Abstract
Inactivating mutations in the breast cancer susceptibility gene BRCA2 cause gross chromosomal rearrangements. Chromosome structural instability in the absence of BRCA2 is thought to result from defective homology-directed DNA repair. Here, we show that BRCA2 links the fidelity of telomere maintenance with genetic integrity. Absence of BRCA2 resulted in signs of dysfunctional telomeres, such as telomere shortening, erosions, and end fusions in proliferating mouse fibroblasts. BRCA2 localized to the telomeres in S phase in an ATR-dependent manner, and its absence resulted in the accumulation of common fragile sites, particularly at the G-rich lagging strand, and increased the telomere sister chromatid exchange in unchallenged cells. The incidence of common fragile sites and telomere sister chromatid exchange increased markedly after treatment with replication inhibitors. Congruently, telomere-induced foci were frequently observed in the absence of Brca2, denoting activation of the DNA damage response and abnormal chromosome end joining. These telomere end fusions constituted a significant portion of chromosome aberrations in Brca2-deficient cells. Our results suggest that BRCA2 is required for telomere homeostasis and may be particularly important for the replication of G-rich telomeric lagging strands.
Keywords: Cancer, Chromosomes, DNA Replication, Mouse Genetics, Telomeres, BRCA2, Genetic Instability, Telomere Erosion, Telomere Fragility
Introduction
The inheritance of one mutant allele of BRCA2 predisposes carriers to early onset breast cancer through loss of heterozygosity; thus, BRCA2 is a tumor suppressor (1, 2). Recently, it has been shown that BRCA2 heterozygosity also promotes KrasG12D-driven carcinogenesis (3), indicating that mutation of BRCA2 is critical for both the initiation and progression of cancer.
A truncated Brca2 allele (Brca2Tr) in mice causes embryonic lethality and growth retardation due to accumulation of DNA double-stranded breaks (DSBs)3 and consequent checkpoint activation (4). Metaphase chromosome spreads of the mouse embryonic fibroblasts (MEFs) from Brca2Tr/Tr mice display chromatid, chromosome breaks, and radial structured chromosomes, strongly indicating that DSB repair is impaired in Brca2Tr/Tr mice (4). Congruently, molecular and biochemical studies of BRCA2 have revealed that BRCA2 regulates homologous recombination (HR), also called homology-directed repair (HDR) (5), by interacting with the recombinase Rad51, through the BRC repeats in exon 11 (6–8) and the C terminus (6, 9). These studies confirmed the well defined role of BRCA2 as a tumor suppressor and a critical regulator of error-free DNA repair.
HDR begins when a damaged DNA strand invades the undamaged duplex of its sister DNA strand. The damaged strand is then repaired by DNA synthesis using the sister strand as a template. Thus, HDR is an error-free DSB repair pathway that takes place during the S or G2 phases of the cell cycle (10). Notably, HDR is implicated in the repair and rescue of stalled DNA replication forks (11). The inefficient resolution of stalled replication forks that occurs in the absence of BRCA2 greatly contributes to the accumulation of gross chromosomal rearrangements, such as translocations, deletions, inversions, and amplifications (12). Moreover, DNA intermediates at stalled DNA replication forks collapse into double strand breaks in BRCA2-deficient cells (13). Recently, it has been shown that BRCA2 blocks the resection of stalled replication forks by the MRE11 nuclease and that this function requires the RAD51-binding C-terminal region of BRCA2 in a manner that is independent from HDR (14). Collectively, these studies suggest that BRCA2 is crucial for the stabilization of stalled replication forks.
Mammalian telomeres are composed of long arrays of TTAGGG repeats. When cells proliferate, telomere DNA can be lost due to the inability of the DNA replication machinery to duplicate the linear DNA ends. This end replication problem is solved by the reverse transcriptase, telomerase, which adds TTAGGG repeats onto the 3′ ends of chromosomes (15) to compensate for the loss of terminal sequences. In addition to the critical role of telomerase, the DNA replication machinery is required for the maintenance of telomeres in proliferating cells; most of the long TTAGGG repeat at the end of the chromosome is maintained by semi-conservative DNA replication (16).
Interestingly, a recent study has indicated that telomeric repeats impose a challenge to the DNA replication machinery. Replication-dependent defects that resemble the common fragile sites (CFS), which occur when DNA polymerase α is inhibited by aphidicolin (Aph), arise at the telomere (17). The study suggested that telomeres challenge replication fork progression because of TTAGGG repeats forming G-G Hoogsteen base pairs (18) that make the G quadruplex (G4) DNA structures. G4 structures inhibit the progression of the DNA replication machinery through steric hindrance (17). Helicases such as Pif1 (19), FANCJ (20), Bloom syndrome (BLM) (21), and RTEL in mouse (22) are reported to unwind G4 structures and facilitate DNA replication. In Escherichia coli, the RecQ helicase family of proteins is involved in unwinding the G4 structures (23). Consequently, absence of RecQ helicases implicated in the removal of G4 secondary structures provokes the increase of telomeric CFSs (17). From these, it is anticipated that proteins associated with telomere G4 structures will participate in the homeostasis of telomeres.
We observed end fusions while analyzing the chromosomes of Brca2-deficient cells. Therefore, we asked if telomere erosions contributed to the chromosome aberrations in BRCA2-related tumorigenesis. If so, involvement of BRCA2 in telomere maintenance would constitute an important tumor suppressor function, linking telomere homeostasis to the prevention of cancer. Therefore, we set out to determine whether BRCA2 is involved in telomere maintenance.
EXPERIMENTAL PROCEDURES
Plasmids, siRNA, and Antibodies
The plasmid used for generating telomere probes, pSXneo270T2AG3, was a gift from Dr. Titia De Lange (The Rockefeller University, New York). Synthetic siRNA for BRCA2 (GAA GAA CAA UAU CCU ACU ATT), GFP (GUU CAG CGU GUC CGG CGA GTT), ATM (AAC AUA CUA CUC AAA GAC AUU), ATR-1 (AAC CUC CGU GAU GUU GCU UGA), or ATR-2 (AAG CCA AGA CAA AUU CUG UGU) were purchased from Bioneer Co. (Daejeon, Korea).
Sheep polyclonal antibodies specific to mouse Brca2 were generated by injection of recombinant mouse Brca2 protein (3,107–3,303 amino acids) purified from E. coli (National Blood Transfusion Service, UK). Rabbit polyclonal antibodies specific to human BRCA2 were generated by injection of a peptide, corresponding to 1,382–1,395 amino acids of human BRCA2. The following antibodies were purchased: anti-actin (Santa Cruz Biotechnology; AC-15); anti-BrdU 3D4 (BD Biosciences; 555627); anti-FLAG M2 (Sigma; F3165); anti-TRF1 (Abcam; ab1423); anti-TRF2 (NOVUS; NB100–2577); anti-ATM (Abcam; ab78); and anti-ATR (Santa Cruz Biotechnology; N-19).
Mouse Breeding, Generation of MEFs, Adenoviral Infection, and Cell Culture
Brca2 conditional knock-out mice (Brca2f11) (24) were from Dr. Anton Berns (NCI, The Netherlands). Mice were housed in a semi-conventional (virus antibody-free) facility, and all mouse experiments were approved by the Institutional Animal Care and Use Committee of Seoul National University. We strictly followed the Seoul National University guidelines, policies, and regulations for the Care and Use of Laboratory Animals.
The MEFs were isolated from E13.5 embryos, cultured in DMEM supplemented with 15% FBS in 5% CO2-humidified incubator. Cre recombinase was introduced with 100 multiplicities of infection of Cre-expressing adenovirus (Ad-Cre).
Telomere FISH, CO-FISH, and Quantitative-FISH to Analyze Telomere Lengths
MEFs were incubated with 0.1 mg/ml colcemid (Sigma) for 4 h at 37 °C, fixed in methanol/acetic acid (3:1), and processed for metaphase chromosome spreads (25). When required, 0.2 μm Aph was added for 16 h before cells were subjected to colcemid treatment. Chromosomes were stained using Cy3-OO-(CCCTAA)3-peptide nucleic acid (PNA) probe (Panagene) and DAPI.
For chromosome orientation FISH (CO-FISH) experiments, MEFs were treated with 10 mm BrdU/BrdC (3:1) for 16 h and incubated with colcemid for the last 4 h. Subsequent steps followed the previously described protocol (26), using Cy3-OO-(TTAGGG)3 and FAM-OO-(CCCTAA)3-PNA probes. Fluorescent micrograph images were acquired using an AxioCam MRm camera on a Zeiss Observer Z1 inverted microscope with ×40/0.6 NA lens (Zeiss).
