Background: ChR2 is a light-gated ion channel allowing fast non-invasive control of cell membrane potential.
Results: We combined bioinformatic modeling and electrophysiology to infer structure/function details on ChR2.
Conclusion: We show a complete structural model of the channel, describe the ion-conducting pathway and identify key residues involved in ionic permeability and in photoactivation.
Significance: These results expand our knowledge on the structural determinants of ChR2.
Keywords: Biophysics, Calcium Channels, Protein Structure, Rhodopsin, Sodium Channels, Channelrhodopsin-2, Light-activated Channels, Optogenetics, Structure Modeling
Abstract
Channelrhodopsin-2 (ChR2) is a light-gated cation channel widely used as a biotechnological tool to control membrane depolarization in various cell types and tissues. Although several ChR2 variants with modified properties have been generated, the structural determinants of the protein function are largely unresolved. We used bioinformatic modeling of the ChR2 structure to identify the putative cationic pathway within the channel, which is formed by a system of inner cavities that are uniquely present in this microbial rhodopsin. Site-directed mutagenesis combined with patch clamp analysis in HeLa cells was used to determine key residues involved in ChR2 conductance and selectivity. Among them, Gln-56 is important for ion conductance, whereas Ser-63, Thr-250, and Asn-258 are previously unrecognized residues involved in ion selectivity and photocurrent kinetics. This study widens the current structural information on ChR2 and can assist in the design of new improved variants for specific biological applications.
Introduction
Channelrhodopsins are light-gated ion channels that form the phototactic machinery of the unicellular alga Chlamydomonas reinhardtii (1). They are seven-transmembrane domains proteins and contain the light-isomerizable chromophore all-trans-retinal covalently bound to the protein via a protonated Schiff base.
Channelrhodopsin-2 (ChR2)3 is a light-activated cation channel (2, 3), which can be used to control with millisecond resolution Na+ permeability of the cell membrane. Exogenous expression of ChR2 has been exploited to achieve non-invasive control of membrane potential in neuronal cells in the intact brain (4–9) and, more recently, in cardiac cells and tissue (10, 11). A number of new channelrhodopsin variants have recently been generated with the aim to modify spectral properties of the photoprotein, ionic conductance, as well as efficiency in membrane localization and protein expression (12, 13). For instance, introduction of H134R mutation in the ChR2 sequence has yielded a variant with increased photocurrent amplitude (14), and further mutation of the Glu-123 residue to threonine led to faster off-kinetics (15). Other investigators have identified and modified those residues involved in ChR2 photocycle (15–17). Recently, the E123T/T159C double mutant that combines both large photocurrents and accelerated photocycle was generated (18).
To allow the production of new variants with characteristics suited for improved biological applications, it is essential to identify the residues involved in the basic functions of ChR2, such as the photocurrent kinetics and ionic selectivity. The identification of the determinants of ion conductance and selectivity is of great interest as it would allow the design of mutations able to optimize ChR2 properties in accordance with the application of interest and allow the widening of ChR2 application fields.
To get a deeper understanding of the ChR2 structure and of the mechanism of cation conductance, we have developed a bioinformatic model of ChR2 from C. reinhardtii by threading and homology modeling. This allowed us to identify two chambers that are part of the ion pathway inside the channel. The identification of putative important residues for ion conductance and selectivity was validated by patch clamp analysis of HeLa cells expressing the ChR2 mutants. A single point mutation (Q56E) of residues exposed in these two chambers allowed us to decrease conductance to Na+, the main ChR2-permeating ion. Three variants with a single amino acid mutation displayed a different Ca2+-to-Na+ conductance ratio (S63D, T250E, and N258D) and faster off-kinetics (T250E). These results support the model and identify residues along the cation pathway, thus adding information for engineering new variants with different ion selectivity and photocurrent kinetics.
EXPERIMENTAL PROCEDURES
Molecular Biology and Expression of ChR2 and Its Variants in HeLa Cells
PCR-based site directed mutagenesis was performed with oligonucleotides carrying the specific mutation using Pfu DNA polymerase (Stratagene, La Jolla, CA). All constructs were verified by sequencing.
HeLa cells devoid of connexins (K. Willecke, University of Bonn, Germany) (19) were used to minimize electrical noise and avoid electrical coupling between cells. Cells were grown in DMEM supplemented with 10% FBS, 2 mm glutamine, and 1% penicillin/streptomycin under a 5% CO2 atmosphere at 37 °C. Transient transfection of cells was obtained with Lipofectamine (Invitrogen, Paisley, UK) reagent following the manufacturer's instructions. Cells were used for experiments 40–72 h after transfection, when the largest fraction of the protein reached the plasma membrane. Transfected cells were identified based on their fluorescence when imaged using a red fluorescence cube (excitation, 515 ± 35 nm; emission, 590 long pass).
To estimate protein expression, cells were fixed with 4% paraformaldehyde 48 h after transfection and imaged with an inverted confocal microscope (TCS SP5, Leica Mikrosysteme Vertrieb GmbH) equipped with oil immersion objectives (Leica 63×, 1.4 NA). Cells transfected with ChR2(H134R)-mCherry and respective mutants were excited by the 561-mm laser line, and emission was collected between 570 and 660 nm. For intracellular Ca2+ imaging, Fluo-4 (1 μm, Molecular Probes, Invitrogen) was loaded at 37 °C for 30 min. Cells were then washed and transferred into an extracellular solution consisting of 80 mm Ca2+, 5 mm Na+, 3 mm KCl, 135 mm N-methyl-d-glucamine, 10 mm Hepes, 20 mm glucose, adjusted at pH = 7.4. Fluo-4 and ChR2 were excited with 100-ms pulses of 490 ± 20 nm, and the emitted light was detected with a CCD camera at 510 ± 20 nm.
