Abstract
The three types of interferon (IFNs) are essential for immunity against at least some viruses in the mouse model of experimental infections, type I IFNs displaying the broadest and strongest anti-viral activity. Consistently, human genetic studies have shown that type II IFN is largely redundant for immunity against viruses in the course of natural infections. The precise contributions of human type I and III IFNs remain undefined. However, various inborn errors of anti-viral IFN immunity have been described, which can result in either broad or narrow immunological and viral phenotypes. The broad disorders impair the response to (STAT1, TYK2) or the production of at least type I and type III IFNs following multiple stimuli (NEMO), resulting in multiple viral infections at various sites, including herpes simplex encephalitis (HSE). The narrow disorders impair exclusively (TLR3) or mostly (UNC-93B, TRIF, TRAF3) the TLR3-dependent induction of type I and III IFNs, leading to HSE in apparently otherwise healthy individuals. These recent discoveries highlight the importance of human type I and III IFNs in protective immunity against viruses, including the TLR3-IFN pathway in protection against HSE.
Introduction
Since the first descriptions of interferon (IFN) as a factor interfering with virus replication [1, 2], our understanding of IFNs has significantly improved leading to the current appreciation of these cytokines in various biological functions including the induction of an antiviral state [3–5]. There are three types of IFNs classified by on their nucleotide sequence, chromosomal location, and receptor specificity [6]. Human type I IFNs, first discovered in 1957, are found as a cluster on chromosome 9 and are comprised of twelve IFN-α, one IFN-β, one IFN-ω, one IFN-ε, and one IFN-κ that utilize the IFN-α receptors 1 and 2 (IFN-αR1 and 2) [6–8] (J. Manry et al, unpublished). Mouse studies have demonstrated the essential role of type I IFNs, via IFNAR1 or IFNAR2 knockout mice [9], and of IFN-β, via IFNB1 knockout mice, in a wide range of viral infections [10, 11]. Type II IFN discovered in 1965 is represented only by IFN-γ, which is encoded on chromosome 12 and uses the distinct IFN-γR1 and 2 receptors [12, 13]. To an even greater degree than in the mouse model [13, 14], human IFN-γ plays an important role as a ‘macrophage-activating factor’ rather than an ‘IFN’ [15] as patients lacking either receptor chain suffer mostly from intra-macrophagic bacterial infections, mycobacterial diseases in particular [16, 17]. Type III IFNs discovered in 2003, including IFN-λ1 (IL-29), IFN-λ2 (IL28B), and IFN-λ3 (IL28A), are encoded on human chromosome 19 and signal through the IFN-λR1 and IL-10RB receptors [18–20]. IFN-λ was shown to protect against some viral infections in mice [21, 22]. Descriptions of autoimmune diseases as a result of enhanced type I IFN immunity [23] and mycobacterial diseases as a result of decreased type II IFN immunity [16] have been reviewed elsewhere. We will herein review the known human inborn errors of immunity leading to reduced type I and/or III IFN immunity, associated with viral diseases [22].
Defects in antiviral IFNs - Broad Defects
The first inborn errors of anti-viral IFNs have been deciphered via the clinical investigation of children with herpes simplex virus (HSV)-1 encephalitis (HSE). HSE is a rare and potentially fatal infection of the central nervous system (CNS) affecting about two to four per 1,000,000 individuals per year [24–26]. Unlike the asymptomatic or benign (herpes labialis) presentation of HSV-1 infection, in the vast majority of the infected population (adult seropositivity is as high as >85%), HSE can result in up to 70% mortality if left untreated [27, 28]. Treatment with acyclovir has significantly improved survival rates however 35–62% of patients suffer life long neurological sequellae of varying severity [29]. There are two peaks of incidence, one occurring in childhood between the ages of 6 months to 3 years during HSV-1 primary infection, and another later in life (>50yrs) probably due to viral reactivation from latency [29]. In a French epidemiological survey of HSE patients it was suggested that HSE may have a strong Mendelian genetic basis, despite being sporadic, as up to 14% of the children were born to consanguineous parents [29]. HSV-1 is thought to remain localized in the CNS as systemic spread of the virus during infection has only rarely been observed [30]. There have been no reported associations of cases with a particular strain of HSV-1, suggesting that variation in virus virulence is not a major determinant of disease [31, 32]. Hence, the pathogenesis of HSE has remained elusive until the description of HSE in patients with IFN deficiencies. We will first focus on three unrelated single gene disorders underlying susceptibly to multiple viral infections, including HSE, owing to their broad impairments of IFN immunity (Table 1 and Figure 1).
Table 1.
Human inborn errors of antiviral IFNs
| Deficiency | Viral Infections | IFN-inducing pathways* | IFNs Response* | ||||
|---|---|---|---|---|---|---|---|
| HSE | Other | TLR3 | TLR7/8 | TLR9 | Cytosolic dsRNA | ||
| STAT1 | yes | CMV, EBV, HSV, Molluscum, Parainfluenza II, Polio III, RSV, VZV | + | + | + | NT | − |
| TYK2 | no | CMV, HSV Molluscum | NT | NT | NT | NT | − |
| NEMO | yes | Adenovirus, CMV, HSV | − | NT | NT | +/− | + |
| UNC-93B | yes | − | − | − | − | + | + |
| TLR3 | yes | Coxsackie B | − | + | + | + | + |
| TRIF | yes | − | − | + | + | +/− | + |
| TRAF3 | yes | − | − | − | − | − | + |
Type I/III IFNs production (TLR3 and intracellular dsRNA pathways) and response tested in patients’ SV40-fibroblasts. TLR7/8/9-induced type I/III IFNs tested in patients’ peripheral blood mononuclear cells.
NT = not tested; “+” = normal; “−“ = impaired; “+/−“ = partial impairment.
HSV = cutaneous HSV infections
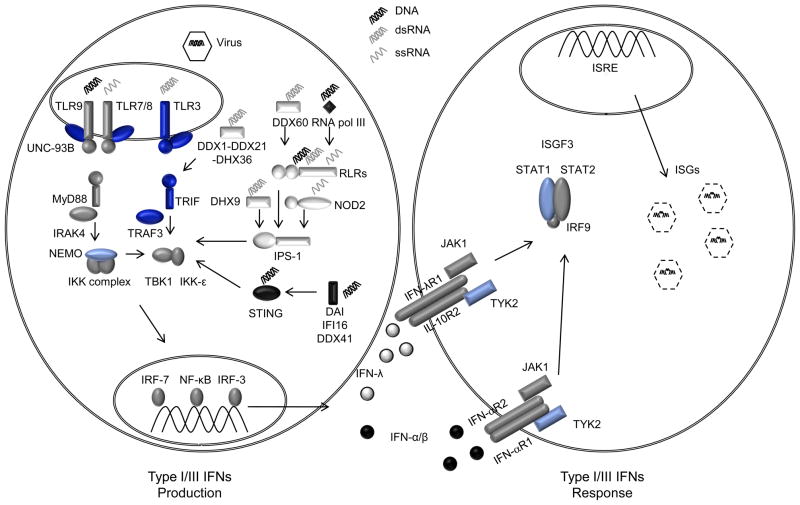
Figure 1. Human deficiencies of types I and III IFNs.
Following viral infection, immune receptors engage in the recognition of viral nucleic acids via the endosomal TLRs (TLR3, TLR7/8, TLR9), the cytosolic RLRs (RIGI, MDA5, LGP2), RNA-detecting DExD helicases (DDX1-DDX21-DHX36; DDX60, DHX9), NLRs (NOD2) or DNA receptors (DAI, IFI16, DDX41, RNA polymerase III, STING) (left panel). This in turn triggers various signaling pathways leading to the activation of the transcription factors NF-κB, IRF3 and IRF7 leading to the production of the antiviral IFNs, IFN-α/β and –λ. These IFNs are detected by their respective receptors: IFN-α/β by the heterodimers of IFN-αR1 and IFN-αR2, and IFN-λ by the heterodimers of IFN-λR1 and IL-10R2. This triggers the formation of the ISGF3 transcription factor complex that binds the IFN-stimulated response element (ISRE) resulting in the induction of numerous IFN stimulated genes (ISGs) initiating an antiviral response leading to the destruction of the virus (right panel). Mutations in NEMO, STAT1 or TYK2 are associated with multiple viral infections (in pale blue) while mutations in UNC93B1, TLR3, TRIF or TRAF3 are strictly associated with HSE. RNA molecules are shown in gray where as DNA is shown in black.