Quantitative-FISH (Q-FISH) on chromosome metaphase spreads was performed as described previously (27), in a blinded fashion. Metaphase spreads from MEFs were hybridized with Cy3-OO-(CCCTAA)3-PNA probe, and images were acquired using CoolSnap HQ cooled CCD camera on DeltaVision. Telomere fluorescence intensity was analyzed using TFL-Telo software (Peter Lansdorp, University of British Columbia, Canada). For statistical analysis, we used SPSS software.
Telomere Oligonucleotide Ligation Assay (T-OLA)
The T-OLA analysis was performed as described previously (28, 29). Briefly, 24 h after Ad-Cre infection, Brca2f11/f11 MEFs were synchronized at G1/S by thymidine-aphidicolin block (30). Cells were then released to S phase by changing to fresh medium for 4 h. 3 μg of genomic DNA was hybridized with 0.5 pmol of γ-32P-end-labeled nucleotide of the sequence (CCCTAA)4 or (TTAGGG)4. Hybridization was performed overnight at 50 °C, followed by ligation with 20 units of TaqDNA ligase (New England Biolab) at 50 °C for 5 h. Then the samples were precipitated, resuspended, and denatured in a formamide loading buffer. The samples were separated on 6% acrylamide, 6 m urea gels. Images were obtained by exposure to FLA7000 (FUJIFILM). 10 ng of T-OLA product was used for the quantitative PCR of the genomic GAPDH gene for loading control.
In-gel Hybridization Assay
In gel hybridization to measure telomeric G-overhang was carried out as described previously (31). Briefly, Brca2f11/f11 MEF genomic DNA was digested with MboI and separated by pulsed field gel electrophoresis and hybridized with (CCCTAA)4 to determine the overhang signal. The gel was denatured and rehybridized with the same probe to determine the total telomeric DNA signals. The overhang signal was normalized to the total telomeric DNA signal.
Telomere-ChIP
HeLa_CFLAP-BRCA2_hTert was generated by introducing hTert by retroviral transfer of pBabe_hTert into HeLa_CFLAP-BRCA2 cells (32). HeLa_CFLAP BRCA2_hTert cells were subjected to double thymidine block. The cells were then released into the cell cycle by changing to fresh medium. Aliquots were collected at the indicated time points after release.
In experiments using KU55933 (20 μm) or CGK733 (20 μm), the drugs were added for 1 h before cell lysis. ATM and ATR knockdown experiments were performed as described previously (25); cell lysis was performed 24 h post-siRNA transfection. Telomere-ChIP assays were performed as described previously (34) with the indicated antibodies. Cell cycle profiles were determined by FACS analysis.
RESULTS
Telomere Dysfunction in Brca2-deficient MEFs
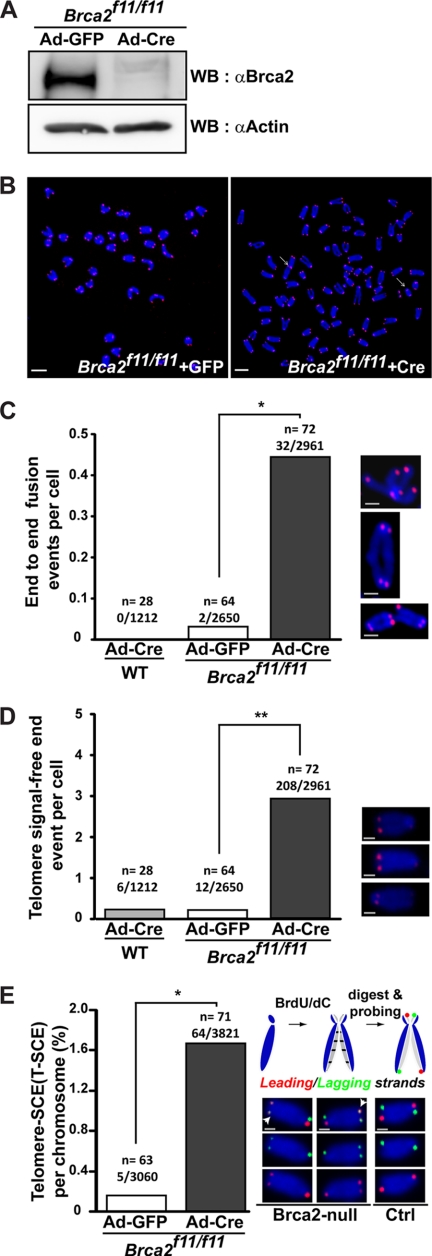
To assess whether BRCA2 is associated with telomere maintenance, we first examined for telomere aberrations in the absence of BRCA2. For this, we used MEFs isolated from Brca2 conditional knock-out mice, Brca2f11/f11 (24), and utilized the Cre-loxP system. After expression of Cre recombinase by infection of Ad-Cre, exon 11 of Brca2 was deleted, as was indicated by the disappearance of intact Brca2 protein in the Western blot, 48 h post-infection (Fig. 1A). Telomeres were examined by telomere-FISH in the presence (Ad-GFP) or absence (Ad-Cre) of Brca2, 5 days post-infection of Brca2f11/f11 MEFs. MEFs depleted of Brca2 displayed telomere end fusions (0.44 events/cell; Fig. 1, B and C); chromosomal and chromatid fusions were observed at similar frequencies (0.22 and 0.22 events/cell each).
FIGURE 1.
Telomere attrition manifested as telomeric end-to-end fusions, telomere signal-free ends, and anaphase bridges in the absence of Brca2. A, Western blot analysis to assess the efficiency of Cre recombinase after Ad-Cre infection in Brca2f11/f11 MEFs. Lysates for Western blots (WB) were collected at the same time as telomere (T)-FISH in B. Brca2 disappears when Cre is expressed. Same blot was reprobed with anti-actin antibodies for normalization. B, Brca2f11/f11 MEFs or wild-type MEFs were infected with Ad-GFP or Ad-Cre. Five days post-infection, MEFs were subjected to T-FISH analysis. At least 28 metaphases from two independent MEFs of the same siblings were analyzed. Representative images of T-FISH in Brca2f11/f11 MEFs infected with Ad-GFP or Ad-Cre are shown. White arrows, chromosomes with end-to-end fusion or chromatid fusion. Scale bars, 5 μm. C, frequency of telomere end-to-end fusion events. Bar graphs indicate the frequency of telomere fusions in all chromosomes analyzed. x/y, total number of chromosomes with end-end fusion/number of chromosomes analyzed; n, number of cells analyzed. Enlarged images are shown at right. Scale bars, 1 μm. D, frequency of telomere signal-free ends. Bar graphs indicate the frequency of telomere SFEs in all chromosomes analyzed. x/y, total number of chromosomes with SFEs/number of chromosomes analyzed; n, number of cells analyzed. Representative images are shown at right. Scale bars, 1 μm. E, T-SCE measured in the presence (Ad-GFP) or absence (Ad-Cre) of Brca2. CO-FISH assay was performed 5 days post-adenoviral infection. Wild-type cells display lagging (green) and the leading (red) strand in a diagonal orientation in control (schematic diagram above, see also “Experimental Procedures”). Leading and/or the lagging strands appearing in the nondiagonal orientation are judged to result from the telomere sister chromatid exchanges and were observed in Brca2-null MEFs. T-SCEs are marked with white arrowheads. The results are from three independent experiments. x/y, total number of T-SCEs/number of chromosomes analyzed; n, number of cells analyzed. Representative images are shown at right. Scale bars, 1 μm (*, p < 0.05; **, p < 0.01; t test).
In addition, telomeres without detectable signal in telomere-FISH, called signal-free ends (SFEs), were frequently observed in Brca2-depleted MEFs (2.89 events/cell; Fig. 1D), indicating the loss of telomeric repeats in the absence of Brca2. As critically shortened telomeres can induce exchanges between leading and lagging strand telomeres through recombination, the incidence of T-SCE (35, 36) was measured by CO-FISH. The result showed that the absence of Brca2 provoked aberrant recombination (T-SCE at 1.67% incidence per chromosome), whereas the control MEFs barely showed any T-SCE (Fig. 1E). Interestingly, high T-SCE is one of the features of alternative lengthening of telomeres (ALT) used to maintain telomeres in cancer cells independent from telomerase (37, 38). That T-SCE took place in Brca2-deficient MEFs suggests the interesting possibility that Brca2 depletion could induce aberrant recombination leading to ALT, if the cells were permitted to grow further.