Electrophysiology
Whole cell patch clamp experiments were performed at room temperature (∼23 °C) on single cells visualized with an inverted microscope (Olympus IX50, Tokyo, Japan). Photocurrents were activated with a 500-ms pulse of 470-nm light and recorded using an EPC-7 amplifier (HEKA Electronik, Lambrecht, Germany) and the Axon Instruments pClamp10 software. Data were sampled at 10 kHz.
Patch pipettes were prepared by pulling borosilicate glass capillaries (1.5-mm outer diameter and 1.16-mm inner diameter, Harvard Apparatus Ltd.) using a micropipette puller (Narishige). Pipette resistance was 2.3–3.5 megaohms when filled with intracellular solution.
Extracellular solutions contained (in mm): 145 NaCl, 3 KCl, 5 N-methyl-d-glucamine, 10 Hepes, 20 Glucose; pH was adjusted to 7.35 with HCl (solution 1). Ca2+ photocurrents were recorded in (in mm): 3 KCl, 135 N-methyl-d-glucamine, 10 CaCl2, 10 Hepes, 20 glucose; pH was adjusted to 7.35 with HCl (solution 2). Intracellular solution was (in mm): 120 CsCl, 10 triethanolamine-Cl, 10 Hepes, 10 EGTA, 4 MgATP, 0.1 NaGTP; pH was adjusted to 7.2 with CsOH.
Light stimulation was provided by a 100-watt mercury lamp equipped with a mechanical shutter (Uniblitz, Vincent Associates, Rochester, NY) with the light filtered through a 480 ± 10-nm band-pass filter (Chroma, Bellows Falls, VT) and reflected off a mirror to the specimen through a 40×, 1.4 NA oil immersion objective (Olympus). This resulted in light power at the sample plan of 0.45 milliwatt/mm2. ChR2 activation spectra were acquired using a monochromator (Polychrome IV, Till Photonics GmbH) triggered via the D/A port of the Digidata interface driven by pClamp 10 (Axon Instruments).
Structure Modeling
ChR2 1–315 models were obtained using the Protein Homology/analogY Recognition Engine (Phyre) Server (20) and the Swiss-Model server (21). The models are based on the following templates: 1m0kA (model 1, 7.0 × 10−26), 1xioA (model 2, 6.2 × 10−27), 1h2sA (model 3, 1.3 × 10−26), and 1h2sA (model 4, 2.0 × 10−44). Retinal was added in the final models by juxtaposition. The Protein3Dfit server was used for structural superposition (22), and the PyMOL viewer was used for visualization (Schrödinger LLC, Portland, OR) (23). The models underwent energy minimization and a short molecular dynamics simulation (100 ps) with constrained α-carbon position to allow the side chain to relax. Both energy minimization and molecular dynamics studies were performed using the Amber94 force field (24) and the Gromacs molecular dynamics package (25). Energy minimization was performed in vacuo, whereas for molecular dynamics, we solvated the proteins using an explicit solvent model (TIP3) and an ion concentration of 0.15 m NaCl. The system was then simulated under periodic boundary conditions at 300 K and 1 atm using the Berendsen thermostat and barostat (26). To investigate the effect of the R120A mutation, we performed unrestrained molecular dynamics for model 2 and for the same model in which Arg-120 was mutated into an alanine. The dynamics of the two systems were followed for 1 ns to let the side chains relax, without the restraint on the α-carbon positions. The simulation conditions were the same as the equilibration described above.
RESULTS
ChR2 Bioinformatic Models
To investigate the structural features of ChR2, we developed four models of the protein by both threading and homology modeling of the fragment 1–315 of ChR2(H134R) from C. reinhardtii. ChR2 models 1, 2, and 3 were obtained by the Phyre Server (20), and model 4 was obtained by the Swiss-Model server (21). In all models, only the central part of the sequence is represented (residues 52–273 in models 1, 2, and 3 and residues 56–263 in model 4), resulting in the classic rhodopsin fold based on seven-transmembrane antiparallel α helices, predicted to have an extracellular N terminus and an intracellular C terminus (supplemental Fig. S1, A and B). Residues composing the transmembrane helices are indicated in supplemental Table S1.
The loops connecting such helices are short (≤10 amino acids) except for the α2-α3 loop, which in most models is up to 16 residues long. This extended loop, which includes a short helix in model 2, is located on the extracellular side of the membrane, on the same side as the N-terminal extracellular region (the first 50 residues at the N-terminal are not modeled). The α2-α3 loop and the N terminus are rich in hydrophobic residues. In HR, a similar structure is present that has been proposed to function as a regulator of the ion flux (6). Although there is general agreement on the residues being part of the last five TM helices of ChR2, sequence homology of helices 1 and 2 with other rhodopsins is low. We took into account structural information from homology and threading modeling to obtain a structure-based alignment of these two helices (supplemental Fig. S2).
