Background: HRG-1-related proteins have low sequence homology but are functionally conserved.
Results: Conserved residues within the transmembrane domain and the C terminus dictate the function of HRG-1-related proteins.
Conclusion: Heme transport is mediated by topologically conserved residues implying an evolutionary ancient transport mechanism.
Significance: Understanding the mechanism of HRG-1-related protein function will provide novel therapeutic insights into human hematological disorders and parasites, which rely on host heme for survival.
Keywords: C. elegans, Heme, Iron metabolism, Transporters, Yeast metabolism, Nematode, Tetrapyrrole
Abstract
Caenorhabditis elegans and human HRG-1-related proteins are conserved, membrane-bound permeases that bind and translocate heme in metazoan cells via a currently uncharacterized mechanism. Here, we show that cellular import of heme by HRG-1-related proteins from worms and humans requires strategically located amino acids that are topologically conserved across species. We exploit a heme synthesis-defective Saccharomyces cerevisiae mutant to model the heme auxotrophy of C. elegans and demonstrate that, under heme-deplete conditions, the endosomal CeHRG-1 requires both a specific histidine in the predicted second transmembrane domain (TMD2) and the FARKY motif in the C terminus tail for heme transport. By contrast, the plasma membrane CeHRG-4 transports heme by utilizing a histidine in the exoplasmic (E2) loop and the FARKY motif. Optimal activity under heme-limiting conditions, however, requires histidine in the E2 loop of CeHRG-1 and tyrosine in TMD2 of CeHRG-4. An analogous system exists in humans, because mutation of the synonymous histidine in TMD2 of hHRG-1 eliminates heme transport activity, implying an evolutionary conserved heme transport mechanism that predates vertebrate origins. Our results support a model in which heme is translocated across membranes facilitated by conserved amino acids positioned on the exoplasmic, cytoplasmic, and transmembrane regions of HRG-1-related proteins. These findings may provide a framework for understanding the structural basis of heme transport in eukaryotes and human parasites, which rely on host heme for survival.
Introduction
Heme (iron protoporphyrin IX) is a macrocycle that not only plays an essential role as a prosthetic group in proteins but is an essential nutrient for parasites and is the major dietary iron source for humans (1, 2). Because hemes are essential but cytotoxic when present in excess, organisms must maintain appropriate levels of cellular heme. One means to accomplish this task is to regulate the synthesis and degradation of heme (3–5). Another would be to regulate the trafficking and compartmentalization of heme (1). Although significant strides have been made in understanding the mechanisms and regulation of heme synthesis and degradation in eukaryotes, how heme is transported, trafficked, and compartmentalized within the cell remains poorly understood (1). Studies of heme transporters in bacteria have provided the molecular underpinnings into protein function (6, 7), but these transporters do not have identifiable homologs in animals.
By utilizing the free-living roundworm Caenorhabditis elegans, we previously identified HRG-12-related proteins, membrane-bound transporters that import heme (8). HRG-1-related proteins have four paralogs in C. elegans, CeHRG-1, CeHRG-4, CeHRG-5, and CeHRG-6, which may function redundantly to ensure that the worm, a heme auxotroph, acquires an adequate supply of heme to sustain its growth and development (9, 10). Mammals, by contrast, have a single homolog, which is only ≈20% identical to CeHRG-1. Previous studies have demonstrated that both worm and human HRG-1 bind and transport heme and that knockdown of hrg-1 in zebrafish embryos results in severe anemia with concomitant defects in yolk tube extension and brain formation; these are phenotypes that are fully rescued by CeHRG-1 (8). These studies predict that HRG-1-related proteins are essential for vertebrate development and have conserved functions across metazoa. To gain mechanistic insights into the heme transport function of HRG-1-related proteins, we conducted a structure-function analysis of CeHRG-1 and CeHRG-4 by exploiting yeast mutants that are unable to synthesize heme. Our studies reveal that HRG-1-related proteins transport heme across membranes through the coordinated actions of residues that are topologically conserved in the worm and human proteins. Our results imply that the mechanism for heme import employed by HRG-1-related proteins is ancient and predates vertebrate origins.
EXPERIMENTAL PROCEDURES
Yeast Strains and Growth Media
The Saccharomyces cerevisiae strains used in this study were derived from W303 and YPH499 background (supplemental Table S1). The hem1Δ(6D) and OPY102 strains were constructed as described elsewhere (11, 12). To construct hem1Δfre1Δfre2ΔMET3-FRE1, plasmid pRS314-MET3-FRE1 was linearized with NdeI and integrated into the TRP1 locus of OPY102. Cells were maintained in YPD (yeast extract-peptone-dextrose) or appropriate synthetic complete (SC) media supplemented with 250 μm δ-aminolevulinic acid (ALA) (Frontier Scientific) (13).
Plasmid Construction and Site-directed Mutagenesis
To make yeast expression plasmids, the open reading frame (ORF) of CeHRG-4, CeHRG-1, hHRG-1, and their hemagglutinin (HA) epitope-tagged version were amplified by PCR from pCDNA3.1(+)Zeo-CeHRG-4-HA, pCDNA3.1(+)Zeo-CeHRG-1-HA, and pCDNA3.1(+)Zeo-hHRG-1-HA using gene-specific primers containing BamHI and XbaI sites, digested, and ligated to pYES-DEST52 vector (Invitrogen) digested with the same enzymes. Codon-optimized hHRG-1 ORF for yeast expression (yhHRG-1) was synthesized by Genscript (OptimumGene) and subcloned into pYES-DEST52 vector as described above. The ORF of hFLVCR2 (GenBankTM accession number BC019087) was PCR-amplified from human FLVCR2 cDNA in pOTB7 (Clone ID 4866427, Open Biosystems) using gene-specific primers and cloned into pYES-DEST52 vector as described above. Site-directed mutagenesis was performed on pCDNA3.1(+)Zeo-CeHRG-4-HA and pCDNA3.1(+)Zeo-CeHRG-1-HA using the QuikChange XL site-directed mutagenesis kit following the manufacturer's instructions (Stratagene). The sequences of primers used for generating mutations are listed in supplemental Table S2. The coding sequence of mutant proteins was sequenced and subcloned into pYES-DEST52 as described above.
Spot Growth Assay
The plasmids for expressing wild-type and mutant HRG-1-related proteins were transformed into strain hem1Δ (6D) using the lithium acetate method (14). Transformants were selected on 2% w/v glucose SC (-Ura) plates supplemented with 250 μm ALA. Five or six colonies of each transformation were picked and restreaked onto 2% w/v raffinose SC (-Ura) plates supplemented with 250 μm ALA to deplete glucose for 48 h. Prior to spotting, cells were cultivated in 2% w/v raffinose SC (-Ura) medium for 18 h to deplete heme. Cells were then suspended in water to an A600 of 0.2. 10 μl of 10-fold serial dilutions of each transformant was spotted onto 2% w/v raffinose SC (-Ura) plates supplemented with either 0.4% w/v glucose and 250 μm ALA (positive control), or 0.4% w/v galactose and different concentrations of hemin (no hemin addition as negative control), incubated at 30 °C for 3 days before imaging.
Ferrireductase Assay
The strain hem1Δ fre1Δ fre2ΔMET3-FRE1 was used for ferrireductase assay. Yeast transformation and selection were performed as described above using respective SC auxotrophic medium supplemented with 250 μm ALA. After being depleted for heme in 2% w/v raffinose SC (-Ura, -Trp, +Met) medium for 12 h, cells were suspended in 2% w/v raffinose SC (-Ura, -Trp, -Met) medium supplemented with 0.4% w/v galactose, 0.1 mm Na2S, and different concentrations of hemin to an A600 of 0.3, cultivated in 96-well plates at 30 °C, 225 rpm for 16 h, and subjected to ferrireductase assay (15). Cells were washed with washing buffer (2% BSA, 0.1% Tween 20 in 2× PBS) three to four times to remove residual hemin in the medium, washed twice with reaction buffer (5% glucose, 0.05 m sodium citrate buffer, pH 6.5), and then suspended in reaction buffer while the A600 was determined using a plate reader (PerkinElmer, Optima). An equal volume of assay buffer (2 mm bathophenanthroline disulfonate, 2 mm FeCl3 in reaction buffer) was added to the cells (T = 0 min) and incubated at 30 °C in the dark until a red color was developed. A535 and A610 were determined, and the ferrireductase activity (nanomoles/106cells/min) was calculated in Equation 1.
