ETOC: TM9/Phg1 proteins are essential for cellular adhesion in many systems, from Dictyostelium to human cells, yet their exact role remains unknown. We demonstrate that TM9 proteins participate in adhesion in Dictyostelium cells by controlling the surface levels of SibA adhesion molecules, notably by influencing their sorting in the endocytic pathway.
Abstract
TM9 proteins form a family of conserved proteins with nine transmembrane domains essential for cellular adhesion in many biological systems, but their exact role in this process remains unknown. In this study, we found that genetic inactivation of the TM9 protein Phg1A dramatically decreases the surface levels of the SibA adhesion molecule in Dictyostelium amoebae. This is due to a decrease in sibA mRNA levels, in SibA protein stability, and in SibA targeting to the cell surface. A similar phenotype was observed in cells devoid of SadA, a protein that does not belong to the TM9 family but also exhibits nine transmembrane domains and is essential for cellular adhesion. A contact site A (csA)-SibA chimeric protein comprising only the transmembrane and cytosolic domains of SibA and the extracellular domain of the Dictyostelium surface protein csA also showed reduced stability and relocalization to endocytic compartments in phg1A knockout cells. These results indicate that TM9 proteins participate in cell adhesion by controlling the levels of adhesion proteins present at the cell surface.
INTRODUCTION
Cell–substrate adhesion is essential in many biological processes, such as development, lymphocyte migration, and metastasic dissemination (Hood and Cheresh, 2002). Attachment of phagocytic cells to particles is also necessary to allow their subsequent engulfment. In one of the best-studied examples, cell surface integrins bind to the extracellular matrix and connect it with the actin cytoskeleton and to a complex network of cytosolic proteins. This system is at play during the migration of fibroblasts in the connective tissue, as well as during integrin-dependent phagocytosis (e.g., in opsonized microorganisms) by macrophages (Cougoule et al., 2004).
The amoeba Dictyostelium discoideum is a widely used model system for studying cellular adhesion and phagocytosis. This professional phagocyte feeds upon microorganisms in the soil, and its phagocytic machinery resembles that of mammalian phagocytes (Cosson and Soldati, 2008). Remarkably, Sib proteins, which perform as adhesion molecules in Dictyostelium, present features also found in mammlian integrin β chains, notably an extracellular Von Willebrandt A domain, a glycine-rich transmembrane domain, and a highly conserved cytosolic domain interacting with talin (Cornillon et al., 2006). Genetic inactivation of sibA or sibC causes a partial loss of cellular adhesion to certain substrates and particles, indicating the redundant roles of these two molecules in cell adhesion (Cornillon et al., 2008). Genetic analysis in Dictyostelium also uncovered the essential role of two additional membrane proteins in adhesion, Phg1A (Cornillon et al., 2000) and SadA (Fey et al., 2002). Both phg1A and sadA knockout cells are strongly defective for cellular adhesion and phagocytosis, but as detailed in this paper and elsewhere, the precise role of these proteins in cellular adhesion has yet to be elucidated.
Phg1A is a member of a family of three proteins in Dictyostelium, named Phg1A, -B, and -C, and characterized by the presence of nine transmembrane domains and a high degree of sequence conservation (Benghezal et al., 2003). There are four related gene products in humans (TM9SF1, -2, -3, and -4), three in Saccharomyces cerevisiae (TMN1, -2, and -3; Froquet et al., 2008), and three in Drosophila (Bergeret et al., 2008). The term TM9SF (for transmembrane 9 superfamily) has been widely used, but such a small family of closely related gene products is more adequately described as the TM9 family. Very little is known about the function of TM9/Phg1 proteins in different organisms. Genetic inactivation of members of this family causes cell adhesion defects in organisms as distinct as Dictyostelium (Cornillon et al., 2000), S. cerevisiae (Froquet et al., 2008), and Drosophila (Bergeret et al., 2008). In humans, TM9SF4 (the orthologue of Phg1A) is overexpressed in metastatic melanoma cells, in which it is essential for cannibal phagocytic activity (Lozupone et al., 2009). The ability of some metastatic cells to phagocytose nonmalignant cells and to feed upon them may contribute to increased cell survival and the metastatic process (Lozupone et al., 2009). Phg1A was found in endosomal compartments in S. cerevisiae (Singer-Kruger et al., 1993) and in Dictyostelium (Cornillon et al., 2000), as well as in human cells (Schimmoller et al., 1998; Bagshaw et al., 2005), but the mechanism linking Phg1 proteins to adhesion remains unelucidated to date.
The situation is even less clear for SadA (Fey et al., 2002): there is no identified orthologue of this protein in non-Dictyostelium organisms. Like Phg1 proteins, it features a large N-terminal domain plus a C-terminal region with nine putative transmembrane domains, and could therefore be classified topologically as a TM9 protein, but it does not show any significant sequence homology with TM9/Phg1 proteins. Although genetic inactivation of SadA results in a loss of cellular adhesion, the exact role of SadA in cellular adhesion is not established.
In this study, we show that Phg1A and SadA play a role in cell adhesion in Dictyostelium by controlling the levels of SibA adhesion molecules at the cell surface, an effect achieved by influencing the level of sibA transcripts, as well as the intracellular transport and stability of the SibA protein.
RESULTS
SibA, phg1A, and sadA mutant cells exhibit similar adhesion defects
To compare the adhesion defects observed in sibA, sadA, and phg1A knockout cells, we measured their ability to phagocytose various particles. For this, wild-type or isogenic mutant cells were incubated in medium containing fluorescent particles (latex beads or bacteria), and the internalized material was quantified by flow cytometry. As shown previously (Cornillon et al., 2000), specific defects in adhesion produce defects in phagocytosis that are limited to a subset of particles. Compared with wild-type cells, phg1A, sadA, and sibA mutant cells exhibited a strong defect for phagocytosis of latex beads, a more attenuated defect for phagocytosis of Escherichia coli bacteria, and essentially unaffected phagocytosis of Klebsiella pneumoniae bacteria (Figure 1). The defects observed were quantitatively less pronounced in sibA than in phg1A or sadA mutant cells, potentially reflecting the residual activity of SibC in sibA knockout cells (Cornillon et al., 2008). Little or no reduction was seen in fluid-phase macropinocytosis (Figure 1), a process mechanistically similar to phagocytosis, but independent of cellular adhesion. The fact that genetic inactivation of PHG1A or SADA creates adhesion defects qualitatively similar to those observed in sibA knockout cells suggests that Phg1A and SadA may regulate function or expression of SibA.
