Abstract
Toxoplasma gondii ME49 is an obligatory intracellular apicomplexa parasite that causes toxoplasmosis in humans, domesticated and wild animals. Waterborne outbreaks of acute toxoplasmosis worldwide reinforce the transmission of Toxoplasma gondii ME49 to humans through contaminated water and may have a greater epidemiological impact than previously believed. In the quest for drug and vaccine target identification subtractive genomics involving subtraction between the host and pathogen genome has been implemented for enlisting essential pathogen specific proteins. Using this approach, our analysis on both human and Toxoplasma gondii ME49 reveals that out of 7987 protein coding sequences of the pathogen, 950 represent essential non human-homologous proteins. Subcellular localization prediction & comparative-biochemical pathway analysis of these essential proteins gives a list of apicoplast-associated proteins having unique pathogen-specific metabolic pathway. These apicoplast-associated enzymes involved in fatty acid biosynthesis pathway of Toxoplasma gondii ME49, may be used as potential drug targets, as the pathway is vital for the protozoan's survival. Structure prediction of drug target proteins was done using fold based recognition method. Screening of the functional inhibitors against these novel targets may result in discovery of novel therapeutic compounds that can be effective against Toxoplasma gondii ME49.
Abbreviations
DEG - Database of Essential Gene, KEGG - Kyoto Encyclopaedia of Genes and Genomes, KAAS - KEGG Automated Annotation Server, PFP - Protein Function Prediction, COG - Cluster of Orthologous Genes.
Keywords: Toxoplasma gondii ME49, Essential gene, Drug targets, Subtractive genomics, Pathway analysis
Background:
Toxoplasma gondii is arguably the most successful parasitic organism in the world. It can infect all warm-blooded animals, including people, and causes a wide spectrum of disease severity in the different host species, ranging from acute fatal disease in marsupials and marine mammals through to very mild clinical signs in other hosts. Toxoplasma gondii ME49 is an obligatory intracellular Apicomplexa parasite, with a complex life cycle, that causes toxoplasmosis in humans, domesticated and wild animals[1].Toxoplasmic encephalitis (TE) caused by Toxoplasma gondii ME49 remains an important human disease, particularly in immuno-suppressed individuals . Murine models employing the ME49 strain of Toxoplasma gondii have been developed and used to study the pathogenesis and therapy of TE[2] [3] [4]. The ME49 strain of Toxoplasma gondii is of typeII, which has been reported as the type most frequently associated with human disease. Infection is usually asymptomatic in people with a normal immune function and occasionally results in eye involvement. Toxoplasmosis can cause severe disease in foetuses of acutely infected pregnant woman, immuno-compromised individuals (AIDS) and therapeutically immuno-suppressed patients, as cancer or transplant recipients. Toxoplasma gondii ME49 is a protozoan parasite that is uniquely adapted for penetrating and surviving within a wide range of host cells. This parasite invades mammalian cells by an active actin-dependent mechanism, and after entry establishes a vacuole with the assistance of products secreted by the parasite's apical organelles [5].
One of the unique features of Toxoplasma gondii ME49 is the presence of a non-photosynthetic plastid called apicoplast (6-15) containing its own 35-kilobase genome, which is essential for parasite survival. Antibiotics that apparently target plastid protein synthesis specifically block parasite replication [9, 10]. It has been postulated that the apicomplexan plastid was acquired by endosymbiosis of an alga [6, 7, 11]. The plastid is bounded by four membranes, and its genome shows substantial sequence similarity to algal and other chloroplast DNA [7, 8, 9, 11]. Although the metabolic functions of this degenerate plastid remain largely unknown, it is possible that it provides a site for fatty acid biosynthesis [14, 15]. Recent studied has revealed directly linkage between apicoplast function and parasite survival, validating this intriguing organelle as an effective target for parasiticidal drug design.
In quest for developing new potent drug candidates against Toxoplasma gondii cell-based drug development strategy has been in use in past decade. In cell-based drug development, researchers attempt to create drugs that kill a pathogen without necessarily understanding the details of how the drugs work. In contrast, target-based drug development entails the search for compounds that act on a specific intracellular target―often a protein known or suspected to be required for survival of the pathogen. The latter approach to drug development has been facilitated greatly by the sequencing of many pathogen genomes.
