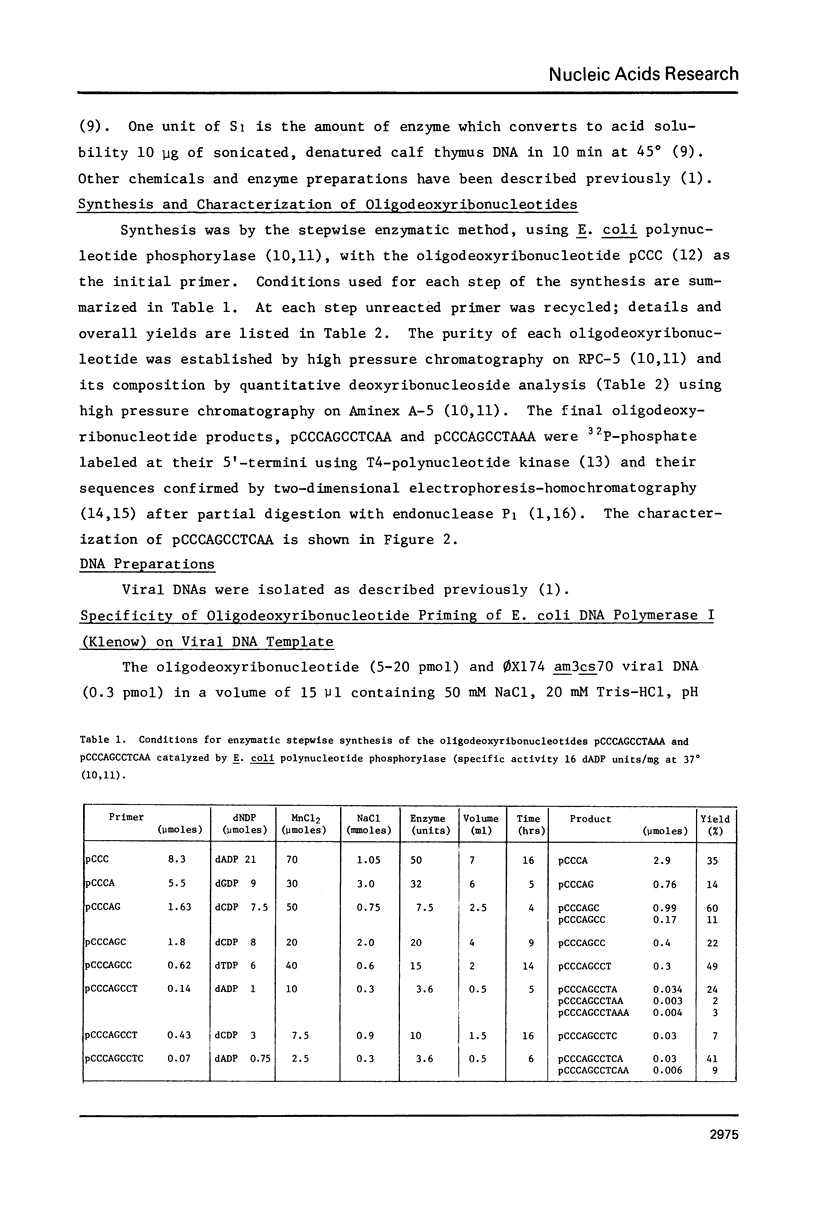
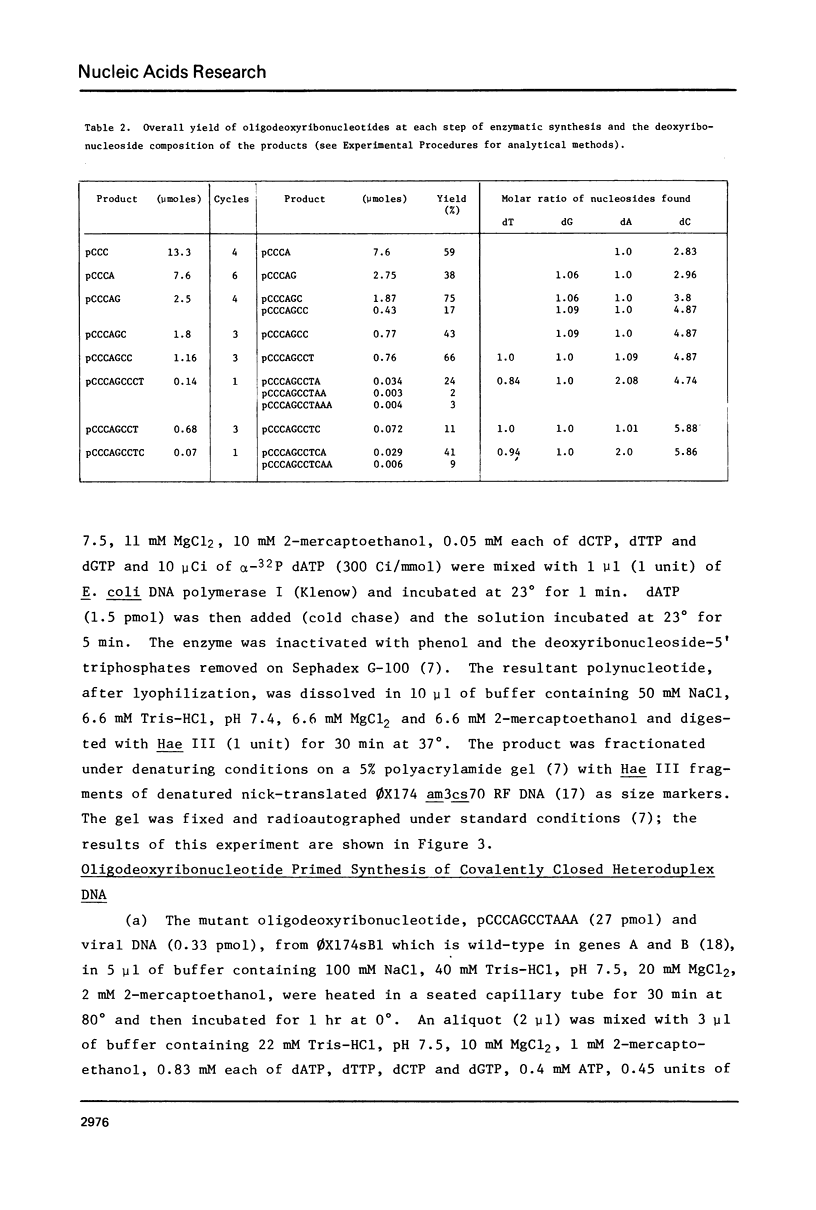
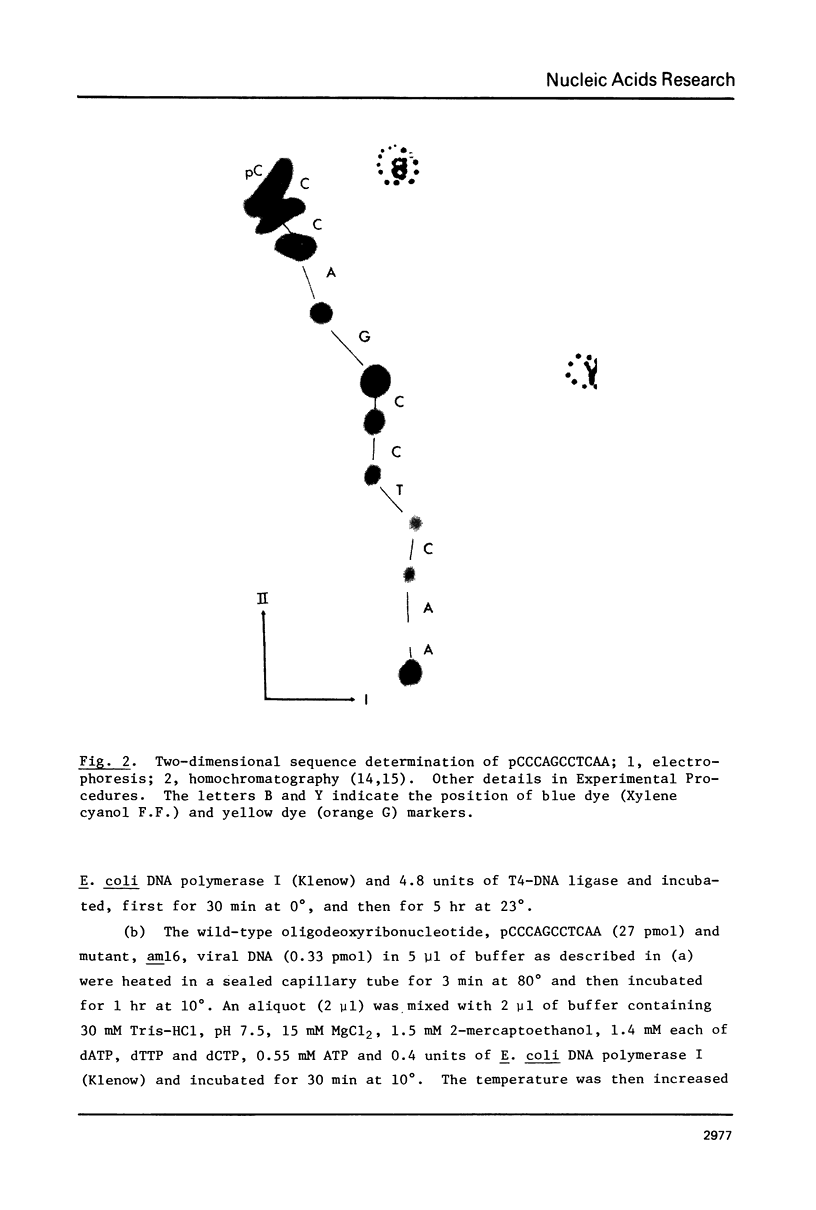
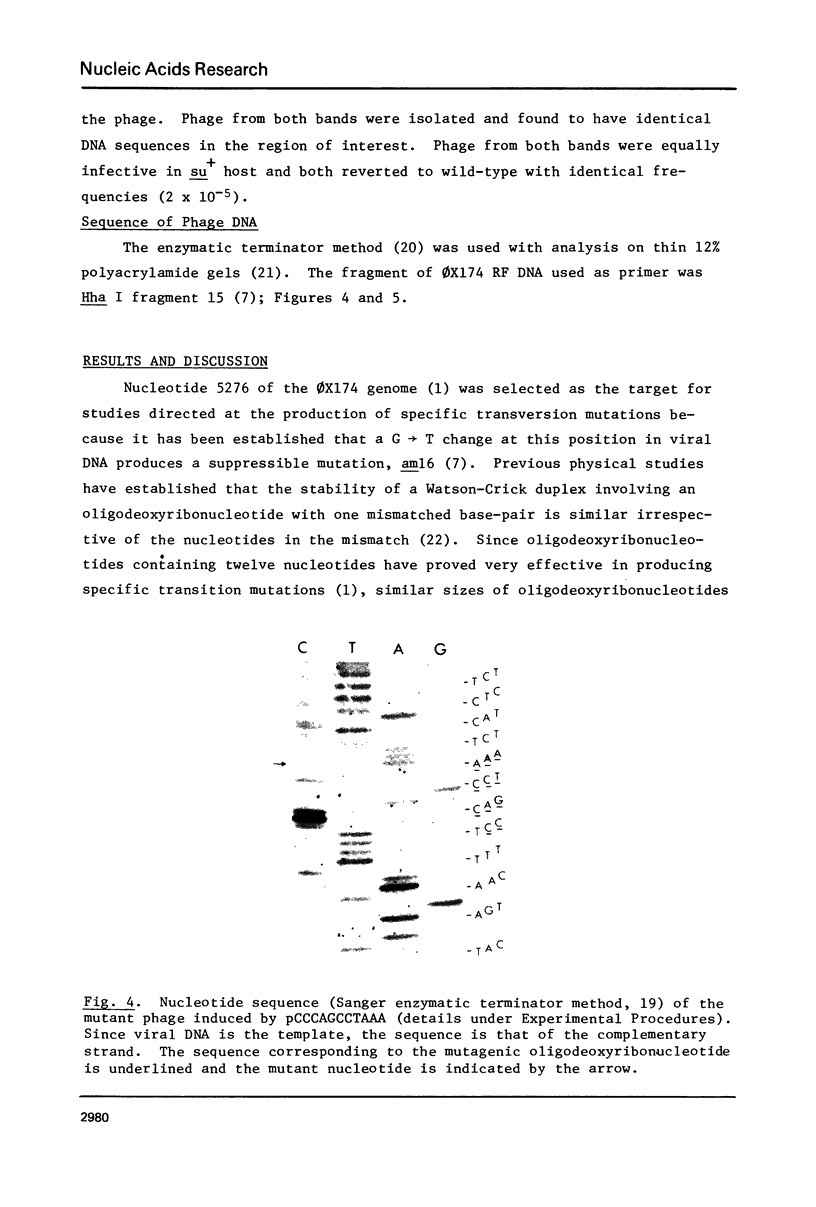
The oligodeoxyribonucleotides, pCCCAGCCTCAA, which is complementary to nucleotides 5274--4284 of bacteriophage phi X174 viral DNA , and pCCCAGCCTAAA, which corresponds to the same sequence with a C leads to A change at the ninth nucleotide, were synthesized enzymatically. The second of these oligonucleotides was used as a primer for E. coli DNA polymerase I, from which the 5'-exonculease has been removed by proteolysis (Klenow enzyme), on wild-type phi X174 viral DNA template. After ligation, this yielded closed circular heteroduplex DNA with a G, A mismatch at nucleotide 5276. Transfection of E. coli spheroplasts with the heteroduplex DNA produced phage mutated at this nucleotide (G leads to T in the viral DNA) with high efficiency (13%). The mutant DNA, which corresponds to the gene B mutant am16, was reverted (T leads to G) by the wild type oligonucleotide with an efficiency of 19%. The nucleotide changes were established by sequence determination of the mutated viral DNA using the enzymatic terminator method. The production of specific transversion mutations, together with a previous demonstration of specific transition mutations (1), established that short enzymatically synthesized oligodeoxyribonucleotides can be used to induce any class of single nucleotide replacement with high efficiency and thus provide a powerful tool for specific genetic manipulations in circular genomes like that of phi X174.