Background: The RanBP2 internal repeat domain (IR1-M-IR2) catalyzes SUMO E3 ligase activity and binds SUMO1-RanGAP1/UBC9 at the nuclear pore complex.
Results: Biochemistry and structures of RanBP2/SUMO-RanGAP1/UBC9 are presented.
Conclusion: IR1 protects RanGAP1-SUMO1/UBC9 and functions as the primary E3 ligase of RanBP2, whereas IR2 interacts with SUMO1 to promote weaker SUMO1-specific E3 ligase activity.
Significance: RanBP2/SUMO interactions provide insight to SUMO isoform specificity.
Keywords: E3 Ubiquitin Ligase, Nuclear Pore, SUMO, Ubiquitin-conjugating Enzyme (Ubc), X-ray Crystallography, Localization, SUMO E2, SUMO E3 ligase, SUMO Interaction Motif
Abstract
The RanBP2 nucleoporin contains an internal repeat domain (IR1-M-IR2) that catalyzes E3 ligase activity and forms a stable complex with SUMO-modified RanGAP1 and UBC9 at the nuclear pore complex. RanBP2 exhibits specificity for SUMO1 as RanGAP1-SUMO1/UBC9 forms a more stable complex with RanBP2 compared with RanGAP1-SUMO2 that results in greater protection of RanGAP-SUMO1 from proteases. The IR1-M-IR2 SUMO E3 ligase activity also shows a similar preference for SUMO1. We utilized deletions and domain swap constructs in protease protection assays and automodification assays to define RanBP2 domains responsible for RanGAP1-SUMO1 protection and SUMO1-specific E3 ligase activity. Our data suggest that elements in both IR1 and IR2 exhibit specificity for SUMO1. IR1 protects RanGAP1-SUMO1/UBC9 and functions as the primary E3 ligase of RanBP2, whereas IR2 retains the ability to interact with SUMO1 to promote SUMO1-specific E3 ligase activity. To determine the structural basis for SUMO1 specificity, a hybrid IR1 construct and IR1 were used to determine three new structures for complexes containing UBC9 with RanGAP1-SUMO1/2. These structures show more extensive contacts among SUMO, UBC9, and RanBP2 in complexes containing SUMO1 compared with SUMO2 and suggest that differences in SUMO specificity may be achieved through these subtle conformational differences.
Introduction
Post-translational modification by members of the small ubiquitin-related modifier (SUMO)3 protein family regulates multiple cellular processes, including nuclear transport, transcription, chromosome segregation, and DNA repair (1, 2). Conjugation of SUMO to substrates occurs through an enzymatic cascade composed of an E1-activating enzyme (SAE1/UBA2), an E2-conjugating enzyme (UBC9), and E3 protein ligases that results in formation of an isopeptide bond between the C terminus of SUMO and the ϵ-amino group of a substrate lysine residue (3). The SUMO E2, UBC9, is able to specifically recognize and conjugate SUMO to substrates containing a SUMO consensus motif (ΨKXD/E), although E3s often aid in conjugation through (i) recruitment of the E2∼SUMO (where ∼ indicates a thioester linkage) and substrate into a complex to promote specificity and (ii) stimulation of SUMO discharge from the E2 to the substrate (4).
SUMO E3 protein ligases include members of the Siz/PIAS family which contain an SP-RING domain and are thought to function like ubiquitin RING E3s by binding the substrate and by binding and activating E2∼SUMO for SUMO discharge (5–7). A second and distinct class of SUMO E3 is represented singularly by RanBP2 (also termed Nup358), a component of the cytoplasmic fibrils of the nuclear pore complex (NPC). RanBP2 is required to localize SUMO1-modified RanGAP1 and UBC9 in a complex at the NPC to facilitate nucleocytoplasmic trafficking by promoting the Ran GTPase cycle (8–10). In addition to its role in binding RanGAP1-SUMO1/UBC9, the RanBP2 IR1-M-IR2 domain can promote SUMO conjugation in vitro and in vivo. Furthermore, the internal repeats IR1 and IR2 each possess E3 activity in vitro (11–14).
Previous structural work demonstrated that IR1 contains five motifs that are required to bind RanGAP1-SUMO1/UBC9 (14). Motif I is a SUMO-interacting motif (SIM) that forms an anti-parallel β-strand with SUMO β-strand 2, establishing contacts to SUMO β-strand 2 and SUMO α-helix 1. Motif II is composed of an α-helix and coil that makes contacts to both SUMO and UBC9, whereas motifs III–V wrap around and form additional interactions with UBC9. In the absence of RanGAP1-SUMO1/UBC9, IR1 can bind an E2∼SUMO thioester and coordinate it in an optimal orientation to promote catalysis.
RanGAP1 was the first SUMO substrate to be identified and is preferentially modified by SUMO1 in vivo (15–17). This is not because SUMO2 does not modify RanGAP1, but rather because RanGAP1-SUMO1 and UBC9 form a more stable complex with RanBP2 that is better protected from the deconjugating activities of SUMO proteases (18). The preference for SUMO1 is also observed in its E3 ligase activity, as previous studies demonstrated that the E3 ligase domains of RanBP2 exhibit higher ligase activity with SUMO1 compared with SUMO2 under single and multiple turnover conditions with a number of model substrates (11, 14).
Although it is clear that the E3 ligase domain of RanBP2 displays a preference for SUMO1, it is not yet known how this specificity is achieved. In addition, it is unclear whether IR1 and IR2 function together or separately in E3 ligase and binding activities of RanBP2. To define better the RanBP2 domains responsible for RanGAP1-SUMO1 protection and SUMO1-specific E3 ligase activity, we utilized IR1-M-IR2 deletions and domain swap constructs in protease protection assays and automodification assays. Our results support a model in which SUMO1 specificity is achieved by both internal repeats of RanBP2. IR1 protects RanGAP1-SUMO1/UBC9 and functions as the primary E3 ligase of RanBP2, whereas IR2 retains the ability to interact with SUMO1 to promote SUMO1-specific E3 ligase activity. Domain swaps further suggest that a hybrid IR1 containing IR2 motif II can promote SUMO1-specific modification of a model substrate. To determine the structural basis for SUMO1 specificity, IR1 and a hybrid IR1 construct containing IR2 motif II were used to crystallize and determine three new structures for complexes containing UBC9 with either RanGAP1-SUMO1 or RanGAP1-SUMO2. These structures show more extensive contacts among SUMO, UBC9, and RanBP2 in complexes containing SUMO1 compared with SUMO2.
