Background: The osteogenic Runt-related (RUNX) transcription factor Runx2 is frequently elevated in osseous and non-osseous tumor cells.
Results: Genomic RUNX2 target genes involved in motility were identified; RUNX2 depletion reduces cell motility in osteosarcoma cells.
Conclusion: RUNX2 regulates cell motility and adhesion in osteosarcoma cells.
Significance: RUNX2 may also control migration of normal osteoblasts and/or tumor cells.
Keywords: Adhesion, Cell Migration, Cell Motility, Chromatin Immunoprecipitation (ChIP), Collagen, Gene Expression, Transcription, Focal Adhesion, Osteosarcoma, RUNX2
Abstract
Runt-related transcription factors (RUNX1, RUNX2, and RUNX3) are key lineage-specific regulators of progenitor cell growth and differentiation but also function pathologically as cancer genes that contribute to tumorigenesis. RUNX2 attenuates growth and stimulates maturation of osteoblasts during bone formation but is also robustly expressed in a subset of osteosarcomas, as well as in metastatic breast and prostate tumors. To assess the biological function of RUNX2 in osteosarcoma cells, we examined human genomic promoter interactions for RUNX2 using chromatin immunoprecipitation (ChIP)-microarray analysis in SAOS-2 cells. Promoter binding of both RUNX2 and RNA polymerase II was compared with gene expression profiles of cells in which RUNX2 was depleted by RNA interference. Many RUNX2-bound loci (1550 of 2339 total) exhibit promoter occupancy by RNA polymerase II and contain the RUNX consensus motif 5′-((T/A/C)G(T/A/C)GG(T/G). Gene ontology analysis indicates that RUNX2 controls components of multiple signaling pathways (e.g. WNT, TGFβ, TNFα, and interleukins), as well as genes linked to cell motility and adhesion (e.g. the focal adhesion-related genes FAK/PTK2 and TLN1). Our results reveal that siRNA depletion of RUNX2, PTK2, or TLN1 diminishes motility of U2OS osteosarcoma cells. Thus, RUNX2 binding to diverse gene loci may support the biological properties of osteosarcoma cells.
Introduction
Runt-related (RUNX) transcription factors have emerged as potent gene regulators that are associated with both tissue development and oncogenesis. RUNX proteins control cell fate by regulating cell growth and differentiation of progenitor cells in different lineages (1–3). Deregulation of the function or expression of these factors is causally linked to distinct cancer phenotypes (4–8). Null mutations in Runt-related transcription factors (RUNX1/AML1, RUNX2/CBFA1, and RUNX3/PEBP2αC) cause major lineage-specific defects during mammalian development, although both loss- and gain-of-function mutations have been pathologically associated with cancer. For example, RUNX1 is frequently rearranged in acute myelogenous leukemia (1, 2), and null mutations in mice abolish definitive hematopoiesis during fetal development (9–11). Silencing of RUNX3 gene expression contributes to the etiology of carcinomas in multiple tissues (4, 5), and loss-of-function mutations cause alterations in gastrointestinal and neuronal tissues during postnatal development (12–14). Ectopic activation of RUNX2 upon retroviral integration contributes to T cell lymphomas in a Myc-dependent mouse model (15–18). Furthermore, genetic mutations in RUNX2 are linked to cleidocranial dysplasia in human patients (19–22). Mutations that abolish DNA binding and/or transcriptional functions or mutations that generate dosage insufficiency of RUNX2 in mouse models result in skeletal malformations at least in part due to a maturational arrest (23–26). Such RUNX2 mutations also result in growth deregulation in osteoblasts and embryonic fibroblasts (27–32). Although RUNX3 is silenced during tumorigenesis, unscheduled expression of RUNX1 and RUNX2 has been observed in several major cancer types (e.g. breast and prostate) (1–8), indicating that the latter two proteins play active roles in tumor etiology.
Cell autonomous effects in tumors that exhibit altered RUNX function or expression are attributable to gene regulatory functions of RUNX proteins. RUNX2 is endogenously expressed during the cell cycle in normal osteoblasts and expressed at increased levels upon cessation of growth and subsequent maturation of osteoblasts (27, 28, 33). Although RUNX2 is a natural suppressor of normal osteoblast proliferation, it is aberrantly expressed at elevated levels in a subset of cells derived from patients with osteosarcoma, a pediatric disease that is prevalent in adolescent patients (34–37). The increased levels of RUNX2 suggest that its growth-suppressive potential may be bypassed, thus permitting expression of its putative oncogenic functions in osteosarcoma. An extensive but incomplete catalog of RUNX target genes expressed in osteoblasts, as well as in osteosarcoma, breast, and prostate tumor cells, has emerged (7, 31, 38–52). These genes generally alter pathways linked to cell proliferation and survival, as well as other cellular activities required for tumorigenesis or cancer metastasis. However, a comprehensive assessment of gene regulatory networks controlled by RUNX proteins in specific tumors is necessary.
In this study, we have analyzed the genomic function of RUNX2 in osteosarcoma cells to gain insight into molecular pathways that are perturbed in bone cancer. We examined loci that are directly bound and controlled by RUNX2 using whole genome chromatin immunoprecipitations (ChIPs) for RUNX2 combined with genome-wide promoter microarrays (ChIP-on-chip), as well as gene expression profiling of cells depleted of RUNX2 using siRNAs. Our results reveal that RUNX2 controls genes and networks that are related to cell migration and adhesion, as well as other programs in osteosarcoma cells.
EXPERIMENTAL PROCEDURES
ChIP Assays
ChIP assays were performed with SAOS-2 cells that were grown in McCoy's medium (Thermo Scientific, Logan, UT) supplemented with 15% fetal bovine serum (FBS), penicillin/streptomycin, and l-glutamine (all from Invitrogen, Grand Island, NY). SAOS-2 cells were grown to ∼80% confluence and were cross-linked for 10 min in culture medium at room temperature with 1% formaldehyde solution. Fresh formaldehyde stock solution contained 50 mm HEPES-KOH, pH 7.5, 100 mm NaCl, 1 mm EDTA, 0.5 mm EGTA, and 11% formaldehyde. Cross-linking was terminated by incubation of cells with 0.125 m glycine solution for 5 min. Cells were washed twice with 1× PBS, placed on ice, and harvested using a cell scraper in PBS with protease inhibitors (Complete, Roche Diagnostics, Indianapolis, IN). Cells were collected by centrifugation at 4 °C, rapidly frozen in liquid nitrogen, and stored at −80 °C. Cell pellets were thawed on ice before each use.
ChIP was performed using previously published protocols (53–55). In brief, cells were resuspended in Lysis Buffer 1 (50 mm HEPES-KOH, pH 7.5, 140 mm NaCl, 1 mm EDTA, 10% glycerol, 0.5% Nonidet P-40, 0.25% Triton X-100, 1× protease inhibitors) for 10 min, collected by low speed centrifugation, and resuspended in Lysis Buffer 2 (10 mm Tris-HCl, pH 8.0, 200 mm NaCl, 1 mm EDTA, 0.5 mm EGTA, 1× protease inhibitors) for 10 min at room temperature. After the second centrifugation step, pellets were resuspended in 3 ml of Lysis Buffer 3 (10 mm Tris-HCl, pH 8.0, 100 mm NaCl, 1 mm EDTA, 0.5 mm EGTA, 0.1% sodium deoxycholate, 0.5% N-lauroylsarcosine, 1× protease inhibitors), and chromatin was sonicated to an average length of ∼500 bp. For SAOS-2 cells, sonication was optimized to 16 pulses of 30 s with intervals of 30 s using a Sonic Dismembrator (model 550, Thermo Fisher Scientific, Waltham, MA), while samples were maintained on ice at 4 °C. Triton X-100 was added (300 μl) to the sonicated lysate, and cellular debris was removed by microcentrifugation at 16,000 × g. Supernatants were adjusted with Sonication Buffer to the equivalent of ∼1.5 × 107 cells per ChIP sample. Input genomic DNA was saved separately and stored at −20 °C until further use.
Dynal beads (protein G, 100 μl/ChIP, Invitrogen) were washed with blocking solution (PBS, 0.5% BSA, filtered) and precoated overnight with anti-RUNX2 antibodies (M70, 12 μg, Santa Cruz Biotechnology, Santa Cruz, CA), polymerase II antibody (8WG16, 10 μl, Covance, Princeton, NJ), or IgG (rabbit polyclonal, 12 μg, Santa Cruz Biotechnology) as a negative control. After three washes with blocking solution, 1 ml of chromatin was added, and suspensions were rotated for a minimum of 8 h at 4 °C. Beads were collected by magnetic attraction using a Magnarack (Invitrogen) and resuspended in 1 ml of RIPA buffer (50 mm HEPES-KOH, pH 7.5, 500 mm LiCl, 1 mm EDTA, 1.0% Nonidet P-40, 0.7% sodium deoxycholate). This rinse cycle was repeated five times. Samples were then subjected to a single wash with 1 ml of TE buffer (10 mm Tris-HCl, pH 8.0, 1 mm EDTA) and recovered by low speed centrifugation at 900 × g for 3 min. Beads were resuspended in 210 μl of elution buffer (50 mm Tris-HCl, pH 8.0, 10 mm EDTA, 1.0% SDS) and incubated at 65 °C for 25 min with intermittent agitation (vortex) at ∼2-min intervals to elute chromatin. Beads were recovered by centrifugation for 1 min at 16,000 × g at room temperature, and 200 μl was transferred to a new 1.5-ml tube. At this step, 20 μl (2%) of the input sample was diluted with 180 μl of elution buffer and processed in parallel with the ChIP samples.
Cross-linking was reversed by incubating the suspensions for 12 h at 65 °C in an oven. The next day, samples were incubated for 2 h at 37 °C in the presence of RNase A (0.2 mg/ml in 200 μl of TE buffer) and subsequently for an additional 2 h at 55 °C in the presence of proteinase K (0.2 mg/ml). DNA was purified using phenol/chloroform/isoamyl alcohol (United States Biochemical, Cleveland, OH) extraction with phase separation in Heavy Phase Lock tubes (Eppendorf, Hamburg, Germany) followed by DNA precipitation with ethanol using standard procedures. DNA pellets were resuspended in 20 μl of 10 mm Tris-HCl, pH 8.0, and DNA concentrations of input samples were measured using a Nanodrop instrument (Thermo Fisher Scientific) (note: DNA concentrations of ChIP samples are below the level of detection). The DNA size range and efficiency of DNA fragmentation were evaluated by ethidium bromide staining of input samples using a 1.5% agarose gel (average DNA fragment size ∼500 bp).