FIGURE 1:
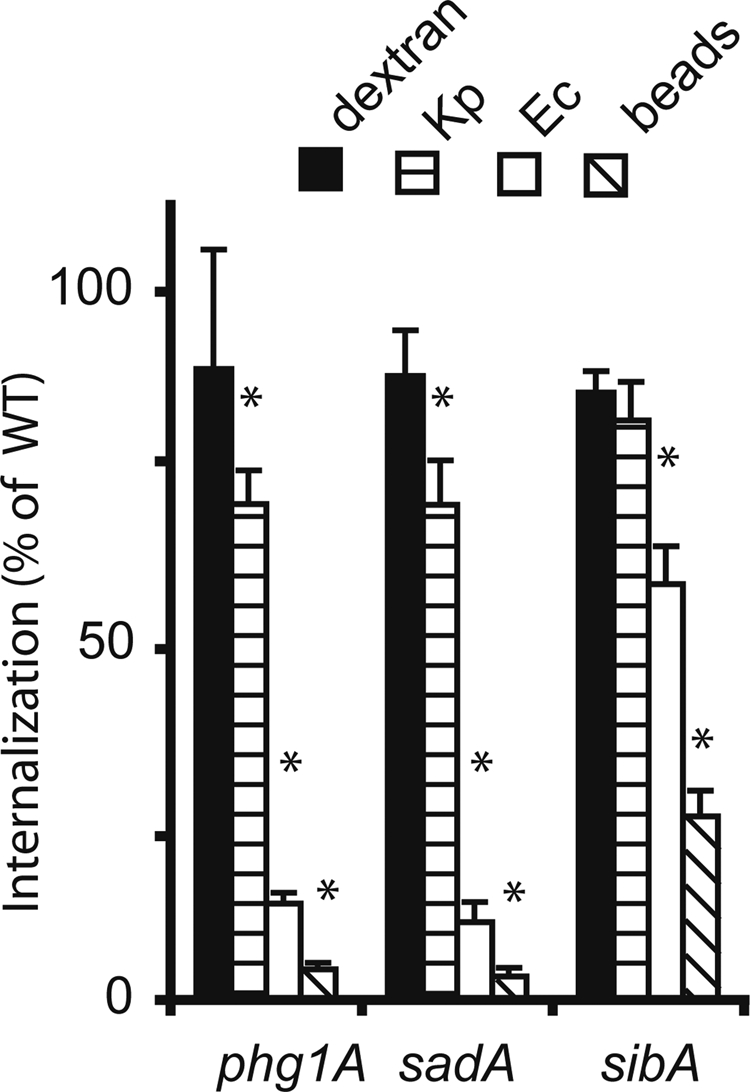
Phagocytosis defects in phg1A, sadA, and sibA knockout cells. Wild-type or mutant Dictyostelium cells were incubated for 20 min in HL5 medium containing either fluorescent phagocytic particles (latex beads [beads], K. pneumoniae [Kp], or E. coli [Ec] bacteria) or a fluorescent fluid-phase marker (dextran). The internalized fluorescence was analyzed by flow cytometry and expressed as a percentage of internalization in wild-type cells. Qualitatively, phg1A, sadA, and sibA mutants presented similar phenotypes compared with wild-type cells: virtually normal uptake of fluid phase and of K. pneumoniae, diminished phagocytosis of E. coli, and minimal phagocytosis of latex beads. The average and SEM of at least four experiments are indicated. *, significantly different from wild-type (p < 0.05 with Student's t test).
Phg1A and SadA control surface levels of SibA
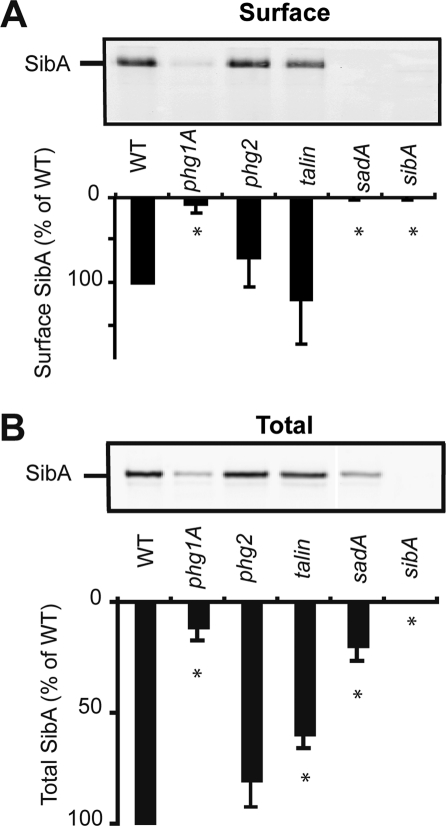
To assess the possible involvement of Phg1A and SadA in controlling surface levels of SibA, we biotinylated the surface of wild-type or mutant cells, immunoprecipitated SibA, and revealed it with horseradish peroxidase (HRP)-coupled avidin. As in sibA knockout cells, SibA was virtually absent from the surface of phg1A and sadA knockout cells (Figure 2A). The surface levels of SibA were not significantly affected in talin or phg2 mutant cells compared with wild-type cells (Figure 2A).
FIGURE 2:
Phg1A and SadA control surface expression of SibA. (A) To assess the presence of SibA at the cell surface, we surface-biotinylated and then lysed wild-type and mutant cells. SibA was purified by immunoprecipitation, migrated on a 7% polyacrylamide gel, and revealed with HRP-coupled avidin. While SibA was readily detectable at the surface of wild-type, talin, or phg2 cells, it was virtually absent from the surface of sibA, phg1A, and sadA mutant cells. (B) To determine the total cellular amount of SibA, we separated cellular proteins by electrophoresis and revealed SibA with a specific antibody. (lane 10): The amount of SibA was quantified and expressed as the percentage of the amount observed in wild-type cells. The average and SEM of four independent experiments are indicated. *, significantly different from wild-type (p < 0.05 with Student's t test).
To assess the total amount of SibA, we used gel electrophoresis to separate whole-cell lysates, transferred them to nitrocellulose, and revealed SibA with anti-SibA antibodies. The total amount of SibA in both phg1A and sadA mutant cells was significantly reduced compared with wild-type cells, whereas only relatively minor differences were seen in talin or phg2 mutant cells (Figure 2B). These results suggest the absence of SibA from the surface of phg1A and sadA mutant cells is at least partly due to a decrease in the total amount of cellular SibA. In addition, the SibA surface-to-total ratio was markedly lower in phg1A and in sadA mutant cells (0.27 and 0.0, respectively) than in wild-type cells (1.0), suggesting the intracellular distribution of SibA may be affected in phg1A and sadA mutant cells.
To further test whether loss of SibA accounted for the adhesion defect observed in phg1A mutant cells, we assessed the effect of temperature on cellular levels of SibA. Previous experiments have shown that the adhesion defect of phg1A mutant cells is significantly increased at 26°C compared with 20°C (Benghezal et al., 2003). We observed that the levels of SibA were indeed significantly lower at 26°C than at 20°C (Figure 3). It was also shown previously that phg1B mutant cells exhibit a (modest) adhesion defect only at 26°C. In agreement with this, cellular levels of SibA in phg1B mutant cells were not affected at 20°C, but were significantly lower at 26°C (Figure 3).