EXPERIMENTAL PROCEDURES
Cloning, Expression, and Protein Purification
Preparation of human E1 (SAE1/UBA2), E2 (UBC9), SUMO1 (1–97), SUMO2 (1–93), SUMO1 (18–97), SUMO2 (14–93), RanGAP1 (419–578), IR1-M-IR2 (RanBP2 residues 2631–2771), IR1 (RanBP2 residues 2631–2695), IR2 (RanBP2 residues 2709–2771), and the p53 tetramerization domain have been described previously (14, 19–22). Additional RanBP2 constructs (RanBP2-IR, IR1-M-IR1SIM-IR2II–V, IR2SIM-IR1II–V-M-IR2, ΔSIMIR1-M-IR2, IR1-M-ΔSIMIR2, ΔSIMIR1-M-ΔSIM-IR2, IR2SIM-IR1, IR2SIM-IR2II-IR1III–V, IR1SIM-IR2II-IR1III–V, and IR1SIM-IR2; see Fig. 1) were amplified by PCR and cloned into a pSmt3 vector. SENP1(419–644) was cloned into pET-28b (Novagen). Vectors were transformed into Escherichia coli BL21(DE3)-CP cells, grown at 37 °C to an A600 of 0.8, induced with 1 mm isopropyl 1-thio-β-d-galactopyranoside for 4 h at 30 °C, harvested, and suspended in buffer containing 50 mm Tris-HCl, pH 8.0, 20% w/v sucrose, 350 mm NaCl, 20 mm imidazole, 0.1% IGEPAL, 1 mm PMSF, 10 μg/ml DNase, and 1 mm β-mercaptoethanol prior to sonication and centrifugation. Proteins were purified by nickel-nitrilotriacetic acid-agarose resin (Qiagen) and gel filtration (Superdex75; GE Healthcare). Purified proteins were stored in 20 mm Tris, pH 8.0, 350 mm NaCl, 1 mm β-mercaptoethanol, flash frozen, and stored at −80 °C. RanGAP1-SUMO1 and RanGAP1-SUMO2 were prepared and combined with UBC9 and RanBP2 constructs, purified by gel filtration (Superdex200; GE Healthcare) and concentrated to 10 mg/ml in 20 mm Tris, pH 8.0, 100 mm NaCl, 1 mm β-mercaptoethanol.
FIGURE 1.
RanBP2 constructs. A, amino acid alignment of the internal repeat domain of RanBP2. A schematic representation of IR1 (magenta), M (gray), and IR2 (pink) is displayed below the amino acid sequence of RanBP2. Regions corresponding to the motif I SIM, motif II, and motifs III–V are shown above the IR1 sequence. Asterisks indicating identical amino acid positions between IR1 and IR2 are displayed above the IR2 sequence. B, IR1-M-IR2 constructs. Schematics of IR1-M-IR2 constructs used in this study are color-coded as above. C, IR constructs. Schematics of individual internal repeat domain constructs are shown.
Biochemical Assays
Protease protection assays were carried out in buffer containing 20 mm Tris, pH 8.0, 150 mm NaCl, 0.1% Tween 20, and 2 mm DTT. Reactions with 8 μm RanGAP1-SUMO, 20 μm UBC9, and 20 μm RanBP2 construct were preincubated for 10 min at room temperature, SENP1 was added, and reactions were incubated for 20 min at 37 °C. Samples were quenched by the addition of SDS-loading buffer, separated by SDS-PAGE (12% BisTris gels and MOPS running buffer; Invitrogen) and stained with SYPRO Ruby (Bio-Rad). Gels were scanned using an FLA-5000 (Fujifilm), and images were processed and quantified using MultiGauge v2.02 (Fujifilm). Data were fit to an exponential decay curve (y = y0 + ae−bx) using SigmaPlot (Systat Software, Inc.).
Automodification assays were performed in buffer containing 20 mm HEPES, pH 7.5, 5 mm MgCl2, 0.1% Tween 20, 1 mm DTT. Reactions containing 1 μm SUMO, 100 nm E1, 100 nm E2, and 100 nm RanBP2 construct were initiated by addition of 1 mm ATP and incubated at 37 °C. For assays in the presence of excess RanGAP1-SUMO and UBC9, a mixture containing both RanGAP1-SUMO and UBC9 was preincubated for 10 min at room temperature and added to the reaction at a final concentration of 250 nm before the addition of ATP. Samples were quenched by addition of SDS-loading buffer, separated by SDS-PAGE (4–12% BisTris gels and MES running buffer; Invitrogen), and analyzed by Western blotting with polyclonal antibodies against SUMO1 (Boston Biochem) and SUMO2 (Sigma).
SUMO conjugation assays under multiple turnover conditions with individual IR domain constructs were performed in buffer containing 20 mm HEPES, pH 7.5, 5 mm MgCl2, 0.1% Tween 20, and 1 mm DTT. Reactions included 200 nm E1, 200 nm E2, 100 nm RanBP2 construct, 8 μm p53, 15 μm SUMO. Conjugation assays performed in the absence of RanBP2 or with IR1-M-IR2 constructs were performed in the same buffer, but reactions included 100 nm E1, 100 nm E2, 100 nm RanBP2 construct, 8 μm p53, and 5 μm SUMO. Reactions were initiated by the addition of 1 mm ATP, and samples were quenched by addition of SDS-loading buffer and were separated by SDS-PAGE (4–12% BisTris gels and MES running buffer). Gels were stained with SYPRO Ruby, scanned using an FLA-5000, and images were processed and quantified using MultiGauge v2.02.
Crystallographic Analysis
Crystals of SUMO1-RanGAP1-UBC9-IR1SIM-IR2II-IR1III–V were obtained at 6 °C by hanging-drop vapor diffusion against a well solution containing 22% PEG 4000, 100 mm HEPES, pH 7.5, 400 mm ammonium citrate, and 2% isopropyl alcohol. These crystals were cryoprotected with 12% ethylene glycol and diffracted to 2.3 Å. Crystals of the SUMO2-RanGAP1-UBC9-IR1SIM-IR2II-IR1III–V complex were obtained at 6 °C by hanging-drop vapor diffusion with a well solution containing 14% PEG4000, 100 mm sodium citrate, pH 6.0, and 200 mm ammonium acetate. These crystals were cryoprotected with 16% ethylene glycol and diffracted to 2.6 Å. Crystals of SUMO2-RanGAP1-UBC9-IR1 were obtained at 18 °C by hanging-drop vapor diffusion against a well solution containing 16% PEG4000, 100 mm sodium citrate, pH 5.0, and 200 mm ammonium acetate. These crystals were cryoprotected with 12% ethylene glycol and diffracted to 2.6 Å. Data sets were collected at the Advanced Photon Source (Argonne, IL) at NE-CAT beamline 24-IDC. Diffraction data were processed using HKL-2000 (23) and CCP4 (24). After molecular replacement using a model derived from the SUMO1-RanGAP1-UBC9-IR1 structure (PDB 1Z5S), atomic models were built into the density using COOT (25) and refined with Refmac (24), CNS (26), and Phenix (27).
