Abstract
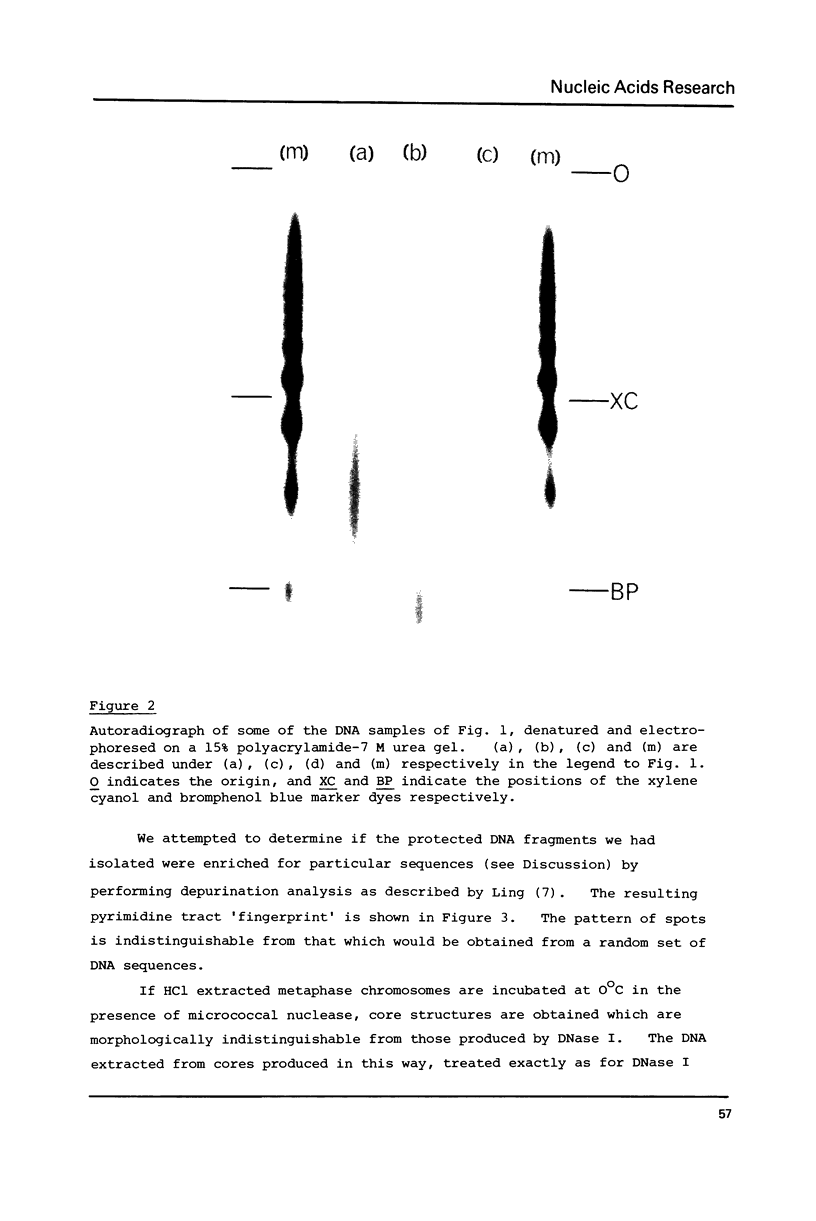
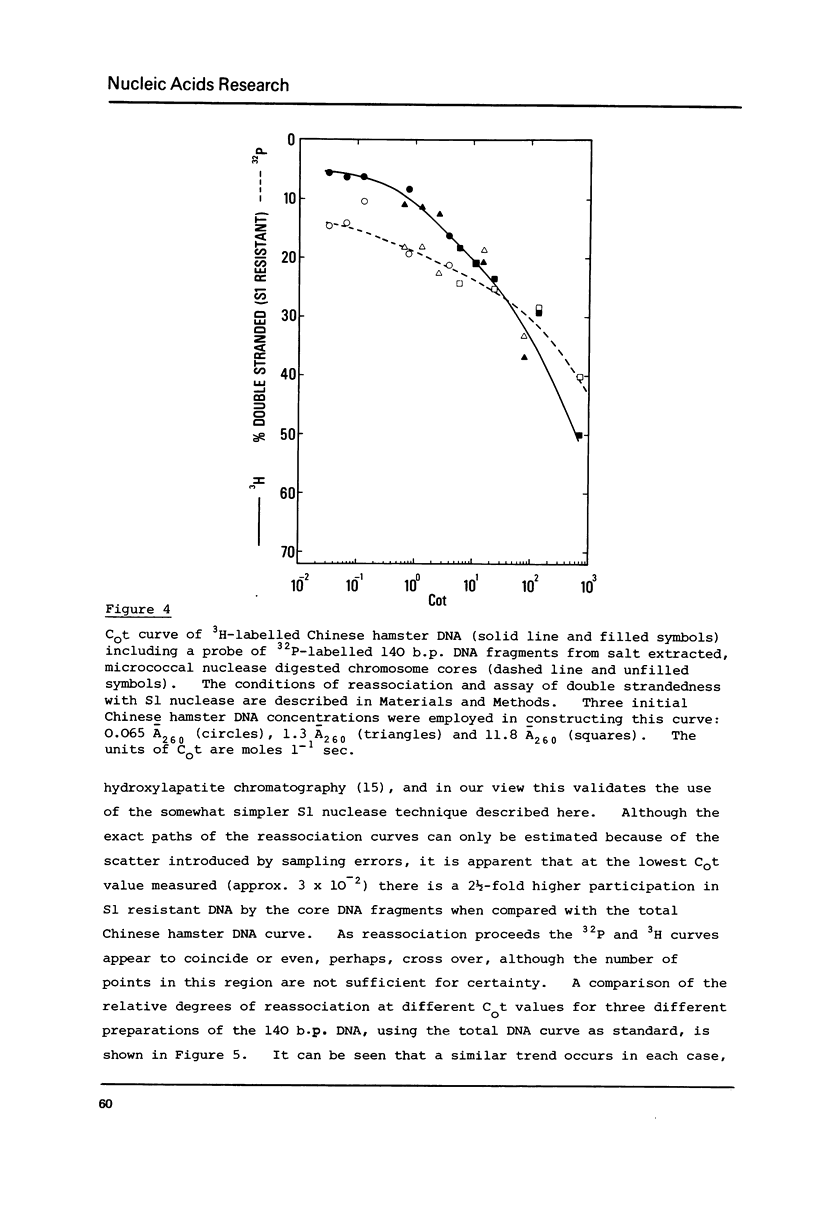
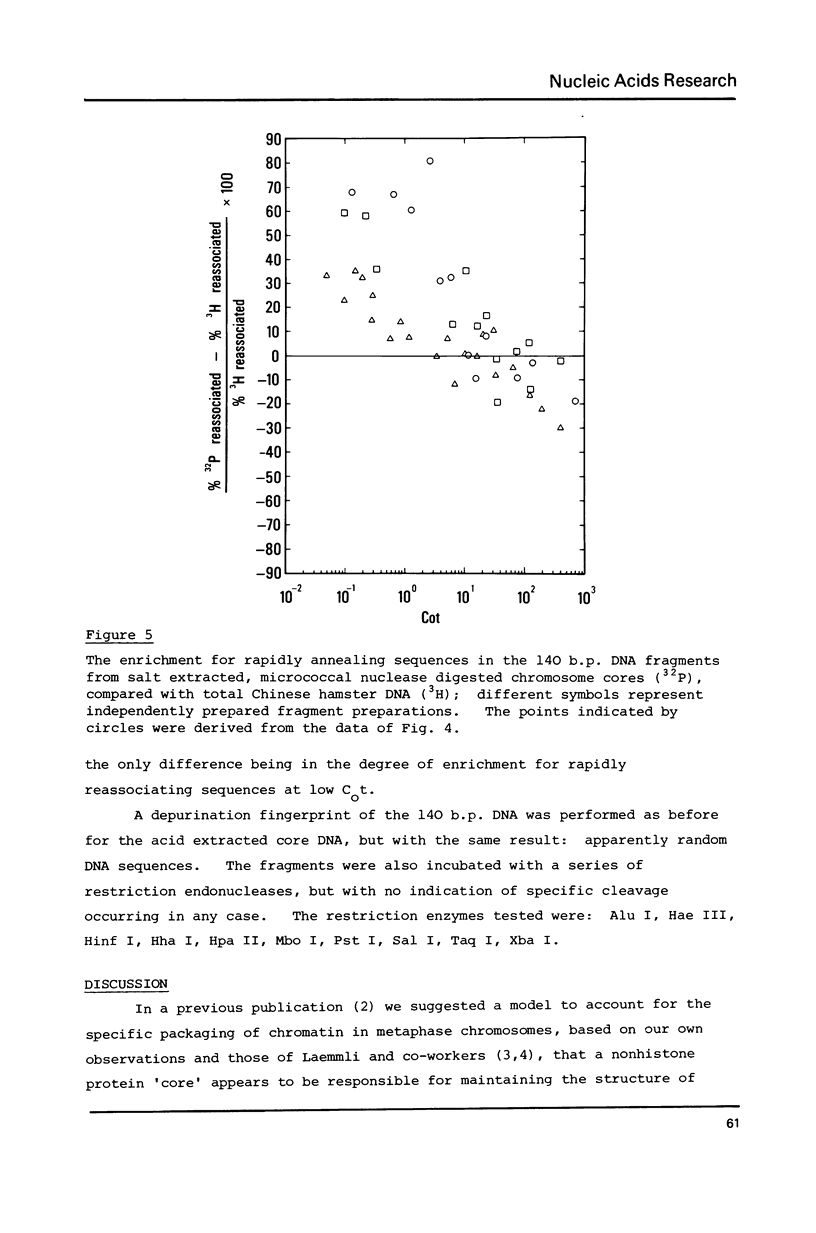
A small proportion (0.1-0.5%) of the total DNA content of native Chinese hamster metaphase chromosomes is protected from nucleolytic degradation following the removal of histones by extraction with either 0.2 N HCl or 2 M NaCl, and remains attached to the nonhistone protein core. Acid extraction followed by DNase I digestion leads to small fragments of 10-30 bases. Salt extraction followed by micrococcal nuclease digestion gives approx. 140 b.p. fragments which are undistinguishable in size from nucleosome core DNA fragments. Furthermore, DNase I treatment of salt extracted chromosomes gives DNA fragments containing single strands which are multiples of 10 bases in length, again characteristic of the nucleosome structure. Reassociation kinetics using the 32P-labelled 140 b.p. fragments as probes suggests they are enriched for rapidly reassociating sequences.
