ETOC: Ribosome synthesis is a multistep process initiated in the nucleolus with the transcription of a precursor rRNA that is subjected to a series of modification and processing steps to generate the mature rRNA. In this paper, we describe a novel 60S ribosome biogenesis complex associating with LAS1L that controls rRNA processing and synthesis of the 28S rRNA.
Abstract
The coordination of RNA polymerase I transcription with pre-rRNA processing, preribosomal particle assembly, and nuclear export is a finely tuned process requiring the concerted actions of a number of accessory factors. However, the exact functions of some of these proteins and how they assemble in subcomplexes remain poorly defined. LAS1L was first described as a nucleolar protein required for maturation of the 60S preribosomal subunit. In this paper, we demonstrate that LAS1L interacts with PELP1, TEX10, and WDR18, the mammalian homologues of the budding yeast Rix1 complex, along with NOL9 and SENP3, to form a novel nucleolar complex that cofractionates with the 60S preribosomal subunit. Depletion of LAS1L-associated proteins results in a p53-dependent G1 arrest and leads to defects in processing of the pre-rRNA internal transcribed spacer 2 region. We further show that the nucleolar localization of this complex requires active RNA polymerase I transcription and the small ubiquitin-like modifier–specific protease SENP3. Taken together, our data identify a novel mammalian complex required for 60S ribosomal subunit synthesis, providing further insight into the intricate, yet poorly described, process of ribosome biogenesis in higher eukaryotes.
INTRODUCTION
Actively dividing cells must increase their rate of protein synthesis to sustain the growth associated with proliferation. As ribosomes are the main effectors of translation, their synthesis underlies a cell's ability to grow and divide (Lempiainen and Shore, 2009). Eukaryotic ribosome biogenesis is initiated in the nucleolus as RNA polymerase I (Pol I) transcribes a 47S (35S in yeast) pre-rRNA. This 47S pre-rRNA is subjected to a series of cotranscriptional and posttranscriptional modifications involving the collaborative actions of more than 200 trans-acting factors to form what will become the mature 28S (25S in yeast), 18S, and 5.8S rRNAs (Hadjiolova et al., 1993; Kopp et al., 2007; Kos and Tollervey, 2010; Panse and Johnson, 2010). The 18S rRNA and small ribosomal proteins transcribed by RNA polymerase II (Pol II) are incorporated into the pre-40S ribosomal subunit (Panse and Johnson, 2010). The 28S and 5.8S rRNAs, along with the 5S rRNA transcribed by RNA polymerase III (Pol III) and large ribosomal proteins, are incorporated into the pre-60S ribosomal subunit (Panse and Johnson, 2010). These preribosomal subunits are then subjected to further modifications, as each subunit is exported separately into the cytoplasm to undergo the final maturation steps before translation is initiated and fully competent ribosomes are formed (Panse and Johnson, 2010).
Ribosome biogenesis relies on a balance among transcription, processing, and export of the ribosomal particles; it is tightly coupled to cell cycle progression. Defects in any of these steps ultimately leads to nucleolar stress, followed by a G1 cell cycle arrest dependent on the stabilization of the tumor suppressor p53 (Deisenroth and Zhang, 2010). Interestingly, proteins involved in tumorigenesis also control ribosome biogenesis at various stages. The protooncogene c-Myc, as well as mTOR, both positively regulate ribosome biogenesis via increased transcription of all three RNA polymerases (Schlosser et al., 2003; Mayer et al., 2004; Poortinga et al., 2004; Arabi et al., 2005; Grandori et al., 2005; Gomez-Roman et al., 2006; Mayer and Grummt, 2006; van Riggelen et al., 2010). Alternatively, several tumor suppressor proteins have been shown to have a repressive effect on Pol I (Cavanaugh et al., 1995; Budde and Grummt, 1999; Zhai and Comai, 2000; Zhang et al., 2005). Nucleophosmin (NPM1), a nucleolar protein required for rRNA processing, is also often found overexpressed, mutated, or deleted in various types of cancer (Tanaka et al., 1992; Nozawa et al., 1996; Shields et al., 1997; Skaar et al., 1998; Subong et al., 1999; Mendes-da-Silva et al., 2000; Tsui et al., 2004; Falini et al., 2005; Grisendi et al., 2006). Together these observations suggest that ribosome biogenesis represents an important pathway that could be targeted to stop the growth of cancer cells.
Recently a panoply of large-scale studies in Saccharomyces cerevisiae have led to the purification of various preribosomal particles and the identification of a considerable number of proteins involved in the maturation and export of both the 60S and 40S ribosomal subunits (Harnpicharnchai et al., 2001; Nissan et al., 2002; Peng et al., 2003; Schafer et al., 2003; Krogan et al., 2004; Kressler et al., 2008; Li et al., 2009). Many of the factors identified have putative mammalian homologues, yet most of these proteins remain to be characterized in higher eukaryotes. Although the processing of rRNA requires several proteins, there are few complexes that have been identified in this process in mammals. The mammalian Pes1-Bop1-WDR12 complex was shown to be critical for the processing of the 32S pre-rRNA into the mature 28S rRNA (Strezoska et al., 2000, 2002; Lapik et al., 2004; Holzel et al., 2005, 2010; Grimm et al., 2006; Rohrmoser et al., 2007). Recently the nucleolar small ubiquitin-like modifier (SUMO)-specific proteases SENP3 and SENP5 were shown to be required for efficient rRNA processing via their interaction with the nucleolar chaperone NPM1, indicating a possible role for SUMOylation in the regulation of ribosome synthesis (Yun et al., 2008).
We recently identified LAS1L as a nucleolar protein required for the synthesis of the 60S ribosomal subunit and maturation of the 28S rRNA (Castle et al., 2010). Depletion of LAS1L results in accumulation of the 32S pre-rRNA intermediate, indicating a pre-rRNA processing defect of the internal transcribed spacer-2 (ITS-2), a region between the 5.8S and 28S rRNAs in the pre-RNA transcript (Castle et al., 2010). Interestingly, reduction of LAS1L levels also resulted in nucleolar disruption and increased levels of the tumor suppressor p53 and its transcriptional target p21, resulting in a cell cycle arrest in G1 phase (Castle et al., 2010). The stabilization of p53 suggests that LAS1L plays an important role in controlling ribosome biogenesis and nucleolar integrity.
To further define the function of LAS1L, we sought to identify interacting proteins using a proteomic approach. In this study, we show that LAS1L forms a protein complex with PELP1, TEX10, NOL9, SENP3, and WDR18. Our data demonstrate that all the components of the LAS1L complex cosediment with the pre-60S ribosomal particle and are required for proper processing of the 32S rRNA intermediate. Moreover, depletion of LAS1L-associated complex proteins resulted in a G1 arrest through stabilization of p53, suggesting their importance in regulating nucleolar function. Finally, we found that LAS1L and PELP1 are SUMOylated in an SENP3-dependent manner and that depletion of the SUMO-specific protease SENP3 induces relocalization of LAS1L and PELP1 from the nucleolus to the nucleoplasm, suggesting a role for SUMOylation in regulating the functions of LAS1L and PELP1. These findings demonstrate that the LAS1L complex is critical for ribosome biogenesis and cell proliferation and provide additional evidence that multiple protein complexes have distinct functions in the synthesis of eukaryotic ribosomes.
RESULTS
Identifying LAS1L-interacting proteins
Las1p was originally described as an essential protein required for cell proliferation in S. cerevisiae (Doseff and Arndt, 1995). Our previous studies have described the human homologue of Las1p, LAS1L, as a nucleolar protein required for processing of the rRNA ITS-2 region and synthesis of the 60S ribosomal particle (Castle et al., 2010).
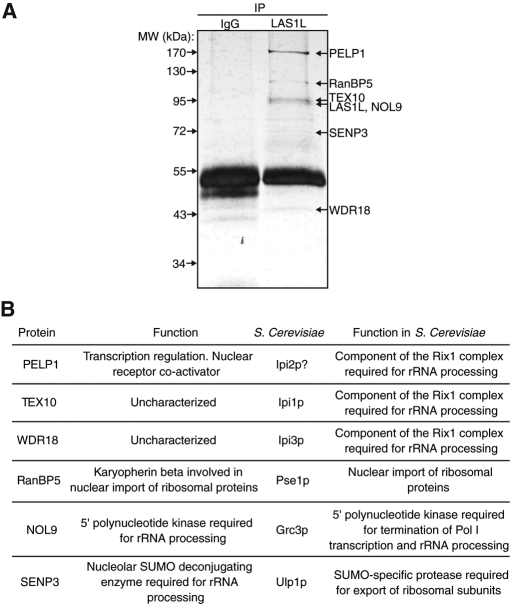
To gain insight into the role of LAS1L in ribosome biogenesis, we performed a proteomic analysis of immunoprecipitated endogenous LAS1L complexes purified from HEK 293T cells (Figure 1A). Six proteins associated with LAS1L were identified by liquid chromatography–tandem mass spectrometry (Figure 1, A and B). We confirmed that all these interactions are positive in HEK 293T, HCT116 (unpublished data), and MCF7 (unpublished data) cells by coimmunoprecipitation of endogenous LAS1L with PELP1, RanBP5, TEX10, NOL9, SENP3, and WDR18, using commercially available antibodies or antibodies that we generated (WDR18; Figure 2A). Specificity of each antibody was verified by RNA interference (RNAi) depletion, which was followed by Western blot analysis (Supplemental Figure S1, A and B). Figure 1B compiles the known function of the human proteins found, as well as their putative S. cerevisiae homologues. TEX10 and WDR18 are uncharacterized proteins that share sequence homology with budding yeast Ipi1 and Ipi3, respectively (Finkbeiner et al., 2011). In budding yeast, Ipi1p and Ipi3p with Rix1p are essential components of the Rix1 complex required for rRNA ITS-2 processing and export of the 60S ribosomal subunit (Bassler et al., 2001; Galani et al., 2004; Krogan et al., 2004). PELP1 was first characterized as a nuclear receptor coactivator and a regulator of transcription often overexpressed in cancer (Vadlamudi et al., 2001, 2004; Nair et al., 2010). Interestingly, PELP1 shares some sequence homology with Rix1p and, therefore, with TEX10 and WDR18, represents the mammalian counterpart of the budding yeast Rix1 complex (Finkbeiner et al., 2011). RanBP5 is a karyopherin-β protein that has been shown to mediate nuclear import of ribosomal proteins (Jakel and Gorlich, 1998; Chou et al., 2010). NOL9 was recently described as a 5′-polynucleotide kinase required for processing of the ITS-2 region of rRNA (Heindl and Martinez, 2010), while SENP3 is a nucleolar SUMO-specific protease shown to be required for both ITS-2 pre-rRNA processing and SUMO deconjugation of ribosomal proteins and NPM1 (Gong and Yeh, 2006; Haindl et al., 2008; Yun et al., 2008). These results indicate that LAS1L interacts with proteins known to be implicated in transcriptional regulation (PELP1) and control of ribosome biogenesis (NOL9, RanBP5, and SENP3).
FIGURE 1:
Isolation of LAS1L-associated proteins. (A) LAS1L was affinity-purified from HEK 293T cell lysate using an anti-LAS1L antibody coupled to protein G Sepharose beads. Normal rabbit IgG was used as negative control. LAS1L-associated complexes were separated on SDS–PAGE and silver-stained and bands present only in the LAS1L IP lane were cut out, digested with trypsin, and analyzed by mass spectrometry. Identified proteins are marked with arrows. (B) The proteins identified in (A) and their corresponding S. cerevisiae putative homologues (Rout et al., 1997; Jakel and Gorlich, 1998; Bassler et al., 2001; Vadlamudi et al., 2001, 2004; Peng et al., 2003; Galani et al., 2004; Krogan et al., 2004; Gong and Yeh, 2006; Panse et al., 2006; Nair et al., 2007; Haindl et al., 2008; Yun et al., 2008; Braglia et al., 2010; Chou et al., 2010; Heindl and Martinez, 2010; Finkbeiner et al., 2011).
