Abstract
Human immunodeficiency virus type-1 (HIV-1) encodes a polypeptide called Gag that is able to form virus-like particles in vitro in the absence of any cellular or viral constituents. During the late phase of the HIV-1 infection, Gag polyproteins are transported to the plasma membrane (PM) for assembly. In the past two decades, in vivo, in vitro, and structural studies have shown that Gag trafficking and targeting to the PM are orchestrated events that are dependent on multiple factors including cellular proteins and specific membrane lipids. The matrix (MA) domain of Gag has been the focus of these studies as it appears to be engaged in multiple intracellular interactions that are suggested to be critical for virus assembly and replication. The interaction between Gag and the PM is perhaps the most understood. It is now established that the ultimate localization of Gag on punctate sites on the PM is mediated by specific interactions between the MA domain of Gag and phosphatidylinositol-4,5-bisphosphate [PI(4,5)P2], a minor lipid localized on the inner leaflet of the PM. Structure-based studies revealed that binding of PI(4,5)P2 to MA induces minor conformational changes, leading to exposure of the myristyl (myr) group. Exposure of the myr group is also triggered by binding of calmodulin, enhanced by factors that promote protein self-association like the capsid domain of Gag, and is modulated by pH. Despite the steady progress in defining both the viral and cellular determinants of retroviral assembly and release, Gag’s intracellular interactions and trafficking to its assembly sites in the infected cell are poorly understood. In this review, we summarize the current understanding of the structural and functional role of MA in HIV replication.
Keywords: assembly, Gag, matrix, myristyl, NMR, trafficking, plasma membrane
Human immunodeficiency virus type-1 (HIV-1), the causative agent of AIDS, is blamed for over 34 million deaths and is poised to claim over two million lives a year in the absence of efficient therapeutic intervention (UNAIDS report 2011). HIV-1 replication is strongly dependent on the cellular machinery to produce progeny virus. Since its discovery, efforts have focused on the development of effective vaccines and drugs that target different stages of the HIV lifecycle. Combinations of drugs that target protease, reverse transcriptase, integrase, or virus fusion form the basis of current antiretroviral therapy (ART; Greene et al., 2008). However, the genetic diversity and the ability of the virus to mutate to evade drug treatment remains a threat to the future success of ART. The discovery of cellular factors that participate in HIV-1 replication pathways has provided new insights into the molecular basis of virus–host cell interactions. Recent studies have identified hundreds of cellular factors that are involved in HIV replication (Brass et al., 2008; König et al., 2008; Zhou et al., 2008; Jäger et al., 2011; Lever and Jeang, 2011). Drugs that target the essential interactions of viral factors with host-proteins have the potential to not only complement current therapy, but may also overcome the problem of viral escape. As host-factors are stable and not diverse, the mutational capacity of the virus that usually allows for drug escape is thus restricted by the requirement to remain compatible to an invariant host-factor. Elucidation of the molecular interactions between the host cell and HIV are important for understanding the virus replication and the subsequent cytopathogenesis in the infected cell, which will aid in the development of more efficient antiviral drugs.
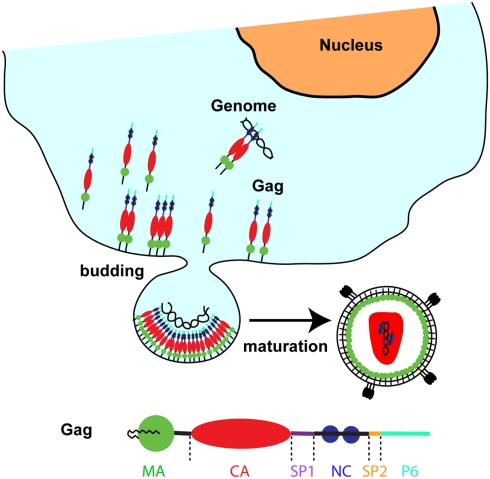
Retroviral genomes encode a polyprotein called Gag (Figure 1) that contains all the viral elements required for virus assembly and is capable of forming virus-like particles (VLPs) in vitro in the absence of viral and cellular constituents (Gheysen et al., 1989; Campbell and Rein, 1999; Campbell et al., 2001a). Subsequent to their synthesis, Gag proteins are targeted to the plasma membrane (PM) for assembly and budding (Adamson and Freed, 2007; Chu et al., 2010). During or shortly after budding, the virally encoded protease cleaves off Gag proteins at five positions into matrix (MA), capsid (CA), nucleocapsid (NC), spacer peptide 1 (SP1), spacer peptide 2 (SP2), and P6, which rearrange to form mature and infectious virions (Figure 1; Turner and Summers, 1999; Adamson and Freed, 2007; Ganser-Pornillos et al., 2008; Briggs and Kräusslich, 2011).
Figure 1.
Schematic representation showing the late phase of HIV replication involving Gag targeting and assembly on the plasma membrane.
It is widely accepted that HIV-1 Gag budding and assembly occur predominantly on the PM (Figure 1; Hermida-Matsumoto and Resh, 2000; Jouvenet et al., 2006, 2008; Finzi et al., 2007; Li et al., 2007; Welsch et al., 2007; Gousset et al., 2008; Joshi et al., 2009a; Ono, 2009, 2010). Membrane binding is mediated by Gag’s N-terminal myristoylated MA domain [myr(+)MA; Adamson and Freed, 2007; Ganser-Pornillos et al., 2008; Ono, 2009; Chukkapalli and Ono, 2011; Hamard-Peron and Muriaux, 2011]. The myristyl (myr) group functions in concert with a group of conserved basic residues to facilitate membrane anchoring and assembly of Gag. Mutations that either block myristoylation or disrupt the basic patch lead to inefficient Gag targeting to the PM, resulting in dramatically reduced virus production (Bryant and Ratner, 1990; Freed et al., 1994; Spearman et al., 1997; Hermida-Matsumoto and Resh, 2000; Ono et al., 2000b).
Role of the MA protein in HIV replication has been extensively examined. In this review, we focus on the functional role of MA in the late phase of infection. In the last few years, efforts have been focused on the identification of structural requirements that enable efficient Gag-membrane association. Success in producing the first retroviral matrix myristoylated protein for structural studies (Tang et al., 2004) afforded invaluable insight into understanding specific factors that regulate the myr switch and the potential implication in virus assembly.
Intrinsic Factors that Modulate the myr Switch in HIV-1 MA
In vitro studies have established that Gag binds membranes more efficiently than the isolated MA protein (Zhou and Resh, 1996; Spearman et al., 1997; Hermida-Matsumoto and Resh, 1999; Ono and Freed, 1999; Paillart and Gottlinger, 1999; Bouamr et al., 2003). This led to the hypothesis that the myr group is exposed in HIV-1 Gag and sequestered in the MA protein, which has come to be known the “myristyl switch mechanism” (Spearman et al., 1997; Hermida-Matsumoto and Resh, 1999; Ono and Freed, 1999; Bouamr et al., 2003). However, pioneering NMR structural studies by the Summers lab confirmed that the myr group can adopt sequestered [myr(s)] and exposed [myr(e)] conformations even in the isolated MA protein (Tang et al., 2004). Equilibrium data revealed that while myr(+)MA resides in monomer–trimer equilibrium, the myr(−)MA protein maintains the monomeric character in solution under all conditions (Tang et al., 2004). In addition, exposure of the myr group is coupled with protein trimerization (Tang et al., 2004). The myr switch was found to be sensitive to protein concentration, which led to the proposition of the “entropic switch mechanism” (Tang et al., 2004). Additionally, inclusion of the CA domain increases exposure of the myr group, which indicates that exposure of the myr group is dependent on Gag multimerization (Tang et al., 2004). Most recently, we have shown that myr exposure is also modulated by pH (Fledderman et al., 2010). Since these factors (protein concentration, multimerization, and pH) are intrinsically confined to the MA protein, we call them the “internal modulators.”
Gag Multimerization, a Trigger for myr Exposure and a Requirement for Efficient Membrane Association
As mentioned above, previous studies revealed that exposure of the myr group is coupled with protein trimerization and is enhanced by factors that promote protein self-association, such as increasing protein concentration or inclusion of the CA domain (Tang et al., 2004). HIV-1 Gag multimerization is directly linked to efficient membrane binding and is required for particle formation and viral infectivity (Burniston et al., 1999; Ono et al., 2000a; Lindwasser and Resh, 2001; Li et al., 2007). The oligomerization properties of HIV-1 Gag, MA, and MACA constructs have been extensively studied (Morikawa et al., 1998, 2000; Tang et al., 2004; Alfadhli et al., 2007, 2009a,b; Dalton et al., 2007; Datta et al., 2007b; Li et al., 2007; Saad et al., 2007a,b; Dou et al., 2009; Hogue et al., 2009). The CA domain contains a dimerization site in the C terminus and is known to play a central role in mediating Gag–Gag interactions (Li et al., 2000; Joshi et al., 2006; Ganser-Pornillos et al., 2007, 2008; Wright et al., 2007; Briggs et al., 2009; Hogue et al., 2009; Chukkapalli and Ono, 2011). Other domains such as SP1 and NC also play important roles in Gag multimerization and assembly (Dawson and Yu, 1998; Burniston et al., 1999; Campbell and Rein, 1999; Cimarelli et al., 2000; Morikawa et al., 2000; Muriaux et al., 2001; Wright et al., 2007; Briggs et al., 2009). Altogether, despite the lack of a detailed structural mechanism these findings indicate that myr exposure is enhanced through synergistic intermolecular interactions between Gag subdomains that cooperatively promote assembly.
Myristyl Exposure is Modulated by pH
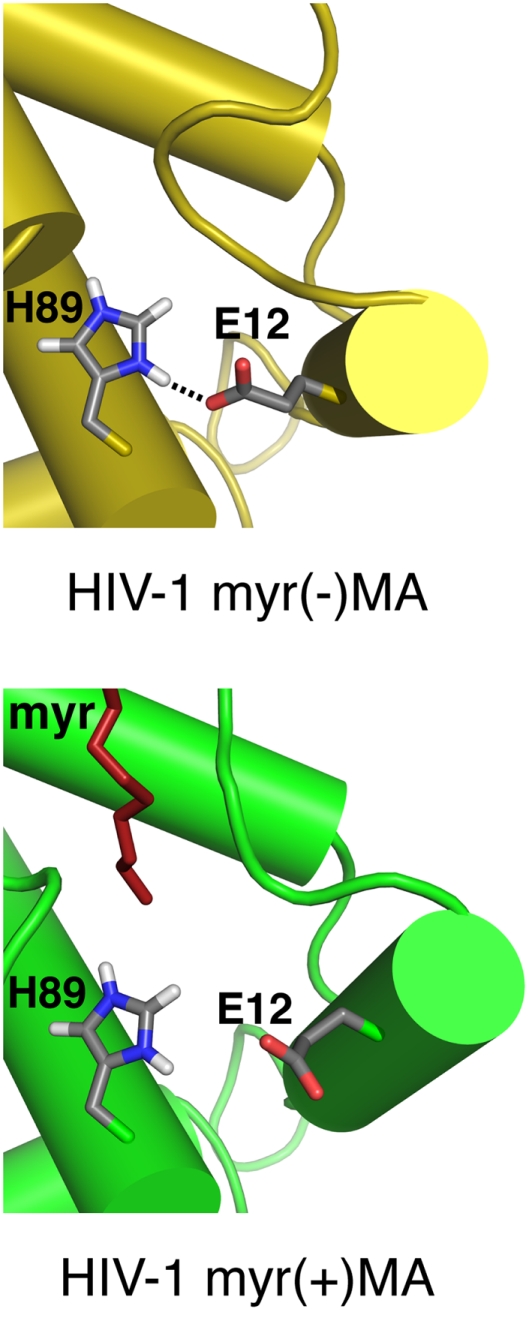
One of the key characteristics of the HIV-1 myr(+)MA is the presence of the myr group in two energetically stable states: exposed and sequestered (Tang et al., 2004). In solution, these two states are at equilibrium. However, it seems that subtle changes in sample conditions such as protein concentration can perturb the equilibrium state (Tang et al., 2004). Analysis of the HIV-1 myr(+)MA structure revealed that the imidazole ring of His-89, which is highly conserved among all strains of HIV-1, HIV-2, and simian immunodeficiency virus (SIV; Los Alamos National Laboratory, http://www.hiv.lanl.gov), forms a salt bridge with Glu-12(COO−) in HIV-1 myr(−)MA but not in the myr(+)MA protein. Histidine residues play important roles in biological activity as they are frequently included in the active sites of enzymes and contribute to protein stability and function. The intrinsic pKa of the histidine imidazole group, which varies depending on the degree of burial or exposure within proteins (Edgcomb and Murphy, 2002), typically lies between 6.0 and 7.0 (Liu et al., 1997; Edgcomb and Murphy, 2002). Previous studies have shown that mutation of His-89 led to targeting of Gag to intracellular compartments and severely reduced virus production (Freed et al., 1994). We have recently shown that deprotonation of the His-89 imidazole ring in myr(+)MA destabilizes the salt bridge formed between His-89(Hδ2) and Glu-12(COO−; Figure 2), leading to tight sequestration of the myr group and a shift in the equilibrium from trimer to monomer (Fledderman et al., 2010). By using NMR methods, we calculated the pKa of the histidine imidazole group in both myr(−)MA and myr(+)MA to be 6.5 (Fledderman et al., 2010). Furthermore, we have shown that oligomerization of a Gag-like construct containing MACA is also pH-dependent. Increasing the pH to 8 inhibited the formation of trimers for MA and MACA (Figure 3; Fledderman et al., 2010). These findings indicate that trimerization of Gag in vitro, although enhanced by CA, is solely driven by the MA domain and tightly regulated by pH. Based on these findings, we suggested that changes in pH might have an effect on the affinity of MA and Gag binding to membranes.
Figure 2.
The imidazole ring of His-89 forms a salt bridge with Glu-12(COO−) in HIV-1 myr(−)MA but not in myr(+)MA.
Figure 3.
Schematic representation showing multimerization events of Gag and MA proteins as a function of pH. Reprinted with permission from (Fledderman et al., 2010). Copyright {2010} American Chemical Society.