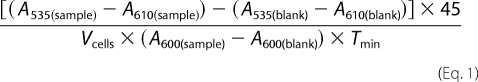 |
β-Galactosidase Reporter Assay
Reporter genes were cloned into plasmid pRS314m-CYC1-HIS3 by replacing HIS3 through in vivo recombination in yeast. The plasmids for wild-type and mutant HRG-1-related proteins expression were co-transformed into strain hem1Δ(6D) with pRS314m-CYC1-LacZ. Transformants selection was performed as described above using appropriate SC auxotrophic medium supplemented with 250 μm ALA. Cells were depleted for heme in 2% w/v raffinose SC (-Ura, -Trp) medium for 12 h, and then were suspended in 10 ml of 2% w/v raffinose SC (-Ura, -Trp) medium supplemented with 0.4% w/v galactose and different concentrations of hemin to an A600 of 0.1. Cell culture was cultivated at 30 °C, 225 rpm for 12 h and subjected to β-galactosidase assay as described elsewhere (16). β-Galactosidase activities were normalized to total protein concentration for comparison.
Immunoblotting
For protein extraction, yeast transformants were resuspended in lysis buffer (1% sodium dodecyl sulfate (SDS), 8 m urea, 10 mm Tris-HCl, pH 8.0, 10 mm EDTA) with protease inhibitors (1 mm phenylmethylsulfonyl fluoride, 4 mm benzamidine, 2 μg/ml leupeptin, and 1 μg/ml pepstatin) and 0.5-mm acid-washed glass beads. Cells were heated at 65 °C for 10 min and disrupted using FastPrep-24 (MPBio) for 3 × 30 s at the 6.5 m/s setting. Cell lysates were collected, and the total protein concentration was quantified with Bradford reagent (Bio-Rad). Protein samples were separated on 12% SDS-PAGE gel and transferred to a nitrocellulose membrane (Bio-Rad). For immunoblotting, the membranes were incubated with rabbit anti-HA (Sigma) as primary antibody at a 1:5,000 dilution for 16 h at 4 °C, followed by HRP-conjugated goat anti-rabbit antibody at a 1:10,000 dilution for 1 h at room temperature. The membranes were re-probed with anti-PGK1 (1:5,000, Invitrogen) as loading control. The signal was detected by using SuperSignal chemiluminescence reagents (Thermo Scientific) in the Gel Documentation system (Bio-Rad).
Immunofluorescence
Yeast transformants were cultivated in 2% w/v raffinose SC (-Ura) medium supplemented with 0.4% w/v galactose and 250 μm ALA to mid-log phase, fixed with 4% formaldehyde for 1 h at room temperature. Immunofluorescence microscopy was performed as described elsewhere (16). Images were taken using a DMIRE2 epifluorescence microscope (Leica) connected to a Retiga 1300 cooled mono 12-bit camera.
Worm Culture and Strains
C. elegans strains were maintained on nematode growth medium (NGM) agar plates seeded with OP50 bacteria at 20 °C. Worm synchronization and cross were performed as described elsewhere (17). The deletion strains Δhrg-1 (tm3199) and Δhrg-4 (tm2994) were obtained from the National Bioresource Project (Japan). The deletion strains were outcrossed eight times with the N2 Bristol strain before further study. The Δhrg-1Δhrg-4 double deletion strain was generated by crossing Δhrg-1 with Δhrg-4; progenies that were homozygous mutants and their wild-type brood mates were picked by single-worm PCR genotyping. For single-worm PCR, individual worms were lysed in 5 μl of lysis buffer (50 mm KCl, 10 mm Tris-HCl, 2.5 mm MgCl2, 0.45% Nonidet P-40, 0.45% Tween 20, 0.01% gelatin, and 1 mg/ml freshly prepared proteinase K) by freezing and heating (2 h at −80 °C, 1 h at 65 °C, and 30 min at 95 °C). Worm lysates were subjected to PCR reactions with primers CeHRG-1_del_f and CeHRG-1_del_b to detect a 650-bp fragment of hrg-1 in the Δhrg-1 strain, and with primers CeHRG-4_del_f and CeHRG-4_del_b to detect a 450-bp fragment of hrg-4 in the Δhrg-4 strain (supplemental Table S2).
PCR Genotyping of Worm Deletion Strains
For genomic DNA extraction, synchronized late-L4 worms grown at 4 μm hemin chloride in mCeHR-2 medium were harvested by centrifugation at 800 × g for 5 min. The worm pellet was washed with M9 buffer and lysed in disruption buffer (0.2 m NaCl, 0.1 m Tris, pH 8.0, 0.05 m EDTA, 0.5% SDS, 200 μg/ml proteinase K) by incubating at 65 °C for 1 h. Worm genomic DNA was then isolated by phenol:chloroform extraction and ethanol precipitation and was dissolved in sterile deionized water. For RNA isolation, worms were grown as described for genomic DNA extraction. Worm pellets were transferred into tubes with the lysing matrix C (MPBio) and disrupted using FastPrep-24 (MPBio) for 60 s at the 6.5 m/s setting. Total RNA was extracted using TRIzol reagent (Invitrogen), treated with RNase-free DNase for 1 h at 37 °C, and purified using the RNeasy kit (Qiagen). 2 μg of total RNA was used to synthesize first strand cDNA using a Superscript II first strand cDNA synthesis kit (Invitrogen). Primers CeHRG-1_del_f and CeHRG-1_del_b were used for hrg-1 deletion genotyping, and primers CeHRG-4_del_f and CeHRG-4_del_b were used for hrg-4 deletion genotyping, as described above.
Generation of Transgenic Worms
The translational fusion plasmids Phrg-4::CeHRG-4::YFP and Phrg-1::CeHRG-1::GFP were constructed using the multisite Gateway system (Invitrogen). The 3 kb upstream of hrg-4 and of hrg-1 gene was used as putative hrg-4 promoter (Phrg-4) and putative hrg-1 promoter (Phrg-1), respectively. The promoter, coding region, and 3′-UTR of all the rescue plasmids were sequenced before further analysis. The transgenic worms were generated by microparticle bombardment (18). 10 μg of the rescue plasmids was mixed with 5 μg of unc-119 rescue plasmid pDP#MM016B and co-bombarded into unc-119 worms using the PDS-1000 particle delivery system (Bio-Rad). At least two transgenic lines of each construct were analyzed for all the experiments.
Worm ZnMP Uptake Assay
Worms were harvested at L4 stage from NGM plates, washed twice with M9 buffer, and then transferred to 12-well plates. Approximately 100 worms/well were inoculated in mCeHR-2 medium supplemented with 1.5 μm hemin chloride and 10 μm zinc mesoporphyrin (ZnMP) for 16 h with continuous shaking at 20 °C. Images with a z resolution of 1 μm were acquired in a laser-scanning confocal microscope (LSM 510) with argon (458 nm and 488 nm) and He-Ne (543 nm and 633 nm) lasers (Zeiss).
Worm Growth Assay on RP523 Escherichia coli
The heme-deficient E. coli strain RP523 (19) was grown overnight in Luria-Bertani medium supplemented with 1 μm hemin and diluted at a ratio of 1:6 into fresh LB medium supplemented with different concentrations of hemin, cultivated for 5.5 h, heat-inactivated at 65 °C for 5 min, measured for A600, and diluted to a A600 = 0.5 with corresponding LB medium. Each 35-mm NGM plate was seeded with 200 μl of cell culture.