STAT1
Signal transducer and activator of transcription-1 (STAT-1) is a protein involved in the transduction of cellular responses to IFN-α/β, -λ and γ, and IL-27, via the formation of two transcription factor complexes: the interferon stimulated gamma factor 3 (ISGF3) composed of STAT1-STAT2-p48/IRF9 trimers and the gamma activated factor (GAF) comprised of STAT1 homodimers [33, 34]. Heterozygous loss-of-function germ-line mutations in STAT1 impairing STAT-1 phosphorylation or DNA binding activity underlie selective impairment of the IFN-γ dependent GAF responses but intact IFN-α/β-dependent ISGF3 responses [35–37]. These patients suffer from autosomal dominant (AD) mycobacterial disease due to the impaired IFN-γ responses. First described in 2003, complete autosomal recessive (AR) mutations in STAT1 lead to a complete loss of STAT1 protein expression and consequently have no STAT1-dependent responses to IFN-γ, IFN-α/β, and –λ, leading to both mycobacterial and numerous viral diseases (Table 1) [38–41]. Two of these patients died of viral disease, one of confirmed HSE due to abolished responses to IFN-α/β and/or -λ. A milder form of partial AR STAT1 deficiency has also been reported in several patients [42, 43]. More recently, heterozygous gain-of-function mutations in STAT1 have been described in patients suffering from AD chronic mucocutaneous candidiasis (CMC). These mutations are loss-of-dephosphorylation, gain-of-function, and enhanced cellular responses to IFN-α/β, -λ and γ, and/or IL-27 are probably responsible for the poor development of IL-17-producing T cells, which are essential for muco-cutaneous immunity against Candida albicans [44–46]. These findings explain the fungal infections and the lack of mycobacterial or viral diseases in these patients. Overall, the various types of STAT1 mutations allowed a fine dissection of the role of STAT1-dependent IFN immunity in host response, demonstrating in particular an essential role of IFN-α/β- and/or -λ-dependent ISGF3 immunity to control various viral infections, including HSV1 infection in the CNS.
TYK2
Shortly after the description of STAT1-deficient patients with viral infections, TYK2 deficiency was reported in 2006 in a single individual with multiple viral infections and surprisingly no HSE. TYK2 is a kinase that is constitutively associated with various cytokine receptors, including IFN-αR1, in which case it is activated upon ligand-induced IFN-αR1 and IFNαR2 dimerization, allowing for the formation of ISGF3 [47]. Mouse TYK2 was initially shown to be essential for IFN-α/β signalling [48, 49] but since been shown to be involved in other cytokine response pathways including those of IL-6 and IL-12 [50, 51]. The patient with complete deficiency in TYK2 [52] displayed a broad clinical phenotype including susceptibility to Staphylococci, atopic dermatitis, high serum IgE, similar to hyper-IgE patients [53, 54] perhaps due to reduced IL-6 and/or IL-10 responses. He also displayed infections by Mycobacteria and Salmonella, reminiscent of patients with Mendelian susceptibility to mycobacterial disease (MSMD) [16], which was attributed to his poor cellular responses to IL-12 and subsequent impairment to upregulate IFN-γ. Finally he also suffered from recurrent cutaneous HSV-1 and Molluscum contagiosum infections, probably owing to his poor cellular response to IFN-α/β. Unlike the AD STAT1-deficient patients, the viral diseases manifested in the TYK2 patient were mild and limited to cutaneous infections, the reasons for which remain unclear and may reflect residual, TYK2-independent cellular responses to IFNs. The description of other TYK2 patients will be helpful in better delineating the immunological and viral phenotypes associated with this defect. In any case, TYK2 deficiency impairs at least IFN-α/β responses and confers predisposition to at least cutaneous viral diseases.
NEMO
Nuclear factor-κB (NF-κB) essential moderator (NEMO), also known as IKK-γ, is a regulatory subunit of the IKK complex activating the canonical NF-κB signalling pathway, and operating downstream of multiple receptors including TNFRs, TCR, BCR, IL-1Rs, TLRs and the RIGI/IPS-1 pathways [55, 56]. Mutations in NEMO cause a very wide spectrum of disease ranging from incontinentia pigmenti causing in-utero lethality in hemizygous males and ectodermal abnormalities in heterozygous females [57], to various forms of X-linked recessive forms of anhidrotic ectodermal dysplasia with immunodeficiency (EDA-ID) [55, 58]. Children with EDA-ID usually suffer from multiple infections by various pathogens including pyogenic bacteria, fungi, mycobacteria, and viruses [55, 58]. Pneumococcal infections are the most common infections they suffer, probably due to impaired TLR and IL-1R responses [59–61]. Mycobacterial diseases probably occur due to an impaired CD40-IL-12 pathway, leading to impaired IFN-γ mediated immunity [62]. Systemic or intestinal cytomegalovirus (CMV), adenoviral and cutaneous HSV-1 infections are the most common viral manifestations of disease (Table 1) [55, 63, 64].. Interestingly, a patient with a hypomorphic mutation in NEMO developed recurrent HSE, which led to his death [63, 65]. NEMO, unlike the previous two deficiencies, is involved in the induction of, as opposed to the response to, type I and III IFNs following cell stimulation by various receptors. A recent report has demonstrated NEMO to be involved in IRF3 activation following viral infections [56]. Of course NF-κB activation is also important to trigger the transcription of IFNs [66–69]. This dual action of NEMO potentially explains viral susceptibility in patients carrying mutations in NEMO, via impaired type I and/or III IFN production. Fibroblasts from the HSE patient with a hypomorphic frameshift mutation in NEMO (110_111insC), showed impaired IFN-α/β and -λ production following viral infection or Toll-like receptor 3 (TLR3) stimulation [70]. TLR3 recognizes dsRNA of viral origin, furthermore inborn errors of TLR3 immunity underlie HSE in other patients (see below), hence this defined a plausible molecular mechanism for HSE, at least in this patient [70]. In fact 8 additional patients with different mutations in NEMO showed varying degrees of impaired IFN-α/β, -λ production but no HSE despite being seropositive for HSV-1. Hence, all NEMO patients might be potentially prone to HSE however there is incomplete clinical penetrance [70]. Whether the mutations in NEMO or other factors, genetic or environmental, account for the observed rarity of HSE in patients with NEMO mutations is unclear. The specific molecular basis of viral infections other than HSE in NEMO patients also remains elusive. In any case, the role of NEMO in regulating the induction of type I and III IFNs responses is probably associated with viral diseases and the TLR3-IFN pathway with HSE in particular.
Defects in anti-viral IFNs- Narrow Defects
NEMO, STAT1, and TYK2 deficiencies all have in common broad immunological defects affecting not only type I and III IFNs but also other cytokines. They thus confer a broad infectious phenotype including viral, bacterial as well as fungal infections. It is no surprise that these patients are susceptible to multiple viral infections as their defects lie where multiple antiviral pathways converge: STAT1 and TYK2 downstream of type I and III IFN receptors and NEMO downstream of the numerous IFN-inducing receptors. In recent years, a growing number of receptors that induce antiviral IFNs have been identified, including TLRs, retinoic acid-inducible gene-I (RIG)-like receptors (RLRs), DExD/H helicases, nucleotide-binding oligomerization domain (NOD)-like receptors, and various DNA receptors (Figure 1). Ten TLRs have been described in humans, each stimulated by different agonists [71]. TLR3, TLR7/8 and TLR9 in particular are intracellular (endosomal) and involved in the sensing of extracellular and endosomal nucleic acids, dsRNA, ssRNA and dsDNA respectively, leading to type I and III IFNs production [72, 73]. RLRs consist of RIG-I, and melanoma differentiation-associated gene (MDA)-5, and laboratory of genetics and physiology-2 (LGP2) that recognize cytosolic RNA and DNA inducing IFNs through its specific adaptor, interferon-β promoter stimulator (IPS)-1 (also known as MAVS, VISA or Cardif) [74–77]. NOD2 has also been shown to recognize ssRNA and produce type I IFNs in an IPS1-dependent manner [78]. Recent reports have revealed DExD/H helicases to be involved in dsRNA sensing either in a TRIF-dependent (DDX1-DDX21-DHX36) [79] or in a RLR-dependent or IPS-1-dependent manner (DDX60 and DHX9 respectively) [80, 81]. The DNA sensor RNA polymerase III indirectly induces IFNs by transcribing AT rich DNA into uncapped 5′ triphosphate–bearing RNA which then signals via the RLR-pathway [82, 83]. Finally, DNA-dependent activator of IFN-regulatory factors (DAI or DLM1/ZBP1), a PYHIN (pyrin and HIN200 domain–containing proteins; also known as p200 or HIN200 proteins) protein interferon gamma-inducible protein 16 (IFI16), and another DExD/H helicase DDX41 have all been implicated in cytoplasmic recognition of intracellular DNA, leading to type I IFN production [84–87]. They signal through stimulator of interferon genes or STING (MITA, ERIS, MPYS or TMEM173) which has itself been shown to detect cyclic di-GMP, and the kinases TANK-binding kinase 1 (TBK1) and IKK-ε [86, 88–91]. The antiviral nature of these receptors have been studied primarily in the context of mouse models of experimental infection or in vitro [72, 74, 78–80, 84, 85, 87, 92, 93]. This shed light on their function in host defence in experimental conditions in vivo, which appeared to be relatively broad for each receptor; however their respective role in natural immunity remained elusive. Recent developments in understanding antiviral immunity in natura, thanks to human studies, have revealed the crucial importance of the TLR3-IFN pathway in protection against the development of HSE. Here we review the few known inborn errors of antiviral IFNs that are associated with HSE (Table 1 and Figure 1). The function of other IFN-inducing pathways, like that of the many IFN-inducible genes, remains largely unknown.