Earlier studies have also shown that mutation of His-89 and other residues in the vicinity led to targeting of Gag to intracellular compartments and severely reduced virus production (Freed et al., 1994). We recently found that the internal Gag puncta observed with H89G mutation colocalized extensively with CD63, indicating multivesicular bodies (MVB) localization (Fledderman et al., 2010). These results indicate that His-89 is essential for proper targeting of Gag to the PM and efficient particle formation in HeLa cells. Structural studies have shown that substitution of His-89 destabilized the tertiary structure, triggering myr exposure, and promoting high-order protein self-association (Fledderman et al., 2010).
Altogether, structural studies revealed that MA acts as a “pH sensor” and that, at least in vitro, myr exposure is regulated by pH. A potential role of pH variations in subcellular localization and targeting of Gag has not yet been established. Intracellular pH often fluctuates (6.3–7.8) in response to cell growth, development, and apoptosis (Schuldiner and Rozengurt, 1982; Moolenaar et al., 1983; Gottlieb et al., 1995). Alterations in the PM function can also be induced by cytopathic viruses including HIV-1 (Makutonina et al., 1996). A significant decrease in pH from 7.2 to as low as 6.0 was observed in cells infected by HIV-1 (Makutonina et al., 1996). Despite the in vitro evidence for pH effect on the myr switch, it has yet to be established whether manipulation of intracellular pH can alter kinetic pathways and sites or efficiency of Gag assembly.
MA Residues that Regulate the myr Switch Mechanism and/or Gag Assembly Sites
Mutagenesis studies in the past two decades have identified several residues in the MA domain that can alter Gag targeting and assembly (Bryant and Ratner, 1990; Freed et al., 1994; Zhou et al., 1994; Ono et al., 1997, 2000b; Ono and Freed, 1999, 2004; Paillart and Gottlinger, 1999; Joshi et al., 2009a; Chukkapalli et al., 2010). Several of these residues are located in the N-terminus of MA and are considered critical for proper membrane selection (Freed et al., 1994; Ono et al., 1997; Ono and Freed, 1999; Paillart and Gottlinger, 1999). More specifically, V7R, L8A, and L8I mutations give rise to a phenotype similar to that observed for the unmyristoylated protein without blocking myristoylation. These mutants, which display a hazy, non-punctate staining pattern typical of HIV-1 Gag mutants that are defective in membrane binding, cause a significant increase in cytosolic localization of Gag and severely inhibit virus assembly and release (Freed et al., 1994; Ono et al., 1997; Ono and Freed, 1999; Paillart and Gottlinger, 1999). In order to identify the structural basis for weak membrane association and diminished assembly of Gag V7R, L8A and L8I mutants, NMR, and analytical centrifugation studies have been performed on MA V7R, L8A, and L8I mutants (Saad et al., 2007b). NMR structures revealed that the backbone atoms of these mutants are very similar to the coordinates of the wild-type MA protein, indicating that single mutations in the N-terminal loop had no effect on the globular fold of the protein (Saad et al., 2007b). These studies, however, have shown that these mutations shut off the myr switch and lead to complete sequestration of the myr group (Saad et al., 2007b). The main differences between the structures are localized within the disordered loop formed by residues Gly-2–Ser-9. For example, in the V7R MA structure the side chain of Arg-7 makes a salt bridge with the side chain of Glu-52, and the methylene groups of Arg-7 contribute to the hydrophobic interactions with the methylene groups of the myristate (Saad et al., 2007b). For L8A, the less bulky methyl group of Ala-8 creates a better cavity for the myristate group, which packs closer to Trp-16. Thus, structural changes involve only residues located near the mutation site, which may have only average intrinsic conformational mobility. The sensitivity of the myristyl switch equilibrium to subtle molecular changes is evident in the conservative substitution of Leu-8 by Ile, which is sufficient to block myristate exposure under conditions in which exposure is normally highly favored (Saad et al., 2007b). Altogether, in vivo, biochemical, and structural studies confirm that even conserved structural changes in the N-terminus of MA can lead to major effects on the myr switch, which in turn abrogate Gag binding to the PM and inhibit virus assembly.
Interestingly, the phenotype observed for mutants in the N-terminus is very different from that observed for mutants in the basic domain (e.g., K30E and K32E) or between residues 85 and 89, which have been shown to associate specifically with intracellular compartments like MVBs (Freed et al., 1994; Ono et al., 1997, 2000b; Ono and Freed, 2004; Joshi et al., 2009a). While the structural effect of K30E and K32E mutations on the myr switch is not yet known, we predict that substitutions between residues 85 and 89 may directly perturb the myr switch equilibrium. As mentioned above, H89G substitution in myr(+)MA led to formation of protein aggregates as a result of exposure of the myr group (Fledderman et al., 2010). Thus, it appears that sequestration or lack of myr group favors cytosolic localization rather than MVB association. Although targeting of Gag K30E/K32E mutant to MVB compartments led to severely defective virus particle production in HeLa cells (Ono et al., 2000b; Ono and Freed, 2004), recent evidence revealed that it can still support highly efficient assembly and release in T cells (Joshi et al., 2009a). It was suggested that under some circumstances, late endosomal compartments serve as productive sites for HIV assembly in some cell types (Joshi et al., 2009a).
Role of Phosphatidylinositol-4,5-Bisphosphate [PI(4,5)P2] in HIV-1 Gag Targeting, Membrane Binding, and Virus Assembly
It is well established that assembly of retroviral Gag proteins occur predominantly on the PM (Hermida-Matsumoto and Resh, 2000; Jouvenet et al., 2006, 2008; Finzi et al., 2007; Li et al., 2007; Welsch et al., 2007; Gousset et al., 2008; Joshi et al., 2009a; Ono, 2009, 2010). Membrane selection appears to be critical for productive virus production. Rein and co-workers have shown that Gag constructs assemble in vitro into VLPs with diameters significantly smaller (25–30 nm) than those of authentic particles, whereas particles assembled in the presence of reticulocyte lysates exhibit normal diameters (∼100 nm; Campbell et al., 2001a). Further studies identified inositol pentaphosphate (IP5) as the active agent that promotes normal particle morphology and is capable of conferring resistance to disassembly in RNase, NaCl, and trypsin assays. Other inositol phosphates and IP derivatives, including inositol hexakisphosphate (IP6) and PI(4,5)P2 (Figure 4A) had similar influence on in vitro Gag assembly, and mutagenesis studies suggested that they promote HIV-1 particle assembly by interacting with the MA domain of Gag (Campbell et al., 2001a; Crist et al., 2009). A Gag protein lacking residues 16–99 in its MA domain lost the requirement for inositol phosphates in assembly in vitro. Further exploration of the binding properties of IP6 to a Gag construct lacking the myr group and the p6 domain [myr(−)GagΔp6] revealed that both MA and NC domains of Gag contribute to IP6 binding (Datta et al., 2007b). Equilibrium studies have shown that myr(−)GagΔp6 undergoes an equilibrium switch from monomer–dimer in the absence of IP6 to monomer–trimer upon binding to IP6. However, myr(−)GagΔp6 with a mutation in the dimer interface of CA is monomeric in the presence of IP6, suggesting that the “dimer interface” is essential for the trimeric interaction (Datta et al., 2007b). These findings supported by hydrodynamic and small-angle neutron scattering (SANS) data led to the suggestion that the participation of both MA and NC domains of Gag in IP6 interaction is evidence for a folded conformation of Gag in solution, with its ends close to each other in the three-dimensional space (Datta et al., 2007a).
Figure 4.
(A) Representation of the PI(4,5)P2 molecule (phosphate groups are shown as yellow balls), and (B) membrane binding model predicted from the structural studies; the exposed 1′-fatty acid chains (yellow) and myristyl groups (green) project from a highly basic surface (basic side chains shown in blue) in a manner expected to synergistically promote membrane binding. [(B) is reprinted from Saad et al., 2006; Copyright, Michael F. Summers, used with permission)].
Studies by Freed, Ono, and co-workers thereafter have provided compelling evidence that the ultimate localization of HIV-1 Gag on the PM is critically dependent on PI(4,5)P2 (Ono et al., 2004; Chukkapalli et al., 2008, 2010; Chukkapalli and Ono, 2011). PI(4,5)P2 is a membrane marker for proteins that are specifically targeted to the PM (Behnia and Munro, 2005; McLaughlin and Murray, 2005). Depletion of PI(4,5)P2 by overexpression of 5-phosphatase IV (5ptase IV) led to accumulation of Gag at the membranes of late endosomes and MVBs, inhibited HIV assembly, and severely reduced virus production (Ono et al., 2004). Likewise, overexpression of a constitutively active form of Arf6 (Arf6/Q67L), which induces the formation of PI(4,5)P2-enriched endosomal structures led to retargeting of Gag to these vesicles and severely reduced virus production. Sensitivity of Gag localization and assembly to PI(4,5)P2 manipulation was lost upon substitution of the MA domain with the N-terminus of Fyn kinase [Fyn(10)ΔMA/delNC], which is not affected by 5ptaseIV overexpression. Altogether, these findings supported the hypothesis that interactions between MA and PI(4,5)P2 are essential for proper Gag targeting and membrane binding.
NMR studies have been utilized to identify how MA interacts with PI(4,5)P2 (Saad et al., 2006). NMR chemical shift perturbation data obtained for the MA protein revealed that residues Arg-22, Lys-26, Lys-27, His-33, Glu-73, Leu-75, and Ser-77 were the most affected upon binding of soluble analogs of PI(4,5)P2 (Saad et al., 2006). These residues reside on a cleft formed by helices II and V, and a β hairpin (β-II-V cleft). Although these studies marked the first structural evidence for direct interactions between HIV-1 MA and a cellular constituent, the two most intriguing observations were the PI(4,5)P2 binding mode and the effect of binding on the myr switch. The three-dimensional structure of the MA-PI(4,5)P2 complex revealed that the acyl chain attached to the 2′-position of the glycerol packs within the β-II–V cleft against the side chains of Leu-21, Lys-27, Tyr-29, His-33, Trp-36, and Ser-77, while the phosphoinositide head group packs against Leu-21 and Lys-27, burying the 2′-fatty acid chain (Figure 4A). The 1′-acyl chain, however, is exposed to solvent with PI(4,5)P2 molecule adopting an “extended lipid” conformation. Binding of PI(4,5)P2 to MA induces a minor conformational change entailing a repositioning of helix I and destabilization of the hydrophobic cavity, leading to myr exposure. These results indicate that PI(4,5)P2 acts as both a trigger of the myr switch and as a membrane anchor and suggest a potential mechanism for targeting Gag to membrane rafts (Figure 4B; Saad et al., 2006). Interestingly, the proposed membrane binding mode is very similar to the “extended lipid” conformation predicted in the phospholipid–cytochrome c model (Kinnunen et al., 1994; Rytömaa and Kinnunen, 1995; Tuominen et al., 2002).
Of particular note, our studies represent the first evidence that PI(4,5)P2 can adopt an extended conformation. The hydrophobic cleft that interacts with the 2′-acyl chain of PI(4,5)P2 analogs with short chains (four- and eight-carbon atoms) is also capable of accommodating longer fatty acids without inducing any major changes in the protein structure. A model of the trimeric PI(4,5)P2:myr(+)MA–membrane complex constructed by using 18- and 20-carbon 1′- and 2′-acyl chains is shown in Figure 4B. Native PI(4,5)P2 forms micelles in aqueous solution, making it virtually impossible to study its interaction with MA by NMR methods (Janmey et al., 1987). Difficulties in characterizing the binding of native PI(4,5)P2 with the MA protein have only been overcome upon encapsulating the protein in reverse micelles (Valentine et al., 2010). Titration of MA with native PI(4,5)P2 in reverse micelles has led to significant chemical shift changes in a subset of amide signals. Mapping of the chemical shift changes on the MA surface indicates a binding mode similar to that of the soluble analogs with short chains.
Binding of native PI(4,5)P2 to myr(−)GagΔp6 has been characterized by mass spectrometric protein footprinting (Shkriabai et al., 2006). NHS-biotin modification approach was used to identify lysine residues in Gag that are exposed to solvent or those that are protected from biotinylation due to direct Gag-PI(4,5)P2 contacts. Data revealed that among 21 surface Lys residues readily modified in free Gag, only MA K30 and K32 are protected in the Gag-PI(4,5)P2 complex. Although NMR and mass spectrometry methods provide evidence for direct binding of native PI(4,5)P2 to MA, the exact binding interface, and a high-resolution structural model have yet to be determined. Based on the NMR data obtained for MA in reverse micelles, our best guess is that the 20-carbon 2′-acyl chain of PI(4,5)P2 is sequestered by the protein.
Confocal microscopy data revealed that Gag molecules assemble at punctate sites on the PM (Hermida-Matsumoto and Resh, 2000). What are these punctate sites and why does Gag select these sites? To address these questions, we recall a few previous observations that could support the structural studies and the MA–PI(4,5)P2 binding mode as well as the overall association with the PM. First, there is evidence that punctate sites comprise lipid raft microdomains (Aloia et al., 1993; Nguyen and Hildreth, 2000; Campbell et al., 2001b; Ono and Freed, 2001, 2005; Ding et al., 2003; Holm et al., 2003; Waheed and Freed, 2009). Lipid rafts are typically liquid-ordered membrane structures that contain elevated levels of cholesterol and sphingolipids with saturated fatty acids (Brown and London, 1997, 1998; Melkonian et al., 1999; Zacharias et al., 2002; Ono and Freed, 2005; Waheed and Freed, 2009). Proteins that associate with lipid rafts generally contain two saturated acyl chains or are anchored by adaptor molecules that contain two saturated chains (for example, glycosylphosphatidylinositol-anchored proteins; Brown and London, 2000; Ono and Freed, 2005). Because PI(4,5)P2 contains stearate (18-carbon saturated acyl chain) at the 1′-position and arachidonate (20-carbon acyl chain with four non-conjugated double bonds) at the 2′-position, sequestration of the 2′-chain is likely to reduce the affinity of PI(4,5)P2 for fluid regions of the membrane and promote its association with rafts. Since previous studies also suggest that PI(4,5)P2 molecules are homogeneously dispersed within the PM of quiescent cells but they can colocalize with lipid rafts upon stimulation (Golub and Caroni, 2005), sequestration of the 2′-acyl chain by MA may suggest a potential mechanism for the lateral targeting of PI(4,5)P2:Gag complexes to lipid raft microdomains (Saad et al., 2006). Second, the proposed mechanism for Gag binding to membrane is supported by the finding that substitution of the saturated myr group of HIV-1 Gag by unsaturated lipids reduces the affinity of Gag for rafts and inhibits particle assembly (Lindwasser and Resh, 2002).