DNA was amplified by adapting the standard protocol for whole genome amplification using the GenomePlex WGA2 kit (Sigma) as described previously (55). Briefly, the initial random fragmentation step was eliminated, and DNA from 10 μl of ChIP sample or from 10 ng of input chromatin was amplified using 22 PCR cycles. DNA was purified using the Qiaquick PCR purification kit (Qiagen Sciences, Beverly, MA) and resuspended in 30 μl of nuclease-free water. Final DNA concentrations were measured using the nanodrop device. ChIP DNA was analyzed with quantitative PCR using positive and negative controls, both before and after whole genome amplification. ChIP enrichment was determined using a 7300 sequence detection system (Applied Biosystems, Foster City, CA) with SYBR Green chemistry. The ChIP signal was normalized to the input sample and total DNA content. ChIP qPCR3 primers were designed in the peak regions of human gene promoters, preferably around genomic locations where potential RUNX2-binding elements were located (sequences for the ChIP PCR primers are summarized in supplemental Table S1). Only samples that met stringent quality criteria based on multiple positive and negative controls (e.g. expected immunoprecipitation and amplification of known RUNX2-responsive genes but not unrelated genes based on ChIP-qPCR; uniform whole genome amplification of different genomic segments) were submitted for Nimblegen ChIP-on-chip analysis (Roche NimbleGen Inc., Roche Applied Science).
ChIP-on-chip Assays
Amplicons were labeled with Cy5 (ChIP sample) and Cy3 (input sample) by Roche Nimblegen (Roche Applied Science) and hybridized to human HG18 Refseq promoter arrays that cover on average 2200 bp upstream and 500 bp downstream with a median probe spacing of 100 bp. In brief, raw data of the fluorescence intensities were obtained from scanned images of the oligonucleotide tiling arrays using NimbleScan 2.3 extraction software (Nimblegen Systems). For each spot on the array, log2 ratios of the Cy5-labeled test sample versus the Cy3-labeled reference (input) sample were calculated. The biweight mean of this log2 ratio was subtracted from each data point. Peak search analysis was performed with NimbleScan 2.3 software, and log2 ratios and false discovery rates were calculated for every peak (supplemental Table S2).
The ChIP-chip samples were validated using several control experiments. First, we examined the enrichment on the arrays of selected positive and negative control promoters by ChIP-qPCR before and after whole genome amplification to ensure that the positive controls were enriched and that negative controls were not enriched. Second, we performed arrays using two independent ChIP samples that revealed the reproducibility of the data sets. Third, the overlapping peak regions for different ChIP samples were visually inspected and found to be very reproducible in biological replicates. Fourth, a subset of targets identified on the arrays was validated using ChIP-qPCR assays with primers near RUNX motifs in peak regions or outside peak regions (negative controls).
Motif Analysis
Peak regions that were consistently positive (i.e. exhibiting peak overlap) in duplicate ChIP-on-chip experiments were analyzed in both the sense and antisense directions for the occurrence of the RUNX consensus motif 5′-(T/A/C)G(C/T)GGT or the related variant RUNX motifs 5′-TGTGGG and 5′-TGAGGT that are known to bind RUNX2 in vitro (29, 56). Motif searches were conducted within a 500-bp sequence centered on the region of peak overlap (i.e. 250 bp on each side) in duplicate experiments (supplemental Table S3). Aligned peak regions were also examined for co-regulatory elements in the vicinity of RUNX motifs using Clover, JASPAR, and TRANSFAC databases (57–61), but these analyses did not yield definitive evidence of co-motif enrichment.
RNA Interference
Osteosarcoma cells (SAOS-2 or U2OS) were seeded in 6-well plates and transfected the next day at 30–40% confluency with different oligonucleotides using OligofectamineTM reagent (Invitrogen) in 1 ml of Opti-MEM (a reduced serum medium from Invitrogen) according to the manufacturer's instructions. After 4 h, 0.5 ml of fresh culture medium containing 3× concentrated FBS was added. Cells were harvested 48 h (Western blot or qPCR) or 72 h (gene expression profiling) after transfection (40).
For gene expression profiling, three different small interfering RNA duplexes directed against human RUNX2 (siRUNX2) mRNA were purchased from Qiagen, indicated as siRUNX2 A, B, and C. The target sequences were as follows: siRUNX2A, 5′-CTC TGC ACC AAG TCC TTT T dTT-3′; siRUNX2B, 5′-AAT GCC TCT GCT GTT ATG AAA-3′, and siRUNX2C, 5′-AAG GTT CAA CGA TCT GAG ATT-3′), and oligonucleotides were used at 50 nm. Control cells were transfected with siRNA duplexes specific for CAT, GFP, or nonsilencing siRNA (Qiagen, Inc.) using the same concentrations and vehicle alone as control (40).
For Western blots and RT-qPCR validation studies, siRUNX2B and siRUNX2C oligos were used, or siRUNX2 oligos from Dharmacon (on-target plus smartpool J-012665-00). Control cells were treated with nonsilencing oligos from Qiagen (target sequence 5′-AAT TCT CCG AAC GTG TCA CGT-3′) or Dharmacon (on-target plus siControl nontargeting pool D-001810-10).
Gene Expression Profiling and Analysis
Affymetrix microarray analyses (Hu-U133Plus2 chips) were performed using RNA samples isolated from asynchronous SAOS-2 cells that were treated with RUNX2 siRNA and nonsilencing siRNAs. RNA samples were processed essentially as described previously (40, 62, 63). Signals from each microarray were analyzed, normalized, and converted to a numerical output using the Affymetrix GeneChip software. The data generated by the different arrays were globally scaled to an average intensity of 1500. The average expression value for each gene across the arrays was used to normalize the mRNA intensities. Adjusted data were subjected to further analysis using Cluster (63, 64).
Gene expression values from the arrays were calculated from raw CEL data using the method of Li and Wong (62). Raw data from the Hu-U133Plus2 chips were normalized and processed using dChip. Low and negative values were truncated upward to a uniform value of 150, and genes that had at least one P designation were used for further analysis. For a given gene, the mean expression value xt (log units) for three independent siRNA experiments for RUNX2 (oligos A–C) was compared with the mean gene expression xc (log units) of the three nonsilencing negative controls (siCAT, siGFP, and nonsilencing) using a cumulative distribution function, where s is the S.D. (log units) of the samples (19). A t test was applied for comparisons, and a p value ≤ 0.05 was considered significant. Because RUNX2 oligo C is much more efficient than A and B, we also performed a Z-test, in which oligo C was compared with the three nonsilencing oligos to identify genes that are modulated when cells are most depleted of RUNX2 (supplemental Table S4).
RNA Extraction and Real Time Quantitative PCR
Total RNA was purified using TRIzol (Invitrogen) and subjected to DNase I digestion prior to cDNA preparation using the qScript cDNA synthesis kit (Quanta). Relative quantification of amplified DNA was determined using a 7300 sequence detection system (Roche Diagnostics). Gene expression was monitored using real time primer pairs with SYBR Green detection (Applied Biosystems) (see supplemental Table S1), except for RUNX1 primers that used a Taqman probe for detection (Applied Biosystems; catalog number Hs00231079_m1). The relative mRNA expression was calculated with the ΔΔCT method. All primers used in the study were very carefully selected for maximal amplification efficiency (>90%), and all dissociation curves displayed one single peak. For qPCR array analysis, multiple genes (>50 genes with two primer pairs each) were analyzed with the linear regression method (LinRegPCR quantitative PCR data analysis program (version 11.0) (65).
Western Blot Analysis
Cells were lysed in RIPA buffer (Boston Bioproducts) and 2× SDS sample buffer supplemented with protease inhibitors (Complete, EDTA-free, Roche Diagnostics) and MG132 (Calbiochem). Lysates were fractionated in a 10% acrylamide gel and subjected to immunoblotting (Bio-Rad system). Immunoblots were incubated for 1 h with the following primary antibodies: RUNX2 (mouse monoclonal, MBL) or anti-CDK2 (rabbit M2, Santa Cruz Biotechnology). Peroxidase-labeled goat anti-mouse or goat anti-rabbit secondary antibodies (Santa Cruz Biotechnology) were used to visualize bands with enhanced chemiluminescence (ECL) chemistry (PerkinElmer Life Sciences) on BioMax film (Eastman Kodak Co.).
In vitro wound healing assays were carried out with U2OS cells that were pretreated with siRNAs for RUNX2 or representative RUNX2 targets associated with cell migration. Cells were grown until 80% confluence and scratched with a 200-μl pipette tip to create a cell-free area (“wound”). Cells were washed to remove unattached cells, incubated for 24 h, and examined by light microscopy. Three fields for each treatment in three independent experiments were imaged and quantified using ImageJ.
Migration and Invasion Assay
U2OS human osteosarcoma cells were depleted of RUNX2 using siRNAs as described above. At 48 h after transfection, cells were harvested using trypsin and counted in medium containing FBS. Cells were washed once with growth medium without supplements, collected by centrifugation, and resuspended with medium containing 0.1% BSA (fraction V, Sigma).
For trans-well invasion assays, the cell concentration was adjusted to 5 × 104 cell/ml, and the cell suspension was introduced into Matrigel invasion chambers or control plates without Matrigel (BD Biosciences). Before seeding, Matrigel plates were rehydrated for 2 h with warm incomplete medium at 37 °C, and normal growth medium containing FBS (0.75 ml) was added to the lower wells. Cells were incubated for 24 h to permit migration and invasion, and cells were removed from the upper membrane surface using cotton-tipped swabs. Cells that migrated to the lower surface of the membrane were fixed and stained with the Hema-3 stain set (Fisher). Hematoxylin and eosin-stained cells were photographed and counted. For each experimental condition, we calculated the average of four fields per well to cover nearly the entire well.
