Protein disulfide isomerases (PDIs) are conserved chaperone-like proteins that play an essential role during protein folding and in some cases during degradation. Substrate-specific effects of PDI family members occur during the ER-associated degradation of diverse substrates in yeast and mammalian cells.
Abstract
ER-associated degradation (ERAD) rids the early secretory pathway of misfolded or misprocessed proteins. Some members of the protein disulfide isomerase (PDI) family appear to facilitate ERAD substrate selection and retrotranslocation, but a thorough characterization of PDIs during the degradation of diverse substrates has not been undertaken, in part because there are 20 PDI family members in mammals. PDIs can also exhibit disulfide redox, isomerization, and/or chaperone activity, but which of these activities is required for the ERAD of different substrate classes is unknown. We therefore examined the fates of unique substrates in yeast, which expresses five PDIs. Through the use of a yeast expression system for apolipoprotein B (ApoB), which is disulfide rich, we discovered that Pdi1 interacts with ApoB and facilitates degradation through its chaperone activity. In contrast, Pdi1's redox activity was required for the ERAD of CPY* (a misfolded version of carboxypeptidase Y that has five disulfide bonds). The ERAD of another substrate, the alpha subunit of the epithelial sodium channel, was Pdi1 independent. Distinct effects of mammalian PDI homologues on ApoB degradation were then observed in hepatic cells. These data indicate that PDIs contribute to the ERAD of proteins through different mechanisms and that PDI diversity is critical to recognize the spectrum of potential ERAD substrates.
INTRODUCTION
Apolipoprotein B (ApoB) is a large, amphipathic protein that is produced in two isoforms in mammals. ApoB100 (∼540 kDa), synthesized in hepatic cells, is the predominant structural protein in very low density lipoproteins (VLDLs) and low-density lipoproteins (LDLs). A shorter isoform, ApoB48 (containing the amino-terminal 48% of ApoB100) is expressed in enterocytes and assembles into chylomicrons in all mammalian species and into VLDLs in rodents (Rutledge et al., 2010). Collectively, VLDLs, LDLs, and chylomicrons transport cholesterol and triglycerides through the bloodstream and deliver these essential metabolites to every tissue (Fisher and Ginsberg, 2002; Brodsky and Fisher, 2008; Rutledge et al., 2010). The amount of endogenously produced cholesterol must be tightly regulated since either excess or shortage of this sterol can lead to disease. For example, excess cholesterol can accumulate in coronary arteries and contribute to the development of atherosclerosis (Kannel et al., 1971; Brodsky and Fisher, 2008), the primary cause of coronary artery disease. Conversely, hypobetalipoproteinemia is a disease characterized by decreased plasma concentrations of ApoB and results in nutrient malabsorption, ataxia, and neuromuscular degeneration; the most common form of hypobetalipoproteinemia results from a premature stop codon in ApoB (Linton et al., 1993; Whitfield et al., 2004). Because the amount of secreted ApoB that circulates in the bloodstream correlates directly with serum cholesterol levels (Crooke et al., 2005; Zimmermann et al., 2006), a better understanding of ApoB secretion is vital for the treatment and prevention of hypobetalipoproteinemia and coronary artery disease.
ApoB secretion is tightly controlled, in large part through regulated degradation, an important component of which is accomplished via the endoplasmic reticulum–associated degradation (ERAD) pathway (Fisher and Ginsberg, 2002; Brodsky and Fisher, 2008; Rutledge et al., 2010). ERAD involves four major steps: selection of a misfolded protein, retrotranslocation of the protein from the ER to the cytosol, substrate ubiquitination, and proteasome mediated degradation (Meusser et al., 2005; Vembar and Brodsky, 2008; Xie and Ng, 2010). The ERAD of ApoB is unique in that it is metabolically controlled by lipid availability and occurs cotranslationally (Dixon et al., 1991; Benoist and Grand-Perret, 1997; Zhou et al., 1998). When lipids are abundant, ApoB is cotranslationally translocated into the ER, where it becomes N-glycosylated (Harazono et al., 2005), acquires eight disulfide bonds (Yang et al., 1990; Burch and Herscovitz, 2000), and is lipid loaded via the action of microsomal triglyceride transfer protein (MTP) complex. The complex contains an M subunit and an ER chaperone/enzyme known as protein disulfide isomerase (PDI; Hussain et al., 2003), which is the founding member of a group of 20 PDI family members in mammals. Upon sufficient lipid loading, ApoB assembles into a pre-VLDL and traffics to the Golgi, where it undergoes further maturation and is secreted (Tran et al., 2002; Olofsson and Boren, 2005; Gusarova et al., 2007; Siddiqi et al., 2010). However, when the intracellular concentrations of phospholipids and triacylglycerols are depleted, ApoB is selected for ERAD, ubiquitinated by the E3 ligase gp78, retrotranslocated, and degraded by the proteasome (Dixon et al., 1991; Yeung et al., 1996; Fisher et al., 1997; Liao et al., 1998; Pariyarath et al., 2001; Liang et al., 2003). Because ERAD results in lower intracellular and secreted levels of ApoB, it follows that a decline in ApoB production will decrease serum lipid and cholesterol levels. Thus we have sought to better define the ERAD pathway for ApoB, an effort that may provide novel therapeutic targets to treat cardiovascular and other, related diseases.
Disulfide bonds, such as those found in ApoB, are formed within the oxidizing environment of the ER by PDI family members (Kleizen and Braakman, 2004; Anelli and Sitia, 2008; Hatahet and Ruddock, 2009; Brodsky and Skach, 2011; Bulleid and Ellgaard, 2011). PDIs are identified by the presence of one or more thioredoxin-like motifs (Cys-X-X-Cys) and can catalyze the oxidation, reduction, and/or isomerization of disulfide bonds. Along with catalytic redox or isomerase activity, some PDI family members also possess chaperone activity due to the presence of a hydrophobic substrate-binding domain (Cai et al., 1994; Song and Wang, 1995; Klappa et al., 1998; Gillece et al., 1999; Ellgaard and Ruddock, 2005; Tian et al., 2006, 2008; Hatahet and Ruddock, 2009). As noted earlier, in mammals there are 20 PDI family members, 1 of which, known as PDI, associates with the M subunit to form the MTP complex. However, with few exceptions (Hatahet and Ruddock, 2009; Jessop et al., 2009; Park et al., 2009; Rutkevich et al., 2010; Brodsky and Skach, 2011), specific functions for each of these proteins have not been assigned. In contrast, in the yeast Saccharomyces cerevisiae there are five PDI family members, which are encoded by the PDI1, MPD1, MPD2, EUG1, and EPS1 genes. PDI1 is the only essential gene among this group (Farquhar et al., 1991). It contains two active-site, thioredoxin-like domains (denoted the a and a′ active sites), each with a CXXC motif and a substrate-binding domain in the b′ domain (Holst et al., 1997; Gillece et al., 1999; Tian et al., 2008). Although PDI1 is essential, the expression of Mpd1 from the PDI1 promoter supports cell viability in pdi1Δ yeast (Norgaard et al., 2001).
Select examples exist in which a clear link between PDI family members and ERAD or an ERAD-like phenomenon has been established. For example, in yeast the chaperone activity of Pdi1p is required for the recognition and degradation of an ERAD substrate, pro-α factor (pαF), which lacks disulfide bonds (Gillece et al., 1999), and Eps1 appears to help target a misfolded membrane protein, Pma1-D378N, for ERAD (Wang and Chang, 2003). In mammalian cells, ERdj5, a protein that contains both a J domain that interacts with BiP and six thioredoxin repeat motifs, may reduce disulfide bonds to facilitate ERAD substrate retrotranslocation (Ushioda et al., 2008; Hagiwara et al., 2011). Furthermore, some PDIs bind to select bacterial toxins and viruses prior to their retrotranslocation and delivery to the cytoplasm in infected cells (Magnuson et al., 2005; Forster et al., 2006; Schelhaas et al., 2007; Rainey-Barger et al., 2009; Moore et al., 2010; Taylor et al., 2011).
We previously reported on an in vitro assay that monitors the proteasome-dependent degradation of ER membrane-embedded ApoB48 in the presence of yeast cytosol (Gusarova et al., 2001). Although cytosol prepared from wild-type yeast supported ApoB degradation, lysates from Hsp70 or Hsp90 mutant strains were degradation-deficient. The Hsp70 and Hsp90-dependent degradation of ApoB was also observed in hepatic cells (Fisher et al., 1997; Gusarova et al., 2001). We then developed a yeast ApoB expression system that used a truncated form of the protein, ApoB29, which still associates with lipids in hepatic cells, and we identified the Hsp110 chaperone as an ApoB-stabilizing factor. Here, too, the effect of the human Hsp110 homologue on ApoB was recapitulated in hepatic cells (Hrizo et al., 2007). These data indicate that the yeast system provides a powerful model to isolate and characterize components that play important roles in ApoB biogenesis.
As mentioned, mammalian PDI is one component of the MTP complex that helps load lipids onto ApoB (Hussain et al., 2003), but it is unknown whether PDI targets the protein for ERAD under lipid-poor conditions. Two other mammalian PDI family members, ERp57 and ERp72, can be coprecipitated with ApoB (Adeli et al., 1997; Linnik and Herscovitz, 1998; Hussain et al., 2003; Zhang and Herscovitz, 2003), but their roles during ApoB biogenesis are also unknown. Using both yeast and mammalian cell systems, we report on the specific action of PDI family members during the ERAD of ApoB. We also show that the degradation of ApoB and CPY* (a misfolded version of carboxypeptidase Y that has five disulfide bonds), which is a soluble ERAD substrate, require different Pdi1-encoded functions in yeast: ApoB uses Pdi1's chaperone activity, whereas CPY* uses the thioredoxin-like motifs. We then discovered that Pdi1 is dispensable for the turnover of another ERAD substrate, the alpha subunit of the epithelial sodium channel (α-ENaC), and with the exception of Mpd1, the genes encoding the remaining three PDI relatives can be ablated without an effect on the ERAD of any of the substrates examined in this study. Finally, we found that two human PDI family members also facilitate the ERAD of ApoB in hepatic cells, although PDI—which is a component of the MTP complex—enhanced ApoB secretion. These data demonstrate the diverse PDI activities that are required during ERAD and that different PDI family members can act either to fold or degrade ApoB.
RESULTS
Pdi1 is the major contributor to ERAD in yeast
Disulfide bond formation is critical for ApoB maturation and secretion (Yang et al., 1989, 1990; Burch and Herscovitz, 2000). In addition, as described earlier, PDI is a component of the MTP complex, and two PDI family members, ERp57 and ERp72, have been found associated with ApoB (Adeli et al., 1997; Linnik and Herscovitz, 1998; Hussain et al., 2003; Zhang and Herscovitz, 2003). To perform a systematic analysis of the roles of PDIs in the ERAD of ApoB, we first examined ApoB stability in the yeast S. cerevisiae in which the genes encoding the nonessential PDIs were deleted. Our yeast expression system for ApoB was previously used to define several of the requirements for the ERAD of ApoB in hepatic cells (Hrizo et al., 2007). Because the wild-type yeast strain W303 provided a better readout for the extent of ApoB degradation than the BY4742 strain (our unpublished data), we constructed all of our mutants and performed each of the following experiments in this background (Supplemental Table S1).