Gag-PI(4,5)P2 Interactions in Other Retroviruses
In the past few years, efforts have been focused on whether Gag targeting and virus assembly in other retroviruses are also dependent on PI(4,5)P2. Subsequent to our studies on HIV-1, we discovered that HIV-2 MA binds specifically to PI(4,5)P2 in a manner identical to that of HIV-1 (Saad et al., 2008). However, in HIV-2 MA the myr switch is less sensitive to PI(4,5)P2 binding as the myr group is found to be sequestered in the complex. The subcellular site of HIV-2 Gag assembly is also regulated by PI(4,5)P2. Like HIV-1, overexpression of 5ptaseIV or Arf6/Q67L also significantly reduced levels of released HIV-2 virions, indicating that the production of HIV-2 particles is inhibited by PI(4,5)P2 perturbation (Saad et al., 2008).
In most retroviruses, membrane anchoring is strongly dependent on the myr group. However, Gag proteins of some retroviruses, notably alpha retroviruses [the prototype of which is Rous sarcoma virus (RSV)] and the lentivirus equine infectious anemia virus (EIAV), lack myr modification, and Gag targeting and binding to the PM is mainly mediated by electrostatic interactions (Erdie and Wills, 1990; Provitera et al., 2000; Dalton et al., 2005). For example, the association of RSV MA protein with liposomes of defined composition is electrostatic in nature and is dependent on the presence of a biologically relevant concentration of negatively charged lipids like phosphatidylserine (Dalton et al., 2005). In addition, MA-PI(4,5)P2 interactions have been detected in Moloney murine leukemia virus (MMLV) and Mason-Pfizer monkey virus (MPMV; Stansell et al., 2007; Chan et al., 2008; Chen et al., 2008; Hamard-Peron et al., 2010). As has been observed for HIV-1 and HIV-2 MA, NMR-based studies have shown that a soluble analog of PI(4,5)P2 interacts directly with EIAV MA, suggesting a potential role of this lipid in EIAV Gag assembly (Chen et al., 2008). However, a most recent study investigating the role of PI(4,5)P2 in assembly of EIAV show that Gag was detected on the PM and in compartments enriched in phosphatidylinositol 3,5-biphosphate [PI(3,5)P2; Fernandes et al., 2011]. NMR studies have shown that EIAV MA binds to phosphatidylinositol 3-phosphate [PI(3)P] with a higher affinity than that of PI(4,5)P2 (Fernandes et al., 2011). Treatment of cells with YM201636, a kinase inhibitor that blocks production of PI(3,5)P2 from PI(3)P led to Gag colocalization with aberrant compartments and inhibited VLP release (Fernandes et al., 2011). In contrast to HIV-1, release of EIAV VLPs was not significantly diminished by coexpression with 5ptase IV. Taken together, these results indicate that membrane targeting in EIAV proceeds via a mechanism that is probably distinct from that in HIV-1 or HIV-2. On the other hand, when compared to HIV-1 and HIV-2 recent studies have shown that human T-lymphotropic virus type (HTLV-1) and RSV Gag proteins have either lower or no requirement for PI(4,5)P2 for membrane association (Chan et al., 2011; Inlora et al., 2011).
Role of Gag in Envelope (Env) Incorporation
Retroviral Env protein is a glycosylated polyprotein synthesized as intact gp160 precursor that is subsequently cleaved by cellular proteases to form a complex of a surface unit (gp120) and transmembrane domain (gp41). During virus assembly, the gp120/gp41 complex is incorporated in the lipid bilayer of the nascent virions. Despite the progress made in understanding the structural details of Env and its components as well as the trafficking pathway of Env polyprotein, the mechanism by which Env is incorporated into virus particles is not completely understood. The Gag protein has long been implicated in Env incorporation based on the proposed interaction between the MA domain and the cytoplasmic (CT) domain of gp41 (Yu et al., 1992; Dorfman et al., 1994; Freed and Martin, 1995, 1996; Mammano et al., 1995). However, importance of this interaction is controversial since in many cell lines (HeLa, 293T, CV-1, and COS), deletion of the CT domain has virtually no effect on Env incorporation (Wilk et al., 1992; Freed and Martin, 1995, 1996; Murakami and Freed, 2000). Checkley et al. (2011) have recently reviewed the current understanding of HIV-1 Env trafficking and incorporation into virions. Several models have been proposed to explain Env incorporation into virus particles (Checkley et al., 2011):
-
(i)
“Passive” model: Env is expressed on the host PM and subsequently incorporated into budding virions. Support of this model stems from the observation that host membrane proteins are abundantly incorporated into retrovirus particles (Lusso et al., 1990; Arthur et al., 1992; Ott, 2008).
-
(ii)
Direct Gag–Env interaction: Genetic data revealed that the MA domain of Gag is required for Env incorporation (Yu et al., 1992; Dorfman et al., 1994; Freed and Martin, 1995, 1996). Mutations in or deletion of the MA domain of Gag decreased Env incorporation (Freed and Martin, 1995, 1996; Mammano et al., 1995; Reil et al., 1998; Brandano and Stevenson, 2012), while deletion of the gp41 CT domain reversed the Env incorporation defect dictated by MA mutations (Freed and Martin, 1995, 1996; Mammano et al., 1995), or MA deletion (Reil et al., 1998). Altogether, these studies suggest that MA is required for full-length Env incorporation whereas Env–MA interaction is apparently not required for incorporation of truncated Env. The requirement of the MA and CT domains for Gag–gp41 interactions appears to be supported by a few studies albeit evidence for direct interactions is still weak. For example, previous studies have shown that HIV-1 Env is localized to the basolateral surface of polarized epithelial cells (Owens and Compans, 1989). Gag has been shown to bud in a non-polarized manner in these cells when Env was not expressed. Budding, however, was restricted to the basolateral surface upon coexpression of Gag and Env (Owens et al., 1991). Mutations in MA or CT truncation undermined the ability of Env to direct basolateral budding of Gag (Lodge et al., 1994). Although these studies suggest a cross-talk between the MA and CT domains, they do not necessarily indicate a direct interaction (Checkley et al., 2011).
-
(iii)
Gag–Env cotargeting: As mentioned above, the absence of compelling evidence for direct interactions between Gag and gp41 may suggest that a cellular constituent like a protein or membrane acts as a bridge or a docking platform for the Gag–Env complex. A role of a cellular factor(s) in Env incorporation is perhaps supported by the results showing that the requirement of CT domain is strongly dependent on the cell type (permissive cells like HeLa, COS, and 293T; Wilk et al., 1992; Akari et al., 2000; Murakami and Freed, 2000). In non-permissive cell lines (e.g., T cells and macrophages), removal of gp41 CT led to ∼10-fold decrease in Env incorporation (Murakami and Freed, 2000). The identity of cellular factors required for Env incorporation in permissive and non-permissive cells have yet to be determined.
-
(iv)
Indirect Gag–Env interaction: Since it is not clear whether there is a direct interaction between the MA domain of Gag and gp41 CT, it is conceivable that the interaction is mediated by cellular protein(s). Among the gp41 CT-interacting proteins are the adaptor protein complexes AP-1 and AP-2. The AP complexes are heterotetramers that mediate both the recruitment of clathrin to membranes and the recognition of sorting signals within the cytosolic tails of transmembrane cargo molecules (Robinson and Bonifacino, 2001; Boehm and Bonifacino, 2002; Nakatsu and Ohno, 2003). While AP-1 has been shown to regulate the subcellular localization of Env via binding to a dileucine motif in gp41 CT (Berlioz-Torrent et al., 1999; Byland et al., 2007; Wyss et al., 2011), AP-2 has been shown to bind to the gp41 CT domain and drive clathrin-mediated endocytosis of Env from the PM (Ohno et al., 1997; Boge et al., 1998). Interestingly, AP-1 and AP-2 has been shown to bind to the MA domain of Gag (see below; Batonick et al., 2005; Camus et al., 2007). Other proteins like the 47-kDa tail interacting protein (TIP47) and calmodulin (CaM) have been proposed to play some role in Env incorporation (discussed in more details below).
Potential Role of MA in Genomic RNA Packaging
Although MA’s role in HIV-1 Gag targeting and assembly on the PM is the best understood, there is mounting evidence that it also plays a role in retroviral genome packaging. A recent review has shed light on the current understanding of the functional role of MA in genome packaging (Parent and Gudleski, 2011). Since the MA protein is usually associated to the lipid bilayer and on the opposite end from the NC domain, the biological importance for the proposed MA–RNA interactions is controversial and has been under scrutiny for a long time. Interest in assessing the functional role of MA in genome packaging stems from earlier studies on the interactions between HIV-1 MA and other retroviruses (e.g., RSV and bovine leukemia virus) with single-stranded and double-stranded DNA and RNA (Leis et al., 1978; Darlix and Spahr, 1982; Méric and Spahr, 1986; Steeg and Vogt, 1990; Katoh et al., 1991; Ott et al., 2005; Alfadhli et al., 2009b; Cai et al., 2010; Ramalingam et al., 2011). It has been shown that MA binds directly to an RNA molecule that is highly homologous to a fragment of the pol domain (Purohit et al., 2001). The interaction interface of MA is confined to the basic domain; substitutions in the basic residues led to weakened affinity to RNA in vitro and a delay in virus replication in vivo (Purohit et al., 2001). However, it has yet to be established whether MA binds directly to the packaging signal (ψ) of the RNA genome.
Most recent studies by Akira Ono and co-workers revealed that the highly basic region (HBR) in the N-terminus of MA not only contributes to binding of PI(4,5)P2 to Gag, but is also capable of binding to RNA (Chukkapalli et al., 2010). These results are consistent with in vitro and NMR studies (Purohit et al., 2001; Hearps et al., 2008; Alfadhli et al., 2011). Interestingly, binding of RNA to HBR was found to inhibit association of Gag to membrane liposomes in the absence of PI(4,5)P2 (Chukkapalli et al., 2010). As a consequence, RNA was considered as a negative regulator of Gag-membrane binding. Thus, the interplay between Gag, PI(4,5)P2, and RNA appears to be an important aspect of Gag assembly that has to be examined further.
The Missing Link: HIV-1 Gag en route to the Plasma Membrane?
Great progress has been made in defining both the viral and cellular determinants of HIV-1 assembly and release (Chu et al., 2010). However, the trafficking pathway used by Gag to reach its assembly sites in the infected cell is poorly understood. There is mounting evidence that HIV-1 Gag interacts with several cellular proteins during the replication cycle. These include CaM (Radding et al., 2000), the human adaptor protein complexes AP-1, AP-2, and AP-3 (Batonick et al., 2005; Dong et al., 2005; Camus et al., 2007), TIP47 (Lopez-Vergès et al., 2006), Golgi-localized gammaear containing Arf-binding protein (GGA; Joshi et al., 2008, 2009b), ADP ribosylation factor (Arf; Joshi et al., 2008), the suppressor of cytokine signaling 1 (SOCS1; Ryo et al., 2008; Nishi et al., 2009), a lymphoid specific Src kinase (Lck; Strasner et al., 2008), N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE; Joshi et al., 2011), Filamin A (Cooper et al., 2011), vacuolar protein sorting-associated protein 18 (Vps18; Tomita et al., 2011), Mon2 (Tomita et al., 2011), and Lyric (Engeland et al., 2011). The majority of these proteins have been implicated in retroviral assembly. Since we are focusing on the MA protein, we will summarize the current understanding of the proposed interactions between the MA domain of Gag and a small selection of the proteins described above including CaM, AP-3, TIP47, SOCS1, and Lyric.
Potential Role of CaM in HIV Replication
CaM is a ubiquitous calcium-binding protein expressed in all eukaryotic cells. It binds to and regulates different protein targets, thereby affecting many different cellular functions (Osawa et al., 1999; Chin and Means, 2000; Hoeflich and Ikura, 2002; Vetter and Leclerc, 2003; Yamniuk and Vogel, 2004; Ishida and Vogel, 2006). It binds more than 100 proteins “specifically” (Rogers and Strehler, 1996) and is found in different subcellular locations, including the cytoplasm, within organelles, or associated with the plasma or organelle membranes (Osawa et al., 1999; Chin and Means, 2000; Hoeflich and Ikura, 2002; Vetter and Leclerc, 2003; Yamniuk and Vogel, 2004; Ishida and Vogel, 2006). CaM is perhaps one of the most characterized cellular proteins and its interactions with proteins have been extensively studied (Chattopadhyaya et al., 1992; Finn et al., 1995; Chin and Means, 2000; Fallon and Quiocho, 2003). Binding of calcium to CaM triggers a major conformational change and helical rearrangement, enabling it to bind to specific proteins for a specific response.