FIGURE 3:
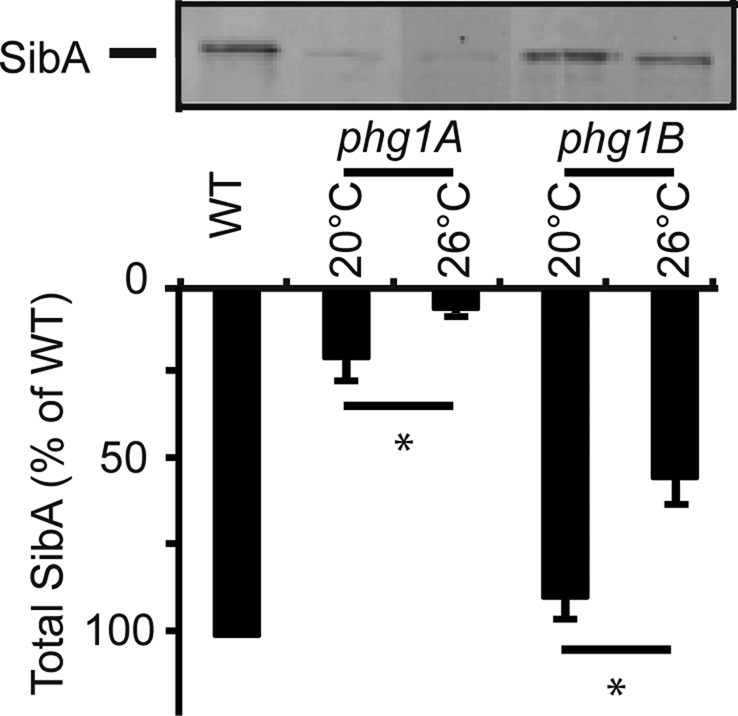
Phg1A and Phg1B control cellular levels of SibA in a temperature-dependent manner. Wild-type or mutant (phg1A or phg1B) cells were grown at the indicated temperature for 24 h. The total cellular amount of SibA was determined by Western blot, as described in the legend to Figure 2. The average and SEM of four independent experiments are indicated. *, significantly different from wild-type (p < 0.05 with Student's t test).
Phg1A and SadA control production and stability of SibA
Decreased cellular levels of SibA could be due to a decrease in its production, to an increased turnover, or to a combination of both. To evaluate these two possibilities, we first measured by quantitative reverse transcriptase PCR (qRT-PCR) the level of sibA mRNA in various cell lines to provide an indication of sibA mRNA expression level. We observed a statistically significant decrease in the amount of sibA mRNA in phg1A mutant cells relative to wild-type cells (37% of wild-type level) (Figure 4). Less prominent and not statistically significant alterations in sibA mRNA levels were observed in sadA mutant cells (72% of wild-type, p > 0.1) and in other mutant cells analyzed (Figure 4).
FIGURE 4:
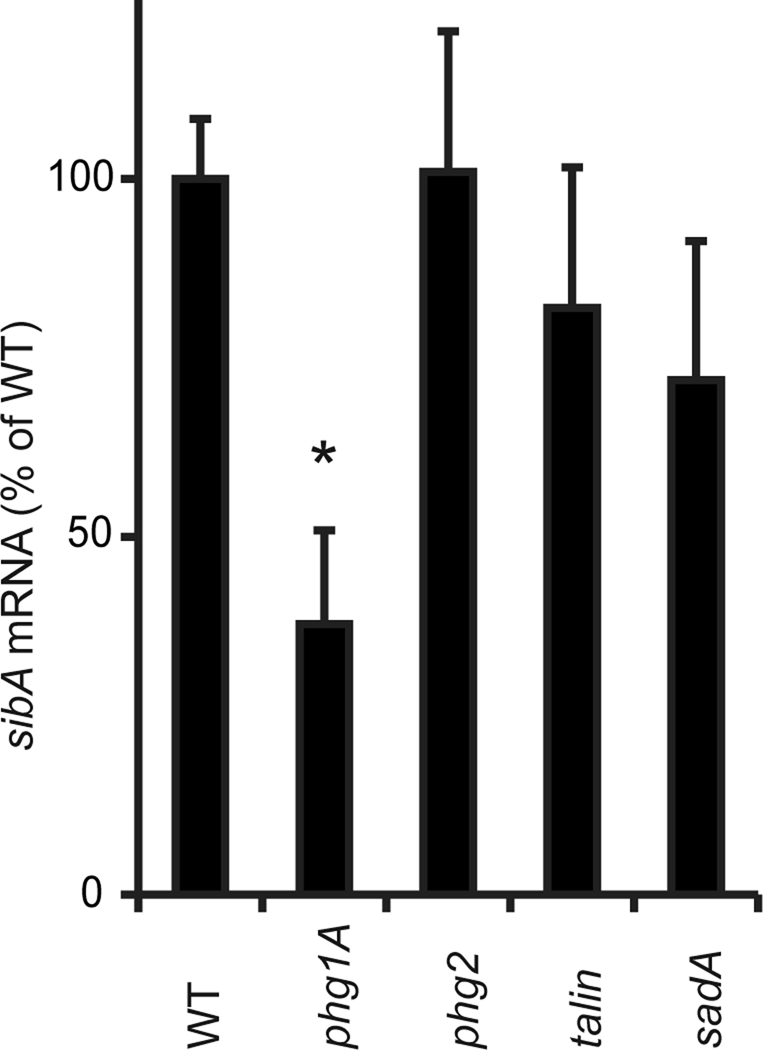
SibA mRNA levels are decreased in phg1A mutant cells. The amount of sibA mRNA was determined in mutant cells by RT-PCR and expressed as a percentage of the level measured in wild-type cells. The average and SEM of three independent experiments are indicated. *, significantly different from wild-type (p < 0.05 with Student's t test).
To measure the stability of the SibA protein, we incubated cells in the presence of cycloheximide to inhibit protein synthesis. At various times, aliquots of the cells were collected, and the remaining SibA was revealed by immunoblotting (Figure 5). In wild-type cells, SibA was relatively stable (70% remaining after 2 h of chase). In contrast, SibA was more rapidly degraded in phg1A mutant cells, and even more so in sadA mutant cells (37 and 13% remaining after 2 h of chase, respectively; Figure 5).