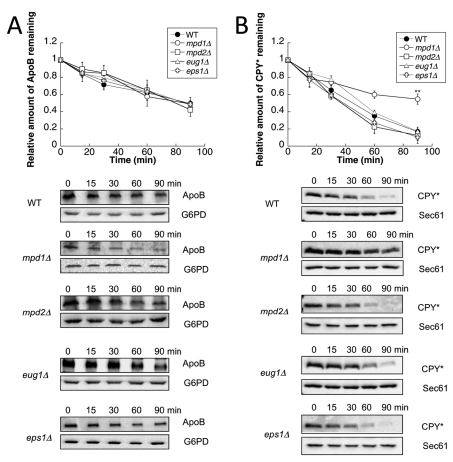
The gene encoding Pdi1 is essential (Farquhar et al., 1991), so to assess whether any of the other PDI family members were important for the degradation of ApoB, mpd1Δ, mpd2Δ, eug1Δ, or eps1Δ cells were transformed with the ApoB expression vector, and the degradation rate was measured using a cycloheximide chase assay, as described in Materials and Methods. For these studies, we again chose to express ApoB29 from an inducible reporter; ApoB29 is the shortest ApoB isoform whose degradation is metabolically controlled (Wang et al., 1994; Segrest et al., 2001). ApoB29 also contains seven of the eight disulfide bonds found in full-length ApoB (Yang et al., 1990; Harazono et al., 2005). In each mutant, we found that ApoB was degraded at wild-type levels (Figure 1A). We also measured the ERAD of a well-characterized substrate, CPY* (Wolf and Fink, 1975; Finger et al., 1993). In contrast to ApoB, CPY* was stabilized when MPD1 was deleted, although no stabilization was evident when the other nonessential PDIs were deleted (Figure 1B). In addition, we examined the degradation of pαf, a yeast ERAD substrate that lacks cysteines and that was previously shown to be selected by Pdi1 for degradation (Gillece et al., 1999), as well as α-ENaC, a mammalian protein with seven disulfide bonds whose proteasome- and chaperone-dependent degradation were characterized in yeast (Kashlan et al., 2007; Buck et al., 2010). We found that deletion of the nonessential PDIs also had no effect on the ERAD of these proteins (Supplemental Figure S1). Combined with previous data demonstrating that Eps1 is required for the ERAD of a mutant form of Pma1 (Wang and Chang, 2003), these results indicate substrate specificity among the nonessential PDI family members.
FIGURE 1:
Mpd1 contributes to the ERAD of CPY* but not ApoB29. (A) Cycloheximide chase reactions were performed as described in Materials and Methods in wild-type (●), mpd1Δ (○), mpd2Δ (□), eug1Δ (Δ), or eps1Δ (†) yeast strains expressing ApoB29 (A) or CPY* (B). Chase reactions were performed at 30°C, and lysates were immunoblotted with anti-HA antibody. Anti-Sec61 antiserum was used as a loading control for chase reactions monitoring CPY* turnover, and anti-G6PD antiserum was used as a loading control for chase reactions measuring ApoB29 degradation. Top, quantitative data. Bottom, representative images. Data represent the means of four to six experiments, ± SEM. The lack of visible error bars indicates that the SEM is less than the size of the symbol. **p < 0.01.
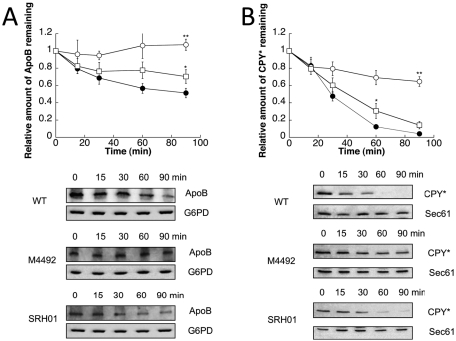
We next asked whether yeast Pdi1 facilitates the ERAD of ApoB. Because previous work indicated that Mpd1 overexpression from the PDI1 promoter supported the growth of a pdi1Δ mutant (Norgaard et al., 2001), we examined ApoB degradation in pdi1Δmpd2Δeug1Δeps1Δ cells in which Mpd1 is the only PDI family member expressed (strain M4492). In this strain, ApoB was completely stabilized (Figure 2A, open circles). We also examined the degradation of ApoB in a strain in which Pdi1 is the only yeast PDI family member expressed (strain SRH01). In this yeast, ApoB degradation was mostly restored (Figure 2A, open squares). We then measured the degradation of CPY* in these strains, since Pdi1 facilitates the ERAD of CPY* (Gillece et al., 1999; Sakoh-Nakatogawa et al., 2009). As was evident for ApoB, CPY* was also significantly stabilized when Mpd1 is the only PDI family member expressed, and protein degradation was again mostly restored when only Pdi1 was expressed (Figure 2B). Because there was a residual degradation defect when ApoB and CPY* turnover was assessed in SRH01, we suggest that at least one of the other PDI family members contributes during the process of substrate selection and/or retrotranslocation.
FIGURE 2:
The ERAD of ApoB29 and CPY* is slowed by the loss of PDI1. Cycloheximide chase reactions were performed as described in Materials and Methods in wild-type (●), M4492 (pdi1Δmpd1Δmpd2Δeug1Δeps1Δ [MPD1]) (○), or SRH01 (pdi1Δmpd1Δmpd2Δeug1Δeps1Δ [PDI1]) (□) yeast strains expressing ApoB29 (A) or CPY* (B). Chase reactions were performed at 30°C, and lysates were immunoblotted with anti-HA antibody. Anti-Sec61 antiserum was used as a loading control for chase reactions monitoring CPY* turnover, and anti-G6PD antiserum was used as a loading control for chase reactions measuring ApoB29 degradation. Top, quantitative data. Bottom, representative blots. Data represent the means of four to six experiments, ± SEM. The lack of visible error bars indicates that the SEM is less than the size of the symbol. *p 0.05, **p < 0.01.
The chaperone activity of Pdi1 was previously found to be important for pαf ERAD (Gillece et al., 1999), but the ability of Mpd1 to support the degradation of this substrate as the only expressed PDI family member has not been investigated. We observed measurable stabilization of pαf when only Mpd1 was expressed, but degradation was robust when only Pdi1 was expressed (Supplemental Figure S2A). Of interest, although α-ENaC has seven disulfide bonds that are important for protein folding and function (Firsov et al., 1999; Kashlan et al., 2011), an ERAD defect was absent when Mpd1 was expressed (Supplemental Figure S2B). These data suggest that Pdi1 is not required for the ERAD of all substrates. Normally, ENaC functions as a heterotrimer composed of three subunits—α, β, and γ (Snyder, 2002; Jasti et al., 2007)—but in this expression system α-ENaC is an orphan subunit. Therefore the “decision” to select α-ENaC for degradation may be distinct from other substrates in which intermolecular folding events or protein assembly are monitored. Together these results suggest that Pdi1 function is not imperative for the ERAD of all substrates, and, consistent with previous data (Gillece et al., 1999; Wang and Chang, 2003), our data indicate that some substrates rely in part on the function of other PDI family members during ER quality control.
ApoB and CPY* associate with Pdi1, and their degradation requires either Pdi1's chaperone activity or the thioredoxin-like motifs
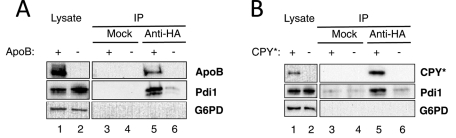
We next determined whether Pdi1 directly associates with ApoB and CPY*. To this end, we used a method in which the ability of Pdi1 to form a mixed disulfide with the ER lumenal protein Htm1/Mnl1 was assessed (Sakoh-Nakatogawa et al., 2009). In this protocol, yeast cells expressing ApoB are spheroplasted and lysed, and then proteins are precipitated under conditions in which preformed disulfide bonds are trapped (see Materials and Methods). We therefore precipitated ApoB under these conditions and asked whether Pdi1 bound to this substrate after we added reductant and performed SDS–PAGE. When ApoB was immunoprecipitated, Pdi1 was also present, but an abundant cytosolic protein (glucose-6-phosphate dehydrogenase [G6PD]) that contains a single cysteine was absent (Figure 3A, lane 5). We note that a minor amount of Pdi1 associated with resin when strains lacked ApoB (lane 6), but neither ApoB nor PDI was precipitated using unconjugated resin, indicating a specific association between Pdi1 and ApoB. Consistent with Pdi1 having a direct effect on the degradation of CPY*, we also observed a Pdi1–CPY* interaction (Figure 3B, lane 5). A prominent association between these proteins was absent in precipitations from lysates that lacked the substrate (lane 6) or in mock precipitations (lanes 3 and 4), although here too a small amount of Pdi1 appears to nonspecifically associate with the resin. Nevertheless, these results strongly suggest that ApoB and CPY* form mixed disulfides with Pdi1.
FIGURE 3:
Pdi1 physically interacts with ApoB29 and CPY*. Native immunoprecipitation reactions were performed using anti-HA resin or unconjugated Sepharose (Mock), using lysates from wild-type yeast strains expressing ApoB29 (A) or CPY* (B). In both cases, cell lysates were also examined that contained a vector control (–) in place of the ApoB29 or CPY* (+) expression vector. A total of 1% of the input for the precipitation was also examined (Lysate). After precipitation and SDS–PAGE, the indicated proteins were examined by immunoblot analysis.
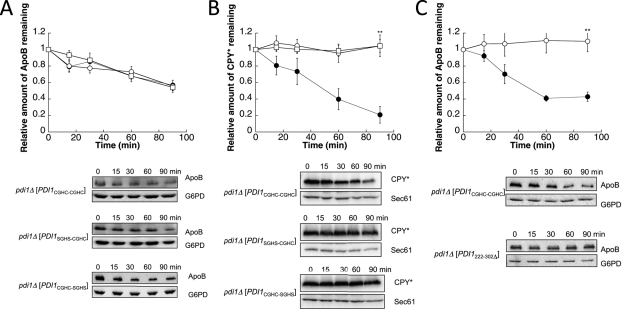
Because Pdi1 interacts with ApoB and facilitates its degradation, we next investigated which Pdi1-embedded function was important for ApoB ERAD. We first asked whether the a or a′ thioredoxin-like active site was necessary. To this end, we used strains in which PDI1 had been deleted but the cells expressed wild-type PDI1 on a plasmid (PDI1CGHC-CGHC), PDI1 on a plasmid with both cysteines in the a active site mutated to serines (PDI1SGHS-CGHC), or PDI1 on a plasmid with both cysteines in the a′ active site mutated to serines (PDI1CGHC-SGHS). ApoB degradation was then measured by cycloheximide chase analysis. When either active site's cysteines were mutated, ApoB was degraded at wild-type levels (Figure 4A). In contrast, when either active site was mutated, CPY* was completely stable (Figure 4B), indicating that both thioredoxin-like motifs—and their redox activities—are necessary for CPY* but not ApoB degradation.
FIGURE 4:
The chaperone-like activity and the oxidoreductase activity of Pdi1 respectively facilitate the ERAD of ApoB29 and CPY*. Cycloheximide chase reactions were performed as described in Materials and Methods in pdi1Δ [PDI1CGHC-CGHC] (●), pdi1Δ [PDI1SGHS-CGHC] (○), or pdi1Δ [PDI1CGHC-SGHS] (□) yeast strains expressing ApoB29 (A) or CPY* (B). Cycloheximide chase reactions were also performed in pdi1Δ [PDI1CGHC-CGHC] (●) or pdi1Δ [PDI1222-302Δ] (○) yeast strains expressing ApoB29 (C). Chase reactions were conducted at 30°C, and lysates were immunoblotted with anti-HA antibody. Anti-Sec61 antiserum was used as a loading control for chase reactions monitoring CPY*, and anti-G6PD antiserum was used as a loading control for chase reactions measuring ApoB29 degradation. Top, quantitative data. Bottom, representative blots. Data represent the means of five to seven experiments, ± SEM. The lack of visible error bars indicates that the SEM is less than the size of the symbol. **p < 0.01.
Another interpretation of the data presented in Figure 4A is that the a and a′ sites function redundantly to support ApoB degradation; however, it is believed that the two sites are not equivalent (Holst et al., 1997; Hatahet and Ruddock, 2009; Wang et al., 2009; Vitu et al., 2010), and at least one of the thioredoxin motifs must be capable of forming a thiolate to support cell viability (Laboissiere et al., 1995; Chivers et al., 1996). A more likely possibility is that Pdi1's chaperone activity, which mediates Pdi1's ability to bind some peptides in the ER (Klappa et al., 1998; Gillece et al., 1999), is necessary for ApoB degradation. To test this hypothesis, we used strains in which PDI1 was deleted but the cells expressed either wild-type PDI1 on a plasmid or PDI1 with its b′ chaperone domain deleted (PDI1222-302Δ). As shown in Figure 4C, ApoB was completely stable when Pdi1's chaperone domain was absent. To determine whether the chaperone mutant strain supported the ERAD of other substrates, CPY* degradation was measured. Consistent with previous results (Gillece et al., 1999), no CPY* degradation defect was observed when the Pdi1 chaperone domain was mutated (Supplemental Figure S3A). We also confirmed that the mutant Pdi1 protein was expressed to the same level as wild-type Pdi1 and remained stable over the 90-min chase period (Supplemental Figure S3B). Together these results indicate distinct requirements for Pdi1-embedded activities during ERAD: some substrates (α-ENaC) are Pdi1 independent, some (ApoB and pαF) require the protein's chaperone activity, and others (CPY*) require both of the enzyme's thioredoxin-like motifs.