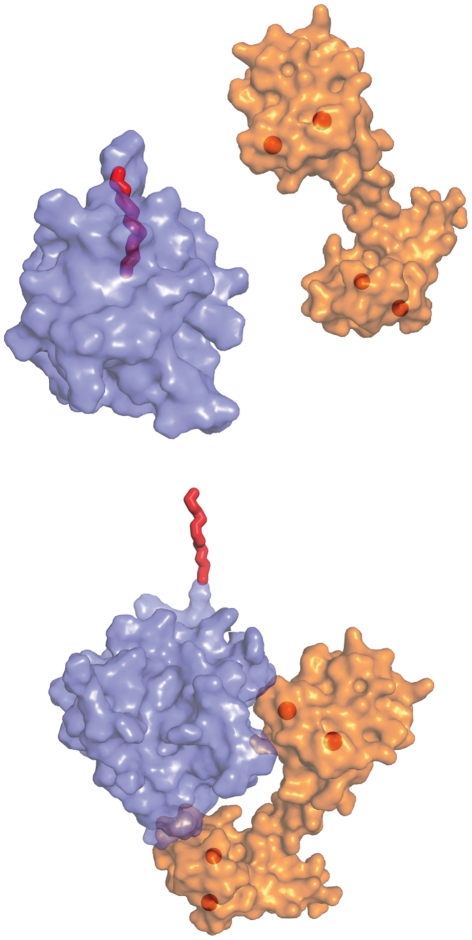
Subcellular distribution of CaM in HIV-infected cells has been shown to be distinct from that observed in uninfected cells (Radding et al., 1996). In vivo and in vitro studies revealed that CaM interacts with HIV-1 Gag, Nef, and gp160 proteins (Towler et al., 1988; Srinivas et al., 1993; Radding et al., 2000; Hayashi et al., 2002; Matsubara et al., 2005). Pioneering studies by Hunter and co-workers have shown that HIV-1 Gag can interact with CaM in a calcium-dependent manner (Radding et al., 2000). Gag and CaM colocalize in a diffuse pattern in the cytoplasm. Investigation of the underlying mechanism of Gag–CaM interactions has gained some momentum recently. Our lab (Ghanam et al., 2010) and others (Chow et al., 2010) have shown that CaM binds directly to the MA protein with 1:1 stoichiometry in a Ca2+-dependent manner. We have also established that: (i) CaM-binding to myr(+)MA induces a structural change that triggers myr exposure (Figure 5), (ii) the myr group is not involved in CaM-binding, and (iii) CaM–MA interaction is hydrophobic and entropically driven, while electrostatic interactions appear to be inconsequential (Ghanam et al., 2010).
Figure 5.
CaM-binding to MA triggers myr exposure. Surface representation of CaM (orange) and myr(+)MA (slate) with the myr group (red) sequestered and exposed in the free and bound states, respectively. Spheres represent Ca2+ ions.
Radding et al. (2000) have shown that peptides from the N-terminus of MA bind to CaM with variable affinities and in different modes. These studies have prompted others to conduct SAXS experiments to elucidate the global features of CaM bound to short peptides derived from the MA protein (Izumi et al., 2008). SAXS studies conducted on CaM complexed with full-length MA shed light on the global features of the complex (Chow et al., 2010). However, SAXS methods cannot provide details on the complex interface and cannot identify specific residues that are important for the formation and stabilization of the complex. We have utilized NMR, biochemical, and biophysical methods to elucidate the precise molecular mechanism of MA–CaM interactions (Samal et al., 2011). In this work, we have (i) identified the interaction interface of CaM bound to short peptides derived from the MA protein, (ii) shown that MA peptides bind to CaM with variable affinities and diverse modes, and (iii) identified the minimal CaM-binding domain of MA as residues 8–43 (MA-8–43).
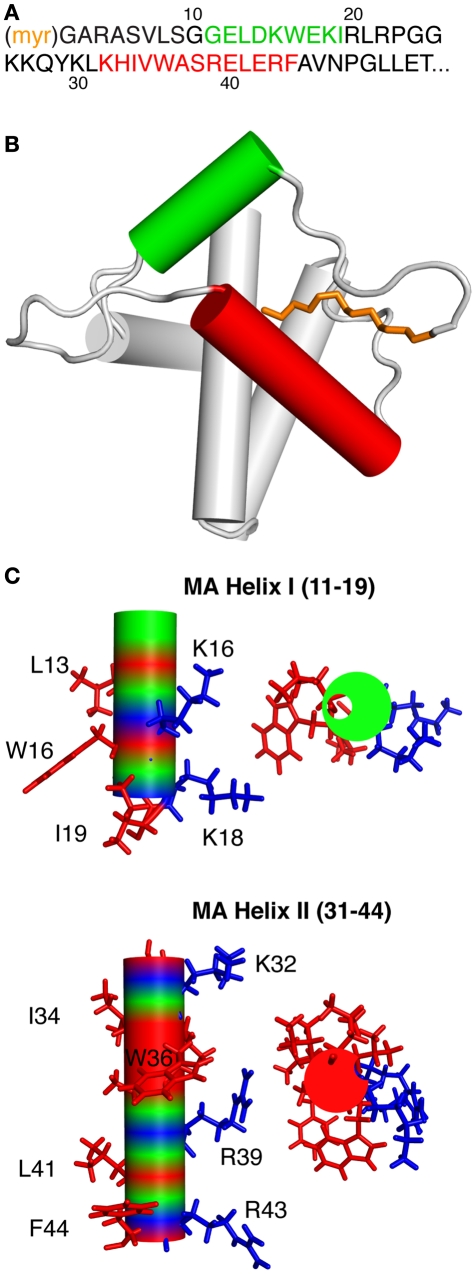
A typical CaM-binding peptide is 15–30 residues long with amphipathic character and a propensity to form an α-helix (Ishida and Vogel, 2006), features that helices I and II of MA possess (Figure 6). The 36-residue long MA-8–43 peptide is perhaps one of the largest CaM-binding motifs to be discovered. The fact that MA-8–43 forms two helices is another striking feature of this peptide since almost all known CaM-target proteins usually possess a domain that forms only one α-helix. In many of the classical CaM-binding targets, hydrophobic residues usually occupy conserved positions at 1–5–10 or 1–8–14, which point to one face of the helix (Ishida and Vogel, 2006). Although these patterns are typically found in many CaM-binding proteins, numerous unclassified examples were also identified (Ishida and Vogel, 2006). By using a web-based tool to identify a sequence pattern with potential CaM-binding sites (Yap et al., 2000), the region between Arg22 and Glu40 (Figure 6) scored the highest. Scores are based on several criteria including hydropathy, α-helical propensity, residue weight and charge, hydrophobic residue content, helical class, and occurrence of particular residues. Arg22–Glu40 represents the majority of helix II. However, this peptide does not contain a motif that matches any of the known patterns of CaM-binding sites. Thus, MA-8–43 is considered a novel and unclassified CaM-binding sequence that is substantially different (in length and composition) from all known CaM-binding domains. These findings confirm the enormous versatility of CaM-target complex formation and suggest a novel CaM-binding mode that requires engagement of hydrophobic residues from both helices I and II of MA (Ghanam et al., 2010; Samal et al., 2011).
Figure 6.
(A) Amino acid sequence of the N-terminus of MA. (B) Cartoon representation of the NMR structure (2H3I) of myr(+)MA highlighting helices I and II in green and red, respectively. Myr group is shown in orange. (C) Amphipathic character of helices I and II with hydrophobic and basic residues shown in red and blue, respectively, “A modified version of this figure was published in the Journal of Biological Chemistry. Samal et al., 2011; The American Society for Biochemistry and Molecular Biology.”
The exact functional role of CaM in HIV replication is not known. The observation that CaM colocalized with Gag in the cytoplasm of the infected cell may suggest a role of CaM in the (pre)assembly stage. Could it be that CaM facilitates Gag transport to the PM? This is a reasonable hypothesis given the fact that one of the major functions of CaM is shuttling proteins to the PM. However, this hypothesis has yet to be tested. One interesting characteristic of CaM is its preferential binding to myristoylated proteins. Membrane association and dissociation of myristoylated proteins are often regulated by intracellular Ca2+ signaling through a “Ca2+-myr switch” such as recoverin (Tanaka et al., 1995), neurocalcin (Faurobert et al., 1996), and S-modulin (Matsuda et al., 1998). As a consequence, intracellular localization from the cytosol to the membrane is dependent on Ca2+ binding (Braunewell and Gundelfinger, 1999). Other myristoylated proteins including myristoylated alanine-rich C kinase substrate (MARCKS), brain-specific protein kinase C substrate (CAP-23/NAP-22) and HIV-1 Nef interact with CaM via the myr group to facilitate their intracellular localization and membrane targeting (Matsubara et al., 2003, 2004, 2005). Our structural studies were the first of their kind to be conducted on binding of CaM to a native myristoylated protein and the first to show the subsequent exposure of the myr group (Figure 5). The finding that the myr group is not “grabbed” by CaM may suggest a novel functional role of CaM in the trafficking and/or targeting of Gag to the PM for assembly.
The functional role of CaM in the HIV replication cycle could be rather complex since interactions with gp160 and Nef have also been detected. Expression of the HIV-1 gp160 protein has led to a marked increase in CaM distribution (Radding et al., 1996). The CaM-binding region in gp160 was identified to be a helical peptide in gp41. Deletion of this region led to diminished virus infectivity (Srinivas et al., 1993; Radding et al., 1996). On the other hand, mutations in the cytoplasmic domain of gp41 eliminated co-immunoprecipitation of Env with calmodulin but had no significant effect on viral production or Env expression (Micoli et al., 2006). Confocal microscopy data confirmed the elevated level of CaM distribution and also showed that CaM colocalizes with the gp160 protein (Radding et al., 1996). Thus, the interplay between gp160, CaM, and Gag could be important in HIV replication. Although it is reasonable to hypothesize that HIV–CaM protein interactions may lead to disruption of CaM-dependent cell-signaling pathways and contribute to immune dysfunction during HIV pathogenesis, the exact functional role of CaM in HIV replication has yet to be examined.
Role of the Adaptor Protein Complexes in Gag Trafficking
To understand the critical steps of the molecular mechanism of HIV assembly, it is vital to identify the cellular machinery, its components, and how these components interact with each other and/or the HIV proteins. Although it is widely accepted that in most cell types HIV-1 Gag budding and assembly occur on the PM (Hermida-Matsumoto and Resh, 2000; Jouvenet et al., 2006, 2008; Finzi et al., 2007; Li et al., 2007; Welsch et al., 2007; Gousset et al., 2008; Joshi et al., 2009a; Ono, 2009, 2010), several research groups have shown that late endosomal compartments can serve as major assembly sites (Nguyen et al., 2003; Nydegger et al., 2003; Pelchen-Matthews et al., 2003; Sherer et al., 2003; Grigorov et al., 2006). To further explore the assembly pathway in late endosomal compartments, efforts focused on cellular proteins and protein complexes that typically play roles in protein sorting to and from the endosomal pathway. Several of these factors, which are implicated in Gag trafficking and assembly have been discovered. These include AP-1, AP-2, and AP-3 (Batonick et al., 2005; Dong et al., 2005; Camus et al., 2007). AP-3 appears to be involved in the sorting of a subset of transmembrane proteins targeted to lysosomes and lysosome-related organelles. Studies by Spearman et al. (1997) indicate that HIV-1 Gag binds to the δ subunit of the AP-3 complex (Dong et al., 2005). The AP-3 binding domain in Gag is suggested to be within the N-terminal region of MA (helix I; Figure 6). Gag constructs lacking the MA domain failed to bind and recruit the AP-3 complex. In addition, particle formation by WT Gag was severely diminished by a dominant negative AP-3 δ fragment and by siRNA depletion of AP-3, indicating that interaction between Gag and AP-3 is essential for normal productive trafficking pathway of Gag in the cell (Dong et al., 2005). The trafficking of Gag to CD63-positive intracellular compartments was also eliminated when Gag–AP-3 interaction was disrupted.
In addition to binding to Gag, the cytoplasmic tail of gp41 contains a tyrosine-based signal that has been shown to be active in promoting clathrin-mediated endocytosis through interactions with the μ subunit of AP-2 (Boge et al., 1998; Berlioz-Torrent et al., 1999). This observation has led to the hypothesis that the Env protein is internalized to sorting/recycling endosome compartment where it becomes associated with Gag (Dong et al., 2005). The dual role of AP complexes in Env and Gag trafficking pathways and compelling evidence for direct physical interactions between MA and AP-3 have yet to be determined.
TIP47: Role in Gag Localization and Env Incorporation
TIP47 is an essential protein for endosome-to-TGN retrograde transport of mannose-6-phosphate receptors, has been shown to form a ternary complex by bridging between the Env and Gag proteins (Lopez-Vergès et al., 2006). The interaction between Gag and TIP47 is mediated by the MA domain. Mutations in the MA domain of Gag that abrogate TIP47 binding were found to inhibit Env incorporation, Env–Gag colocalization, and virus infectivity (Lopez-Vergès et al., 2006). Silencing of TIP47 was found to impair Env incorporation and infectivity and abolished co-immunoprecipitation of Gag and Env. Based on these findings, it was suggested that TIP47 acts as a connector between Gag and Env during assembly (Lopez-Vergès et al., 2006). Interestingly, several point mutations and deletions within the first 23 residues of MA resulted in loss of TIP47 binding and thus Env incorporation. Among these mutations were W16A and E17A. Mutations in helix I of MA (L13, W16, E17, and K18) have been shown to block Env incorporation (Dorfman et al., 1994; Freed and Martin, 1996; Davis et al., 2006), which led to the suggestion that MA interacts directly with the Env protein. Altogether, studies on TIP47 suggest a functional role of this protein in Env incorporation and colocalization of Gag and Env. However, role of TIP47 in HIV biology remains unclear as studies conducted in other laboratories have failed to support any role of TIP47 in particle production or virus replication (Checkley et al., 2011).
Gag–SOCS1 Interactions
SOCS1, a member of negative regulators of cytokine signaling (Yoshimura et al., 2007) has been recently shown to be an inducible host-factor during HIV-1 infection and to regulate the late stages of the virus replication pathway (Ryo et al., 2008; Nishi et al., 2009). SOCS1 interacts with the MA and NC regions of Gag to facilitate its intracellular trafficking and stability. The depletion of SOCS1 caused a severe reduction in Gag trafficking and assembly (Ryo et al., 2008). It has been also shown that SOCS1 is involved in regulating Gag trafficking via the microtubule-dependent cellular machinery (Nishi et al., 2009).
Genetically, SOCS family members have a central SH2 domain and a conserved SOCS box (Yoshimura et al., 2007). Structure–function analyses revealed that the SH2 domain is critical for efficient binding to its substrates (Narazaki et al., 1998). Ryo et al. (2008) reinforced these findings by showing that a mutant lacking the SH2 domain does not bind HIV-1 Gag, whereas an N-terminus or a SOCS1 box deletion did not affect binding to Gag in 293T cells. Thus, elucidation of the structural requirements of Gag–SOCS1 interactions will provide insights into the functional role of SOCS1 in Gag trafficking and stability throughout the HIV replication cycle.