FIGURE 5:
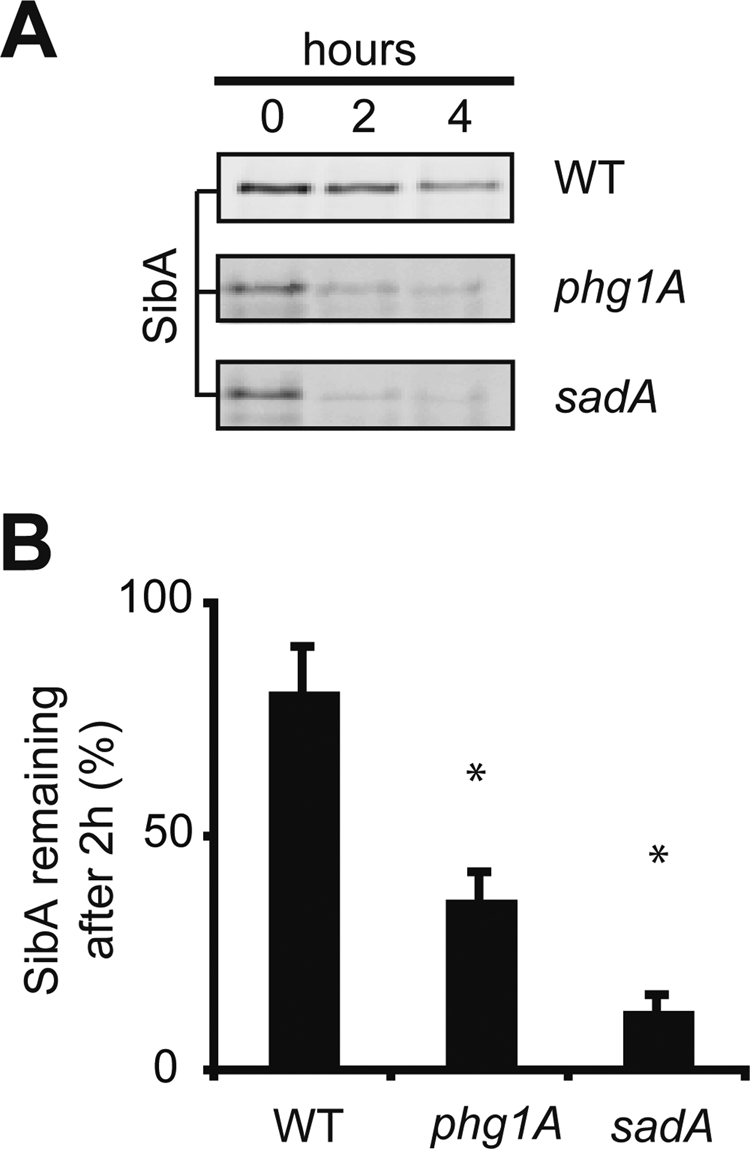
The SibA protein is less stable in phg1A and sadA mutant cells than in wild-type cells. (A) Wild-type and mutant cells were incubated in the presence of cycloheximide for 0, 2, or 4 h and then lysed; the remaining SibA was detected by Western blotting after gel electrophoresis. To obtain equivalent signals at time zero, 106 cells were loaded per lane for phg1A and sadA mutant cells and 105 for wild-type cells. (B) The signals were quantified to determine the percentage of SibA remaining after 2 h of chase. The average and SEM of six independent experiments are indicated. *, significantly different from wild-type (p < 0.05 with Student's t test).
These results indicate that Phg1A and SadA control the total cellular amount of SibA by influencing its turnover and, at least in the case of Phg1A, its production.
Phg1A controls the sorting of SibA in the endocytic pathway
The decreased stability of SibA in phg1A and sadA mutant cells may be indicative of perturbed intracellular targeting. Endogenous SibA is present in such relatively small amounts in Dictyostelium cells that it is not detectable by immunofluorescence. To circumvent this limitation, we constructed a plasmid encoding a chimeric protein (csA-SibA) composed of the extracellular domain of the Dictyostelium adhesion molecule contact site A (csA) linked to the transmembrane and cytosolic domains of SibA (Figure 6A). We then introduced this plasmid in wild-type or phg1A mutant cells. SadA mutant cells were not analyzed further in this study, because they proved difficult to grow and to transfect, due to marked defects in their actin cytoskeleton (Fey et al., 2002).
FIGURE 6:
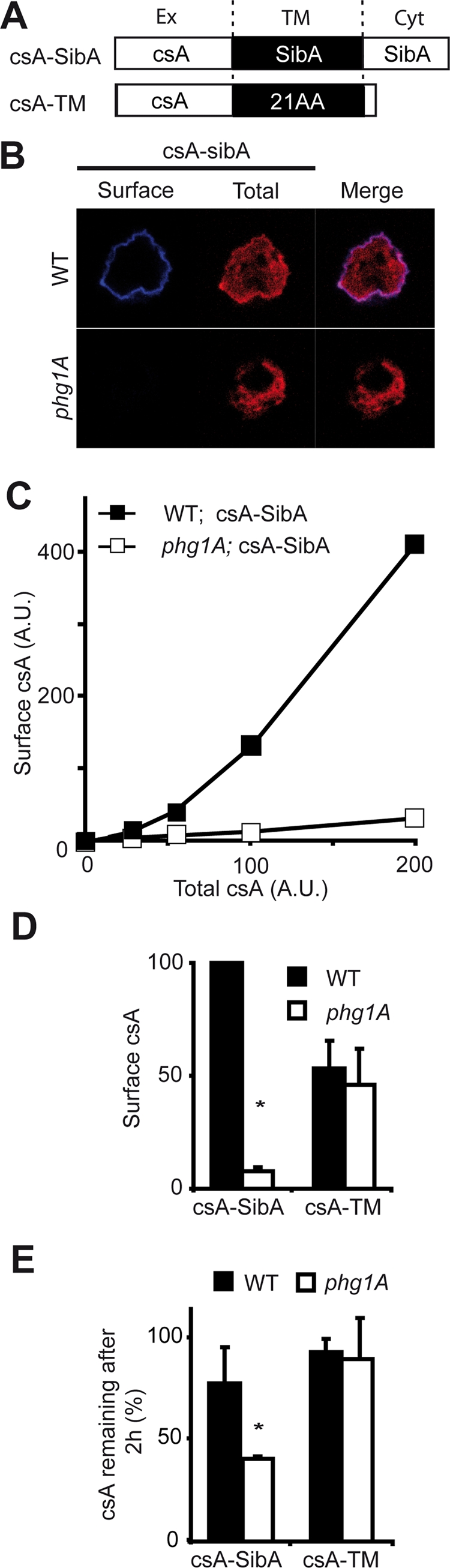
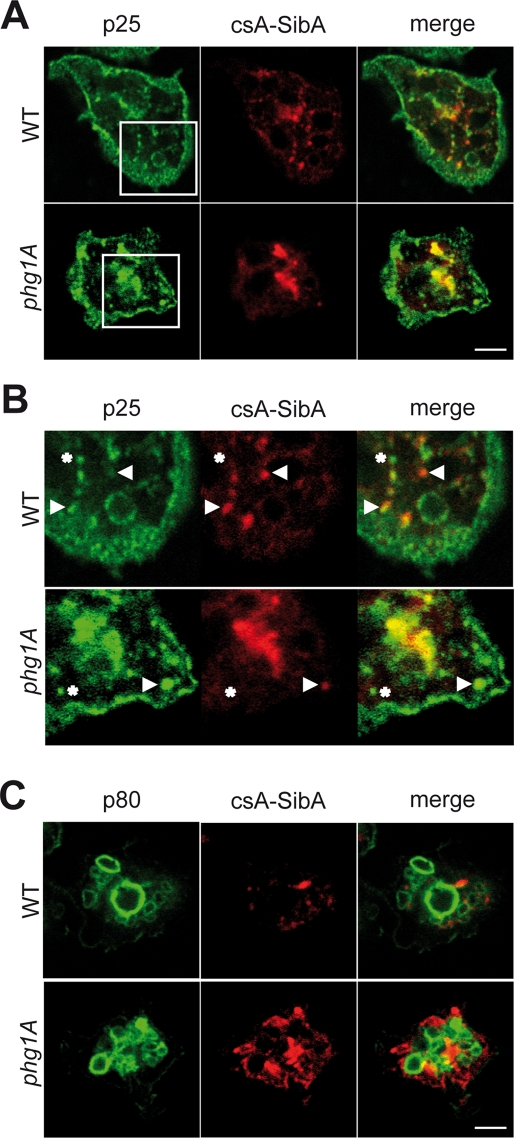
Intracellular sequestration of csA-SibA in phg1A mutant cells. (A) A chimeric protein comprising the csA extracellular domain fused to the transmembrane and cytosolic domains of SibA (csA-SibA) was expressed in wild-type and in phg1A mutant cells. As a control, a csA protein fused to a classical hydrophobic transmembrane domain was used (csA-TM). (B) csA-SibA was labeled by immunofluorescence before (Surface, blue) and after (Total, red) permeabilization in transfected wild-type and phg1A mutant cells, using an antibody specific for the csA extracellular domain. The pictures presented were obtained with a confocal microscope in consecutive sequence and with identical settings. (C) The surface and total csA was labeled as described in (B) for wild-type and phg1A mutant cells expressing csA-SibA. The fluorescence was quantified by flow cytometry. (D) The average and SEM of seven independent flow cytometry experiments (described in C) are represented in this bar graph. The surface csA value used was that corresponding to a total csA expression of 200 in (C). (E) Stability of csA-SibA and csA-TM in wild-type and phg1A mutant cells. The stability of the protein was determined during a chase in the presence of cycloheximide, as described in the legend to Figure 5. CsA-SibA was less stable in phg1A mutant cells than in wild-type cells, while the stability of csA-TM was comparable in both cells. The average and SEM of four independent experiments are indicated. *, significantly different from wild-type (p < 0.05 with Student's t test).