RESULTS
Activities of RanBP2 IR1-M-IR2
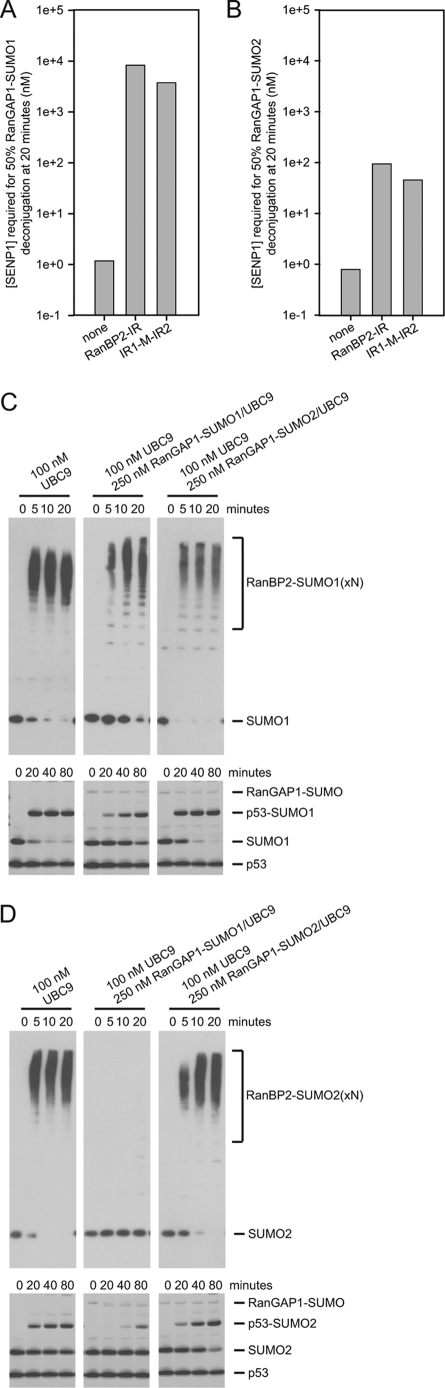
To confirm previous results with respect to preferential protection of RanGAP1-SUMO1 by RanBP2 (18), we utilized a RanBP2 construct from residues 2611 to 2794 that encompassed the E3 ligase domain of RanBP2, RanBP2-IR, and a smaller construct from 2631 to 2771 containing both internal repeat domains, IR1-M-IR2 (Fig. 1, A and B). Reactions containing 20 μm RanBP2 construct and 20 μm UBC9 were preincubated with either 8 μm RanGAP1-SUMO1 or 8 μm RanGAP1-SUMO2 for 10 min to allow for complex formation. The SUMO protease SENP1 was added at increasing concentrations to determine how much SENP1 is required to deconjugate 50% of RanGAP1-SUMO after 20 min (supplemental Figs. 1 and 2).
7000-fold and 3200-fold more SENP1 was required to achieve 50% deconjugation of RanGAP-SUMO1/UBC9 in the presence of RanBP2-IR and IR1-M-IR2 (8.3 μm and 3.8 μm, respectively) compared with RanGAP1-SUMO1/UBC9 alone (1.2 nm) (Fig. 2A). In similar reactions with RanGAP-SUMO2, however, 86-fold less SENP1 is required to achieve the same level of deconjugation (94 nm for RanBP2-IR and 45 nm for IR1-M-IR2) (Fig. 2B). The difference in protection of RanGAP1-SUMO1 and RanGAP1-SUMO2 is not due to preference of the protease for SUMO1-modified substrate as SENP1 does not exhibit SUMO isoform specificity in deconjugation assays (28), and protease protection assays with RanGAP1-SUMO1/UBC9 and RanGAP1-SUMO2/UBC9 in the absence of RanBP2 require similar SENP1 concentrations (1.2 nm and 0.8 nm, respectively). These results confirm previous findings with respect the ability of RanBP2 to preferentially bind and protect RanGAP1-SUMO1 (18).
FIGURE 2.
Activities of RanBP2 IR1-M-IR2. A and B, bar graphs indicating the concentration of SENP1 required to deconjugate 50% of RanGAP-SUMO in the presence of UBC9 and indicated RanBP2 constructs for RanGAP-SUMO1 (A) and RanGAP-SUMO2 (B) as interpolated using data obtained from three independent experiments (supplemental Figs. 1 and 2). C, time course of IR1-M-IR2 automodification with SUMO1 (upper) and p53 modification with SUMO1 in the presence of IR1-M-IR2 (lower). Reactions were performed in the presence of the specified concentrations of UBC9 and RanGAP-SUMO/UBC9. Automodification was detected by Western blotting with SUMO1 antibody. p53 conjugation was detected by SYPRO staining. D, same as C, except that reactions were performed with SUMO2 and automodification of IR1-M-IR2 with SUMO2 was detected by Western blotting with SUMO2 antibody.
RanBP2 E3 ligase activity was monitored in automodification assays as reported previously (12–13). In our assays, IR1-M-IR2 is modified by SUMO1 and SUMO2 in reactions with E1, ATP/Mg, and UBC9 (Fig. 2, C and D). Our previous work suggested that RanGAP1-SUMO/UBC9 is a structural mimetic of UBC9∼SUMO because the same elements required to bind RanGAP1-SUMO/UBC9 are required to activate the UBC9∼SUMO thioester in E3 ligase assays with other substrates (14). In reactions with UBC9 and SUMO1, automodification was inhibited but not abolished by excess RanGAP1-SUMO1/UBC9, but not by excess RanGAP1-SUMO2/UBC9 (Fig. 2C). In reactions with UBC9 and SUMO2, automodification was undetectable in the presence of excess RanGAP1-SUMO1/UBC9 and partially inhibited by RanGAP1-SUMO2/UBC9 (Fig. 2D). A similar pattern of inhibition was observed when RanGAP1-SUMO1/UBC9 and RanGAP1-SUMO2/UBC9 were added to conjugation assays using p53 as a substrate (Fig. 2, C and D) This suggests that RanBP2 has a distinct preference for complexes containing SUMO1 and that RanGAP1-SUMO1/UBC9 and RanGAP1-SUMO2/UBC9 effectively compete with UBC9∼SUMO1 and UBC9∼SUMO2 for binding, respectively. Whereas RanGAP1-SUMO2/UBC9 is unable to compete with UBC9∼SUMO1, RanGAP1-SUMO1/UBC9 appears to prevent interaction between UBC9∼SUMO2 and RanBP2. The preference observed for SUMO1 in E3 ligase assays is also evident in protease protection assays using RanGAP1-SUMO1/UBC9 and RanGAP1-SUMO2/UBC9.