Lyric–Gag Interactions
Lyric is a ubiquitously expressed 64 kDa protein with several isoforms due to differential splicing, which differ in their intracellular localization, and is normally involved in various signaling pathways (Thirkettle et al., 2009). Previous studies have shown that Lyric is an HIV-inducible gene that is involved in various signaling pathways and acts in a positive-feedback loop promoting HIV replication (Thirkettle et al., 2009). A recent study has identified Lyric as an HIV-1 Gag-interacting protein. Endogenous Lyric has been shown to be incorporated in HIV-1 virions (Engeland et al., 2011). Both MA and NC appear to be required for Lyric binding. However, deleting NC and fusing MA with a GCN4 motif to drive trimerization led to the same binding affinity to Lyric. Thus, it is possible that Lyric binding to Gag is mostly dependent on the MA protein but also requires multimerization. Gag-interacting region in the Lyric protein has been mapped to a region spanning residues 101–289. Expression of this region increased Gag expression and viral infectivity. Interestingly, the observation of Gag–Lyric interactions in MMLV and EIAV (Engeland et al., 2011) suggests that Lyric’s role in the replication cycle is probably a conserved feature among retroviruses. Despite the experimental evidence for Gag–Lyric interaction, the precise functional role for Lyric has yet to be established.
Summary
Several cellular factors have been proposed to interact directly with the MA domain to facilitate Gag trafficking and assembly. However, molecular interactions have yet to be validated and characterized in most cases. Despite the tremendous progress achieved in understanding the structural properties of Gag and its domains, high-resolution structures of their complexes with cellular partners are very limited. The discovery of new cellular factors that bind to Gag or Gag components (Brass et al., 2008; Jäger et al., 2011) should pave the way for further structural investigation and hopefully will lead to identification of novel mechanisms and/or pathways critical for virus assembly. Such knowledge should facilitate the development of better approaches for the treatment of HIV.
Nomenclature
MA mutants are named based on the myr group being designated as residue 1. In contrast, studies that performed the characterization of the mutants discussed here considered the N-terminal Gly of the myristoylated protein as residue 1 (Freed et al., 1994; Ono and Freed, 1999, 2004; Ono et al., 2000a; Joshi et al., 2009a; Chukkapalli et al., 2010). Thus, the mutant names are offset by one residue.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Thanks to our laboratory members for the invaluable comments on the manuscript. Work from our laboratory described in this article is supported by the National Institutes of Health (1R01AI087101) and intramural funding from the Center for AIDS Research and Comprehensive Cancer Center at the University of Alabama at Birmingham.
References
- Adamson C. S., Freed E. O. (2007). Human immunodeficiency virus type 1 assembly, release and maturation. Adv. Pharmacol. 55, 347–387 10.1016/S1054-3589(07)55010-6 [DOI] [PubMed] [Google Scholar]
- Akari H., Fukumori T., Adachi A. (2000). Cell-dependent requirement of human immunodeficiency virus type 1 gp41 cytoplasmic tail for Env incorporation into virions. J. Virol. 74, 4891–4893 10.1128/JVI.74.6.2907-2912.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfadhli A., Barklis R. L., Barklis E. (2009a). HIV-1 matrix organizes as a hexamer of trimers on membranes containing phosphatidylinositol-(4,5)-bisphosphate. Virology 387, 466–472 10.1016/j.virol.2009.02.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfadhli A., Still A., Barklis E. (2009b). Analysis of human immunodeficiency virus type 1 matrix binding to membranes and nucleic acids. J. Virol. 83, 12196–12203 10.1128/JVI.01197-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfadhli A., Huseby D., Kapit E., Colman D., Barklis E. (2007). Human immunodeficiency virus type 1 matrix protein assembles on membranes as a hexamer. J. Virol. 81, 1472–1478 10.1128/JVI.02122-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfadhli A., McNett H., Tsagli S., Bächinger H. P., Peyton D. H., Barklis E. (2011). HIV-1 matrix protein binding to RNA. J. Mol. Biol. 410, 653–666 10.1016/j.jmb.2011.04.063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aloia R. C., Tian H., Jensen F. C. (1993). Lipid composition and fluidity of the human immunodeficiency virus envelope and host cell plasma membranes. Proc. Natl. Acad. Sci. U.S.A. 90, 5181–5185 10.1073/pnas.90.11.5181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arthur L. O., Bess J. W., Jr., Sowder R. C. I., Benveniste R. E., Mann D. L., Chermann J.-C., Henderson L. E. (1992). Cellular proteins bound to immunodeficiency virus: implications for pathogenesis and vaccines. Science 258, 1935–1938 10.1126/science.1470916 [DOI] [PubMed] [Google Scholar]
- Batonick M., Favre M., Boge M., Spearman P., Höning S., Thali M. (2005). Interaction of HIV-1 Gag with the clathrin-associated adaptor AP-2. Virology 342, 190–200 10.1016/j.virol.2005.08.001 [DOI] [PubMed] [Google Scholar]
- Behnia R., Munro S. (2005). Organelle identity and the signposts for membrane traffic. Nature 438, 597–604 10.1038/nature04397 [DOI] [PubMed] [Google Scholar]
- Berlioz-Torrent C., Shacklett B. L., Erdtmann L., Delamarre L., Bouchaert I., Sonigo P., Dokhelar M. C., Benarous R. (1999). Interactions of the cytoplasmic domains of human and simian retroviral transmembrane proteins with components of the clathrin adaptor complexes modulate intracellular and cell surface expression of envelope glycoproteins. J. Virol. 73, 1350–1361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm M., Bonifacino J. S. (2002). Genetic analyses of adaptin function from yeast to mammals. Gene 286, 175–186 10.1016/S0378-1119(02)00422-5 [DOI] [PubMed] [Google Scholar]
- Boge M., Wyss S., Bonifacino J. S., Thali M. (1998). A membrane-proximal tyrosine-based signal mediates internalization of the HIV-1 envelope glycoprotein via interaction with the AP-2 clathrin adaptor. J. Biol. Chem. 273, 15773–15778 10.1074/jbc.273.25.15773 [DOI] [PubMed] [Google Scholar]
- Bouamr F., Scarlata S., Carter C. A. (2003). Role of myristylation in HIV-1 Gag assembly. Biochemistry 42, 6408–6417 10.1021/bi020692z [DOI] [PubMed] [Google Scholar]
- Brandano L., Stevenson M. (2012). A highly conserved residue in the C-terminal helix of HIV-1 matrix is required for envelope incorporation into virus particles. J. Virol. 86, 2347–2359 10.1128/JVI.06047-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brass A. L., Dykxhoorn D. M., Benita Y., Yan N., Engelman A., Xavier R. J., Lieberman J., Elledge S. J. (2008). Identification of host proteins required for HIV infection through a functional genomic screen. Science 319, 921–926 10.1126/science.1152725 [DOI] [PubMed] [Google Scholar]
- Braunewell K. H., Gundelfinger E. D. (1999). Intracellular neuronal calcium sensor proteins: a family of EF-hand calcium-binding proteins in search of a function. Cell Tissue Res. 295, 1–12 10.1007/s004410051207 [DOI] [PubMed] [Google Scholar]
- Briggs J. A. G., Kräusslich H.-G. (2011). The molecular architecture of HIV. J. Mol. Biol. 410, 491–500 10.1016/j.jmb.2011.04.021 [DOI] [PubMed] [Google Scholar]
- Briggs J. A. G., Riches J. D., Glass B., Bartonova V., Zanetti G., Kräusslich H.-G. (2009). Structure and assembly of immature HIV. Proc. Natl. Acad. Sci. U.S.A. 106, 11090–11095 10.1073/pnas.0903535106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown D. A., London E. (1997). Structure of detergent-resistant membrane domains: does phase separation occur in biological membranes? Biochem. Biophys. Res. Commun. 240, 1–7 10.1006/bbrc.1997.7575 [DOI] [PubMed] [Google Scholar]
- Brown D. A., London E. (1998). Structure and origin of ordered lipid domains in biological membranes. J. Membr. Biol. 164, 103–114 10.1007/s002329900397 [DOI] [PubMed] [Google Scholar]
- Brown D. A., London E. (2000). Structure and function of sphingolipid- and cholesterol-rich membrane rafts. J. Biol. Chem. 275, 17221–17224 10.1074/jbc.275.18.13441 [DOI] [PubMed] [Google Scholar]
- Bryant M., Ratner L. (1990). Myristoylation-dependent replication and assembly of human immunodeficiency virus 1. Proc. Natl. Acad. Sci. U.S.A. 87, 523–527 10.1073/pnas.87.2.523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burniston M. T., Cimarelli A., Colgan J., Curtis S. P., Luban J. (1999). Human immunodeficiency virus type 1 Gag polyprotein multimerization requires the nucleocapsid domain and RNA and is promoted by the capsid-dimer interface and the basic region of matrix protein. J. Virol. 73, 8527–8540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byland R., Vance P. J., Hoxie J. A., Marsh M. (2007). A conserved dileucine motif mediates clathrin and AP-2-dependent endocytosis of the HIV-1 envelope protein. Mol. Biol. Cell 18, 414–425 10.1091/mbc.E06-06-0535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai M., Huang Y., Craigie R., Clore G. M. (2010). Structural basis of the association of HIV-1 matrix protein with DNA. PLoS ONE 5, e15675. 10.1371/journal.pone.0015675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell S., Fisher R. J., Towler E. M., Fox S., Issaq H. J., Wolfe T., Phillips L. R., Rein A. (2001a). Modulation of HIV-like particle assembly in vitro by inositol phosphates. Proc. Natl. Acad. Sci. U.S.A. 98, 10875–10879 10.1073/pnas.191224698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell S. M., Crowe S. M., Mak J. (2001b). Lipid rafts and HIV-1: from viral entry to assembly of progeny virions. J. Clin. Virol. 22, 217–227 10.1016/S1386-6532(01)00193-7 [DOI] [PubMed] [Google Scholar]
- Campbell S., Rein A. (1999). In vitro assembly properties of human immunodeficiency virus type 1 Gag protein lacking the p6 domain. J. Virol. 73, 2270–2279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camus G., Segura-Morales C., Molle D., Lopez-Vergès S., Begon-Pescia C., Cazevieille C., Schu P., Bertrand E., Berlioz-Torrent C., Basyuk E. (2007). The clathrin adaptor complex AP-1 binds HIV-1 and MLV Gag and facilitates their budding. Mol. Biol. Cell 18, 3193–3203 10.1091/mbc.E06-12-1147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J., Dick R. A., Vogt V. M. (2011). Rous sarcoma virus Gag has no specific requirement for phosphatidylinositol-(4,5)-bisphosphate for plasma membrane association in vivo or for liposome interaction in vitro. J. Virol. 85, 10851–10860 10.1128/JVI.00760-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan R., Uchil P. D., Jin J., Shui G., Ott D. E., Mothes W., Wenk M. R. (2008). Retroviruses human immunodeficiency virus and murine leukemia virus are enriched in phosphoinositides. J. Virol. 82, 11228–11238 10.1128/JVI.00438-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattopadhyaya R., Meador W. E., Means A. R., Quiocho F. A. (1992). Calmodulin structure refined at 1.7 Å resolution. J. Mol. Biol. 228, 1177–1192 10.1016/0022-2836(92)90324-D [DOI] [PubMed] [Google Scholar]
- Checkley M. A., Luttge B. G., Freed E. O. (2011). HIV-1 envelope glycoprotein biosynthesis, trafficking, and incorporation. J. Mol. Biol. 410, 582–608 10.1016/j.jmb.2011.04.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K., Bachtiar I., Piszczek G., Bouamr F., Carter C., Tjandra N. (2008). Solution NMR characterization of oligomerization and dynamics of equine infectious anemia virus matrix protein and its interaction with PIP2. Biochemistry 47, 1928–1937 10.1021/bi800566u [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin D., Means A. R. (2000). Calmodulin: a prototypical calcium sensor. Trends Cell Biol. 10, 322–328 10.1016/S0962-8924(00)01800-6 [DOI] [PubMed] [Google Scholar]
- Chow J. Y. H., Jeffries C. M., Kwan A. H., Guss J. M., Trewhella J. (2010). Calmodulin disrupts the structure of the HIV-1 MA protein. J. Mol. Biol. 400, 702–714 10.1016/j.jmb.2010.05.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu H., Wang J. J., Spearman P. (2010). Human immunodeficiency virus type-1 Gag and host vesicular trafficking pathways. Curr. Top. Microbiol. Immunol. 339, 67–84 10.1007/978-3-642-02175-6_4 [DOI] [PubMed] [Google Scholar]