Synchronized L1 larvae of worms grown on OP50-seeded NGM plates were transferred onto NGM plates seeded with RP523 grown with 50 μm hemin and incubated at 20 °C for 4 days (P0). The P0 worms were bleached when they reached gravid stage. Forty synchronized F1 L1 larvae were transferred onto each RP523 plate and inoculated at 20 °C for 3–5 days. Five F1 gravid worms were placed onto each new appropriate RP523 plate and allowed to lay eggs (F2) for 12 h. The growth of F1 and F2 worms was recorded by differential interference contrast imaging and COPAS BioSort (Union Biometrica, Holliston, MA) when the wild-type worms reached young adult stage. For COPAS BioSort, about 200 worms for each sample were analyzed for both length (time of flight) and optical density (extinction).
Statistical Analysis
Statistical significance was calculated by using one-way analysis of variance with the Student-Newman-Keuls multiple comparison test in INSTAT version 3.01 (GraphPad, San Diego, CA). Data values were presented as mean ± S.E. A p value of <0.05 was considered as significant.
RESULTS
CeHRG-1 and CeHRG-4 Are Both Essential for Heme Homeostasis in C. elegans
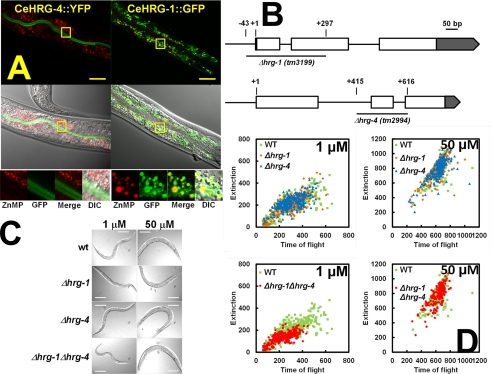
To determine the in vivo localization of HRG-1-related proteins, we generated transgenic worms that expressed CeHRG-1 and CeHRG-4 tagged at the C terminus with GFP. Both genes were expressed in the worm intestine with CeHRG-4 localized to the apical plasma membrane and CeHRG-1 primarily localized within an intracellular vesicular compartment (Fig. 1A). Metabolic labeling with the fluorescent heme analog ZnMP revealed that CeHRG-1 localized to vesicular membranes containing ZnMP fluorescence. These localization studies are consistent with our previous genetic knockdown experiments in worms, which showed that ZnMP accumulated in vesicles when CeHRG-1 was depleted while RNAi of CeHRG-4 reduced ZnMP entry into the intestine.
FIGURE 1.
Functional characterization of HRG-1-related proteins in C. elegans. A, transgenic worms stably expressing CeHRG-4::YFP or CeHRG-1::GFP translational fusions were cultivated in mCeHR-2 medium with 1.5 μm heme and 10 μm ZnMP for 16 h with constant shaking. The localization of GFP variants (green) and ZnMP (red) was examined using a Zeiss 710 confocal microscope. Boxed regions in the upper images have been enlarged for clarity and are shown at the bottom. B, schematic representation of Δhrg-1 (tm3199) and Δhrg-4 (tm2994). Open boxes represent open reading frames and gray boxes indicate untranslated regions. “+1” indicates the transcription start site. The deleted regions are represented by underscoring with the start and end indicated above. C, wild-type, Δhrg-1, Δhrg-4, and Δhrg-1Δhrg-4 worms were synchronized, placed on NGM plates seeded with RP523 bacteria grown at 50 μm hemin (P0), synchronized again, and transferred to RP523 plates of low (1 μm) and high (50 μm) hemin (F1) and follow the growth for two subsequent generations. Representative images of F2 progeny are shown. Scale bar, 50 μm. D, the sizes of worms in C were measured by COPAS BioSort. Each dot represents an individual worm: wild-type (green), Δhrg-1 (orange), Δhrg-4 (blue), and Δhrg-1Δhrg-4 (red). Time of flight and Extinction indicate the length and the optical density of worms, respectively.
To delineate the function of HRG-1-related proteins, we analyzed worms with a genetic deletion in hrg-1 and hrg-4. As shown in Fig. 1B, Δhrg-1 (tm3199) contains a 341-bp deletion, which results in the loss of exon 1 (including 43 bp upstream of the ATG start codon), intron 1, and a portion of exon 2. By contrast, Δhrg-4 (tm2994) has a 202-bp deletion, which eliminates part of intron 1, exon 2, and intron 2. PCR amplification of the genomic DNA from Δhrg-1 and Δhrg-4 confirmed the presence of these deletions (supplemental Fig. S1). Analysis by RT-PCR from RNA extracted from Δhrg-1 strain revealed no discernable product, although a fragment of ≈450 bp was amplified in RNA samples obtained from the Δhrg-4 deletion strain (supplemental Fig. S1A). DNA sequencing of the 450-bp product revealed that a truncated CeHRG-4 protein, containing the first two TMDs and six additional amino acids followed by several stop codons, could be synthesized if the Δhrg-4 mRNA were to be translated (supplemental Fig. S1B). Together, these results suggest that the Δhrg-1 strain is a null mutant, whereas the Δhrg-4 strain could be a hypomorph.
Because CeHRG-1 and CeHRG-4 are highly up-regulated when worms are grown with low environmental heme, we analyzed the deletion mutants for heme-dependent phenotypes (8, 20). Neither Δhrg-1 nor Δhrg-4 showed any obvious phenotypes compared with wild-type broodmates when fed with RP523, a mutant E. coli strain that does not synthesize heme (21). Feeding worms RP523 cultivated in different concentrations of heme permits external control of heme levels within the worm via dietary manipulations. Because the C. elegans genome contains multiple paralogs of HRG-1-related proteins that may function redundantly, we generated a Δhrg-1Δhrg-4 double mutant strain. Unlike the single mutants, the Δhrg-1Δhrg-4 mutant worms were significantly growth-retarded in low heme corresponding to a delay of up to three larval stages during development (Fig. 1, C and D). Most importantly, this growth delay was fully suppressed when double mutant worms were fed a heme-replete diet (Fig. 1, C and D). These genetic studies affirm that both hrg-1 and hrg-4 are essential for heme homeostasis in C. elegans when worms are maintained under minimal dietary heme conditions. Moreover, the additive phenotypes observed in the Δhrg-1Δhrg-4 double mutants indicate that the aberrant mRNA generated in Δhrg-4 worms has little or no function.
Expression of HRG-1-related Proteins in S. cerevisiae hem1Δ Strain
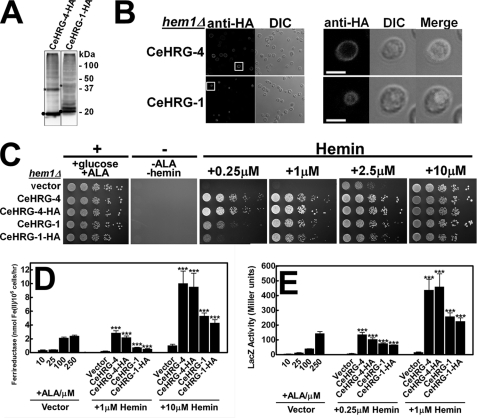
Our genetic investigations of C. elegans clearly indicate that CeHRG-1 and CeHRG-4 function in a concerted manner to maintain heme homeostasis in the worm. The presence of multiple paralogs of HRG-1-related proteins in C. elegans hinders a systematic investigation of the mechanisms employed by individual HRG-1-related proteins within the animal. To circumvent this complication, we sought to recapitulate the heme auxotrophy of C. elegans in a simpler, defined system. We exploited S. cerevisiae, because this single-cell eukaryote does not contain HRG-1-related protein homologs and utilizes exogenous heme poorly even in the absence of endogenous heme synthesis (12, 22). A forward genetic screen in yeast had recently identified the porphyrin transporter Pug1p using sensitive, functional assays established to assess heme transport activity in hem1Δ strain, a yeast mutant that is deficient in HEM1, which encodes for 5-aminolevulinate synthase, the first enzyme of the heme synthesis pathway (12). HRG-1-related proteins tagged at the C terminus with an HA epitope was expressed in hem1Δ under the control of the inducible GAL1 promoter. When both CeHRG-1 and CeHRG-4 are expressed in yeast, their proteins migrated at the predicted molecular weights, similar to our earlier expression analysis in mammalian cell lines (Fig. 2A) (8). Indirect immunofluorescence microscopy indicated that CeHRG-4-HA localized primarily on the plasma membrane, whereas CeHRG-1-HA was found on the yeast vacuolar membrane, a membrane compartment equivalent to the mammalian lysosome (Fig. 2B). These studies are consistent with expression studies of CeHRG-1 and CeHRG-4 proteins in worms (Fig. 1B) and mammalian cells (8).