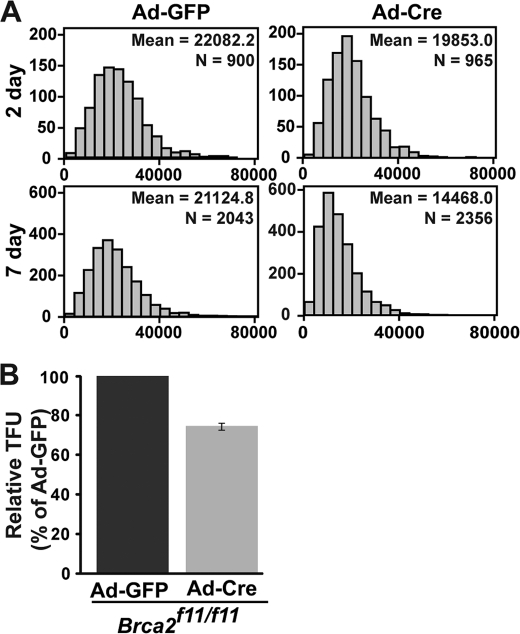
Chromosomal end fusions and SFEs indicated that telomere shortening occurred in the Brca2-deficient cells. To verify the telomere shortening, we measured the length of telomeres by Q-FISH in the presence and absence of Brca2. Q-FISH analysis was conducted in Brca2f11/f11 MEFs 2 and 7 days post-infection of Ad-GFP or Ad-Cre adenovirus. The mean telomere lengths in Brca2-deleted MEFs were ∼25% shorter than those of control MEFs (Fig. 2, A and B, Ad-Cre), confirming that telomere repeats are subject to shortening when Brca2 is depleted.
FIGURE 2.
Telomere shortening in Brca2-depleted MEFs. Brca2f11/f11 MEFs were analyzed for their overall telomere length after infection with Ad-GFP or Ad-Cre by measuring fluorescent telomere signals with Q-FISH (TFL-Telo program). A, telomeres shorten in Brca2-deficient MEFs in a passage-dependent manner. Ad-Cre-infected Brca2f11/f11 MEFs display a significant degree of telomere shortening after Cre expression, compared with control (Ad-GFP). x axis, relative fluorescence units; y axis, number of chromosomes. N, number of chromosomes analyzed. Mean value of relative fluorescence units are also indicated. Data were analyzed using SPSS software. B, relative intensity of the telomere signals in Ad-GFP- or Ad-Cre-infected MEFs. Chromosomes from at least 60 different cells were analyzed. TFU, telomere fluorescence unit. Bars represent the relative fluorescence intensity over control MEFs (Ad-GFP), measured at day 7 post-viral infection in Brca2f11/f11 MEFs. Error bar represents the mean ± S.D.
DNA Damage Response at Telomeres of Brca2-deficient MEFs
Unprotected telomeres, either due to the disruption of T-loops by depleting shelterin proteins, such as TRF2 (39), or to critical telomere shortening (40–42), are recognized as sites of DNA damage. Markers of DSBs, γ-H2AX or 53BP1 (43), colocalize with damaged telomeres and constitute telomere dysfunction-induced foci (TIFs) (39). The presence of these TIFs is the signature of telomere erosions in cells.
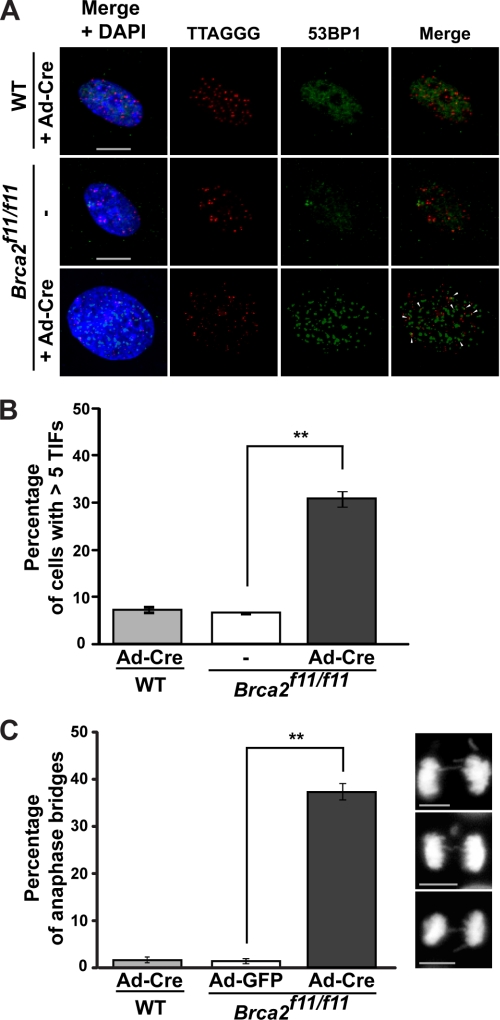
As Brca2-deficient MEFs exhibited shortened telomeres and telomere erosion, we analyzed the incidence of TIFs in these cells. Cells with more than five TIFs were counted as positive in this assay. The results show that >30% of Brca2-depleted MEFs displayed TIFs (4–5-fold increase compared with control; Fig. 3, A and B). None of the shelterin proteins tested directly associated with BRCA2 (data not shown); therefore, the TIFs in Brca2-deficient cells are less likely to result from disruption of the shelterin complex or the T-loop (44). Of note, there was a significant shortening of telomeres associated with the depletion of Brca2. These results suggest that excessive loss of telomeric DNA may have induced the DNA damage response at telomeres, resulting in chromosome end fusions in Brca2-deficient MEFs.
FIGURE 3.
Increase in telomeric DNA damage in the absence of Brca2. A, TIF increase in Brca2-null MEFs. TIFs were analyzed using anti-53BP1 antibody (green) and PNA-conjugated telomere probe (CCCTAA, red) in Brca2f11/f11 MEFs 5 days post-infection with Ad-Cre. Wild-type MEFs infected with Ad-Cre were used for control. TIFs appeared as merged yellow foci and are marked with arrows. B, quantification of the TIFs shown in A. Bar graphs indicate the percentage of cells with >5 TIFs. Scale bar, 5 μm. C, quantification of anaphase bridges after Cre expression in wild-type or Brca2f11/f11 MEFs. MEFs were grown on coverslips and fixed with 4% paraformaldehyde, and the DNA was counterstained with DAPI. Two experiments were performed independently and scored in a blind manner. Images were acquired using AxioCam MRm camera on Zeiss Observer Z1 inverted microscope with ×40/0.6 NA lens (Zeiss). Bar graphs indicate the percentage of cells displaying anaphase bridges 5 days post-adenoviral infection. At least 63 cells in anaphase were analyzed. Error bars, standard deviation. Scale bars, 5 μm (**, p < 0.01; t test).
Anaphase bridges form when chromosomes of two ends of a dicentric chromosome, as a result of end-to-end fusions, are pulled to opposite directions by the spindle apparatus during anaphase (16). We analyzed the formation and incidence of anaphase bridges in Brca2f11/f11 MEFs. In Brca2-deficient MEFs (+Cre in Brca2f11/f11), 37% of cells displayed anaphase bridges, whereas wild-type MEFs infected with Ad-Cre or Brca2f11/f11 MEFs with Ad-GFP infection barely did (Fig. 3C). Because anaphase bridges lead to breakage-fusion-bridge cycles (45), these events can result in chromosomal translocations in BRCA2-deficient cells and contribute to genetic instability.
Telomere Fragility and Replication Stress in Brca2-deficient Cells
How might BRCA2 deficiency cause telomere aberrations? It is known that telomeres are difficult to replicate because their repetitive arrays of guanosine-rich DNA sequences can form G quadruplexes and hamper the progression of DNA replication machinery (46). Even the T-loops that protect the telomere end from resection can inhibit the passage of replication machinery. Although hypothetical and done in vitro, recent data suggest that the presence of triple helices, four-way junctions, and D-loops at the telomeres can inhibit the progression of the replication machinery as well. Together, these studies suggest that telomeres are susceptible to replication fork stalling (16, 47). However, several lines of evidence suggest that BRCA2 is essential for stabilizing and preventing the degradation of stalled replication forks (13, 14) and that chromosome gaps and breaks in metaphase chromosome spreads, the so-called CFSs, occur in BRCA2-deficient cells as a result of defective replication (48). These considerations prompted us to investigate whether Brca2 might help to maintain telomere homeostasis through its role in stabilizing stalled replication forks.