Activities of RanBP2 IR1 and IR2
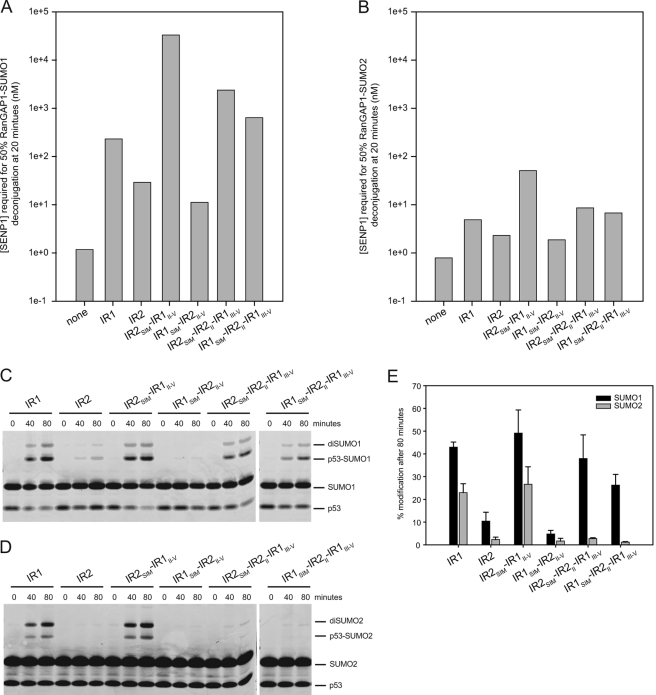
Constructs corresponding either to IR1 or IR2 were utilized in protease protection and E3 ligase assays to further dissect elements within RanBP2 that contribute SUMO1 specificity (Fig. 1C). Similar to results obtained for IR1-M-IR2 which exhibited an 83-fold preference for RanGAP1-SUMO1 over RanGAP1-SUMO2, both IR1 and IR2 protected RanGAP1-SUMO1 47-fold and 13-fold better than RanGAP1-SUMO2, respectively (Fig. 3, A and B), although IR1-M-IR2 protected RanGAP1-SUMO1 16-fold better than IR1, and IR1 protected RanGAP1-SUMO1 8-fold better than IR2. In summary, both IR1 and IR2 exhibit a preference for SUMO1, but IR1 appears to form a more stable complex with RanGAP1-SUMO1/UBC9 than IR2. These data are consistent with previous work showing that IR1, and not IR2, is able to interact with UBC9 in GST pulldown experiments (11).
FIGURE 3.
Activities of RanBP2 IR1 and IR2. A and B, bar graphs indicating the concentration of SENP1 required to deconjugate 50% of RanGAP-SUMO in the presence of UBC9 and indicated RanBP2 constructs for RanGAP-SUMO1 (A) and RanGAP-SUMO2 (B) as interpolated using data obtained from three independent experiments (supplemental Figs. 1 and 2). C, time course of SUMO1 conjugation to p53 with the specified RanBP2 construct. D, same as C, except that conjugation assays were performed with SUMO2. E, bar chart displaying quantitation of the conjugation assays shown in C and D. Assays were performed in triplicate. Error bars represent ± 1 S.D.
Previous work from our laboratory demonstrated that deletion of the motif I SIM from either IR1 or IR2 abrogated or greatly diminished their E3 ligase activity (14). Furthermore, deletion of IR1 motifs III–V diminished its E3 ligase activity to levels similar to that observed for IR2. Because the IR1/SUMO1-RanGAP1/UBC9 structure showed that motifs I and II contact SUMO (motif I) and the SUMO/UBC9 interface (motif II), these data suggest that motifs I and II are critical for E3 ligase activity and that differences in primary sequence between motifs III–IV in IR1 and IR2 (18% identity) may account for the inability of IR2 to engage UBC9 fully (Fig. 1A).
The preference exhibited by IR1 and IR2 for SUMO1 has been suggested previously based on E3 ligase activity with the substrates Sp100 (11) and p53 (14). We confirmed these results using IR1 and IR2 in conjugation assays with the tetramerization domain of p53 as a model substrate (Fig. 3, C–E). IR1 was able to conjugate both SUMO1 and SUMO2, with a slight preference for SUMO1 (2-fold). Although IR2 is less active than IR1, IR2 exhibits a 4-fold preference for SUMO1 in conjugation assays, indicating that IR2 retains SUMO1-specific E3 ligase activity. It should be noted that di-SUMO chains form in these reactions conducted under conditions of multiple turnover, and these chains form more readily with SUMO2 due to a SUMO consensus site near its N terminus (29).
SUMO isoform-specific SIMs have been reported in the literature (30–33), and IR1 of RanBP2 is known to bind SUMO primarily through its SIM (motif I). We posited that the SIM may be responsible for the higher affinity interaction between RanGAP1-SUMO1 and IR1 (Fig. 3, A and B) which has also been reported previously using RanBP2 (18). To test whether the SIMs of IR1 or IR2 contribute differentially to SUMO1 specificity, chimeras were made in which the SIMs of IR1 and IR2 were swapped (Fig. 1C). Like IR1 and IR2, IR1 motifs II–V with an IR2 SIM (IR2SIM-IR1II–V) and IR2 with an IR1 SIM (IR1SIM-IR2II–V) required relatively low levels of SENP1 to deconjugate RanGAP1-SUMO2 (51 and 2 nm, respectively) (Fig. 3B). In contrast, the IR2SIM-IR1II–V hybrid protected RanGAP1-SUMO1 144-fold better relative than IR1 and 9-fold better than IR1-M-IR2 whereas the IR1SIM-IR2II–V hybrid was 3-fold less able to protect RanGAP1-SUMO1 compared with IR2 (Fig. 3A). This suggests that the IR2 SIM interacts better with SUMO1 especially when placed in the context of IR1 motifs II–V. Because the IR2 SIM exhibited a preference for SUMO1, we tested whether IR2 motif II could contribute further to SUMO1 specificity because motif II is well conserved between IR1 and IR2 and because IR1 motif II interacts with SUMO1 in the context of an IR1 complex (14). IR1 hybrids containing IR2 motif II (IR1SIM-IR2II-IR1III–V) or the IR2 SIM and IR2 motif II (IR2SIM-IR2II-IR1III–V) displayed 3-fold and 10-fold enhanced protection, respectively, of RanGAP1-SUMO1 relative to IR1 in protease protection assays (Fig. 3A).
In conjugation assays, the IR2SIM-IR1II–V hybrid exhibited a 2-fold preference for SUMO1 over SUMO2 similar to that observed for IR1, whereas IR2 exhibited a 4-fold preference for SUMO1 over SUMO2 (Fig. 3E). When IR1 hybrids containing IR2 motif II were used in conjugation assays, E3 ligase activity was clearly distinct from IR1, IR2, or the IR2SIM-IR1II–V hybrid (Fig. 3, C–E). Both IR2SIM-IR2II-IR1III–V and IR1SIM-IR2II-IR1III–V hybrids maintained E3 ligase activities in SUMO1 conjugation assays to levels similar to that observed for IR1, but each was less able to promote conjugation using SUMO2. In other words, IR2SIM-IR2II-IR1III–V and IR1SIM-IR2II-IR1III–V exhibited 14-fold and 23-fold specificity for SUMO1 in E3 ligase assays, suggesting a role for IR2 motif II in discriminating between SUMO1 and SUMO2 in these assays.