Sequence alignment and structure superimposition with HR, BR, and a third light-driven proton pump with known structure, archaerhodopsin (AR) (27, 28) (Protein Data Bank accession numbers: 3a7k, 2zzl, and 2ei4, respectively), were used to investigate the structural features of ChR2. Superposition of the four ChR2 models with HR, BR, and AR allowed us to identify a structure corresponding to the retinal binding pocket previously identified in the other rhodopsins. In our models, Lys-257 results in the correct position to be covalently bound to the retinal forming a Schiff base, which is in line with current evidence. The aromatic residues surrounding the retinal are Trp-124, Phe-178, Tyr-184, Trp-223, Phe-226, and Phe-230 (supplemental Table S2), showing a high degree of homology with the corresponding residues in HR (29). The proton donor and acceptors for the retinal Schiff base were reported to be Arg-134, Glu-123, and Asp-253, respectively (16). Accordingly, in our model, Arg-134 and Glu-123 are located on opposite sides with respect to the retinal. The first is located toward the intracellular part, about 11.6 Å from the Schiff base, whereas the second is located toward the protein extracellular side, at about 4.1 Å from the Schiff base. The other residues described to be involved in the ChR2 photocycle are Cys-128 and Asp-156 (16, 30–32); in our model, they point toward the retinal pocket with their sulfur and oxygen atoms at a relative distance of 2.6 Å, compatible with formation of a hydrogen bond.
Inner Chamber System and Ion Selectivity Filter in ChR2
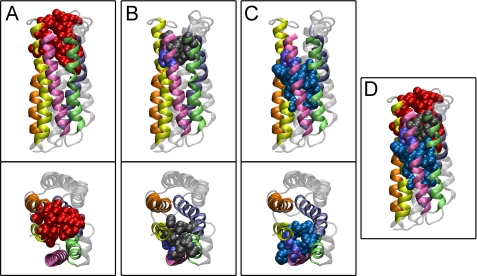
Internal water-filled cavities have been described in BR and microbial rhodopsins (33), and a system of inner chambers determines the ion pathway in ion-conducting rhodopsin (29). In our ChR2 models, we identified a system of inner chambers in the extracellular half of the protein. Three spatially conserved chambers, hereafter named chambers A, B, and C, were present in the ChR2 models (Fig. 1). Among these, only chamber A (located toward the extracellular side) is also present in HR, AR, and BR. By contrast, chambers B and C seem to be a specific feature of ChR2 because only partial superposition with HR, AR, and BR chambers is found (not shown). The inner surface of chambers B and C is formed by both polar/charged residues, whose spatial position is well conserved in the different simulations, and hydrophobic residues whose identity and position are less well defined. This accounts for the different shape and size of the chambers among the models. A list of the residues that form the surface of chambers B and C in the corresponding models can be found in Table 1. The structural features of the inner chambers and their position within ChR2 suggest that they are likely to form the cationic pathway. Our model is consistent with a role of the negatively charged and polar side chains of the two consecutive chambers B and C in coordinating and progressively dehydrating the ion while it proceeds along the channel cavity. Therefore, we designed a set of point mutations in chambers B and C (Fig. 2) to change the geometry and the charge distribution of the chambers. In the first set of mutants, we aimed at increasing the net negative charge of the residues facing the inner chambers to allow dehydration of ions with higher charge density (small divalent cations, i.e. Ca2+) than Na+. Therefore, selected residues were mutated to either Asp or Glu to change as little as possible the bulk of the side chain. In the case of the mutant A59S, we sought to modify the geometry of the coordination site, adding a further polar contact.
FIGURE 1.
Inner chamber system in ChR2 according to molecular modeling. Spatially conserved chambers in ChR2 bioinformatic model 2 are shown. A–C, chamber A (A), chamber B (B), and chamber C (C) with their side and top view. Helices surrounding the chambers are colored in mauve (helix 1), green (helix 2), blue (helix 3), orange (helix 6), and yellow (helix 7). In D, superposition of all chambers is shown.
TABLE 1.
Residues forming the inner surface of chambers B and C in ChR2 bioinformatic models
*, residues shared by all ChR2 models. Single-letter amino acid codes were used.
FIGURE 2.
Position of mutated residues in ChR2 model 2. Point mutations of residues shared by all ChR2 bioinformatic models in chambers B (gray) and C (cyan) were performed. All residues belong to helix 1 (mauve) and helix 7 (yellow). A, side view. B, enlargement of A. C, top view.
Functional Characterization of ChR2 Mutants
ChR2 mutants tagged with the fluorescent mCherry protein fused at the C terminus were expressed in HeLa cells and tested by whole cell voltage clamp. Photocurrents were evoked by a 500-ms pulse of blue light (480 ± 10 nm, 0.45 milliwatt/mm2) and measured at different membrane potentials (20-mV steps from −120 to 20 mV). In Fig. 3A, the typical photocurrent of ChR2(H134R)-mCherry (hereafter designated as WT) in solution 1 (see “Experimental Procedures”) at −120 mV is shown. Confocal microscopy analysis revealed that all mutants were preferentially distributed to the plasma membrane (Fig. 3B). Quantification of mCherry fluorescence intensity in the cell contour confirmed that these values were not significantly different among ChR2 mutants (not shown). To exclude that differences in photocurrent amplitudes could be caused by shifts in the maximum absorbance wavelength of the mutants as compared with the WT, activation spectra were recorded by exciting cells from 390 to 590 nm with a monochromator. Differences in the activation spectra were modest, and the peak activation wavelength was included within the excitation window of the light used for photocurrent measurements for all the mutants (supplemental Fig. S3).