Yeast expressing only Mpd1 or specific mutant forms of Pdi1 are dithiothreitol sensitive
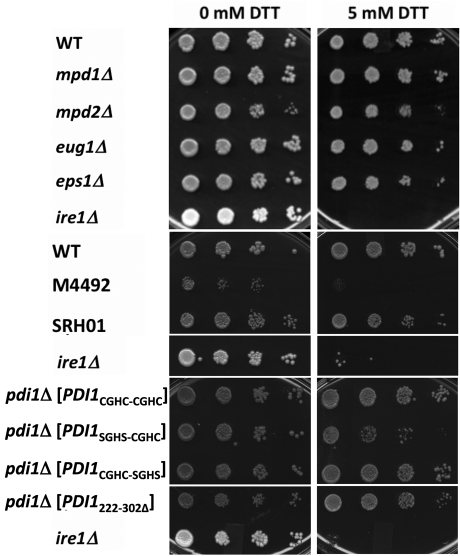
We hypothesized that if the chaperone mutant strain (pdi1Δ[PDI1222-302Δ]) was more sensitive to ER stress and less viable, then the stabilization of ApoB observed in these Pdi1222-302Δ-expressing yeast could have resulted from a nonspecific effect on induction of ER stress and compromised cell growth. Therefore the sensitivities of all of the examined mutants to an ER stress-inducing agent, dithiothreitol (DTT), were measured. We first found that the strains with a single deletion of any of the individual nonessential PDIs were DTT insensitive. In contrast, and as a control, ire1Δ yeast, which are unable to induce the unfolded protein response (Cox and Walter, 1996), were inviable when plated on DTT (Figure 5). Strain M4492 (pdi1Δmpd1Δmpd2Δeug1Δeps1Δ [MPD1]) was significantly sicker than wild-type cells on rich media and was extremely DTT sensitive; however, when Pdi1 was the only PDI family member expressed (strain SRH01, pdi1Δmpd1Δmpd2Δeug1Δeps1Δ [PDI1]), DTT sensitivity was absent (Figure 5). In accordance with previous data (Holst et al., 1997), we also found that mutation of the a active site in Pdi1 (pdi1Δ[PDI1SGHS-CGHC]) led to modest DTT sensitivity. Notably, the b′ chaperone mutant strain (pdi1Δ[PDI1222-302Δ]) was DTT insensitive. These results suggest that ApoB stabilization in the Pdi1222-302Δ chaperone-defective strain does not result from unmitigated stress and poor cell growth.
FIGURE 5:
Strains with deletions of or mutations in the PDI family members exhibit varying sensitivities to the reducing agent DTT. The indicated yeast strains were grown in selective medium to mid log phase. Serial dilutions of the cells were spot plated onto medium containing either 0 mM or 5 mM DTT, as indicated, and grown for 2 d at 30°C. The data are representative of several independent trials, and in liquid culture the doubling times for the single deletions in the absence of DTT were essentially identical.
ApoB degradation is α-mannosidase–like lectin independent
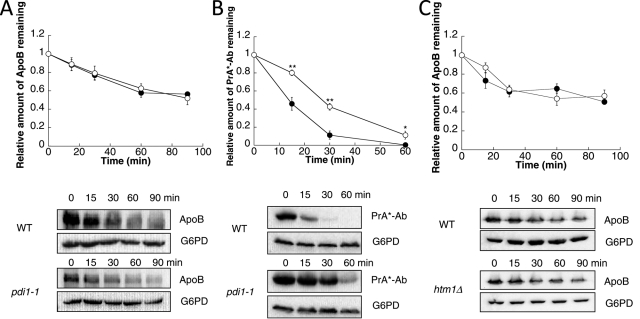
Pdi1 interacts with and is required for the oxidation of an intermolecular disulfide bond in Htm1/Mnl1 (Clerc et al., 2009; Sakoh-Nakatogawa et al., 2009), which is the ER degradation-enhancing α-mannosidase–like lectin (EDEM) homologue in yeast. As in mammals, Htm1 recognizes misfolded glycoproteins and targets them for degradation (Jakob et al., 2001; Nakatsukasa et al., 2001). Recently a mutation in PDI1, pdi1-1, was identified in a screen for yeast that require a functioning unfolded protein response for viability (Gauss et al., 2011). The pdi1-1 mutation harbors a leucine in place of a proline in the center of the b′ domain (amino acid 313) and disrupts the interaction of Pdi1 with Htm1. This, in turn, affects the ERAD of select substrates. Formally, the delayed degradation of ApoB, a glycoprotein, in the PDI1222-302Δ strain (Figure 4C) might have arisen from an indirect effect on Htm1 activity.
To address this possibility, we asked whether ApoB degradation was altered in the pdi1-1 mutant. ApoB was degraded at wild-type levels in the pdi1-1 strain (Figure 6A). As a control for this experiment, PrA*-Ab, a mutated version of the vacuolar protein proteinase A that requires Htm1 (Finger et al., 1993; Spear and Ng, 2005; Kanehara et al., 2010), was stabilized in pdi1-1 yeast (Figure 6B). These data suggest that ApoB degradation is Htm1 independent and that the effect of the PDI1222-302Δ mutant on ApoB (Figure 4C) was not due to a secondary effect via disrupted association between Htm1 and Pdi1. Consistent with this hypothesis, we also failed to observe an effect on ApoB stability in htm1Δ strains compared with wild-type yeast (Figure 6C). Further, as shown earlier, ApoB was degraded at wild-type levels in pdi1Δ [PDI1 SGHS-CGHC] and pdi1Δ [PDI1 CGHC-SGHS] strains (Figure 4A). These mutants were previously reported to ablate the function of Htm1 during ERAD (Sakoh-Nakatogawa et al., 2009). Overall, these data indicate that the yeast EDEM homologue is dispensable for ApoB turnover and confirm that ApoB degradation requires Pdi1's chaperone activity.
FIGURE 6:
The EDEM homologue Htm1 does not play a role in the degradation of ApoB. Cycloheximide chase reactions were performed as described in Materials and Methods in wild-type W303a (●) or KKY415 (pdi1-1) (○) yeast strains expressing ApoB29 (A) or PrA*-Ab (B). Cycloheximide chase reactions were also performed in wild-type SEY6210 (●) or htm1Δ (○) yeast strains expressing ApoB29 (C). Chase reactions were performed at 30°C, and lysates were immunoblotted with anti-HA antibody. Anti-G6PD antiserum was used as a loading control. Data represent the means of four to six experiments, ± SEM. The lack of visible error bars indicates that the SEM is less than the size of the symbol. *p < 0.05, **p < 0.01.
Defects in chaperone function can affect the biochemical properties of an ERAD substrate in yeast (Nishikawa et al., 2001; Kabani et al., 2003). Because the chaperone activity of Pdi1 was required for maximal ApoB turnover, we were curious to see whether ApoB's conformation was altered in the b′ mutant strain. As a readout for ApoB's conformation in the yeast ER, we chose to measure the reactivity of any free cysteines in ApoB to the thiol-modifying reagent maleimide-PEG5000 after ApoB isolation and reduction. Interestingly, the amount of ApoB that could be recovered after this analysis was performed in the pdi1Δ[PDI1222-302Δ] strain—but not in the wild-type strain—was significantly reduced (data not shown). These results suggest that when the chaperone domain of Pdi1 is absent, the conformation of ApoB may be altered.
Distinct PDI family members contribute differently to ApoB biogenesis in hepatic cells
To assess whether the knowledge acquired from the use of the yeast system could inform us as to which mammalian chaperones might similarly mediate ApoB quality control, we examined the contributions of select mammalian PDI family members on ApoB biogenesis in hepatic cells. Although there are only five yeast PDI family members, mammals express 20 PDI-like proteins (Hatahet and Ruddock, 2009). To identify which of these PDIs to examine, we first performed a BLAST search with yeast Pdi1 against the human protein database. The three best hits were ERp72, PDI, and ERp57 (our unpublished data). Another criterion for this study was to ascertain which PDIs exhibited overlapping expression patterns with ApoB. PDI, ERp57, and ERp72 are all expressed in the liver (Marcus et al., 1996; also see the Human Protein Reference Database, www.hprd.org/), but the fourth- and fifth-best hits from the BLAST search, PDIp and PDILT, respectively, are not expressed in this tissue. Furthermore, PDI, ERp57, and ERp72 have a similar domain organization to Pdi1 (a-b-b′-a′), although ERp72 has an additional thioredoxin-like active site (a°-a-b-b′-a′; Hatahet and Ruddock, 2009). The crystal structures of yeast Pdi1 and ERp57 have been solved (Tian et al., 2006, 2008; Dong et al., 2009) and share broad overall similarity, having domains organized in a U-shaped conformation, although ERp57 has a more twisted conformation than Pdi1 (Dong et al., 2009). Of interest, PDI and ERp72 are also able to support the growth of pdi1Δ yeast strains (Gunther et al., 1993).
Because Pdi1's b′ domain was required to facilitate ApoB ERAD in yeast (Figure 4C), we also wanted to examine mammalian PDI family members that harbor substrate-binding domains. PDI, ERp57, and ERp72 all have substrate-binding domains, and specific substrates have been identified for each of these proteins. The chaperone activity of PDI is necessary to refold proinsulin (Winter et al., 2002) and lysozyme (Puig and Gilbert, 1994). ERp57 specifically associates with lectins through a binding site in its b′ domain (Russell et al., 2004), and a range of substrates have been identified for ERp57 in vivo, including, laminin, collagen, and clusterin (Jessop et al., 2009). Similarly, ERp72 is found within chaperone complexes and interacts with thrombospondin (Kuznetsov et al., 1997) and thyroglobulin (Menon et al., 2007), but, it is unknown whether these interactions occur through the b′ substrate-binding domain. Nevertheless, PDI, ERp57, and ERp72 have previously been shown to precipitate with ApoB, and through its integration into the MTP complex, PDI interacts with ApoB during lipid loading (Adeli et al., 1997; Linnik and Herscovitz, 1998; Zhang and Herscovitz, 2003). However, the roles of PDI, ERp57, or ERp72 as regulators of ApoB degradation in the mammalian ER have not been examined.
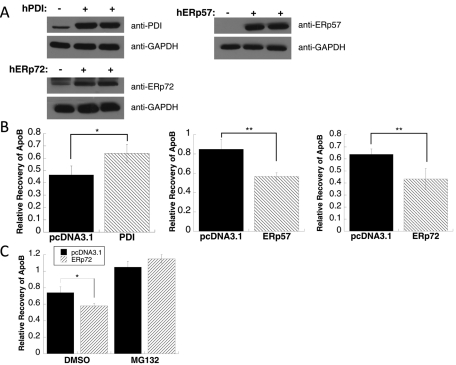
To determine whether PDI, ERp57, or ERp72 contributes to ApoB ERAD, we overexpressed each protein in rat hepatoma McA-RH7777 cells, an established cell line for studying ApoB biogenesis (Tanabe et al., 1989). First, to monitor whether each protein could be overexpressed, cells were assayed after they were transfected with a vector control or vectors engineered for the transient expression of human PDI, ERp57, or ERp72. After 48 h the cells were broken and the lysates were subjected to SDS–PAGE and probed with anti-PDI, anti-ERp57, or anti-ERp72 antibodies. When duplicate samples were analyzed and the signals were quantified relative to a loading control, PDI and ERp72 were overexpressed by approximately sevenfold and sixfold, respectively (Figure 7A), and—assuming similar antibody avidity—the quantity of overexpressed ERp57 appears comparable to that of PDI and ERp72 when overexpressed (Figure 7A).