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barrell B. G., Air G. M., Hutchison C. A., 3rd Overlapping genes in bacteriophage phiX174. Nature. 1976 Nov 4;264(5581):34–41. doi: 10.1038/264034a0. [DOI] [PubMed] [Google Scholar]
- Brown N. L., Smith M. DNA sequence of a region of the phi X174 genome coding for a ribosome binding site. Nature. 1977 Feb 24;265(5596):695–698. doi: 10.1038/265695a0. [DOI] [PubMed] [Google Scholar]
- Brown N. L., Smith M. The sequence of a region of bacteriophage phiX174 DNA coding for parts of genes A and B. J Mol Biol. 1977 Oct 15;116(1):1–28. doi: 10.1016/0022-2836(77)90115-2. [DOI] [PubMed] [Google Scholar]
- Brownlee G. G., Sanger F. Chromatography of 32P-labelled oligonucleotides on thin layers of DEAE-cellulose. Eur J Biochem. 1969 Dec;11(2):395–399. doi: 10.1111/j.1432-1033.1969.tb00786.x. [DOI] [PubMed] [Google Scholar]
- Brutlag D., Atkinson M. R., Setlow P., Kornberg A. An active fragment of DNA polymerase produced by proteolytic cleavage. Biochem Biophys Res Commun. 1969 Dec 4;37(6):982–989. doi: 10.1016/0006-291x(69)90228-9. [DOI] [PubMed] [Google Scholar]
- Gillam S., Jahnke P., Smith M. Enzymatic synthesis of oligodeoxyribonucleotides of defined sequence. J Biol Chem. 1978 Apr 25;253(8):2532–2539. [PubMed] [Google Scholar]