FIGURE 3.
Photocurrents of ChR2(H134R)-mCherry and variants in Na+- and Ca2+-based extracellular solutions. A, typical photocurrent of ChR2(H134R)-mCherry in HeLa cells measured at −120 mV upon excitation with a 500-ms light pulse (480 nm, black bar) in extracellular solution 1 (in mm, 145 NaCl, 3 KCl, 5 N-methyl-d-glucamine, 10 Hepes, 20 glucose; pH 7.35). On the right, the IV relationship from −120 mV to 20 mV in 20 mV steps is shown (n = 9). B, expression of ChR2(H134R)-mCherry (WT) and variants in HeLa cells as assessed by confocal microscopy. C and D, inward photocurrents at −120 mV in extracellular solution 1 (C) and solution 2 (D) (in mm, 10 CaCl2, 3 KCl, 135 N-methyl-d-glucamine, 10 Hepes, 20 glucose; pH 7.35). *, p < 0.05, unpaired two-tailed t test. pF, picofarads. E, ratio between photocurrent peaks in solution 1 and 2. *, p < 0.002, unpaired two-tailed t test. F, Fluo-4 measurement of intracellular Ca2+ in HeLa cells transfected with the ChR2 WT and S63D mutant upon 100-ms pulses of 490-nm light. A significant increase in intracellular Ca2+ in ChR2-S63D as compared with WT-ChR2 expressing cells was detected. Error bars in panels A and C–F indicate S.E.
Mutations of conserved residues of chamber B (Gln-56) and between chambers B and C (Thr-250 and Ala-59) were designed in the hypothesis that ion dehydration is a step in ion transport, thus affecting ion selectivity. Among all mutations in those residues facing either chamber B or C, Q56E significantly reduced both Na+ and Ca2+ currents at −120 mV (Fig. 3, C and D). Mutation T250E caused a dramatic reduction in Ca2+ photocurrent, whereas T250D did not cause any effect in both Ca2+ and Na+ currents.
As residue Thr-250 is located between chambers B and C, this may suggest the presence of a dimension filter between these two cavities, which would be in line with the effect of increasing the steric hindrance of the side chain (i.e. from Asp to Glu) on Ca2+ currents. This would also support that Thr-250 faces the pore.
The conserved residues of chamber C, Ser-63 and Asn-258, were also mutated into Asp. The analysis of currents evoked by light in an extracellular solution with either Na+ or Ca2+ as the main ion able to permeate the channel (solutions 1 and 2, respectively, see “Experimental Procedures”) revealed that although the monovalent Na+ currents were unchanged in both mutants, Ca2+ current was increased by about 65% in S63D mutant (Fig. 3D). When considering the Ca2+/Na+ current ratio, both S63D and N258D mutants showed a relative increase in Ca2+ conductance (Fig. 3E) (Ca2+/Na+: S63D versus H134R = 1.39, n = 10; N258D versus H134R = 1.32, n = 12). These data suggest that chamber C may function as the Ca2+ coordination site because increasing the negative charge in this chamber leads to a higher Ca2+/Na+ current ratio. An effect of the mutations on the open probability of the channel is not likely as this would affect the amplitude of Na+ currents. This may not hold for Q56E mutant, for which further studies would be required. It has been shown that ChR2 Ca2+ photocurrents reach saturation at high Ca2+ concentration (>40 mm) (34), suggesting the presence of a Ca2+ binding site in the channel. Our data indicate that the Ca2+ binding site might reside in chamber C.
To better address the permeability to Ca2+ ions, we transfected HeLa cells with one of the mutants that display enhanced Ca2+/Na+ current ratio, ChR2-S63D-mCherry, and loaded them with the Ca2+ indicator Fluo-4. The excitation wavelength used for Fluo-4 imaging (490 ± 20 nm) allows simultaneous image acquisition and photoactivation, In an extracellular solution containing 80 mm Ca2+, we measured a significant increase in intracellular Ca2+ in S63D-expressing as compared with WT-expressing cells (S63D:1.55 ± 0.02 WT:1.27 ± 0.01; n = 24 (S63D) and 57 (WT); p < 0.001)(Fig. 3F).
To investigate whether point mutations performed also affected the photocurrent kinetics, the opening rate (τON), the transition from peak to stationary current (τDES), and the closing rate (τOFF) after light was switched off were estimated (Table 2). T250E substitution lead to significantly faster off-kinetics, similar to that of previously published ultrafast ChR2 variants, whereas N258D mutation induced a slower closing rate. Both mutants that showed a higher Ca2+/Na+ ratio (S63D and N258D) also displayed a slower transition from the peak current to the stationary state. The summary of the kinetic properties of all the mutants is reported in Table 2.
TABLE 2.