To identify apicoplast associated drug targets we have implemented the concept of subtractive genomics approach in which the subtraction dataset between the host and pathogen genome provides information of pathogen-specific essential proteins. This approach has been used successfully in recent times to identify essential genes in Pseudomonas aeruginosa. We have used the same methodology to analyse the whole genome sequence of the Toxoplasma gondii ME49. The differences in the proteins of the host and the pathogen can be effectively used for designing a drug specifically targeting the pathogen.
Methodology:
The process-flow of the methodology is displayed in Figure 1.
Figure 1.
Flowchart of the methodology used. CD-HIT = Cluster Database at High Identity with Tolerance; NR Set = Non-Redundant Set; BLASTP = Basic Local Alignment Search Tool Protein; DEG = Database of Essential Genes; PFP Server = Protein Function Prediction Server; KEGG = Kyoto Encyclopedia of Genes and Genomes; KAAS = KEGG Automatic Annotation Server;
Retrieval of Proteomes of Host and Pathogen:
The complete proteome of Toxoplasma gondii ME49 & Homo sapiens was retrieved from NCBI. Eukaryotic essential protein sequences were retrieved from the database of essential genes (DEG) [16] from its location (http://tubic.tju.edu.cn/deg/) by manual annotation.
Identification of Essential Proteins in Toxoplasma gondii ME49:
Toxoplasma gondii ME49 proteome was purged at 20% threshold using CD-HIT [17] to exclude paralogs or duplicates for further analysis. The set of proteins obtained were subjected to Blastp against Homo sapiens proteome with the expectation value (Evalue) cut-off of 10-4. The protein sequences of Toxoplasma gondii ME49 showing no significant similarity with Homo sapiens proteome was retrieved manually. Blastp analysis was performed for the non homologous protein sequences of Toxoplasma gondii ME49 against database of essential proteins retrieved from DEG with E-value cut off score of 10-10 and 30% identity. A minimum bit-score cut-off of 100 was used to screen out proteins that appeared to represent essential genes. The protein sequences obtained are non homologous essential proteins of Toxoplasma gondii ME49.
Sub Cellular Localization Prediction:
Sub-cellular localization analysis of the essential proteins has been done by Proteome Analyst Specialized Sub Cellular Localization Server v2.5 (PA-SUB) [18] to identify the location of the essential proteins in different cellular organelles.
Metabolic Pathway Analysis:
Metabolic pathway analysis of the essential proteins of Toxoplasma gondii ME49 was done by KEGG Automatic Annotation Server (KAAS) [19]. Comparative analysis of the metabolic pathways of the host and pathogen was performed by using Kyoto Encyclopaedia of Genes and Genomes (KEGG) pathway database [20] to trace out essential proteins involved in pathogen specific metabolic pathways for the identification of potential drug targets.
Protein Function Prediction:
Protein function prediction was done using PFP Automated Protein Function Prediction Server (http://kiharalab.org/web/pfp.php) to find the function of uncharacterised essential protein [21].
Cluster of Orthologous Genes:
Essential proteins were manually segregated into different categories as per the norms of Cluster of Orthologous Genes (http://www.ncbi.nlm.nih.gov/COG/).
Structure prediction of drug targets:
Crystal structures for the enzymes in apicoplast involved in fat metabolism are not available in Protein data Bank therefore modelling of the target proteins were performed using different structure prediction methods. Homology modelling did not give good results because of low sequence identity between target and template sequence (less than 20%).For getting better prediction accuracy fold based prediction using Phyre2 server [22] was implemented. Protein threading or fold recognition is a method of protein modelling which is used to model those proteins which have the same fold as proteins of known structures, but do not have homologous proteins with known structure. Energy minimization of generated 3D-model was done through GROMACS (OPLS force field) by using Steepest Descent and Conjugate Gradient Algorithms. Parameters like covalent bond distances and angles, stereo-chemical validation, atom nomenclature were validated using RAMPAGE and overall quality factor of non-bonded interactions between different atoms types were measured by ERRAT program. RMSD (root-mean-square deviation) and RMSF (Root Mean Square Fluctuation) was calculated for modelled structures.