RESULTS
Genome-wide Occupancy of RUNX2 at Gene Loci in Osteosarcoma Cells
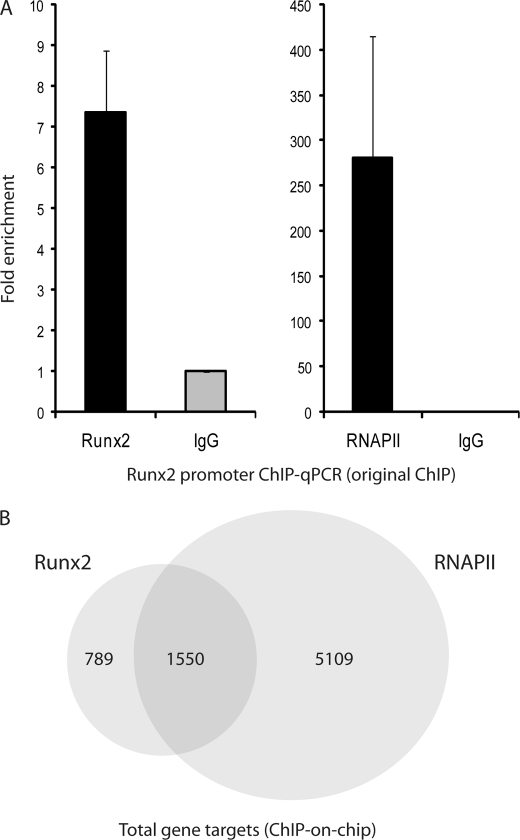
We performed two independent biological replicates of ChIP-on-chip experiments for RUNX2 in SAOS-2 human osteosarcoma cells that express high levels of RUNX2, compared with U2OS cells and normal osteoblastic cells, thus facilitating technical execution. We also carried out RNA polymerase II (RNAPII) ChIPs to distinguish between genes that are being (or poised to be) transcribed and those that are not transcribed. For each sample, we initially examined RUNX2 and RNAPII binding to the RUNX2 promoter, which is known to be autoregulated (66). The average enrichment of the two independent samples as determined by ChIP-qPCR was ∼7–8-fold for RUNX2 antibodies and ∼300-fold for RNAPII compared with the IgG controls (Fig. 1A), thus prevalidating our samples for genome-wide ChIP analysis.
FIGURE 1.
RUNX2 and RNAPII ChIP-on-chip. A, average occupancy by RUNX2 and RNAPII is shown from two independent original ChIP samples with ChIP-qPCR. B, total number of genes with RUNX2 and RNAPII promoter occupancy in two independent ChIP samples. 1550 gene targets were both bound by RUNX2 and RNAPII.
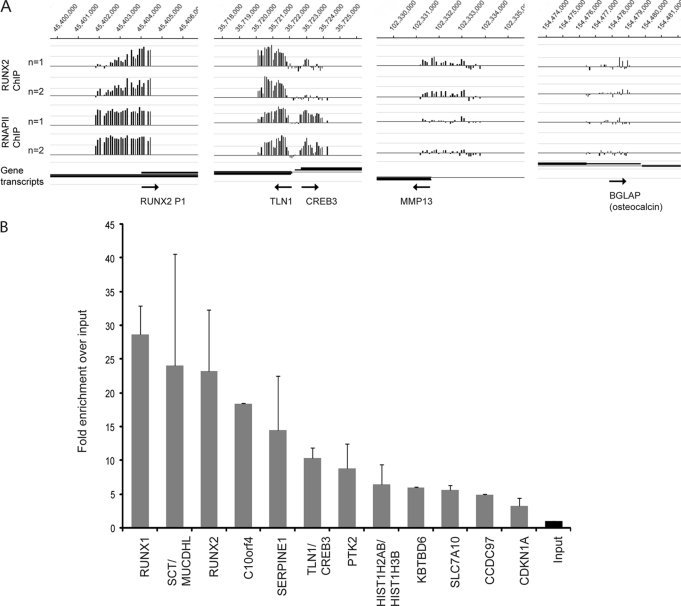
Duplicate ChIP-on-chip experiments with NimbleGen promoter tiling arrays revealed 2265 reproducible peaks for RUNX2 binding that are located adjacent to 2339 unique genes (Fig. 1B and supplemental Table S2). Of these genes, 1550 were also occupied by RNAPII (Fig. 1B). Corroborating the prevalidation assays, we find that the RUNX2 P1 promoter itself and the established RUNX2 target gene MMP13 are both occupied in duplicate arrays (Fig. 2A). However, no RUNX2 binding is observed for the osteocalcin (BGLAP) gene, a classical RUNX2 target that is not expressed in SAOS-2 cells as reviewed by Rodan and Noda (67). Our analyses revealed many new potential target genes such as Talin1 (TLN1) and cAMP-responsive element-binding protein 3 (CREB3) that are controlled by a shared intergenic regulatory region (Fig. 2A).
FIGURE 2.
RUNX2 occupancy in promoter regions identified with ChIP-on-chip experiments. A, in two independent experiments (n = 1 and n = 2), RUNX2 enrichment is high on its own P1 promoter, as well as on the shared TLN1-CREB3 promoter region. Enrichment is relatively low on the known RUNX2 target gene MMP13, and there is no significant enrichment on BGLAP (osteocalcin) that is not expressed in SAOS-2 cells. B, validation of RUNX2 targets with ChIP-qPCR. Averages are shown from two independent ChIP samples. Primers were designed around Runx motifs observed in peak regions identified in A (see supplemental Table S1 for exact locations within the peak). The fold enrichments observed in A and B are not directly comparable because the tiles in the Nimblegen promoter array do not match the amplicons of the qPCR primers.
Post-validation of the ChIP-on-chip results using qPCR and ChIP DNA was analyzed for enrichment of RUNX2-bound promoter segments (Fig. 2B). As negative controls, we used primers that amplify genomic DNA adjacent to RUNX2 binding regions (“peaks”) and exhibit negligible occupancy in NimbleGen arrays (data not shown). As a positive control, binding of RUNX2 to the RUNX2 P1 promoter on the arrays was post-validated by qPCR analysis (Fig. 2, A and B). A number of genes exhibiting robust RUNX2 binding were further characterized by qPCR analysis and represent novel targets, including RUNX1, PTK2, C10orf4, and SERPINE1, as well as the SCT-MUCDHL and TLN1-CREB3 gene pairs. Identification of RUNX1 as a RUNX2 target is consistent with functional cross-regulation between these related transcription factors in other biological contexts.4 SCT and CREB3 are both linked to cell signaling, whereas MUCDHL, SERPINE1, TLN1, and PTK2 are all linked to cell adhesion and/or migration (see below). C10orf4 is an anonymous gene that is not well characterized. Our results suggest that ∼9–10% of genes in the human genome (i.e. ∼25,000 genes present on the array) may be controlled by RUNX2 in osteosarcoma cells, although RUNX2 is bound to ∼23% of all genes that are associated with RNAPII (n = 6659 genes) (Fig. 1B). Furthermore, binding of RUNX2 to genes, whether or not they are co-occupied by RNAPII, is consistent with the bi-functional role of RUNX2 in gene activation and suppression.
Genome-wide Analyses of RUNX2 Recognition Motifs and Co-regulatory Elements
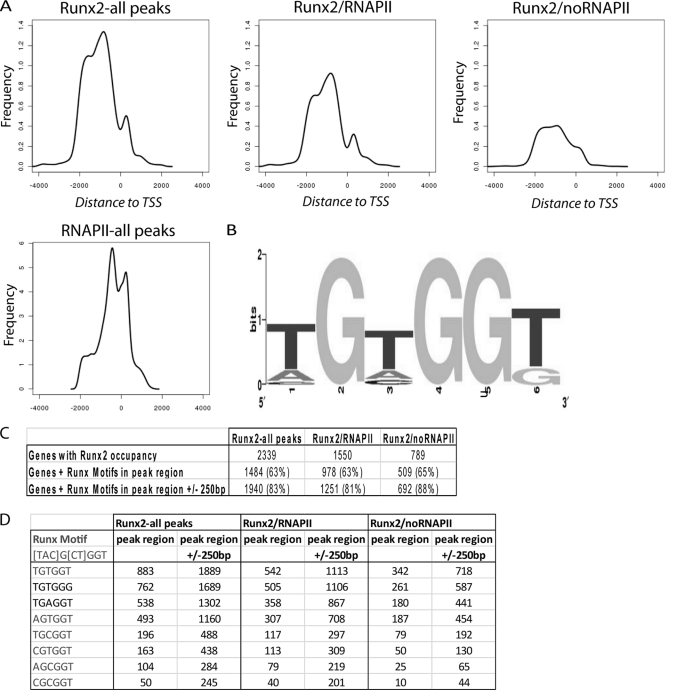
The recognition motif of Runt-related transcription factors was previously established by binding site selection for RUNX1/AML1 (68, 69). We performed motif analysis to refine this recognition element and to establish its relative location at a genome-wide level as it occurs within the first 2 kb of selected target genes (i.e. the promoter segments that are represented on the NimbleGen arrays). We analyzed the distribution of RUNX2 binding in all 2265 peak regions that were adjacent to 2339 genes, relative to the transcription start site (TSS) and relative to RNAPII binding. RNAPII is concentrated near the TSS as expected. RUNX2 generally binds distal to RNAPII at ∼800 bp upstream of the TSS, but RUNX2 binding is also enriched at ∼300 bp downstream of the TSS (Fig. 3A).
FIGURE 3.
Average distance of RUNX2 and RNAPII binding relative to the TSS and RUNX2 motif analysis. A, frequency of RUNX2 binding is maximal at ∼800 bp upstream of the TSS on average and relatively low at the TSS. When RNAPII-positive promoters are included (left and middle panel), there is an enrichment in RUNX2 binding at ∼300 bp downstream of the TSS, which is absent in promoters of genes that are RNAPII-negative. The average RNAPII binding is proximal to the maximal average RUNX2 binding. B, a RUNX2 motif was generated from the consensus motif present in all peak regions that aligned in duplicate experiments. The motif was generated with weblogo.berkeley.edu. C, frequency of detected Runx motifs in peak regions of genes in relation to co-occupancy with RNA polymerase II. D, summary of specific Runx-binding motifs and their occurrence in peak regions (left columns) or a 500-bp interval (± 250 bp) centered on the peak region (right columns).