As a control, we constructed and expressed in wild-type and phg1A knockout cells a second chimeric csA protein anchored in the membrane by a classical 21-amino acid hydrophobic transmembrane domain followed by a very short cytosolic domain (csA-TM; Figure 6A).
csA-SibA was detected by immunofluorescence at the cell surface, as well as in intracellular compartments in wild-type cells (Figure 6B). In contrast, surface csA-SibA was undetectable in phg1A mutant cells (Figure 6B). To confirm this result in a more quantitative manner, we analyzed labeled cells by flow cytometry to simultaneously measure the levels of csA protein at the surface and inside individual cells. This analysis confirmed that phg1A knockout cells presented at their surface a much smaller portion of total csA-SibA than wild-type cells (Figure 6C). We observed a very similar behavior for a csA-SibC chimera in which the csA extracellular domain was fused to the transmembrane and cytosolic domains of SibC: a greater amount was expressed at the surface of wild-type than of phg1A mutant cells (respectively: 81 ± 19 and 18 ± 2; n = 6; p = 0.04). In contrast, csA-TM was expressed at similar levels at the surface of both cell types (Figure 6D).
Finally, like SibA, csA-SibA was less stable in phg1A mutant cells than in wild-type cells, as revealed by chase experiments in the presence of cycloheximide (Figure 6E). Taken together these results indicate that csA-SibA, like SibA, is excluded from the cell surface and presumably targeted for intracellular degradation in phg1A mutant cells.
To determine where phg1A-dependent intracellular sorting occurred, we assessed with immunofluorescence the localization of the csA-SibA chimera in wild-type and in phg1A mutant cells (Figure 7). We detected csA-SibA in small compartments in both cell types, sometimes grouped in a juxtanuclear region, a disposition reminiscent of previously characterized early endocytic compartments (Charette et al., 2006). Indeed, all compartments containing csA-SibA also contained p25, a marker present at the cell surface, as well as in early and recycling endosomes in Dictyostelium (Charette et al., 2006; Figure 7, A and B). However, some of the p25-positive compartments did not contain detectable levels of csA-SibA, suggesting csA-SibA was restricted to a subpopulation of early and recycling endosomes. CsA-SibA was not detected in more mature lysosomal compartments labeled with an anti-p80 antibody in either wild-type (Figure 7C) or phg1A mutant cells (unpublished data). As shown previously, p80 is present at the cell surface and in lysosomes, but virtually absent from recycling endosomes (Ravanel et al., 2001). Note that in these experiments, permeabilization resulted in weaker cell surface staining not readily detectable by immunofluorescence. These experiments indicate that in both wild-type and phg1A knockout cells, csA-SibA is present at the cell surface and in early endocytic compartments, but that cell surface levels of csA-SibA are much lower in phg1A knockout cells than in wild-type cells.
FIGURE 7:
Intracellular csA-SibA is localized in early endocytic compartments. Wild-type and phg1A mutant cells expressing csA-SibA were fixed and permeabilized, and intracellular csA-SibA was detected with a rabbit antibody recognizing the SibA cytosolic domain, and a fluorescent secondary antibody (red). In the same cells, p25 (A and B) or p80 (C) were detected with specific mouse antibodies (green). (B) An enlarged portion of (A). In both wild-type and phg1A cells, csA-SibA was present in intracellular compartments positive for p25 (arrowheads) but not for p80. Some p25-positive compartments did not contain csA-SibA (asterisks), suggesting that csA-SibA is sorted differently from p25 in the early endocytic pathway. Note that in these experiments, surface csA-SibA is difficult to detect due to cell permeabilization.
Absence of SibA-associated proteins
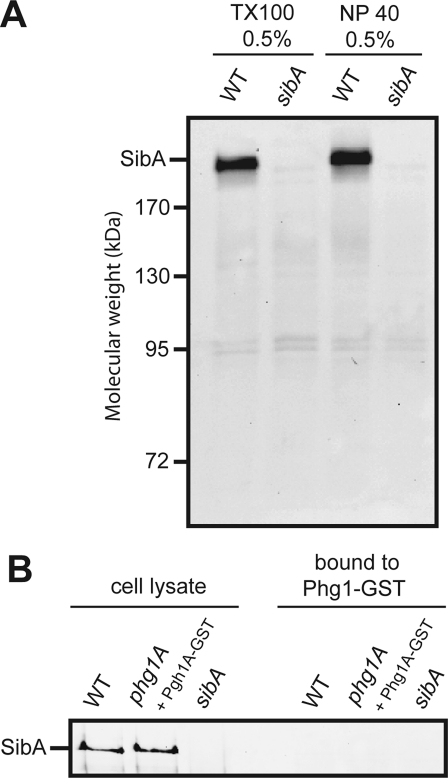
Phg1A may control the sorting and stability of SibA via direct interactions or via a more general effect on sorting and transport in the endocytic pathway. To distinguish between these two possibilities, we designed two distinct experiments to detect direct interaction between Phg1A and SibA. First, we labeled cell surface proteins with biotin, lysed the cells, immunoprecipitated SibA, and probed for coprecipitated biotinylated proteins with HRP-coupled avidin (Figure 8A). No associated proteins were detected, which suggests that there may not be a stable association of SibA with another chain (equivalent to the α chain of integrins) or with Phg1A or SadA. This method, however, would not detect an association of Phg1A with SibA occurring in an intracellular compartment. Therefore, in a second experiment, we used cells expressing Phg1A tagged with a glutathione S-transferase (GST) at its C-terminus, a fusion protein that was shown previously to fully complement the defects of a phg1A mutant cell (Benghezal et al., 2003). We then purified Phg1A-GST using a glutathione-coupled resin, and attempted to detect associated SibA by Western blotting (Figure 8B). We did not detect any SibA coprecipitated with Phg1A-GST, even when material precipitated from 30 × 106 cells was loaded on a single lane. Although these results suggest that the Phg1A and SibA proteins do not associate stably in living cells, they do not rule out a transient or an unstable interaction between these two proteins.