FIGURE 7:
Overexpression of ERp57 or ERp72 leads to an increase in ApoB100 ERAD. (A) McArdle-RH7777 cells were transfected with pcDNA3.1 lacking an insert (–) or containing human (h) PDI, ERp57, or ERp72. Equal amounts of cell lysates were analyzed by immunoblotting with an anti-PDI antibody, anti-ERp57 antibody, or anti-ERp72 antibody. (B) Following a metabolic labeling reaction, a 60-min chase was performed as described in Materials and Methods in McArdle-RH7777 cells transfected with a vector control (pcDNA3.1 lacking an insert) or containing the PDI, ERp57, or ERp72 genes. The bars indicating “Relative Recovery of ApoB” indicate the amount of ApoB-precipitable material recovered from cell lysates and secreted into the medium at the completion of the chase divided by the amount of ApoB-precipitable material recovered after 15 min of chase (this prolonged period is required to complete the synthesis of ApoB (Gusarova et al., 2001)). Data represent the means of three experiments, ± SEM. *p < 0.05, **p < 0.01. (C) Pulse-chase reactions were performed as in B, except that either DMSO or the proteasome inhibitor MG132 was added to the cells prior to the start of the metabolic labeling. Data represent the means of three experiments, ± SEM. *p < 0.05.
We hypothesized that if PDI played a strong MTP complex–independent role in mammalian cells during ApoB degradation, then its overexpression would result in decreased recovery of ApoB (Wang et al., 1997). However, if PDI's only role in ApoB biogenesis was through the MTP complex, we predicted that increased recovery of ApoB would be evident because the MTP complex promotes ApoB secretion (Gordon et al., 1994; Leiper et al., 1994; Haghpassand et al., 1996) and escape from ERAD. No change in ApoB recovery could indicate that a prodegradative role of PDI was balanced by the stabilizing role of the MTP complex. When the amount of radiolabeled ApoB recovered after a 60-min chase in PDI-overexpressing cells was compared with the control, a 37% overall increase in ApoB recovery was evident (Figure 7B, left). These results suggest that the expression of greater amounts of PDI improve MTP complex function and augment ApoB transport and escape from ERAD. As controls for this experiment, we also assessed the secretion of albumin, as described (Gusarova et al., 2007; Hrizo et al., 2007), but found that its biogenesis was unaffected by PDI overexpression (data not shown). In addition, we asked whether PDI overexpression enhanced ApoB secretion because it inhibited ERAD. However, we found that PDI overexpression, regardless of whether the proteasome was active or not (i.e., in the presence of MG-132), led to an increase in the amount of secreted ApoB (Supplemental Figure 4).
If yeast Pdi1 is required for ApoB degradation (Figures 2 and 4), why is the closest homologue in human cells, PDI, not required for ERAD? Instead, human PDI overexpression stabilized the protein and increased its recovery in this experiment. The simplest explanation for this result is that yeast lack the MTP complex, which is required to load lipids onto maturing, ApoB-containing chylomicrons and VLDLs in the ER. In mammalian cells, the loss of MTP function—brought about either by genetic means or through the use of small-molecule inhibitors—is accompanied by increased ApoB degradation (Jamil et al., 1996; Benoist and Grand-Perret, 1997; Zimmermann et al., 2006). Thus our yeast ApoB model most closely resembles the lipid- or MTP complex–deficient state. We conclude that Pdi1 is a prodegradative factor for ApoB in yeast, and yet, when complexed with the M subunit in mammals, the closest Pdi1 homologue, PDI, promotes ApoB folding and secretion.
On the basis of our yeast data, we also hypothesized that overexpression of ERp57 and ERp72 would lead to increased ApoB degradation. As predicted, when either ERp57 or ERp72 was overexpressed, ∼33% less ApoB was recovered compared with the vector control (Figure 7B, middle and right). To confirm that ApoB degradation was proteasome mediated and that the effect of ERp57 and ERp72 was via the ERAD pathway and not an alternate degradative system (Pan et al., 2008), we assessed the impact of a proteasome inhibitor, MG132, on ApoB recovery when ERp72 was overexpressed. In this experiment, in the presence of DMSO, less ApoB was again recovered when ERp72 was overexpressed compared with the vector control, but when cells were treated with MG132, ApoB recovery in both the mock and ERp72-overexpressing cells rose (Figure 7C). Overall, these data indicate that PDI is primarily an ApoB-stabilizing chaperone in mammalian cells, most likely through its function as a component of the MTP complex, but that ERp57 and ERp72 facilitate ApoB ERAD. More generally, these results support the use of the yeast ApoB expression system as a means to identify components that play diverse roles during the ERAD of this protein in mammals.
DISCUSSION
ApoB is targeted for degradation unlike any other known ERAD substrate. Under lipid-poor conditions, ApoB is cotranslationally selected for degradation by the Hsp70 and Hsp90 molecular chaperones, as well as a J domain–containing cochaperone, P58IPK. The apolipoprotein is then ubiquitinated by gp78 while translocon embedded and is retrotranslocated into the cytosol through the Sec61 translocon by the action of the AAA-ATPase p97. Ultimately, this immature form of ApoB is captured and degraded by the proteasome (Dixon et al., 1991; Yeung et al., 1996; Benoist and Grand-Perret, 1997; Fisher et al., 1997, 2008; Yao et al., 1997; Liao et al., 1998; Mitchell et al., 1998; Gusarova et al., 2001; Pariyarath et al., 2001; Liang et al., 2003; Oyadomari et al., 2006). Given these unique attributes, and because of the profound link between ApoB secretion and human disease, we sought to identify and characterize other factors that affect the decision between stabilizing and degrading ApoB.
In this study, we show for the first time that PDI family members play opposing roles during the degradation of an ERAD substrate in mammalian cells. Specifically, we find that ERp57 and ERp72 facilitate the proteasome-mediated degradation of ApoB, but PDI helps stabilize ApoB in hepatic cells, an effect most likely brought about by its membership in the MTP complex. In yeast, which lack the MTP complex, we discovered that Pdi1 facilitates the ERAD of ApoB. We also discovered that the chaperone activity of ApoB is required for degradation. In contrast, the degradation of another substrate (i.e., CPY*) depends on Pdi1's thioredoxin motifs, and destruction of yet another ERAD substrate (i.e., α-ENaC) is PDI independent. These data highlight the complex and unique actions of PDI family members during ERAD.
One surprising aspect of this study is that ApoB forms a disulfide cross-linked species with Pdi1 in the yeast ER (Figure 3). Thus one might predict that the redox or isomerization activity—but not the chaperone activity—of Pdi1 might have been most important for ERAD. It was unexpected that mutating the cysteine residues in the a or a′ site had no effect on the rate of ApoB degradation, although CPY* degradation was blocked. There are two explanations for these data. First, there may be two pools of ApoB: One that has formed cross-links and one that is en route for degradation, an event that may require a noncovalent, chaperone-like association between ApoB and Pdi1. In fact, we note that the ERAD of ApoB is incomplete when chase reactions are conducted in yeast (see, e.g., Figure 1), and so perhaps the remaining, stable material represents the cross-linked pool. Second, Pdi1's two activities, as a redox enzyme and as a chaperone (LaMantia and Lennarz, 1993; Gillece et al., 1999), might provide unique functions during ApoB biogenesis. For example, Pdi1 might initially form disulfide bonds with ApoB, which could represent an attempt by Pdi1 and other chaperones to assist protein folding. However, because yeast lack the MTP complex, the chaperone activity of Pdi1 then becomes essential to maintain ApoB's retrotranslocation competence prior to degradation. Consistent with this view, we find that ApoB can cross-link to Pdi1 even if the chaperone domain or the a or a′ active sites have been mutated; CPY* also cross-links to each Pdi1 species (Supplemental Figure 5). In this model, however, it remains mysterious how the generated disulfide bonds in ApoB might be broken prior to retrotranslocation. Yeast lack ERdj5, which has been proposed to perform this function (Ushioda et al., 2008). So, one is left with the scenario that a yet-to-be identified enzyme is capable of breaking disulfides in the yeast ER. Alternatively, the reduction of disulfide bonds might be unnecessary for Sec61-dependent retrotranslocation and proteasome degradation. It is worth noting that polypeptide “loops” can be inserted into the Sec61 translocon (Skach, 2009). Disulfide-bonded substrates and circular substrates can also be degraded by the proteasome (Lee et al., 2002; Liu et al., 2003). In addition, large, folded domains have been shown to retrotranslocate across the ER membrane (Fiebiger et al., 2002; Tirosh et al., 2003; Schelhaas et al., 2007).
ApoB maturation requires the formation of specific disulfide bonds, and mutations in cysteines that form disulfide bonds in ApoB diminish VLDL assembly and secretion (Huang and Shelness, 1997; Tran et al., 1998; DeLozier et al., 2001). Mutations in the M subunit of the MTP complex, which contains PDI, can also prevent ApoB maturation, a phenomena that results in abetalipoproteinemia (Narcisi et al., 1995; Ohashi et al., 2000). Because we observed measurable effects of PDI, ERp57, and ERp72 overexpression on the amount of secreted ApoB, our results may be relevant to explain the relative severity of human diseases such as hypobetalipoproteinemia and atherosclerosis. In most cases, the factor(s) that lead to differences in the circulating lipoprotein levels in the population are mysterious, but it is plausible that variations in the amount or activities of components that control ApoB maturation may be at play. More generally, polymorphisms in ApoB itself are known to contribute to altered levels of secreted VLDLs, as specific nonsense codons in APOB cause hypobetalipoproteinemia (Linton et al., 1993; Whitfield et al., 2004). Overall, we hope that these ongoing studies will provide therapeutic candidates that may be targeted to prevent the catastrophic effects of diseases related to ApoB-containing lipoproteins, in particular atherosclerosis, which is the leading cause of death in Western societies.
MATERIALS AND METHODS
Yeast strains, strain construction, and growth assays
Yeast strains were grown at 26°C using standard conditions for growth, media preparation, and transformation unless otherwise noted (Adams et al., 1997). A complete list of yeast strains used in this study is given in Supplemental Table S1.
To create the PDI single-deletion strains (mpd1Δ, mpd2Δ, eug1Δ, and eps1Δ), PCR-mediated gene disruption was used, using the W303 background (Brachmann et al., 1998). In each case, the KanMX cassette was amplified from the pFA6a-KanMX6 plasmid using primers containing 20 nucleotides of homology to the KanMX cassette (underlined) and 40 nucleotides of homology flanking the gene to be disrupted. To amplify KanMX for the disruption of MPD1 the following primers were used: (forward) 5′-TCC ACT TAA CAC AAT TAG GAG AGA CAA AAT TTG ACA TAT AAG ATT GTA CTG AGA GTG CAC-3′ and (reverse) 5′-TGT GTT TAA TTA GAT AAT CAT TGA ATG AGG AAA CGT ACC ACT GTG CGG TAT TTC ACA CCG-3′. To amplify KanMX for the disruption of MPD2 the following primers were used: (forward) 5′-GTC TAG TGC AAG TAC GTC GGC AAA GTA AAA CAC AAA GGA GAG ATT GTA CTG AGA GTG CAC-3′ and (reverse) 5′-TCG GTA TTC GTA AAG TAA AAG ACA GAG CGA AGC TTA TGT TCT GTG CGG TAT TTC ACA CCG-3′. To amplify KanMX for the disruption of EUG1 the following primers were used: (forward) 5′-ATA TGG CAA TCT CCC AAC AAG CAC CCG CTC ATA TAA TAC CAG ATT GTA CTG AGA GTG CAC-3′ and (reverse) 5′-AGA TGT TAA AAA TGT GCA TTA TAT ATG CTT TAT TTA TTG ACT GTG CGG TAT TTC ACA CCG-3′. To amplify KanMX for the disruption of EPS1 the following primers were used: (forward) 5′-AAA AAT ACT ATC TAT AAA AAC TAG CTG TAA GGC AGC AGC CAG ATT GTA CTG AGA GTG CAC-3′ and (reverse) 5′-AGA TAT CAG CAT TCT TTT ATT TTT ATA ACT ACT TAA GCG TCT GTG CGG TAT TTC ACA CCG-3′. The deletions of MPD1, MPD2, EUG1, and EPS1 were confirmed by PCR.
Strain SRH01 was created using a plasmid shuffle. Briefly, plasmid pSG01 (see later discussion) was transformed into strain M4492, kindly provided by the Schmitt lab (Saarland University, Saarbrücken, Germany). Transformants were plated on medium containing 5- fluoroanthranilic acid, which selects for cells that have lost the TRP1 gene. Therefore the MPD1-TRP plasmid from strain M4492 was lost and replaced with a plasmid in which the expression of Pdi1 was driven from its own promoter.
DTT sensitivity was determined by growing cells overnight at 26°C to logarithmic phase in the appropriate selective medium. The number of cells in each culture was normalized. and a dilution series from each culture (1:10, 1:100, 1:1000) was plated on yeast extract/peptone/dextrose solid media (pH 5.5) either containing or lacking a final concentration of 5 mM DTT. The plates were incubated at 30°C for 2 d.