We addressed whether RUNX2-binding motifs are enriched in promoters with RUNX2 occupancy by assessing the presence of the consensus RUNX motif 5′-(T/A/C)G(C/T)GGT (68) in peak regions. Motif analysis was performed using peak regions that overlap in duplicate experiments (i.e. alignment of a minimum of one tile). We found that ∼83% of all overlapping peak regions contained RUNX motifs within 250 bp from the region of overlap, and 65% of these motifs were located within the region of peak overlap (Fig. 3, B and C). The motifs 5′-TGTGGT and 5′-AGTGGT, which are a perfect match with the consensus, were present with the highest and fourth highest frequency, respectively (Fig. 3D). Interestingly, the one-mismatch motifs 5′-TGTGGG and 5′-TGAGGT, which are known to bind RUNX2 based on in vitro protein/DNA interaction assays (29, 56), were the second and third most frequent, although the motif 5′-CGCGGT was the least frequent. Our analysis establishes a consensus motif (5′-(T/A/C)G(T/A/C)GG(T/G)) in which the subsequence (T/A)G(T/A)GG(T/G) accounts for the four most prevalent RUNX2-binding elements that together encompass 80–90% of all empirically determined binding sites (Fig. 3D). The genome-wide RUNX2 consensus motif is in good agreement with the RUNX1/AML1 motif established by binding site selection (68), consistent with the high evolutionary conservation of the Runt homology DNA binding domain.
Most peak regions (>80%) encompass one to five copies of this genome-wide RUNX2 consensus motif. A limited number of gene promoters (∼24) contain a larger number of RUNX motifs (≥12) (e.g. NALP4, DRD1IP, DPCR1, RUNX1, ARSA, TRIM28, LTBP1, and HCCS) (supplemental Table S3). For example, the promoter for the NALP4 gene contains ∼81 RUNX motifs near the peak region. This gene and others that contain many RUNX motifs in their promoters (e.g. NALP4, DPCR1, and MMEL1) are RNAPII-negative. However, there is no clear correlation between the number of motifs and transcriptional status (i.e. whether a gene is activated or repressed by RUNX2 as inferred from RNAPII binding) (supplemental Fig. S1).
Pathway Analysis for RUNX2 Target Genes
To identify RUNX2-dependent regulatory networks that support its postulated pathological activity in osteosarcoma cells, we carried out gene ontology analysis using GeneSpring, Ingenuity, and David 2.0 annotation programs (70). Target genes that exhibit the most robust binding in ChIP-on-chip analyses (i.e. genes with highest average log2 ratios; n = 1000) were selected for these analyses. Based on GeneSpring analysis, RUNX2 occupies genes that support cell signaling by a number of extracellular ligands (e.g. Wnt, TNFα, TGFβ, EGF receptor, Notch, Hedgehog, and A6B4 integrin) (supplemental Table S4). A6B4 integrin and TGFBR are of particular interest because these pathways are linked to cell adhesion and Smad signaling, respectively, which are both mechanistically associated with the biological functions of RUNX2 (supplemental Fig. S2) (3). Ingenuity pathway analysis revealed that RUNX2 controls networks related to general cellular functions (e.g. protein synthesis, cellular assembly and organization, cell morphology, as well as molecular transport and amino acid metabolism) (supplemental Table S5). Furthermore, RUNX2 target genes are associated with, for example, endocrine disorders, immunological diseases, cancer, cell cycle, cellular movement, tumor morphology, and embryonic development. DAVID 2.0 analysis identified a large number of zinc ion-binding proteins (e.g. zinc finger transcription factors), as well as other transcription factors (e.g. ETS-related proteins) and co-regulatory factors (e.g. SMAD4 and WW domain proteins) (supplemental Table S6). Taken together, these analyses indicate that RUNX2 controls multiple distinct cellular functions in osteosarcoma cells.
Genome-wide Responsiveness of Target Genes to Modulation by RUNX2 siRNA
To understand which genes are most responsive to modulations in Runx2 levels, we determined expression values of genes using Affymetrix cDNA microarray profiling in actively proliferating SAOS-2 cells that were treated with or without siRUNX2. SAOS-2 cells were treated with three distinct siRNAs for RUNX2 and three nonsilencing oligos. Analysis of these triplicate datasets identified ∼140 genes that exhibit statistically significant changes in expression (p < 0.05) and ∼80 genes that trended toward significance (0.05 < p < 0.07) by the Student's t test (supplemental Table S7 and data not shown). Differences in the efficacies of the three siRNAs in reducing RUNX2 levels generate statistical variation that prevents positive identification of responsive target genes. Therefore, we also determined gene expression values for the most effective siRunx2 oligo compared with the results for the three nonsilencing oligos. This analysis identified ∼488 genes that are modulated by 2-fold (statistical significance determined using a Z-test) (supplemental Table S8).
We compared genes directly bound by RUNX2 (identified by ChIP-on-chip analysis) with genes that display changes in expression upon RUNX2 depletion. The inherent difficulty of this comparison is that genes that are most tightly bound are least responsive to modest reductions in RUNX2 levels (see “Discussion”). Indeed, of the ∼220 genes that responded to changes in RUNX2 gene expression (using a t test), only 25 genes were also identified by RUNX2 promoter binding. Just 10 of these genes exhibit a greater than 1.4-fold change in expression upon RUNX2 depletion, beyond RUNX2 itself (see below). Similarly, of the ∼488 genes that change by 2-fold (using a Z-test) (supplemental Table S8), only 67 were also discovered by ChIP-on-chip analysis (data not shown). The low representation of genes that are robustly modulated based on expression profiling and also detected by ChIP-on-chip analysis indicates that direct identification of RUNX2 targets by ChIP-on-chip does not necessarily predict responsiveness of genes to modulation of RUNX2 levels by siRNA knockdown (see “Discussion”).
Of the 10 genes that are modulated >1.4-fold respond to siRUNX2 depletion and exhibit promoter occupancy by RUNX2, two genes exhibit decreased expression as follows: the planar cell polarity related gene PRICKLE1 and the RNA polymerase III regulatory factor SNAPC1 (supplemental Table S9). The expression of a third RUNX2-bound gene, TGFBR2, also increases with siRUNX2, but this gene does not associate with RNAPII (i.e. in untreated control cells). This result suggests that TGFBR2 may be both directly and indirectly controlled by RUNX2. We identified five genes that are negatively regulated by RUNX2, based on clearly increased expression (>1.7-fold) with siRUNX2. Of these, three genes are transcribed or poised for transcription and suppressed by RUNX2 based on the presence of both RUNX2 and RNAPII (i.e. RUNX1, RBBP4, and COL5A1). Regulation of RUNX1 by RUNX2 reflects cross-regulation that complements auto-regulatory mechanisms for RUNX genes (66). RBBP4 interacts with the RUNX2-binding proteins pRB and HDAC3, which are both linked to gene repression (71), suggesting that RUNX2 participates in intricate transcriptional inhibitory networks. Two Runx2-bound genes (i.e. IGFS4 and PSCD2) do not associate with RNAPII and are thus transcriptionally inactive, perhaps due to active repression by RUNX2. We conclude that RUNX2 interacts with genes that are actively transcribed or poised for transcription in osteosarcoma cells, as well as silent genes.
RUNX2 Regulates Genes Involved in Cell Adhesion and Motility in Osteosarcoma Cells and Breast Cancer Cells
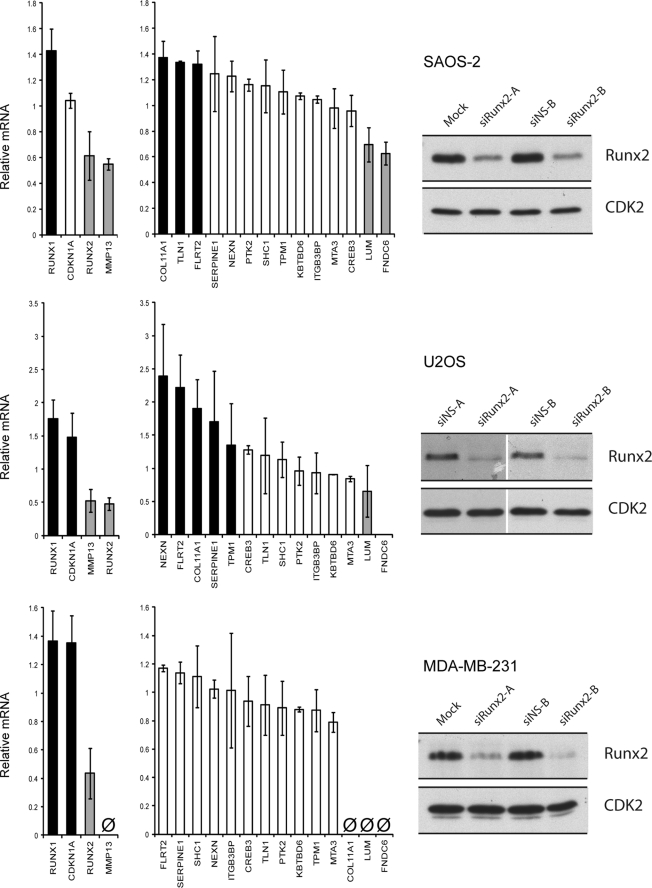
Many genes that strongly interact with RUNX2 have functions in cell motility and/or adhesion (Figs. 4–6). For example, two prominent target genes (i.e. appearing at the top of supplemental Table S2) are related to FAK function, i.e. FAK/PTK2 and Talin 1 (TLN1). We first examined these two genes and other representative targets by RT-qPCR analysis using mRNA isolated from cells treated with or without siRUNX2. We used SAOS-2 and U2OS osteosarcoma cells, which express endogenous RUNX2, as well as MDA-MB-231 breast cancer cells that ectopically express RUNX2 (47).
FIGURE 4.
RUNX2 regulates genes that play a role in cell motility. RUNX2 knockdown with two different siRunx2 oligonucleotides (48 h) in SAOS-2, U2OS, and MDA-MB-231 cells affects several genes that are involved in motility and adhesion of cells as identified with ChIP-on-chip. The bar graphs (middle panels) show average effects of the two different siRNA oligos compared with two negative nonsilencing RNAs on gene expression as measured by RT-qPCR. The errors in technical replicates within the same biological sample were negligible and not indicated. Black and gray bars indicate effects of more than 30%; white bars indicate no significant effects; Ø indicates no expression. RUNX2 knockdown at the protein level (left panels) is shown by Western blot analysis using lysates from SAOS-2, U2OS, and MDA-MB-231 cells treated with or without siRUNX2. Western blots were quantified using ImageJ (imagej.nih.gov) and revealed a >70% reduction in RUNX2 levels in all cell lines.
FIGURE 5.