- Gillam S., Rottman F., Jahnke P., Smith M. Enzymatic synthesis of oligonucleotides of defined sequence: synthesis of a segment of yeast iso-1-cytochrome c gene. Proc Natl Acad Sci U S A. 1977 Jan;74(1):96–100. doi: 10.1073/pnas.74.1.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillam S., Waterman K., Smith M. The base-pairing specificity of cellulose-pdT9. Nucleic Acids Res. 1975 May;2(5):625–634. doi: 10.1093/nar/2.5.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulian M., Kornberg A., Sinsheimer R. L. Enzymatic synthesis of DNA, XXIV. Synthesis of infectious phage phi-X174 DNA. Proc Natl Acad Sci U S A. 1967 Dec;58(6):2321–2328. doi: 10.1073/pnas.58.6.2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gschwender H. H., Hofschneider P. H. Lysis inhibition of phi-X174-, M12-, and Q-beta-infected Escherichia coli bacteria by magnesium ions. Biochim Biophys Acta. 1969 Oct 22;190(2):454–459. doi: 10.1016/0005-2787(69)90094-x. [DOI] [PubMed] [Google Scholar]
- Hutchison C. A., 3rd, Phillips S., Edgell M. H., Gillam S., Jahnke P., Smith M. Mutagenesis at a specific position in a DNA sequence. J Biol Chem. 1978 Sep 25;253(18):6551–6560. [PubMed] [Google Scholar]
- Jay E., Bambara R., Padmanabhan R., Wu R. DNA sequence analysis: a general, simple and rapid method for sequencing large oligodeoxyribonucleotide fragments by mapping. Nucleic Acids Res. 1974 Mar;1(3):331–353. doi: 10.1093/nar/1.3.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klenow H., Henningsen I. Selective elimination of the exonuclease activity of the deoxyribonucleic acid polymerase from Escherichia coli B by limited proteolysis. Proc Natl Acad Sci U S A. 1970 Jan;65(1):168–175. doi: 10.1073/pnas.65.1.