FIGURE 2.
Improved hemin utilization in HRG-1-related proteins overexpressing yeast strain. A, immunoblotting was performed on transformants overexpressing C-terminal HA epitope-tagged CeHRG-4 or CeHRG-1 in the hem1(6D) strain. Predicted molecular masses of HRG-1-related proteins were detected as indicated by filled circles. Asterisks indicated putative dimers (15 μg/lane). B, the hem1Δ(6D) strain transformed with CeHRG-4-HA or CeHRG-1-HA were inoculated in SC medium supplemented with 250 μm ALA and subjected to indirect immunofluorescence microscopy. Rabbit anti-HA was used as the primary antibody, and an AlexaFluo568-conjugated goat anti-rabbit antibody was used as the secondary antibody. For clarity, the boxed regions in the left images have been enlarged to show the intracellular localization of HRG-1-related proteins. Scale bar, 5 μm. C, the hem1Δ(6D) strain transformed with empty vectors pYes-DEST52, CeHRG-4, and CeHRG-1, and their HA-tagged versions was cultivated overnight in SC medium without ALA and spotted in serial dilutions on SC plates supplemented with the indicated concentration of hemin (positive control: +0.4% glucose, +250 μm ALA, negative control: -ALA, -hemin). Plates were incubated at 30 °C for 3 days prior to imaging. D, the hem1Δfre1Δfre2ΔMET3-FRE1 strain was transformed with empty vector pYes-DEST52 and plasmids for HRG-1-related proteins expression. Transformants were cultivated in SC medium without ALA for 12 h, washed, and grown in SC medium supplemented with the indicated concentrations of hemin for 16 h. Cells were then washed and subjected to ferric reductase activity assay. E, the hem1Δ(6D) strain co-transformed with pCYC1-LacZ and empty vector pYes-DEST52 or plasmids for expressing HRG-1-related proteins were cultivated in SC medium without ALA for 12 h, washed, and grown in SC medium supplemented with indicated concentration of hemin for 12 h. Cells were then collected and lysed, and β-galactosidase activity was measured. Error bars represent the ± S.E. from three independent experiments. ***, p < 0.001.
CeHRG-1 and CeHRG-4 Mediate Heme Transport in Yeast
A hem1Δ yeast mutant is unable to grow unless it is supplemented with either ALA, the product of the 5-aminolevulinate synthase enzyme, or an excess of exogenous heme (supplemented as hemin chloride) in the growth medium. We used three independent assays established in hem1Δ yeast mutants (12) as follows: (a) First, we determined the rescue of growth of hem1Δ strain transformed with a plasmid expressing HRG-1-related proteins and grown on agar plates containing varying concentrations of heme (Fig. 2C). The degree of growth rescue and the heme concentration indicate relative uptake of heme by HRG-1-related proteins. (b) Second, we measured the activity of a heme-dependent enzyme, ferric reductase (15, 23), as an indicator of intracellular heme concentrations in a hem1Δfre1Δfre2ΔMET3-FRE1 strain (Fig. 2D). This strain lacks both native genes for FRE1 and FRE2, the major surface reductases that are sensitive to regulation by iron and copper, but has one of these ferric reductases (Fre1p) present under the control of the inducible MET3 promoter. Thus, any perturbations in cellular heme levels due to expression of the HRG-1-related proteins are reflected in a concomitant change in the ferric reductase enzyme activity (15, 18). (c) Third, we measured changes in the regulatory pools of heme by determining β-galactosidase activity from a CYC1::lacZ promoter-reporter fusion (Fig. 2E). Here, lacZ expression is dependent on Hap1–5, a transcription complex that binds heme and activates CYC1 expression in response to cellular changes in heme (24, 25). Thus, the level of β-galactosidase activity in CYC1::lacZ-expressing HRG-1-related proteins reflects the amount of intracellular heme available to activate the CYC1 promoter.
The hem1Δ strain transformed with pYES-DEST52 vector alone did not grow unless supplemented with 2.5–10 μm heme. By contrast, cells overexpressing CeHRG-1 and CeHRG-4, either with or without an HA epitope tag, showed a significant improvement in growth at both 0.25 μm and 1 μm heme (Fig. 2C). The imported heme was incorporated into cellular hemoproteins, because yeast expressing CeHRG-4 and CeHRG-1 showed >10-fold and 5-fold increases in ferrireductase activity, respectively (Fig. 2D). Furthermore, cells expressing HRG-1-related proteins had elevated cytoplasmic heme, because CeHRG-4 and CeHRG-1 expression increased the activity of CYC1::LacZ reporter by 10- to 20-fold (Fig. 2E). In addition, CeHRG-5 and CeHRG-6 also significantly and consistently enhanced the heme-dependent growth of the hem1Δ strain confirming our in silico observations that these putative paralogs transport heme. (supplemental Fig. S2). Together, these studies unequivocally demonstrate that multiple HRG-1-related proteins transport heme in the worm and lend validity to the use of the yeast as a heterologous system to dissect the mechanism of HRG-1-related proteins activity.
Identification of Conserved Heme-binding Ligands in HRG-1-related Proteins
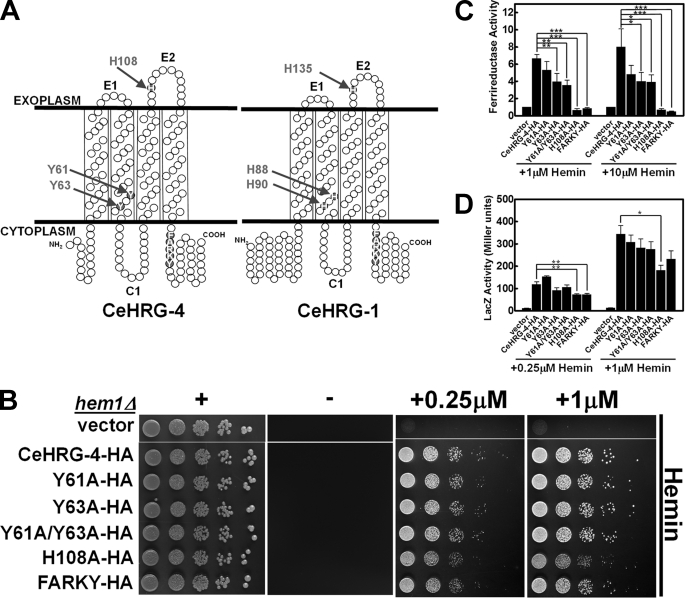
Although multiple sequence alignments reveal only 27% identity and 41% similarity between CeHRG-1 and CeHRG-4, an examination of the two protein sequences using membrane topology modeling revealed highly conserved amino acids that are located within the protein's predicted secondary structure that may function as axial ligands (His, Tyr, or Cys) for heme (Fig. 3A). CeHRG-1 and CeHRG-4 are predicted to contain four transmembrane domains (TMDs) and both the N and C termini reside in the cytoplasm (8). Conserved residues in CeHRG-1 include His-88 and His-90 in TMD2, His-135 in the second predicted exoplasmic (E2) loop, and in the C terminus a FARKY motif, a cluster of aromatic and basic amino acids that could interact and orient heme side chains (Fig. 3A and supplemental Fig. S3). It is noteworthy that the potential His heme ligands in the TMD2 of CeHRG-1 are substituted with Tyr (Tyr-61 and Tyr-63) in CeHRG-4 (Fig. 3A). Tyr, which preferentially binds ferric iron, forms heme ligands with a lower redox potential than His, and the coordination stabilizes heme and prevents it from carrying out oxidative chemistry (26, 27).