Kinetic properties of ChR2(H134R)-mCherry and respective mutant variants. Opening rates (τON), closing rates (τOFF), and desensitization rates (τDES) were estimated from photocurrents of at least six cells. Values represent means ± S.E. * = p < 0.01, unpaired t test. Imax is the largest peak current, and Iplateau is the current at the steady state level
| IPlateau/IMax | τON | τOFF | τDES | |
|---|---|---|---|---|
| ms | ms | ms | ||
| WT | 75.8 ± 4.8 | 2.17 ± 0.26 | 14.94 ± 1.24 | 9.18 ± 0.64 |
| Q56E | 71.1 ± 4.3 | 3.78 ± 0.61* | 14.56 ± 0.46 | 11.75 ± 0.06 |
| A59S | 71.3 ± 3.3 | 1.56 ± 0.14 | 12.76 ± 1.08 | 8.26 ± 1.45 |
| S63D | 83.3 ± 2.2* | 1.64 ± 0.10 | 17.26 ± 0.90 | 15.27 ± 1.06* |
| T250D | 71.1 ± 3.2 | 2.44 ± 0.25 | 12.72 ± 0.48 | 9.89 ± 0.97 |
| T250E | 68.4 ± 4.0* | 2.42 ± 0.18 | 8.21 ± 0.70* | 11.83 ± 1.85 |
| N258D | 82.7 ± 2.4* | 2.29 ± 0.16 | 27.42 ± 0.96* | 25.83 ± 3.07* |
Role of Counterion System in ChR2 Photoactivation
As suggested by sequence similarity and functional data, the activation mechanism of ChR2 is similar to other microbial rhodopsins, and our bioinformatic model is in agreement with this concept. In BR, the proton transfer occurs in an extended hydrogen-bonded complex containing the two negatively charged Asp-85 and Asp-212, two positively charged groups, Lys-216 (the Schiff base) and Arg-82, and coordinated water (35). In our ChR2 models, the corresponding residues are predicted to be Glu-123, Asp-253, Lys-257 (the Schiff base), and Arg-120, respectively.
We used molecular dynamics simulations to include water in our model and explore equilibrium fluctuations of the side chains. Quite intriguingly, after 1 ns, the side chain of Arg-120 faces chamber B and obstructs the cation pathway (Fig. 4, A and B) as corresponding basic residues in BR and HR do (33). Arg-120 is found in a position in between the counterions Glu-123 and Asp-253, at a relative distance of 7.4 and 4.6 Å, respectively (Fig. 4C). This is consistent with the structure of BR, in which these 4 residues and a centrally coordinated water molecule form a quadrupole (36).
FIGURE 4.
Role of ARS-120 in the counterion system. A and B, side chain of residue Arg-120 (in red) obstructs cation pathway (represented by chambers B and C, in cyan), as shown for ChR2 model 2 after a 1-ns molecular dynamics simulation (A, side view; B, top view). C, enlargement of the Schiff base region, with the key residues forming the hydrogen bond network. Arg-120 is found in a position in between the counterions Glu-123 and Asp-253, at a relative distance of 7.4 and 4.6 Å, respectively. D, R120A mutation caused a 10-fold reduction in photocurrent amplitude. In the graphs, currents at −120 mV in solution 1 are shown, n = 10). pF, picofarads. Error bars in indicate S.D.
To test whether Arg-120 is involved in the mechanism of photoactivation, we substituted the arginine with a non-protonable alanine (R120A). Energy minimization of the ChR2 R120A model demonstrated that this mutation does not alter the structure of the helices and protein stability and that its position did not change upon molecular dynamics simulation. Photocurrent of R120A mutant was compared with that of the wild type ChR2 in a subset of cells with comparable expression levels at the plasma membrane. We found that R120A mutation caused a 10-fold reduction in photocurrent amplitude (Fig. 4D).
DISCUSSION
In this study, we used a combination of bioinformatic modeling, molecular dynamics simulations, and site-directed mutagenesis to gain information on structure-function relationship in ChR2. Bioinformatic structure prediction and structural superposition of ChR2 with BR, AR, and HR, other microbial rhodopsins with ion conductance, allowed us to identify the putative ion pathway within the channel. In ChR2, this is formed by a series of three consecutive chambers made by residues belonging to helices 1–4 and 7. Among these, only chamber A (located toward the extracellular side) is also present in HR, AR, and BR. By contrast, chambers B and C are a specific feature of ChR2. Internal water-filled cavities have been described in BR and microbial rhodopsins (33), and a system of inner chambers determines the ion pathway in ion-conducting rhodopsin (29).
Mutagenesis of residues predicted to be exposed in chambers B and C caused alterations in conductance to Na+ (Q56E) or relative Ca2+ or Na+ conductance (S63D, T250E, and N258D), supporting that these residues participate in the pore formation. It has been reported that only dehydrated cations can permeate the “selective filter” of ChR2 (3). Our structural modeling of the ion conduction pathway is consistent with a role of the negatively charged and polar side chains of the two consecutive chambers B and C in coordinating and progressively dehydrating the ion while it proceeds along the channel cavities. The experimental evidence suggests that adding further polar residues to chamber B has a negative or no effect on ion current. On the contrary, increasing the net negative charge exposed in chamber C results in a higher Ca2+/Na+ current ratio. Given that Ca2+ has a higher charge density and bigger hydration shell than Na+, this may indicate that a limiting step in the dehydration process occurs mostly in chamber C.
To our knowledge, most of the ChR2 mutations have been engineered so far on the basis of sequence homology to BR, whose crystal structure is known in detail, and have targeted regions close to the retinal binding pocket (helix 2 and 3). This has generated a number of mutants with increased photocurrent and altered kinetics. Among these, only L132C mutation affected the relative permeability to Ca2+ and Na+ (37). Our analysis explores a new region of the protein, suggesting new determinants for ion selectivity and permeability (Ser-63 and Asn-258).