Given the weaker interactions between IR2 and UBC9, it is likely that the ability of IR2 to protect RanGAP1-SUMO1 and to promote SUMO1-specific E3 ligase activity relies more heavily on the IR2 SIM and motif II, which show a clear preference for SUMO1 within the context of IR1. This claim is supported not only by the decreased protection of RanGAP1-SUMO1 observed for IR1SIM-IR2II–V, but also by the behavior of the SIM swap constructs in conjugation assays. Whereas the IR2SIM-IR1II–V hybrid maintains the ability to act as an E3 with SUMO1 and SUMO2, E3 activity with SUMO1 is nearly abolished in the IR1SIM-IR2II–V hybrid (Fig. 3, C–E).
Structures with IR1 and IR1SIM-IR2II-IR1III–V
The structure of IR1 was previously determined in complex with RanGAP1-SUMO1/UBC9 at 3.0 Å resolution (14). To determine whether structural differences could explain the IR1 preference for SUMO1 we crystallized and determined the structure of IR1 in complex with RanGAP1-SUMO2/UBC9 at 2.6 Å resolution. Our biochemical data suggest that the IR2 SIM and motif II exhibit properties distinct from IR1, so attempts were made to determine structures of IR2 and the IR2SIM-IR2II-IR1III–V hybrid in complex with RanGAP1-SUMO1/UBC9 or RanGAP1-SUMO2/UBC9. Although those efforts unsuccessful, we did obtain crystals of the IR1SIM-IR2II-IR1III–V hybrid, containing IR2 motif II, in complex with RanGAP1-SUMO1/UBC9 at 2.3 Å resolution and RanGAP1-SUMO2/UBC9 at 2.6 Å resolution. Structures were solved by molecular replacement using the SUMO1-RanGAP1/UBC9 complex as a starting model as obtained from the SUMO1-RanGAP1/UBC9/IR1 complex (Table 1).
TABLE 1.
Crystallographic data statistics
| Crystal parameters | RanGAP1-SUMO2, UBC9, IR1 | RanGAP1-SUMO1, UBC9, IR1-IR2(MII) | RanGAP1-SUMO2, UBC9, IR1-IR2(MII) |
|---|---|---|---|
| Data collection | |||
| Source | APS 24IDC | APS 24IDC | APS 24IDC |
| Wavelength (Å) | 0.9795 | 0.9792 | 0.9792 |
| Space group | P3221 | C2221 | P3221 |
| Cell dimensions | |||
| a, b, c (Å) | 155.41, 155.41, 57.93 | 136.65, 199.19, 63.41 | 151.35, 151.35, 57.44 |
| α, β, γ (°) | 90, 90, 120 | 90, 90, 90 | 90, 90, 120 |
| Resolution (Å) | 25-2.6 (2.69-2.60)a | 35-2.3 (2.38-2.30) | 35-2.6 (2.69-2.60) |
| Completeness (%) | 99.2 (98.0) | 99.5 (99.4) | 99.5 (99.0) |
| Number of reflections | 24,830 | 38,980 | 23,329 |
| Redundancy | 8.4 (4.3) | 3.8 (3.6) | 4.8 (3.5) |
| Rmergeb (%) | 4.0 (35.6) | 5.7 (29.4) | 6.7 (43.9) |
| <I>/σ (I) | 17.7 (1.8) | 14.1 (5.6) | 14.3 (3.5) |
| Refinement statistics | |||
| Resolution (Å) | 24.41–2.60 (2.70–2.60) | 33.98–2.29 (2.35–2.29) | 34.78–2.60 (2.71–2.60) |
| Number of reflections | 24,821 (1,263) | 38,956 (1,949) | 23,317 (1,198) |
| Rworkc/Rfree (5% of data) | 0.211 (0.359)/0.264 (0.400) | 0.192 (0.247)/0.232 (0.301) | 0.204 (0.297)/0.251 (0.366) |
| Number of atoms | 3,652 | 3,955 | 3,769 |
| Protein | 3,610 | 3,612 | 3,626 |
| Water | 42 | 343 | 143 |
| B-factors (Å) | 79.1 | 38.9 | 53.3 |
| Protein | 79.3 | 38.7 | 55.8 |
| Water | 64.9 | 40.3 | 42.6 |
| r.m.s.d.d | |||
| Bond lengths (Å) | 0.009 | 0.008 | 0.009 |
| Bond angles (°) | 1.18 | 1.19 | 1.19 |
| Ramachandran plote | |||
| Core (%) | 95.5 | 96.5 | 96.2 |
| Allowed (%) | 4.3 | 3.3 | 3.5 |
| Generously allowed (%) | 0.3 | 0.3 | 0.3 |
| Disallowed region (%) | 0.0 | 0.0 | 0.0 |
| Molprobityf | |||
| Favored (%) | 97.5 | 98.6 | 98.9 |
| Allowed (%) | 2.5 | 1.4 | 1.1 |
| Outliers (%) | 0.0 | 0.0 | 0.0 |
| Clash score | 93th percentile | 97th percentile | 93th percentile |
| Molprobity score | 82th percentile | 95th percentile | 88th percentile |
| PDB code | 3UIN | 3UIP | 3UIO |
a Data in parentheses indicate statistics for data in the highest resolution bin.
b Rmerge = Σhkl Σi|I(hkl)i − <I(hkl)>|/Σhkl Σi <I(hkl)i>.
c Rwork = Σhkl Fo(hkl) − Fc(hkl)|/Σhkl |Fo(hkl)|, where Fo and Fc are observed and calculated structure factors, respectively.
d Root mean square difference.
e Calculated with the program PROCHECK.
f Calculated with the program MolProbity (40).
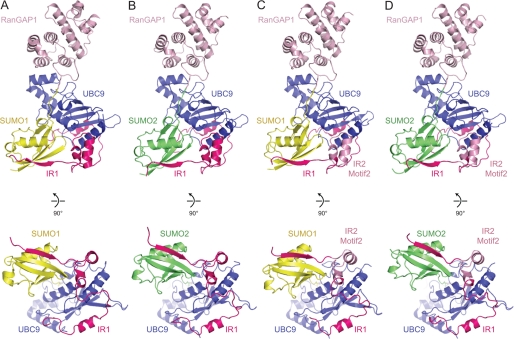
The three new structures share overall architectural similarities to that observed previously for IR1 in complex with RanGAP1-SUMO1/UBC9, including contacts between UBC9 and RanGAP1 and most contacts observed between RanBP2 elements that interact with SUMO and UBC9 (Fig. 4). We speculated originally that RanBP2 binds RanGAP1-SUMO1/UBC9 by trapping the conjugation product without release of UBC9 and that the conformation observed for UBC9 and SUMO likely resembles the conformation of UBC9∼SUMO in its activated state just prior to discharge. These claims are now supported by recent studies in the ubiquitin pathway (34–36).
FIGURE 4.