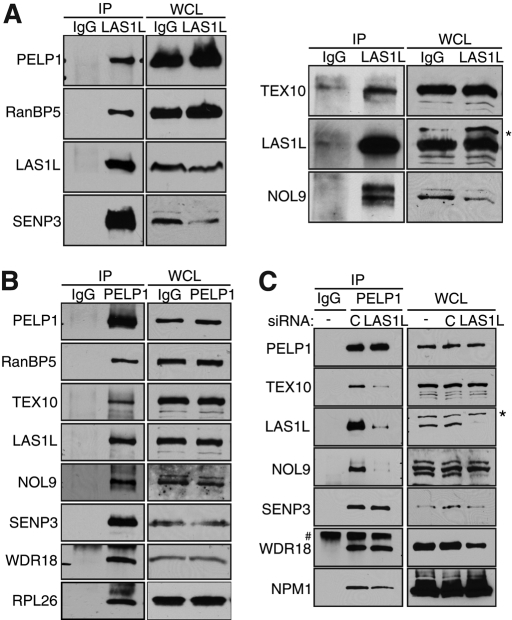
FIGURE 2:
LAS1L associates with PELP1, TEX10, WDR18, RanBP5, NOL9, and SENP3. (A) LAS1L was immunoprecipitated (IP) with an anti-LAS1L–specific antibody from HEK 293T cell lysates. Associated proteins were separated on SDS–PAGE and analyzed by Western blotting with specific antibodies (indicated on the left). Normal rabbit IgG was used as negative control. WCL, whole-cell lysate; *, presence of an unspecific band. (B) PELP1 was immunoprecipitated (IP) with an anti-PELP1 antibody from HEK 293T cell lysates. Associated proteins were separated on SDS–PAGE and analyzed by Western blotting with specific antibodies (indicated on the left). Normal rabbit IgG was used as negative control. WCL, whole-cell lysate. (C) HEK 293T cells were transfected with nontargeting control (represented by the letter “C”) or LAS1L siRNA for 48 h. Cells were lysed and immunoprecipitated with an anti-PELP1 antibody. Proteins associating with PELP1 in the absence of LAS1L were separated on SDS–PAGE and analyzed by Western blotting with specific antibodies (indicated on the left). Normal rabbit IgG was used as negative control. WCL, whole-cell lysate; *, presence of an unspecific band; #, an IgG band.
To evaluate whether LAS1L is part of two different complexes, one involved in regulating transcription and another involved in ribosome synthesis, we tested whether PELP1 could interact with the other LAS1L-interacting proteins. Coimmunoprecipitation experiments using an anti-PELP1 antibody revealed that endogenous PELP1 also interacts with RanBP5, TEX10, WDR18, NOL9, SENP3, and LAS1L, as well as the large ribosomal protein RPL26 (Figure 2B). Mass spectrometry analysis of PELP1-purified complexes confirmed the interaction of PELP1 with the LAS1L-associated proteins, as well as with NPM1 (Figure 2B; unpublished data). Small interfering RNA (siRNA) down-regulation of LAS1L indicated that PELP1 interaction with both NOL9 and TEX10 was dependent on the presence of LAS1L (Figure 2C), suggesting that LAS1L and PELP1 could be part of the same complex. Based on these observations, we propose that all of these proteins could constitute a large complex involved in pre-rRNA processing and the synthesis of mature ribosomes.
Depletion of LAS1L-interacting proteins induces a p53-dependent nucleolar stress
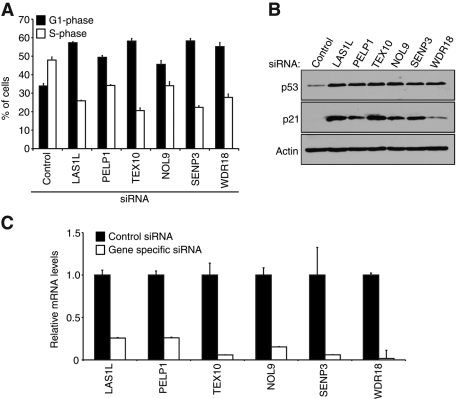
We previously demonstrated that depletion of LAS1L resulted in a nucleolar stress characterized by a G1-phase arrest and stabilization of the tumor suppressor p53 (Castle et al., 2010). To evaluate whether LAS1L-associated proteins are also required for nucleolar integrity, we depleted HCT116 colon carcinoma cells of complex proteins by RNAi, as determined by quantitative reverse transcriptase PCR (qRT-PCR) analysis (Figure 3C). Cells were then labeled with bromodeoxyuridine (BrdU), stained with propidium iodide (PI), and subjected to fluorescence-activated cell sorter (FACS) analysis. Depletion of all the identified LAS1L-interacting proteins resulted in a 15–24% increase in the number of cells in G1 phase and a decrease in the S-phase population, suggesting an arrest in G1 phase (Figure 3A). To further resolve the nature of this G1 arrest, we subjected lysates of cells from the same experiment to Western blot analysis. In accordance with the FACS analysis, reduction of all proteins in the complex resulted in stabilization of the tumor suppressor p53 and increased levels of its transcriptional target p21 (Figure 3B), suggesting the cell cycle arrest is due to increased levels of these proteins. These results demonstrate that LAS1L-associated proteins are required for cell division, and the stabilization of p53 suggests that their depletion results in a nucleolar stress.
FIGURE 3:
Depletion of LAS1L-interacting proteins induces a p53-dependent G1 cell cycle arrest. (A) Cell cycle profiles of HCT116 cells transfected with control, LAS1L, PELP1, TEX10, NOL9, SENP3, and WDR18 siRNA. Seventy-two hours after transfection the cells were pulse-labeled with BrdU for 30 min, stained with propidium iodide, and analyzed by flow cytometry to determine the percentage of cells in G1 and S phase. Error bars indicate SD from triplicate experiments. (B) Western blot analysis of the siRNA-transfected cells from (A) with specific antibodies against p53, p21, and β-actin. (C) Total RNA was extracted from the siRNA-transfected cells from panel (A), and knockdowns were confirmed by qRT-PCR with gene-specific primers. Relative mRNA levels for each gene-specific primer were normalized to β-actin. Error bars indicate SD from triplicate experiments.
LAS1L-interacting proteins localize to the nucleolus
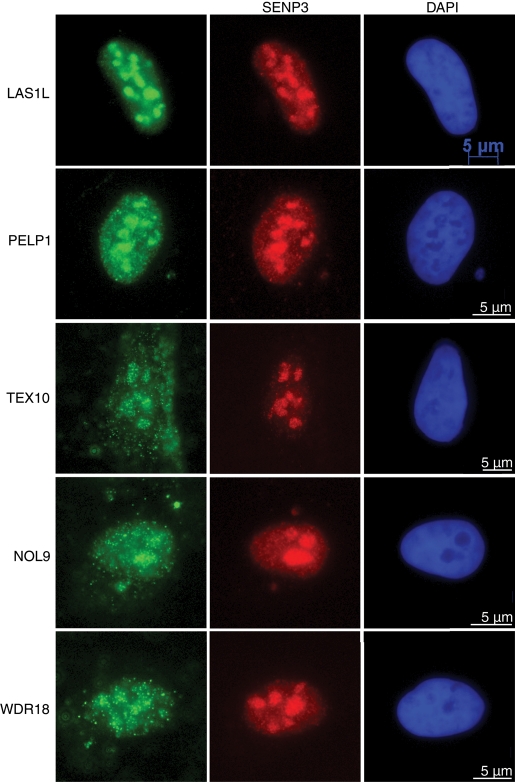
In our previous studies, we observed the subcellular localization of LAS1L to be predominantly nucleolar (Castle et al., 2010). To further characterize the other proteins in complex with LAS1L, we performed immunofluorescence analysis in U2OS osteosarcoma cells with specific antibodies. LAS1L, PELP1, TEX10, NOL9, and WDR18 showed nucleolar localization at endogenous levels, as determined by colocalization with the known nucleolar protein SENP3 (Figure 4). Because ribosome biogenesis takes place in the nucleolus (Boisvert et al., 2007), these results further support a role for LAS1L and its interacting proteins in that process.
FIGURE 4:
LAS1L-associated proteins localize to the nucleolus. Immunofluorescence analysis of U2OS cells with complex protein-specific antibodies. Cells were preextracted with 0.1% Triton, fixed, and immunostained with anti-LAS1L, PELP1, TEX10, NOL9, and WDR18 antibodies (green). Colocalization with SENP3 was confirmed using an anti-SENP3 antibody (red). DNA was visualized by staining with Hoechst 33342 (blue). Scale is representative of all three panels.
LAS1L and its associated proteins cofractionate with pre-60S particles
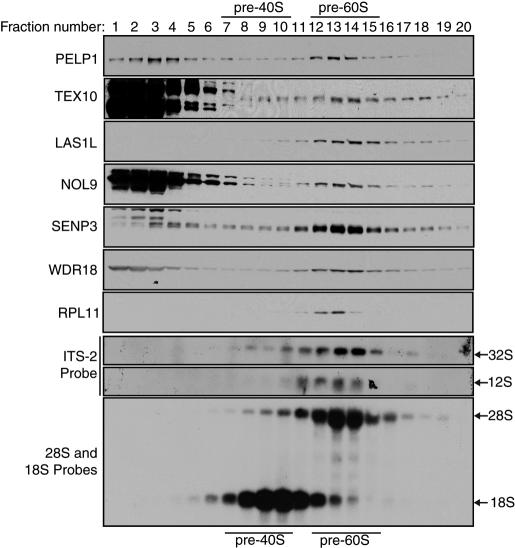
LAS1L is required for formation of the 60S preribosomal subunit, and depletion of LAS1L results in a distinct loss of the 60S, but not the 40S, preribosomal subunit (Castle et al., 2010). To further assess the likelihood that LAS1L and its interacting proteins are factors that associate with the maturing 60S ribosomal particle, we subjected HCT116 nuclear extracts to sucrose gradient fractionation experiments. LAS1L, PELP1, TEX10, NOL9, SENP3, and WDR18 all cofractionated with the pre-60S particle, as demonstrated by cofractionation with the large ribosomal protein RPL11 and Northern blot analysis with the 28S and 18S probes (Figure 5). Moreover, all proteins in the complex were found to be enriched in fractions 13 and 14, the same fractions containing the 32S and 12S rRNA intermediates, as shown by Northern blot analysis with an ITS-2 probe (Figure 5). This evidence suggests that the LAS1L-interacting proteins could possibly play a role in the processing of the rRNA ITS-2 region, which is found in the 32S pre-rRNA; this processing role is similar to the one we previously demonstrated for LAS1L (Castle et al., 2010).
FIGURE 5:
LAS1L-interacting proteins cosediment with pre-60S ribosomal particles. Nuclear extracts from HCT116 cells were fractionated by centrifugation on a 10–30% sucrose gradient. Fractions were collected, and the optical density was measured at 260 nm (A260). The positions of the pre-40S and pre-60S native subunits are indicated. Presence of the LAS1L complex proteins in each fraction were confirmed by Western blotting using specific antibodies (indicated on the left). Total RNA from each fraction was extracted and separated on a 1.2% formaldehyde gel and analyzed by Northern blotting with ITS-2, 28S, and 18S probes, as indicated. Arrows indicate the positions of the 32S, 28S, 18S, and 12S rRNAs.
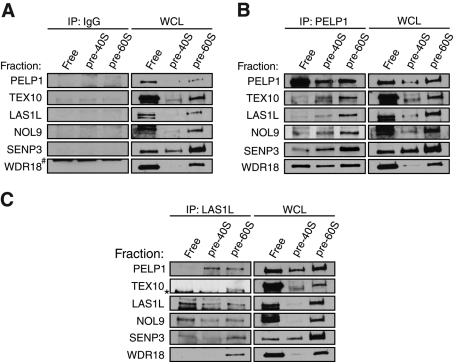
Although all of the LAS1L-interacting proteins cofractionate with pre-60S particles, it is clear that a major population of some proteins in the complex is present in the fractions at the top of the gradient (Figure 5). It is therefore possible that the LAS1L-associated proteins could exist in separate complexes when not associated with the pre-60S particle. To further investigate this possibility, we performed sucrose gradient fractionation, and fractions corresponding to free nuclear proteins and smaller protein complexes (fractions 1, 2, and 3), pre-40S (fractions 8, 9, and 10), and pre-60S (fractions 12, 13, and 14) ribosomal particles were pooled separately. These pooled fractions were then subjected to coimmunoprecipitation experiments with antibodies specific for LAS1L or PELP1. Consistent with the sucrose gradient cofractionation experiments, all of the LAS1L-accociated proteins coprecipitated with LAS1L and PELP1 in the fractions corresponding to pre-60S ribosomal particles (Figure 6, B and C). However, only NOL9 was found associated with LAS1L in the fractions corresponding to free proteins and smaller complexes (Figure 6C). Furthermore, only WDR18 was found to interact with PELP1 in the free protein fractions (Figure 6B). Collectively, these data indicate that all of the proteins form a complex when they are on the pre-60S particle but are part of separate complexes when not associated with preribosomal particles.