In previous models of ChR2, several glutamic acid residues in helix B (Glu-82, Glu-83, Glu-90, Glu-97, and Glu-101) were predicted (i) to be part of the water-containing cavity allowing ions to cross the channel and (ii) to point toward the outer part of the ring formed by the seven-transmembrane helices (38), suggesting that the functional ChR2 may work as a trimer. However, no evidence in favor of the trimeric unit was provided in other studies (39, 40).
In addition, ChR2 was recently shown to form dimers rather than trimers, and the arrangement of the monomers does not seem to support this latter model (41). However, these negatively charged residues represent a striking feature of helix 2 in our alignment as well (supplemental Fig. S2), and at least some of those were shown to affect the photocurrent (42, 43).
In our models, despite a linear arrangement of their Cα, residues Glu-82, Glu-83, and Glu-90 are located on the surface of the protein, pointing toward the outside of the monomer, whereas Glu-101 points to the interior of the ring of helices. The exact position of the side chain of Glu-97 is uncertain as in different models, it bends over either the ring of helices or the external surface. One possible explanation might be that helix 2 can exist in different degrees of rotation, which could represent different conformational states of the protein during the photocycle.
The mechanism of functioning of ChR2 is similar to other microbial rhodopsins. ChR2 has been proposed to function as a proton pump, with a proton “leak” suggested as a basis of its ion channel behavior (44).
As mutation R120A almost entirely abolished the photocurrent, and given the position of this residue between the putative counterions Glu-123 and Asp-253, it can be speculated that electrostatic interactions of the charged side chain of Arg-120 take part in the mechanism of photoactivation. This would be in line with the general proton-pumping mechanism of rhodopsins, as recently described using BR as a model (33).
In BR, deprotonation of the Schiff base Lys-216 (Lys-257 in ChR2) upon photon absorption breaks its interaction with the counterions Asp-85 and Asp-212 (Glu-123 and Asp-253 in ChR2). This event breaks the salt bridge between Asp-85 and Arg-82 (Arg-120 in ChR2), and this latter in turn moves toward Glu-194/Glu-204 (Glu-235/Ser-245 in ChR2), and by this mechanism, a proton is released to the bulk.
The homology of the structural features of BR and ChR2 in the Schiff base region of the retinal binding site further supports the concept of a similar molecular basis of the photoactivation mechanism. Therefore, we propose that the proton-pumping mechanism in ChR2 proceeds by displacement of the side chain of Arg-120 following protonation of the Schiff base counterion.
In BR, electrostatic interaction between the Schiff base and the surrounding residues in the binding pocket affects the retinal absorption spectrum. It is thus tempting to speculate that mutagenesis in the residues surrounding the Schiff base region may result in spectrally shifted variants.
In conclusion, we show a complete structural model of ChR2, describe the ion-conducting pathway, and identify novel key residues involved in ionic permeability and in the photoactivation mechanism. These results expand our current knowledge on the structural determinants of ChR2 function and direct further biotechnological efforts to generate new variants with specific biophysical properties (i.e. higher Ca2+ conductance, higher Na+ specificity, faster/slower kinetics). Notably, most of our mutants were obtained by targeting previously unrecognized regions regulating ChR2 function. Possible combination with existing variants may therefore be used to tune ChR2 function to specific applications.
Supplementary Material
Acknowledgments
We thank Prof. Peter Hegemann (Humboldt University of Berlin, Germany) and Prof. Tullio Pozzan (University of Padova, Italy) for helpful discussion and Prof. Karl Deisseroth (Stanford University, Stanford, CA) for the ChR2(H134R)-mCherry construct. We are indebted to Dr. Andrea Lelli for the technical help.
This work was supported by funding from the European Community Seventh Framework Program FP7/2007-2013 under Grant Agreement HEALTH-F2-2009-241526, EUTrigTreat (to M. M.).

This article contains supplemental Tables S1 and S2 and Figs. S1–S3.
- ChR2
- channelrhodopsin-2
- BR
- bacteriorhodopsin
- AR
- archaerhodopsin
- HR
- halorhodopsin.