Discussion:
Toxoplasma gondii ME49 proteome was found to contain 7987 functional & structural proteins. The protein which had conserved region in Human proteome detected by BLASTp was excluded for further analysis. The non homologous proteins when analysed with BLASTp in DEG (Database of Essential Genes) gave a list of 950 putative uncharacterised essential proteins. The 950 essential proteins were grouped into 12 classes according to COGs functional classification. The subcellular localization prediction for the 950 essential proteins of Toxoplasma gondii ME49 was done by using Proteome Analyst Specialized Sub Cellular Localization Server v2.5 (PA-SUB) to locate the apicoplast associated proteins. Sub-cellular localization prediction gave the following set of proteins: Apicoplast: 31; Cytoplasm: 396; Periplasm: 3; Mitochondria: 149; Nucleus:567; Extracellularenvironment:49; Plasmamembrane:152;Peroxisome:4;ER:3;Golgiapparatus:5; Cytoskeleton:4; Outer membrane: 8 and Lysosome:1. Out of 131 proteins located in apicoplast which are mentioned above, 75 are Miscellaneous Enzymes and two of the enzymes glutamate dehydrogenase & aspartate amino transferase play an important role in the fatty acid biosynthesis pathway.
The KEGG-KAAS analysis identified the biochemical pathways of the apicoplast associated proteins. Comparative analysis of biochemical pathways of apicoplast associated proteins of Toxoplasma gondii ME49 with the pathways of Homo sapiens gives 18 proteins which are involved in pathogen-specific metabolic pathways. The complete list of the apicoplast associated proteins having unique pathways is given in . Table 1 (See supplementary material) has 18 drug targets which are active in pathogen-specific metabolic pathways. Out of those 18 targets, 16 are engaged in wide-ranging metabolic processes and only two are involved in apicoplast-specific fatty acid metabolic reactions. Apicoplast-associated fatty acid metabolism is very unique to the Toxoplasma gondii and any alteration in the direction of this metabolism cascade will be fatal to the pathogen & therefore they have good prospects for targeting of new drugs. Thus we have considered these proteins for further structural & functional analysis. The two drug targets identified are Long chain fatty acid CoA ligase & Succinate- semi aldehyde dehydrogenase (Figure 2. (a, b)).
Figure 2.
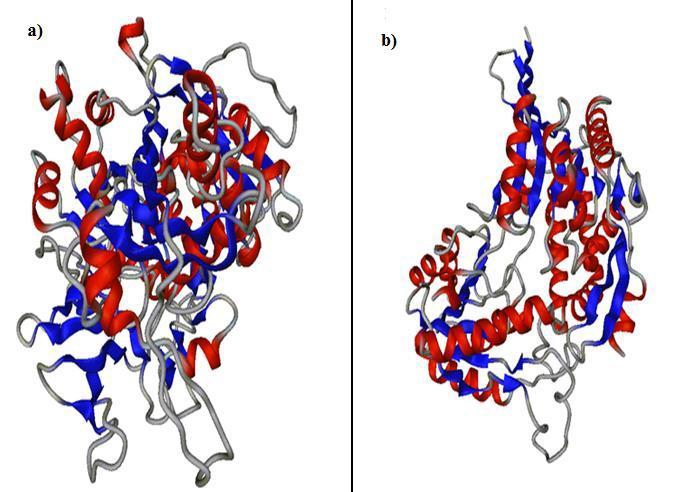
(a) Modeled structure of Modeled structure of Longchain fatty acid CoA ligase (b) Modeled structure of Succinate semi-aldehyde dehydrogenase. The structures have been viewed using PPG (Protein Picture Generator http://bioserv.rpbs.jussieu.fr/PPG) and DINO (Visualizing Structural Biology (2002) http://www.dino3d.org)
Drugs designed for these targets can have potent anti-microbial activity against the Toxoplasma gondii ME49.X ray crystallography or NMR derived structures of these two proteins were not available in RCSB-PDB so computationalapproach of structure prediction was the only alternative. The alignment of these two proteins with RCSB-PDB templates gave less than 18% identity so homology modelling based approach was not a good choice for such a low-alignment of targettemplate.