UNC-93B
In 2006, the first genetic etiology for isolated HSE was reported in two children with AR UNC-93B-deficicency [94]. UNC-93B is an endoplasmic reticulum (ER) protein that contributes to the translocation of TLRs (TLR3, 7, 8 and 9) from the ER to endolysosomes [95, 96]. The four human intracellular TLRs rarely tolerate missense or nonsense mutations suggesting that they are under strong purifying selection, unlike surface-expressed TLRs [97]. TLR7 and TLR8 are primarily involved in detection of ssRNAs, TLR9 in the detection of dsDNA and TLR3 of dsRNA, which is produced during the replication of almost all viruses including HSV-1 [71, 98]. Mouse TLR7 and TLR9 have been shown to play an important role in pDCs, contributing to a significant amount of type I IFN produced by these cells [98], however human TLR7, TLR8, and TLR9 are largely redundant in host defence against viruses, as revealed by the lack of any detectable viral diseases in patients with IRAK4 and MyD88 deficiency [59, 60, 99–101]. UNC-93B was first discovered in an N-ethyl-N-nitrosourea (ENU) mutagenesis screen, underlying the 3d mouse mutant phenotype which displayed abolished responses to TLR3, 7 and 9 agonists as well as susceptibly to murine CMV (MCMV) infection [102]. Human UNC-93B deficiency was reported shortly after, in two unrelated HSE patients homozygous for null mutations in UNC93B1 [94]. Similar to the 3d mouse, the patients’ peripheral blood mononuclear cells (PBMCs) failed to produce type I and III IFNs in response to TLR3, TLR7, TLR8, and TLR9 agonists. The patients’ fibroblasts also showed no production of type I or III IFNs (specifically IFN-β and IFN-λ1/3) following stimulation with the TLR3 agonist polyinosinic-polycytidylic acid (poly(I:C)) or following infection with vesicular stomatitis virus (VSV) or HSV-1. This viral susceptibility was complemented by the addition of recombinant IFN-α2b, suggesting the lack of IFN production was responsible for sensitivity to viral infection and demonstrating that IFN response was normal. Despite multiple TLRs affected, UNC-93B-deficient patients suffered only from HSE with no systemic or cutaneous dissemination of HSV-1, nor did they present with other severe viral infections. Hence, UNC-93B-deficiency, in combination with STAT1 and NEMO deficiencies, hinted at the essential requirement of UNC-93B-dependent IFN production for protective immunity against HSV-1 primary infection in the CNS, at least in some children. Moreover, the contribution of TLR7, TLR8 and TLR9 to disease, if any, is probably small as IRAK4- and MyD88-deficient patients, who display impaired responses to all TLRs except TLR3 do not develop HSE upon infection by HSV-1, nor any other severe viral diseases but typically present with pyogenic bacterial infections [59–61, 100]. It followed then that our attention should turn to human TLR3 as the receptor mediating the necessary protection against HSV-1 infection in the CNS.
TLR3
Shortly after the discovery of AR UNC-93B deficiency, AD TLR3 deficiency was discovered in two unrelated HSE patients carrying the same heterozygous missense mutation affecting the ectodomain of TLR3 [103]. The patients’ fibroblasts displayed dominant negative properties for poly(I:C) induced type I and III IFNs production, consistent with impaired but not abolished responses to poly(I:C) [103]. Similarly, the patients’ cells were more susceptible to VSV and HSV-1 infections as assessed by IFN production, viral replication, and virus-induced cell death. The addition of recombinant IFN-α2b to the patients’ cells reversed the phenotype to control levels such that the patients’ cells were able to control viral induced cell mortality and replication. However, not all TLR3 expressing cells were found to have impaired poly(I:C) responses, which can be attributed to other dsRNA sensors, or residual TLR3 signalling, or both. Interestingly, another patient heterozygous for the same TLR3 mutation was subsequently reported as having coxsackie B virus myocarditis [104], suggesting that TLR3 mutations may underlie susceptibility to other viral diseases. We have also more recently identified an AR form of TLR3 deficiency, in a patient compound heterozygous for two null TLR3 alleles [105] whose fibroblasts displayed abolished responses to TLR3 whereas the patient’s PBMCs, monocytes, and monocyte derived macrophages responded to poly(I:C) normally. Genome-wide transcriptome analysis using cells from this patient revealed that TLR3 is largely redundant for responses to dsRNA in leukocytes, where other dsRNA sensors may play a more significant role. Whereas the normal resistance to most viral infections in patients with AD TLR3 may result from residual TLR3 cellular responses, the occurrence of childhood HSE in an otherwise healthy adult with complete TLR3 deficiency suggests that TLR3 is largely redundant in host defence, against viruses in particular. The redundancy of TLR3 in leukocytes probably explains the narrow clinical vulnerability. Conversely, the development of HSE suggests that resident cells in the CNS, like fibroblasts, rely on TLR3 to respond to poly(I:C) and HSV-1.
TRIF
The confirmation that mutations in the TLR3 pathway underlie HSE also came recently from the discovery of Toll/IL1R (TIR) domain-containing adaptor inducing IFN-β or TRIF deficient patients [106]. TRIF, also known as TIR domain containing adaptor molecule 1 (TICAM-1), is an adaptor protein serving as the sole adaptor to TLR3 and as an alternative adaptor via TRIF-related adaptor molecule (TRAM) to TLR4 [107, 108]. Once activated by agonist-induced TLR3 dimerization (or via TLR4-TRAM), this cytosolic protein homo-oligomerizes to form a platform from which all downstream signalling events occur, ultimately resulting in type I and III IFNs and proinflammatory cytokine production [109, 110]. A recent report described an additional role for TRIF engaging in detection of cytosolic dsRNA through the DExD/H-box helicase complex, DDX1-DDX21-DHX36 [79]. TRIF-deficient mice show an abolished response to poly(I:C), resistance to endotoxic shock, and are susceptible to MCMV and vaccinia virus infections [107, 108]. Two HSE patients with TRIF deficiency were identified, one with a homozygous nonsense mutation, resulting in complete AR TRIF deficiency, and another with a heterozygous missense mutation, leading to partial AD TRIF deficiency. Similar to TLR3-deficient patients, TRIF-deficient patients’ fibroblasts did not produce IFNs after stimulation with poly(I:C) and showed increased susceptibility to VSV and HSV-1 infections, highlighting the importance of the TLR3-TRIF-IFN pathway in protection against HSE. In addition, as the AR TRIF patient was a loss-of expression, loss-of-function mutation, the patient’s cells also displayed impaired responses to both transfected poly(I:C) and LPS in terms of IFN induction, due to TRIF’s role in the DExD/H helicase pathway and the TLR4 pathways respectively. These observations suggest that the human TRIF-dependent TLR4 and DExD/H helicase pathways are largely redundant for host defence, as this patient only suffered from HSE which can be attributed solely to the unresponsive TLR3 pathway. The impairment of these other pathways had no clinical consequence, or at least not for the moment and their role in other diseases, viral illnesses in particular, cannot be entirely excluded as the AR TRIF patient is still relatively young.