REFERENCES
- 1. Foster K. W., Saranak J., Patel N., Zarilli G., Okabe M., Kline T., Nakanishi K. (1984) A rhodopsin is the functional photoreceptor for phototaxis in the unicellular eukaryote Chlamydomonas. Nature 311, 756–759 [DOI] [PubMed] [Google Scholar]
- 2. Nagel G., Ollig D., Fuhrmann M., Kateriya S., Musti A. M., Bamberg E., Hegemann P. (2002) Channelrhodopsin-1: a light-gated proton channel in green algae. Science 296, 2395–2398 [DOI] [PubMed] [Google Scholar]
- 3. Nagel G., Szellas T., Huhn W., Kateriya S., Adeishvili N., Berthold P., Ollig D., Hegemann P., Bamberg E. (2003) Channelrhodopsin-2, a directly light-gated cation-selective membrane channel. Proc. Natl. Acad. Sci. U.S.A. 100, 13940–13945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Boyden E. S., Zhang F., Bamberg E., Nagel G., Deisseroth K. (2005) Millisecond-timescale, genetically targeted optical control of neural activity. Nat. Neurosci. 8, 1263–1268 [DOI] [PubMed] [Google Scholar]
- 5. Zhang F., Wang L. P., Brauner M., Liewald J. F., Kay K., Watzke N., Wood P. G., Bamberg E., Nagel G., Gottschalk A., Deisseroth K. (2007) Multimodal fast optical interrogation of neural circuitry. Nature 446, 633–639 [DOI] [PubMed] [Google Scholar]
- 6. Arenkiel B. R., Peca J., Davison I. G., Feliciano C., Deisseroth K., Augustine G. J., Ehlers M. D., Feng G. (2007) In vivo light-induced activation of neural circuitry in transgenic mice expressing channelrhodopsin-2. Neuron 54, 205–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhang F., Wang L. P., Boyden E. S., Deisseroth K. (2006) Channelrhodopsin-2 and optical control of excitable cells. Nat. Methods 3, 785–792 [DOI] [PubMed] [Google Scholar]
- 8. Ishizuka T., Kakuda M., Araki R., Yawo H. (2006) Kinetic evaluation of photosensitivity in genetically engineered neurons expressing green algae light-gated channels. Neurosci. Res. 54, 85–94 [DOI] [PubMed] [Google Scholar]
- 9. Li X., Gutierrez D. V., Hanson M. G., Han J., Mark M. D., Chiel H., Hegemann P., Landmesser L. T., Herlitze S. (2005) Fast noninvasive activation and inhibition of neural and network activity by vertebrate rhodopsin and green algae channelrhodopsin. Proc. Natl. Acad. Sci. U.S.A. 102, 17816–17821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bruegmann T., Malan D., Hesse M., Beiert T., Fuegemann C. J., Fleischmann B. K., Sasse P. (2010) Optogenetic control of heart muscle in vitro and in vivo. Nat. Methods 7, 897–900 [DOI] [PubMed] [Google Scholar]
- 11. Hofmann B., Maybeck V., Eick S., Meffert S., Ingebrandt S., Wood P., Bamberg E., Offenhäusser A. (2010) Light-induced stimulation and delay of cardiac activity. Lab Chip 10, 2588–2596 [DOI] [PubMed] [Google Scholar]
- 12. Hegemann P., Möglich A. (2011) Channelrhodopsin engineering and exploration of new optogenetic tools. Nat. Methods 8, 39–42 [DOI] [PubMed] [Google Scholar]
- 13. Deisseroth K. (2011) Optogenetics. Nat. Methods 8, 26–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nagel G., Brauner M., Liewald J. F., Adeishvili N., Bamberg E., Gottschalk A. (2005) Light activation of channelrhodopsin-2 in excitable cells of Caenorhabditis elegans triggers rapid behavioral responses. Curr. Biol. 15, 2279–2284 [DOI] [PubMed] [Google Scholar]
- 15. Gunaydin L. A., Yizhar O., Berndt A., Sohal V. S., Deisseroth K., Hegemann P. (2010) Ultrafast optogenetic control. Nat. Neurosci. 13, 387–392 [DOI] [PubMed] [Google Scholar]
- 16. Berndt A., Yizhar O., Gunaydin L. A., Hegemann P., Deisseroth K. (2009) Bi-stable neural state switches. Nat. Neurosci. 12, 229–234 [DOI] [PubMed] [Google Scholar]
- 17. Stehfest K., Ritter E., Berndt A., Bartl F., Hegemann P. (2010) The branched photocycle of the slow-cycling channelrhodopsin-2 mutant C128T. J. Mol. Biol. 398, 690–702 [DOI] [PubMed] [Google Scholar]
- 18. Berndt A., Schoenenberger P., Mattis J., Tye K. M., Deisseroth K., Hegemann P., Oertner T. G. (2011) High-efficiency channelrhodopsins for fast neuronal stimulation at low light levels. Proc. Natl. Acad. Sci. U.S.A. 108, 7595–7600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Beltramello M., Bicego M., Piazza V., Ciubotaru C. D., Mammano F., D'Andrea P. (2003) Permeability and gating properties of human connexins 26 and 30 expressed in HeLa cells. Biochem. Biophys. Res. Commun. 305, 1024–1033 [DOI] [PubMed] [Google Scholar]
- 20. Kelley L. A., Sternberg M. J. (2009) Protein structure prediction on the web: a case study using the Phyre server. Nat. Protoc. 4, 363–371 [DOI] [PubMed] [Google Scholar]
- 21. Arnold K., Bordoli L., Kopp J., Schwede T. (2006) The SWISS-MODEL workspace: a web-based environment for protein structure homology modeling. Bioinformatics 22, 195–201 [DOI] [PubMed] [Google Scholar]
- 22. Lessel U., Schomburg D. (1994) Similarities between protein three-dimensional structures. Protein Eng. 7, 1175–1187 [DOI] [PubMed] [Google Scholar]