Structures of RanGAP1-SUMO/UBC9/RanBP2 complexes. Schematics represent complexes containing RanGAP1-SUMO1/UBC9/IR1 (A; PDB 1Z5S), RanGAP1-SUMO2/UBC9/IR1 (B), RanGAP1-SUMO1/UBC9/IR1SIM-IR2II-IR1III–V (C), and RanGAP1-SUMO2/UBC9/IR1SIM-IR2II-IR1III–V (D). SUMO1, SUMO2, RanGAP1, UBC9, IR1, and IR2 motif II are displayed in yellow, green, light pink, blue, magenta, and pink, respectively. Lys524 of RanGAP1 and the SUMO C-terminal glycine (Gly97 for SUMO1, Gly93 for SUMO2) and are in stick representation. A rotated view of each complex is shown below each panel to highlight the position of SUMO and the IR1 SIM with respect to UBC9. Structural figures were generated with PyMOL (38).
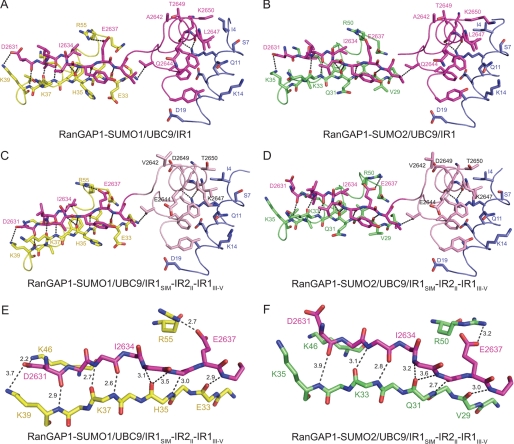
The improved resolution for the new RanGAP1-SUMO/UBC9/RanBP2 complexes warrants further discussion. The interface between RanGAP1 and UBC9 is similar to that observed in the presence of SUMO at 3.0 Å (14) and in the absence of SUMO at 2.5 Å (21) and 1.8 Å resolution (37). An extensive network of interactions between the SUMO1 C-terminal diglycine motif, UBC9, and the RanGAP1 lysine residue to which SUMO is attached was noted previously based on the 3.0 Å structure of IR1 in complex with RanGAP1-SUMO1/UBC9. Of particular interest is the arrangement of “catalytic” groups around the E2 active site cysteine, the C-terminal glycine, and substrate lysine side chain (Fig. 5). Although oxidized in the 2.3 Å structure, the cysteine Sγ is 4.0 Å and 4.3 Å from the RanGAP1 Lys524 Nϵ and SUMO1 Gly97 carbonyl carbon. The SUMO1 Gly97 carbonyl oxygen is well defined by electron density and is 3.0 Å from UBC9 Asn85 Nδ in a position that supports its role in organizing the thioester in the proper geometry for attack by the incoming Lys nucleophile. In addition, the UBC9 Asp127 carbonyl oxygen is 2.9 Å from the SUMO1 Gly97 carbonyl carbon. Very little solvent was observed in the previous structure due to the lower resolution, but in the new structures it is now evident that no solvent exists within this network of interactions, suggesting a highly complementary surface that encompasses the SUMO C-terminal diglycine motif, the UBC9 catalytic site, and the RanGAP1 lysine side chain. Analogous contacts are observed in each of the complexes presented in this study.
FIGURE 5.
Active site of the RanGAP1-SUMO1/UBC9/IR1SIM-IR2II-IR1III–V complex. A, stereo view of the E2 active site in complex with RanGAP1-SUMO1, shown in schematic and stick representation. Residues are labeled, and potential hydrogen bonds are indicated by dashed lines. SUMO, RanGAP1, and UBC9 are colored yellow, light pink, and blue, respectively. B, alternate view of the isopeptide linkage with simulated annealing omit map contoured to 2.0 σ at 2.3 Å.
Structural Basis for SUMO1 Specificity
No gross structural differences were observed that could definitively explain why IR1 or its derivatives display a preference for SUMO1, and amino acids that differ between motif II of IR1 and motif II of IR2 do not make contact with SUMO and UBC9. However, alignment of UBC9 in each of the complexes (root mean square difference of 0.48 Å for SUMO2-RanGAP1/UBC9/IR1, 0.80 Å for SUMO1-RanGAP1/UBC9/IR1SIM-IR2II-IR1III–V, and 0.41 Å for SUMO1-RanGAP1/UBC9/IR1SIM-IR2II-IR1III–V, when aligned with SUMO1-RanGAP1/UBC9/IR1) (38) revealed that SUMO1 was shifted closer to UBC9 in both our previous structure (14) and the current structures compared with the those containing SUMO2 (Fig. 4). This shift results in slightly greater total buried surface areas between UBC9 and SUMO1 (110–130 Å2), suggesting that RanBP2 may organize UBC9 and SUMO in subtly different ways depending on the SUMO isoform present.
Further inspection of the structures revealed other differences with respect to RanBP2 contacts to SUMO1 or SUMO2 that may account for some of the observed SUMO1 specificity. In surface complementarity calculations using SC (24), slightly higher complementarity was observed between SUMO1 and RanBP2 compared with complexes containing SUMO2 (0.730 average for SUMO1 complexes and 0.696 average for SUMO2 complexes). In addition, the N terminus of the IR1 SIM is positioned further away from SUMO in SUMO2-containing complexes (Fig. 6). This results in an average distance of 3.4 Å between backbone contacts of IR1 Asp2631-Leu2633 and SUMO2 Lys35-Lys33 compared with 3.0 Å with SUMO1 Lys39-Lys37 and, in the case of SUMO2-RanGAP1/UBC9/IR1SIM-IR2II-IR1III–V, loss of contacts between the IR1 Asp2631 and SUMO2 Lys35 and Lys46 side chains. In other words, the number and distance of hydrogen bonds between the IR1 SIM and SUMO1 were greater and closer, respectively, compared with structures containing SUMO2 (Fig. 6). This presumably accounts for the slightly higher buried surface area between RanBP2 and SUMO1 compared with complexes containing SUMO2. In all, complexes containing SUMO1 have ∼300 Å2 or 13% greater additional buried surface area between SUMO1/UBC9 and RanBP2/SUMO1 compared with those complexes containing SUMO2.
FIGURE 6.
SUMO/SIM interactions in RanGAP1-SUMO/UBC9/RanBP2 complexes. A–D, ribbon and stick representation of interactions between RanBP2 SIM and motif II with SUMO and UBC9 in RanGAP1-SUMO1/UBC9/IR1 (A; PDB 1Z5S), RanGAP1-SUMO2/UBC9/IR1 (B), RanGAP1-SUMO1/UBC9/IR1SIM-IR2II-IR1III–V (C), and RanGAP1-SUMO2/UBC9/IR1SIM-IR2II-IR1III–V (D). SUMO β-strand 2 and SUMO α-helix 1 are colored yellow and green for SUMO1 and SUMO2, respectively. The IR1 SIM and motif II, IR2 motif II, and UBC9 are colored magenta, pink, and blue, respectively. Hydrogen bonding interactions are indicated by dashed lines. E and F, close-up view of hydrogen bonds between SUMO and the IR1 SIM for RanGAP1-SUMO1/UBC9/IR1SIM-IR2II-IR1III–V (E) and RanGAP1-SUMO2/UBC9/IR1SIM-IR2II-IR1III–V (F).