FIGURE 8:
No detectable association between Phg1A and SibA. (A) Cell surface proteins were biotinylated in wild-type or sibA mutant cells. Cells were lysed in the presence of either Triton X-100 or Nonidet P40, and SibA was immunoprecipitated. Proteins precipitated from 107 cells were separated on a polyacrylamide gel and revealed with HRP-coupled avidin. No protein specifically coprecipitating with SibA was detected. (B) Phg1A-GST was expressed in phg1A mutant cells and precipitated in the presence of 0.5% Triton X-100. The proteins were separated by electrophoresis and SibA was revealed with a specific antibody. As a control, wild-type or sibA cells were used. Cellular lysates (0.5 × 106 cells), as well as precipitated samples (107 cells), were analyzed. No detectable SibA was coprecipitated with Phg1A-GST.
DISCUSSION
The results presented in this study indicate that Phg1A and SadA participate in cellular adhesion by controlling the levels of SibA adhesion molecules present at the cell surface. Phg1A and SadA achieve this effect by controlling to various extents the levels of sibA mRNA, the stability of the SibA protein, and its targeting to the cell surface. In the absence of Phg1A, SibA is less abundantly produced, less stable, and excluded from the cell surface. A similar pattern is observed in sadA mutant cells, in which the stability and surface targeting of SibA are strongly defective. By establishing connections among Phg1A, SadA, and SibA, these results provide a more complete picture of the adhesion machinery in Dictyostelium, one that is centered on the function of SibA adhesion molecules. Indeed, the major gene products shown to be essential for efficient adhesion now have a clear link with SibA: talin and myosin VII form a complex (Gebbie et al., 2004; Tuxworth et al., 2005) that binds the cytosolic domain of SibA (Cornillon et al., 2006), while Phg1A and SadA control expression of SibA at the cell surface (this study).
It is surprising to observe that genetic inactivation of sadA or phg1A would yield such similar phenotypes in mutant cells, since the corresponding proteins show no clear homology. On the other hand, both proteins share the same general topology with a large N-terminal extracellular domain and nine C-terminal transmembrane domains. Although the classical definition of TM9 proteins, based on sequence conservation (Schimmoller et al., 1998), would not include SadA, it is possible that more subtle structural features are conserved between Phg1A and SadA proteins. A more advanced knowledge of the structure and function of TM9 proteins may allow us in the future to delineate an extended TM9 superfamily that would include SadA and possibly other proteins. As our knowledge of TM9 proteins increases, we will presumably discover whether the structural and functional similarities between SadA and Phg1A are significant or merely coincidental.
The overall organization of the adhesion machinery in Dictyostelium and in mammalian cells is remarkably similar. SibA proteins and integrin β chains also share a number of structural and functional features (Cornillon et al., 2006). It is thus tempting to speculate that TM9 proteins in mammalian cells may also control surface expression of proteins involved in cellular adhesion, particularly integrins. In addition, Phg1A has also been implicated in the control of adhesion in yeast cells (Froquet et al., 2008) and Drosophila phagocytic cells (Bergeret et al., 2008), which use very different sets of adhesion molecules. We speculate that a connection will be established between TM9/Phg1 proteins and surface expression of key adhesion molecules in these various systems.
MATERIALS AND METHODS
Cell culture and mutant cells
D. discoideum strains were grown in HL5 medium at 23°C and subcultured twice a week to maintain a maximal density of 106 cells/ml. All mutant strains used in this study were derived from the subclone DH1–10 (Cornillon et al., 2000) of the parental axenic Dictyostelium strain DH1 (Caterina et al., 1994). For simplicity, DH1–10 cells are referred to as wild-type cells. The phg1A (Cornillon et al., 2000), sibA (Cornillon et al., 2006), and talin (Gebbie et al., 2004) mutant strains have been described previously. The sadA knockout mutants were generated by using the knockout plasmids previously described (Fey et al., 2002). In addition to their adhesion defects, sadA knockout mutants presented a huge cytokinesis defect (unpublished data), as described previously (Fey et al., 2002).
Expression of csA chimera
The extracellular domain of csA was amplified from genomic DNA using the two following oligonucleotides: GATACAGTTAACAAAATGAAATTTTTATTAGTATTGATAATATTATA and CTTTTCGGTACCAGTTTCTTCTGGTGTGCTGGTTG. The PCR fragment was digested with HpaI and KpnI and cloned into prepSC3 (a pDXA-3C plasmid with a modified HindIII-HpaI-KpnI-NotI-XbaI multiple-cloning site [ Manstein et al., 1995]) digested with HpaI-KpnI, creating the csA-0 intermediate construct. The csA-TM construct was obtained by amplifying the sequence coding for the transmembrane domain of CD1b-TM3 (21 aa; Mercanti et al., 2010) and subcloning it in csA-0, using the KpnI and XbaI sites. The amino acid sequence of the transmembrane domain of csA-TM is: ESDSIVLAIIVPSLLLLLCLALLWYMRRRSM.
To construct csA-SibA, we amplified the transmembrane and cytoplasmic domains of SibA (from Tyr-1850 to stop codon) by PCR and cloned them into csA-0.
Internalization assays
Phagocytosis and fluid-phase uptake were determined by shaking the cells for 20 min at 21°C in HL5 medium with either 1-μm-diameter Fluoresbrite YG carboxylate microspheres (Polysciences, Warrington, PA), rhodamine-labeled K. pneumoniae (Cornillon et al., 2000), rhodamine-labeled E. coli, or 10 μg/ml of Alexa Fluor 647–labeled dextran (Molecular Probes, Eugene, OR), and measuring the internalized fluorescence in a fluorescence-activated cell sorter (FACSCalibur, Becton Dickinson, San Jose, CA) after two washes with HL5 containing 0.1% sodium azide, as described previously (Cornillon et al., 2000).