- 23. Lill M. A., Danielson M. L. (2011) Computer-aided drug design platform using PyMOL. J. Comput. Aided Mol. Des. 25, 13–19 [DOI] [PubMed] [Google Scholar]
- 24. Cornell W. D., Cieplak P., Bayly C. I., Gould I. R., Merz K. M., Ferguson D. M., Spellmeyer D. C., Fox T., Caldwell J. W., Kollman P. A. (1995) A second generation force field for the simulation of proteins, nucleic acids, and organic molecules. J. Am. Chem. Soc. 117, 5179–5197 [Google Scholar]
- 25. Hess B., Kutzner C., van der Spoel D., Lindahl E. (2008) Gromacs 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation. J. Chem. Theory Comput. 4, 435–447 [DOI] [PubMed] [Google Scholar]
- 26. Berendsen H. J., Postma J. P., van Gunsteren W. F., DiNola A., Haak J. R. (1984) Molecular dynamics with coupling to an external bath. J. Chem. Phys. 81, 3684–3690 [Google Scholar]
- 27. Enami N., Yoshimura K., Murakami M., Okumura H., Ihara K., Kouyama T. (2006) Crystal structures of archaerhodopsin-1 and -2: common structural motif in archaeal light-driven proton pumps. J. Mol. Biol. 358, 675–685 [DOI] [PubMed] [Google Scholar]
- 28. Yoshimura K., Kouyama T. (2008) Structural role of bacterioruberin in the trimeric structure of archaerhodopsin-2. J. Mol. Biol. 375, 1267–1281 [DOI] [PubMed] [Google Scholar]
- 29. Kouyama T., Kanada S., Takeguchi Y., Narusawa A., Murakami M., Ihara K. (2010) Crystal structure of the light-driven chloride pump halorhodopsin from Natronomonas pharaonis. J. Mol. Biol. 396, 564–579 [DOI] [PubMed] [Google Scholar]
- 30. Nack M., Radu I., Gossing M., Bamann C., Bamberg E., von Mollard G. F., Heberle J. (2010) The DC gate in channelrhodopsin-2: crucial hydrogen bonding interaction between Cys-128 and Asp-156. Photochem. Photobiol. Sci. 9, 194–198 [DOI] [PubMed] [Google Scholar]
- 31. Radu I., Bamann C., Nack M., Nagel G., Bamberg E., Heberle J. (2009) Conformational changes of channelrhodopsin-2. J. Am. Chem. Soc. 131, 7313–7319 [DOI] [PubMed] [Google Scholar]
- 32. Bamann C., Gueta R., Kleinlogel S., Nagel G., Bamberg E. (2010) Structural guidance of the photocycle of channelrhodopsin-2 by an interhelical hydrogen bond. Biochemistry 49, 267–278 [DOI] [PubMed] [Google Scholar]
- 33. Wolf S., Freier E., Potschies M., Hofmann E., Gerwert K. (2010) Directional proton transfer in membrane proteins achieved through protonated protein-bound water molecules: a proton diode. Angew Chem. Int. Ed Engl. 49, 6889–6893 [DOI] [PubMed] [Google Scholar]
- 34. Tsunoda S. P., Hegemann P. (2009) Glu-87 of channelrhodopsin-1 causes pH-dependent color tuning and fast photocurrent inactivation. Photochem. Photobiol. 85, 564–569 [DOI] [PubMed] [Google Scholar]
- 35. Brown L. S., Váró G., Hatanaka M., Sasaki J., Kandori H., Maeda A., Friedman N., Sheves M., Nedleman R., Lanyi J. K. (1995) The complex extracellular domain regulates the deprotonation and reprotonation of the retinal Schiff base during the bacteriorhodopsin photocycle. Biochemistry 34, 12903–12911 [DOI] [PubMed] [Google Scholar]
- 36. Dér A., Száraz S., Tóth-Boconádi R., Tokaji Z., Keszthelyi L., Stoeckenius W. (1991) Alternative translocation of protons and halide ions by bacteriorhodopsin. Proc. Natl. Acad. Sci. U.S.A. 88, 4751–4755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kleinlogel S., Feldbauer K., Dempski R. E., Fotis H., Wood P. G., Bamann C., Bamberg E. (2011) Ultra light-sensitive and fast neuronal activation with the Ca2+-permeable channelrhodopsin CatCh. Nat. Neurosci. 14, 513–518 [DOI] [PubMed] [Google Scholar]
- 38. Hegemann P., Tsunoda S. (2007) Light tools for neuroscience: channelrhodopsin and light-activated enzymes. Cell Sci. Rev. 3, 108–123 [Google Scholar]
- 39. Hegemann P., Ehlenbeck S., Gradmann D. (2005) Multiple photocycles of channelrhodopsin. Biophys. J. 89, 3911–3918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Petreanu L., Huber D., Sobczyk A., Svoboda K. (2007) Channelrhodopsin-2-assisted circuit mapping of long-range callosal projections. Nat. Neurosci. 10, 663–668 [DOI] [PubMed] [Google Scholar]
- 41. Müller M., Bamann C., Bamberg E., Kühlbrandt W. (2011) Projection structure of channelrhodopsin-2 at 6 Å resolution by electron crystallography. J. Mol. Biol. 414, 86–95 [DOI] [PubMed] [Google Scholar]
- 42. Ruffert K., Himmel B., Lall D., Bamann C., Bamberg E., Betz H., Eulenburg V. (2011) Glutamate residue 90 in the predicted transmembrane domain 2 is crucial for cation flux through channelrhodopsin 2. Biochem. Biophys. Res. Commun. 410, 737–743 [DOI] [PubMed] [Google Scholar]
- 43. Sugiyama Y., Wang H., Hikima T., Sato M., Kuroda J., Takahashi T., Ishizuka T., Yawo H. (2009) Photocurrent attenuation by a single polar-to-nonpolar point mutation of channelrhodopsin-2. Photochem. Photobiol. Sci. 8, 328–336 [DOI] [PubMed] [Google Scholar]
- 44. Feldbauer K., Zimmermann D., Pintschovius V., Spitz J., Bamann C., Bamberg E. (2009) Channelrhodopsin-2 is a leaky proton pump. Proc. Natl. Acad. Sci. U.S.A. 106, 12317–12322 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.