TRAF3
Discovered prior to TRIF deficiency, TNF-receptor associated factor 3 (TRAF3) deficiency was surprising at first glance because of the broad involvement of TRAF3 in both various IFN-inducing pathways and multiple TNFR superfamily responsive pathways [111, 112]. TRAF3 is an adaptor protein found downstream of the TNF receptors [111, 112], and IFN-inducing receptors including TLR3, TLR7, TLR8, TLR9, and the cytosolic dsRNA receptors RIG-I/MDA5 [112, 113]. Surprisingly, we discovered AD TRAF3 deficiency in a patient with HSE, carrying a de novo missense allele, which is loss-of-expression, loss-of-function, and dominant-negative, resulting in impaired but not abolished TLR3-mediated induction of IFNs and increased susceptibility to viral infections in heterozygous cells from the HSE patient [114]. Consistent with the loss-of-function observed in the TLR3-IFN mediated pathway, the TRAF3 heterozygous cells displayed impaired but not abolished function for all other TRAF3-dependent pathways, including those downstream from TNF receptors such as CD40, BAFF, and LT, and IFN-inducing receptors such as TLR7/8, and the cytosolic dsRNA pathways. At odds with the prediction and observation that human TRAF3 plays such a broad role, the patient only suffered from HSE. It is surprising that this patient has not displayed any other infectious or immunological diseases given the broad impact of the heterozygous mutation in various cell types. This suggests that the residual signalling threshold below that of predisposition to disease varies among the numerous TRAF3-dependent pathways. In other words, impaired TRAF3-dependent responses to TLR3 predisposed to HSE, whereas impaired TRAF3-dependent responses to other receptors remained clinically silent. A complete defect would be predicted to have broader clinical consequences. We cannot however exclude that such clinical consequences may appear later in life in the HSE patient. So far, TLR3-independent TRAF3-dependent IFNs (due to some residual function of TRAF3 in the patients’ cells) and/or TRAF3-independent IFN production probably protected the patient against other viral infections. In any case, this experiment of Nature demonstrated that the human TLR3-and TRAF3-dependent induction of IFNs is essential for protection against HSV-1 in the CNS.
Conclusion
Broad inborn errors of antiviral IFNs, as shown by NEMO, STAT1, and TYK2 deficiencies, result in multiple viral infections in different tissues including systemic or intestinal CMV, cutaneous Molluscum, cutaneous herpes infections as well as HSE. Indeed, these three molecules are critical for IFN immunity, controlling cellular production of all three types of IFNs in response to the stimulation of various signalling pathways (NEMO) or cellular responses to most anti-viral IFNs, not to mention other cytokines that may also contribute to anti-viral immunity (STAT1 and TYK2). In contrast, narrow inborn errors of antiviral IFNs are thus far limited to one particular viral phenotype, HSE, due to the alteration of one particular IFN-inducing pathway, controlled by TLR3. The discoveries of UNC93B, TLR3, TRIF and TRAF3 deficiencies which all have in common a defect in the TLR3-IFN pathway strongly suggest that this mechanism is at the heart of resistance against HSV-1 primary infection in the CNS, at least in some children. Not in all children, however, as incomplete clinical penetrance was observed for all single-gene inborn errors of TLR3 immunity for which at least one relative of the proband was also genetically affected, explaining the paradoxically sporadic nature of genetically determined HSE. Multiple factors may contribute to incomplete clinical penetrance, whether environmental (infectious virus nature and amount), or host factors (genetic and even epigenetic, as suggested by the early-onset) [29].
In any case, inborn errors of TLR3-dependent IFN immunity predispose to HSE, at least in some children. TLR3 is highly expressed in the CNS, and is capable of recognizing dsRNA intermediates produced during the life cycle of HSV-1, which with rabies virus is one of the few viruses that infect the human CNS via a neurotropic route, not crossing the blood-brain barrier [92, 115, 116]. Moreover, children with HSE are not prone to HSV-1 infections at other sites. Hence, it is tempting to speculate that TLR3 acts as a predominant dsRNA sensor in the CNS, controlling CNS-intrinsic immunity against HSV-1 in particular via its control of antiviral IFN. The selective pressure exerted by HSV-1 and other neurotropic viruses, such as rabies virus, may account for the strong signatures of purifying selection documented for human TLR3 and TRIF (e.g. missense and nonsense mutations were rarely tolerated), suggesting that they played a crucial role in the survival of mankind [97, 117]. In this context, the identification of UNC93B, TLR3, TRIF, and TRAF3 deficiencies underlying HSE suggests that other genes controlling this particular pathway may be mutated in other children with HSE. The intriguing observation of a patient with viral myocarditis and AD TLR3 deficiency [104] however raises the possibility that the infectious phenotype associated with inborn errors of TLR3 immunity may be narrow in individual patients but broader at the population level, consistent with the incomplete clinical penetrance documented for both HSE and viral myocarditis.
Naturally the role of the other IFN-inducing viral sensors in host defence comes into question, as well as that of the diverse IFNs and IFN-inducible antiviral target genes. Why are there so many genes at each of these three levels? Which viral diseases would occur in individuals with inborn errors of one or another component? Reports of rare non-synonymous sequence variants in RIGI and MDA5 have been reported, several with proven defect in function but with no clear viral association reported yet [118, 119]. This suggests that these receptors might be largely redundant in anti-viral immunity. No such mutations associated with a loss of function have been reported yet for any of the other receptors. Deleterious mutations in these genes very possibly may have clinical consequences that have not yet been brought to light. It would be interesting to study these genes from an evolutionary genetic perspective and determine which ones are under purifying selective pressure [120]. A clinical genetic approach similar to that followed for HSE may also be fruitful. We hypothesize that other inborn errors of IFN immunity may underlie severe viral infections other than HSE, such as severe influenza, myocarditis, or hepatitis [121, 122]. As some of these infections show some degree of tissue specificity (like HSE restricted to the CNS), it is possible that a particular IFN-related pathway may be devoted to a specific virus or tissue, or both, in a non-redundant manner. Consistent with this notion, recent studies have associated sequence variants in IFN-λ2 with hepatitis C infection treatment response and spontaneous viral clearance in humans [123–126]. Each IFN-inducing pathway, or each IFN type and subtype, or each anti-viral IFN-inducible gene, may have selectively evolved towards protection against a particular viral infection, the diversity of IFN-inducing, IFNs, and IFN-inducible genes reflecting the diversity of viruses and tissues.