Plasmids
The plasmids used in this study are shown in Supplemental Table S2. To assess the degradation of ApoB29 in yeast, pSLW1-B29 was used (Hrizo et al., 2007). To monitor the degradation of CPY*, we used pRS316CPY*-3HA, which was kindly provided by the Weissman lab (University of California, San Francisco, San Francisco, CA; Bhamidipati et al., 2005).
Plasmids pBH1464, pBH1852, pBH1630, and pCT37 (Holst et al., 1997) were generous gifts from the Schmitt lab (Saarland University). Plasmid pSG01 was constructed by subcloning a PstI–BamHI fragment from plasmid pBH1464 into the same sites in pRS314.
To study the degradation of pαF, we used pSM36 (Kim et al., 2005). To monitor the degradation of α-ENaC in the nonessential PDI deletion strains, we used plasmid pRS426GPD ENaC-HA (Buck et al., 2010). To measure the degradation of α-ENaC in strains M4492 and SRH01 (Supplemental Table S1) a methionine-repressible α-ENaC plasmid was constructed. To this end, an EcoRI–XhoI fragment corresponding to the coding sequence of α-ENaC and the hemagglutinin (HA) epitope tag from plasmid pRS426GPD α-ENaC-HA was subcloned into the same sites in pRS426MET25 (Mumberg et al., 1994) and was named pRS426MET25 α-ENaC-HA. To assess the degradation of PrA*-Ab–HA, plasmid pKK223 (Kanehara et al., 2010) was kindly provided by the Ng lab (National University of Singapore, Singapore).
Assays to measure the degradation of ERAD substrates in yeast
To assess the degradation of ApoB29 in yeast, cells transformed with pSLW1-B29 were grown to logarithmic phase (OD600 = 0.4–1.0) overnight at 26°C in synthetic complete medium lacking uracil but supplemented with glucose to a 2% final concentration. The cells were harvested and resuspended in complete medium supplemented with galactose to a final concentration of 2% and were grown for 5 h at 26°C to obtain maximal expression of ApoB29. The cycloheximide chase analysis was performed at 30°C as previously described (Hrizo et al., 2007). Total protein was precipitated as described (Zhang et al., 2001) and immediately resolved by SDS–PAGE before immunoblot analysis. ApoB29 was detected by using an anti-HA-horseradish peroxidase (HRP)–conjugated (clone 3F10; Roche, Indianapolis, IN) antibody. Immunoblots were also probed with anti-G6PD (Sigma-Aldrich, St. Louis, MO) antiserum as a loading control. The G6PD primary antibody was decorated with donkey HRP-conjugated anti–rabbit immunoglobulin G (IgG) secondary antibody (GE Healthcare, Waukesha, WI). The Supersignal West Femto Chemiluminescent Substrate (Pierce, Rockford, IL) was used for the detection of anti-HA ApoB29 in immunoblots, and the Supersignal West Pico Chemiluminescent Substrate (Pierce) was used for the detection of anti-G6PD in immunoblots. The signals were quantified using a Kodak 440CF Image Station and the associated Kodak 1D software (Eastman Kodak, Rochester, NY).
To monitor the degradation of CPY*, cells expressing pRS316CPY*-3HA were grown in the appropriate selective media overnight at 26°C to logarithmic phase (OD600 = 0.4–1.0). Cycloheximide chase at 30°C, protein precipitation, and SDS–PAGE were performed as previously described (Tran et al., 2011). Immunoblots were probed with an anti-HA-HRP-conjugated (clone 3F10; Roche) antibody to detect CPY* and with anti-Sec61 antiserum (Stirling et al., 1992), which served as a loading control. The anti-Sec61 primary antibody was probed with donkey HRP-conjugated anti–rabbit IgG secondary antibody. The Supersignal West Pico Chemiluminescent Substrate was used to detect anti-HA CPY* and immunoblots were also probed with anti-Sec61. The resulting signals were quantified as described.
The degradation of the α subunit of ENaC (α-ENaC) was assessed by introducing plasmid pRS426GPD α–ENaC-HA (see earlier discussion and Supplemental Table S2) into the recipient strains that lacked single, nonessential PDI family members. The cells were grown in selective medium overnight at 26°C to logarithmic phase, and cycloheximide chases, protein precipitation, and SDS–PAGE were performed as described (Buck et al., 2010). α-ENaC degradation in strains M4492 and SRH01 was determined by introducing plasmid pRS426MET25 α–ENaC-HA (see earlier discussion), which is engineered for the methionine-repressible expression of α-ENaC. The cells were grown at 26°C in selective medium with 2 mM methionine overnight to logarithmic phase. To obtain maximal expression of α-ENaC, cells were harvested and resuspended to an initial concentration of 0.5 A600/ml in selective medium lacking methionine and were grown for another 1.5 h. Cycloheximide chases, protein precipitation, and SDS–PAGE were performed as published (Buck et al., 2010). Immunoblots were probed with an anti-HA-HRP-conjugated antibody (clone 3F10; Roche) to detect α-ENaC, and with anti-G6PD antiserum, which served as a loading control. The anti-G6PD primary antibody was probed with donkey HRP-conjugated anti–rabbit IgG secondary antibody. The Supersignal West Pico Chemiluminescent Substrate was used to detect the anti–HA-α-ENaC and anti-G6PD antibodies on the immunoblots, and the signals were quantified as described.
The ERAD of pαF was measured using plasmid pSM36 (see Supplemental Table S2 and earlier discussion). In brief, cells were grown in selective medium at 26°C overnight to logarithmic phase and were harvested and resuspended to 0.5 A600/ml. Next the cells were preincubated at 30°C with vigorous shaking for 10 min, and then protein synthesis was stopped by the addition of cycloheximide to a final concentration of 50 μg/ml. At the indicated time points, 1 ml of cells was harvested and frozen in liquid nitrogen. Total protein was precipitated as described (Zhang et al., 2001) and was resolved by SDS–PAGE, followed by Western blot analysis. Immunoblots were probed with an anti-HA-HRP-conjugated (clone 3F10; Roche) antibody to detect pαf and with anti-G6PD antiserum, which served as a loading control. The anti-G6PD primary antibody was probed with donkey HRP-conjugated anti–rabbit IgG secondary antibody. The Supersignal West Pico Chemiluminescent Substrate was used to detect the bound antibodies, and the signals were quantified as described.
To monitor the degradation of PrA*-Ab, cells expressing pKK223 were grown in the appropriate selective media overnight at 26°C to logarithmic phase (OD600 = 0.4–1.0). A cycloheximide chase at 30°C, protein precipitation, and SDS–PAGE were performed as described. Immunoblots were probed with an anti-HA-HRP-conjugated antibody to detect PrA*-Ab and with anti-G6PD antiserum, which served as a loading control. The anti-G6PD primary antibody was probed with donkey HRP-conjugated anti–rabbit IgG secondary antibody (GE Healthcare). The Supersignal West Pico Chemiluminescent Substrate was used for detection of anti-HA PrA*-Ab and anti-G6PD immunoblots, and the signals were quantified as described.
Coimmunoprecipitation assays
Disulfide-conjugated proteins were immunoprecipitated using a previously published protocol with minor modifications (Sakoh-Nakatogawa et al., 2009). In brief, spheroplasts were prepared from cells expressing ApoB29 or CPY*, as described (McCracken and Brodsky, 1996) and were resuspended in buffer containing 1.2 M sorbitol for 30 min at 30°C. Proteins were precipitated with 10% trichloroacetic acid (TCA), centrifuged at 15,000 × g for 5 min at 4°C, and resuspended in 2% SDS, 20 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES)–KOH, pH 7.4, 50 mM NaCl, and 35 mM iodoacetamide with a protease inhibitor cocktail. The solution was incubated at 75°C for 5 min, and insoluble material was removed by centrifugation at 15,000 × g for 5 min at 4°C. The supernatant was diluted 10-fold with 20 mM HEPES-KOH, pH 7.4, and 50 mM NaCl with protease inhibitors and incubated with anti-HA-conjugated resin (Roche) or Sepharose 6B resin (Sigma-Aldrich) as a negative control for 3 h at room temperature. The immunoprecipitates were washed three times with 20 mM HEPES-KOH, pH 7.4, and 50 mM NaCl and eluted in sample buffer prepared with 120 mM of freshly added DTT for 3 min at 75°C for SDS–PAGE, as described. Prior to the immunoprecipitation, 1% of the lysate was retained and loaded as a control. The samples were immunoblotted with anti-Pdi1 antibody (a kind gift from V. Denic, Harvard University, Cambridge, MA), and the primary antibody was probed with donkey HRP-conjugated anti–rabbit IgG secondary. The Supersignal West Pico Chemiluminescent Substrate was used for detection. ApoB was detected as described.
Cysteine modification assays
Cysteines were modified by the addition of maleimide-PEG5000 using a previously published protocol (Tetsch et al., 2011) that we adapted for yeast. Briefly, spheroplasts were prepared from cells expressing ApoB29 and were resuspended in 20 mM HEPES, pH 6.8, 150 mM KOAc, 5 mM MgOAc, and 1.2 M sorbitol. Iodoacetamide was added to a final concentration of 10 mM, and cells were incubated at 30°C for 15 min. Following incubation, proteins were precipitated by the addition of TCA to a final concentration of 10% and were incubated on ice for 30 min. The cells were centrifuged (16,000 × g, 4°C, 15 min) and the pellets were resuspended in denaturing buffer (6 M urea, 200 mM Tris-HCl, pH 8.5, 10 mM EDTA, 0.5% SDS) containing 1 mM DTT and were incubated at 37°C for 1 h. The samples were then divided into two tubes, and again proteins were precipitated with 10% TCA on ice for 30 min. Following centrifugation (15 min, 4°C, 16, 000 × g) and removal of residual TCA, the pellets were resuspended in either denaturing buffer (see earlier discussion) or denaturing buffer with 10 mM maleimide-PEG5000 (Layson Bio, Arab, AL). Samples were incubated at 37°C for 1 h. TCA was added again to a final concentration of 10%, and the samples were kept on ice for 15 min before centrifugation (15 min, 4°C, 16,000 × g). After removal of all TCA, pellets were resuspended in denaturing buffer, and 2× nondenaturing sample buffer was added. Samples were heated to 75°C for 5 min and subjected to SDS–PAGE and immunoblotting. Pdi1 and ApoB were detected as described above.
PDI, ERp57, and ERp72 expression and ApoB secretion and degradation in rat hepatoma cells
The genes encoding human PDI, ERp57, and ERp72 cDNA were obtained from Open Biosystems (Huntsville, AL; MHS1011-98054157, MHS1011-98054265, and MHS1011-75436 respectively). The cDNA was amplified using standard PCR conditions and the following primers, which contained a Kozak consensus sequence (in bold) and KpnI or XbaI restriction sites (underlined). PDI was amplified using the primers (forward) 5′-G TAC GGT ACC ACC GCC ACC ATG CTG CGC CGC GCT CTG CTG-3′ and (reverse) 5′-G TAC TCT AGA TTA CAG TTC ATC TTT CAC AGC-3′. ERp57 was amplified using the primers (forward) 5′-G TAC GGT ACC ACC GCC ACC ATG CGC CTC CGC CGC CTA GCG-3′ and (reverse) 5′-G TAC TCT AGA TTA GAG ATC CTC CTG TGC CTT C-3′. ERp72 was amplified using the primers (forward) 5′-G TAC GGT ACC ACC GCC ACC ATG AGG CCC CGG AAA GCC TTC CTG-3′ and (reverse) 5′-G TAC TCT AGA TTA TCA AAG CTC TTC CTT GGT CCT G-3′. The engineered restriction sites at the 5′ and 3′ ends allowed for subcloning into the pcDNA3.1 vector. High-fidelity amplification and cloning was confirmed by sequence analysis.