FIGURE 6:
LAS1L and NOL9 interact with the mammalian Rix1 complex on pre-60S ribosomal particles. Nuclear extracts from HCT116 cells were fractionated by centrifugation on a 10–30% sucrose gradient. Fractions were collected, and the optical density was measured at 260 nm (A260). Based on the A260 profile, fractions corresponding to free nuclear proteins (1, 2, and 3), pre-40S ribosomal particles (8, 9, and 10), and pre-60S ribosomal particles (12, 13, and 14) were then combined and immunoprecipitated (IP) with rabbit IgG (A) as a negative control or with PELP1 (B) or LAS1L (C) antibodies. Associated proteins were separated on SDS–PAGE and analyzed by Western blotting with specific antibodies (indicated on the left). WCL, whole-cell lysate; *, presence of an unspecific band; #, an IgG band.
LAS1L-interacting proteins are required for pre-rRNA processing
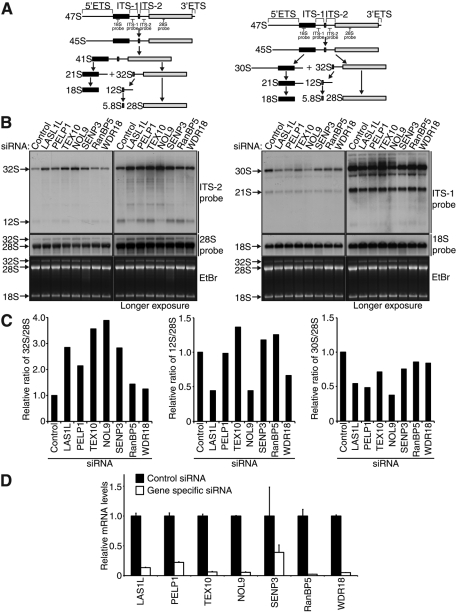
The 47S pre-rRNA contains four regions that must be modified, processed and removed from pre-rRNA to form the mature 18S, 5.8S, and 28S rRNAs (Hadjiolova et al., 1993). The two interior regions are known as the internal transcribed spacers (ITSs), with ITS-1 located between the 18S and 5.8S rRNAs and ITS-2 situated between the 5.8S and 28S rRNAs (Figure 7A). The 47S precursor is also bounded on each end by the external transcribed spacer (ETS) regions, the 5′ETS and the 3′ETS. Each ETS and ITS region is processed through a series of exonucleolytic and endonucleolytic reactions in order to form the mature rRNAs (Figure 7A; Kressler et al., 2010; Panse and Johnson, 2010), and depletion of proteins involved in these steps results in accumulation of pre-rRNA intermediates. To examine the possibility that the LAS1L-associated proteins are required for pre-rRNA processing, we treated HCT116 cells with control or gene-specific siRNA, and knockdowns were assessed by qPCR analysis (Figure 7C). Total rRNA was harvested and subjected to Northern blot analysis using probes specific for the ITS-1 and ITS-2 regions. Abrogation of LAS1L-interacting proteins resulted in a notable increase in the level of the 32S rRNA intermediate, with approximately two to fourfold increases in the 32S/28S ratio occurring in cells depleted of LAS1L, PELP1, TEX10, NOL9, and SENP3 (Figure 7, B, C, and D). A slight decrease in the levels of the 30S pre-rRNA was also observed (Figure 7, B, C, and D). Interestingly, although the 32S precursor is directly upstream of the 12S precursor (Figure 7A), only depletion of LAS1L, NOL9, and to a lesser extent WDR18 resulted in a loss of the 12S intermediate (Figure 7, B and C). Taken together, these data demonstrate that depletion of LAS1L and its associated factors results in processing defects of pre-rRNA intermediates, especially of those found in the 60S preribosomal subunit, further supporting a role for these proteins in pre-rRNA processing.
FIGURE 7:
LAS1L-associated proteins are required for proper processing of ITS-2. (A) Schematic representation of the primary 47S rRNA transcript and the two major processing pathways with rRNA intermediates, as indicated (adapted from Hadjiolova et al., 1993). (B) Northern blot analysis of total RNA from HCT116 cells transfected with control, LAS1L, PELP1, TEX10, NOL9, SENP3, RanBP5, and WDR18 siRNA. Seventy-two hours after transfection, equal amounts of total RNA were hybridized with specific probes for ITS-1, ITS-2, 28S, and 18S rRNA intermediates (indicated on the right). The right panel shows longer exposure times for each probe. The positions of the specific probes used for Northern blot analysis are indicated on the schematic in (A). (C) The 32S/28S, 12S/28S, and 30S/28S ratios were determined by quantification of the relative band intensities of the 32S, 30S, and 28S rRNA intermediates in the lower exposures and the 12S rRNA intermediate in the longer exposure from (B). Intensities were normalized to the control siRNA-treated sample. (D) Knockdowns were confirmed by qRT-PCR with gene-specific primers. Relative mRNA levels for each gene-specific primer were normalized to β-actin. Error bars indicate SD from triplicate experiments.
Nucleolar localization of LAS1L interactors requires active Pol I transcription
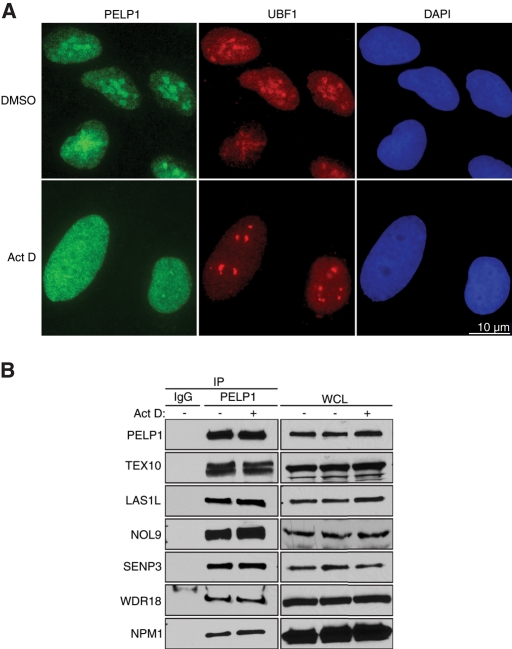
The rate-limiting step in ribosome biogenesis is thought to be the transcription of tandem rDNA genes by Pol I, and inhibition of rRNA transcription has been shown to cause relocalization out of the nucleolus of proteins involved in ribosome biogenesis (Huang et al., 2005; Zhang et al., 2009). To analyze whether nucleolar localization of LAS1L-associated proteins requires Pol I transcription, we treated U2OS cells with a low concentration of actinomycin D to inhibit Pol I, but not Pol II, transcription. The subcellular localization of endogenous PELP1 was then determined by immunofluorescence microscopy, and upstream binding factor 1 (UBF1), a transcription factor required for Pol I transcription, was used as a nucleolar marker. PELP1 showed a predominantly nucleolar localization when treated with dimethyl sulfoxide (DMSO) as a control (Figure 8A). However, when U2OS cells were treated with actinomycin D, the localization of PELP1 was determined to be mainly nuclear (Figure 8A). Coimmunoprecipitation experiments using an antibody specific for endogenous PELP1 indicated that the complex containing LAS1L remains associated with PELP1 in the absence of Pol I transcription (Figure 8B).
FIGURE 8:
PELP1 requires active Pol I transcription for nucleolar localization. (A) U2OS cells were treated with either DMSO or 20 nM actinomycin D for 2 h. Cells were fixed and immunostained with an anti-PELP1 antibody (green) and an anti-UBF1 antibody (red). DNA was visualized by staining with Hoechst 33342 (blue). Scale is representative of both panels. (B) Cells were treated with DMSO (−) or actinomycin D (+) as in (A), and lysates were immunoprecipitated (IP) with an anti-PELP1 antibody. Associated proteins were separated on SDS–PAGE and analyzed by Western blotting with specific antibodies (indicated on the left). Normal rabbit IgG was used as negative control. WCL, whole-cell lysate.
We showed that PELP1 also coprecipitates with NPM1 (Figure 2C), a nucleolar chaperone that associates with SENP3 and is required for pre-rRNA processing (Itahana et al., 2003; Haindl et al., 2008; Yun et al., 2008). NPM1 has been shown to modulate the stability and nucleolar localization of several of its binding partners, including SENP3 (Valdez et al., 1994; Li et al., 1996; Yun et al., 2008). Interestingly, NPM1 remained associated with PELP1 irrespective of Pol I transcription status (Figure 8B), indicating that the nuclear relocalization is not due to loss of interaction with NPM1. These observations demonstrate that the complex assembles independent of Pol I transcription, yet localizes to the nucleolus only when transcription of the rDNA gene is active.
Nucleolar localization of the LAS1L complex requires association with SENP3 and NPM1
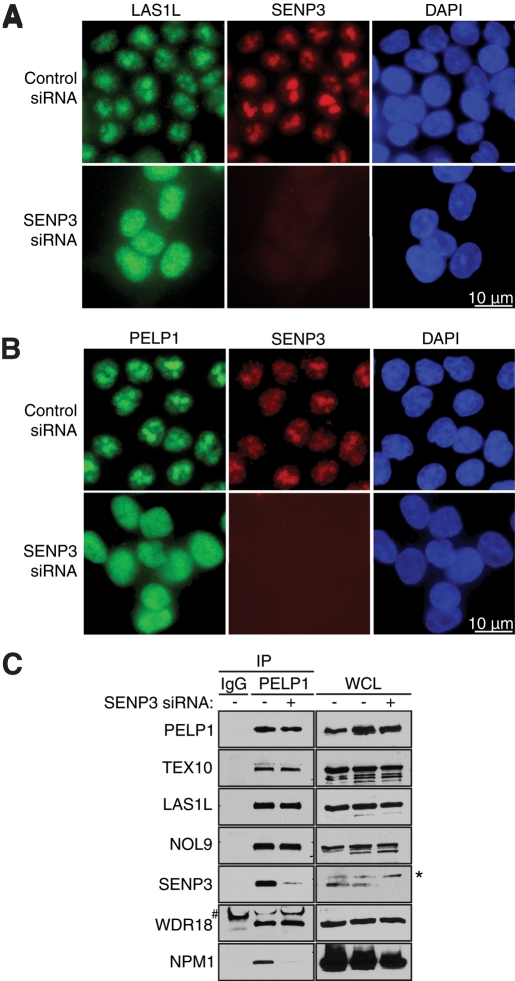
It is well established that posttranslational SUMO modifications can affect subcellular localization of proteins (Geiss-Friedlander and Melchior, 2007). Since the SUMO-specific protease SENP3 interacts with LAS1L and PELP1 (Figure 2, A and C), we sought to determine whether SENP3 is required for localization of LAS1L and PELP1 to the nucleolar compartment. HCT116 cells were treated with control or SENP3 siRNA, and cells were analyzed by immunofluorescence microscopy with antibodies specific for LAS1L, PELP1, and SENP3. In control siRNA-treated cells, LAS1L and PELP1 exhibited a strong nucleolar staining (Figure 9, A and B). In contrast, when SENP3 protein level was depleted by RNAi treatment, LAS1L and PELP1 redistributed to the nucleoplasm (Figure 9, A and B), indicating that SENP3 is important for this subcellular localization. We next evaluated whether SENP3 was required for the interaction of LAS1L and its associated factors. Immunoprecipitation experiments revealed that the proteins of the complex could still interact with PELP1 in the absence of SENP3 (Figure 9C), suggesting that nucleolar localization is not necessary for complex formation.