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klenow H., Overgaard-Hansen K., Patkar S. A. Proteolytic cleavage fo native DNA polymerase into two different catalytic fragments. Influence of assay condtions on the change of exonuclease activity and polymerase activity accompanying cleavage. Eur J Biochem. 1971 Oct 14;22(3):371–381. doi: 10.1111/j.1432-1033.1971.tb01554.x. [DOI] [PubMed] [Google Scholar]
- Lautenberger J. A., Kan N. C., Lackey D., Linn S., Edgell M. H., Hutchison C. A., 3rd Recognition site of Escherichia coli B restriction enzyme on phi XsB1 and simian virus 40 DNAs: an interrupted sequence. Proc Natl Acad Sci U S A. 1978 May;75(5):2271–2275. doi: 10.1073/pnas.75.5.2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T., Jeffrey A., Kleid D. G. Nucleotide sequence of the rightward operator of phage lambda. Proc Natl Acad Sci U S A. 1975 Mar;72(3):1184–1188. doi: 10.1073/pnas.72.3.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panet A., van de Sande J. H., Loewen P. C., Khorana H. G., Raae A. J., Lillehaug J. R., Kleppe K. Physical characterization and simultaneous purification of bacteriophage T4 induced polynucleotide kinase, polynucleotide ligase, and deoxyribonucleic acid polymerase. Biochemistry. 1973 Dec 4;12(25):5045–5050. doi: 10.1021/bi00749a003. [DOI] [PubMed] [Google Scholar]