Rat hepatoma McA-RH7777 cells (American Type Culture Collection, Manassas, VA; CRL-1601) were cultured at 37°C in DMEM supplemented with 10% fetal bovine serum, 10% horse serum, l-glutamine, and penicillin/streptomycin. Cells were transfected with 2 μg of pcDNA3.1 vector (control), pcDNA3.1-hPDI expression vector, pcDNA3.1-hERp57 expression vector, or pcDNA3.1-hERp72 expression vector, prepared as described, using FuGENE HD transfection reagent (Roche) according to the manufacturer's specifications. A total of 24 h after the initial transfection a second transfection with 2 μg of DNA was performed. A pulse-chase analysis was performed 48 h after the second transfection, as previously described (Gusarova et al., 2001). The proteasome dependence of ApoB100 degradation and secretion was performed as described (Meex et al., 2011) with either DMSO or the proteasomal inhibitor MG132 (Z-Leu-Leu-Leu-al; Sigma, St. Louis, MO) added to a final concentration of 25μM.
Supplementary Material
Acknowledgments
We thank Chris Bahur, Jessica Coblentz, Vlad Denic, Rebecca Hughey, Davis Ng, Karin Römisch, Manfred Schmitt, and Jonathan Weissman for technical assistance, advice, and/or reagents. This work was supported by National Institutes of Health Grants HL58541 to E.A.F. and J.L.B. and GM75061 and DK65161 to J.L.B. We also acknowledge the Core Facilities of the Pittsburgh Center for Kidney Research, Pittsburgh, PA (DK79307).
Abbreviations used:
- ApoB
apolipoprotein B
- CPY*
carboxypeptidase Y*
- ERAD
endoplasmic reticulum–associated degradation
- LDL
low-density lipoproteins
- PDI
protein disulfide isomerase
- VLDL
very low-density lipoproteins
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-08-0704) on December 21, 2011.
REFERENCES
- Adams A, Gottschling DE, Kaiser CA, Stearns T. Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1997. pp. 59–63. [Google Scholar]
- Adeli K, Macri J, Mohammadi A, Kito M, Urade R, Cavallo D. Apolipoprotein B is intracellularly associated with an ER-60 protease homologue in HepG2 cells. J Biol Chem. 1997;272:22489–22494. doi: 10.1074/jbc.272.36.22489. [DOI] [PubMed] [Google Scholar]
- Anelli T, Sitia R. Protein quality control in the early secretory pathway. EMBO J. 2008;27:315–327. doi: 10.1038/sj.emboj.7601974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benoist F, Grand-Perret T. Co-translational degradation of apolipoprotein B100 by the proteasome is prevented by microsomal triglyceride transfer protein: synchronized translation studies on Hepg2 cells treated with an inhibitor of microsomal triglyceride transfer protein. J Biol Chem. 1997;272:20435–20442. doi: 10.1074/jbc.272.33.20435. [DOI] [PubMed] [Google Scholar]
- Bhamidipati A, Denic V, Quan EM, Weissman JS. Exploration of the topological requirements of ERAD identifies Yos9p as a lectin sensor of misfolded glycoproteins in the ER lumen. Mol Cell. 2005;19:741–751. doi: 10.1016/j.molcel.2005.07.027. [DOI] [PubMed] [Google Scholar]
- Brachmann CB, Davies A, Cost GJ, Caputo E, Li J, Hieter P, Boeke JD. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Brodsky J, Skach W. Protein folding and quality control in the endoplasmic reticulum: recent lessons from yeast and mammalian cell systems. Curr Opin Cell Biol. 2011;23:464–475. doi: 10.1016/j.ceb.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky JL, Fisher EA. The many intersecting pathways underlying apolipoprotein B secretion and degradation. Trends Endocrinol Metab. 2008;19:254–259. doi: 10.1016/j.tem.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck TM, Kolb AR, Boyd CR, Kleyman TR, Brodsky JL. The endoplasmic reticulum-associated degradation of the epithelial sodium channel requires a unique complement of molecular chaperones. Mol Biol Cell. 2010;21:1047–1058. doi: 10.1091/mbc.E09-11-0944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulleid NJ, Ellgaard L. Multiple ways to make disulfides. Trends Biochem Sci. 2011;36:485–492. doi: 10.1016/j.tibs.2011.05.004. [DOI] [PubMed] [Google Scholar]
- Burch WL, Herscovitz H. Disulfide bonds are required for folding and secretion of apolipoprotein B regardless of its lipidation state. J Biol Chem. 2000;275:16267–16274. doi: 10.1074/jbc.M000446200. [DOI] [PubMed] [Google Scholar]
- Cai H, Wang CC, Tsou CL. Chaperone-like activity of protein disulfide isomerase in the refolding of a protein with no disulfide bonds. J Biol Chem. 1994;269:24550–24552. [PubMed] [Google Scholar]
- Chivers PT, Laboissiere MC, Raines RT. The CXXC motif: imperatives for the formation of native disulfide bonds in the cell. EMBO J. 1996;15:2659–2667. [PMC free article] [PubMed] [Google Scholar]
- Clerc S, Hirsch C, Oggier DM, Deprez P, Jakob C, Sommer T, Aebi M. Htm1 protein generates the N-glycan signal for glycoprotein degradation in the endoplasmic reticulum. J Cell Biol. 2009;184:159–172. doi: 10.1083/jcb.200809198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox JS, Walter P. A novel mechanism for regulating activity of a transcription factor that controls the unfolded protein response. Cell. 1996;87:391–404. doi: 10.1016/s0092-8674(00)81360-4. [DOI] [PubMed] [Google Scholar]
- Crooke RM, Graham MJ, Lemonidis KM, Whipple CP, Koo S, Perera RJ. An apolipoprotein B antisense oligonucleotide lowers LDL cholesterol in hyperlipidemic mice without causing hepatic steatosis. J Lipid Res. 2005;46:872–884. doi: 10.1194/jlr.M400492-JLR200. [DOI] [PubMed] [Google Scholar]
- DeLozier JA, Parks JS, Shelness GS. Vesicle-binding properties of wild-type and cysteine mutant forms of alpha(1) domain of apolipoprotein B. J Lipid Res. 2001;42:399–406. [PubMed] [Google Scholar]
- Dixon JL, Furukawa S, Ginsberg HN. Oleate stimulates secretion of apolipoprotein B-containing lipoproteins from Hep G2 cells by inhibiting early intracellular degradation of apolipoprotein B. J Biol Chem. 1991;266:5080–5086. [PubMed] [Google Scholar]
- Dong G, Wearsch PA, Peaper DR, Cresswell P, Reinisch KM. Insights into MHC class I peptide loading from the structure of the tapasin-ERp57 thiol oxidoreductase heterodimer. Immunity. 2009;30:21–32. doi: 10.1016/j.immuni.2008.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellgaard L, Ruddock L. The human disulfide isomerase family: substrate interactions and functional properties. EMBO J. 2005;6:28–32. doi: 10.1038/sj.embor.7400311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farquhar R, Honey N, Murant SJ, Bossier P, Schultz L, Montgomery D, Ellis RW, Freedman RB, Tuite MF. Protein disulfide isomerase is essential for viability in Saccharomyces cerevisiae. Gene. 1991;108:81–89. doi: 10.1016/0378-1119(91)90490-3. [DOI] [PubMed] [Google Scholar]
- Fiebiger E, Story C, Ploegh HL, Tortorella D. Visualization of the ER-to-cytosol dislocation reaction of a type I membrane protein. EMBO J. 2002;21:1041–1053. doi: 10.1093/emboj/21.5.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finger A, Knop M, Wolf DH. Analysis of two mutated vacuolar proteins reveals a degradation pathway in the endoplasmic reticulum or a related compartment of yeast. Eur J Biochem. 1993;218:565–574. doi: 10.1111/j.1432-1033.1993.tb18410.x. [DOI] [PubMed] [Google Scholar]
- Firsov D, Robert-Nicoud M, Gruender S, Schild L, Rossier BC. Mutational analysis of cysteine-rich domains of the epithelium sodium channel (ENaC). Identification of cysteines essential for channel expression at the cell surface. J Biol Chem. 1999;274:2743–2749. doi: 10.1074/jbc.274.5.2743. [DOI] [PubMed] [Google Scholar]
- Fisher EA, Ginsberg HN. Complexity in the secretory pathway: the assembly and secretion of apolipoprotein B-containing lipoproteins. J Biol Chem. 2002;277:17377–17380. doi: 10.1074/jbc.R100068200. [DOI] [PubMed] [Google Scholar]
- Fisher EA, Lapierre LR, Junkins RD, McLeod RS. The AAA-ATPase p97 facilitates degradation of apolipoprotein B by the ubiquitin-proteasome pathway. J Lipid Res. 2008;49:2149–2160. doi: 10.1194/jlr.M800108-JLR200. [DOI] [PubMed] [Google Scholar]
- Fisher EA, Zhou M, Mitchell DM, Wu X, Omura S, Wang H, Goldberg AL, Ginsberg HN. The degradation of apolipoprotein B100 is mediated by the ubiquitin-proteasome pathway and involves heat shock protein 70. J Biol Chem. 1997;272:20427–20434. doi: 10.1074/jbc.272.33.20427. [DOI] [PubMed] [Google Scholar]
- Forster ML, Sivick K, Park YN, Arvan P, Lencer WI, Tsai B. Protein disulfide isomerase–like proteins play opposing roles during retrotranslocation. J Cell Biol. 2006;173:853–859. doi: 10.1083/jcb.200602046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauss R, Kanehara K, Carvalho P, Ng DT, Aebi M. A complex of pdi1p and the mannosidase htm1p initiates clearance of unfolded glycoproteins from the endoplasmic reticulum. Mol Cell. 2011;42:782–793. doi: 10.1016/j.molcel.2011.04.027. [DOI] [PubMed] [Google Scholar]
- Gillece P, Luz JM, Lennarz WJ, de la Cruz FJ, Romisch K. Export of a cysteine-free misfolded secretory protein from the endoplasmic reticulum for degradation requires interaction with protein disulfide isomerase. J Cell Biol. 1999;147:1443–1456. doi: 10.1083/jcb.147.7.1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon DA, Jamil H, Sharp D, Mullaney D, Yao Z, Gregg RE, Wetterau J. Secretion of apolipoprotein B-containing lipoproteins from HeLa cells is dependent on expression of the microsomal triglyceride transfer protein and is regulated by lipid availability. Proc Natl Acad Sci USA. 1994;91:7628–7632. doi: 10.1073/pnas.91.16.7628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunther R, Srinivasan M, Haugejorden S, Green M, Ehbrecht IM, Kuntzel H. Functional replacement of the Saccharomyces cerevisiae Trg1/Pdi1 protein by members of the mammalian protein disulfide isomerase family. J Biol Chem. 1993;268:7728–7732. [PubMed] [Google Scholar]
- Gusarova V, Caplan AJ, Brodsky JL, Fisher EA. Apoprotein B degradation is promoted by the molecular chaperones hsp90 and hsp70. J Biol Chem. 2001;276:24891–24900. doi: 10.1074/jbc.M100633200. [DOI] [PubMed] [Google Scholar]
- Gusarova V, Seo J, Sullivan ML, Watkins SC, Brodsky JL, Fisher EA. Golgi-associated maturation of very low density lipoproteins involves conformational changes in apolipoprotein B, but is not dependent on apolipoprotein E. J Biol Chem. 2007;282:19453–19462. doi: 10.1074/jbc.M700475200. [DOI] [PubMed] [Google Scholar]
- Haghpassand M, Wilder D, Moberly JB. Inhibition of apolipoprotein B and triglyceride secretion in human hepatoma cells (HepG2) J Lipid Res. 1996;37:1468–1480. [PubMed] [Google Scholar]
- Hagiwara M, Maegawa K, Suzuki M, Ushioda R, Araki K, Matsumoto Y, Hoseki J, Nagata K, Inaba K. Structural basis of an ERAD pathway mediated by the ER-resident protein disulfide reductase ERdj5. Mol Cell. 2011;41:432–444. doi: 10.1016/j.molcel.2011.01.021. [DOI] [PubMed] [Google Scholar]
- Harazono A, Kawasaki N, Kawanishi T, Hayakawa T. Site-specific glycosylation analysis of human apolipoprotein B100 using LC/ESI MS/MS. Glycobiology. 2005;15:447–462. doi: 10.1093/glycob/cwi033. [DOI] [PubMed] [Google Scholar]