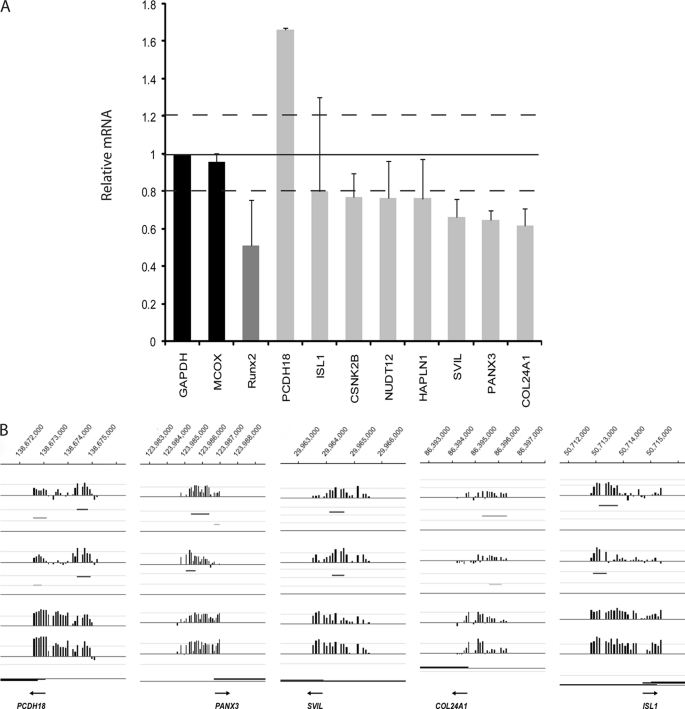
RUNX2 target genes that play a role in cell motility and adhesion as identified using a qPCR array. A, qPCR array was developed for ∼50 different genes. The genes that showed more than 20% up- or down-regulation (on average) are displayed in this graph. RUNX2 knockdown (48 h) in SAOS-2 cells affects several genes that are involved in cell motility and adhesion. Averages are shown from two different siRunx2 oligonucleotides versus the average of mock transfection and nonsilencing RNA. Expression levels were analyzed using a qPCR array with GAPDH as internal control. Black bars, negative controls; dark gray bar, RUNX2 down-regulation; light gray bars, new potential target genes that are affected more than 20% (indicated with dotted lines) compared with GAPDH upon treatment with siRUNX2 (“smart pool”) oligos. B, RUNX2 occupancy on gene promoters of a selection of genes that are siRNA-responsive. Occupancy of independent duplicate experiments are shown (n = 1 and n = 2). Significant peak regions are shown in different shades of gray, based on level of significance.
FIGURE 6.
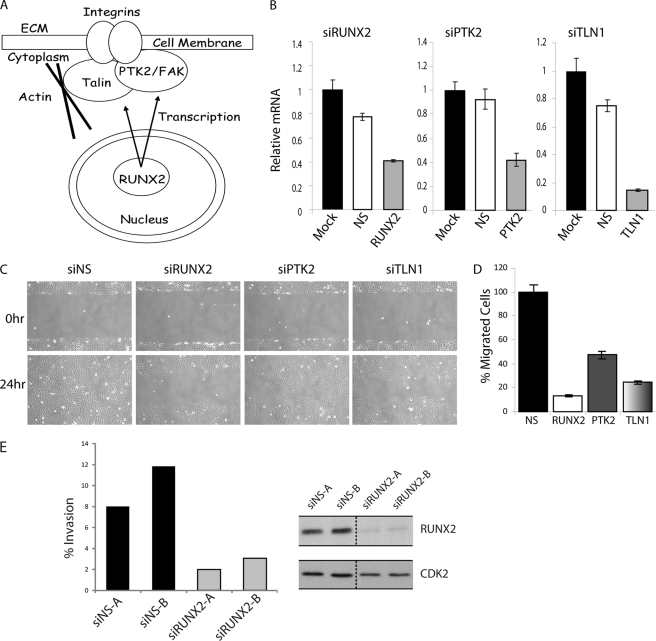
RUNX2 down-regulation inhibits motility of U2OS osteosarcoma cells. A, diagram showing the transcriptional regulation of two proteins (PTK2/FAK and TLN) that function together at focal adhesions at the cell surface. B, RT-qPCR analysis of RUNX2, PTK2, and TLN1 knockdown by siRNA (48 h) in U2OS cells, nonsilencing siRNA (NS) was used as control. Error bars represent S.D. values of triplicate measurements. C, depletion of RUNX2 and two representative target genes delays migration of U2OS cells in wound healing assays. Cell migration was analyzed by phase contrast microscopy over a 24-h time course. Representative images of wound healing at 0 and 24 h after scratch are presented. D, difference in cell migration and wound healing was quantified as the percentage of wound healing compared with control siRNA. Wound healing was quantified using ImageJ (rsbweb.nih.gov). Values represent averages (± S.D.) of three independent measurements along the wound scratch. Data are representative of three independent experiments. E, invasion of U2OS cells is impaired when RUNX2 is down-regulated (48 h) in a transwell culture assay with different oligos siRUNX2-A and -B. Right panels, RUNX2 down-regulation at the protein level. Graphs depict the number of invaded cells as a percentage of the migrated cells, based on four separate cell counts that covered almost the entire well. The averages and S.D. values of these cell counts show only minimal technical variation and are not shown. Biological variation is evidenced by the values obtained for two different siRNAs for specific depletion of Runx2 (siRUNX2-A and -B) and two negative nonsilencing controls (siNS-A and siNS-B). One of two representative experiments with similar results is shown.
Control experiments yielded the desired result that RUNX2 mRNA and protein were each significantly down-regulated with two distinct siRNA oligos in all three cell lines (∼40–60% mRNA and >70% protein) (Fig. 4). Using RT-qPCR, we also validated several genes that are related to the FAK pathway (NEXN, TPM1, and SHC1), as well as other genes that play a role in migration/adhesion targets by RT-qPCR validation (Figs. 4 and 5). The expression of several motility-related genes (e.g. COL11A1 and FLRT2) is increased by more than 30% in response to siRUNX2 in SAOS-2 cells, suggesting that these genes are normally repressed by RUNX2. In MDA-MB-231 cells, the response of these same genes to siRUNX2 was more modest (Fig. 4). Five genes that exhibit altered expression in RT-qPCR arrays upon RUNX2 depletion (PCDH18, PANX3, SVIL, COL24A1, and ISL1) exhibit clear RUNX2 occupancy in duplicate ChIP-on-chip experiments (Fig. 5).
ChIP-on-chip analysis identified a number of proteins associated with the FAK pathway (for example, PTK2, TLN1, NEXN, TPM1, and SHC1). This finding suggests that RUNX2 controls cellular functions linked to motility and adhesion. PTK2 and TLN1 are among the 10 most prominent RUNX2 target genes (by peak ratio), and both PTK2 and TLN1 are bound by RNAPII (Fig. 2 and supplemental Table S2 and data not shown). PTK and TLN1 are functionally related genes that form a complex at focal adhesions to connect integrins with the actin cytoskeleton (Fig. 6A) (72, 73). Therefore, these two proteins are in principle very attractive candidate target genes that may contribute to the predicted role of RUNX2 in cell adhesion and motility in osteosarcoma cells.
In testing this hypothesis, we encountered two major technical obstacles. First, standard SAOS-2 cells are neither motile nor invasive (supplemental Fig. S3 and data not shown). Hence, RUNX2 binding to components of the FAK pathway (e.g. TLN1 and PTK2) is not sufficient to generate a mobile cell. Therefore, we investigated the function of RUNX2 and its targets PTK2 and TLN1 in U2OS cells, which are intrinsically capable of migrating in cell culture.
Second, RUNX2 depletion in SAOS-2, U2OS, or MDA-MB-231 cells has modest if any effects on TLN1 or PTK2 as established by RT-qPCR (Fig. 4). Also, neither TLN1 nor PTK2 was discovered as a Runx2-responsive gene by Affymetrix profiling using SAOS-2 cells (see supplemental Tables S7 and S8). Because of the lack of effects on the respective mRNAs, RUNX2 depletion is not likely to change the protein levels of either TLN1 or PTK2. Therefore, we tested the functions of these two RUNX2 targets directly using siRNAs for PTK2 and TLN1 in U2OS cells in parallel with studies using siRNA for RUNX2.
Cell motility and invasion studies were performed with in vitro wound healing (“scratch”) assays and trans-well systems (Fig. 6, B–E). RUNX2 siRNA only reduces RUNX2 mRNA by 60% (Fig. 6B), but RUNX2 protein is decreased to barely detectable levels as determined by immunoblot analysis (Fig. 6E). Thus, we have obtained a robust knockdown of Runx2 that is expected to achieve a biological effect.
Cell motility is strongly reduced upon RUNX2 depletion and to a lesser extent upon depletion of either TLN1 or PTK2 using siRNAs (Fig. 6, B–D). Furthermore, studies that monitor migration through Matrigel revealed that RUNX2 depletion results in considerable inhibition of U2OS cell invasion (Fig. 6E). The latter experiments were carried out with two different RUNX2 siRNAs. We obtained consistent results thus ruling out siRNA off-target effects. The combined results obtained with siRUNX2, siPTK2, and siTLN1 (Fig. 6) corroborate the generalized concept that RUNX2 regulates motility and is bound to a broad spectrum of motility-related genes. However, because RUNX2 depletion does not appreciably change PTK2 or TLN1 expression, these two genes cannot account for the observed siRUNX2-dependent changes in cell motility. We propose that other key target genes that are less tightly bound by RUNX2 may become rate-limiting for cell motility when RUNX2 gene expression is inhibited.
DISCUSSION
Aberrant expression of RUNX proteins has been linked to pathological events in cancer cells. For example, increased expression of RUNX2 is observed in both osseous (e.g. osteosarcoma) and nonosseous cancers (e.g. breast and prostate) (7, 8, 34–36), indicating that RUNX2 may have an oncogenic function in tumor etiology. In this study, we have examined physiological targets of RUNX2 in osteosarcoma cells where the protein is endogenously overexpressed relative to normal osteoblastic cells. We analyzed genome-wide interactions of RUNX2 and RNAPII with gene promoters and also performed gene expression profiling in cells depleted of RUNX2. One key finding is that RUNX2 is bound to a large number (>2,000) of genes that are either actively transcribed or poised for expression (∼60–70%) based on co-interactions of the same genes with RNAPII. RUNX2 also interacts with genes that appear to be inactive (∼30–40%) as reflected by the absence of RNAPII. The global observation that RUNX2 binds to both active and inactive genes in osteosarcoma cells is consistent with the general concept that RUNX2 is a bi-functional regulator that can activate or repress gene transcription depending on promoter context and cellular milieu.