- Razin A., Hirose T., Itakura K., Riggs A. D. Efficient correction of a mutation by use of chemically synthesized DNA. Proc Natl Acad Sci U S A. 1978 Sep;75(9):4268–4270. doi: 10.1073/pnas.75.9.4268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R., Friedmann T., Air G. M., Barrell B. G., Brown N. L., Fiddes J. C., Hutchison C. A., 3rd, Slocombe P. M., Smith M. The nucleotide sequence of bacteriophage phiX174. J Mol Biol. 1978 Oct 25;125(2):225–246. doi: 10.1016/0022-2836(78)90346-7. [DOI] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R. The use of thin acrylamide gels for DNA sequencing. FEBS Lett. 1978 Mar 1;87(1):107–110. doi: 10.1016/0014-5793(78)80145-8. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt V. M. Purification and further properties of single-strand-specific nuclease from Aspergillus oryzae. Eur J Biochem. 1973 Feb 15;33(1):192–200. doi: 10.1111/j.1432-1033.1973.tb02669.x. [DOI] [PubMed] [Google Scholar]
- Weisbeek P. J., Van Arkel G. A. On the origin of bacteriophage phiX174 replicative form DNA replication. Virology. 1976 Jul 1;72(1):72–79. doi: 10.1016/0042-6822(76)90312-3. [DOI] [PubMed] [Google Scholar]
- Weymouth L. A., Loeb L. A. Mutagenesis during in vitro DNA synthesis. Proc Natl Acad Sci U S A. 1978 Apr;75(4):1924–1928. doi: 10.1073/pnas.75.4.1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Sande J. H., Kleppe K., Khorana H. G. Reversal of bacteriophage T4 induced polynucleotide kinase action. Biochemistry. 1973 Dec 4;12(25):5050–5055. doi: 10.1021/bi00749a004. [DOI] [PubMed] [Google Scholar]