- Hatahet F, Ruddock LW. Protein disulfide isomerase: a critical evaluation of its function in disulfide bond formation. Antioxid Redox Signal. 2009;11:2807–2850. doi: 10.1089/ars.2009.2466. [DOI] [PubMed] [Google Scholar]
- Holst B, Tachibana C, Winther JR. Active site mutations in yeast protein disulfide isomerase cause dithiothreitol sensitivity and a reduced rate of protein folding in the endoplasmic reticulum. J Cell Biol. 1997;138:1229–1238. doi: 10.1083/jcb.138.6.1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrizo SL, Gusarova V, Habiel DM, Goeckeler JL, Fisher EA, Brodsky JL. The Hsp110 molecular chaperone stabilizes apolipoprotein B from endoplasmic associated degradation (ERAD) J Biol Chem. 2007;282:32665–32675. doi: 10.1074/jbc.M705216200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang XF, Shelness GS. Identification of cysteine pairs within the amino-terminal 5% of apolipoprotein B essential for hepatic lipoprotein assembly and secretion. J Biol Chem. 1997;272:31872–31876. doi: 10.1074/jbc.272.50.31872. [DOI] [PubMed] [Google Scholar]
- Hussain MM, Shi J, Dreizen P. Microsomal triglyceride transfer protein and its role in apoB-lipoprotein assembly. J Lipid Res. 2003;44:22–32. doi: 10.1194/jlr.r200014-jlr200. [DOI] [PubMed] [Google Scholar]
- Jakob CA, Bodmer D, Spirig U, Battig P, Marcil A, Dignard D, Bergeron JJ, Thomas DY, Aebi M. Htm1p, a mannosidase-like protein, is involved in glycoprotein degradation in yeast. EMBO Rep. 2001;2:423–430. doi: 10.1093/embo-reports/kve089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamil H, et al. An inhibitor of the microsomal triglyceride transfer protein inhibits apoB secretion from HepG2 cells. Proc Natl Acad Sci USA. 1996;93:11991–11995. doi: 10.1073/pnas.93.21.11991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasti J, Furukawa H, Gonzales EB, Gouaux E. Structure of acid-sensing ion channel 1 at 1.9 A resolution and low pH. Nature. 2007;449:316–323. doi: 10.1038/nature06163. [DOI] [PubMed] [Google Scholar]
- Jessop CE, Watkins RH, Simmons JJ, Tasab M, Bulleid NJ. Protein disulfide isomerase family members show distinct substrate specificity: P5 is targeted to BiP client proteins. J Cell Sci. 2009;122:4287–4295. doi: 10.1242/jcs.059154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabani M, Kelley SS, Morrow MW, Montgomery DL, Sivendran R, Rose MD, Gierasch LM, Brodsky JL. Dependence of endoplasmic reticulum-associated degradation on the peptide binding domain and concentration of BiP. Mol Biol Cell. 2003;14:3437–3448. doi: 10.1091/mbc.E02-12-0847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehara K, Xie W, Ng DT. Modularity of the Hrd1 ERAD complex underlies its diverse client range. J Cell Biol. 2010;188:707–716. doi: 10.1083/jcb.200907055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannel WB, Castelli WP, Gordon T, McNamara PM. Serum cholesterol, lipoproteins, and the risk of coronary heart disease. The Framingham Study. Ann Intern Med. 1971;74:1–12. doi: 10.7326/0003-4819-74-1-1. [DOI] [PubMed] [Google Scholar]
- Kashlan OB, Adelman JL, Okumura S, Blobner BM, Zuzek Z, Hughey RP, Kleyman TR, Grabe M. Constraint-based, homology model of the extracellular domain of the epithelial Na+ channel alpha subunit reveals a mechanism of channel activation by proteases. J Biol Chem. 2011;286:649–660. doi: 10.1074/jbc.M110.167098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashlan OB, Mueller GM, Qamar MZ, Poland PA, Ahner A, Rubenstein RC, Hughey RP, Brodsky JL, Kleyman TR. Small heat shock protein alphaA-crystallin regulates epithelial sodium channel expression. J Biol Chem. 2007;282:28149–28156. doi: 10.1074/jbc.M703409200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W, Spear ED, Ng DT. Yos9p detects and targets misfolded glycoproteins for ER-associated degradation. Mol Cell. 2005;19:753–764. doi: 10.1016/j.molcel.2005.08.010. [DOI] [PubMed] [Google Scholar]
- Klappa P, Ruddock LW, Darby NJ, Freedman RB. The b′ domain provides the principal peptide-binding site of protein disulfide isomerase but all domains contribute to binding of misfolded proteins. EMBO J. 1998;17:927–935. doi: 10.1093/emboj/17.4.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleizen B, Braakman I. Protein folding and quality control in the endoplasmic reticulum. Curr Opin Cell Biol. 2004;16:343–349. doi: 10.1016/j.ceb.2004.06.012. [DOI] [PubMed] [Google Scholar]
- Kuznetsov G, Chen LB, Nigam SK. Multiple molecular chaperones complex with misfolded large oligomeric glycoproteins in the endoplasmic reticulum. J Biol Chem. 1997;272:3057–3063. doi: 10.1074/jbc.272.5.3057. [DOI] [PubMed] [Google Scholar]
- Laboissiere MC, Sturley SL, Raines RT. The essential function of protein-disulfide isomerase is to unscramble non-native disulfide bonds. J Biol Chem. 1995;270:28006–28009. doi: 10.1074/jbc.270.47.28006. [DOI] [PubMed] [Google Scholar]
- LaMantia ML, Lennarz WJ. The essential function of yeast protein disulfide isomerase does not reside in its isomerase activity. Cell. 1993;74:899–908. doi: 10.1016/0092-8674(93)90469-7. [DOI] [PubMed] [Google Scholar]
- Lee C, Prakash S, Matouschek A. Concurrent translocation of multiple polypeptide chains through the proteasomal degradation channel. J Biol Chem. 2002;277:34760–34765. doi: 10.1074/jbc.M204750200. [DOI] [PubMed] [Google Scholar]
- Leiper JM, Bayliss JD, Pease RJ, Brett DJ, Scott J, Shoulders CC. Microsomal triglyceride transfer protein, the abetalipoproteinemia gene product, mediates the secretion of apolipoprotein B-containing lipoproteins from heterologous cells. J Biol Chem. 1994;269:21951–21954. [PubMed] [Google Scholar]
- Liang JS, Kim T, Fang S, Yamaguchi J, Weissman AM, Fisher EA, Ginsberg HN. Overexpression of the tumor autocrine motility factor receptor Gp78, a ubiquitin protein ligase, results in increased ubiquitinylation and decreased secretion of apolipoprotein B100 in HepG2 Cells. J Biol Chem. 2003;278:23984–23988. doi: 10.1074/jbc.M302683200. [DOI] [PubMed] [Google Scholar]
- Liao W, Yeung SC, Chan L. Proteasome-mediated degradation of apolipoprotein B targets both nascent peptides cotranslationally before translocation and full-length apolipoprotein B after translocation into the endoplasmic reticulum. J Biol Chem. 1998;273:27225–27230. doi: 10.1074/jbc.273.42.27225. [DOI] [PubMed] [Google Scholar]
- Linnik K, Herscovitz H. Multiple molecular chaperones interact with apolipoprotein B during its maturation. J Biol Chem. 1998;273:21368–21373. doi: 10.1074/jbc.273.33.21368. [DOI] [PubMed] [Google Scholar]
- Linton MF, Farese RVJ, Young SG. Familial hypolipoproteinemia. J Lipid Res. 1993;34:521–541. [PubMed] [Google Scholar]
- Liu CW, Corboy MJ, DeMartino GN, Thomas PJ. Endoproteolytic activity of the proteasome. Science. 2003;299:408–411. doi: 10.1126/science.1079293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnuson B, Rainey EK, Benjamin T, Baryshev M, Mkrtchian S, Tsai B. ERp29 triggers a conformational change in polyomavirus to stimulate membrane binding. Mol Cell. 2005;20:289–300. doi: 10.1016/j.molcel.2005.08.034. [DOI] [PubMed] [Google Scholar]
- Marcus N, Shaffer D, Farrar P, Green M. Tissue distribution of three members of the murine protein disulfide isomerase (PDI) family. Biochim Biophys Acta. 1996;1309:253–260. doi: 10.1016/s0167-4781(96)00133-9. [DOI] [PubMed] [Google Scholar]
- McCracken AA, Brodsky JL. Assembly of ER-associated protein degradation in vitro: dependence on cytosol, calnexin, and ATP. J Cell Biol. 1996;132:291–298. doi: 10.1083/jcb.132.3.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meex SJ, Andreo U, Sparks JD, Fisher EA. Huh-7 or HepG2 cells: which is the better model for studying human apolipoprotein-B100 assembly and secretion. J Lipid Res. 2011;52:152–158. doi: 10.1194/jlr.D008888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon S, Lee J, Abplanalp WA, Yoo SE, Agui T, Furudate S, Kim PS, Arvan P. Oxidoreductase interactions include a role for ERp72 engagement with mutant thyroglobulin from the rdw/rdw rat dwarf. J Biol Chem. 2007;282:6183–6191. doi: 10.1074/jbc.M608863200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meusser B, Hirsch C, Jarosch E, Sommer T. ERAD: the long road to destruction. Nat Cell Biol. 2005;7:766–772. doi: 10.1038/ncb0805-766. [DOI] [PubMed] [Google Scholar]
- Mitchell DM, Zhou M, Pariyarath R, Wang H, Aitchison JD, Ginsberg HN, Fisher EA. Apoprotein B100 has a prolonged interaction with the translocon during which its lipidation and translocation change from dependence on the microsomal triglyceride transfer protein to independence. Proc Natl Acad Sci USA. 1998;95:14733–14738. doi: 10.1073/pnas.95.25.14733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore P, Bernardi KM, Tsai B. The Ero1alpha-PDI redox cycle regulates retro-translocation of cholera toxin. Mol Biol Cell. 2010;21:1305–1313. doi: 10.1091/mbc.E09-09-0826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumberg D, Muller R, Funk M. Regulatable promoters of Saccharomyces cerevisiae: comparison of transcriptional activity and their use for heterologous expression. Nucleic Acids Res. 1994;22:5767–5768. doi: 10.1093/nar/22.25.5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatsukasa K, Nishikawa S, Hosokawa N, Nagata K, Endo T. Mnl1p, an alpha -mannosidase-like protein in yeast Saccharomyces cerevisiae, is required for endoplasmic reticulum-associated degradation of glycoproteins. J Biol Chem. 2001;276:8635–8638. doi: 10.1074/jbc.C100023200. [DOI] [PubMed] [Google Scholar]
- Narcisi TM, et al. Mutations of the microsomal triglyceride-transfer-protein gene in abetalipoproteinemia. Am J Hum Genet. 1995;57:1298–1310. [PMC free article] [PubMed] [Google Scholar]
- Nishikawa SI, Fewell SW, Kato Y, Brodsky JL, Endo T. Molecular chaperones in the yeast endoplasmic reticulum maintain the solubility of proteins for retrotranslocation and degradation. J Cell Biol. 2001;153:1061–1070. doi: 10.1083/jcb.153.5.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norgaard P, Westphal V, Tachibana C, Alsoe L, Holst B, Winther JR. Functional differences in yeast protein disulfide isomerases. J Cell Biol. 2001;152:553–562. doi: 10.1083/jcb.152.3.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi K, et al. Novel mutations in the microsomal triglyceride transfer protein gene causing abetalipoproteinemia. J Lipid Res. 2000;41:1199–1204. [PubMed] [Google Scholar]
- Olofsson SO, Boren J. Apolipoprotein B: a clinically important apolipoprotein which assembles atherogenic lipoproteins and promotes the development of atherosclerosis. J Intern Med. 2005;258:395–410. doi: 10.1111/j.1365-2796.2005.01556.x. [DOI] [PubMed] [Google Scholar]
- Oyadomari S, et al. Cotranslocational degradation protects the stressed endoplasmic reticulum from protein overload. Cell. 2006;126:727–739. doi: 10.1016/j.cell.2006.06.051. [DOI] [PubMed] [Google Scholar]