Previous studies with osteogenic and osteosarcoma cells, as well as breast and prostate tumor cells, have identified a number of individual genes that respond to changes in RUNX2 protein levels (7, 31, 38–52). For example, in breast cancer cells, RUNX2 controls matrix metalloproteinases (e.g. MMP-9 and MMP-13) and VEGF that may be pathologically linked to key steps in cancer metastasis (74). However, few of the previously identified RUNX2 target genes characterized in different cell types appear to be controlled by RUNX2 in SAOS-2 osteosarcoma cells. Thus, the RUNX2 target genes identified in this study represent a distinct cell context-dependent subset of the total cohort of RUNX2 target genes.
Pathway analysis of RUNX2 target genes indicates that RUNX2 controls multiple regulatory networks. As expected from its well known role as a regulator of osteoblast growth and differentiation, RUNX2 regulates genes that support bone cell growth and survival, as well as lineage commitment and differentiation (i.e. WNT, TFGβ, TNFα, and EGF signaling). However, the most interesting discovery of this study is that RUNX2 also controls pathways that are broadly related to cell adhesion and motility. Our results establish that depletion of RUNX2 or the RUNX2 target genes TLN1 or PTK2 decreases motility of U2OS cells. We note that these genes do not affect motility of SAOS-2 cells, because these cells are relatively immobile. Our finding that RUNX2 may have functions in cell adhesion and motility of mobile osteosarcoma cells complements earlier findings obtained with both loss-of function and gain-of-function experiments in murine models (e.g. Runx2 null mouse embryonic fibroblasts and murine T cell lymphoma cells (14–18)), as well as human breast and prostate cancer cells (45–47, 75–78).
Our study combined ChIP data with gene expression profiling results of SAOS-2 cells treated with RUNX2 siRNA. The results revealed sets of RUNX2 target genes that are either down- or up-regulated by RUNX2 depletion, corroborating the bi-functional role of RUNX2 in gene regulation in osteosarcoma cells. Triplicate Affymetrix gene expression experiments with siRUNX2- treated cells identified multiple genes (between ∼200 and ∼1000) that were significantly modulated (>1.4-fold), but only a small subset (<20) of these genes were also occupied by RUNX2. We also observed that a number of RUNX2-bound genes are not responsive to RUNX2 depletion, although RUNX2 occupancy is clearly and reproducibly evident in separate experiments. The observation that tightly bound target genes are not necessarily the most responsive to modulations in RUNX2 levels may have several molecular explanations (reviewed in Ref. 79). For example, reduction of RUNX2 levels is more likely to affect genes to which RUNX2 binds with low affinity, because RUNX2 may only vacate high affinity genes upon complete loss of RUNX2 expression.
Two of the more fascinating genes controlled by RUNX2 are the focal adhesion kinase PTK2/FAK and TLN1. Both proteins are part of a complex that connects integrins with the actin cytoskeleton (72, 73). Consistent with the important role of these proteins and other RUNX2-responsive genes in cell adhesion and motility, knockdown of RUNX2, TLN1, or PTK2/FAK alters motility of U2OS cells. Because RUNX2 depletion only minimally affects the mRNA levels of either TLN1 or PTK2 (presumably because these genes are very tightly bound by RUNX2), the observed effects of RUNX2 on cell motility reflect broader involvement of RUNX2 in regulating expression programs supporting cellular movement.
In summary, RUNX2 interacts with the promoters of a cohort of motility genes in SAOS-2 cells. These interactions are not sufficient to generate a mobile cell phenotype, because SAOS-2 cells are not particularly motile. Depletion of RUNX2, TLN1, and PTK2 affects motility of U2OS but not SAOS-2 cells. These findings indicate that RUNX2 is important for movement, but this effect clearly differs among osteosarcoma cell lines and thus depends on biological context. Finally, although the focal adhesion-related RUNX2 target genes TLN1 and PTK2 control cell movement in U2OS cells, RUNX2 becomes rate-limiting at levels that do not yet affect TLN1 or PTK2 gene expression. Therefore, it appears that RUNX2 can control cell movement through alternate molecular pathways, perhaps independent of TLN1 or PTK2. We conclude that elevation of RUNX2 levels in osteosarcoma cells supports its binding to diverse gene loci, which may be linked to the pathology of this pediatric bone cancer.
Acknowledgments
We thank Srivatsan Padmanabhan, Sandhya Pande, Viktor Teplyuk, Nadiya Teplyuk, Jitesh Pratap, S. Kaleem Zaidi, Mei Xu, Akhter Ali, and Mohammad Hassan for stimulating discussions. We also thank Charlene Baron for digital imaging advice, Judy Rask for assistance with manuscript preparation, and Phyllis Spatrick from the University of Massachusetts Genomics Core for assistance with Affymetrix analysis.
This work was supported, in whole or in part, by National Institutes of Health Grants AR49069, CA082834, and AR048818, as well as from contract grant sponsor FONDAP 15090007.

This article contains supplemental Figs. S1–S3 and Tables S1–S9.
A. J. van Wijnen, G. S. Stein, J. B. Lian, and J. L. Stein, unpublished observations.
- qPCR
- quantitative PCR
- RNAP
- RNA polymerase II
- oligo
- oligonucleotide
- TSS
- transcription start site
- FAK
- focal adhesion kinase.
REFERENCES
- 1. Wang C. Q., Jacob B., Nah G. S., Osato M. (2010) Runx family genes, niche, and stem cell quiescence. Blood Cells Mol. Dis. 44, 275–286 [DOI] [PubMed] [Google Scholar]
- 2. Niebuhr B., Fischer M., Täger M., Cammenga J., Stocking C. (2008) Gatekeeper function of the RUNX1 transcription factor in acute leukemia. Blood Cells Mol. Dis. 40, 211–218 [DOI] [PubMed] [Google Scholar]
- 3. Lian J. B., Stein G. S., Javed A., van Wijnen A. J., Stein J. L., Montecino M., Hassan M. Q., Gaur T., Lengner C. J., Young D. W. (2006) Networks and hubs for the transcriptional control of osteoblastogenesis. Rev. Endocr. Metab. Disord. 7, 1–16 [DOI] [PubMed] [Google Scholar]
- 4. Chuang L. S., Ito Y. (2010) RUNX3 is multifunctional in carcinogenesis of multiple solid tumors. Oncogene 29, 2605–2615 [DOI] [PubMed] [Google Scholar]
- 5. Subramaniam M. M., Chan J. Y., Yeoh K. G., Quek T., Ito K., Salto-Tellez M. (2009) Molecular pathology of RUNX3 in human carcinogenesis. Biochim. Biophys. Acta 1796, 315–331 [DOI] [PubMed] [Google Scholar]
- 6. Kilbey A., Terry A., Cameron E. R., Neil J. C. (2008) Oncogene-induced senescence. An essential role for Runx. Cell Cycle 7, 2333–2340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pratap J., Lian J. B., Javed A., Barnes G. L., van Wijnen A. J., Stein J. L., Stein G. S. (2006) Regulatory roles of Runx2 in metastatic tumor and cancer cell interactions with bone. Cancer Metastasis Rev. 25, 589–600 [DOI] [PubMed] [Google Scholar]
- 8. Blyth K., Cameron E. R., Neil J. C. (2005) The RUNX genes. Gain or loss of function in cancer. Nat. Rev. Cancer 5, 376–387 [DOI] [PubMed] [Google Scholar]
- 9. Okuda T., van Deursen J., Hiebert S. W., Grosveld G., Downing J. R. (1996) AML1, the target of multiple chromosomal translocations in human leukemia, is essential for normal fetal liver hematopoiesis. Cell 84, 321–330 [DOI] [PubMed] [Google Scholar]
- 10. Wang Q., Stacy T., Binder M., Marin-Padilla M., Sharpe A. H., Speck N. A. (1996) Disruption of the Cbfa2 gene causes necrosis and hemorrhaging in the central nervous system and blocks definitive hematopoiesis. Proc. Natl. Acad. Sci. U.S.A. 93, 3444–3449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dowdy C. R., Xie R., Frederick D., Hussain S., Zaidi S. K., Vradii D., Javed A., Li X., Jones S. N., Lian J. B., van Wijnen A. J., Stein J. L., Stein G. S. (2010) Definitive hematopoiesis requires Runx1 C-terminal mediated subnuclear targeting and transactivation. Hum. Mol. Genet. 19, 1048–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Li Q. L., Ito K., Sakakura C., Fukamachi H., Inoue K., Chi X. Z., Lee K. Y., Nomura S., Lee C. W., Han S. B., Kim H. M., Kim W. J., Yamamoto H., Yamashita N., Yano T., Ikeda T., Itohara S., Inazawa J., Abe T., Hagiwara A., Yamagishi H., Ooe A., Kaneda A., Sugimura T., Ushijima T., Bae S. C., Ito Y. (2002) Causal relationship between the loss of RUNX3 expression and gastric cancer. Cell 109, 113–124 [DOI] [PubMed] [Google Scholar]
- 13. Inoue K., Ozaki S., Shiga T., Ito K., Masuda T., Okado N., Iseda T., Kawaguchi S., Ogawa M., Bae S. C., Yamashita N., Itohara S., Kudo N., Ito Y. (2002) Runx3 controls the axonal projection of proprioceptive dorsal root ganglion neurons. Nat. Neurosci. 5, 946–954 [DOI] [PubMed] [Google Scholar]
- 14. Woolf E., Xiao C., Fainaru O., Lotem J., Rosen D., Negreanu V., Bernstein Y., Goldenberg D., Brenner O., Berke G., Levanon D., Groner Y. (2003) Runx3 and Runx1 are required for CD8 T cell development during thymopoiesis. Proc. Natl. Acad. Sci. U.S.A. 100, 7731–7736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Stewart M., Terry A., O'Hara M., Cameron E., Onions D., Neil J. C. (1996) til-1. A novel proviral insertion locus for Moloney murine leukemia virus in lymphomas of CD2-myc transgenic mice. J. Gen. Virol. 77, 443–446 [DOI] [PubMed] [Google Scholar]