Consistent with SIM contacts to SUMO, previous studies demonstrated that SUMO isoform preference could be swapped in SUMO1 hybrids that contain the SUMO2 α-helix and β-sheet (18). Based on our structures containing SUMO1 and SUMO2, we attempted to pinpoint those SUMO amino acid side chains that are responsible for these differences. Inspection of the surface suggested a notable difference between SUMO1 (Val38) and SUMO2 (Ile34) (supplemental Fig. 3). The absence of isoleucine in SUMO1 creates a deeper hydrophobic pocket that might account for the ability of the IR1 SIM to approach SUMO1 more closely compared with SUMO2, although this hypothesis remains to be tested.
SIM Functions in IR1-M-IR2
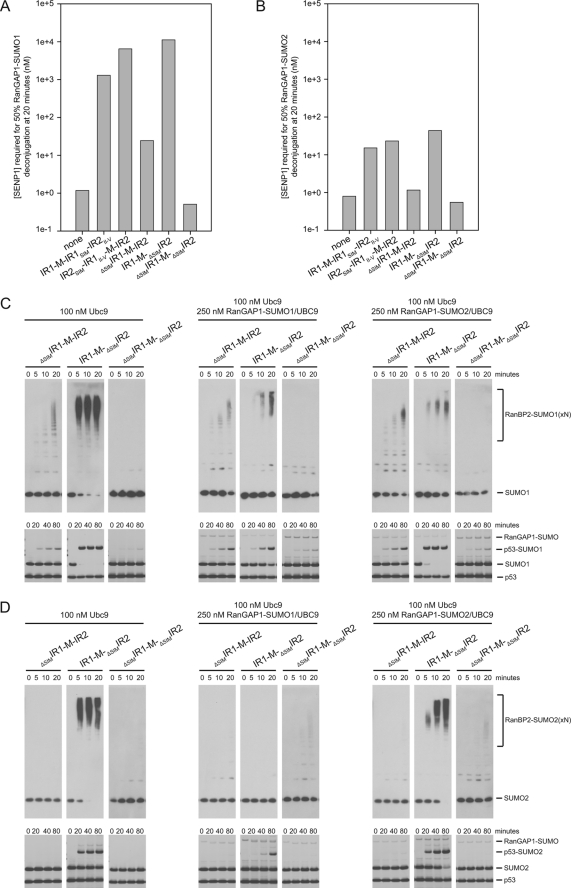
The IR1-M-IR2 SIMs appear to play a vital role in mediating SUMO binding, so we swapped and/or deleted the SIMs to assess their role in protease protection and E3 ligase assays in the context of IR1-M-IR2. Constructs were designed for IR1-M-IR2 to replace the IR2 SIM with the IR1 SIM (IR1-M-IR1SIM-IR2II–V) and to replace the IR1 SIM with the IR2 SIM (IR2SIM-IR1II–V-M-IR2) (Fig. 1B). The activities of these SIM swap constructs correlate well with our results for the individual IR domains. IR1-M-IR2 containing two IR2 SIMs increased protection of RanGAP1-SUMO1 2-fold relative to the wild-type IR1-M-IR2 and 5-fold better than an IR1-M-IR2 construct with two IR1 SIMs (Fig. 7A).
FIGURE 7.
SIM contributions to IR1-M-IR2 activities. A and B, bar graphs indicating concentration of SENP1 required to deconjugate 50% of RanGAP-SUMO in the presence of UBC9 and indicated RanBP2 constructs for RanGAP-SUMO1 (A) and RanGAP-SUMO2 (B) as interpolated using data obtained from three independent experiments (supplemental Figs. 1 and 2). C, time course of automodification with SUMO1 (upper) and p53 modification with SUMO1 (lower) with the indicated IR1-M-IR2 constructs. Reactions were performed in the presence of the specified concentrations of UBC9 and RanGAP-SUMO/UBC9. Automodification was detected by Western blotting with SUMO1 antibody. p53 conjugation was detected by SYPRO staining. D, same as C, except that reactions were performed with SUMO2, and automodification was detected by Western blotting with SUMO2 antibody.
The data presented thus far point to IR1 as the major site for RanGAP1-SUMO1/UBC9 and UBC9∼SUMO1 binding. We tested this by creating IR1-M-IR2 constructs containing SIM deletions. A construct lacking the IR1 SIM (ΔSIMIR1-M-IR2) behaves similarly to IR2 alone and is less able to protect RanGAP1-SUMO1 and RanGAP1-SUMO2 (153-fold and 39-fold, respectively) (Fig. 7, A and B). This suggests that IR1 motifs II–V do not work in trans with the IR2 SIM within ΔSIMIR1-M-IR2. In contrast, IR1-M-ΔSIMIR2 protects RanGAP1-SUMO1 3-fold better than wild-type IR1-M-IR2 and requires a 256-fold higher concentration of SENP1 to deconjugate RanGAP1-SUMO1 relative to RanGAP-SUMO2 (Fig. 7, A and B). Furthermore, each of the constructs containing a SIM followed by IR1 motifs II–V was better able to protect RanGAP1-SUMO1, whereas constructs containing a SIM followed by IR2 motifs II–V were less able to protect RanGAP1-SUMO1. This suggests that IR1 is the site of stable complex binding and that IR2, although still able to interact with SUMO1 via its SIM and perhaps motif II, is less effective at engaging UBC9 as evidenced by its lower E3 activity and inability to protect RanGAP1-SUMO/UBC9 complexes.
It is interesting that IR1-M-ΔSIMIR2 appears slightly better at protecting RanGAP1-SUMO1 compared with IR1-M-IR2 (3-fold). Although speculative, this may be due to RanGAP1-SUMO1/UBC9 sampling two binding sites when both IR1 and IR2 SIMs are present. In IR1-M-IR2, RanGAP1-SUMO1/UBC9 is best protected when in complex with IR1 elements, but less protected from SENP1 when it interacts with the IR2 SIM due to weaker interactions between IR2 motifs II–V and UBC9. In IR1-M-ΔSIMIR2 RanGAP1-SUMO1/UBC9 is more stable because it is funneled to IR1 where it forms a complex that is better protected from SENP1 activities.