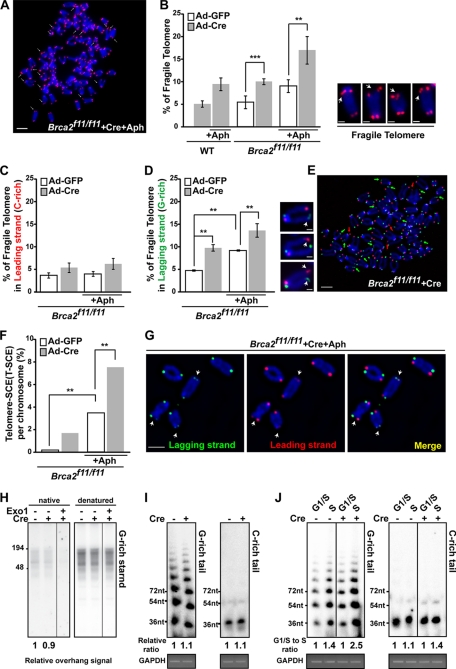
We first determined whether telomere fragility was increased after BRCA2 depletion. To analyze the CFSs at telomeres, telomere-FISH was performed and examined under the fluorescence microscope. Broken telomeric signals and multiple telomeric signals, which reflect defects in telomere DNA synthesis (17), were counted with or without treatment with Aph, a DNA polymerase α inhibitor (Fig. 4, A and B). CFSs are induced under partial inhibition of DNA polymerase α (49, 50). The result showed that even in the absence of Aph treatment, Brca2-deficient MEFs exhibited CFSs comparable with the level measured when controls (wild-type and Brca2f11/f11 infected with Ad-GFP MEFs) were treated with Aph (∼9%; Fig. 4B). Clearly, depletion of Brca2 led to a marked increase of fragile telomeres. Upon Aph treatment, CFSs in Brca2-depleted MEFs increased to 2-fold more compared with untreated and ∼3-fold more compared with the untreated wild-type control (∼18%; Fig. 4B). Thus, telomere fragility is increased by BRCA2 depletion in unchallenged cells as well as cells exposed to replication inhibitors.
FIGURE 4.
Replication block at the telomeres in the absence of Brca2. A, Brca2f11/f11 MEFs, infected with Ad-Cre, were subjected to telomere FISH after Aph treatment. Arrows depict chromosomes with fragile telomere. Bar, 5 μm. B, Brca2-deficient MEFs display marked increase of fragile telomeres. Brca2f11/f11 MEFs, infected with Ad-GFP or Ad-Cre, were subjected to telomere FISH after Aph treatment and analyzed. Wild-type MEFs were used as controls. Bars represent the percentage of fragile telomeres in total number of chromosomes analyzed, 5 days post-adenoviral infection. Representative images of telomere fragility are shown at the right. Scale bar, 1 μm. C and D, comparison of fragile telomeres in leading and lagging strands after Brca2 depletion. Brca2f11/f11 MEFs infected with Ad-GFP or Ad-Cre were subjected to CO-FISH after Aph treatment. Bars represent the percentage of fragile telomeres in leading or lagging strand, analyzed 2.5 days post-adenoviral infection. Representative images of telomere fragility are shown at the right. Scale bar, 1 μm. E, telomere fragility in lagging strands is higher compared with the leading strand in Brca2-deficient MEFs. Green arrowheads, fragile telomeres in lagging strand. Red arrowhead, fragile telomeres in leading strand. Scale bar, 5 μm. F, T-SCE measured in the presence (Ad-GFP) or absence (Ad-Cre) of Brca2 with/without Aph treatment. G, Brca2-deficient MEFs treated with Aph display a marked increase in T-SCE. Note the arrowheads, which indicate T-SCE. Scale bar, 5 μm. H, in-gel hybridization in a native gel and a denatured gel, using (CCCTAA)4 probe. DNA was extracted from Brca2f11/f11 MEFs, after infection with Ad-GFP or Ad-Cre. Telomere overhangs hybridize with the probe under the native condition (left), as exonuclease I (Exo1) treatment abolishes the radioactivity. Total telomere signals are obtained in the denatured condition. The relative radioactivity of telomere overhangs was obtained (marked as relative ratio) by normalizing the intensities of the hybridized radioactivity from the native condition to the denatured. Signals of control MEFs were set to 1. I, T-OLA analysis (28) of Brca2f11/f11 MEFs with oligonucleotides complementary to the G-rich tail (CCCTAA)3 or the C-rich tail (TTAGGG)3 2.5 days post-Ad-GFP (−Cre) or Ad-Cre (+Cre) infection. Relative hybridization intensity and length to the control are marked. GAPDH PCR product is shown for normalization of genomic DNA employed. J, Brca2f11/f11 MEFs infected with Ad-GFP or Ad-Cre were synchronized in G1/S by thymidine-aphidicolin block and then released into S phase progression by incubating in fresh media for 4 h. T-OLA assay was performed 2.5 days post-adenoviral infection. Relative intensity and length of T-OLA product in each lane are marked; the intensity of T-OLA product in G1/S in each setting is set to 1. PCR product of GAPDH is included for control. Error bars, standard deviation (**, p < 0.01; ***, p < 0.001; t test).
We next considered whether BRCA2 might suppress telomere fragility through its role in stabilizing stalled DNA replication forks (13, 14). Although it has been suggested that BRCA2 is required for telomere homeostasis by loading Rad51 at stalled replication forks to promote repair or restart (51), more recent evidence suggests for the role of BRCA2 in preventing stalled replication fork collapse (13, 47), which is distinct from its function in HDR and DSB repair (14). Furthermore, Rad51 was shown to protect newly synthesized DNA from Mre11-dependent degradation (52).
Indeed, it is known that in E. coli DNA repeats on the lagging strand template can adopt unusual DNA structures when it becomes single-stranded in a region between Okazaki fragments during DNA replication, causing preferential instability of repeats on the lagging strand (53, 54). To test if fragile telomeres induced by Brca2 depletion display this replication-associated strand preference, we used chromosome orientation-FISH (CO-FISH), which enables us to distinguish lagging strand (G-rich, Fig. 4, C–E, green) from leading strand (C-rich, red). Even in control MEFs, treatment with 0.2 μm Aph led to the increase of lagging strand-specific fragile telomere (2-fold, ∼8%; Fig. 4D), which indicates that G-rich lagging strands are more susceptible to CFSs (compare Fig. 4, C and D). Treatment with Aph in the absence of Brca2 further increased the G-rich lagging strand telomere fragility (∼13%; Fig. 4, D and E). Moreover, T-SCEs were frequently observed in Aph treated MEFs (Fig. 4F). Also, T-SCE in Brca2-depleted MEFs was increased by 2-fold upon Aph treatment (7.3%; Fig. 4, F and G), suggesting that the replication problems in G-rich lagging strand telomeres are linked to the marked increase of T-SCE in the absence of Brca2.
As the G-rich telomere lagging strands displayed an increase in fragility, we asked whether the length and amount of G-rich single strands would differ in the absence of Brca2. This was assessed by Southern hybridization of telomere DNA in the native and denatured conditions. Incubation with exonuclease I was included in the native condition to confirm that the hybridized signals were single-stranded telomeres. The result showed that the absence of Brca2 did not significantly alter the length of G-rich telomere single strand repeat arrays (Fig. 4H).
To corroborate the result, the length of the single-stranded DNA at the telomeres was measured and compared by T-OLA (29, 55, 56). In this assay, γ-32P-labeled oligonucleotides complementary to the telomeric overhang or single-stranded DNA at the telomere are annealed to the nondenatured genomic DNA and ligated. The length of the ligated product and the hybridization signal intensity are the indications for the length and amount of telomeric overhangs and the single-stranded telomeric DNA. Brca2f11/f11 MEFs infected with Ad-GFP (−Cre) or Ad-Cre (+Cre) were blocked in G1/S phase and released into cell cycle progression. Then the genomic DNA was extracted from G1/S-arrested or S phase cells and subjected to T-OLA analysis.