Acknowledgments
We thank the members of the Laboratory of Human Genetics of Infectious Diseases, in particular Laurent Abel, Capucine Picard, Anne Puel and Stéphanie Boisson-Dupuis for helpful discussions and Lazaro Lorenzo for technical assistance. We thank the children and their families who took part in the studies referred to in this review. V. Sancho-Shimizu was supported by the Marie Curie Intra-European Fellowship 2008–2010. J.-L. Casanova was an international scholar of the Howard Hughes Medical Institute from 2005 to 2008. Research on anti-viral immunity in the Laboratory of Human Genetics of Infectious Diseases is supported by grants from the Agence national de la Recherche ANR-08-MNP-014, The Rockefeller University Center for Clinical and Translational Science grant number 5UL1RR024143, the National Institute of Allergy and Infectious Diseases grant number 1R01AI088364, the St. Giles Foundation, the Thrasher Research Fund, the March of Dimes, the Jeffrey Modell Foundation, Talecris Biotherapeutics, and The Rockefeller University.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Isaacs A, Lindenmann J. Virus interference. I. The interferon. Proc R Soc Lond B Biol Sci. 1957;147(927):258–67. doi: 10.1098/rspb.1957.0048. [DOI] [PubMed] [Google Scholar]
- 2.Nagano Y, Kojima Y, Sawai Y. Immunity and interference in vaccinia; inhibition of skin infection by inactivated virus. C R Seances Soc Biol Fil. 1954;148(7–8):750–2. [PubMed] [Google Scholar]
- 3.Gresser I. The antitumor effects of interferon: a personal history. Biochimie. 2007;89(6–7):723–8. doi: 10.1016/j.biochi.2007.03.005. [DOI] [PubMed] [Google Scholar]
- 4.Jouanguy E, Zhang SY, Chapgier A, Sancho-Shimizu V, Puel A, Picard C, Boisson-Dupuis S, Abel L, Casanova JL. Human primary immunodeficiencies of type I interferons. Biochimie. 2007;89(6–7):878–83. doi: 10.1016/j.biochi.2007.04.016. [DOI] [PubMed] [Google Scholar]
- 5.Gresser I. Wherefore interferon? J Leukoc Biol. 1997;61(5):567–74. doi: 10.1002/jlb.61.5.567. [DOI] [PubMed] [Google Scholar]
- 6.Pestka S, Krause CD, Walter MR. Interferons, interferon-like cytokines, and their receptors. Immunol Rev. 2004;202:8–32. doi: 10.1111/j.0105-2896.2004.00204.x. [DOI] [PubMed] [Google Scholar]
- 7.Genin P, Vaccaro A, Civas A. The role of differential expression of human interferon--a genes in antiviral immunity. Cytokine Growth Factor Rev. 2009;20(4):283–95. doi: 10.1016/j.cytogfr.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 8.Thomas C, et al. Structural Linkage between Ligand Discrimination and Receptor Activation by Type I Interferons. Cell. 2011;146(4):621–32. doi: 10.1016/j.cell.2011.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bogdan C. The function of type I interferons in antimicrobial immunity. Curr Opin Immunol. 2000;12(4):419–24. doi: 10.1016/s0952-7915(00)00111-4. [DOI] [PubMed] [Google Scholar]
- 10.Deonarain R, Alcami A, Alexiou M, Dallman MJ, Gewert DR, Porter AC. Impaired antiviral response and alpha/beta interferon induction in mice lacking beta interferon. J Virol. 2000;74(7):3404–9. doi: 10.1128/jvi.74.7.3404-3409.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Trinchieri G. Type I interferon: friend or foe? J Exp Med. 2010;207(10):2053–63. doi: 10.1084/jem.20101664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bach EA, Aguet M, Schreiber RD. The IFN gamma receptor: a paradigm for cytokine receptor signaling. Annu Rev Immunol. 1997;15:563–91. doi: 10.1146/annurev.immunol.15.1.563. [DOI] [PubMed] [Google Scholar]
- 13.Vilcek J. Fifty years of interferon research: aiming at a moving target. Immunity. 2006;25(3):343–8. doi: 10.1016/j.immuni.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 14.van den Broek MF, Muller U, Huang S, Zinkernagel RM, Aguet M. Immune defence in mice lacking type I and/or type II interferon receptors. Immunol Rev. 1995;148:5–18. doi: 10.1111/j.1600-065x.1995.tb00090.x. [DOI] [PubMed] [Google Scholar]
- 15.Nathan CF, Murray HW, Wiebe ME, Rubin BY. Identification of interferon-gamma as the lymphokine that activates human macrophage oxidative metabolism and antimicrobial activity. J Exp Med. 1983;158(3):670–89. doi: 10.1084/jem.158.3.670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Filipe-Santos O, et al. Inborn errors of IL-12/23- and IFN-gamma-mediated immunity: molecular, cellular, and clinical features. Semin Immunol. 2006;18(6):347–61. doi: 10.1016/j.smim.2006.07.010. [DOI] [PubMed] [Google Scholar]
- 17.Zhang SY, et al. Inborn errors of interferon (IFN)-mediated immunity in humans: insights into the respective roles of IFN-alpha/beta, IFN-gamma, and IFN-lambda in host defense. Immunol Rev. 2008;226:29–40. doi: 10.1111/j.1600-065X.2008.00698.x. [DOI] [PubMed] [Google Scholar]
- 18.Sheppard P, et al. IL-28, IL-29 and their class II cytokine receptor IL-28R. Nat Immunol. 2003;4(1):63–8. doi: 10.1038/ni873. [DOI] [PubMed] [Google Scholar]
- 19.Kotenko SV, et al. IFN-lambdas mediate antiviral protection through a distinct class II cytokine receptor complex. Nat Immunol. 2003;4(1):69–77. doi: 10.1038/ni875. [DOI] [PubMed] [Google Scholar]
- 20.Uze G, Monneron D. IL-28 and IL-29: newcomers to the interferon family. Biochimie. 2007;89(6–7):729–34. doi: 10.1016/j.biochi.2007.01.008. [DOI] [PubMed] [Google Scholar]
- 21.Ank N, West H, Bartholdy C, Eriksson K, Thomsen AR, Paludan SR. Lambda interferon (IFN-lambda), a type III IFN, is induced by viruses and IFNs and displays potent antiviral activity against select virus infections in vivo. J Virol. 2006;80(9):4501–9. doi: 10.1128/JVI.80.9.4501-4509.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kotenko SV. IFN-lambdas. Curr Opin Immunol. 2011 doi: 10.1016/j.coi.2011.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Crow YJ. Type I interferonpathies: a novel set of inborn errors of immunity. Ann N Y Acad Sci. 2011 doi: 10.1111/j.1749-6632.2011.06220.x. In Press. [DOI] [PubMed] [Google Scholar]
- 24.Najioullah F, Bosshard S, Thouvenot D, Boibieux A, Menager B, Biron F, Aymard M, Lina B. Diagnosis and surveillance of herpes simplex virus infection of the central nervous system. J Med Virol. 2000;61(4):468–73. doi: 10.1002/1096-9071(200008)61:4<468::aid-jmv9>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 25.Puchhammer-Stockl E, Presterl E, Croy C, Aberle S, Popow-Kraupp T, Kundi M, Hofmann H, Wenninger U, Godl I. Screening for possible failure of herpes simplex virus PCR in cerebrospinal fluid for the diagnosis of herpes simplex encephalitis. J Med Virol. 2001;64(4):531–6. doi: 10.1002/jmv.1082. [DOI] [PubMed] [Google Scholar]
- 26.Whitley RJ. Herpes Simplex Virus in Children. Curr Treat Options Neurol. 2002;4(3):231–237. doi: 10.1007/s11940-002-0040-2. [DOI] [PubMed] [Google Scholar]
- 27.Whitley RJ, Lakeman F. Herpes simplex virus infections of the central nervous system: therapeutic and diagnostic considerations. Clin Infect Dis. 1995;20(2):414–20. doi: 10.1093/clinids/20.2.414. [DOI] [PubMed] [Google Scholar]
- 28.Sancho-Shimizu V, Zhang SY, Abel L, Tardieu M, Rozenberg F, Jouanguy E, Casanova JL. Genetic susceptibility to herpes simplex virus 1 encephalitis in mice and humans. Curr Opin Allergy Clin Immunol. 2007;7(6):495–505. doi: 10.1097/ACI.0b013e3282f151d2. [DOI] [PubMed] [Google Scholar]
- 29*.Abel L, et al. Age-dependent Mendelian predisposition to herpes simplex virus type 1 encephalitis in childhood. J Pediatr. 2010;157(4):623–9. 629, e1. doi: 10.1016/j.jpeds.2010.04.020. The first genetic epidemiological survey of herpes simplex encephalitis in children from a French cohort. [DOI] [PubMed] [Google Scholar]
- 30.Studahl M, Skoldenberg B. Herpes simplex encephalitis and other neurological syndromes caused by herpes simplex virus-1. In: Studahl M, Cinque P, Bergstrom T, editors. Herpes simplex viruses. New York: Taylot & Francis; 2006. pp. 275–316. [Google Scholar]