FIGURE 9:
SENP3 is necessary for LAS1L and PELP1 nucleolar localization. (A) HCT116 cells were transfected with control or SENP3 siRNA for 72 h. Cells were fixed and immunostained with anti-LAS1L (green) and anti-SENP3 (red) antibodies. DNA was visualized by staining with Hoechst 33342 (blue). Scale is representative of all panels. (B) HCT116 cells were transfected with control or SENP3 siRNA for 72 h. Cells were fixed and immunostained with anti-PELP1 (green) and anti-SENP3 (red) antibodies. DNA was visualized by staining with Hoechst 33342 (blue). Scale is representative of all panels. (C) Cells were transfected with a Control (−) or SENP3 (+) siRNA for 72 h. Lysates were then immunoprecipitated with rabbit IgG (as negative control) or PELP1 antibody. Coprecipitating proteins were separated on SDS–PAGE and analyzed by Western blotting using specific antibodies (indicated on the left). WCL, whole-cell lysate; *, presence of an unspecific band; #, an IgG band.
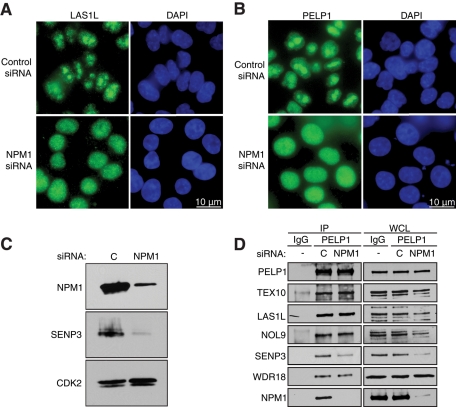
We observed that depletion of SENP3 resulted in abrogation of the interaction between PELP1 and NPM1 (Figure 9C), suggesting that this protein could be required for the nucleolar localization of the LAS1L interactors as well. To test this hypothesis, we treated HCT116 cells with control or NPM1 siRNA, and the levels of NPM1 were determined by Western blot analysis (Figure 10C). The subcellular localization of endogenous LAS1L and PELP1 was then determined using immunofluorescence microscopy. Cells treated with control siRNA revealed strong nucleolar localization for both LAS1L and PELP1 (Figure 10, A and B). In contrast, cells treated with NPM1 siRNA resulted in a relocalization of LAS1L and PELP1 from the nucleolus to the nucleus (Figure 10, A and B). Similar to what we observed upon depletion of SENP3, treatment with NPM1 siRNA did not affect formation of the complex (Figure 10D). Interestingly, the depletion of NPM1 also resulted in decreased total levels of SENP3 and SENP3 bound to PELP1 (Figure 10C), which is consistent with previous studies showing that NPM1 regulates SENP3 protein stability (Yun et al., 2008). Another report has demonstrated that SENP3 is involved in deconjugating SUMO-2 from NPM1, a process critical for efficient 28S rRNA maturation (Haindl et al., 2008). It is therefore possible that SUMOylated NPM1 is unable to bind and stabilize the complex in the nucleolus. Taken together, these data indicate that the nucleolar localization of LAS1L and PELP1 relies on the presence of both SENP3 and NPM1, and suggests a role for regulation of ribosome biogenesis by SUMOylation.
FIGURE 10:
NPM1 is required for LAS1L and PELP1 nucleolar localization. (A) HCT116 cells were transfected with control or NPM1 siRNA for 48 h. Subcellular localization of LAS1L was determined by immunofluorescence analysis using a LAS1L antibody (green). DNA was visualized by staining with Hoechst 33342 (blue). Scale is representative of all panels. (B) HCT116 cells were transfected with control or NPM1 siRNA for 48 h. Subcellular localization of PELP1 was determined by immunofluorescence analysis using a PELP1 antibody (green). DNA was visualized by staining with Hoechst 33342 (blue). Scale is representative of all panels. (C) Knockdowns of NPM1 for (A) and (B) were confirmed by Western blotting using specific antibodies (indicated on the left). An anti-CDK2 antibody was used as loading control. (D) Cells were transfected with a control (“C”) or NPM1 siRNA for 72 h. Lysates were then immunoprecipitated with rabbit IgG (as negative control) or PELP1 antibody. Coprecipitating proteins were separated on SDS–PAGE and analyzed by Western blotting using specific antibodies (indicated on the left). WCL, whole-cell lysate.
LAS1L and PELP1 are modified by SUMO in an SENP3-dependent manner
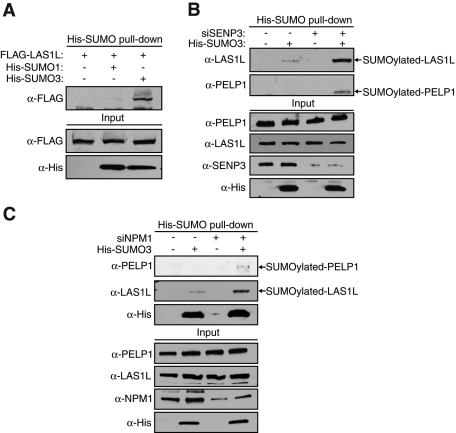
Because the SUMOylation status of a protein can affect its subcellular localization (Geiss-Friedlander and Melchior, 2007) and treatment with SENP3 siRNA results in relocalization of both LAS1L and PELP1 (Figure 9, A and B), we sought to determine whether these proteins are directly modified by SUMO. HEK 293T cells were transfected with FLAG-LAS1L and His-tagged SUMO-1 or SUMO-3, and pulldowns were performed on cellular extracts using Ni-nitrolotriacetic acid (Ni-NTA) beads. FLAG-LAS1L was precipitated with His-SUMO-3 and only slightly with His-SUMO-1, suggesting that LAS1L is modified more specifically by SUMO-3 (Figure 11A). To test whether the SUMOylation status of LAS1L is modulated by SENP3, we transfected HEK 293T cells with SENP3 siRNA followed by His-SUMO-3, and pulldowns were performed on cellular lysates with Ni-NTA beads. Endogenous LAS1L was found to be modified by SUMO-3, and this SUMOylation increased upon depletion of SENP3 (Figure 11B). Furthermore, we were also able to identify endogenous PELP1 as being modified by SUMO-3 upon depletion of SENP3 (Figure 11B). In support of these findings, Finkbeiner and coworkers also identified PELP1 and LAS1L as being modified by SUMO (Finkbeiner et al., 2011). Because we also observed a relocalization of LAS1L and PELP1 from the nucleolus to the nucleoplasm upon depletion of NPM1, we sought to determine whether or not these proteins showed increased SUMOylation with abrogation of NPM1. Indeed, similar to what we observed upon SENP3 depletion, knockdown of NPM1 by RNAi also resulted in increased levels of SUMOylated endogenous LAS1L and PELP1 (Figure 11C). Taken together, these observations suggest that both SENP3 and NPM1 are required for nucleolar localization of LAS1L and PELP1. These data support the previously described links between NPM1 and SENP3 and further suggest that modification of LAS1L and PELP1 by SUMO regulates their subcellular localization (Haindl et al., 2008; Kuo et al., 2008; Yun et al., 2008).
FIGURE 11:
LAS1L and PELP1 are modified by SUMO in an SENP3-dependent manner. (A) HEK 293T cells were transfected with FLAG-LAS1L (+) plus either empty vector (−) or plasmids expressing 6xHis-tagged SUMO-1 or SUMO-3 (+). Forty-eight hours after transfection, SUMOylated proteins were pulled down from cell lysates using Ni-NTA agarose beads. Eluates were analyzed on SDS–PAGE and by Western blotting with the indicated antibodies. (B) HEK 293T cells were transfected with either control (−) or SENP3 (+) siRNA. The next day, cells were transfected with empty vector (−) or a 6xHis-tagged SUMO-3 (+) expressing plasmid. Forty-eight hours after transfection, pulldowns and protein analyses were performed as in (A). (C) HEK 293T cells were transfected with either control (−) or NPM1 (+) siRNA. The next day, cells were transfected with empty vector (−) or a plasmid expressing 6xHis-tagged SUMO-3 (+). Forty-eight hours after transfection, pulldowns and protein analyses were performed as in (A).
DISCUSSION
Synthesis of ribosomal subunits is a multistep process requiring the coordinated activity of more than 170 factors recently identified by large-scale studies in budding yeast (Harnpicharnchai et al., 2001; Nissan et al., 2002; Peng et al., 2003; Schafer et al., 2003; Krogan et al., 2004; Kressler et al., 2008; Li et al., 2009). Although most of these factors are conserved throughout evolution, little is known about their roles in mammalian systems. In this study, we present evidence that LAS1L is part of a novel ribosome biogenesis complex consisting of PELP1, TEX10, WDR18, NOL9, and SENP3 (Figure 2, A and B). We demonstrate that all these proteins localize to the nucleolus (Figure 4) and cosediment with the pre-60S ribosomal particle (Figure 5) and that they are required for efficient processing of the 28S rRNA (Figure 7, B and D).
Recently Finkbeiner et al. (2011) determined that PELP1, TEX10, and WDR18 interact together as part of a complex sharing sequence homology with the Rix1 complex (Ipi1p, Rix1p, and Ipi3p) required for ribosome biogenesis in S. cerevisiae (Finkbeiner et al., 2011). Indeed, TEX10 contains a conserved N-terminal Ipi1 domain and is ∼20% identical to Ipi1p (Finkbeiner et al., 2011). Likewise, WDR18 and Ipi3p are ∼25% identical, and both proteins contain multiple WD40 repeats (Finkbeiner et al., 2011). At its N-terminus, PELP1 displays a Rix1 homology region, and both PELP1 and Rix1 share a C-terminal region rich in acidic residues (Finkbeiner et al., 2011). In mammals, PELP1 has been characterized as a transcriptional regulator capable of modulating the functions of diverse nuclear receptors, such as the estrogen (ERα and ERβ), progesterone, and glucocorticoid receptors (Vadlamudi et al., 2001, 2004; Nair et al., 2004; Kayahara et al., 2008). Our study demonstrates that PELP1 localizes to the nucleus and nucleolus and is important for efficient synthesis of the 28S rRNA (Figures 4 and 7). Interestingly, PELP1 was found to bind RNA and interact with components of the mRNA splicing machinery (Nair et al., 2006), and a recent study has shown that PELP1 can associate with the rDNA promoter (Gonugunta et al., 2011). It is possible that, depending on its subcellular compartmentalization, PELP1 regulates either Pol I- or Pol II-specific RNA-processing events.
The Rix1 complex in S. cerevisiae associates with a late pre-60S particle and has been indicated by multiple groups as being required for pre-rRNA processing (Galani et al., 2004; Krogan et al., 2004; Nissan et al., 2004). This complex is thought to be involved in the later steps of pre-rRNA processing that occur after cleavage of the ITS-2 region, and it has been shown to preferentially bind the 7S rRNA (Krogan et al., 2004). We demonstrate that the human homologues of the Rix1 complex (PELP1, TEX10, and WDR18) cofractionate with a particle that contains the 32S pre-rRNA (Figure 5), suggesting it could associate with the 60S subunit before cleavage of the ITS-2 occurs. Furthermore, depletion of PELP1, TEX10, and WDR18 showed an increase in the 32S intermediate (Figure 7, B and D), suggesting they are also required for the processing of this earlier step. Another possibility is that the Rix1 complex is required in mammals for other ITS-2–processing factors, such as LAS1L and NOL9, to be recruited to the particle. Indeed, we observed that depletion of PELP1, TEX10, or WDR18 leads to a dramatic decrease in LAS1L protein stability (Figure S1A). Whether the Rix1 complex participates directly in the modification of pre-rRNAs or is simply required for the stabilization of other rRNA-processing factors remains to be determined. The possibility also remains that the Rix1 complex functions in a different manner in mammals than in yeast.