As expected, G-rich tails were significantly longer compared with C-rich tails (29); the absence of Brca2 was associated with slightly longer G-rich tails compared with control (Fig. 4I). Because the single-stranded telomeric DNA is expected to be predominantly generated in S phase, we assessed the length and amount of G tails and C tails in G1/S and S phase, respectively, and compared them in the presence or absence of Brca2. Interestingly, G-rich tails increased to 2.5-fold in S phase of Brca2-depleted cells (Fig. 4J, +Cre), whereas the control cells displayed 1.4-fold increase in S phase (Fig. 4J, −Cre), suggesting that the increase of G-rich overhang in the absence of Brca2 is associated with the resection of nascent daughter strand of the G-rich lagging strands during telomere replication (52). These results suggest that BRCA2 has a role in facilitating G-rich lagging strand synthesis at telomeres, by blocking stalled replication fork degradation (14) or by helping the replication to overcome obstacles on the single-stranded templates, such as G4 structures. Regardless of this distinction, it will be interesting in future work to explore the link between BRCA2 and G4 structures formed in the G-rich lagging strand (TTAGGG repeats) in the telomere.
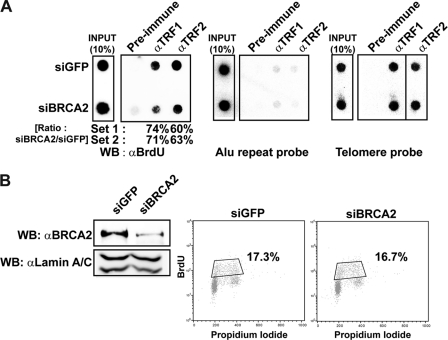
To confirm the involvement of BRCA2 in telomere replication, the ability to incorporate BrdU into telomere repeats was measured and compared in the presence and absence of BRCA2. HeLa cells were transfected with control or siRNA against BRCA2, then subjected to six rounds of BrdU pulse (30 min) and chase (3.5 h), 24 h post-siRNA transfection. Cells were then subjected to telomere-ChIP using antibodies to TRF1 or TRF2. Telomere ChIP products were subjected to Western blot with anti-BrdU antibody to be analyzed for BrdU incorporation in telomeres. The results show that telomere DNA synthesis, as reflected by BrdU incorporation measured with densitometry, was reduced to ∼60–70% in BRCA2-depleted cells (siBRCA2; Fig. 5A, left panel). The assay was normalized because similar amounts of telomere ChIP products were employed in the assay (Fig. 5A, middle and right panels).
FIGURE 5.
Processing of telomere replication is hampered by the absence of BRCA2. A, HeLa cells were transfected with siRNA for GFP (siGFP) or BRCA2 (siBRCA2). The cells were then pulsed with BrdU for 30 min and chased for 3 h and 30 min. This cycle was repeated six times. Lysates were incubated with anti-TRF1 or -TRF2 antibodies and subjected to telomere-ChIP. The blot was immunoblotted with anti-BrdU antibodies to assess the amount of replicated telomeric DNAs. ChIP assays were also performed with Alu or telomere probes. The results from two different sets of experiments are marked below the left panel. The values in percentage depict the ratio of BrdU incorporation in BRCA2-depleted cells (siBRCA2) compared with control (siGFP). B, flow cytometry analysis of cell cycle progression with or without BRCA2 depletion in HeLa cells. The percentage of cells in S phase is boxed and marked (right panel). Simultaneously, lysates were prepared for Western blot (WB) to assess the efficiency of siRNA against BRCA2. Same blot was probed with anti-lamin A/C for normalization (left panel).
The absence of BRCA2 can affect the cell cycle, such that the cells may arrest in G2/M due to accumulation of unrepaired DSBs, particularly upon ectopic DNA damage (4). To rule out this secondary effect from possible cell cycle arrest, we assessed the BrdU incorporation at the telomere, 48 h post-siRNA transfection. At this time point, it is assumed that BRCA2 depletion has not accumulated enough DNA damage to affect the cell cycle progression. Nevertheless, we assessed the cell cycle profile in this experimental setting by analyzing the global BrdU incorporation and propidium iodide staining; the result showed that 48 h post-siRNA transfection, S phase global DNA replication was not measurably affected (Fig. 5B), thus suggesting that the inefficiency of the BrdU incorporation at telomeres resulted primarily from a telomere-specific defect in DNA synthesis.
ATR Pathway Acts Upstream of BRCA2
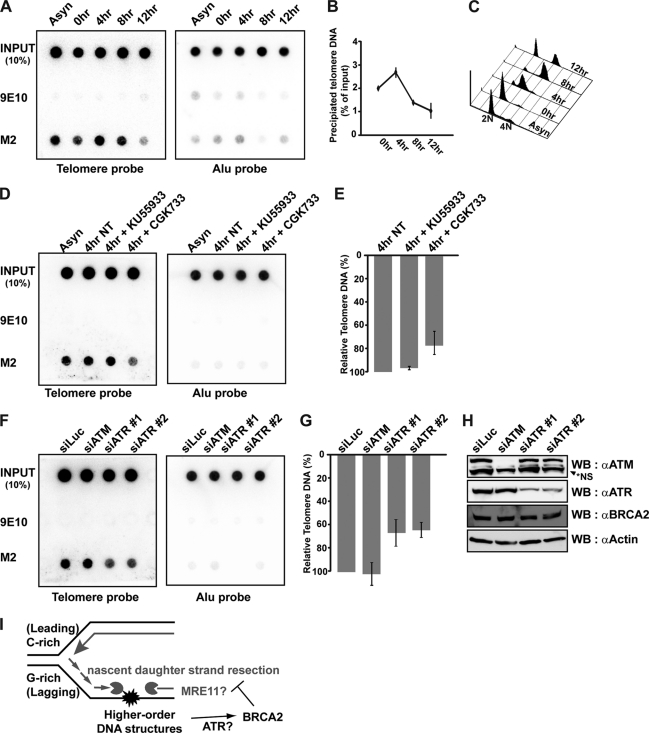
We next asked whether BRCA2 is capable of binding to telomeres. For this, we took advantage of HeLa cells expressing BAC-encoded full-length BRCA2, tagged with GFP and FLAG (32). Because HeLa cells carry a short telomere (<5 kb), telomerase (hTERT) was introduced into these cells to elongate the telomeres and facilitate the telomere-ChIP assay (57). Cells were arrested in G1/S by double thymidine block and released into the cell cycle by changing to fresh medium. Lysates were collected and subjected to telomere-ChIP assays at the indicated time points after release. Simultaneously, aliquots of cells were analyzed for their cell cycle stage by flow cytometry (Fig. 6, A–C). The result revealed that BRCA2 bound to telomeres in S phase; binding was observed in asynchronous cells, at 0 h, peaking at 4 h from thymidine release (Fig. 6B), which corresponded to S phase (Fig. 6C).
FIGURE 6.
BRCA2 localizes to telomeres during S phase in an ATR-dependent manner. A, HeLa_CFLAP-BRCA2_hTert cells were synchronized at the G1/S boundary by double thymidine block and then released into the cell cycle. Cell lysates were prepared at the indicated time points after thymidine release and subjected to telomere ChIP with anti-FLAG antibody to detect BRCA2 at the telomere (M2). Nonrelated monoclonal 9E10 was employed to control for nonspecific binding. B, percentage of precipitated telomere DNA was measured as a ratio of input signals and marked as dotted graphs at each time point of analysis. C, cell cycle profiles for each time point were measured by FACS analysis. D, HeLa_CFLAP-BRCA2_hTert cells were treated or left alone with 20 μm KU55933 (inhibitor specific for ATM) or 20 μm CGK733 (inhibits ATM and ATR) for 1 and 4 h post-thymidine release. Lysates were also subjected to telomere ChIP with anti-FLAG (M2) to detect BRCA2 bound to telomere. Negative control, 9E10. E and G, bars represent the relative amount of telomere DNA bound to BRCA2 measured by densitometry. The signals were normalized to the input controls. NT, not treated. F, HeLa_CFLAP-BRCA2_hTert cells transfected with indicated siRNAs. Cell lysates were prepared 24 h post-transfection. Telomere ChIP assays were performed with anti-FLAG (M2, BRCA2) and 9E10. Error bars, standard deviation. H, Western blot (WB) analysis to assess the efficiency of siRNAs employed in the assay in F and G. NS, not specific. I, model for the proposed function of BRCA2 in the telomere maintenance. At telomeres, obstacles for elongating DNA polymerase form more frequently at the lagging strands, because G-rich parental strands exposed as single strands between Okazaki fragments can adopt higher order structures such as G-quadruplex. BRCA2 prevents the nascent strand at an aberrant fork from MRE11-dependent uncontrolled resection. In the absence of BRCA2, the resection of daughter strands would be more frequent at lagging strands, resulting in the marked increase of telomere fragility, telomere shortening, and erosion.