- Chukkapalli V., Hogue I. B., Boyko V., Hu W.-S., Ono A. (2008). Interaction between HIV-1 Gag matrix domain and phosphatidylinositol-(4,5)-bisphosphate is essential for efficient Gag-membrane binding. J. Virol. 82, 2405–2417 10.1128/JVI.01614-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chukkapalli V., Oh S. J., Ono A. (2010). Opposing mechanisms involving RNA and lipids regulate HIV-1 Gag membrane binding through the highly basic region of the matrix domain. Proc. Natl. Acad. Sci. U.S.A. 107, 1600–1605 10.1073/pnas.0908661107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chukkapalli V., Ono A. (2011). Molecular determinants that regulate plasma membrane association of HIV-1 Gag. J. Mol. Biol. 410, 512–524 10.1016/j.jmb.2011.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimarelli A., Sandin S., Hoglund S., Luban J. (2000). Basic residues in human immunodeficiency virus type 1 nucleocapsid promote virion assembly via interaction with RNA. J. Virol. 74, 3046–3057 10.1128/JVI.74.15.6734-6740.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper J., Liu L., Woodruff E. A., Taylor H. E., Goodwin J. S., D’Aquila R. T., Spearman P., Hildreth J. E. K., Dong X. (2011). Filamin A protein interacts with human immunodeficiency virus type 1 Gag protein and contributes to productive particle assembly. J. Biol. Chem. 286, 28498–28510 10.1074/jbc.M111.239053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crist R. M., Datta S. A. K., Stephen A. G., Soheilian F., Mirro J., Fisher R. J., Nagashima K., Rein A. (2009). Assembly properties of human immunodeficiency virus type 1 Gag-leucine zipper chimeras: implications for retrovirus assembly. J. Virol. 83, 2216–2225 10.1128/JVI.02031-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalton A. K., Ako-Adjei D., Murray P. S., Murray D., Vogt M. V. (2007). Electrostatic interactions drive membrane association of the human immunodeficiency virus type 1 Gag MA domain. J. Virol. 81, 6434–6445 10.1128/JVI.02757-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalton A. K., Murray P. S., Murray D., Vogt V. M. (2005). Biochemical characterization of Rous sarcoma virus MA protein interaction with membranes. J. Virol. 79, 6227–6238 10.1128/JVI.79.10.6227-6238.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darlix J. L., Spahr P. F. (1982). Binding sites of viral protein P19 onto Rous sarcoma virus RNA and possible controls of viral functions. J. Mol. Biol. 160, 147–161 10.1016/0022-2836(82)90172-3 [DOI] [PubMed] [Google Scholar]
- Datta S. A. K., Curtis J. E., Ratcliff W., Clark P. K., Crist R. M., Lebowitz J., Krueger S., Rein A. (2007a). Conformation of the HIV-1 Gag protein in solution. J. Mol. Biol. 365, 812–824 10.1016/j.jmb.2006.10.072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta S. A. K., Zhao Z., Clark P. K., Tarasov S., Alexandratos J. N., Campbell S. J., Kvaratskhelia M., Lebowitz J., Rein A. (2007b). Interactions between HIV-1 Gag molecules in solution: an inositol phosphate-mediated switch. J. Mol. Biol. 365, 799–811 10.1016/j.jmb.2006.10.072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis M. R., Jiang J., Zhou J., Freed E. O., Aiken C. (2006). A mutation in the human immunodeficiency virus type 1 Gag protein destabilizes the interaction of the envelope protein subunits gp120 and gp41. J. Virol. 80, 2405–2417 10.1128/JVI.80.3.1290-1301.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson L., Yu X. F. (1998). The role of nucleocapsid of HIV-1 in virus assembly. Virology 251, 141–157 10.1006/viro.1998.9374 [DOI] [PubMed] [Google Scholar]
- Ding L., Derdowski A., Wang J.-J., Spearman P. (2003). Independent segregation of human immunodeficiency virus type 1 Gag protein complexes and lipid rafts. J. Virol. 77, 1916–1926 10.1128/JVI.77.20.11027-11039.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X., Li H., Derdowski A., Ding L., Burnett A., Chen X., Peters T. R., Dermody T. S., Woodruff E., Wang J.-J., Spearman P. (2005). AP-3 directs the intracellular trafficking of HIV-1 Gag and plays a key role in particle assembly. Cell 120, 663–674 10.1016/j.cell.2004.12.023 [DOI] [PubMed] [Google Scholar]
- Dorfman T., Mammano F., Haseltine W. A., Göttlinger H. G. (1994). Role of the matrix protein in the virion association of the human immunodeficiency virus type 1 envelope glycoprotein. J. Virol. 68, 1689–1696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou J., Wang J.-J., Chen X., Li H., Ding L., Spearman P. (2009). Characterization of a myristoylated, monomeric HIV Gag protein. Virology 387, 341–352 10.1016/j.virol.2009.02.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgcomb S. P., Murphy K. P. (2002). Variability in the pKa of histidine side-chains correlates with burial within proteins. Proteins 49, 1–6 10.1002/prot.10177 [DOI] [PubMed] [Google Scholar]
- Engeland C. E., Oberwinkler H., Schümann M., Krause E., Müller G. A., Kräusslich H.-G. (2011). The cellular protein lyric interacts with HIV-1 Gag. J. Virol. 85, 13322–13332 10.1128/JVI.00174-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdie C. R., Wills J. W. (1990). Myristoylation of Rous sarcoma virus Gag protein does not prevent replication in avian cells. J. Virol. 64, 5204–5208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fallon J. L., Quiocho F. A. (2003). A closed compact structure of native Ca+ 2-calmodulin. Structure 11, 1303–1307 10.1016/j.str.2003.09.004 [DOI] [PubMed] [Google Scholar]
- Faurobert E., Chen C.-K., Hurley J. B., Teng D. H.-F. (1996). Drosophila neurocalcin, a fatty acylated, Ca-binding protein that associates with membranes and inhibits in vitro phosphorylation of bovine rhodopsin. J. Biol. Chem. 271, 10256–10262 10.1074/jbc.271.17.10256 [DOI] [PubMed] [Google Scholar]
- Fernandes F., Chen K., Ehrlich L. S., Jin J., Chen M. H., Medina G. N., Symons M., Montelaro R., Donaldson J., Tjandra N., Carter C. A. (2011). Phosphoinositides direct equine infectious anemia virus Gag trafficking and release. Traffic 12, 438–451 10.1111/j.1600-0854.2010.01153.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn B. E., Evenäs J., Drakenberg T., Waltho J. P., Thulin E., Forsén S. (1995). Calcium-induced structural changes and domain autonomy in calmodulin. Nat. Struct. Biol. 2, 777–783 10.1038/nsb0995-777 [DOI] [PubMed] [Google Scholar]
- Finzi A., Orthwein A., Mercier J., Cohen E. A. (2007). Productive human immunodeficiency virus type 1 assembly takes place at the plasma membrane. J. Virol. 81, 7476–7490 10.1128/JVI.00308-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fledderman E. L., Fujii K., Ghanam R. H., Waki K., Prevelige P. E., Freed E. O., Saad J. S. (2010). Myristate exposure in the HIV-1 matrix protein is modulated by ph. Biochemistry 49, 9551–9562 10.1021/bi101245j [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freed E. O., Martin A. M. (1996). Domains of the human immonodeficiency virus type 1 matrix and gp41 cytoplasmic tail required for envelope incorporation into virions. J. Virol. 70, 341–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freed E. O., Orenstein J. M., Buckler-White A. J., Martin M. A. (1994). Single amino acid changes in the human immunodeficiency virus type 1 matrix protein block virus particle production. J. Virol. 68, 5311–5320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freed O. E., Martin A. M. (1995). Virion incorporation of envelope glycoproteins with long but not short cytoplasmic tails is blocked by specific, single amino acid substitutions in the human immunodeficiency virus type 1 matrix. J. Virol. 69, 1984–1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganser-Pornillos B. K., Cheng A., Yeager M. (2007). Structure of full-length HIV-1 CA: a model for the mature capsid lattice. Cell 131, 70–79 10.1016/j.cell.2007.08.018 [DOI] [PubMed] [Google Scholar]
- Ganser-Pornillos B. K., Yeager M., Sundquist W. I. (2008). The structural biology of HIV assembly. Curr. Opin. Struct. Biol. 18, 203–217 10.1016/j.sbi.2008.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghanam R. H., Fernandez T. F., Fledderman E. L., Saad J. S. (2010). Binding of calmodulin to the HIV-1 matrix protein triggers myristate exposure. J. Biol. Chem. 285, 41911–41920 10.1074/jbc.M110.179093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gheysen D., Jacobs E., De Foresta F., Thiriart C., Francotte M., Thines D., De Wilde M. (1989). Assembly and release of HIV-1 precursor Pr55gag virus-like particles from recombinant baculovirus-infected insect cells. Cell 59, 103–112 10.1016/0092-8674(89)90873-8 [DOI] [PubMed] [Google Scholar]
- Golub T., Caroni P. (2005). PI(4,5)P2-dependent microdomain assemblies capture microtubules to promote and control leading edge motility. J. Cell Biol. 169, 151–162 10.1083/jcb.200407058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottlieb R. A., Giesing H. A., Zhu J. Y., Engler R. L., Babior B. M. (1995). Cell acidification in apoptosis: granulocyte colony-stimulating factor delays programmed cell death in neutrophils by up-regulating the vacuolar H+-ATPase. Proc. Natl. Acad. Sci. U.S.A. 92, 5965–5968 10.1073/pnas.92.13.5965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gousset K., Ablan S. D., Coren L. V., Ono A., Soheilian F., Nagashima K., Ott D. E., Freed E. O. (2008). Real-time visualization of HIV-1 GAG trafficking in infected macrophages. PLoS Pathog. 4, e1000015. 10.1371/journal.ppat.1000015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene W. C., Debyser Z., Ikeda Y., Freed E. O., Stephens E., Yonemoto W., Buckheit R. W., Esté J. A., Cihlar T. (2008). Novel targets for HIV therapy. Antiviral Res. 80, 251–265 10.1016/j.antiviral.2008.08.003 [DOI] [PubMed] [Google Scholar]
- Grigorov B., Arcanger F., Roingeard P., Darlix J. L., Muriaux D. (2006). Assembly of infectious HIV-1 in human epithelial and T-lymphoblastic cell lines. J. Mol. Biol. 359, 848–862 10.1016/j.jmb.2006.04.017 [DOI] [PubMed] [Google Scholar]
- Hamard-Peron E., Juilliard F., Saad J. S., Roy C., Roingeard P., Summers M. F., Darlix J. L., Picart C., Muriaux D. (2010). Targeting of MuLV Gag to the plasma membrane is mediated by PI(4,5)P2/PS and a polybasic region in the Matrix. J. Virol. 84, 503–515 10.1128/JVI.01134-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamard-Peron E., Muriaux D. (2011). Retroviral matrix and lipids, the intimate interactions. Retrovirology 8, 15. 10.1186/1742-4690-8-S2-O15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi N., Matsubara M., Jinbo Y., Titani K., Izumi Y., Matsushima N. (2002). Nef of HIV-1 interacts directly with calcium-bound calmodulin. Protein Sci. 11, 529–537 10.1110/ps.23702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hearps A. C., Wagstaff K. M., Piller S. C., Jans D. A. (2008). The N-terminal basic domain of the HIV-1 matrix protein does not contain a conventional nuclear localization sequence but is required for DNA binding and protein self-association. Biochemistry 47, 2199–2210 10.1021/bi701360j [DOI] [PubMed] [Google Scholar]
- Hermida-Matsumoto L., Resh M. D. (1999). Human immunodeficiency virus type 1 protease triggers a myristoyl switch that modulates membrane binding fo Pr55gag and p17MA. J. Virol. 73, 1902–1908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermida-Matsumoto L., Resh M. D. (2000). Localization of human immunodeficiency virus type 1 Gag and Env at the plasma membrane by confocal imagine. J. Virol. 74, 8670–8679 10.1128/JVI.74.18.8670-8679.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeflich K. P., Ikura M. (2002). Calmodulin in action: diversity in target recognition and activation mechanisms. Cell 108, 739–742 10.1016/S0092-8674(02)00682-7 [DOI] [PubMed] [Google Scholar]
- Hogue I. B., Hoppe A., Ono A. (2009). Quantitative FRET microscopy analysis of HIV-1 Gag-Gag interaction: the relative contributions of CA and NC domains, and membrane binding. J. Virol. 83, 7322–7336 10.1128/JVI.02545-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm K., Weclewicz K., Hewson R., Suomalainen M. (2003). Human immunodeficiency virus type 1 assembly and lipid rafts: Pr55gag associates with membrane domains that are largely resistant to Brij98 but sensitive to Triton X-100. J. Virol. 77, 4805–4817 10.1128/JVI.77.8.4805-4817.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inlora J., Chukkapalli V., Derse D., Ono A. (2011). Gag localization and virus-like particle release mediated by the matrix domain of human T-lymphotropic virus type 1 Gag are less dependent on phosphatidylinositol-(4,5)-bisphosphate than those mediated by the matrix domain of HIV-1 Gag. J. Virol. 85, 3802–3810 10.1128/JVI.02383-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida H., Vogel H. J. (2006). Protein-peptide interaction studies demonstrate the versatility of calmodulin target protein binding. Protein Pept. Lett. 13, 455–465 10.2174/092986606776819600 [DOI] [PubMed] [Google Scholar]
- Izumi Y., Watanabe H., Watanabe N., Aoyama A., Jinbo J., Hayashi N. (2008). Solution X-ray scattering reveals a novel structure of calmodulin complexed with a binding domain peptide from the HIV-1 matrix protein p17. Biochemistry 47, 7158–7166 10.1021/bi702416b [DOI] [PubMed] [Google Scholar]
- Jäger S., Cimermancic P., Gulbahce N., Johnson J. R., McGovern K. E., Clarke S. C., Shales M., Mercenne G., Pache L., Li K., Hernandez H., Jang G. M., Roth S. L., Akiva E., Marlett J., Stephens M., D’Orso I., Fernandes J., Fahey M., Mahon C., O’Donoghue A. J., Todorovic A., Morris J. H., Maltby D. A., Alber T., Cagney G., Bushman F. D., Young J. A., Chanda S. K., Sundquist W. I., Kortemme T., Hernandez R. D., Craik C. S., Burlingame A., Sali A., Frankel A. D., Krogan N. J. (2011). Global landscape of HIV-human protein complexes. Nature 481, 365–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janmey P. A., Iida K., Yin H. L., Stossel T. P. (1987). Polyphosphoinositide micelles and polyphosphoinositide-containing vesicles dissociate endogenous gelsolin-actin complexes and promote actin assembly from the fast-growing end of actin filaments blocked by gelsolin. J. Biol. Chem. 262, 12228–12236 [PubMed] [Google Scholar]