For structures which have low identity of template-target alignment, fold- recognition based structure prediction is preferred. Structure prediction using Phyre2 (Protein Homology/analogY Recognition Engine V 2.0) server was employed as described under Methods. (Table 2, see supplementary material) represents the summary of Phyre2 results. The Ramachandran plot analysis (by RAMPAGE) of the model structure from Phyre2 server showed that > 90% of the residues is in favoured and additional allowed regions. It signifies the good quality of our model. Active site prediction using Q-Site Finder (http://www.modelling.leeds.ac.uk/qsitefinder) predicts pocket of Long chain fatty acid CoA ligase (having site volume of 994 Cubic A0) and Succinate- semi aldehyde dehydrogenase (having site volume of 618 Cubic A0) which can be used as potential inhibitory sites (Figure 2 (a, b)).As apicoplast is unique to the protozoa the apicoplast associated essential proteins may be attractive drug targets for Toxoplasma gondii ME49. Thus, further investigations on the predicted apicoplast associated essential proteins are required to verify the reliability of the data.
Conclusion:
The present study has thus led to the identification of several apicoplast associated proteins that can be targeted for effective drug design against Toxoplasma gondii ME49 .The drugs developed against these will be specific to the pathogen, and therefore less or non-toxic to the host. Virtual screening against these novel targets might be useful in the discovery of novel therapeutic compounds against Toxoplasma gondii ME49.This research thus draw attention to novel means of finding new generation of drug targets for drug resistant microbes.
Supplementary material
Acknowledgments
The authors express gratitude for the technical support from Department of Biotechnology, PES Institute of Technology, Bangalore & Institute of Computational Biology, Bangalore.
Footnotes
Citation:Saremy et al, Bioinformation 7(8): 379-383 (2011)
References
- 1.ACS Guimaraes, et al. Rev Inst Med Trop São Paulo. 1993;35:479. doi: 10.1590/s0036-46651993000600001. [DOI] [PubMed] [Google Scholar]
- 2.FG Araujo, et al. Antimicrob Agents Chemother. 1997;41:2137. doi: 10.1128/aac.41.10.2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.FG Araujo, et al. Int J Antimicrob Agents. 1992;4:153. doi: 10.1016/0924-8579(92)90002-9. [DOI] [PubMed] [Google Scholar]
- 4.DG Mordue, et al. J Immunol. 2001;167:4574. doi: 10.4049/jimmunol.167.8.4574. [DOI] [PubMed] [Google Scholar]
- 5.I Coppens, et al. Expert Rev Mol. 2001;15:1. [Google Scholar]
- 6.JHP Hackstein, et al. Parasitology Res. 1995;81:207. [Google Scholar]
- 7.DH Williamson, et al. Mol Gen Genet. 1994;243:249. doi: 10.1007/BF00280323. [DOI] [PubMed] [Google Scholar]
- 8.Fadden Mc, et al. Nature. 1996;381:482. [Google Scholar]
- 9.RF Waller, et al. Proc Natl Acad Sci. 1998;95:12352. [Google Scholar]
- 10.RJM Wilson, et al. Mol Biol Rev. 1997;61:1. [Google Scholar]
- 11.S Kohler, et al. Nature. 1997;275:1485. [Google Scholar]
- 12.ME Fichera, et al. Nature. 1997;390:407. [Google Scholar]
- 13.Beckers, et al. J Clin Invest. 1995;95(1):367–76. doi: 10.1172/JCI117665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pfefferkorn, et al. Antimicrob Agents Chemother. 1992;36:1091. doi: 10.1128/aac.36.5.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.F Roberts, et al. Nature. 1998;393:80. [Google Scholar]
- 16.R Zhang, et al. Nucleic Acids Res. 2009;37:455. [Google Scholar]
- 17.W Li, et al. Bioinformatics. 2006;22:1658. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- 18.Z Lu, et al. Bioinformatics. 2004;20:547. [Google Scholar]
- 19.Y Moriya, et al. Nucleic Acids Res. 2007;35:182. [Google Scholar]
- 20.M Kanehisa, et al. Nucleic Acids Res. 2000;28:27. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hawkins Troy, et al. Protein Sci. 2006;15:1550. doi: 10.1110/ps.062153506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kelley LA, et al. Nat Protoc. 2009;4:363. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



