Abstract
Computer programs for the analysis of data from techniques frequently used in nucleic acids research are described. In addition to calculating non-linear, least-squares solutions to equations describing these systems, the programs allow for data editing, normalization, plotting and storage, and are flexible and simple to use. Typical applications of the programs are described.
Full text
PDF








Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Applequist J., Damle V. Theory of the effects of concentration and chain length on helix-coil equilibria in two-stranded nucleic acids. J Chem Phys. 1963 Nov 15;39(10):2719–2721. doi: 10.1063/1.1734089. [DOI] [PubMed] [Google Scholar]
- Azbel M. Y. DNA sequencing and melting curve. Proc Natl Acad Sci U S A. 1979 Jan;76(1):101–105. doi: 10.1073/pnas.76.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakke A. C., Wu J. R., Bonner J. Chromatin structure in the cellular slime mold Dictyostelium discoideum. Proc Natl Acad Sci U S A. 1978 Feb;75(2):705–709. doi: 10.1073/pnas.75.2.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonner J., Wallace R. B., Sargent T. D., Murphy R. F., Dube S. K. The expressed portion of eukaryotic chromatin. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 2):851–857. doi: 10.1101/sqb.1978.042.01.086. [DOI] [PubMed] [Google Scholar]
- Garrard W. T., Pearson W. R., Wake S. K., Bonner J. Stoichiometry of chromatin proteins. Biochem Biophys Res Commun. 1974 May 7;58(1):50–57. doi: 10.1016/0006-291x(74)90889-4. [DOI] [PubMed] [Google Scholar]
- Gottesfeld J. M., Murphy R. F., Bonner J. Structure of transcriptionally active chromatin. Proc Natl Acad Sci U S A. 1975 Nov;72(11):4404–4408. doi: 10.1073/pnas.72.11.4404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy R. F., Bonner J. Alkaline extraction of non-histone proteins from rat liver chromatin. Biochim Biophys Acta. 1975 Sep 9;405(1):62–66. doi: 10.1016/0005-2795(75)90314-1. [DOI] [PubMed] [Google Scholar]
- Murphy R. F., Wallace R. B., Bonner J. Altered nucleosome spacing in newly replicated chromatin from Friend leukemia cells. Proc Natl Acad Sci U S A. 1978 Dec;75(12):5903–5907. doi: 10.1073/pnas.75.12.5903. [DOI] [PMC free article] [PubMed] [Google Scholar]
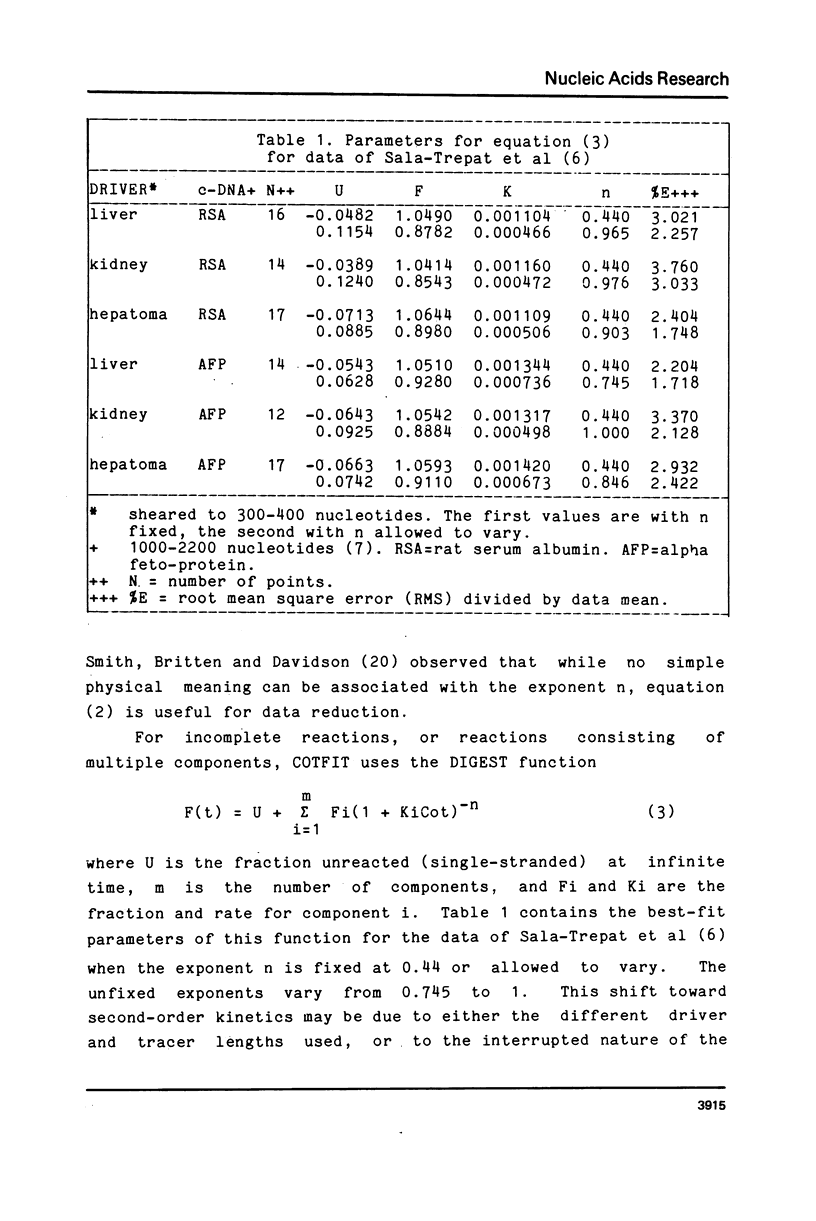
- Pearson W. R., Davidson E. H., Britten R. J. A program for least squares analysis of reassociation and hybridization data. Nucleic Acids Res. 1977 Jun;4(6):1727–1737. doi: 10.1093/nar/4.6.1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sala-Trepat J. M., Sargent T. D., Sell S., Bonner J. alpha-Fetoprotein and albumin genes of rats: no evidence for amplification-deletion or rearrangement in rat liver carcinogenesis. Proc Natl Acad Sci U S A. 1979 Feb;76(2):695–699. doi: 10.1073/pnas.76.2.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M. J., Britten R. J., Davidson E. H. Studies on nucleic acid reassociation kinetics: reactivity of single-stranded tails in DNA-DNA renaturation. Proc Natl Acad Sci U S A. 1975 Dec;72(12):4805–4809. doi: 10.1073/pnas.72.12.4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace R. B., Dube S. K., Bonner J. Localization of the globin gene in the template active fraction of chromatin of Friend leukemia cells. Science. 1977 Dec 16;198(4322):1166–1168. doi: 10.1126/science.270812. [DOI] [PubMed] [Google Scholar]
- Wallace R. B., Sargent T. D., Murphy R. F., Bonner J. Physical properties of chemically acetylated rat liver chromatin. Proc Natl Acad Sci U S A. 1977 Aug;74(8):3244–3248. doi: 10.1073/pnas.74.8.3244. [DOI] [PMC free article] [PubMed] [Google Scholar]