We have shown compelling lines of evidence that BRCA2 is required for telomere homeostasis. The absence of BRCA2 resulted in an increase in stalled replication forks at telomeres, shortening of telomeres, and end-to-end fusions. We have also shown that TIFs increase in the absence of BRCA2, indicating a DNA damage response at telomeres.
Damage in DNA is sensed by PI3K family proteins, ATM and ATR, depending on the type of DNA damage; ATM senses DSBs (58), and ATR recognizes single strand break and stalled replication forks (59). The two kinases function separately or in concert, depending on the type of damage, and coordinate the DNA damage response, the repair, and cell cycle regulation (60–62). Notably, the maintenance of telomeres requires both ATM and ATR pathways (63, 64). The ATR machinery is recruited to telomeres before the completion of telomere replication, consistent with its role in DNA replication, and the ATM and HR machinery is required for telomere-specific structure after replication (63).
Taking all this information into account, we asked whether the binding of BRCA2 to telomeres in S phase was ATM- or ATR-dependent. For this, two inhibitors, CGK733 and KU55933, were used. CGK733 can inhibit the activity of ATM and also ATR (65, 66), and a moderate concentration of KU55933 (20 μm) preferentially inhibits the ATM pathway, but not ATR, in vivo (67). In our experimental system, we found that 20 μm KU55933 specifically blocked ATM activity, because it inhibited phosphorylation of Chk2 (68) upon irradiation, but not after treatment of hydroxyurea (supplemental Fig. S1, A and B). In comparison, 20 μm CGK733 treatment effectively inhibited the phosphorylation of Chk1, and thus ATR inhibition (33, 65, 69–71), upon hydroxyurea treatment (supplemental Fig. S1A). However, the same concentration was not effective toward ATM inhibition upon irradiation in HeLa_CFLAP_BRCA2_hTert cells (supplemental Fig. S1B). Taking this information into account, HeLa_CFLAP-BRCA2_hTert cells were synchronized by double thymidine block and released into the cell cycle. Four hours after release, the time when cells are enriched in S phase (Fig. 6C), cells were challenged with CGK733 or KU55933 for 1 h. The result showed that the binding of BRCA2 to telomeres in S phase, as assayed by telomere-ChIP, was reduced to ∼68% in cells treated with 20 μm CGK733 (Fig. 6, D and E). By comparison, 20 μm KU55933 treatment did not interfere with BRCA2 binding to telomeres.
As these inhibitors can be argued for their specificity to ATM or ATR, HeLa_CFLAP-BRCA2_hTert cells were transfected with siRNAs against ATM or ATR and assessed for the binding of BRCA2 to telomeres. Upon knockdown expression of ATR (siATR), BRCA2 binding to telomeres was reduced to two-thirds (∼60%) (Fig. 6G), whereas ATM knockdown (siATM) had little effect (Fig. 6, F–H), confirming that BRCA2 binding to telomeres requires ATR.
The result is consistent with the data showing that the absence of BRCA2 resulted in problems in G-rich telomere replication. Collectively, our results show that BRCA2 binds to telomeres in S phase, and suggest that it is involved in telomere maintenance likely through inhibition of the degradation of stalled replication forks, particularly in the G-rich lagging strands (Fig. 6I).
DISCUSSION
Maintaining the fidelity of telomere replication is an important function of proliferating cells but not in differentiated nondividing cells. Hence, yin and yang sides of telomere maintenance exist in terms of tumorigenesis; the end replication problem of telomeres induces cellular senescence, which in fact is a preventive mechanism for tumorigenesis in vivo. However, homeostasis of telomere length is crucial for the genetic integrity of proliferating cells because critically shortened telomeres are susceptible to unwanted telomere sister chromatid exchanges and end fusions that lead to gross chromosomal rearrangements, contributing to chromosome instability.
Examples of the connection between telomere erosion and human cancers are the cancer-prone genetic diseases Bloom syndrome and Werner syndrome. Genes encoding BLM and WRN, the two genes mutated in Bloom syndrome and Werner syndrome, respectively, are RecQ helicases thought to be involved in the unwinding of secondary structures; hence, they are critically required for telomere replication (33). The absence of BLM or WRN results in fragile telomeres and telomere erosion (17).
Here, we show that BRCA2, a gene mutated in familial breast cancer with chromosome instability, localizes to telomeres in S phase of the cell cycle, and we suggest that it is required for the prevention of stalled replication fork collapse in telomeres. Although BLM and WRN are helicases that may unwind secondary structures, BRCA2 does not possess any helicase activity. However, the absence of BRCA2 led to the accumulation of G-rich single strand telomere repeats, suggesting that its absence precipitates problems in telomere lagging strand synthesis.
As has been shown by previous studies, BRCA2 is required for the maintenance of stalled replication forks, preventing them from breakdown or collapse. Because G4 structures cause steric hindrance of the progression of replication forks, the requirement of BRCA2 may be maximized at the lagging strand of the replicating telomeres. The results in Fig. 5 support this notion; DNA synthesis at telomeres was more affected (Fig. 5A) than global DNA synthesis (Fig. 5B) in the first couple of cell divisions after BRCA2 depletion.
While preparing this manuscript, Badie et al. (51) reported that BRCA2 loads Rad51 onto telomeres, thereby fulfilling the need of the HR machinery to restart replication. These observations are consistent with ours and confirm the role of BRCA2 in telomere maintenance. However, we have further shown that the TTAGGG repeats of lagging strand synthesis face problems if BRCA2 is depleted, suggesting a link between BRCA2 and obstacles to telomere lagging strand replication, such as G4 secondary structures. The fact that ATR, which is involved in checkpoint activation in response to replication stress, is required for BRCA2 to bind to telomeres in S phase (Fig. 6) supports the notion that BRCA2 function is critical to prevent degradation of stalled replication forks at telomeres caused by problems in G-rich lagging strand synthesis (Fig. 6I). This speculation is consistent with the recent report suggesting that BRCA2 inhibits MRE11-dependent degradation of stalled replication forks, which is independent from DSB repair (14).
Interestingly, T-SCE and anaphase bridges increased in the Brca2-depleted MEFs (Figs. 1 and 3). These results suggest an interesting possibility that BRCA2 may be involved in the suppression of ALT. Whether BRCA2 is indeed involved in the suppression of ALT is an important question for understanding the basis of genetic instability and tumor initiation in ALT-type cancers and also the cancers with BRCA2 mutations.
Supplementary Material
Acknowledgments
HeLa_CFLAP_BRCA2 was a gift from M. Petronczki (Claire Hall Laboratories, London, United Kingdom). We are grateful to Dr. Titia de Lange (The Rockefeller University, New York) for providing telomere DNA constructs. Imaging and flow cytometric analyses were supported by Grant 2005-0048590 from the Research Center for Functional Cellulomics, Seoul National University (Seoul, Korea).
This work was supported by Basic Science Research Program Grant 2011-0018630 and National Research Foundation of Korea Grants 2009-0093927 and 2010-0026104.

This article contains supplemental Fig. S1.
- DSB
- double-stranded break
- MEF
- mouse embryonic fibroblast
- HR
- homologous recombination
- HDR
- homology-directed repair
- CFS
- common fragile sites
- PNA
- peptide nucleic acid
- CO-FISH
- chromosome orientation fluorescence in situ hybridization
- Q-FISH
- quantitative fluorescence in situ hybridization
- SFE
- signal-free end
- T-SCE
- telomere sister chromatid exchange
- ALT
- alternative lengthening of telomeres
- TIF
- telomere dysfunction-induced foci
- Aph
- aphidicolin
- ATR
- ataxia telangiectasia and rad3 related
- ATM
- ataxia telangiectasia mutated
- hTert
- human Tert
- T-OLA
- telomere-oligonucleotide ligation assay.