- Joshi A., Ablan S. D., Soheilian F., Nagashima K., Freed E. O. (2009a). Evidence that productive human immunodeficiency virus type 1 assembly can occur in an intracellular compartment. J. Virol. 83, 5375–5387 10.1128/JVI.00109-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi A., Nagashima K., Freed E. O. (2009b). Defects in cellular sorting and retroviral assembly induced by GGA overexpression. BMC Cell Biol. 10, 72. 10.1186/1471-2121-10-72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi A., Garg H., Ablan S. D., Freed E. O. (2011). Evidence of a role for soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE) machinery in HIV-1 assembly and release. J. Biol. Chem. 286, 29861–28971 10.1074/jbc.M111.241521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi A., Garg H., Nagashima K., Bonifacino J. S., Freed E. O. (2008). GGA and Arf proteins modulate retrovirus assembly and release. Mol. Cell 30, 227–238 10.1016/j.molcel.2008.03.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi A., Nagashima K., Freed E. O. (2006). Mutation of dileucine-like motifs in the human immunodeficiency virus type 1 capsid disrupts virus assembly, Gag-Gag interactions, Gag-membrane binding, and virion maturation. J. Virol. 80, 7939–7951 10.1128/JVI.00355-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouvenet N., Bieniasz P. D., Simon S. M. (2008). Imaging the biogenesis of individual HIV-1 virions in live cells. Nature 454, 236–240 10.1038/nature06998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouvenet N., Neil S. J. D., Bess C., Johnson M. C., Virgen C. A., Simon S. M., Bieniasz P. D. (2006). Plasma membrane is the site of productive HIV-1 particle assembly. PLoS Biol. 4, e435. 10.1371/journal.pbio.0040435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh I., Kyushiki H., Sakamoto Y., Ikawa Y., Yoshinaka Y. (1991). Bovine leukemia virus matrix-associated protein MA(p15): further processing and formation of a specific complex with the dimer of the 5′-terminal genomic rna fragment. J. Virol. 65, 6845–6855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinnunen P. K., Kõiv A., Lehtonen J. Y., Rytömaa M., Mustonen P. (1994). Lipid dynamics and peripheral interactions of proteins with membrane surfaces. Chem. Phys. Lipids 73, 181–207 10.1016/0009-3084(94)90170-8 [DOI] [PubMed] [Google Scholar]
- König R., Zhou Y., Elleder D., Diamond T. L., Bonamy G. M. C., Irelan J. T., Chiang C.-Y., Tu B. P., De Jesus P. D., Lilley C. E., Seidel S., Opaluch A. M., Caldwell J. S., Weitzman M. D., Kuhen K. L., Bandyopadhyay S., Ideker T., Orth A. P., Miraglia L. J., Bushman F. D., Young J. A., Chanda S. K. (2008). Global analysis of host-pathogen interactions that regulate early-stage HIV-1 replication. Cell 135, 49–60 10.1016/j.cell.2008.07.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leis J. P., McGinnis J., Green R. W. (1978). Rous sarcoma virus p19 binds to specific double-stranded regions of viral RNA: effect of p19 on cleavage of viral RNA by RNase III. Virology 84, 87–98 10.1016/0042-6822(78)90220-9 [DOI] [PubMed] [Google Scholar]
- Lever A. M. L., Jeang K.-T. (2011). Insights into cellular factors that regulate HIV-1 replication in human cells. Biochemistry 50, 920–931 10.1021/bi101805f [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Dou J., Ding L., Spearman P. (2007). Myristoylation is required for human immunodeficiency virus type 1 Gag-Gag multimerization in mammalian cells. J. Virol. 81, 12899–12910 10.1128/JVI.00529-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Hill C. P., Sundquist W. I., Finch J. T. (2000). Image reconstructions of helical assemblies of the HIV-1 CA protein. Nature 407, 409–413 10.1038/35030177 [DOI] [PubMed] [Google Scholar]
- Lindwasser O. W., Resh M. D. (2001). Multimerization of human immunodeficiency virus type 1 Gag promotes its localization to barges, raft-like membrane microdomains. J. Virol. 75, 7913–7924 10.1128/JVI.75.17.7913-7924.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindwasser O. W., Resh M. D. (2002). Myristoylation as a target for inhibiting HIV assembly: unsaturated fatty acids block viral budding. Proc. Natl. Acad. Sci. U.S.A. 99, 13037–13042 10.1073/pnas.212409999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T., Ryan M., Dahlquist F. W., Griffith O. H. (1997). Determination of pKa values of the histidine side chains of phosphatidylinositol-specific phospholipase C from Bacillus cereus by NMR spectroscopy and site-directed mutagenesis. Protein Sci. 6, 1937–1944 10.1002/pro.5560060914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodge R., Gottlinger H., Gabuzda D., Cohen E. A., Lemay G. (1994). The intracytoplasmic domain of gp41 mediates polarized budding of human immunodeficiency virus type 1 in MDCK cells. J. Virol. 68, 4857–4861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Vergès S., Camus G., Blot G., Beauvoir R., Benarous R., Berlioz-Torrent C. (2006). Tail-interacting protein TIP47 is a connector between Gag and Env and is required for Env incorporation into HIV-1 virions. Proc. Natl. Acad. Sci. U.S.A. 103, 14947–14952 10.1073/pnas.0602941103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lusso P., Di Marzo Veronese F., Ensoli B., Franchini G., Jemma C., Derocco S. E., Kalyanaraman V. S., Gallo R. C. (1990). Expanded HIV-1 cellular tropism by phenotypic mixing with murine endogenous retroviruses. Science 247, 848–852 10.1126/science.2305256 [DOI] [PubMed] [Google Scholar]
- Makutonina A., Voss T. G., Plymale D. R., Fermin C. D., Norris C. H., Vigh S., Garry R. F. (1996). Human immunodeficiency virus infection of T-lymphoblastoid cells reduces intracellular pH. J. Virol. 70, 7049–7055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mammano F., Kondo E., Sodroski J., Bukovsky A., Gottlinger H. G. (1995). Rescue of human immunodeficiency virus type I matrix protein mutants by envelope glycoproteins with short cytoplasmic domains. J. Virol. 69, 3824–3830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubara M., Jing T., Kawamura K., Shimojo N., Titani K., Hashimoto K., Hayashi N. (2005). Myristoyl moiety of HIV Nef is involved in regulation of the interaction with calmodulin in vivo. Protein Sci. 14, 494–503 10.1110/ps.04969605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubara M., Nakatsu T., Kato H., Taniguchi H. (2004). Crystal structure of a myristoylated CAP-23/NAP-22 N-terminal domain complexed with Ca2+/calmodulin. EMBO J. 23, 712–718 10.1038/sj.emboj.7600093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubara M., Titani K., Taniguchi H., Hayashi N. (2003). Direct involvement of protein myristoylation in myristoylated alanine-rich C kinase substrate (MARCKS)-calmodulin interaction. J. Biol. Chem. 278, 48898–48902 10.1074/jbc.M305488200 [DOI] [PubMed] [Google Scholar]
- Matsuda S., Hisatomi O., Ishino T., Kobayashi Y., Tokunaga F. (1998). The role of calcium-binding sites in S-modulin function. J. Biol. Chem. 273, 20223–20227 10.1074/jbc.273.20.12378 [DOI] [PubMed] [Google Scholar]
- McLaughlin S., Murray D. (2005). Plasma membrane phosphoinositide organization by protein electrostatics. Nature 438, 605–611 10.1038/nature04398 [DOI] [PubMed] [Google Scholar]
- Melkonian K. A., Ostermeyer A. G., Chen J. Z., Roth M. G., Brown D. A. (1999). Role of lipid modifications in targeting proteins to detergent-resistant membrane rafts. J. Biol. Chem. 274, 3910–3917 10.1074/jbc.274.6.3910 [DOI] [PubMed] [Google Scholar]
- Méric C., Spahr P.-F. (1986). Rous sarcoma virus nucleic acid-binding protein p12 is necessary for viral 70S RNA dimer formation and packaging. J. Virol. 60, 450–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micoli K. J., Mamaeva O., Piller S. C., Barker J. L., Pan G., Hunter E., McDonald J. M. (2006). Point mutations in the C-terminus of HIV-1 gp160 reduce apoptosis and calmodulin binding without affecting viral replication. Virology 344, 468–479 10.1016/j.virol.2005.08.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moolenaar W. H., Tsien R. Y., Van Der Saag P. T., De Laat S. W. (1983). Na+/H+ exchange and cytoplasmic pH in the action of growth factors in human fibroblasts. Nature 304, 645–648 10.1038/304645a0 [DOI] [PubMed] [Google Scholar]
- Morikawa Y., Hockley D. J., Nermut M. V., Jones I. M. (2000). Roles of matrix, p2, and N-terminal myristoylation in human immunodeficiency virus type 1 Gag assembly. J. Virol. 74, 16–23 10.1128/JVI.74.1.16-23.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morikawa Y., Zhang W.-H., Hockley D. J., Nermut M. V., Jones I. M. (1998). Detection of a trimeric human immunodeficiency virus type 1 Gag intermediate is dependent on sequences in the matrix protein, p17. J. Virol. 72, 7659–7663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami T., Freed E. O. (2000). The long cytoplasmic tail of gp41 is required in a cell type-dependent manner for HIV-1 envelope glycoprotein incorporation into virions. Proc. Natl. Acad. Sci. U.S.A. 97, 343–348 10.1073/pnas.97.7.3579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muriaux D., Mirro J., Harvin D., Rein A. (2001). RNA is a structural element in retrovirus particles. Proc. Natl. Acad. Sci. U.S.A. 98, 5246–5251 10.1073/pnas.091000398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatsu F., Ohno H. (2003). Adaptor protein complexes as the key regulators of protein sorting in the post-Golgi network. Cell Struct. Funct. 28, 419–429 10.1247/csf.28.419 [DOI] [PubMed] [Google Scholar]
- Narazaki M., Fujimoto M., Matsumoto T., Morita Y., Saito H., Kajita T., Yoshizaki K., Naka T., Kishimoto T. (1998). Three distinct domains of SSI-1/SOCS-1/JAB protein are required for its suppression of interleukin 6 signaling. Proc. Natl. Acad. Sci. U.S.A. 95, 13130–13134 10.1073/pnas.95.22.13130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen D. G., Booth A., Gould S. J., Hildreth J. E. (2003). Evidence that HIV budding in primary macrophages occurs through the exosome release pathway. J. Biol. Chem. 278, 52347–52354 10.1074/jbc.M308770200 [DOI] [PubMed] [Google Scholar]
- Nguyen D. H., Hildreth J. E. (2000). Evidence for budding of human immunodeficiency virus type 1 selectively from glycolipid-enriched membrane lipid rafts. J. Virol. 74, 3264–3272 10.1128/JVI.74.12.5495-5501.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishi M., Ryo A., Tsurutani N., Ohba K., Sawasaki T., Morishita R., Perrem K., Aoki I., Morikawa Y., Yamamoto N. (2009). Requirement for microtubule integrity in the SOCS1-mediated intracellular dynamics of HIV-1 Gag. FEBS Lett. 583, 1243–1250 10.1016/j.febslet.2009.03.041 [DOI] [PubMed] [Google Scholar]
- Nydegger S., Foti M., Derdowski A., Spearman P., Thali M. (2003). HIV-1 egress is gated through late endosomal membranes. Traffic 4, 902–910 10.1046/j.1600-0854.2003.00145.x [DOI] [PubMed] [Google Scholar]
- Ohno H., Aguilar R. C., Fournier M. C., Hennecke S., Cosson P., Bonifacino J. S. (1997). Interaction of endocytic signals from the HIV-1 envelope glycoprotein complex with members of the adaptor medium chain family. Virology 238, 305–315 10.1006/viro.1997.8839 [DOI] [PubMed] [Google Scholar]
- Ono A. (2009). HIV-1 assembly at the plasma membrane: Gag trafficking and localization. Future Virol. 4, 241–257 10.2217/fvl.09.4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono A. (2010). Relationships between plasma membrane microdomains and HIV-1 assembly. Biol. Cell 102, 335–350 10.1042/BC20090165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono A., Ablan S. D., Lockett S. J., Nagashima K., Freed E. O. (2004). Phosphatidylinositol (4,5) bisphosphate regulates HIV-1 Gag targeting to the plasma membrane. Proc. Natl. Acad. Sci. U.S.A. 101, 14889–14894 10.1073/pnas.0405816101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono A., Demirov D., Freed E. O. (2000a). Relationship between human immunodeficiency virus type-1 Gag multimerization and membrane binding. J. Virol. 74, 5142–5150 10.1128/JVI.74.11.5142-5150.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono A., Orenstein J. M., Freed E. O. (2000b). Role of the Gag matrix domain in targeting human immunodeficiency virus type 1 assembly. J. Virol. 74, 2855–2866 10.1128/JVI.74.11.5142-5150.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono A., Freed E. O. (1999). Binding of human immunodeficiency virus type 1 Gag to membrane: role of the matrix amino terminus. J. Virol. 73, 4136–4144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono A., Freed E. O. (2001). Plasma membrane rafts play a critical role in HIV-1 assembly and release. Proc. Natl. Acad. Sci. U.S.A. 98, 13925–13930 10.1073/pnas.241320298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono A., Freed E. O. (2004). Cell-type-dependent tageting of human immunodeficiency virus type 1 assembly to the plasma membrane and the multivesicular body. J. Virol. 78, 1552–1563 10.1128/JVI.78.3.1552-1563.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono A., Freed E. O. (2005). Role of lipid rafts in virus replication. Adv. Virus Res. 64, 311–358 10.1016/S0065-3527(05)64010-9 [DOI] [PubMed] [Google Scholar]
- Ono A., Huang M., Freed E. O. (1997). Characterization of human immunodeficiency virus type 1 matrix revertants: effects on virus assembly, Gag processing, and Env incorporation into virions. J. Virol. 71, 4409–4418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osawa M., Tokumitsu H., Swindells M. B., Kurihara H., Orita M., Shibanuma T., Furuya T., Ikura M. (1999). A novel target recognition revealed by calmodulin in complex with Ca2+-calmodulin-dependent kinase kinase. Nat. Struct. Biol. 6, 819–824 10.1038/12271 [DOI] [PubMed] [Google Scholar]
- Ott D. E. (2008). Cellular proteins detected in HIV-1. Rev. Med. Virol. 18, 159–175 10.1002/rmv.570 [DOI] [PubMed] [Google Scholar]