Full text
PDF















Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adolph K. W., Cheng S. M., Laemmli U. K. Role of nonhistone proteins in metaphase chromosome structure. Cell. 1977 Nov;12(3):805–816. doi: 10.1016/0092-8674(77)90279-3. [DOI] [PubMed] [Google Scholar]
- Allet B., Jeppesen P. G., Katagiri K. J., Delius H. Mapping the DNA fragments produced by cleavage by lambda DNA with endonuclease RI. Nature. 1973 Jan 12;241(5385):120–123. doi: 10.1038/241120a0. [DOI] [PubMed] [Google Scholar]
- Benz R. D., Burki H. J. The distribution of moderately repeated DNA sequences among Chinese hamster chromosomes. Exp Cell Res. 1978 Mar 1;112(1):155–165. doi: 10.1016/0014-4827(78)90536-0. [DOI] [PubMed] [Google Scholar]
- Britten R. J., Kohne D. E. Repeated sequences in DNA. Hundreds of thousands of copies of DNA sequences have been incorporated into the genomes of higher organisms. Science. 1968 Aug 9;161(3841):529–540. doi: 10.1126/science.161.3841.529. [DOI] [PubMed] [Google Scholar]
- Gilbert W., Maxam A. The nucleotide sequence of the lac operator. Proc Natl Acad Sci U S A. 1973 Dec;70(12):3581–3584. doi: 10.1073/pnas.70.12.3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeppesen P. G., Bankier A. T., Sanders L. Non-histone proteins and the structure of metaphase chromosomes. Exp Cell Res. 1978 Sep;115(2):293–302. doi: 10.1016/0014-4827(78)90284-7. [DOI] [PubMed] [Google Scholar]
- Jeppesen P. G., Sanders L., Slocombe P. M. A restriction cleavage map of phiX 174 DNA by pulse-chase labelling using E. coli DNA polymerase. Nucleic Acids Res. 1976 May;3(5):1323–1329. doi: 10.1093/nar/3.5.1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling V. Fractionation and sequences of the large pyrimidine oligonucleotides from bacteriophage fd DNA. J Mol Biol. 1972 Feb 28;64(1):87–102. doi: 10.1016/0022-2836(72)90322-1. [DOI] [PubMed] [Google Scholar]
- Noll M. Internal structure of the chromatin subunit. Nucleic Acids Res. 1974 Nov;1(11):1573–1578. doi: 10.1093/nar/1.11.1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oudet P., Gross-Bellard M., Chambon P. Electron microscopic and biochemical evidence that chromatin structure is a repeating unit. Cell. 1975 Apr;4(4):281–300. doi: 10.1016/0092-8674(75)90149-x. [DOI] [PubMed] [Google Scholar]
- Paulson J. R., Laemmli U. K. The structure of histone-depleted metaphase chromosomes. Cell. 1977 Nov;12(3):817–828. doi: 10.1016/0092-8674(77)90280-x. [DOI] [PubMed] [Google Scholar]
- Razin S. V., Mantieva V. L., Georgiev G. P. DNA adjacent to attachment points of deoxyribonucleoprotein fibril to chromosomal axial structure is enriched in reiterated base sequences. Nucleic Acids Res. 1978 Dec;5(12):4737–4751. doi: 10.1093/nar/5.12.4737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw B. R., Herman T. M., Kovacic R. T., Beaudreau G. S., Van Holde K. E. Analysis of subunit organization in chicken erythrocyte chromatin. Proc Natl Acad Sci U S A. 1976 Feb;73(2):505–509. doi: 10.1073/pnas.73.2.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton W. D. A crude nuclease preparation suitable for use in DNA reassociation experiments. Biochim Biophys Acta. 1971 Jul 29;240(4):522–531. doi: 10.1016/0005-2787(71)90709-x. [DOI] [PubMed] [Google Scholar]
- WALKER P. M., MCLAREN A. SPECIFIC DUPLEX FORMATION IN VITRO OF MAMMALIAN DNA. J Mol Biol. 1965 Jun;12:394–409. doi: 10.1016/s0022-2836(65)80263-7. [DOI] [PubMed] [Google Scholar]