FIGURE 3.
Heme transport analysis of wild-type and mutant CeHRG-4. A, putative topology of CeHRG-4 and CeHRG-1 predicted by TMHMM 2.0 and SOSUI. The N and C termini are cytoplasmic, C1 is the cytoplasmic loop, and E1 and E2 are the exoplasmic loops. Potential heme interacting residues are shaded in red. B, the hem1Δ(6D) strain was transformed with empty vector pYes-DEST52, CeHRG-4-HA, and its mutants. A spot growth assay were performed with the indicated conditions. Plates were incubated at 30 °C for 3 days prior to imaging. C, the hem1Δfre1Δfre2ΔMET3-FRE1 strain was transformed with empty vector pYes-DEST52, CeHRG-4, and its mutants. A ferric reductase assay was performed with the indicated concentrations of hemin. Ferric reductase activity (nanomoles/106cells/min) was measured and normalized to the activity of vector. D, the hem1Δ(6D) strain co-transformed with pCYC1-LacZ and empty vector pYes-DEST52, CeHRG-4, or its mutants. β-Galactosidase activity was measured. Error bars represent the ±S.E. from three independent experiments. *, p < 0.05; **, p < 0.01; and ***, p < 0.001.
CeHRG-4 Function Requires a Histidine in the E2 Loop and the FARKY Motif in the C Terminus
To examine the role of specific residues for CeHRG-4 functions, we replaced Tyr-61, Tyr-63, His-108, and FARKY with Ala (Fig. 3A and supplemental Fig. S3). Immunoblotting and fluorescence microscopy studies revealed comparable expression and localization of the mutant and wild-type proteins (supplemental Fig. S4). Heme-dependent growth of transformed hem1Δ strain showed reduced growth only for the H108A mutant (Fig. 3B). By contrast, ferrireductase assays revealed a significant reduction in the activity for Tyr-63, His-108, and FARKY mutants, although only mutations in His-108 and the FARKY motif resulted in severe attenuation (Fig. 3C). Although β-galactosidase assays confirmed that substitutions of His-108 and FARKY with Ala in CeHRG-4 significantly decreased cytoplasmic heme levels at low heme, the Y63A mutant had consistently reduced activity but was statistically not significant (Fig. 3D). We found that the Y61A mutant did not alter the activity of heme-dependent reporters in any of the assays. The observation that only the H108A mutant showed impaired growth of hem1Δ strain may reflect the reduced sensitivity of the growth assay to residual heme transport activity in other mutants (Fig. 3, compare B with C and D). Collectively, these results strongly implicate His-108 and the FARKY motif, and to a lesser extent Tyr-63, in heme transport by CeHRG-4.
CeHRG-1 Function Requires a Histidine in TMD2 and the C Terminus FARKY Motif
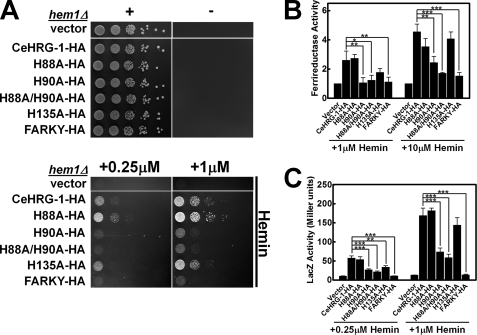
Based on the primary amino acid sequence homology and secondary structure prediction of CeHRG-1 and CeHRG-4, we set out to establish whether select conserved amino acids were essential for heme transport by CeHRG-1 (Fig. 3A and supplemental Fig. S3). Mutations in His-90 and the FARKY motif resulted in a dramatic reduction in growth compared with wild-type CeHRG-1 at low and high heme (Fig. 4A). By contrast, mutating His-135 in the putative E2 loop resulted only in a slight growth delay even though each of the mutants were expressed and localized to the yeast vacuolar membrane (supplemental Fig. S5). Consistent with the growth assays, substitutions of His-90 and FARKY with Ala resulted in significant reduction in ferrireductase and β-galactosidase activities in low and high heme conditions (Fig. 4, B and C). As observed in the growth assay, the H135A mutant showed reduction in ferrireductase and β-galactosidase activities at low heme, which was alleviated by increasing the heme concentrations. As observed for the Y61A mutant of CeHRG-4, the equivalent H88A mutant showed no obvious defects indicating that this ligand does not compensate in the H90A mutant. Our results suggest that, under low heme deplete conditions, CeHRG-1 predominantly requires His-90 in the predicted TMD2 and the C terminus FARKY motif, and to a lesser extent His-135 in the predicted E2 loop, for heme transport.
FIGURE 4.
Heme transport analysis of wild-type and mutant CeHRG-1. A, the hem1Δ(6D) strain was transformed with empty vector pYes-DEST52, CeHRG-1-HA, and its mutants. A spot growth assay were performed with the indicated conditions. Plates were incubated at 30 °C for 3 days prior to imaging. B, the hem1Δfre1Δfre2ΔMET3-FRE1 strain was transformed with empty vector pYes-DEST52, CeHRG-1, and its mutants. A ferric reductase assay was performed with the indicated concentrations of hemin. Ferric reductase activity (nanomoles/106cells/min) was measured and normalized to the activity of vector. C, the hem1Δ(6D) strain co-transformed with pCYC1-LacZ and empty vector pYes-DEST52, CeHRG-1, or its mutants. β-Galactosidase activity was measured. Error bars represent the ±S.E. from three independent experiments. *, p < 0.05; **, p < 0.01; and ***, p < 0.001.
To determine whether other potential heme-binding ligands (His, Tyr, or Cys) conserved in HRG-1-related proteins may compensate as secondary ligands in the site-directed mutants, we mutagenized His-14 and His-40 in the N terminus, His-41 and Tyr-67 in the E1 loop, and Cys-97 and Cys-127 in a highly conserved CLV motif within TMD3, of CeHRG-4 and CeHRG-1, respectively (supplemental Figs. S1 and S6). Mutating each of these residues to Ala did not significantly alter the function of either CeHRG-1 or CeHRG-4, suggesting the predominant heme ligands in both proteins are the His in the E2 loop, the His or Tyr in TMD2, and FARKY motif in the C terminus.