- 31.Norberg P, Bergstrom T, Rekabdar E, Lindh M, Liljeqvist JA. Phylogenetic analysis of clinical herpes simplex virus type 1 isolates identified three genetic groups and recombinant viruses. J Virol. 2004;78(19):10755–64. doi: 10.1128/JVI.78.19.10755-10764.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bergstrom T. Pathogenesis. In: Sudahl M, Cinque P, Bergstrom T, editors. Herpes Simplex Viruses. New York: Taylor & Francis; 2006. pp. 99–117. [Google Scholar]
- 33.Schroder K, Hertzog PJ, Ravasi T, Hume DA. Interferon-gamma: an overview of signals, mechanisms and functions. J Leukoc Biol. 2004;75(2):163–89. doi: 10.1189/jlb.0603252. [DOI] [PubMed] [Google Scholar]
- 34.van Boxel-Dezaire AH, Rani MR, Stark GR. Complex modulation of cell type-specific signaling in response to type I interferons. Immunity. 2006;25(3):361–72. doi: 10.1016/j.immuni.2006.08.014. [DOI] [PubMed] [Google Scholar]
- 35.Dupuis S, Dargemont C, Fieschi C, Thomassin N, Rosenzweig S, Harris J, Holland SM, Schreiber RD, Casanova JL. Impairment of mycobacterial but not viral immunity by a germline human STAT1 mutation. Science. 2001;293(5528):300–3. doi: 10.1126/science.1061154. [DOI] [PubMed] [Google Scholar]
- 36.Chapgier A, et al. Novel STAT1 alleles in otherwise healthy patients with mycobacterial disease. PLoS Genet. 2006;2(8):e131. doi: 10.1371/journal.pgen.0020131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kristensen IA, Veirum JE, Moller BK, Christiansen M. Novel STAT1 alleles in a patient with impaired resistance to mycobacteria. J Clin Immunol. 2011;31(2):265–71. doi: 10.1007/s10875-010-9480-8. [DOI] [PubMed] [Google Scholar]
- 38.Chapgier A, Wynn RF, Jouanguy E, Filipe-Santos O, Zhang S, Feinberg J, Hawkins K, Casanova JL, Arkwright PD. Human complete Stat-1 deficiency is associated with defective type I and II IFN responses in vitro but immunity to some low virulence viruses in vivo. J Immunol. 2006;176(8):5078–83. doi: 10.4049/jimmunol.176.8.5078. [DOI] [PubMed] [Google Scholar]
- 39.Dupuis S, et al. Impaired response to interferon-alpha/beta and lethal viral disease in human STAT1 deficiency. Nat Genet. 2003;33(3):388–91. doi: 10.1038/ng1097. [DOI] [PubMed] [Google Scholar]
- 40.Vairo D, et al. Severe impairment of IFN-{gamma} and IFN-{alpha} responses in cells of a patient with a novel STAT1 splicing mutation. Blood. 2011;118(7):1806–17. doi: 10.1182/blood-2011-01-330571. [DOI] [PubMed] [Google Scholar]
- 41.Averbuch D, Chapgier A, Boisson-Dupuis S, Casanova JL, Engelhard D. The clinical spectrum of patients with deficiency of Signal Transducer and Activator of Transcription-1. Pediatr Infect Dis J. 2011;30(4):352–5. doi: 10.1097/INF.0b013e3181fdff4a. [DOI] [PubMed] [Google Scholar]
- 42.Kong XF, et al. A novel form of human STAT1 deficiency impairing early but not late responses to interferons. Blood. 2010;116(26):5895–906. doi: 10.1182/blood-2010-04-280586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chapgier A, et al. A partial form of recessive STAT1 deficiency in humans. J Clin Invest. 2009;119(6):1502–14. doi: 10.1172/JCI37083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44*.Liu L, et al. Gain-of-function human STAT1 mutations impair IL-17 immunity and underlie chronic mucocutaneous candidiasis. J Exp Med. 2011;208(8):1635–48. doi: 10.1084/jem.20110958. Identification of gain-of-function mutations in STAT1 predisposing to Candida infections. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van de Veerdonk FL, et al. STAT1 mutations in autosomal dominant chronic mucocutaneous candidiasis. N Engl J Med. 2011;365(1):54–61. doi: 10.1056/NEJMoa1100102. [DOI] [PubMed] [Google Scholar]
- 46.Puel A, et al. Chronic mucocutaneous candidiasis in humans with inborn errors of interleukin-17 immunity. Science. 2011;332(6025):65–8. doi: 10.1126/science.1200439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stark GR, Kerr IM, Williams BR, Silverman RH, Schreiber RD. How cells respond to interferons. Annu Rev Biochem. 1998;67:227–64. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- 48.Velazquez L, Fellous M, Stark GR, Pellegrini S. A protein tyrosine kinase in the interferon alpha/beta signaling pathway. Cell. 1992;70(2):313–22. doi: 10.1016/0092-8674(92)90105-l. [DOI] [PubMed] [Google Scholar]
- 49.Uze G, Schreiber G, Piehler J, Pellegrini S. The receptor of the type I interferon family. Curr Top Microbiol Immunol. 2007;316:71–95. doi: 10.1007/978-3-540-71329-6_5. [DOI] [PubMed] [Google Scholar]
- 50.Bacon CM, McVicar DW, Ortaldo JR, Rees RC, O’Shea JJ, Johnston JA. Interleukin 12 (IL-12) induces tyrosine phosphorylation of JAK2 and TYK2: differential use of Janus family tyrosine kinases by IL-2 and IL-12. J Exp Med. 1995;181(1):399–404. doi: 10.1084/jem.181.1.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stahl N, et al. Association and activation of Jak-Tyk kinases by CNTF-LIF-OSM-IL-6 beta receptor components. Science. 1994;263(5143):92–5. doi: 10.1126/science.8272873. [DOI] [PubMed] [Google Scholar]
- 52.Minegishi Y, et al. Human tyrosine kinase 2 deficiency reveals its requisite roles in multiple cytokine signals involved in innate and acquired immunity. Immunity. 2006;25(5):745–55. doi: 10.1016/j.immuni.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 53.Puel A, et al. Recurrent staphylococcal cellulitis and subcutaneous abscesses in a child with autoantibodies against IL-6. J Immunol. 2008;180(1):647–54. doi: 10.4049/jimmunol.180.1.647. [DOI] [PubMed] [Google Scholar]
- 54.Heimall J, Freeman A, Holland SM. Pathogenesis of hyper IgE syndrome. Clin Rev Allergy Immunol. 2010;38(1):32–8. doi: 10.1007/s12016-009-8134-1. [DOI] [PubMed] [Google Scholar]
- 55.Puel A, Picard C, Ku CL, Smahi A, Casanova JL. Inherited disorders of NF-kappaB-mediated immunity in man. Curr Opin Immunol. 2004;16(1):34–41. doi: 10.1016/j.coi.2003.11.013. [DOI] [PubMed] [Google Scholar]
- 56.Zhao T, Yang L, Sun Q, Arguello M, Ballard DW, Hiscott J, Lin R. The NEMO adaptor bridges the nuclear factor-kappaB and interferon regulatory factor signaling pathways. Nat Immunol. 2007;8(6):592–600. doi: 10.1038/ni1465. [DOI] [PubMed] [Google Scholar]
- 57.Smahi A, Courtois G, Rabia SH, Doffinger R, Bodemer C, Munnich A, Casanova JL, Israel A. The NF-kappaB signalling pathway in human diseases: from incontinentia pigmenti to ectodermal dysplasias and immune-deficiency syndromes. Hum Mol Genet. 2002;11(20):2371–5. doi: 10.1093/hmg/11.20.2371. [DOI] [PubMed] [Google Scholar]
- 58.Ku CL, et al. Inherited disorders of human Toll-like receptor signaling: immunological implications. Immunol Rev. 2005;203:10–20. doi: 10.1111/j.0105-2896.2005.00235.x. [DOI] [PubMed] [Google Scholar]
- 59.von Bernuth H, et al. Pyogenic bacterial infections in humans with MyD88 deficiency. Science. 2008;321(5889):691–6. doi: 10.1126/science.1158298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Picard C, et al. Pyogenic bacterial infections in humans with IRAK-4 deficiency. Science. 2003;299(5615):2076–9. doi: 10.1126/science.1081902. [DOI] [PubMed] [Google Scholar]
- 61*.Casanova JL, Abel L, Quintana-Murci L. Human TLRs and IL-1Rs in host defense: natural insights from evolutionary, epidemiological, and clinical genetics. Annu Rev Immunol. 2011;29:447–91. doi: 10.1146/annurev-immunol-030409-101335. A review of the role of human TLRs and IL-1Rs in host defense, from the agles of evolutionary, epidemiological, and clinical genetics. [DOI] [PubMed] [Google Scholar]