- 16. Stewart M., Terry A., Hu M., O'Hara M., Blyth K., Baxter E., Cameron E., Onions D. E., Neil J. C. (1997) Proviral insertions induce the expression of bone-specific isoforms of PEBP2αA (CBFA1). Evidence for a new myc collaborating oncogene. Proc. Natl. Acad. Sci. U.S.A. 94, 8646–8651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Vaillant F., Blyth K., Andrew L., Neil J. C., Cameron E. R. (2002) Enforced expression of Runx2 perturbs T cell development at a stage coincident with β-selection. J. Immunol. 169, 2866–2874 [DOI] [PubMed] [Google Scholar]
- 18. Blyth K., Vaillant F., Hanlon L., Mackay N., Bell M., Jenkins A., Neil J. C., Cameron E. R. (2006) Runx2 and MYC collaborate in lymphoma development by suppressing apoptotic and growth arrest pathways in vivo. Cancer Res. 66, 2195–2201 [DOI] [PubMed] [Google Scholar]
- 19. Mundlos S., Otto F., Mundlos C., Mulliken J. B., Aylsworth A. S., Albright S., Lindhout D., Cole W. G., Henn W., Knoll J. H., Owen M. J., Mertelsmann R., Zabel B. U., Olsen B. R. (1997) Mutations involving the transcription factor CBFA1 cause cleidocranial dysplasia. Cell 89, 773–779 [DOI] [PubMed] [Google Scholar]
- 20. Ott C. E., Leschik G., Trotier F., Brueton L., Brunner H. G., Brussel W., Guillen-Navarro E., Haase C., Kohlhase J., Kotzot D., Lane A., Lee-Kirsch M. A., Morlot S., Simon M. E., Steichen-Gersdorf E., Tegay D. H., Peters H., Mundlos S., Klopocki E. (2010) Deletions of the RUNX2 gene are present in about 10% of individuals with cleidocranial dysplasia. Hum. Mutat. 31, E1587–E1593 [DOI] [PubMed] [Google Scholar]
- 21. Han M. S., Kim H. J., Wee H. J., Lim K. E., Park N. R., Bae S. C., van Wijnen A. J., Stein J. L., Lian J. B., Stein G. S., Choi J. Y. (2010) The cleidocranial dysplasia-related R131G mutation in the Runt-related transcription factor RUNX2 disrupts binding to DNA but not CBF-β. J. Cell. Biochem. 110, 97–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kim H. J., Nam S. H., Kim H. J., Park H. S., Ryoo H. M., Kim S. Y., Cho T. J., Kim S. G., Bae S. C., Kim I. S., Stein J. L., van Wijnen A. J., Stein G. S., Lian J. B., Choi J. Y. (2006) Four novel RUNX2 mutations including a splice donor site result in the cleidocranial dysplasia phenotype. J. Cell. Physiol. 207, 114–122 [DOI] [PubMed] [Google Scholar]
- 23. Komori T., Yagi H., Nomura S., Yamaguchi A., Sasaki K., Deguchi K., Shimizu Y., Bronson R. T., Gao Y. H., Inada M., Sato M., Okamoto R., Kitamura Y., Yoshiki S., Kishimoto T. (1997) Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell 89, 755–764 [DOI] [PubMed] [Google Scholar]
- 24. Otto F., Thornell A. P., Crompton T., Denzel A., Gilmour K. C., Rosewell I. R., Stamp G. W., Beddington R. S., Mundlos S., Olsen B. R., Selby P. B., Owen M. J. (1997) Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell 89, 765–771 [DOI] [PubMed] [Google Scholar]
- 25. Choi J. Y., Pratap J., Javed A., Zaidi S. K., Xing L., Balint E., Dalamangas S., Boyce B., van Wijnen A. J., Lian J. B., Stein J. L., Jones S. N., Stein G. S. (2001) Subnuclear targeting of Runx/Cbfa/AML factors is essential for tissue-specific differentiation during embryonic development. Proc. Natl. Acad. Sci. U.S.A. 98, 8650–8655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lou Y., Javed A., Hussain S., Colby J., Frederick D., Pratap J., Xie R., Gaur T., van Wijnen A. J., Jones S. N., Stein G. S., Lian J. B., Stein J. L. (2009) A Runx2 threshold for the cleidocranial dysplasia phenotype. Hum. Mol. Genet. 18, 556–568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pratap J., Galindo M., Zaidi S. K., Vradii D., Bhat B. M., Robinson J. A., Choi J. Y., Komori T., Stein J. L., Lian J. B., Stein G. S., van Wijnen A. J. (2003) Cell growth regulatory role of Runx2 during proliferative expansion of preosteoblasts. Cancer Res. 63, 5357–5362 [PubMed] [Google Scholar]
- 28. Galindo M., Pratap J., Young D. W., Hovhannisyan H., Im H. J., Choi J. Y., Lian J. B., Stein J. L., Stein G. S., van Wijnen A. J. (2005) The bone-specific expression of Runx2 oscillates during the cell cycle to support a G1-related antiproliferative function in osteoblasts. J. Biol. Chem. 280, 20274–20285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Young D. W., Hassan M. Q., Pratap J., Galindo M., Zaidi S. K., Lee S. H., Yang X., Xie R., Javed A., Underwood J. M., Furcinitti P., Imbalzano A. N., Penman S., Nickerson J. A., Montecino M. A., Lian J. B., Stein J. L., van Wijnen A. J., Stein G. S. (2007) Mitotic occupancy and lineage-specific transcriptional control of rRNA genes by Runx2. Nature 445, 442–446 [DOI] [PubMed] [Google Scholar]
- 30. Zaidi S. K., Pande S., Pratap J., Gaur T., Grigoriu S., Ali S. A., Stein J. L., Lian J. B., van Wijnen A. J., Stein G. S. (2007) Runx2 deficiency and defective subnuclear targeting bypass senescence to promote immortalization and tumorigenic potential. Proc. Natl. Acad. Sci. U.S.A. 104, 19861–19866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Teplyuk N. M., Galindo M., Teplyuk V. I., Pratap J., Young D. W., Lapointe D., Javed A., Stein J. L., Lian J. B., Stein G. S., van Wijnen A. J. (2008) Runx2 regulates G protein-coupled signaling pathways to control growth of osteoblast progenitors. J. Biol. Chem. 283, 27585–27597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kilbey A., Blyth K., Wotton S., Terry A., Jenkins A., Bell M., Hanlon L., Cameron E. R., Neil J. C. (2007) Runx2 disruption promotes immortalization and confers resistance to oncogene-induced senescence in primary murine fibroblasts. Cancer Res. 67, 11263–11271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Galindo M., Kahler R. A., Teplyuk N. M., Stein J. L., Lian J. B., Stein G. S., Westendorf J. J., van Wijnen A. J. (2007) Cell cycle related modulations in Runx2 protein levels are independent of lymphocyte enhancer-binding factor 1 (Lef1) in proliferating osteoblasts. J. Mol. Histol. 38, 501–506 [DOI] [PubMed] [Google Scholar]
- 34. San Martin I. A., Varela N., Gaete M., Villegas K., Osorio M., Tapia J. C., Antonelli M., Mancilla E. E., Pereira B. P., Nathan S. S., Lian J. B., Stein J. L., Stein G. S., van Wijnen A. J., Galindo M. (2009) Impaired cell cycle regulation of the osteoblast-related heterodimeric transcription factor Runx2-Cbfβ in osteosarcoma cells. J. Cell. Physiol. 221, 560–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pereira B. P., Zhou Y., Gupta A., Leong D. T., Aung K. Z., Ling L., Pho R. W., Galindo M., Salto-Tellez M., Stein G. S., Cool S. M., van Wijnen A. J., Nathan S. S. (2009) Runx2, p53, and pRB status as diagnostic parameters for deregulation of osteoblast growth and differentiation in a new prechemotherapeutic osteosarcoma cell line (OS1). J. Cell. Physiol. 221, 778–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sadikovic B., Thorner P., Chilton-Macneill S., Martin J. W., Cervigne N. K., Squire J., Zielenska M. (2010) Expression analysis of genes associated with human osteosarcoma tumors shows correlation of RUNX2 overexpression with poor response to chemotherapy. BMC Cancer 10, 202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Martin J. W., Yoshimoto M., Ludkovski O., Thorner P. S., Zielenska M., Squire J. A., Nuin P. A. (2010) Analysis of segmental duplications, mouse genome synteny, and recurrent cancer-associated amplicons in human chromosome 6p21-p12. Cytogenet. Genome Res. 128, 199–213 [DOI] [PubMed] [Google Scholar]
- 38. Teplyuk N. M., Zhang Y., Lou Y., Hawse J. R., Hassan M. Q., Teplyuk V. I., Pratap J., Galindo M., Stein J. L., Stein G. S., Lian J. B., van Wijnen A. J. (2009) The osteogenic transcription factor runx2 controls genes involved in sterol/steroid metabolism, including CYP11A1 in osteoblasts. Mol. Endocrinol. 23, 849–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Jeong J. H., Jung Y. K., Kim H. J., Jin J. S., Kim H. N., Kang S. M., Kim S. Y., van Wijnen A. J., Stein J. L., Lian J. B., Stein G. S., Kato S., Choi J. Y. (2010) The gene for aromatase, a rate-limiting enzyme for local estrogen biosynthesis, is a downstream target gene of Runx2 in skeletal tissues. Mol. Cell. Biol. 30, 2365–2375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Young D. W., Hassan M. Q., Yang X. Q., Galindo M., Javed A., Zaidi S. K., Furcinitti P., Lapointe D., Montecino M., Lian J. B., Stein J. L., van Wijnen A. J., Stein G. S. (2007) Mitotic retention of gene expression patterns by the cell fate-determining transcription factor Runx2. Proc. Natl. Acad. Sci. U.S.A. 104, 3189–3194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Westendorf J. J., Zaidi S. K., Cascino J. E., Kahler R., van Wijnen A. J., Lian J. B., Yoshida M., Stein G. S., Li X. (2002) Runx2 (Cbfa1, AML-3) interacts with histone deacetylase 6 and represses the p21(CIP1/WAF1) promoter. Mol. Cell. Biol. 22, 7982–7992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Jensen E. D., Niu L., Caretti G., Nicol S. M., Teplyuk N., Stein G. S., Sartorelli V., van Wijnen A. J., Fuller-Pace F. V., Westendorf J. J. (2008) p68 (Ddx5) interacts with Runx2 and regulates osteoblast differentiation. J. Cell. Biochem. 103, 1438–1451 [DOI] [PubMed] [Google Scholar]
- 43. Teplyuk N. M., Haupt L. M., Ling L., Dombrowski C., Mun F. K., Nathan S. S., Lian J. B., Stein J. L., Stein G. S., Cool S. M., van Wijnen A. J. (2009) The osteogenic transcription factor Runx2 regulates components of the fibroblast growth factor/proteoglycan signaling axis in osteoblasts. J. Cell. Biochem. 107, 144–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Haupt L. M., Murali S., Mun F. K., Teplyuk N., Mei L. F., Stein G. S., van Wijnen A. J., Nurcombe V., Cool S. M. (2009) The heparan sulfate proteoglycan (HSPG) glypican-3 mediates commitment of MC3T3-E1 cells toward osteogenesis. J. Cell. Physiol. 220, 780–791 [DOI] [PubMed] [Google Scholar]