A Conserved Histidine in TMD2 Is Essential for Heme Transport by Human HRG-1
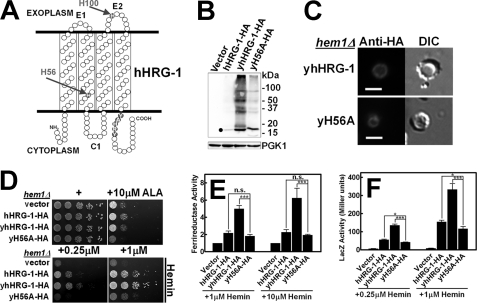
The human homolog of HRG-1-related proteins is only ≈20% identical to CeHRG-1 even though worm and human proteins co-localize to the endolysosomal compartment, and bind and transport heme, implying that these proteins are functional orthologs (8). To verify whether the results of our mechanistic studies of worm HRG-1-related proteins can be extended to humans, we expressed hHRG-1 in hem1Δ yeast. Immunoblotting of lysates from yeast cells expressing hHRG-1-HA revealed hHRG-1 migrating at the predicted molecular weight, albeit the steady-state protein level was significantly lower than CeHRG-1 and CeHRG-4 (Fig. 5B). To optimize the expression of hHRG-1 in yeast, we generated a synthetic open-reading frame (yhHRG-1), which was modified for codon usage bias in S. cerevisiae; the Codon Adaptation Index was upgraded from 0.51 to 0.90 for maximal expression. Steady-state levels for optimized yhHRG-1-HA were at least 10-fold greater than the unoptimized hHRG-1-HA (Fig. 5B). This difference in expression was concomitant with the significantly greater ability of yhHRG-1-HA to rescue the growth of hem1Δ yeast (10,000-fold compared with vector control), stimulate Fre1p enzymatic activity, and induce CYC1::lacZ reporter activity (Fig. 5, D--F). Indirect immunofluorescence microscopy indicated that, similar to CeHRG-1, the human homolog also localized primarily to the yeast vacuole (Fig. 5C).
FIGURE 5.
Heme transport analysis of wild-type and mutant hHRG-1. A, putative topology of hHRG-1 predicted by TMHMM 2.0 and SOSUI. The N and C termini are cytoplasmic, C1 is the cytoplasmic loop, and E1 and E2 are the exoplasmic loops. Potential heme interacting residues are shaded in red. B, immunoblotting was performed on transformants overexpressing C-terminal HA epitope-tagged hHRG-1 or yhHRG-1 and its H56A mutant (yH56A-HA) in the hem1Δ(6D) strain. The predicted molecular mass of hHRG-1 was detected as indicated by filled circles. The endogenous expression level of phosphoglycerate kinase 1 (PGK1) was used as loading control (30 μg of total protein/lane). C, immunofluorescence microscopy results of yhHRG-1-HA and yH56A-HA mutant in the hem1Δ(6D) strain. Rabbit anti-HA was used as the primary antibody, and an AlexaFluo568-conjugated goat anti-rabbit antibody was used as the secondary antibody. Scale bar, 5 μm. D, the hem1Δ(6D) strain was transformed with empty vector pYes-DEST52, hHRG-1 and its mutants. A spot growth assay was performed with the indicated conditions. Plates were incubated at 30 °C for 3 days prior to imaging. E, the hem1Δ fre1Δ fre2Δ MET3-FRE1 strain was transformed with empty vector pYes-DEST52, hHRG-1, and its mutants. A ferric reductase assay was performed with the indicated concentrations of hemin. Ferric reductase activity (nanomoles/106 cells/min) was measured and normalized to the activity of vector. F, the hem1Δ(6D)) strain co-transformed with pCYC1-LacZ and empty vector pYes-DEST52, CeHRG-1, or its mutants. β-Galactosidase activity was measured. Error bars represent the ±S.E. from three independent experiments. *, p < 0.05; **, p < 0.01; and ***, p < 0.001.
Despite low identity at the amino acid sequence level between the human and worm proteins, several features are remarkably conserved in HRG-1-related proteins. Namely, both homologs localized to similar endocytic compartments, showed similar predicted four TMDs, and possess conserved amino acids that were topologically preserved. Potential ligands at the corresponding position in hHRG-1 were His-56 in TMD2, His-100 in the E2 loop, and YAHRY in the C terminus (Fig. 5A). As proof-of-principle, we generated an H56A mutant in hHRG-1. The mutant protein expressed and localized to the vacuole but had significantly reduced function consistently in all three assays irrespective of heme concentrations (Fig. 5, B–F). These results strongly support our proposition that the overall mechanism for heme transport by HRG-1-related proteins is likely conserved between worms and humans.
We next determined whether the assays employed to determine the heme import function of hHRG-1 can be extended to FLVCR2, another transporter recently postulated to be a heme importer in humans cells (28). hFLVCR2 was abundantly expressed in hem1Δ yeast and localized primarily to the plasma membrane. However, hFLCVR2 did not enhance heme-dependent growth hem1Δ yeast even at 10 μm heme (supplemental Fig. S7). These results suggest that, in our assay conditions, hFLVCR2 does not function as a heme importer.
DISCUSSION
Our genetic studies in worms reveal a definitive growth phenotype only when both hrg-1 and hrg-4 are deleted (Fig. 1, C and D). This is not surprising given that worms are solely dependent on environmental heme for growth and development. Results from the hem1Δ yeast growth assays clearly demonstrate that, in addition to CeHRG-1 and CeHRG-4, two additional paralogs exist for heme transport in C. elegans. Thus, multiple backup systems exist in worms that ensure adequate heme is acquired for sustenance and transfer to their progeny (21). From an experimental standpoint, these backup systems, however, occlude a thorough mechanistic analysis of HRG-1-related proteins using the C. elegans model. To recapitulate the heme auxotrophy of worms, we exploited the hem1Δ yeast strain, which is unable to synthesize heme and lacks the ability to utilize exogenous heme at low concentrations. In the current study, we employed independent functional assays in yeast to probe the molecular characteristics of heme transport by HRG-1-related proteins (12). Our results reveal that, despite sharing only 27% identity and localizing to different subcellular compartments, the overall mechanisms for heme transport employed by CeHRG-1 and CeHRG-4 are similar. We attribute the greater activity of CeHRG-4 in utilizing exogenous heme in the yeast reporter assays to its plasma membrane localization. By contrast, although CeHRG-1 predominantly localizes to the vacuolar membrane, it still has significant ability to utilize exogenous heme. It is conceivable that, as observed in mammalian cells (8), a portion of CeHRG-1 traffics to the plasma membrane.
Based on data from our assays, we propose a mechanistic model for heme import by HRG-1-related proteins. A histidine in the E2 loop on the extracellular/luminal side binds and transfers heme to a histidine, or a tyrosine at the synonymous position in CeHRG-4, in TMD2 within the channel. The heme is subsequently translocated to the cytoplasmic side facilitated by the FARKY (YAHRY in humans) motif in the C-terminal tail. The aromatic and positively charged amino acids in this motif may serve as heme ligands and stabilize or orient the vinyl and propionic acid side chains. A precedent for such a mechanism was recently established for the Helicobacter hepaticus CcsBA, a multipass ten-transmembrane channel (29). Heme is transported from the cytoplasm into the bacterial periplasm by CcsBA, a cytochrome c synthetase, by first binding to two histidines, defined as a weak heme binding site within the TMDs, followed by translocation to a periplasmic heme binding region comprising two histidines and a WWD domain (29). This mechanism of heme transfer prevents oxidation of the heme-iron and allows for proper orientation of the heme for covalent bond formation with the reduced apocytochrome. Our previous studies show that HRG-1-related proteins migrate as dimers and trimers on nondenaturing PAGE, raising the possibility that HRG-1-related protein functions as a multimer (8). Thus, optimal transport function may be dependent on the cumulative contribution from individual heme-binding ligands from each subunit. Such a situation is observed with the three-TMD copper transporter Ctr1, which functions as a symmetric trimer to form a channel lined with ligands for copper binding and transport (30).
Based on the degree of functional rescue in the yeast reporter assays for each mutant of the HRG-1-related proteins, we postulate that a hierarchy may exist for binding heme among the ligands. HRG-1-related proteins may contain dual binding sites or heme translocation: a weak low affinity site followed by a high affinity primary substrate binding site. This concept is supported by the recent crystal structure of the LeuT leucine transporter and biochemical characterization of CcsBA heme transporter (29, 31–33). Both proteins possess a low affinity, transiently occupied substrate binding site that works in conjunction with a high affinity primary binding site to promote substrate movement and conformational gating.