Western blot analysis
To detect SibA at the cell surface, we cooled cells to 4°C and then pelleted and washed them twice with ice-cold phosphate-buffered saline (PBS; pH 7.3). The cells were then resuspended and incubated for 30 min in ice-cold PBS containing 0.25 mg/ml sulfo-succinimido-biotin (Molecular Biosciences, Boulder, CO), washed twice with ice-cold PBS containing 40 mM NH4Cl, and then lysed on ice for 15 min in lysis buffer (PBS, 0.5% Triton X-100, 5 mg/ml iodoacetamide, 100 μm phenylmethylsulfonylfluoride, 1.5 μM aprotinin, and 47 μM leupeptin). After the nuclei were pelleted (9300 × g at 4°C for 5 min), the supernatant was first incubated for 15 min with protein G Sepharose beads (GE Healthcare, Sweden) and then for 1 h in the presence of a polyclonal anti-SibA antibody (Cornillon et al., 2006) coupled to protein G Sepharose. Beads were washed five times with PBS containing 0.1% Triton X-100 and resuspended in sample buffer (0.103 g/ml sucrose, 5 × 10−2 M Tris, pH 6.8, 5 × 10−3 M EDTA, 0.5 mg/ml bromophenol blue, 2% SDS). Biotinylated proteins were separated on a 7% polyacrylamide gel, incubated with HRP-coupled avidin (Bio-Rad, Hercules, CA), and revealed by enhanced chemiluminescence (ECL; Amersham Biosciences, GE Healthcare, Waukesha, WI).
To determine the total amount of cellular SibA, we resuspended cell pellets in sample buffer and separated the proteins on a 7% polyacrylamide gel, after which they were transferred to a nitrocellulose membrane (Invitrogen, Carlsbad, CA). The membranes were incubated with a polyclonal anti-SibA antibody (Cornillon et al., 2006) and then with an HRP-coupled donkey anti-rabbit immunoglobulin (Ig) G (Bio-Rad), washed, and revealed by ECL (Amersham Biosciences). The amount of both surface and total cellular SibA protein was evaluated using Quantity One software (Bio-Rad). As a control, serial dilutions of the wild-type sample confirmed the existence of a linear relationship between the measured signal and the amount of protein (unpublished data). Nitrocellulose membranes were stained with Ponceau red to ascertain that similar amounts of proteins were loaded in each lane.
To measure the stability of SibA or csA-SibA proteins, we incubated 2 × 106 Dictyostelium cells for 0, 2, or 4 h in 2 ml HL5 containing 2 mM cycloheximide. At each time point, an aliquot of the cells was collected, pelleted, and resuspended in sample buffer, and the total amount of cellular SibA was revealed by Western blotting, as described above. To obtain equivalent signals at time zero, 10 times more cells were loaded per lane for phg1A and sadA mutant cells than for wild-type cells (106 vs. 105 cells per lane, respectively).
Real-time PCR
As described previously (Cornillon et al., 2008), wild-type or mutant cells were grown in HL5 medium for 3 d to a final density of 106 cells/ml. The cells were harvested, RNAs were purified with a NucleoSpin RNA II kit (Macherey-Nagel, Duren, Germany), and real-time PCR was performed using sibA-specific oligonucleotides (forward: 5′-CCAACTCCGACAGATGGTTCTC-3′; reverse: 5′-AAAGAATTTGTTTTGTTTACTTCATATGCA-3′). Raw Ct values obtained with SDS 2.2 (Applied Biosystems, Bedford, MA) were imported into Excel (Microsoft, Redmond, WA), and normalization factors and fold changes were calculated. The Ig7 gene (mitochondrial large-subunit rRNA) was used to normalize PCRs. The background measured in sibA knockout cells was subtracted from all other results.
Immunofluorescence
To localize intracellular csA-sibA, we performed immunofluorescence on cells attached to glass coverslips. To elicit efficient adhesion of wild-type and phg1A cells, we allowed cells to adhere for 10 min to coverslips in phosphate buffer (2 mM Na2HPO4, 14.7 mM KH2PO4, pH 6.0, supplemented with 0.5% HL5, 100 mM sorbitol, and 100 μM CaCl2). This buffer preserves the morphology of endocytic compartments (Smith et al., 2010), while allowing cells to attach to their substrate in a Phg1A-independent manner (Gebbie et al., 2004). We then performed immunofluorescence on fixed cells as previously described (Mercanti et al., 2006), using a rabbit anti-SibA to detect csA-SibA (Cornillon et al., 2006) and a mouse anti-p25 (H72) to detect p25 (Charette et al., 2006).
To detect csA-TM or csA-SibA by surface immunofluorescence in unfixed cells, we used a mouse monoclonal antibody (41-71-21) directed to the csA extracellular domain (Bertholdt et al., 1985). Since incubations at 4°C were found to affect cell morphology, the cell surface was labeled with this antibody for 1 min at room temperature. Cells were then rinsed twice rapidly and fixed with 4% paraformaldehyde in 100 mM phosphate buffer (pH 7.4), incubated with an anti-mouse antibody coupled to Alexa Fluor 647, and permeabilized with methanol at −20°C. Permeabilized cells were then further incubated sequentially with anti-csA antibody and a rhodamine-coupled anti-mouse antibody to reveal intracellular csA. Cells were observed with a LSM 510 Zeiss confocal microscope (Jena, Germany). Alternatively, levels of csA-sibA and csA-TM at the cell surface and inside the cell were measured by flow cytometry (FACSCalibur) after the cells were detached from coverslips.
Coprecipitation experiments
To detect proteins associated with SibA at the cell surface, we biotinylated proteins present at the cell surface, as described in Western blot analysis. After lysis and immunoprecipitation of SibA, the material precipitated from 107 cells was loaded on each lane of an 8% polyacrylamide gel. After transfer to a nitrocellulose membrane, HRP-coupled avidin was used to reveal proteins coprecipitated with SibA.
In another attempt to detect an association of Phg1A and intracellular SibA, we expressed Phg1A-GST in phg1A mutant cells and purified it on glutathione beads after cell lysis, as previously described (Benghezal et al., 2006). Proteins precipitated from 107 cells were separated on an 8% polyacrylamide gel and SibA was revealed by Western blotting, as described in Western blot analysis.
Supplementary Material
Acknowledgments
This work was supported by grants from the Swiss National Science Foundation (31003A-135789 to P.C.) and from the Doerenkamp-Zbinden and the FENRIV foundations. We thank Prashant Nair for his participation in the initial experiments and Thierry Soldati and Oliver Hartley for critical reading of the manuscript.
Abbreviations used:
- csA
contact site A
- ECL
enhanced chemiluminescence
- GST
glutathione S-transferase
- HRP
horseradish peroxidase
- Ig
immunoglobulin
- PBS
phosphate-buffered saline
- qRT-PCR
quantitative reverse transcriptase PCR
- TM9SF
transmembrane 9 superfamily
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-04-0338) on January 4, 2012.