In this study, we also identified NOL9 as a novel LAS1L-interacting protein (Figures 1A and 2A). NOL9 was recently discovered to be a 5′-polynucleotide kinase that can phosphorylate both single- and double-stranded RNA (Heindl and Martinez, 2010). The pre-rRNA–processing defects seen upon depletion of NOL9 and LAS1L are identical. Endonucleolytic cleavage within the ITS-2 in the 32S pre-rRNA generates the 12S and 28S pre-rRNAs, and the lower levels of 12S pre-rRNA and accrual of the 32S intermediate upon depletion implies that both NOL9 and LAS1L are required for efficient ITS-2 scission (Figure 7, B and C). Depletion of NOL9 results in a 5′ extended 5.8S rRNA intermediate following ITS-1 cleavage (Heindl and Martinez, 2010), suggesting that 5′ phosphorylation of the extended 5.8S is required for subsequent exonucleolytic processing. Indeed, the exonuclease Rat1p (XRN2 in human) is necessary for the 5′-end maturation of the 5.8S and 25S rRNAs following endonucleolytic cleavage (Henry et al., 1994; Geerlings et al., 2000; Xue et al., 2000; Fang et al., 2005; El Hage et al., 2008) and requires a 5′-monophosphate for efficient hydrolysis of substrates (Stevens, 1980). Interestingly, the S. cerevisiae homologues of NOL9 (Grc3p) and LAS1L (Las1p) were both found by tandem affinity purification and mass spectrometry analysis to be interactors of Rai1p, a cofactor for Rat1p (Sydorskyy et al., 2003). Although the role of this interaction has not been explored, it indicates that Grc3p and Las1p might act with Rat1p and Rai1p in rRNA processing. The endonuclease performing the ITS-2 cleavage to generate the 12S and extended 28S rRNAs has yet to be identified both in budding yeast and mammals. LAS1L, with the Rix1 complex (PELP1-WDR18-TEX10), could be responsible for recruiting this endonuclease with NOL9 and XRN2 to the 32S rRNA for efficient cleavage, 5′ phosphorylation, and subsequent exonucleolytic processing.
Although Finkbeiner et al. (2011) previously described the mammalian Rix1 complex and LAS1L as interacting, the exact nature of the association between LAS1L and the proteins in the mammalian Rix1 complex has not been fully explored. Our further analysis of these interactions using sucrose gradient fractionation on nuclear extracts and subsequent coimmunoprecipitation experiments has revealed that LAS1L and the Rix1 complex proteins do indeed interact in the pre-60S fractions (Figure 6, B and C). However, in the fractions corresponding to free nuclear proteins and smaller protein complexes, LAS1L only associates with NOL9 (Figure 6C). Furthermore, PELP1 interacts only with WDR18 in the fractions corresponding to free proteins (Figure 6B). Based on these coimmunoprecipitation experiments, it is therefore likely that LAS1L and NOL9 form a separate complex that does not include PELP1 and WDR18 when LAS1L and NOL9 are not associated with the pre-60S particle. It is also interesting to note that, in our experiments, TEX10 only associates with PELP1 in fractions that correspond to pre-60S ribosomal particles (Figure 6B). One could then conclude from our experiments that the mammalian Rix1 complex could form only on the pre-60S particle. However, further analysis of the maturation of mammalian pre-60S ribosomal particles will be necessary to investigate the temporal association of these proteins with pre-60S particles in mammalian ribosome biogenesis.
Another protein found to interact with LAS1L in this study is RanBP5 (Figure 2A), a karyopherin β that associates with the nuclear pore complex to facilitate the import of both small and large ribosomal proteins (Jakel and Gorlich, 1998; Chou et al., 2010). RanBP5 is homologous with the karyopherin-β proteins Pse1p/Kap121p and Yrb4p/Kap123p in S. cerevisiae. Both Kap121p and Kap123p are responsible not only for the nuclear import of ribosomal proteins (Rout et al., 1997), but also for the export of preribosomal particles (Moy and Silver, 1999; Sydorskyy et al., 2003). Furthermore, Kap121p is required for nuclear import of factors involved in preribosomal particle formation (Leslie et al., 2004; Lebreton et al., 2006). RanBP5 may be involved in importing LAS1L and its interacting proteins to the nucleus, and this could explain the aberrant accumulation of the 32S intermediate observed upon RanBP5 depletion (Figure 7, B and D). Furthermore, it is possible that the defects in rRNA processing seen upon depletion of RanBP5 are a downstream effect resulting from failed nuclear import of ribosomal proteins, which leads to nuclear accumulation of immature preribosomal particles (Figure 7, B and D). Impending investigations will be aimed toward defining a precise role for RanBP5 in the maturation of the 60S preribosomal particles.
Recent studies in budding yeast have suggested that SUMOylation could play an important role in ribosome biogenesis. Analysis of the 40S and 60S preribosomal particles at different maturation stages has shown that early particles are decorated with SUMO (Panse et al., 2006). Moreover, investigation of the SUMO proteome revealed that ribosomal proteins and several processing factors involved in the 40S and 60S synthesis are also modified by SUMOylation (Panse et al., 2006). The nucleolar SUMO-deconjugating enzyme SENP3 was recently shown to catalyze the removal of SUMO conjugates on NPM1, a process required for efficient rRNA processing and synthesis of the 60S ribosomal subunit (Haindl et al., 2008). NPM1 was also shown to be required for stable accumulation of SENP3 in the nucleolus (Kuo et al., 2008; Yun et al., 2008). In this study, we found that SENP3 associates with LAS1L, and both SENP3 and NPM1 interact with PELP1 (Figure 2, A and B). We further describe LAS1L and PELP1 as SUMOylated substrates of SENP3, wherein loss of SENP3 or NPM1 results in accrual of SUMOylated forms of these proteins (Figure 11), and this is supported by previously published work (Finkbeiner et al., 2011). Moreover, we demonstrate that depletion of SENP3 or NPM1 results in a relocalization of LAS1L and PELP1 from the nucleolus with no observed disassembly of the LAS1L complex (Figures 9 and 10), suggesting that nucleolar localization is not required for the complex to assemble. We further observed that induction of a nucleolar stress through treatment with actinomycin D also results in relocalization of the complex from the nucleolus to the nucleoplasm (Figure 8). Previous work has demonstrated that depletion of NPM1 and SENP3 impairs ribosome biogenesis, which could in turn lead to a nucleolar stress (Haindl et al., 2008; Yun et al., 2008). We therefore cannot eliminate the possibility that nucleolar stress in general causes relocalization of LAS1L and PELP1 out of the nucleolus. However, as depletion of SENP3 or NPM1 results in accumulation of SUMOylated LAS1L and PELP1 concomitant with their relocalization from the nucleolus to the nucleoplasm, it is likely that the nucleolar relocalization is due to the accumulation of SUMOylated forms of these proteins. It is also conceivable that SUMOylation of LAS1L and PELP1 is a downstream effect of general nucleolar stress, and it will be interesting to further investigate this possibility. Taken together, our data suggest that SUMOylation partially regulates ribosome biogenesis by modulating the subcellular localization of LAS1L and PELP1. Indeed, studies from Finkbeiner and colleagues demonstrated that expression of a PELP1-SUMO fusion protein results in relocalization of PELP1 from the nucleolus to the nucleoplasm (Finkbeiner et al., 2011). Interestingly, we observed that SENP3 mainly interacts with LAS1L and the Rix1 complex in the fractions corresponding to the 60S particle (Figure 6). SENP3 could participate in a quality control checkpoint serving to limit access of SUMOylated ribosome biogenesis factors to the 60S particle. Whether or not the other proteins in the complex can be SUMOylated remains to be determined, and future experiments will be focused on investigating a role for SUMO in regulating the function of the LAS1L complex.
Increased ribosome biogenesis has been correlated with the rapid growth of cancer cells (Ruggero and Pandolfi, 2003). PELP1 is considered an oncogene and is found overexpressed and mislocalized in a variety of hormonal-responsive tumors (Vadlamudi et al., 2004; Nair et al., 2007; Habashy et al., 2010). We further determined that PELP1 plays a role in ribosome biogenesis in addition to its role as a nuclear receptor coactivator. Interestingly, early studies in the rat pituitary have correlated estrogen treatment with enhanced Pol I transcription and increased amounts of total rRNA (Ying et al., 1996), though no molecular mechanism for this association has been described to date. More recently analysis of the estrogen-signaling transcriptome revealed that estrogen regulates the activity of RNA Pol I (Hah et al., 2011). It will be interesting to determine whether PELP1 provides a link among estrogen signaling, Pol I transcription, and pre-rRNA processing.
MATERIALS AND METHODS
Cell culture and RNA interference
Cells (HEK 293T, HCT116, and U2OS) were cultured at 37°C with 5% CO2 in DMEM high-glucose (Hyclone; ThermoFisher, Lafayette, CO) supplemented with 5% fetal bovine serum (FBS; Hyclone) and penicillin–streptomycin. Synthetic siRNA oligonucleotides (Sigma-Aldrich, St. Louis, MO) were delivered into cells with Lipofectamine 2000 (Invitrogen, Carlsbad, CA) for the HEK 293T cells and Lipofectamine RNAi Max (Invitrogen) for the HCT116 cells. siRNA oligonucleotide (20 nM) was transfected using the reverse-transfection protocol according to the manufacturer's instructions. The following siRNA sequences were used: nontargeting scramble: 5′-GAUCAUACGUGCGAUCAGAdTdT-3′; human LAS1L: 5′-CUGAUACGCUGUAAGCUCUdTdT-3′; human PELP1: 5′-GGAAUGAAGGCUUGUAUGAdTdT-3′; human TEX10: 5′-CUGAUUUGCUUUCUCGGUUdTdT-3′; human WDR18: 5′-GUCUCCUGCCUUCAGUUCAdTdT-3′; human SENP3: 5′-CACCAGGGCUGGAAAGGUUdTdT-3′; human NOL9: 5′-CCACUUUCUCCUUUACAUAdTdT-3′; human RanBP5: 5′-CUCUACAGCUAAGUCUAAAdTdT-3′ human NPM1: 5′- GGACAAGAATCCTTCAAGAdTdT-3′.
Cell cycle analysis
Analysis of cell cycle profiles was performed by measuring DNA content using PI and BrdU labeling, as we described previously (Castle et al., 2010).
Coimmunoprecipitation, immunoblotting, and antibodies
For coimmunoprecipitation experiments, cells were lysed in ELB buffer (50 mM HEPES, pH 7.2, 250 mM NaCl, 2 mM EDTA, 0.5% NP-40) plus protease inhibitors (aprotinin, leupeptin, 4-(2-aminoethyl) benzenesulfonyl fluoride hydrochloride) on ice for 15 min. Lysates were cleared by centrifugation at 22,000 × g for 10 min at 4°C and the supernatant was incubated with 1 μg of antibody for 1 h with rotation at 4°C. Protein G Sepharose beads (Invitrogen) were added for another 1 h with rotation at 4°C. The antibodies used for immunoprecipitations were: normal rabbit immunoglobulin G (IgG; Calbiochem, San Diego, CA), LAS1L (AV34629; Sigma-Aldrich), PELP1 (Bethyl Laboratories, Montgomery, TX). For immunoblotting, cells were lysed in RIPA buffer (25 mM Tris-HCl, pH 7.6, 150 mM NaCl, 1% NP-40, 1% Triton X-100, 1% sodium deoxycholate, and 0.1% SDS) plus protease inhibitors (aprotinin, leupeptin, AEBSF) and phosphatase inhibitor cocktail (ThermoFisher, Lafayette, CO) for 15 min on ice. Lysates were cleared by centrifugation at 22,000 × g for 10 min at 4°C. Protein concentrations were evaluated with the BCA kit (Pierce, Rockford, IL). Proteins were separated by SDS–PAGE and transferred to a nitrocellulose membrane (Bio-Rad, Hercules, CA). The following antibodies were used: LAS1L (Sigma-Aldrich), PELP1 (Bethyl Laboratories, Montgomery, TX), NOL9 (ProteinTech), RanPB5 (Sigma-Aldrich), NPM1 (Santa Cruz Biotechnology, Santa Cruz, CA), TEX10 (ProteinTech), SENP3 (Santa Cruz Biotechnology), NOL9 (Abgent, San Diego, CA), p53 (Santa Cruz Biotechnology), β-actin (Santa Cruz Biotechnology), CDK2 (Santa Cruz Biotechnology), RPL11 (Sigma-Aldrich), RPL26 (Sigma-Aldrich), and p21 (PharMingen, BD Biosciences, San Diego, CA). An anti-WDR18 antibody was raised by immunizing rabbits with the CRKINRDLFDFSTRFITRPAK peptide coupled to keyhole limpet hemocyanin (Open Biosystems, Huntsville, AL).