Site-directed mutation reveals that His-90 in CeHRG-1 (His-56 in hHRG-1) is replaced by a synonymous Tyr-63 in CeHRG-4 in TMD2. Although both His and Tyr are heme-binding ligands, replacing His with a Tyr as a ligand in hemoproteins decreases the reactivity of heme possibly because of the increased anionic character of the phenolic group in Tyr, which preferentially stabilizes oxidized heme (ferric and ferryl-oxo) (26, 27). Primary amino acid sequence alignment of putative HRG-1-related proteins from other species reveals that the His in TMD2 is highly conserved in the majority of species examined. However, in a small number of HRG-1 homologs, the His is substituted by a Tyr, as in the case of the HRG-1 homolog from the human pathogen Leishmania amazonensis, which functions as a heme transporter (not shown). C. elegans is a heme auxotroph and must acquire heme from the environment to sustain growth and development. Under physiological conditions, this soil-dwelling bacteriovorus nematode likely ingests heme from multiple sources to maximize heme utilization through the concerted actions of both CeHRG-1 and CeHRG-4 in the worm intestine. In its natural habitat, CeHRG-4 on the intestinal plasma membrane might encounter oxidized heme more readily than CeHRG-1, which might transport heme derived from the digestion of ingested hemoproteins in the lysosome, an organelle equivalent to the yeast vacuole, a storage compartment for iron, and plausibly, heme (34, 35).
What drives translocation of heme through the HRG-1-related transporters? One possible mechanism could be heme concentration gradients generated by interactions of the transporter with one or more downstream binding partners. In Gram-positive bacteria, heme transfer from IsdA to IsdC, cell wall-anchored proteins in Staphylococcus aureus, is driven by kinetics and transient stereospecific weak “handclasp” interactions (36, 37). This is in stark contrast to Gram-negative bacteria, which utilize high affinity protein-protein interactions for heme transfer (6, 7, 38). Another mechanism could be coupling with an energetically favorable flow or counterflow of ion gradients generated by primary transporters, a feature characteristic of permeases. Indeed, a recent study demonstrated that human HRG-1 associates with the V-ATPase proton pump (39), suggesting the possibility that a proton gradient generated by V-ATPases may drive heme transport, similar to the endocytic calcium transporters in yeast (40).
Is the mechanism for heme transport by HRG-1-related proteins shared with other metazoan heme transporters? FLVCR2, a member of the major facilitator superfamily, was recently reported to import heme in mammalian cells (28). Although we did not observe heme import in our assay conditions, given the high degree of homology between FLVCR2 and the heme effluxer FLVCR1 (41–43), it is possible that FLVCR2 may export heme. Hemopexin facilitates heme export by FLVCR1 by binding to a 69-amino acid peptide (residues 132–201), which contains two exoplasmic loops and two TMDs (44). In silico examination of the membrane topology of FLVCR2 predicts a 12-TMD protein with several strategically placed, evolutionarily conserved, heme-binding ligands in the exoplasmic and cytoplasmic loops and within the TMDs. Genetic studies have unequivocally demonstrated that FLVCR1 is essential for development; FLVCR1-null mice lack definitive erythropoiesis and are embryonic-lethal (45). A structure-function analysis of FLVCR heme transporters will lay the groundwork for establishing whether the overall mechanism for heme transport identified for HRG-1-related proteins is also conserved in FLVCR. Although the physiological role for heme import by hHRG-1 in humans is still unclear, concurrent genetic studies with the mouse equivalent of Hrg-1 will shed significant insight on the biological role of heme transporters in mammalian growth and development.
Supplementary Material
Acknowledgments
We thank H. Dailey for critical discussions and reading of the manuscript, T. Samuel for confocal microscopy of transgenic worms, J. Hanover for use of the COPAS BioSort, and the National Bioresource Project and S. Mitani for the hrg-1 and hrg-4 strains.
This work was supported, in whole or in part, by National Institutes of Health Grants R01DK74797 and R01DK85035 (to I. H.).

This article contains supplemental Figs. S1–S7, Tables S1 and S2, and additional references.
- HRG
- heme-responsive gene
- SC
- synthetic complete
- FLVCR
- feline leukemia virus subgroup C cellular receptor
- TMD
- transmembrane domain
- E2 loop
- the second exoplasmic loop
- FRE
- ferric reductase
- CYC1
- cytochrome c, isoform 1
- Ctr1
- copper transporter 1
- CcsBA
- cytochrome c-type biogenesis proteins
- NGM
- nematode growth medium
- mCeHR
- modified C. elegans habitation and reproduction
- ALA
- δ-aminolevulinic acid
- ZnMP
- zinc mesoporphyrin.
REFERENCES
- 1. Severance S., Hamza I. (2009) Trafficking of heme and porphyrins in metazoa. Chem. Rev. 109, 4596–4616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. West A. R., Oates P. S. (2008) Mechanisms of heme iron absorption. Current questions and controversies. World J. Gastroenterol. 14, 4101–4110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Maines M. D. (2005) The heme oxygenase system. Update 2005. Antioxid. Redox Signal. 7, 1761–1766 [DOI] [PubMed] [Google Scholar]
- 4. Schultz I. J., Chen C., Paw B. H., Hamza I. (2010) Iron and porphyrin trafficking in heme biogenesis. J. Biol. Chem. 285, 26753–26759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Layer G., Reichelt J., Jahn D., Heinz D. W. (2010) Structure and function of enzymes in heme biosynthesis. Protein Sci. 19, 1137–1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tong Y., Guo M. (2009) Bacterial heme-transport proteins and their heme-coordination modes. Arch. Biochem. Biophys. 481, 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wilks A., Burkhard K. A. (2007) Heme and virulence. How bacterial pathogens regulate, transport and utilize heme. Nat. Prod. Rep. 24, 511–522 [DOI] [PubMed] [Google Scholar]
- 8. Rajagopal A., Rao A. U., Amigo J., Tian M., Upadhyay S. K., Hall C., Uhm S., Mathew M. K., Fleming M. D., Paw B. H., Krause M., Hamza I. (2008) Haem homeostasis is regulated by the conserved and concerted functions of HRG-1 proteins. Nature 453, 1127–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rao A. U., Carta L. K., Lesuisse E., Hamza I. (2005) Lack of heme synthesis in a free-living eukaryote. Proc. Natl. Acad. Sci. U.S.A. 102, 4270–4275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Severance S., Rajagopal A., Rao A. U., Cerqueira G. C., Mitreva M., El-Sayed N. M., Krause M., Hamza I. (2010) Genome-wide analysis reveals novel genes essential for heme homeostasis in Caenorhabditis elegans. PLoS Genet. 6, e1001044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Crisp R. J., Pollington A., Galea C., Jaron S., Yamaguchi-Iwai Y., Kaplan J. (2003) Inhibition of heme biosynthesis prevents transcription of iron uptake genes in yeast. J. Biol. Chem. 278, 45499–45506 [DOI] [PubMed] [Google Scholar]
- 12. Protchenko O., Shakoury-Elizeh M., Keane P., Storey J., Androphy R., Philpott C. C. (2008) Role of PUG1 in inducible porphyrin and heme transport in Saccharomyces cerevisiae. Eukaryot. Cell 7, 859–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sherman F. (1991) Getting started with yeast. Methods Enzymol. 194, 3–21 [DOI] [PubMed] [Google Scholar]