REFERENCES
- Bagshaw RD, Mahuran DJ, Callahan JW. A proteomic analysis of lysosomal integral membrane proteins reveals the diverse composition of the organelle. Mol Cell Proteomics. 2005;4:133–143. doi: 10.1074/mcp.M400128-MCP200. [DOI] [PubMed] [Google Scholar]
- Benghezal M, Cornillon S, Gebbie L, Alibaud L, Bruckert F, Letourneur F, Cosson P. Synergistic control of cellular adhesion by transmembrane 9 proteins. Mol Biol Cell. 2003;14:2890–2899. doi: 10.1091/mbc.E02-11-0724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benghezal M, et al. Specific host genes required for the killing of Klebsiella bacteria by phagocytes. Cell Microbiol. 2006;8:139–148. doi: 10.1111/j.1462-5822.2005.00607.x. [DOI] [PubMed] [Google Scholar]
- Bergeret E, Perrin J, Williams M, Grunwald D, Engel E, Thevenon D, Taillebourg E, Bruckert F, Cosson P, Fauvarque MO. TM9SF4 is required for Drosophila cellular immunity via cell adhesion and phagocytosis. J Cell Sci. 2008;121:3325–3334. doi: 10.1242/jcs.030163. [DOI] [PubMed] [Google Scholar]
- Bertholdt G, Stadler J, Bozzaro S, Fichtner B, Gerisch G. Carbohydrate and other epitopes of the contact site A glycoprotein of Dictyostelium discoideum as characterized by monoclonal antibodies. Cell Differ. 1985;16:187–202. doi: 10.1016/0045-6039(85)90516-0. [DOI] [PubMed] [Google Scholar]
- Caterina MJ, Milne JL, Devreotes PN. Mutation of the third intracellular loop of the cAMP receptor, cAR1, of Dictyostelium yields mutants impaired in multiple signaling pathways. J Biol Chem. 1994;269:1523–1532. [PubMed] [Google Scholar]
- Charette SJ, Mercanti V, Letourneur F, Bennett N, Cosson P. A role for adaptor protein-3 complex in the organization of the endocytic pathway in Dictyostelium. Traffic. 2006;7:1528–1538. doi: 10.1111/j.1600-0854.2006.00478.x. [DOI] [PubMed] [Google Scholar]
- Cornillon S, Froquet R, Cosson P. Involvement of Sib proteins in the regulation of cellular adhesion in Dictyostelium discoideum. Eukaryot Cell. 2008;7:1600–1605. doi: 10.1128/EC.00155-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornillon S, Gebbie L, Benghezal M, Nair P, Keller S, Wehrle-Haller B, Charette SJ, Bruckert F, Letourneur F, Cosson P. An adhesion molecule in free-living Dictyostelium amoebae with integrin β features. EMBO Rep. 2006;7:617–621. doi: 10.1038/sj.embor.7400701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornillon S, Pech E, Benghezal M, Ravanel K, Gaynor E, Letourneur F, Bruckert F, Cosson P. Phg1p is a nine-transmembrane protein superfamily member involved in Dictyostelium adhesion and phagocytosis. J Biol Chem. 2000;275:34287–34292. doi: 10.1074/jbc.M006725200. [DOI] [PubMed] [Google Scholar]
- Cosson P, Soldati T. Eat, kill or die: when amoeba meets bacteria. Curr Opin Microbiol. 2008;11:271–276. doi: 10.1016/j.mib.2008.05.005. [DOI] [PubMed] [Google Scholar]
- Cougoule C, Wiedemann A, Lim J, Caron E. Phagocytosis, an alternative model system for the study of cell adhesion. Semin Cell Dev Biol. 2004;15:679–689. doi: 10.1016/j.semcdb.2004.09.001. [DOI] [PubMed] [Google Scholar]
- Fey P, Stephens S, Titus MA, Chisholm RL. SadA, a novel adhesion receptor in Dictyostelium. J Cell Biol. 2002;159:1109–1119. doi: 10.1083/jcb.200206067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froquet R, Cherix N, Birke R, Benghezal M, Cameroni E, Letourneur F, Mosch HU, De Virgilio C, Cosson P. Control of cellular physiology by TM9 proteins in yeast and Dictyostelium. J Biol Chem. 2008;283:6764–6772. doi: 10.1074/jbc.M704484200. [DOI] [PubMed] [Google Scholar]
- Gebbie L, et al. Phg2, a kinase involved in adhesion and focal site modeling in Dictyostelium. Mol Biol Cell. 2004;15:3915–3925. doi: 10.1091/mbc.E03-12-0908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood JD, Cheresh DA. Role of integrins in cell invasion and migration. Nat Rev Cancer. 2002;2:91–100. doi: 10.1038/nrc727. [DOI] [PubMed] [Google Scholar]
- Lozupone F, et al. The human homologue of Dictyostelium discoideum phg1A is expressed by human metastatic melanoma cells. EMBO Rep. 2009;10:1348–1354. doi: 10.1038/embor.2009.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manstein DJ, Schuster HP, Morandini P, Hunt DM. Cloning vectors for the production of proteins in Dictyostelium discoideum. Gene. 1995;162:129–134. doi: 10.1016/0378-1119(95)00351-6. [DOI] [PubMed] [Google Scholar]
- Mercanti V, Charette SJ, Bennett N, Ryckewaert JJ, Letourneur F, Cosson P. Selective membrane exclusion in phagocytic and macropinocytic cups. J Cell Sci. 2006;119:4079–4087. doi: 10.1242/jcs.03190. [DOI] [PubMed] [Google Scholar]
- Mercanti V, Marchetti A, Lelong E, Perez F, Orci L, Cosson P. Transmembrane domains control exclusion of membrane proteins from clathrin-coated pits. J Cell Sci. 2010;123:3329–3335. doi: 10.1242/jcs.073031. [DOI] [PubMed] [Google Scholar]
- Ravanel K, de Chassey B, Cornillon S, Benghezal M, Zulianello L, Gebbie L, Letourneur F, Cosson P. Membrane sorting in the endocytic and phagocytic pathway of Dictyostelium discoideum. Eur J Cell Biol. 2001;80:754–764. doi: 10.1078/0171-9335-00215. [DOI] [PubMed] [Google Scholar]
- Schimmoller F, Diaz E, Muhlbauer B, Pfeffer SR. Characterization of a 76 kDa endosomal, multispanning membrane protein that is highly conserved throughout evolution. Gene. 1998;216:311–318. doi: 10.1016/s0378-1119(98)00349-7. [DOI] [PubMed] [Google Scholar]
- Singer-Kruger B, Frank R, Crausaz F, Riezman H. Partial purification and characterization of early and late endosomes from yeastIdentification of four novel proteins. J Biol Chem. 1993;268:14376–14386. [PubMed] [Google Scholar]
- Smith EW, Lima WC, Charette SJ, Cosson P. Effect of starvation on the endocytic pathway in Dictyostelium cells. Eukaryot Cell. 2010;9:387–392. doi: 10.1128/EC.00285-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuxworth RI, Stephens S, Ryan ZC, Titus MA. Identification of a myosin VII-talin complex. J Biol Chem. 2005;280:26557–26564. doi: 10.1074/jbc.M503699200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.