- 45. Akech J., Wixted J. J., Bedard K., van der Deen M., Hussain S., Guise T. A., van Wijnen A. J., Stein J. L., Languino L. R., Altieri D. C., Pratap J., Keller E., Stein G. S., Lian J. B. (2010) Runx2 association with progression of prostate cancer in patients. Mechanisms mediating bone osteolysis and osteoblastic metastatic lesions. Oncogene 29, 811–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Pratap J., Imbalzano K. M., Underwood J. M., Cohet N., Gokul K., Akech J., van Wijnen A. J., Stein J. L., Imbalzano A. N., Nickerson J. A., Lian J. B., Stein G. S. (2009) Ectopic runx2 expression in mammary epithelial cells disrupts formation of normal acini structure. Implications for breast cancer progression. Cancer Res. 69, 6807–6814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Pratap J., Wixted J. J., Gaur T., Zaidi S. K., Dobson J., Gokul K. D., Hussain S., van Wijnen A. J., Stein J. L., Stein G. S., Lian J. B. (2008) Runx2 transcriptional activation of Indian Hedgehog and a downstream bone metastatic pathway in breast cancer cells. Cancer Res. 68, 7795–7802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Baniwal S. K., Khalid O., Gabet Y., Shah R. R., Purcell D. J., Mav D., Kohn-Gabet A. E., Shi Y., Coetzee G. A., Frenkel B. (2010) Runx2 transcriptome of prostate cancer cells. Insights into invasiveness and bone metastasis. Mol. Cancer 9, 258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Pregizer S., Barski A., Gersbach C. A., García A. J., Frenkel B. (2007) Identification of novel Runx2 targets in osteoblasts. Cell type-specific BMP-dependent regulation of Tram2. J. Cell. Biochem. 102, 1458–1471 [DOI] [PubMed] [Google Scholar]
- 50. Thirunavukkarasu K., Pei Y., Wei T. (2007) Characterization of the human ADAMTS-5 (aggrecanase-2) gene promoter. Mol. Biol. Rep. 34, 225–231 [DOI] [PubMed] [Google Scholar]
- 51. Thirunavukkarasu K., Pei Y., Moore T. L., Wei T., Wang H., Chandrasekhar S. (2007) Regulation of NFATc2 gene expression by the transcription factor Runx2. Mol. Biol. Rep. 34, 1–10 [DOI] [PubMed] [Google Scholar]
- 52. Kilbey A., Terry A., Jenkins A., Borland G., Zhang Q., Wakelam M. J., Cameron E. R., Neil J. C. (2010) Runx regulation of sphingolipid metabolism and survival signaling. Cancer Res. 70, 5860–5869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lee T. I., Johnstone S. E., Young R. A. (2006) Chromatin immunoprecipitation and microarray-based analysis of protein location. Nat. Protoc. 1, 729–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. van der Deen M., Hassan M. Q., Pratap J., Teplyuk N. M., Young D. W., Javed A., Zaidi S. K., Lian J. B., Montecino M., Stein J. L., Stein G. S., van Wijnen A. J. (2008) Chromatin immunoprecipitation assays. Application of ChIP-on-chip for defining dynamic transcriptional mechanisms in bone cells. Methods Mol. Biol. 455, 165–176 [DOI] [PubMed] [Google Scholar]
- 55. O'Geen H., Nicolet C. M., Blahnik K., Green R., Farnham P. J. (2006) Comparison of sample preparation methods for ChIP-chip assays. BioTechniques 41, 577–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Drissi H., Pouliot A., Stein J. L., van Wijnen A. J., Stein G. S., Lian J. B. (2002) Identification of novel protein/DNA interactions within the promoter of the bone-related transcription factor Runx2/Cbfa1. J. Cell. Biochem. 86, 403–412 [DOI] [PubMed] [Google Scholar]
- 57. Frith M. C., Fu Y., Yu L., Chen J. F., Hansen U., Weng Z. (2004) Detection of functional DNA motifs via statistical over-representation. Nucleic Acids Res. 32, 1372–1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. McLeay R. C., Bailey T. L. (2010) Motif enrichment analysis. A unified framework and an evaluation on ChIP data. BMC Bioinformatics 11, 165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Vlieghe D., Sandelin A., De Bleser P. J., Vleminckx K., Wasserman W. W., van Roy F., Lenhard B. (2006) A new generation of JASPAR, the open-access repository for transcription factor binding site profiles. Nucleic Acids Res. 34, D95–D97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Sandelin A., Alkema W., Engström P., Wasserman W. W., Lenhard B. (2004) JASPAR. An open-access database for eukaryotic transcription factor binding profiles. Nucleic Acids Res. 32, D91–D94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Knüppel R., Dietze P., Lehnberg W., Frech K., Wingender E. (1994) TRANSFAC retrieval program. A network model database of eukaryotic transcription regulating sequences and proteins. J. Comput. Biol. 1, 191–198 [DOI] [PubMed] [Google Scholar]
- 62. Li C., Wong W. H. (2001) Model-based analysis of oligonucleotide arrays. Expression index computation and outlier detection. Proc. Natl. Acad. Sci. U.S.A. 98, 31–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Balint E., Lapointe D., Drissi H., van der Meijden C., Young D. W., van Wijnen A. J., Stein J. L., Stein G. S., Lian J. B. (2003) Phenotype discovery by gene expression profiling. Mapping of biological processes linked to BMP-2-mediated osteoblast differentiation. J. Cell. Biochem. 89, 401–426 [DOI] [PubMed] [Google Scholar]
- 64. van der Meijden C. M., Lapointe D. S., Luong M. X., Peric-Hupkes D., Cho B., Stein J. L., van Wijnen A. J., Stein G. S. (2002) Gene profiling of cell cycle progression through S-phase reveals sequential expression of genes required for DNA replication and nucleosome assembly. Cancer Res. 62, 3233–3243 [PubMed] [Google Scholar]
- 65. Ruijter J. M., Ramakers C., Hoogaars W. M., Karlen Y., Bakker O., van den Hoff M. J., Moorman A. F. (2009) Amplification efficiency. Linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res. 37, e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Drissi H., Luc Q., Shakoori R., Chuva De Sousa Lopes S., Choi J. Y., Terry A., Hu M., Jones S., Neil J. C., Lian J. B., Stein J. L., Van Wijnen A. J., Stein G. S. (2000) Transcriptional autoregulation of the bone related CBFA1/RUNX2 gene. J. Cell. Physiol. 184, 341–350 [DOI] [PubMed] [Google Scholar]
- 67. Rodan G. A., Noda M. (1991) Gene expression in osteoblastic cells. Crit. Rev. Eukaryot. Gene Expr. 1, 85–98 [PubMed] [Google Scholar]
- 68. Meyers S., Downing J. R., Hiebert S. W. (1993) Identification of AML-1 and the (8;21) translocation protein (AML-1/ETO) as sequence-specific DNA-binding proteins. The runt homology domain is required for DNA binding and protein-protein interactions. Mol. Cell. Biol. 13, 6336–6345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Otto F., Lübbert M., Stock M. (2003) Upstream and downstream targets of RUNX proteins. J. Cell. Biochem. 89, 9–18 [DOI] [PubMed] [Google Scholar]
- 70. Huang da W., Sherman B. T., Lempicki R. A. (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 [DOI] [PubMed] [Google Scholar]
- 71. Wang C., Fu M., Mani S., Wadler S., Senderowicz A. M., Pestell R. G. (2001) Histone acetylation and the cell-cycle in cancer. Front Biosci. 6, D610–D629 [DOI] [PubMed] [Google Scholar]
- 72. Shattil S. J., Kim C., Ginsberg M. H. (2010) The final steps of integrin activation. The end game. Nat. Rev. Mol. Cell Biol. 11, 288–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Moser M., Legate K. R., Zent R., Fässler R. (2009) The tail of integrins, talin, and kindlins. Science 324, 895–899 [DOI] [PubMed] [Google Scholar]
- 74. Pratap J., Javed A., Languino L. R., van Wijnen A. J., Stein J. L., Stein G. S., Lian J. B. (2005) The Runx2 osteogenic transcription factor regulates matrix metalloproteinase 9 in bone metastatic cancer cells and controls cell invasion. Mol. Cell. Biol. 25, 8581–8591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Leong D. T., Lim J., Goh X., Pratap J., Pereira B. P., Kwok H. S., Nathan S. S., Dobson J. R., Lian J. B., Ito Y., Voorhoeve P. M., Stein G. S., Salto-Tellez M., Cool S. M., van Wijnen A. J. (2010) Cancer-related ectopic expression of the bone-related transcription factor RUNX2 in non-osseous metastatic tumor cells is linked to cell proliferation and motility. Breast Cancer Res. 12, R89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Das K., Leong D. T., Gupta A., Shen L., Putti T., Stein G. S., van Wijnen A. J., Salto-Tellez M. (2009) Positive association between nuclear Runx2 and estrogen-progesterone receptor gene expression characterizes a biological subtype of breast cancer. Eur. J. Cancer 45, 2239–2248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Pratap J., Lian J. B., Stein G. S. (2011) Metastatic bone disease. Role of transcription factors and future targets. Bone 48, 30–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. van der Deen M., Akech J., Wang T., FitzGerald T. J., Altieri D. C., Languino L. R., Lian J. B., van Wijnen A. J., Stein J. L., Stein G. S. (2010) The cancer-related Runx2 protein enhances cell growth and responses to androgen and TGFβ in prostate cancer cells. J. Cell. Biochem. 109, 828–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Farnham P. J. (2009) Insights from genomic profiling of transcription factors. Nat. Rev. Genet. 10, 605–616 [DOI] [PMC free article] [PubMed] [Google Scholar]