- 14. Ito H., Fukuda Y., Murata K., Kimura A. (1983) Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 153, 163–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dancis A., Klausner R. D., Hinnebusch A. G., Barriocanal J. G. (1990) Genetic evidence that ferric reductase is required for iron uptake in Saccharomyces cerevisiae. Mol. Cell. Biol. 10, 2294–2301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Burke D., Dawson D., Stearns T. (2000) Methods in Yeast Genetics, Cold Spring Harbor Press, Cold Spring Harbor [Google Scholar]
- 17. Nass R., Hamza I. (2007) The nematode C. elegans as an animal model to explore toxicology in vivo. Solid and axenic growth culture conditions and compound exposure parameters. In: Current Protocols in Toxicology (Maines M. D., Costa L. G., Hodgson E., Reed D. J., Sipes I. G., eds) John Wiley & Sons, Inc., New York: pp 1.9.1–1.9.17 [DOI] [PubMed] [Google Scholar]
- 18. Georgatsou E., Alexandraki D. (1994) Two distinctly regulated genes are required for ferric reduction, the first step of iron uptake in Saccharomyces cerevisiae. Mol. Cell. Biol. 14, 3065–3073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Li J. M., Umanoff H., Proenca R., Russell C. S., Cosloy S. D. (1988) Cloning of the Escherichia coli K-12 hemB gene. J. Bacteriol. 170, 1021–1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sinclair J., Hamza I. (2010) A novel heme-responsive element mediates transcriptional regulation in Caenorhabditis elegans. J. Biol. Chem. 285, 39536–39543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chen C., Samuel T. K., Sinclair J., Dailey H. A., Hamza I. (2011) An intercellular heme-trafficking protein delivers maternal heme to the embryo during development in C. elegans. Cell 145, 720–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Protchenko O., Rodriguez-Suarez R., Androphy R., Bussey H., Philpott C. C. (2006) A screen for genes of heme uptake identifies the FLC family required for import of FAD into the endoplasmic reticulum. J. Biol. Chem. 281, 21445–21457 [DOI] [PubMed] [Google Scholar]
- 23. Shatwell K. P., Dancis A., Cross A. R., Klausner R. D., Segal A. W. (1996) The FRE1 ferric reductase of Saccharomyces cerevisiae is a cytochrome b similar to that of NADPH oxidase. J. Biol. Chem. 271, 14240–14244 [DOI] [PubMed] [Google Scholar]
- 24. Guarente L., Mason T. (1983) Heme regulates transcription of the CYC1 gene of S. cerevisiae via an upstream activation site. Cell 32, 1279–1286 [DOI] [PubMed] [Google Scholar]
- 25. Hon T., Dodd A., Dirmeier R., Gorman N., Sinclair P. R., Zhang L., Poyton R. O. (2003) A mechanism of oxygen sensing in yeast. Multiple oxygen-responsive steps in the heme biosynthetic pathway affect Hap1 activity. J. Biol. Chem. 278, 50771–50780 [DOI] [PubMed] [Google Scholar]
- 26. Goodwin D. C., Rowlinson S. W., Marnett L. J. (2000) Substitution of tyrosine for the proximal histidine ligand to the heme of prostaglandin endoperoxide synthase 2. Implications for the mechanism of cyclooxygenase activation and catalysis. Biochemistry 39, 5422–5432 [DOI] [PubMed] [Google Scholar]
- 27. Liu Y., Moénne-Loccoz P., Hildebrand D. P., Wilks A., Loehr T. M., Mauk A. G., Ortiz de Montellano P. R. (1999) Replacement of the proximal histidine iron ligand by a cysteine or tyrosine converts heme oxygenase to an oxidase. Biochemistry 38, 3733–3743 [DOI] [PubMed] [Google Scholar]
- 28. Duffy S. P., Shing J., Saraon P., Berger L. C., Eiden M. V., Wilde A., Tailor C. S. (2010) The Fowler syndrome-associated protein FLVCR2 is an importer of heme. Mol. Cell. Biol. 30, 5318–5324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Frawley E. R., Kranz R. G. (2009) CcsBA is a cytochrome c synthetase that also functions in heme transport. Proc. Natl. Acad. Sci. U.S.A. 106, 10201–10206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. De Feo C. J., Aller S. G., Siluvai G. S., Blackburn N. J., Unger V. M. (2009) Three-dimensional structure of the human copper transporter hCTR1. Proc. Natl. Acad. Sci. U.S.A. 106, 4237–4242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Merchant S. S. (2009) His protects heme as it crosses the membrane. Proc. Natl. Acad. Sci. U.S.A. 106, 10069–10070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Diallinas G. (2008) Biochemistry. An almost-complete movie. Science 322, 1644–1645 [DOI] [PubMed] [Google Scholar]
- 33. Singh S. K., Piscitelli C. L., Yamashita A., Gouaux E. (2008) A competitive inhibitor traps LeuT in an open-to-out conformation. Science 322, 1655–1661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Raguzzi F., Lesuisse E., Crichton R. R. (1988) Iron storage in Saccharomyces cerevisiae. FEBS Lett. 231, 253–258 [DOI] [PubMed] [Google Scholar]
- 35. Bode H. P., Dumschat M., Garotti S., Fuhrmann G. F. (1995) Iron sequestration by the yeast vacuole. A study with vacuolar mutants of Saccharomyces cerevisiae. Eur. J. Biochem. 228, 337–342 [PubMed] [Google Scholar]
- 36. Grigg J. C., Ukpabi G., Gaudin C. F., Murphy M. E. (2010) Structural biology of heme binding in the Staphylococcus aureus Isd system. J. Inorg. Biochem. 104, 341–348 [DOI] [PubMed] [Google Scholar]
- 37. Villareal V. A., Spirig T., Robson S. A., Liu M., Lei B., Clubb R. T. (2011) Transient weak protein-protein complexes transfer heme across the cell wall of Staphylococcus aureus. J. Am. Chem. Soc. 133, 14176–14179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Cescau S., Cwerman H., Létoffé S., Delepelaire P., Wandersman C., Biville F. (2007) Heme acquisition by hemophores. Biometals 20, 603–613 [DOI] [PubMed] [Google Scholar]
- 39. O'Callaghan K. M., Ayllon V., O'Keeffe J., Wang Y., Cox O. T., Loughran G., Forgac M., O'Connor R. (2010) Heme-binding protein HRG-1 is induced by insulin-like growth factor I and associates with the vacuolar H+-ATPase to control endosomal pH and receptor trafficking. J. Biol. Chem. 285, 381–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yadav J., Muend S., Zhang Y., Rao R. (2007) A phenomics approach in yeast links proton and calcium pump function in the Golgi. Mol. Biol. Cell 18, 1480–1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Brown J. K., Fung C., Tailor C. S. (2006) Comprehensive mapping of receptor-functioning domains in feline leukemia virus subgroup C receptor FLVCR1. J. Virol. 80, 1742–1751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lipovich L., Hughes A. L., King M. C., Abkowitz J. L., Quigley J. G. (2002) Genomic structure and evolutionary context of the human feline leukemia virus subgroup C receptor (hFLVCR) gene. Evidence for block duplications and de novo gene formation within duplicons of the hFLVCR locus. Gene 286, 203–213 [DOI] [PubMed] [Google Scholar]
- 43. Quigley J. G., Yang Z., Worthington M. T., Phillips J. D., Sabo K. M., Sabath D. E., Berg C. L., Sassa S., Wood B. L., Abkowitz J. L. (2004) Identification of a human heme exporter that is essential for erythropoiesis. Cell 118, 757–766 [DOI] [PubMed] [Google Scholar]
- 44. Yang Z., Philips J. D., Doty R. T., Giraudi P., Ostrow J. D., Tiribelli C., Smith A., Abkowitz J. L. (2010) Kinetics and specificity of feline leukemia virus subgroup C receptor (FLVCR) export function and its dependence on hemopexin. J. Biol. Chem. 285, 28874–28882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Keel S. B., Doty R. T., Yang Z., Quigley J. G., Chen J., Knoblaugh S., Kingsley P. D., De Domenico I., Vaughn M. B., Kaplan J., Palis J., Abkowitz J. L. (2008) A heme export protein is required for red blood cell differentiation and iron homeostasis. Science 319, 825–828 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.