Mass spectrometry analysis
Cells were lysed in ELB buffer (50 mM HEPES, pH 7.2, 250 mM NaCl, 2 mM EDTA, 0.5% NP-40) plus protease inhibitors (aprotinin, leupeptin, AEBSF) on ice for 15 min, and lysates were cleared by centrifugation at 22,000 × g for 10 min at 4°C. The supernatants were incubated with 2 μg of normal rabbit IgG or anti-LAS1L antibody for 1 h with rotation at 4°C. Protein G Sepharose beads were added for another 1 h with rotation at 4°C. Precipitated complexes were eluted in SDS–PAGE sample buffer and separated on a 10% SDS–PAGE. The gel was silver-stained (silver stain kit; Pierce) and proteins present only in the anti-LAS1L lane were cut out, digested with trypsin, and analyzed by mass spectrometry.
Sucrose gradient fractionation and gradient coimmunoprecipitation
Sucrose gradient fractionation was performed according to Pestov et al. (2008). Briefly, cells were harvested and incubated in low salt buffer (10 mM HEPES-NaOH, pH 7.5, 10 mM NaCl, 2 mM MgCl2, 1 mM ethylene glycol tetraacetic acid) for 10 min and lysed by the addition of Igepal CA-630 and sodium deoxycholate to final concentrations of 0.3% and 0.2%, respectively. Nuclei were sedimented by centrifugation at 1000 × g for 5 min and resuspended in sonication buffer (25 mM Tris-HCl, pH 7.5, 100 mM KCl, 1 mM NaF, 2 mM EDTA, 0.05% Igepal CA-630, 1 mM dithiothreitol [DTT]). Protease inhibitors (aprotinin, leupeptin, AEBSF) and RNase inhibitor (100 U/ml; ThermoFisher, Lafayette, CO) were added, and nuclei were disrupted by sonication. The lysate was cleared by centrifugation at 15,000 × g for 15 min and was then separated on a 10–30% (wt/vol) sucrose gradient made in 25 mM Tris-HCl (pH 7.5), 100 mM KCl, 2 mM EDTA, and 1 mM DTT for 3 h at 160,000 × g using a Beckman SW 41 Ti rotor. Equal fractions were collected, and proteins were extracted by trichloroacetic acid precipitation, resuspended in urea sample buffer (62.5 mM Tris-HCl, pH 6.8, 3 M urea, 2% SDS, 0.01% bromophenol blue, 0.1 M DTT), and subjected to SDS–PAGE analysis. RNA was purified from sucrose gradient fractions for subsequent Northern blot analysis, as we previously described (Castle et al., 2010). For coimmunoprecipitation from the sucrose gradient fractions, the gradient fractionation was performed as above. A260 of each fraction was measured, and fractions corresponding to free nuclear proteins, pre-40S, and pre-60S ribosomal particles were pooled. The pooled fractions were brought to a final concentration of 250 mM KCl and 0.5% NP-40 and then incubated in 2 μg antibody for 1 h with rotation at 4°C. Protein G Sepharose beads (Invitrogen) were added for another hour with rotation at 4°C. The antibodies used for immunoprecipitations were: normal rabbit IgG (Calbiochem), LAS1L (AV34629; Sigma-Aldrich), and PELP1 (Bethyl Laboratories). ReliaBLOT (Bethyl Laboratories) was used for immunoblotting with the custom WDR18 antibody to minimize the IgG band.
RNA extractions, Northern blot, and qRT-PCR analysis
Total RNA was extracted with Trizol reagent (Invitrogen). Northern blots were performed as we previously described (Castle et al., 2010). For qRT-PCR analysis, cDNA was synthesized using 1 μg of total RNA with the QuantiTect reverse transcription kit (Qiagen, Valencia, CA). Real-time PCR analysis was performed on a Light Cycler 480 real-time PCR instrument (Mnaimneh et al., 2004). Values were normalized to β-actin mRNA. The primers used for the quantitative PCR are the following: β-actin (5′-GGACTTCGAGCAAGAGATGG-3′, 5′-AGCACTGTGTTGGCGTACAG-3′); LAS1L (5′-CGAAGAGGAGCAGTTTACGG-3′, 5′-GGAAGCCATCATCCAGAAAA-3′); PELP1 (5′-AGCAGCACTGTGTGTCTTGG-3′, 5′-TTCCAATGCTGACTGCTCAC-3′); RanBP5 (5′-TGCCTCTTCTCCTGGAGTGT-3′, 5′-TGCACTTTGCAAAAGAATGC-3′); TEX10 (5′-TTGCAACTTGCTCATCTTGG-3′, 5′-AGAGTCTGCAGGGAGAACCA-3′); NOL9 (5′-CTCCGGATTACCCACTCTGA-3′, 5′-GCCTCTACAGATGCCAAAGC-3′); SENP3 (5′-TTTTGATGCCTCAGCAAGTG-3′, 5′-AATGTCAGCGAGGTGCTTTT-3′); and WDR18 (5′-ACCAGACGGTGAAGCTATGG-3′, 5′-GTGGAAGCTCCTCTCCCTCT-3′).
Immunofluorescence microscopy
Cells grown on coverslips were preextracted with 0.1% Triton in phosphate-buffered saline (PBS) for 1 min at room temperature before fixation with 4% paraformaldehyde at room temperature for 15 min. Cells were then permeabilized with 0.1% Triton for 10 min at room temperature. All samples were blocked with 1% bovine serum albumin for 30 min, washed with PBS, and incubated with the appropriate primary antibody for 1 h. Cells were washed with PBS containing 0.05% Tween-20 and incubated with secondary antibody for 1 h (Alexa Fluor 488 goat anti–rabbit antibody and Alexa Fluor 594 goat anti–mouse antibody; Invitrogen). Cells were washed again with PBS containing 0.05% Tween-20, stained with 4′,6-diamidino-2-phenylindole (Molecular Probes, Invitrogen), and mounted on slides with Vectashield (Vector Laboratories, Burlingame, CA). Fluorescence microscopy was performed on a Zeiss Axioskop 40 fluorescence microscope with a Plan-Apochromat 63×/1.4 numerical aperture, oil-immersion, differential interference contrast objective (Jena, Germany). Images were acquired with an Axiocam MRm camera using the Axiovision Release 4.6 software. All microscopy was performed at room temperature, and all images were prepared in Adobe Photoshop and Adobe Illustrator (San Jose, CA).
SUMOylation assays
Analyses of SUMOylated proteins were performed as described in Tatham et al. (2009). For exogenously expressed LAS1L, HEK 293T cells were transfected with FLAG-LAS1L plus either empty vector or plasmids expressing 6xHis-tagged SUMO-1 (aa 1–97) or SUMO-3 (aa 1–93). For SUMOylation analysis of endogenous LAS1L and PELP1, HEK 293T cells were transfected with either control, SENP3, or NMP1 siRNA using Lipofectamine 2000 (Invitrogen). The next day, cells were transfected with empty vector or a plasmid expressing SUMO-3 (aa 1–93), using Lipofectamine 2000 (Invitrogen). Forty-eight hours after transfection, the cells were harvested and lysed in lysis buffer (6 M guanidine-HCl, 0.1 M sodium phosphate buffer, pH 8, 0.01 M Tris-HCl, pH. 8, 10 mM β-mercaptoethanol, and 5 mM imidazole) with sonication. Ni-NTA agarose beads were added to the cleared lysates and incubated for 2 h at 4°C with rotation. The beads were washed one time with lysis buffer, which was followed by one wash in buffer A (8 M urea, 0.1 M sodium phosphate buffer, pH 8, 0.01 M Tris-HCl, pH. 8, 5 mM β-mercaptoethanol, and 0.1% Triton X-100) and three washes in buffer B (8 M urea, 0.1 M sodium phosphate buffer, pH 6.3, 0.01 M Tris-HCl, pH. 6.3, 5 mM β-mercaptoethanol, and 0.1% Triton X-100). The pulled-down SUMOylated proteins were eluted off the beads with 200 mM imidazole, 5% SDS, 0.15 M Tris-HCl (pH 6.8), 30% glycerol, and 0.72 M β-mercaptoethanol. The eluates were analyzed on SDS–PAGE and Western blotting with anti-FLAG M2 (Sigma-Aldrich), LAS1L (Sigma-Aldrich), PELP1 (Bethyl Laboratories), or 6xHis (Bethyl Laboratories) antibodies.
Nucleolar isolation
Nucleolar isolation was performed according to Muramatsu et al. (1963), with the following changes: briefly, HEK 293T cells were washed in PBS and incubated in buffer A (10 mM HEPES, pH 7.9, 10 mM KCl, 1.5 mM MgCl2, 0.5 mM DTT) for 5 min. Cells were then homogenized in a Dounce tissue homogenizer until more than 90% of the cells burst and then centrifuged at 300 × g for 5 min. Nuclei were resuspended in S1 solution (0.25 M sucrose, 10 mM MgCl2), layered over S2 solution (0.35 M sucrose, 0.5 mM MgCl2), and centrifuged at 1400 × g for 5 min. Nuclei were resuspended in S2 solution and sonicated until there were no intact cells, and nucleoli were readily observed by phase-contrast microscopy. Sonicated extracts were layered over S3 solution (0.88 M sucrose, 0.5 MgCl2) and centrifuged at 3000 × g for 10 min. The nucleolar pellet was resuspended in S2 solution and centrifuged at 1400 × g for 5 min. Immunoblotting was then performed on the nucleolar pellet as described in the Coimmunoprecipitation, immunoblotting, and antibodies section.
Supplementary Material
Acknowledgments
We thank Ghislain Breton for critical review of the manuscript. We are grateful to Victoria Borgianini for her technical assistance.
Abbreviations used:
- BrdU
bromodeoxyuridine
- DMSO
dimethyl sulfoxide
- DTT
dithiothreitol
- ETS
external transcribed spacer
- FACS
fluorescence-activated cell sorter
- IgG
immunoglobulin G
- ITS
internal transcribed spacer
- mTOR
mammalian target of rapamycin;
- Ni-NTA
Ni-nitrolotriacetic acid
- NPM1
nucleophosmin
- PBS
phosphate-buffered saline
- PI
propidium iodide
- Pol I/II/III
RNA polymerase I/II/III
- qRT-PCR
quantitative reverse transcriptase PCR
- RNAi
RNA interference
- siRNA
small interfering RNA
- SUMO
small ubiquitin-like modifier
- UBF1
upstream binding factor 1
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-06-0530) on December 21, 2011.