- 62.Filipe-Santos O, et al. X-linked susceptibility to mycobacteria is caused by mutations in NEMO impairing CD40-dependent IL-12 production. J Exp Med. 2006;203(7):1745–59. doi: 10.1084/jem.20060085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Puel A, et al. The NEMO mutation creating the most-upstream premature stop codon is hypomorphic because of a reinitiation of translation. Am J Hum Genet. 2006;78(4):691–701. doi: 10.1086/501532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Orange JS, Levy O, Brodeur SR, Krzewski K, Roy RM, Niemela JE, Fleisher TA, Bonilla FA, Geha RS. Human nuclear factor kappa B essential modulator mutation can result in immunodeficiency without ectodermal dysplasia. J Allergy Clin Immunol. 2004;114(3):650–6. doi: 10.1016/j.jaci.2004.06.052. [DOI] [PubMed] [Google Scholar]
- 65.Niehues T, et al. Nuclear factor kappaB essential modulator-deficient child with immunodeficiency yet without anhidrotic ectodermal dysplasia. J Allergy Clin Immunol. 2004;114(6):1456–62. doi: 10.1016/j.jaci.2004.08.047. [DOI] [PubMed] [Google Scholar]
- 66.Hiscott J, Alper D, Cohen L, Leblanc JF, Sportza L, Wong A, Xanthoudakis S. Induction of human interferon gene expression is associated with a nuclear factor that interacts with the NF-kappa B site of the human immunodeficiency virus enhancer. J Virol. 1989;63(6):2557–66. doi: 10.1128/jvi.63.6.2557-2566.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Thomson SJ, Goh FG, Banks H, Krausgruber T, Kotenko SV, Foxwell BM, Udalova IA. The role of transposable elements in the regulation of IFN-lambda1 gene expression. Proc Natl Acad Sci U S A. 2009;106(28):11564–9. doi: 10.1073/pnas.0904477106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Visvanathan KV, Goodbourn S. Double-stranded RNA activates binding of NF-kappa B to an inducible element in the human beta-interferon promoter. Embo J. 1989;8(4):1129–38. doi: 10.1002/j.1460-2075.1989.tb03483.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Thanos D, Maniatis T. The high mobility group protein HMG I(Y) is required for NF-kappa B-dependent virus induction of the human IFN-beta gene. Cell. 1992;71(5):777–89. doi: 10.1016/0092-8674(92)90554-p. [DOI] [PubMed] [Google Scholar]
- 70.Audry M, et al. NEMO is a key component of NF-kappaB- and IRF-3-dependent TLR3-mediated immunity to herpes simplex virus. J Allergy Clin Immunol. 2011 doi: 10.1016/j.jaci.2011.04.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124(4):783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 72.Kawai T, Akira S. Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity. 2011;34(5):637–50. doi: 10.1016/j.immuni.2011.05.006. [DOI] [PubMed] [Google Scholar]
- 73.Zhang SY, Jouanguy E, Sancho-Shimizu V, von Bernuth H, Yang K, Abel L, Picard C, Puel A, Casanova JL. Human Toll-like receptor-dependent induction of interferons in protective immunity to viruses. Immunol Rev. 2007;220:225–36. doi: 10.1111/j.1600-065X.2007.00564.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Onoguchi K, Yoneyama M, Fujita T. Retinoic acid-inducible gene-I-like receptors. J Interferon Cytokine Res. 2011;31(1):27–31. doi: 10.1089/jir.2010.0057. [DOI] [PubMed] [Google Scholar]
- 75.Matsumiya T, Imaizumi T, Yoshida H, Satoh K. Antiviral signaling through retinoic acid-inducible gene-I-like receptors. Arch Immunol Ther Exp (Warsz) 2011;59(1):41–8. doi: 10.1007/s00005-010-0107-9. [DOI] [PubMed] [Google Scholar]
- 76.Choi MK, et al. A selective contribution of the RIG-I-like receptor pathway to type I interferon responses activated by cytosolic DNA. Proc Natl Acad Sci U S A. 2009;106(42):17870–5. doi: 10.1073/pnas.0909545106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rehwinkel J, et al. RIG-I detects viral genomic RNA during negative-strand RNA virus infection. Cell. 2010;140(3):397–408. doi: 10.1016/j.cell.2010.01.020. [DOI] [PubMed] [Google Scholar]
- 78.Sabbah A, Chang TH, Harnack R, Frohlich V, Tominaga K, Dube PH, Xiang Y, Bose S. Activation of innate immune antiviral responses by Nod2. Nat Immunol. 2009;10(10):1073–80. doi: 10.1038/ni.1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang Z, Kim T, Bao M, Facchinetti V, Jung SY, Ghaffari AA, Qin J, Cheng G, Liu YJ. DDX1, DDX21, and DHX36 helicases form a complex with the adaptor molecule TRIF to sense dsRNA in dendritic cells. Immunity. 2011;34(6):866–78. doi: 10.1016/j.immuni.2011.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Miyashita M, Oshiumi H, Matsumoto M, Seya T. DDX60, a DEXD/H Box Helicase, Is a Novel Antiviral Factor Promoting RIG-I-Like Receptor-Mediated Signaling. Mol Cell Biol. 2011;31(18):3802–19. doi: 10.1128/MCB.01368-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhang Z, Yuan B, Lu N, Facchinetti V, Liu YJ. DHX9 Pairs with IPS-1 To Sense Double-Stranded RNA in Myeloid Dendritic Cells. J Immunol. 2011 doi: 10.4049/jimmunol.1101307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ablasser A, Bauernfeind F, Hartmann G, Latz E, Fitzgerald KA, Hornung V. RIG-I-dependent sensing of poly(dA:dT) through the induction of an RNA polymerase III-transcribed RNA intermediate. Nat Immunol. 2009;10(10):1065–72. doi: 10.1038/ni.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chiu YH, Macmillan JB, Chen ZJ. RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell. 2009;138(3):576–91. doi: 10.1016/j.cell.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhang Z, Yuan B, Bao M, Lu N, Kim T, Liu YJ. The helicase DDX41 senses intracellular DNA mediated by the adaptor STING in dendritic cells. Nat Immunol. 2011 doi: 10.1038/ni.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Unterholzner L, et al. IFI16 is an innate immune sensor for intracellular DNA. Nat Immunol. 2010;11(11):997–1004. doi: 10.1038/ni.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ishii KJ, et al. TANK-binding kinase-1 delineates innate and adaptive immune responses to DNA vaccines. Nature. 2008;451(7179):725–9. doi: 10.1038/nature06537. [DOI] [PubMed] [Google Scholar]
- 87.Takaoka A, et al. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature. 2007;448(7152):501–5. doi: 10.1038/nature06013. [DOI] [PubMed] [Google Scholar]
- 88.Ishikawa H, Barber GN. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 2008;455(7213):674–8. doi: 10.1038/nature07317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhong B, et al. The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity. 2008;29(4):538–50. doi: 10.1016/j.immuni.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 90.Sun W, et al. ERIS, an endoplasmic reticulum IFN stimulator, activates innate immune signaling through dimerization. Proc Natl Acad Sci U S A. 2009;106(21):8653–8. doi: 10.1073/pnas.0900850106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Burdette DL, Monroe KM, Sotelo-Troha K, Iwig JS, Eckert B, Hyodo M, Hayakawa Y, Vance RE. STING is a direct innate immune sensor of cyclic di-GMP. Nature. 2011 doi: 10.1038/nature10429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Suh HS, Brosnan CF, Lee SC. Toll-like receptors in CNS viral infections. Curr Top Microbiol Immunol. 2009;336:63–81. doi: 10.1007/978-3-642-00549-7_4. [DOI] [PubMed] [Google Scholar]
- 93.Saitoh S, Miyake K. Regulatory molecules required for nucleotide-sensing Toll-like receptors. Immunol Rev. 2009;227(1):32–43. doi: 10.1111/j.1600-065X.2008.00729.x. [DOI] [PubMed] [Google Scholar]
- 94.Casrouge A, et al. Herpes simplex virus encephalitis in human UNC-93B deficiency. Science. 2006;314(5797):308–12. doi: 10.1126/science.1128346. [DOI] [PubMed] [Google Scholar]
- 95.Brinkmann MM, Spooner E, Hoebe K, Beutler B, Ploegh HL, Kim YM. The interaction between the ER membrane protein UNC93B and TLR3, 7, and 9 is crucial for TLR signaling. J Cell Biol. 2007;177(2):265–75. doi: 10.1083/jcb.200612056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kim YM, Brinkmann MM, Paquet ME, Ploegh HL. UNC93B1 delivers nucleotide-sensing toll-like receptors to endolysosomes. Nature. 2008;452(7184):234–8. doi: 10.1038/nature06726. [DOI] [PubMed] [Google Scholar]