REFERENCES
- 1. Tavtigian S. V., Simard J., Rommens J., Couch F., Shattuck-Eidens D., Neuhausen S., Merajver S., Thorlacius S., Offit K., Stoppa-Lyonnet D., Belanger C., Bell R., Berry S., Bogden R., Chen Q., Davis T., Dumont M., Frye C., Hattier T., Jammulapati S., Janecki T., Jiang P., Kehrer R., Leblanc J. F., Mitchell J. T., McArthur-Morrison J., Nguyen K., Peng Y., Samson C., Schroeder M., Snyder S. C., Steele L., Stringfellow M., Stroup C., Swedlund B., Swense J., Teng D., Thomas A., Tran T., Tranchant M., Weaver-Feldhaus J., Wong A. K., Shizuya H., Eyfjord J. E., Cannon-Albright L., Tranchant M., Labrie F., Skolnick M. H., Weber B., Kamb A., Goldgar D. E. (1996) The complete BRCA2 gene and mutations in chromosome 13q-linked kindreds. Nat. Genet. 12, 333–337 [DOI] [PubMed] [Google Scholar]
- 2. Wooster R., Bignell G., Lancaster J., Swift S., Seal S., Mangion J., Collins N., Gregory S., Gumbs C., Micklem G. (1995) Identification of the breast cancer susceptibility gene BRCA2. Nature 378, 789–792 [DOI] [PubMed] [Google Scholar]
- 3. Skoulidis F., Cassidy L. D., Pisupati V., Jonasson J. G., Bjarnason H., Eyfjord J. E., Karreth F. A., Lim M., Barber L. M., Clatworthy S. A., Davies S. E., Olive K. P., Tuveson D. A., Venkitaraman A. R. (2010) Germline Brca2 heterozygosity promotes Kras(G12D)-driven carcinogenesis in a murine model of familial pancreatic cancer. Cancer Cell 18, 499–509 [DOI] [PubMed] [Google Scholar]
- 4. Patel K. J., Yu V. P., Lee H., Corcoran A., Thistlethwaite F. C., Evans M. J., Colledge W. H., Friedman L. S., Ponder B. A., Venkitaraman A. R. (1998) Involvement of Brca2 in DNA repair. Mol. Cell 1, 347–357 [DOI] [PubMed] [Google Scholar]
- 5. Moynahan M. E., Pierce A. J., Jasin M. (2001) BRCA2 is required for homology-directed repair of chromosomal breaks. Mol. Cell 7, 263–272 [DOI] [PubMed] [Google Scholar]
- 6. Pellegrini L., Venkitaraman A. (2004) Emerging functions of BRCA2 in DNA recombination. Trends Biochem. Sci. 29, 310–316 [DOI] [PubMed] [Google Scholar]
- 7. Venkitaraman A. R. (2002) Cancer susceptibility and the functions of BRCA1 and BRCA2. Cell 108, 171–182 [DOI] [PubMed] [Google Scholar]
- 8. West S. C. (2003) Molecular views of recombination proteins and their control. Nat. Rev. Mol. Cell Biol. 4, 435–445 [DOI] [PubMed] [Google Scholar]
- 9. Venkitaraman A. R. (2004) Tracing the network connecting BRCA and Fanconi anaemia proteins. Nat. Rev. Cancer 4, 266–276 [DOI] [PubMed] [Google Scholar]
- 10. Venkitaraman A. R. (2009) Linking the cellular functions of BRCA genes to cancer pathogenesis and treatment. Annu. Rev. Pathol. 4, 461–487 [DOI] [PubMed] [Google Scholar]
- 11. Petermann E., Orta M. L., Issaeva N., Schultz N., Helleday T. (2010) Hydroxyurea-stalled replication forks become progressively inactivated and require two different RAD51-mediated pathways for restart and repair. Mol. Cell 37, 492–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yu V. P., Koehler M., Steinlein C., Schmid M., Hanakahi L. A., van Gool A. J., West S. C., Venkitaraman A. R. (2000) Gross chromosomal rearrangements and genetic exchange between nonhomologous chromosomes following BRCA2 inactivation. Genes Dev. 14, 1400–1406 [PMC free article] [PubMed] [Google Scholar]
- 13. Lomonosov M., Anand S., Sangrithi M., Davies R., Venkitaraman A. R. (2003) Stabilization of stalled DNA replication forks by the BRCA2 breast cancer susceptibility protein. Genes Dev. 17, 3017–3022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Schlacher K., Christ N., Siaud N., Egashira A., Wu H., Jasin M. (2011) Double strand break repair-independent role for BRCA2 in blocking stalled replication fork degradation by MRE11. Cell 145, 529–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Greider C. W., Blackburn E. H. (1985) Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell 43, 405–413 [DOI] [PubMed] [Google Scholar]
- 16. Gilson E., Géli V. (2007) How telomeres are replicated. Nat. Rev. Mol. Cell Biol. 8, 825–838 [DOI] [PubMed] [Google Scholar]
- 17. Sfeir A., Kosiyatrakul S. T., Hockemeyer D., MacRae S. L., Karlseder J., Schildkraut C. L., de Lange T. (2009) Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell 138, 90–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Smith F. W., Feigon J. (1992) Quadruplex structure of Oxytricha telomeric DNA oligonucleotides. Nature 356, 164–168 [DOI] [PubMed] [Google Scholar]
- 19. Paeschke K., Capra J. A., Zakian V. A. (2011) DNA replication through G-quadruplex motifs is promoted by the Saccharomyces cerevisiae Pif1 DNA helicase. Cell 145, 678–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Maizels N. (2008) Genomic stability. FANCJ-dependent G4 DNA repair. Curr. Biol. 18, R613–R614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sun H., Karow J. K., Hickson I. D., Maizels N. (1998) The Bloom's syndrome helicase unwinds G4 DNA. J. Biol. Chem. 273, 27587–27592 [DOI] [PubMed] [Google Scholar]
- 22. Wu Y., Shin-ya K., Brosh R. M., Jr. (2008) FANCJ helicase defective in Fanconia anemia and breast cancer unwinds G-quadruplex DNA to defend genomic stability. Mol. Cell. Biol. 28, 4116–4128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wu X., Maizels N. (2001) Substrate-specific inhibition of RecQ helicase. Nucleic Acids Res. 29, 1765–1771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jonkers J., Meuwissen R., van der Gulden H., Peterse H., van der Valk M., Berns A. (2001) Synergistic tumor suppressor activity of BRCA2 and p53 in a conditional mouse model for breast cancer. Nat. Genet. 29, 418–425 [DOI] [PubMed] [Google Scholar]
- 25. Choi E., Choe H., Min J., Choi J. Y., Kim J., Lee H. (2009) BubR1 acetylation at prometaphase is required for modulating APC/C activity and timing of mitosis. EMBO J. 28, 2077–2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Williams E. S., Bailey S. M. (2009) Chromosome orientation fluorescence in situ hybridization (CO-FISH). Cold Spring Harb. Protoc. pdb.prot5269 [DOI] [PubMed] [Google Scholar]
- 27. Poon S. S., Lansdorp P. M. (2001) Quantitative in situ hybridization (Q-FISH). Curr. Protoc. Cell Biol. Chapter 18, Unit 18.14 [DOI] [PubMed] [Google Scholar]
- 28. Cimino-Reale G., Pascale E., Alvino E., Starace G., D'Ambrosio E. (2003) Long telomeric C-rich 5′-tails in human replicating cells. J. Biol. Chem. 278, 2136–2140 [DOI] [PubMed] [Google Scholar]
- 29. Cimino-Reale G., Pascale E., Battiloro E., Starace G., Verna R., D'Ambrosio E. (2001) The length of telomeric G-rich strand 3′-overhang measured by oligonucleotide ligation assay. Nucleic Acids Res. 29, E35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Dimitrova D. S., Gilbert D. M. (2000) Temporally coordinated assembly and disassembly of replication factories in the absence of DNA synthesis. Nat. Cell Biol. 2, 686–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hemann M. T., Greider C. W. (1999) G-strand overhangs on telomeres in telomerase-deficient mouse cells. Nucleic Acids Res. 27, 3964–3969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lekomtsev S., Guizetti J., Pozniakovsky A., Gerlich D. W., Petronczki M. (2010) Evidence that the tumor-suppressor protein BRCA2 does not regulate cytokinesis in human cells. J. Cell Sci. 123, 1395–1400 [DOI] [PubMed] [Google Scholar]
- 33. Opresko P. L., Mason P. A., Podell E. R., Lei M., Hickson I. D., Cech T. R., Bohr V. A. (2005) POT1 stimulates RecQ helicases WRN and BLM to unwind telomeric DNA substrates. J. Biol. Chem. 280, 32069–32080 [DOI] [PubMed] [Google Scholar]