- Pan M, Maitin V, Parathath S, Andreo U, Lin SX, St Germain C, Yao Z, Maxfield FR, Williams KJ, Fisher EA. Presecretory oxidation, aggregation, and autophagic destruction of apoprotein-B: a pathway for late-stage quality control. Proc Natl Acad Sci USA. 2008;105:5862–5867. doi: 10.1073/pnas.0707460104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pariyarath R, Wang H, Aitchison JD, Ginsberg HN, Welch WJ, Johnson AE, Fisher EA. Co-translational interactions of apoprotein B with the ribosome and translocon during lipoprotein assembly or targeting to the proteasome. J Biol Chem. 2001;276:541–550. doi: 10.1074/jbc.M007944200. [DOI] [PubMed] [Google Scholar]
- Park SW, Zhen G, Verhaeghe C, Nakagami Y, Nguyenvu LT, Barczak AJ, Killeen N, Erle DJ. The protein disulfide isomerase AGR2 is essential for production of intestinal mucus. Proc Natl Acad Sci USA. 2009;106:6950–6955. doi: 10.1073/pnas.0808722106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puig A, Gilbert HF. Protein disulfide isomerase exhibits chaperone and anti-chaperone activity in the oxidative refolding of lysozyme. J Biol Chem. 1994;269:7764–7771. [PubMed] [Google Scholar]
- Rainey-Barger EK, Mkrtchian S, Tsai B. The C-terminal domain of ERp29 mediates polyomavirus binding, unfolding, and infection. J Virol. 2009;83:1483–1491. doi: 10.1128/JVI.02057-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell SJ, Ruddock LW, Salo KE, Oliver JD, Roebuck QP, Llewellyn DH, Roderick HL, Koivunen P, Myllyharju J, High S. The primary substrate binding site in the b′ domain of ERp57 is adapted for endoplasmic reticulum lectin association. J Biol Chem. 2004;279:18861–18869. doi: 10.1074/jbc.M400575200. [DOI] [PubMed] [Google Scholar]
- Rutkevich LA, Cohen-Doyle MF, Brockmeier U, Williams DB. Functional relationship between protein disulfide isomerase family members during the oxidative folding of human secretory proteins. Mol Biol Cell. 2010;21:3093–3105. doi: 10.1091/mbc.E10-04-0356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutledge AC, Su Q, Adeli K. Apolipoprotein B100 biogenesis: a complex array of intracellular mechanisms regulating folding, stability, and lipoprotein assembly. Biochem Cell Biol. 2010;88:251–267. doi: 10.1139/o09-168. [DOI] [PubMed] [Google Scholar]
- Sakoh-Nakatogawa M, Nishikawa S, Endo T. Roles of protein-disulfide isomerase-mediated disulfide bond formation of yeast Mnl1p in endoplasmic reticulum-associated degradation. J Biol Chem. 2009;284:11815–11825. doi: 10.1074/jbc.M900813200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schelhaas M, Malmstrom J, Pelkmans L, Haugstetter J, Ellgaard L, Grunewald K, Helenius A. Simian virus 40 depends on ER protein folding and quality control factors for entry into host cells. Cell. 2007;131:516–529. doi: 10.1016/j.cell.2007.09.038. [DOI] [PubMed] [Google Scholar]
- Segrest JP, Jones MK, De Loof H, Dashti N. Structure of apolipoprotein B-100 in low density lipoproteins. J Lipid Res. 2001;42:1346–1367. [PubMed] [Google Scholar]
- Siddiqi S, Saleem U, Abumrad NA, Davidson NO, Storch J, Siddiqi SA, Mansbach CMI. A novel multiprotein complex is required to generate the prechylomicron transport vesicle from intestinal ER. J Lipid Res. 2010;51:1918–1928. doi: 10.1194/jlr.M005611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skach WR. Cellular mechanisms of membrane protein folding. Nat Struct Mol Biol. 2009;16:606–612. doi: 10.1038/nsmb.1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder PM. The epithelial Na+ channel: cell surface insertion and retrieval in Na+ homeostasis and hypertension. Endocr Rev. 2002;23:258–275. doi: 10.1210/edrv.23.2.0458. [DOI] [PubMed] [Google Scholar]
- Song JL, Wang CC. Chaperone-like activity of protein disulfide-isomerase in the refolding of rhodanese. Eur J Biochem/FEBS. 1995;231:312–316. doi: 10.1111/j.1432-1033.1995.tb20702.x. [DOI] [PubMed] [Google Scholar]
- Spear ED, Ng DT. Single, context-specific glycans can target misfolded glycoproteins for ER-associated degradation. J Cell Biol. 2005;169:73–82. doi: 10.1083/jcb.200411136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirling CJ, Rothblatt J, Hosobuchi M, Deshaies R, Schekman R. Protein translocation mutants defective in the insertion of integral membrane proteins into the endoplasmic reticulum. Mol Biol Cell. 1992;3:129–142. doi: 10.1091/mbc.3.2.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanabe S, Sherman H, Smith L, Yang LA, Fleming R, Hay R. Biogenesis of plasma lipoproteins in rat hepatoma McA-RH7777: importance of diffusion-mediated events during cell growth. In Vitro Cell Dev Biol. 1989;25:1129–1140. doi: 10.1007/BF02621264. [DOI] [PubMed] [Google Scholar]
- Taylor M, Banerjee T, Ray S, Tatulian SA, Teter K. Protein-disulfide isomerase displaces the cholera toxin A1 subunit from the holotoxin without unfolding the A1 subunit. J Biol Chem. 2011;286:22090–22100. doi: 10.1074/jbc.M111.237966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tetsch L, Koller C, Donhofer A, Jung K. Detection and function of an intramolecular disulfide bond in the pH-responsive CadC of Escherichia coli. BMC Microbiol. 2011;11:74. doi: 10.1186/1471-2180-11-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian G, Kober FX, Lewandrowski U, Sickmann A, Lennarz WJ, Schindelin H. The catalytic activity of protein-disulfide isomerase requires a conformationally flexible molecule. J Biol Chem. 2008;283:33630–33640. doi: 10.1074/jbc.M806026200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian G, Xiang S, Noiva R, Lennarz WJ, Schindelin H. The crystal structure of yeast protein disulfide isomerase suggests cooperativity between its active sites. Cell. 2006;124:61–73. doi: 10.1016/j.cell.2005.10.044. [DOI] [PubMed] [Google Scholar]
- Tirosh B, Furman MH, Tortorella D, Ploegh HL. Protein unfolding is not a prerequisite for endoplasmic reticulum-to-cytosol dislocation. J Biol Chem. 2003;278:6664–6672. doi: 10.1074/jbc.M210158200. [DOI] [PubMed] [Google Scholar]
- Tran JR, Tomsic LR, Brodsky JL. A Cdc48p-associated factor modulates endoplasmic reticulum-associated degradation, cell stress, and ubiquitinated protein homeostasis. J Biol Chem. 2011;286:5744–5755. doi: 10.1074/jbc.M110.179259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran K, Boren J, Macri J, Wang Y, McLeod R, Avramoglu RK, Adeli K, Yao Z. Functional analysis of disulfide linkages clustered within the amino terminus of human apolipoprotein B. J Biol Chem. 1998;273:7244–7251. doi: 10.1074/jbc.273.13.7244. [DOI] [PubMed] [Google Scholar]
- Tran K, Thorne-Tjomsland G, DeLong CJ, Cui Z, Shan J, Burton L, Jamieson JC, Yao Z. Intracellular assembly of very low density lipoproteins containing apolipoprotein B100 in rat hepatoma McA-RH7777 cells. J Biol Chem. 2002;277:31187–31200. doi: 10.1074/jbc.M200249200. [DOI] [PubMed] [Google Scholar]
- Ushioda R, Hoseki J, Araki K, Jansen G, Thomas DY, Nagata K. ERdj5 is required as a disulfide reductase for degradation of misfolded proteins in the ER. Science. 2008;321:569–572. doi: 10.1126/science.1159293. [DOI] [PubMed] [Google Scholar]
- Vembar SS, Brodsky JL. One step at a time: endoplasmic reticulum-associated degradation. Nat Rev Mol Cell Biol. 2008;9:944–957. doi: 10.1038/nrm2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitu E, Kim S, Sevier CS, Lutzky O, Heldman N, Bentzur M, Unger T, Yona M, Kaiser CA, Fass D. Oxidative activity of yeast Ero1p on protein disulfide isomerase and related oxidoreductases of the endoplasmic reticulum. J Biol Chem. 2010;285:18155–18165. doi: 10.1074/jbc.M109.064931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Yao Z, Fisher EA. The effects of n-3 fatty acids on the secretion of carboxyl-terminally truncated forms of human apoprotein B. J Biol Chem. 1994;269:18514–18520. [PubMed] [Google Scholar]
- Wang L, Fast DG, Attie AD. The enzymatic and non-enzymatic roles of protein-disulfide isomerase in apolipoprotein B secretion. J Biol Chem. 1997;272:27644–27651. doi: 10.1074/jbc.272.44.27644. [DOI] [PubMed] [Google Scholar]
- Wang L, Li SJ, Sidhu A, Zhu L, Liang Y, Freedman RB, Wang CC. Reconstitution of human Ero1-Lalpha/protein-disulfide isomerase oxidative folding pathway in vitro. Position-dependent differences in role between the a and a′ domains of protein-disulfide isomerase. J Biol Chem. 2009;284:199–206. doi: 10.1074/jbc.M806645200. [DOI] [PubMed] [Google Scholar]
- Wang Q, Chang A. Substrate recognition in ER-associated degradation mediated by Eps1, a member of the protein disulfide isomerase family. EMBO J. 2003;22:3792–3802. doi: 10.1093/emboj/cdg378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield AJ, Barrett PH, van Bockxmeer FM, Burnett JR. Lipid disorders and mutations in the APOB gene. Clin Chem. 2004;50:1725–1732. doi: 10.1373/clinchem.2004.038026. [DOI] [PubMed] [Google Scholar]
- Winter J, Klappa P, Freedman RB, Lilie H, Rudolph R. Catalytic activity and chaperone function of human protein-disulfide isomerase are required for the efficient refolding of proinsulin. J Biol Chem. 2002;277:310–317. doi: 10.1074/jbc.M107832200. [DOI] [PubMed] [Google Scholar]
- Wolf DH, Fink GR. Proteinase C (carboxypeptidase Y) mutant of yeast. J Bacteriol. 1975;123:1150–1156. doi: 10.1128/jb.123.3.1150-1156.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie W, Ng DTW. ERAD substrate recognition in budding yeast. Semin Cell Dev Biol. 2010;21 doi: 10.1016/j.semcdb.2010.02.007. [DOI] [PubMed] [Google Scholar]
- Yang CY, et al. Structure of apolipoprotein B-100 of human low density lipoproteins. Arteriosclerosis. 1989;9:96–108. doi: 10.1161/01.atv.9.1.96. [DOI] [PubMed] [Google Scholar]
- Yang CY, Kim TW, Weng SA, Lee BR, Yang ML, Gotto AM Jr. Isolation and characterization of sulfhydryl and disulfide peptides of human apolipoprotein B-100. Proc Natl Acad Sci USA. 1990;87:5523–5527. doi: 10.1073/pnas.87.14.5523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Z, Tran K, McLeod RS. Intracellular degradation of newly synthesized apolipoprotein B. J Lipid Res. 1997;38:1937–1953. [PubMed] [Google Scholar]
- Yeung SJ, Chen SH, Chan L. Ubiquitin-proteasome pathway mediates intracellular degradation of apolipoprotein B. Biochemistry. 1996;35:13843–13848. doi: 10.1021/bi9618777. [DOI] [PubMed] [Google Scholar]
- Zhang J, Herscovitz H. Nascent lipidated apolipoprotein B is transported to the Golgi as an incompletely folded intermediate as probed by its association with network of endoplasmic reticulum molecular chaperones, GRP94, ERp72, BiP, calreticulin, and cyclophilin B. J Biol Chem. 2003;278:7459–7468. doi: 10.1074/jbc.M207976200. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Nijbroek G, Sullivan ML, McCracken AA, Watkins SC, Michaelis S, Brodsky JL. Hsp70 molecular chaperone facilitates endoplasmic reticulum-associated protein degradation of cystic fibrosis transmembrane conductance regulator in yeast. Mol Biol Cell. 2001;12:1303–1314. doi: 10.1091/mbc.12.5.1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M, Fisher EA, Ginsberg HN. Regulated Co-translational ubiquitination of apolipoprotein B100. A new paradigm for proteasomal degradation of a secretory protein. J Biol Chem. 1998;273:24649–24653. doi: 10.1074/jbc.273.38.24649. [DOI] [PubMed] [Google Scholar]
- Zimmermann TS, et al. RNAi-mediated gene silencing in non-human primates. Nature. 2006;441:11–114. doi: 10.1038/nature04688. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.