- 97*.Barreiro LB, et al. Evolutionary dynamics of human Toll-like receptors and their different contributions to host defense. PLoS Genet. 2009;5(7):e1000562. doi: 10.1371/journal.pgen.1000562. A thorough analysis of the evolutionary contributions of each human TLRs as shaped by history, revealing that endosomal TLRs have evolved under purifying selection. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Blasius AL, Beutler B. Intracellular toll-like receptors. Immunity. 2010;32(3):305–15. doi: 10.1016/j.immuni.2010.03.012. [DOI] [PubMed] [Google Scholar]
- 99.Yang K, et al. Human TLR-7-, -8-, and -9-mediated induction of IFN-alpha/beta and -lambda Is IRAK-4 dependent and redundant for protective immunity to viruses. Immunity. 2005;23(5):465–78. doi: 10.1016/j.immuni.2005.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ku CL, et al. IRAK4 and NEMO mutations in otherwise healthy children with recurrent invasive pneumococcal disease. J Med Genet. 2007;44(1):16–23. doi: 10.1136/jmg.2006.044446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Picard C, et al. Clinical features and outcome of patients with IRAK-4 and MyD88 deficiency. Medicine (Baltimore) 2011;89(6):403–25. doi: 10.1097/MD.0b013e3181fd8ec3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Tabeta K, et al. The Unc93b1 mutation 3d disrupts exogenous antigen presentation and signaling via Toll-like receptors 3, 7 and 9. Nat Immunol. 2006;7(2):156–64. doi: 10.1038/ni1297. [DOI] [PubMed] [Google Scholar]
- 103.Zhang SY, et al. TLR3 deficiency in patients with herpes simplex encephalitis. Science. 2007;317(5844):1522–7. doi: 10.1126/science.1139522. [DOI] [PubMed] [Google Scholar]
- 104.Gorbea C, et al. A role for Toll-like receptor 3 variants in host susceptibility to enteroviral myocarditis and dilated cardiomyopathy. J Biol Chem. 2010;285(30):23208–23. doi: 10.1074/jbc.M109.047464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105*.Guo Y, et al. Herpes simplex virus encephalitis in a patient with complete TLR3 deficiency: TLR3 is otherwise redundant in protective immunity. J Exp Med. 2011;208(10):2083–98. doi: 10.1084/jem.20101568. A report describing an HSE patient with complete TLR3 deficiency. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106*.Sancho-Shimizu V, et al. Herpes simplex encephalitis in patients with autosomal recessive and dominant TRIF deficiency. J Clin Invest. 2011 doi: 10.1172/JCI59259. In press. A report describing the first cases of complete and partial human TRIF deficiency in children with HSE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Yamamoto M, et al. Role of adaptor TRIF in the MyD88-independent toll-like receptor signaling pathway. Science. 2003;301(5633):640–3. doi: 10.1126/science.1087262. [DOI] [PubMed] [Google Scholar]
- 108.Hoebe K, et al. Identification of Lps2 as a key transducer of MyD88-independent TIR signalling. Nature. 2003;424(6950):743–8. doi: 10.1038/nature01889. [DOI] [PubMed] [Google Scholar]
- 109.Funami K, Sasai M, Ohba Y, Oshiumi H, Seya T, Matsumoto M. Spatiotemporal mobilization of Toll/IL-1 receptor domain-containing adaptor molecule-1 in response to dsRNA. J Immunol. 2007;179(10):6867–72. doi: 10.4049/jimmunol.179.10.6867. [DOI] [PubMed] [Google Scholar]
- 110.Funami K, Sasai M, Oshiumi H, Seya T, Matsumoto M. Homo-oligomerization is essential for Toll/interleukin-1 receptor domain-containing adaptor molecule-1-mediated NF-kappaB and interferon regulatory factor-3 activation. J Biol Chem. 2008;283(26):18283–91. doi: 10.1074/jbc.M801013200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Karin M, Gallagher E. TNFR signaling: ubiquitin-conjugated TRAFfic signals control stop-and-go for MAPK signaling complexes. Immunol Rev. 2009;228(1):225–40. doi: 10.1111/j.1600-065X.2008.00755.x. [DOI] [PubMed] [Google Scholar]
- 112.Hacker H, Tseng PH, Karin M. Expanding TRAF function: TRAF3 as a tri-faced immune regulator. Nat Rev Immunol. 2011;11(7):457–68. doi: 10.1038/nri2998. [DOI] [PubMed] [Google Scholar]
- 113.Pietras EM, Saha SK, Cheng G. The interferon response to bacterial and viral infections. J Endotoxin Res. 2006;12(4):246–50. doi: 10.1179/096805106X118799. [DOI] [PubMed] [Google Scholar]
- 114*.Perez de Diego R, et al. Human TRAF3 adaptor molecule deficiency leads to impaired Toll-like receptor 3 response and susceptibility to herpes simplex encephalitis. Immunity. 2010;33(3):400–11. doi: 10.1016/j.immuni.2010.08.014. Identification of a germ-line heterozygous, dominant-negative TRAF3 mutation in an HSE patient. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Prehaud C, Megret F, Lafage M, Lafon M. Virus infection switches TLR-3-positive human neurons to become strong producers of beta interferon. J Virol. 2005;79(20):12893–904. doi: 10.1128/JVI.79.20.12893-12904.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Bsibsi M, Ravid R, Gveric D, van Noort JM. Broad expression of Toll-like receptors in the human central nervous system. J Neuropathol Exp Neurol. 2002;61(11):1013–21. doi: 10.1093/jnen/61.11.1013. [DOI] [PubMed] [Google Scholar]
- 117*.Fornarino S, Laval G, Barreiro LB, Manry J, Vasseur E, Quintana-Murci L. Evolution of the TIR Domain-Containing Adaptors in Humans: Swinging between Constraint and Adaptation. Mol Biol Evol. 2011 doi: 10.1093/molbev/msr137. An analysis of the evolutionary contributions of each human TLR adaptors, demonstrating that MyD88 and TRIF have evolved under purifying selection. [DOI] [PubMed] [Google Scholar]
- 118.Shigemoto T, Kageyama M, Hirai R, Zheng J, Yoneyama M, Fujita T. Identification of loss of function mutations in human genes encoding RIG-I and MDA5: implications for resistance to type I diabetes. J Biol Chem. 2009;284(20):13348–54. doi: 10.1074/jbc.M809449200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Pothlichet J, et al. Study of human RIG-I polymorphisms identifies two variants with an opposite impact on the antiviral immune response. PLoS One. 2009;4(10):e7582. doi: 10.1371/journal.pone.0007582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Quintana-Murci L, Barreiro LB. The role played by natural selection on Mendelian traits in humans. Ann N Y Acad Sci. 2010;1214:1–17. doi: 10.1111/j.1749-6632.2010.05856.x. [DOI] [PubMed] [Google Scholar]
- 121.Alcais A, Quintana-Murci L, Thaler DS, Schurr E, Abel L, Casanova JL. Life-threatening infectious diseases of childhood: single-gene inborn errors of immunity? Ann N Y Acad Sci. 2010;1214:18–33. doi: 10.1111/j.1749-6632.2010.05834.x. [DOI] [PubMed] [Google Scholar]
- 122.Casanova JL, Abel L. Primary immunodeficiencies: a field in its infancy. Science. 2007;317(5838):617–9. doi: 10.1126/science.1142963. [DOI] [PubMed] [Google Scholar]
- 123*.Suppiah V, et al. IL28B is associated with response to chronic hepatitis C interferon-alpha and ribavirin therapy. Nat Genet. 2009;41(10):1100–4. doi: 10.1038/ng.447. First studies to report that sequence variants in IL28B are associated with hepatitis C treatment induced viral clearance. [DOI] [PubMed] [Google Scholar]
- 124*.Ge D, et al. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature. 2009;461(7262):399–401. doi: 10.1038/nature08309. See ref 123. [DOI] [PubMed] [Google Scholar]
- 125*.Tanaka Y, et al. Genome-wide association of IL28B with response to pegylated interferon-alpha and ribavirin therapy for chronic hepatitis C. Nat Genet. 2009;41(10):1105–9. doi: 10.1038/ng.449. See ref 123. [DOI] [PubMed] [Google Scholar]
- 126*.Thomas DL, et al. Genetic variation in IL28B and spontaneous clearance of hepatitis C virus. Nature. 2009;461(7265):798–801. doi: 10.1038/nature08463. First studies to report that sequence variants in IL28B are associated with spontaneous clearance of hepatitis C virus. [DOI] [PMC free article] [PubMed] [Google Scholar]