- 34. Rizzo A., Salvati E., Porru M., D'Angelo C., Stevens M. F., D'Incalci M., Leonetti C., Gilson E., Zupi G., Biroccio A. (2009) Stabilization of quadruplex DNA perturbs telomere replication leading to the activation of an ATR-dependent ATM signaling pathway. Nucleic Acids Res. 37, 5353–5364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wang Y., Erdmann N., Giannone R. J., Wu J., Gomez M., Liu Y. (2005) An increase in telomere sister chromatid exchange in murine embryonic stem cells possessing critically shortened telomeres. Proc. Natl. Acad. Sci. U.S.A. 102, 10256–10260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Morrish T. A., Greider C. W. (2009) Short telomeres initiate telomere recombination in primary and tumor cells. PLoS Genet. 5, e1000357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Londoño-Vallejo J. A., Der-Sarkissian H., Cazes L., Bacchetti S., Reddel R. R. (2004) Alternative lengthening of telomeres is characterized by high rates of telomeric exchange. Cancer Res. 64, 2324–2327 [DOI] [PubMed] [Google Scholar]
- 38. Cesare A. J., Reddel R. R. (2010) Alternative lengthening of telomeres. Models, mechanisms, and implications. Nat. Rev. Genet. 11, 319–330 [DOI] [PubMed] [Google Scholar]
- 39. Takai H., Smogorzewska A., de Lange T. (2003) DNA damage foci at dysfunctional telomeres. Curr. Biol. 13, 1549–1556 [DOI] [PubMed] [Google Scholar]
- 40. IJpma A. S., Greider C. W. (2003) Short telomeres induce a DNA damage response in Saccharomyces cerevisiae. Mol. Biol. Cell 14, 987–1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. d'Adda di Fagagna F., Reaper P. M., Clay-Farrace L., Fiegler H., Carr P., Von Zglinicki T., Saretzki G., Carter N. P., Jackson S. P. (2003) A DNA damage checkpoint response in telomere-initiated senescence. Nature 426, 194–198 [DOI] [PubMed] [Google Scholar]
- 42. Longhese M. P. (2008) DNA damage response at functional and dysfunctional telomeres. Genes Dev. 22, 125–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wang B., Matsuoka S., Carpenter P. B., Elledge S. J. (2002) 53BP1, a mediator of the DNA damage checkpoint. Science 298, 1435–1438 [DOI] [PubMed] [Google Scholar]
- 44. de Lange T. (2005) Shelterin. The protein complex that shapes and safeguards human telomeres. Genes Dev. 19, 2100–2110 [DOI] [PubMed] [Google Scholar]
- 45. Hackett J. A., Greider C. W. (2002) Balancing instability. Dual roles for telomerase and telomere dysfunction in tumorigenesis. Oncogene 21, 619–626 [DOI] [PubMed] [Google Scholar]
- 46. Burge S., Parkinson G. N., Hazel P., Todd A. K., Neidle S. (2006) Quadruplex DNA. Sequence, topology, and structure. Nucleic Acids Res. 34, 5402–5415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Verdun R. E., Karlseder J. (2007) Replication and protection of telomeres. Nature 447, 924–931 [DOI] [PubMed] [Google Scholar]
- 48. Schwartz M., Zlotorynski E., Goldberg M., Ozeri E., Rahat A., le Sage C., Chen B. P., Chen D. J., Agami R., Kerem B. (2005) Homologous recombination and nonhomologous end-joining repair pathways regulate fragile site stability. Genes Dev. 19, 2715–2726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Durkin S. G., Ragland R. L., Arlt M. F., Mulle J. G., Warren S. T., Glover T. W. (2008) Replication stress induces tumor-like microdeletions in FHIT/FRA3B. Proc. Natl. Acad. Sci. U.S.A. 105, 246–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Durkin S. G., Glover T. W. (2007) Chromosome fragile sites. Annu. Rev. Genet. 41, 169–192 [DOI] [PubMed] [Google Scholar]
- 51. Badie S., Escandell J. M., Bouwman P., Carlos A. R., Thanasoula M., Gallardo M. M., Suram A., Jaco I., Benitez J., Herbig U., Blasco M. A., Jonkers J., Tarsounas M. (2010) BRCA2 acts as a RAD51 loader to facilitate telomere replication and capping. Nat. Struct. Mol. Biol. 17, 1461–1469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hashimoto Y., Chaudhuri A. R., Lopes M., Costanzo V. (2010) Rad51 protects nascent DNA from Mre11-dependent degradation and promotes continuous DNA synthesis. Nat. Struct. Mol. Biol. 17, 1305–1311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Trinh T. Q., Sinden R. R. (1991) Preferential DNA secondary structure mutagenesis in the lagging strand of replication in E. coli. Nature 352, 544–547 [DOI] [PubMed] [Google Scholar]
- 54. Rosche W. A., Trinh T. Q., Sinden R. R. (1995) Differential DNA secondary structure-mediated deletion mutation in the leading and lagging strands. J. Bacteriol. 177, 4385–4391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Stewart S. A., Ben-Porath I., Carey V. J., O'Connor B. F., Hahn W. C., Weinberg R. A. (2003) Erosion of the telomeric single strand overhang at replicative senescence. Nat. Genet. 33, 492–496 [DOI] [PubMed] [Google Scholar]
- 56. Yang Q., Zheng Y. L., Harris C. C. (2005) POT1 and TRF2 cooperate to maintain telomeric integrity. Mol. Cell. Biol. 25, 1070–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Baur J. A., Zou Y., Shay J. W., Wright W. E. (2001) Telomere position effect in human cells. Science 292, 2075–2077 [DOI] [PubMed] [Google Scholar]
- 58. Shiloh Y. (2001) ATM (ataxia telangiectasia mutated). Expanding roles in the DNA damage response and cellular homeostasis. Biochem. Soc. Trans. 29, 661–666 [DOI] [PubMed] [Google Scholar]
- 59. Paulsen R. D., Cimprich K. A. (2007) The ATR pathway. Fine-tuning the fork. DNA Repair 6, 953–966 [DOI] [PubMed] [Google Scholar]
- 60. Shiloh Y. (2001) ATM and ATR. Networking cellular responses to DNA damage. Curr. Opin. Genet. Dev. 11, 71–77 [DOI] [PubMed] [Google Scholar]
- 61. McGowan C. H., Russell P. (2004) The DNA damage response. Sensing and signaling. Curr. Opin. Cell Biol. 16, 629–633 [DOI] [PubMed] [Google Scholar]
- 62. Durocher D., Jackson S. P. (2001) DNA-PK, ATM, and ATR as sensors of DNA damage. Variations on a theme? Curr. Opin. Cell Biol. 13, 225–231 [DOI] [PubMed] [Google Scholar]
- 63. Verdun R. E., Karlseder J. (2006) The DNA damage machinery and homologous recombination pathway act consecutively to protect human telomeres. Cell 127, 709–720 [DOI] [PubMed] [Google Scholar]
- 64. Palm W., de Lange T. (2008) How shelterin protects mammalian telomeres. Annu. Rev. Genet. 42, 301–334 [DOI] [PubMed] [Google Scholar]
- 65. Yang X. H., Shiotani B., Classon M., Zou L. (2008) Chk1 and Claspin potentiate PCNA ubiquitination. Genes Dev. 22, 1147–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Alao J. P., Sunnerhagen P. (2009) The ATM and ATR inhibitors CGK733 and caffeine suppress cyclin D1 levels and inhibit cell proliferation. Radiat. Oncol. 4, 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Hickson I., Zhao Y., Richardson C. J., Green S. J., Martin N. M., Orr A. I., Reaper P. M., Jackson S. P., Curtin N. J., Smith G. C. (2004) Identification and characterization of a novel and specific inhibitor of the ataxia-telangiectasia mutated kinase ATM. Cancer Res. 64, 9152–9159 [DOI] [PubMed] [Google Scholar]
- 68. Melchionna R., Chen X. B., Blasina A., McGowan C. H. (2000) Threonine 68 is required for radiation-induced phosphorylation and activation of Cds1. Nat. Cell Biol. 2, 762–765 [DOI] [PubMed] [Google Scholar]
- 69. Liu Q., Guntuku S., Cui X. S., Matsuoka S., Cortez D., Tamai K., Luo G., Carattini-Rivera S., DeMayo F., Bradley A., Donehower L. A., Elledge S. J. (2000) Chk1 is an essential kinase that is regulated by Atr and required for the G(2)/M DNA damage checkpoint. Genes Dev. 14, 1448–1459 [PMC free article] [PubMed] [Google Scholar]
- 70. Zhao H., Piwnica-Worms H. (2001) ATR-mediated checkpoint pathways regulate phosphorylation and activation of human Chk1. Mol. Cell. Biol. 21, 4129–4139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Göhler T., Sabbioneda S., Green C. M., Lehmann A. R. (2011) ATR-mediated phosphorylation of DNA polymerase η is needed for efficient recovery from UV damage. J. Cell Biol. 192, 219–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.