REFERENCES
- Arabi A, et al. c-Myc associates with ribosomal DNA and activates RNA polymerase I transcription. Nat Cell Biol. 2005;7:303–310. doi: 10.1038/ncb1225. [DOI] [PubMed] [Google Scholar]
- Bassler J, Grandi P, Gadal O, Lessmann T, Petfalski E, Tollervey D, Lechner J, Hurt E. Identification of a 60S preribosomal particle that is closely linked to nuclear export. Mol Cell. 2001;8:517–529. doi: 10.1016/s1097-2765(01)00342-2. [DOI] [PubMed] [Google Scholar]
- Boisvert FM, van Koningsbruggen S, Navascues J, Lamond AI. The multifunctional nucleolus. Nat Rev Mol Cell Biol. 2007;8:574–585. doi: 10.1038/nrm2184. [DOI] [PubMed] [Google Scholar]
- Braglia P, Heindl K, Schleiffer A, Martinez J, Proudfoot NJ. Role of the RNA/DNA kinase Grc3 in transcription termination by RNA polymerase I. EMBO Rep. 2010;11:758–764. doi: 10.1038/embor.2010.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budde A, Grummt I. p53 represses ribosomal gene transcription. Oncogene. 1999;18:1119–1124. doi: 10.1038/sj.onc.1202402. [DOI] [PubMed] [Google Scholar]
- Castle CD, Cassimere EK, Lee J, Denicourt C. Las1L is a nucleolar protein required for cell proliferation and ribosome biogenesis. Mol Cell Biol. 2010;30:4404–4414. doi: 10.1128/MCB.00358-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanaugh AH, Hempel WM, Taylor LJ, Rogalsky V, Todorov G, Rothblum LI. Activity of RNA polymerase I transcription factor UBF blocked by Rb gene product. Nature. 1995;374:177–180. doi: 10.1038/374177a0. [DOI] [PubMed] [Google Scholar]
- Chou CW, Tai LR, Kirby R, Lee IF, Lin A. Importin β3 mediates the nuclear import of human ribosomal protein L7 through its interaction with the multifaceted basic clusters of L7. FEBS Lett. 2010;584:4151–4156. doi: 10.1016/j.febslet.2010.08.044. [DOI] [PubMed] [Google Scholar]
- Deisenroth C, Zhang Y. Ribosome biogenesis surveillance: probing the ribosomal protein-Mdm2-p53 pathway. Oncogene. 2010;29:4253–4260. doi: 10.1038/onc.2010.189. [DOI] [PubMed] [Google Scholar]
- Doseff AI, Arndt KT. LAS1 is an essential nuclear protein involved in cell morphogenesis and cell surface growth. Genetics. 1995;141:857–871. doi: 10.1093/genetics/141.3.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Hage A, Koper M, Kufel J, Tollervey D. Efficient termination of transcription by RNA polymerase I requires the 5′ exonuclease Rat1 in yeast. Genes Dev. 2008;22:1069–1081. doi: 10.1101/gad.463708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falini B, et al. Cytoplasmic nucleophosmin in acute myelogenous leukemia with a normal karyotype. N Engl J Med. 2005;352:254–266. doi: 10.1056/NEJMoa041974. [DOI] [PubMed] [Google Scholar]
- Fang F, Phillips S, Butler JS. Rat1p and Rai1p function with the nuclear exosome in the processing and degradation of rRNA precursors. RNA. 2005;11:1571–1578. doi: 10.1261/rna.2900205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkbeiner E, Haindl M, Muller S. The SUMO system controls nucleolar partitioning of a novel mammalian ribosome biogenesis complex. EMBO J. 2011;30:1067–1078. doi: 10.1038/emboj.2011.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galani K, Nissan TA, Petfalski E, Tollervey D, Hurt E. Rea1, a dynein-related nuclear AAA-ATPase, is involved in late rRNA processing and nuclear export of 60 S subunits. J Biol Chem. 2004;279:55411–55418. doi: 10.1074/jbc.M406876200. [DOI] [PubMed] [Google Scholar]
- Geerlings TH, Vos JC, Raue HA. The final step in the formation of 25S rRNA in Saccharomyces cerevisiae is performed by 5′ → 3′ exonucleases. RNA. 2000;6:1698–1703. doi: 10.1017/s1355838200001540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiss-Friedlander R, Melchior F. Concepts in sumoylation: a decade on. Nat Rev Mol Cell Biol. 2007;8:947–956. doi: 10.1038/nrm2293. [DOI] [PubMed] [Google Scholar]
- Gomez-Roman N, et al. Activation by c-Myc of transcription by RNA polymerases I, II and III. Biochem Soc Symp. 2006:141–154. doi: 10.1042/bss0730141. [DOI] [PubMed] [Google Scholar]
- Gong L, Yeh ET. Characterization of a family of nucleolar SUMO-specific proteases with preference for SUMO-2 or SUMO-3. J Biol Chem. 2006;281:15869–15877. doi: 10.1074/jbc.M511658200. [DOI] [PubMed] [Google Scholar]
- Gonugunta VK, Nair BC, Rajhans R, Sareddy GR, Nair SS, Vadlamudi RR. Regulation of rDNA transcription by proto-oncogene PELP1. PLoS One. 2011;6:e21095. doi: 10.1371/journal.pone.0021095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandori C, Gomez-Roman N, Felton-Edkins ZA, Ngouenet C, Galloway DA, Eisenman RN, White RJ. c-Myc binds to human ribosomal DNA and stimulates transcription of rRNA genes by RNA polymerase I. Nat Cell Biol. 2005;7:311–318. doi: 10.1038/ncb1224. [DOI] [PubMed] [Google Scholar]
- Grimm T, Holzel M, Rohrmoser M, Harasim T, Malamoussi A, Gruber-Eber A, Kremmer E, Eick D. Dominant-negative Pes1 mutants inhibit ribosomal RNA processing and cell proliferation via incorporation into the PeBoW-complex. Nucleic Acids Res. 2006;34:3030–3043. doi: 10.1093/nar/gkl378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grisendi S, Mecucci C, Falini B, Pandolfi PP. Nucleophosmin and cancer. Nat Rev Cancer. 2006;6:493–505. doi: 10.1038/nrc1885. [DOI] [PubMed] [Google Scholar]
- Habashy HO, Powe DG, Rakha EA, Ball G, Macmillan RD, Green AR, Ellis IO. The prognostic significance of PELP1 expression in invasive breast cancer with emphasis on the ER-positive luminal-like subtype. Breast Cancer Res Treat. 2010;120:603–612. doi: 10.1007/s10549-009-0419-9. [DOI] [PubMed] [Google Scholar]
- Hadjiolova KV, Nicoloso M, Mazan S, Hadjiolov AA, Bachellerie JP. Alternative pre-rRNA processing pathways in human cells and their alteration by cycloheximide inhibition of protein synthesis. Eur J Biochem. 1993;212:211–215. doi: 10.1111/j.1432-1033.1993.tb17652.x. [DOI] [PubMed] [Google Scholar]
- Hah N, Danko CG, Core L, Waterfall JJ, Siepel A, Lis JT, Kraus WL. A rapid, extensive, and transient transcriptional response to estrogen signaling in breast cancer cells. Cell. 2011;145:622–634. doi: 10.1016/j.cell.2011.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haindl M, Harasim T, Eick D, Muller S. The nucleolar SUMO-specific protease SENP3 reverses SUMO modification of nucleophosmin and is required for rRNA processing. EMBO Rep. 2008;9:273–279. doi: 10.1038/embor.2008.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harnpicharnchai P, et al. Composition and functional characterization of yeast 66S ribosome assembly intermediates. Mol Cell. 2001;8:505–515. doi: 10.1016/s1097-2765(01)00344-6. [DOI] [PubMed] [Google Scholar]
- Heindl K, Martinez J. Nol9 is a novel polynucleotide 5′-kinase involved in ribosomal RNA processing. EMBO J. 2010;29:4161–4171. doi: 10.1038/emboj.2010.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry Y, Wood H, Morrissey JP, Petfalski E, Kearsey S, Tollervey D. The 5′ end of yeast 5.8S rRNA is generated by exonucleases from an upstream cleavage site. EMBO J. 1994;13:2452–2463. doi: 10.1002/j.1460-2075.1994.tb06530.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzel M, Orban M, Hochstatter J, Rohrmoser M, Harasim T, Malamoussi A, Kremmer E, Langst G, Eick D. Defects in 18 S or 28 S rRNA processing activate the p53 pathway. J Biol Chem. 2010;285:6364–6370. doi: 10.1074/jbc.M109.054734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzel M, et al. Mammalian WDR12 is a novel member of the Pes1-Bop1 complex and is required for ribosome biogenesis and cell proliferation. J Cell Biol. 2005;170:367–378. doi: 10.1083/jcb.200501141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang N, Negi S, Szebeni A, Olson MO. Protein NPM3 interacts with the multifunctional nucleolar protein B23/nucleophosmin and inhibits ribosome biogenesis. J Biol Chem. 2005;280:5496–5502. doi: 10.1074/jbc.M407856200. [DOI] [PubMed] [Google Scholar]
- Itahana K, Bhat KP, Jin A, Itahana Y, Hawke D, Kobayashi R, Zhang Y. Tumor suppressor ARF degrades B23, a nucleolar protein involved in ribosome biogenesis and cell proliferation. Mol Cell. 2003;12:1151–1164. doi: 10.1016/s1097-2765(03)00431-3. [DOI] [PubMed] [Google Scholar]
- Jakel S, Gorlich D. Importin β, transportin, RanBP5 and RanBP7 mediate nuclear import of ribosomal proteins in mammalian cells. EMBO J. 1998;17:4491–4502. doi: 10.1093/emboj/17.15.4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayahara M, Ohanian J, Ohanian V, Berry A, Vadlamudi R, Ray DW. MNAR functionally interacts with both NH2- and COOH-terminal GR domains to modulate transactivation. Am J Physiol Endocrinol Metab. 2008;295:E1047–E1055. doi: 10.1152/ajpendo.90429.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopp K, Gasiorowski JZ, Chen D, Gilmore R, Norton JT, Wang C, Leary DJ, Chan EK, Dean DA, Huang S. Pol I transcription and pre-rRNA processing are coordinated in a transcription-dependent manner in mammalian cells. Mol Biol Cell. 2007;18:394–403. doi: 10.1091/mbc.E06-03-0249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kos M, Tollervey D. Yeast pre-rRNA processing and modification occur cotranscriptionally. Mol Cell. 2010;37:809–820. doi: 10.1016/j.molcel.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kressler D, Hurt E, Bassler J. Driving ribosome assembly. Biochim Biophys Acta. 2010;1803:673–683. doi: 10.1016/j.bbamcr.2009.10.009. [DOI] [PubMed] [Google Scholar]
- Kressler D, Roser D, Pertschy B, Hurt E. The AAA ATPase Rix7 powers progression of ribosome biogenesis by stripping Nsa1 from pre-60S particles. J Cell Biol. 2008;181:935–944. doi: 10.1083/jcb.200801181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan NJ, et al. High-definition macromolecular composition of yeast RNA-processing complexes. Mol Cell. 2004;13:225–239. doi: 10.1016/s1097-2765(04)00003-6. [DOI] [PubMed] [Google Scholar]
- Kuo M, den Besten W, Thomas MC, Sherr CJ. Arf-induced turnover of the nucleolar nucleophosmin-associated SUMO-2/3 protease Senp3. Cell Cycle. 2008;7:3378–3387. doi: 10.4161/cc.7.21.6930. [DOI] [PubMed] [Google Scholar]
- Lapik YR, Fernandes CJ, Lau LF, Pestov DG. Physical and functional interaction between Pes1 and Bop1 in mammalian ribosome biogenesis. Mol Cell. 2004;15:17–29. doi: 10.1016/j.molcel.2004.05.020. [DOI] [PubMed] [Google Scholar]
- Lebreton A, Saveanu C, Decourty L, Rain JC, Jacquier A, Fromont-Racine M. A functional network involved in the recycling of nucleocytoplasmic pre-60S factors. J Cell Biol. 2006;173:349–360. doi: 10.1083/jcb.200510080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lempiainen H, Shore D. Growth control and ribosome biogenesis. Curr Opin Cell Biol. 2009;21:855–863. doi: 10.1016/j.ceb.2009.09.002. [DOI] [PubMed] [Google Scholar]
- Leslie DM, Zhang W, Timney BL, Chait BT, Rout MP, Wozniak RW, Aitchison JD. Characterization of karyopherin cargoes reveals unique mechanisms of Kap121p-mediated nuclear import. Mol Cell Biol. 2004;24:8487–8503. doi: 10.1128/MCB.24.19.8487-8503.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YP, Busch RK, Valdez BC, Busch H. C23 interacts with B23, a putative nucleolar-localization-signal-binding protein. Eur J Biochem. 1996;237:153–158. doi: 10.1111/j.1432-1033.1996.0153n.x. [DOI] [PubMed] [Google Scholar]