- Ott D. E., Coren L. V., Gagliardi T. D. (2005). Redundant roles for nucleocapsid and matrix RNA-binding sequences in human immunodeficiency virus type 1 assembly. J. Virol. 79, 13839–13847 10.1128/JVI.79.22.13839-13847.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owens R. J., Compans R. W. (1989). Expression of the human immunodeficiency virus envelope glycoprotein is restricted to basolateral surfaces of polarized epithelial cells. J. Virol. 63, 978–982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owens R. J., Dubay J. W., Hunter E., Compans R. W. (1991). Human immunodeficiency virus envelope protein determines the site of virus release in polarized epithelial cells. Proc. Natl. Acad. Sci. U.S.A. 88, 3987–3991 10.1073/pnas.88.9.3987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paillart J.-C., Gottlinger H. G. (1999). Opposing effects of human immunodeficiency virus type 1 matrix mutations support a myristyl switch model of Gag membrane targeting. J. Virol. 73, 2604–2612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parent L. J., Gudleski N. (2011). Beyond plasma membrane targeting: role of the MA domain of Gag in retroviral genome encapsidation. J. Mol. Biol. 410, 553–564 10.1016/j.jmb.2011.04.072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelchen-Matthews A., Krameer B., Marsh M. (2003). Infectious HIV-1 assembles in late endosomes in primary macrophages. J. Cell Biol. 162, 443–445 10.1083/jcb.200304008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provitera P., Bouamr F., Murray D., Carter C., Scarlata S. (2000). Binding of equine infectious anemia virus matrix protein to membrane bilayers involves multiple interactions. J. Mol. Biol. 296, 887–898 10.1006/jmbi.1999.3482 [DOI] [PubMed] [Google Scholar]
- Purohit P., Dupont S., Stevenson M., Green M. R. (2001). Sequence-specific interaction between HIV-1 matrix protein and viral genomic RNA revealed by in vitro genetic selection. RNA 7, 576–584 10.1017/S1355838201002023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radding W., Pan Z. Q., Hunter E., Johnston P., Williams J. P., McDonald J. M. (1996). Expression of HIV-1 envelope glycoprotein alters cellular calmodulin. Biochem. Biophys. Res. Commun. 218, 192–197 10.1006/bbrc.1996.0034 [DOI] [PubMed] [Google Scholar]
- Radding W., Williams J. P., McKenna M. A., Tummala R., Hunter E., Tytler E. M., McDonald J. M. (2000). Calmodulin and HIV type 1: interactions with Gag and Gag products. AIDS Res. Hum. Retroviruses 16, 1519–1525 10.1089/088922200750006047 [DOI] [PubMed] [Google Scholar]
- Ramalingam D., Duclair S., Datta S. A., Ellington A., Rein A., Prasad V. R. (2011). RNA aptamers directed to human immunodeficiency virus type 1 Gag polyprotein bind to the matrix and nucleocapsid domains and inhibit virus production. J. Virol. 85, 305–314 10.1128/JVI.02626-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reil H., Bukovsky A. A., Gelderblom H. R., Gottlinger H. G. (1998). Efficient HIV-1 replication can occur in the absence of the viral matrix protein. EMBO J. 17, 2699–2708 10.1093/emboj/17.9.2699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M. S., Bonifacino J. S. (2001). Adaptor-related proteins. Curr. Opin. Cell Biol. 13, 444–453 10.1016/S0955-0674(00)00235-0 [DOI] [PubMed] [Google Scholar]
- Rogers M. S., Strehler E. E. (1996). “Calmodulin-like proteins,” in Guidebook to the Calcium-Binding Proteins, eds Celio M. R., Pauls T., Schwaller B. (Oxford: Oxford University Press; ), 34–40 [Google Scholar]
- Ryo A., Tsurutani N., Obha K., Kimura R., Komano J., Nishi M., Soeda H., Hattori S., Perrem K., Yamamoto M., Chiba J., Mimaya J.-I., Yoshimura K., Matsushita S., Honda M., Yoshimura A., Sawasaki T., Aoki I., Morikawa Y., Yamamoto N. (2008). SOCS1 is an inducible host factor during HIV-1 infection and regulates the intracellular trafficking and stability of HIV-1 Gag. Proc. Natl. Acad. Sci. U.S.A. 105, 294–299 10.1073/pnas.0704831105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rytömaa M., Kinnunen P. K. (1995). Reversibility of the binding of cytochrome c to liposomes. Implications for lipid-protein interactions. J. Biol. Chem. 270, 3197–3202 10.1074/jbc.270.7.3197 [DOI] [PubMed] [Google Scholar]
- Saad J. S., Ablan S. D., Ghanam R. H., Kim A., Andrews K., Nagashima K., Soheilian F., Freed E. O., Summers M. F. (2008). Structure of the myristylated HIV-2 MA protein and the role of phosphatidylinositol-(4,5)-bisphosphate in membrane targeting. J. Mol. Biol. 382, 434–447 10.1016/j.jmb.2008.07.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saad J. S., Kim A., Ghanam R. H., Dalton A. K., Vogt M. V., Wu Z., Lu W., Summers M. F. (2007a). Mutations that mimic phosphorylation of the HIV-1 matrix protein do not perturb the myristyl switch. Protein Sci. 16, 1793–1797 10.1110/ps.072987607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saad J. S., Loeliger E., Luncsford P., Liriano M., Tai J., Kim A., Miller J., Joshi A., Freed E. O., Summers M. F. (2007b). Point mutations in the HIV-1 matrix protein turn off the myristyl switch. J. Mol. Biol. 366, 574–585 10.1016/j.jmb.2006.11.068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saad J. S., Miller J., Tai J., Kim A., Ghanam R. H., Summers M. F. (2006). Structural basis for targeting HIV-1 Gag to virus assembly sites on the plasma membrane. Proc. Natl. Acad. Sci. U.S.A. 103, 11364–11369 10.1073/pnas.0602818103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samal A. B., Ghanam R. H., Fernandez T. F., Monroe E. B., Saad J. S. (2011). NMR, biophysical and biochemical studies reveal the minimal calmodulin-binding domain of the HIV-1 matrix protein. J. Biol. Chem. 286, 33533–33543 10.1074/jbc.M111.273623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuldiner S., Rozengurt E. (1982). Na+/H+ antiport in Swiss 3T3 cells: mitogenic stimulation leads to cytoplasmic alkalinization. Proc. Natl. Acad. Sci. U.S.A. 79, 7778–7782 10.1073/pnas.79.24.7778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherer N. M., Lehmann M. J., Jimenez-Soto L. F., Ingmundson A., Horner S. M., Cicchetti G., Allen P. G., Pypaert M., Cunningham J. M., Mothes W. (2003). Visualization of retroviral replication in living cells reveals budding into multivesicular bodies. Traffic 4, 785–801 10.1034/j.1600-0854.2003.00135.x [DOI] [PubMed] [Google Scholar]
- Shkriabai N., Datta S. A., Zhao Z., Hess S., Rein A., Kvaratskhelia M. (2006). Interactions of HIV-1 Gag with assembly cofactors. Biochemistry 45, 4077–4083 10.1021/bi052308e [DOI] [PubMed] [Google Scholar]
- Spearman P., Horton R., Ratner L., Kuli-Zade I. (1997). Membrane binding of human immunodeficiency virus type 1 matrix protein in vivo supports a conformational myristyl switch mechanism. J. Virol. 71, 6582–6592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivas S. K., Srinivas R. V., Anantharamaiah G. M., Compans R. W., Segrest J. P. (1993). Cytosolic domain of the human immunodeficiency virus envelope glycoproteins binds to calmodulin and inhibits calmodulin-regulated proteins. J. Biol. Chem. 268, 22895–22899 [PubMed] [Google Scholar]
- Stansell E., Apkarian R., Haubova S., Diehl W. E., Tytler E. M., Hunter E. (2007). Basic residues in the Mason-Pfizer monkey virus Gag matrix domain regulate intracellular trafficking and capsid-membrane interactions. J. Virol. 81, 8977–8988 10.1128/JVI.00657-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steeg C. M., Vogt V. M. (1990). RNA-binding properties of the matrix protein (p19gag) of avian sarcoma and leukemia viruses. J. Virol. 64, 847–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasner A. B., Natarajan M., Doman T., Key D., August A., Henderson A. J. (2008). The Src kinase Lck facilitates assembly of HIV-1 at the plasma membrane. J. Immunol. 181, 3706–3713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T., Ames J. B., Harvey T. S., Stryer L., Ikura M. (1995). Sequestration of the membrane-targeting myristoyl group of recoverin in the calcium-free state. Nature 376, 444–447 10.1038/376444a0 [DOI] [PubMed] [Google Scholar]
- Tang C., Loeliger E., Luncsford P., Kinde I., Beckett D., Summers M. F. (2004). Entropic switch regulates myristate exposure in the HIV-1 matrix protein. Proc. Natl. Acad. Sci. U.S.A. 101, 517–522 10.1073/pnas.0403100101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thirkettle H. J., Girling J., Warren A. Y., Mills I. G., Sahadevan K., Leung H., Hamdy F., Whitaker H. C., Neal D. E. (2009). LYRIC/AEG-1 is targeted to different subcellular compartments by ubiquitinylation and intrinsic nuclear localization signals. Clin. Cancer Res. 15, 3003–3013 10.1158/1078-0432.CCR-08-2046 [DOI] [PubMed] [Google Scholar]
- Tomita Y., Noda T., Fujii K., Watanabe T., Morikawa Y., Kawaoka Y. (2011). The cellular factors Vps18 and Mon2 are required for efficient production of infectious HIV-1 particles. J. Virol. 85, 5618–5627 10.1128/JVI.02071-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towler D. A., Adams S. P., Eubanks S. R., Towery D. S., Jackson-Machelski E., Glaser L., Gordon J. I. (1988). Myristyl CoA:protein N-myristoyltransferase activities from rat liver and yeast possess overlapping yet distinct peptide substrate specificities. J. Biol. Chem. 263, 1784–1790 [PubMed] [Google Scholar]
- Tuominen E. K., Wallace C. J., Kinnunen P. K. (2002). Phospholipid-cytochrome c interaction: evidence for the extended lipid anchorage. J. Biol. Chem. 277, 8822–8826 10.1074/jbc.M200056200 [DOI] [PubMed] [Google Scholar]
- Turner B. G., Summers M. F. (1999). Structural biology of HIV. J. Mol. Biol. 285, 1–32 10.1006/jmbi.1998.2354 [DOI] [PubMed] [Google Scholar]
- Valentine K. G., Peterson R., Saad J. S., Summers M. F., Xu X., Ames J. B., Wand A. J. (2010). Reverse micelle encapsulation of membrane anchored proteins for solution NMR studies. Structure 18, 9–16 10.1016/j.str.2009.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vetter S. W., Leclerc E. (2003). Novel aspects of calmodulin target recognition and activation. Eur. J. Biochem. 270, 404–414 10.1046/j.1432-1033.2003.03414.x [DOI] [PubMed] [Google Scholar]
- Waheed A. A., Freed E. O. (2009). Lipids and membrane microdomains in HIV-1 replication. Virus Res. 143, 162–176 10.1016/j.virusres.2009.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsch S., Keppler O. T., Habermann A., Allespach I., Krijnse-Locker J., Kräusslich H.-G. (2007). HIV-1 buds predominantly at the plasma membrane of primary human macrophages. PLoS Pathog. 3, e36. 10.1371/journal.ppat.0030036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilk T., Pfeiffer T., Bosch V. (1992). Retained in vitro infectivity and cytopathogenicity of HIV-1 despite truncation of the C-terminal tail of the env gene product. Virology 189, 167–177 10.1016/0042-6822(92)90692-I [DOI] [PubMed] [Google Scholar]
- Wright E. R., Schooler J. B., Ding H. J., Kieffer C., Fillmore C., Sundquist W. I., Jensen G. J. (2007). Electron crytomography of immature HIV-1 virions reveals the structure of the CA and SP1 Gag shells. EMBO J. 26, 2218–2226 10.1038/sj.emboj.7601664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyss S., Berlioz-Torrent C., Boge M., Blot G., Höning S., Benarous R., Thali M. (2011). The highly conserved C-terminal dileucine motif in the cytosolic domain of the human immunodeficiency virus type 1 envelope glycoprotein is critical for its association with the AP-1 clathrin adaptor [correction of adapter]. J. Virol. 75, 2982–2992 10.1128/JVI.75.6.2982-2992.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamniuk A. P., Vogel H. J. (2004). Calmodulin’s flexibility allows for promiscuity in its interactions with target proteins and peptides. Mol. Biotechnol. 27, 33–57 10.1385/MB:27:1:33 [DOI] [PubMed] [Google Scholar]
- Yap K. L., Kim J., Truong K., Sherman M., Yuan T., Ikura M. (2000). Calmodulin target database. J. Struct. Funct. Genomics 1, 8–14 10.1023/A:1011320027914 [DOI] [PubMed] [Google Scholar]
- Yoshimura A., Naka T., Kubo M. (2007). SOCS proteins, cytokine signalling and immune regulation. Nat. Rev. Immunol. 7, 454–465 10.1038/nri2093 [DOI] [PubMed] [Google Scholar]
- Yu X., Yuan X., Matsuda Z., Lee T.-H., Essex M. (1992). The matrix protein of human immunodeficiency virus type I is required for incorporation of viral envelope protein into mature virions. J. Virol. 66, 4966–4971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zacharias D. A., Violin J. D., Newton A. C., Tsien R. Y. (2002). Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science 296, 913–916 10.1126/science.1068539 [DOI] [PubMed] [Google Scholar]
- Zhou H., Xu M., Huang Q., Gates A. T., Zhang X. D., Castle J. C., Stec E., Ferrer M., Strulovici B., Hazuda D. J., Espeseth A. S. (2008). Genome-scale RNAi screen for host factors required for HIV replication. Cell Host Microbe 4, 495–504 10.1016/j.chom.2008.10.004 [DOI] [PubMed] [Google Scholar]
- Zhou W., Parent L. J., Wills J. W., Resh M. D. (1994). Identification of a membrane-binding domain within the amino-terminal region of human immunodeficiency virus type 1 Gag protein which interacts with acidic phospholipids. J. Virol. 68, 2556–2569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W., Resh M. D. (1996). Differential membrane binding of the human immunodeficiency virus type 1 matrix protein. J. Virol. 70, 8540–8548 [DOI] [PMC free article] [PubMed] [Google Scholar]