- Li Z, Lee I, Moradi E, Hung NJ, Johnson AW, Marcotte EM. Rational extension of the ribosome biogenesis pathway using network-guided genetics. PLoS Biol. 2009;7:e1000213. doi: 10.1371/journal.pbio.1000213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer C, Grummt I. Ribosome biogenesis and cell growth: mTOR coordinates transcription by all three classes of nuclear RNA polymerases. Oncogene. 2006;25:6384–6391. doi: 10.1038/sj.onc.1209883. [DOI] [PubMed] [Google Scholar]
- Mayer C, Zhao J, Yuan X, Grummt I. mTOR-dependent activation of the transcription factor TIF-IA links rRNA synthesis to nutrient availability. Genes Dev. 2004;18:423–434. doi: 10.1101/gad.285504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendes-da-Silva P, Moreira A, Duro-da-Costa J, Matias D, Monteiro C. Frequent loss of heterozygosity on chromosome 5 in non-small cell lung carcinoma. Mol Pathol. 2000;53:184–187. doi: 10.1136/mp.53.4.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mnaimneh S, et al. Exploration of essential gene functions via titratable promoter alleles. Cell. 2004;118:31–44. doi: 10.1016/j.cell.2004.06.013. [DOI] [PubMed] [Google Scholar]
- Moy TI, Silver PA. Nuclear export of the small ribosomal subunit requires the ran-GTPase cycle and certain nucleoporins. Genes Dev. 1999;13:2118–2133. doi: 10.1101/gad.13.16.2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muramatsu M, Smetana K, Busch H. Quantitative aspects of isolation of nucleoli of the Walker carcinosarcoma and liver of the rat. Cancer Res. 1963;23:510–518. [PubMed] [Google Scholar]
- Nair SS, Guo Z, Mueller JM, Koochekpour S, Qiu Y, Tekmal RR, Schule R, Kung HJ, Kumar R, Vadlamudi RK. Proline-, glutamic acid-, and leucine-rich protein-1/modulator of nongenomic activity of estrogen receptor enhances androgen receptor functions through LIM-only coactivator, four-and-a-half LIM-only protein 2. Mol Endocrinol. 2007;21:613–624. doi: 10.1210/me.2006-0269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair SS, Mishra SK, Yang Z, Balasenthil S, Kumar R, Vadlamudi RK. Potential role of a novel transcriptional coactivator PELP1 in histone H1 displacement in cancer cells. Cancer Res. 2004;64:6416–6423. doi: 10.1158/0008-5472.CAN-04-1786. [DOI] [PubMed] [Google Scholar]
- Nair SS, Nair BC, Cortez V, Chakravarty D, Metzger E, Schule R, Brann DW, Tekmal RR, Vadlamudi RK. PELP1 is a reader of histone H3 methylation that facilitates oestrogen receptor-α target gene activation by regulating lysine demethylase 1 specificity. EMBO Rep. 2010;11:438–444. doi: 10.1038/embor.2010.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair SS, Rajhans R, Nagpal J, Guider J, Kumar R, Vadlamudi R. PELP1/MNAR interacts with BCAS2: potential role in ER-mediated splicing. Proc Am Assoc Cancer Res. 2006;47:2930. [Google Scholar]
- Nissan TA, Bassler J, Petfalski E, Tollervey D, Hurt E. 60S pre-ribosome formation viewed from assembly in the nucleolus until export to the cytoplasm. EMBO J. 2002;21:5539–5547. doi: 10.1093/emboj/cdf547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissan TA, Galani K, Maco B, Tollervey D, Aebi U, Hurt E. A pre-ribosome with a tadpole-like structure functions in ATP-dependent maturation of 60S subunits. Mol Cell. 2004;15:295–301. doi: 10.1016/j.molcel.2004.06.033. [DOI] [PubMed] [Google Scholar]
- Nozawa Y, Van Belzen N, Van der Made AC, Dinjens WN, Bosman FT. Expression of nucleophosmin/B23 in normal and neoplastic colorectal mucosa. J Pathol. 1996;178:48–52. doi: 10.1002/(SICI)1096-9896(199601)178:1<48::AID-PATH432>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Panse VG, Johnson AW. Maturation of eukaryotic ribosomes: acquisition of functionality. Trends Biochem Sci. 2010;35:260–266. doi: 10.1016/j.tibs.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panse VG, Kressler D, Pauli A, Petfalski E, Gnadig M, Tollervey D, Hurt E. Formation and nuclear export of preribosomes are functionally linked to the small-ubiquitin-related modifier pathway. Traffic. 2006;7:1311–1321. doi: 10.1111/j.1600-0854.2006.00471.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng WT, et al. A panoramic view of yeast noncoding RNA processing. Cell. 2003;113:919–933. doi: 10.1016/s0092-8674(03)00466-5. [DOI] [PubMed] [Google Scholar]
- Pestov DG, Lapik YR, Lau LF. Assays for ribosomal RNA processing and ribosome assembly. Curr Protoc Cell Biol. 2008;Chapter 22 doi: 10.1002/0471143030.cb2211s39. Unit 22.11, DOI: 10.1002/0471143030.cb2211s39. [DOI] [PubMed] [Google Scholar]
- Poortinga G, et al. MAD1 and c-MYC regulate UBF and rDNA transcription during granulocyte differentiation. EMBO J. 2004;23:3325–3335. doi: 10.1038/sj.emboj.7600335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohrmoser M, Holzel M, Grimm T, Malamoussi A, Harasim T, Orban M, Pfisterer I, Gruber-Eber A, Kremmer E, Eick D. Interdependence of Pes1, Bop1, and WDR12 controls nucleolar localization and assembly of the PeBoW complex required for maturation of the 60S ribosomal subunit. Mol Cell Biol. 2007;27:3682–3694. doi: 10.1128/MCB.00172-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rout MP, Blobel G, Aitchison JD. A distinct nuclear import pathway used by ribosomal proteins. Cell. 1997;89:715–725. doi: 10.1016/s0092-8674(00)80254-8. [DOI] [PubMed] [Google Scholar]
- Ruggero D, Pandolfi PP. Does the ribosome translate cancer? Nat Rev Cancer. 2003;3:179–192. doi: 10.1038/nrc1015. [DOI] [PubMed] [Google Scholar]
- Schafer T, Strauss D, Petfalski E, Tollervey D, Hurt E. The path from nucleolar 90S to cytoplasmic 40S pre-ribosomes. EMBO J. 2003;22:1370–1380. doi: 10.1093/emboj/cdg121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlosser I, Holzel M, Murnseer M, Burtscher H, Weidle UH, Eick D. A role for c-Myc in the regulation of ribosomal RNA processing. Nucleic Acids Res. 2003;31:6148–6156. doi: 10.1093/nar/gkg794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shields LB, Gercel-Taylor C, Yashar CM, Wan TC, Katsanis WA, Spinnato JA, Taylor DD. Induction of immune responses to ovarian tumor antigens by multiparity. J Soc Gynecol Investig. 1997;4:298–304. [PubMed] [Google Scholar]
- Skaar TC, Prasad SC, Sharareh S, Lippman ME, Brunner N, Clarke R. Two-dimensional gel electrophoresis analyses identify nucleophosmin as an estrogen regulated protein associated with acquired estrogen-independence in human breast cancer cells. J Steroid Biochem Mol Biol. 1998;67:391–402. doi: 10.1016/s0960-0760(98)00142-3. [DOI] [PubMed] [Google Scholar]
- Stevens A. Purification and characterization of a Saccharomyces cerevisiae exoribonuclease which yields 5′-mononucleotides by a 5′ leads to 3′ mode of hydrolysis. J Biol Chem. 1980;255:3080–3085. [PubMed] [Google Scholar]
- Strezoska Z, Pestov DG, Lau LF. Bop1 is a mouse WD40 repeat nucleolar protein involved in 28S and 5.8S RRNA processing and 60S ribosome biogenesis. Mol Cell Biol. 2000;20:5516–5528. doi: 10.1128/mcb.20.15.5516-5528.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strezoska Z, Pestov DG, Lau LF. Functional inactivation of the mouse nucleolar protein Bop1 inhibits multiple steps in pre-rRNA processing and blocks cell cycle progression. J Biol Chem. 2002;277:29617–29625. doi: 10.1074/jbc.M204381200. [DOI] [PubMed] [Google Scholar]
- Subong EN, Shue MJ, Epstein JI, Briggman JV, Chan PK, Partin AW. Monoclonal antibody to prostate cancer nuclear matrix protein (PRO:4-216) recognizes nucleophosmin/B23. Prostate. 1999;39:298–304. doi: 10.1002/(sici)1097-0045(19990601)39:4<298::aid-pros11>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- Sydorskyy Y, Dilworth DJ, Yi EC, Goodlett DR, Wozniak RW, Aitchison JD. Intersection of the Kap123p-mediated nuclear import and ribosome export pathways. Mol Cell Biol. 2003;23:2042–2054. doi: 10.1128/MCB.23.6.2042-2054.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M, Sasaki H, Kino I, Sugimura T, Terada M. Genes preferentially expressed in embryo stomach are predominantly expressed in gastric cancer. Cancer Res. 1992;52:3372–3377. [PubMed] [Google Scholar]
- Tatham MH, Rodriguez MS, Xirodimas DP, Hay RT. Detection of protein SUMOylation in vivo. Nat Protoc. 2009;4:1363–1371. doi: 10.1038/nprot.2009.128. [DOI] [PubMed] [Google Scholar]
- Tsui KH, Cheng AJ, Chang PL, Pan TL, Yung BY. Association of nucleophosmin/B23 mRNA expression with clinical outcome in patients with bladder carcinoma. Urology. 2004;64:839–844. doi: 10.1016/j.urology.2004.05.020. [DOI] [PubMed] [Google Scholar]
- Vadlamudi RK, Balasenthil S, Broaddus RR, Gustafsson JA, Kumar R. Deregulation of estrogen receptor coactivator proline-, glutamic acid-, and leucine-rich protein-1/modulator of nongenomic activity of estrogen receptor in human endometrial tumors. J Clin Endocrinol Metab. 2004;89:6130–6138. doi: 10.1210/jc.2004-0909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vadlamudi RK, Wang RA, Mazumdar A, Kim Y, Shin J, Sahin A, Kumar R. Molecular cloning and characterization of PELP1, a novel human coregulator of estrogen receptor α. J Biol Chem. 2001;276:38272–38279. doi: 10.1074/jbc.M103783200. [DOI] [PubMed] [Google Scholar]
- Valdez BC, Perlaky L, Henning D, Saijo Y, Chan PK, Busch H. Identification of the nuclear and nucleolar localization signals of the protein p120Interaction with translocation protein B23. J Biol Chem. 1994;269:23776–23783. [PubMed] [Google Scholar]
- van Riggelen J, Yetil A, Felsher DW. MYC as a regulator of ribosome biogenesis and protein synthesis. Nat Rev Cancer. 2010;10:301–309. doi: 10.1038/nrc2819. [DOI] [PubMed] [Google Scholar]
- Xue Y, Bai X, Lee I, Kallstrom G, Ho J, Brown J, Stevens A, Johnson AW. Saccharomyces cerevisiae RAI1 (YGL246c) is homologous to human DOM3Z and encodes a protein that binds the nuclear exoribonuclease Rat1p. Mol Cell Biol. 2000;20:4006–4015. doi: 10.1128/mcb.20.11.4006-4015.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying C, Gregg DW, Gorski J. Estrogen-induced changes in rRNA accumulation and RNA polymerase I activity in the rat pituitary: correlation with pituitary tumor susceptibility. Mol Cell Endocrinol. 1996;118:207–213. doi: 10.1016/0303-7207(96)03786-0. [DOI] [PubMed] [Google Scholar]
- Yun C, Wang Y, Mukhopadhyay D, Backlund P, Kolli N, Yergey A, Wilkinson KD, Dasso M. Nucleolar protein B23/nucleophosmin regulates the vertebrate SUMO pathway through SENP3 and SENP5 proteases. J Cell Biol. 2008;183:589–595. doi: 10.1083/jcb.200807185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai W, Comai L. Repression of RNA polymerase I transcription by the tumor suppressor p53. Mol Cell Biol. 2000;20:5930–5938. doi: 10.1128/mcb.20.16.5930-5938.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Comai L, Johnson DL. PTEN represses RNA polymerase I transcription by disrupting the SL1 complex. Mol Cell Biol. 2005;25:6899–6911. doi: 10.1128/MCB.25.16.6899-6911.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Yang Y, Wu J. B23 interacts with PES1 and is involved in nucleolar localization of PES1. Acta Biochim Biophys Sin (Shanghai) 2009;41:991–997. doi: 10.1093/abbs/gmp096. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.