The results observed in SENP1 protection assays are consistent with results obtained in automodification assays and p53 conjugation assays for these deletion constructs (Fig. 7, C and D). IR1-M-ΔSIMIR2 behaves similarly to IR1-M-IR2 and is inhibited by excess RanGAP1-SUMO1/UBC9. Conversely, ΔSIMIR1-M-IR2 displayed weaker automodification activity that is not inhibited in the presence of excess RanGAP1-SUMO1/UBC9 (Fig. 7C). This supports the idea that RanGAP1-SUMO1/UBC9 binding at IR1 is responsible for inhibition of IR1-M-IR2 E3 ligase activity. Importantly, the weak E3 activity observed for ΔSIMIR1-M-IR2 is dependent on the IR2 SIM, as a construct lacking both SIMs (ΔSIMIR1-M-ΔSIMIR2) displays no automodification activity, shows p53 conjugation activity similar to reactions performed in the absence of RanBP2, and is unable to protect RanGAP1-SUMO1 or RanGAP1-SUMO2 (Fig. 7 and supplemental Fig. 4).
DISCUSSION
The RanBP2 IR1-M-IR2 domain is a SUMO E3 ligase and is responsible for localization of RanGAP1-SUMO1/UBC9 at the NPC. Our data suggest that elements in both IR1 and IR2 exhibit specificity for SUMO1. Most amino acid sequence differences between IR1 and IR2 lie in motifs III–V, and our results suggest that the inability of IR2 to interact with UBC9 underlies its inability to protect RanGAP1-SUMO or catalyze E3 ligase activity compared with IR1. With that said, IR2 contains a SIM and motif II that better differentiate between SUMO isoforms, as they confer added protection to RanGAP1-SUMO1 and higher specificity for SUMO1 versus SUMO2 in conjugation assays when placed in the context of IR1 motifs III–V.
The structural data presented here suggest that differences in SUMO specificity may be achieved through subtle conformational differences observed for SUMO1 and SUMO2 in the RanGAP1-SUMO/UBC9/IR complexes. In addition, we observed higher complementarity and additional interactions between the IR1 SIM and SUMO1 surfaces compared with the complexes containing SUMO2. Although it is rather unsatisfying that these structures did not reveal definitive elements or amino acid side chains that explain RanBP2 preference for SUMO1, it is consistent with rather subtle differences in specificity observed for other SUMO-specific SIMs. Although the noncanonical SIM of CoREST1 interacts exclusively with SUMO2, the SUMO2-specific SIMs of USP25, MCAF1, and K-bZIP display 4–10 differences in isoform preference (30–33). Similarly, the phospho-SIM of Daxx displays an ∼8-fold preference for SUMO1 over SUMO2/3 (39).
In summary, our data suggest that IR1 functions as both the primary receptor for RanGAP1-SUMO1/UBC9 as well as the primary E3 ligase within IR1-M-IR2. Although the IR1 E3 ligase activity is inhibited by excess RanGAP1-SUMO1/UBC9, we posit that IR1-mediated E3 ligase activity is maintained in the presence of RanGAP1-SUMO1/UBC9 because activated UBC9∼SUMO1 and RanGAP1-SUMO1/UBC9 compete directly for IR1 binding, as they are structural mimetics from the perspective of IR1. In contrast, the weaker SUMO1-specific E3 ligase activity of IR2, as observed in ΔSIMIR1-M-IR2, is not inhibited by excess RanGAP1-SUMO1/UBC9. This raises the possibility that when IR1 is engaged by RanGAP1-SUMO1/UBC9, IR2 may retain some function as a weaker SUMO1 specific E3 ligase. These in vitro data suggest distinct and somewhat overlapping roles for IR1 and IR2 at the NPC; however, the functional consequence of these activities will only be revealed once hybrid constructs can be tested in a cellular model and when an intact RanBP2 complex is resolved that contains both RanGAP1-SUMO1/UBC9 and an activated UBC9∼SUMO thioester.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grant GM065872 (to C. D. L.). Use of the Advanced Photon Source was supported by the US Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract W-31-109-Eng-38. Use of the NE-CAT beamline at Sector 24 is based upon research conducted at the Northeastern Collaborative Access Team beamlines of the Advanced Photon Source, which is supported by Award RR-15301 from the National Center for Research Resources at the National Institute of Health.

This article contains supplemental Figs. 1–4.
- SUMO
- small ubiquitin-like modifier
- BisTris
- bis(2-hydroxyethyl)iminotris(hydroxymethyl)methane
- IR
- internal repeat
- NPC
- nuclear pore complex
- PDB
- Protein Data Bank
- SIM
- SUMO-interacting motif.
REFERENCES
- 1. Johnson E. S. (2004) Protein modification by SUMO. Annu. Rev. Biochem. 73, 355–382 [DOI] [PubMed] [Google Scholar]
- 2. Geiss-Friedlander R., Melchior F. (2007) Concepts in sumoylation: a decade on. Nat. Rev. Mol. Cell Biol. 8, 947–956 [DOI] [PubMed] [Google Scholar]
- 3. Capili A. D., Lima C. D. (2007) Taking it step by step: mechanistic insights from structural studies of ubiquitin/ubiquitin-like protein modification pathways. Curr. Opin. Struct. Biol. 17, 726–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gareau J. R., Lima C. D. (2010) The SUMO pathway: emerging mechanisms that shape specificity, conjugation and recognition. Nat. Rev. Mol. Cell Biol. 11, 861–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Reindle A., Belichenko I., Bylebyl G. R., Chen X. L., Gandhi N., Johnson E. S. (2006) Multiple domains in Siz SUMO ligases contribute to substrate selectivity. J. Cell Sci. 119, 4749–4757 [DOI] [PubMed] [Google Scholar]
- 6. Yunus A. A., Lima C. D. (2009) Structure of the Siz/PIAS SUMO E3 ligase Siz1 and determinants required for SUMO modification of PCNA. Mol. Cell 35, 669–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rytinki M. M., Kaikkonen S., Pehkonen P., Jääskeläinen T., Palvimo J. J. (2009) PIAS proteins: pleiotropic interactors associated with SUMO. Cell Mol. Life Sci. 66, 3029–3041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Saitoh H., Pu R., Cavenagh M., Dasso M. (1997) RanBP2 associates with Ubc9p and a modified form of RanGAP1. Proc. Natl. Acad. Sci. U.S.A. 94, 3736–3741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Saitoh H., Sparrow D. B., Shiomi T., Pu R. T., Nishimoto T., Mohun T. J., Dasso M. (1998) Ubc9p and the conjugation of SUMO-1 to RanGAP1 and RanBP2. Curr. Biol. 8, 121–124 [DOI] [PubMed] [Google Scholar]
- 10. Zhang H., Saitoh H., Matunis M. J. (2002) Enzymes of the SUMO modification pathway localize to filaments of the nuclear pore complex. Mol. Cell. Biol. 22, 6498–6508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tatham M. H., Kim S., Jaffray E., Song J., Chen Y., Hay R. T. (2005) Unique binding interactions among Ubc9, SUMO, and RanBP2 reveal a mechanism for SUMO paralog selection. Nat. Struct. Mol. Biol. 12, 67–74 [DOI] [PubMed] [Google Scholar]
- 12. Pichler A., Gast A., Seeler J. S., Dejean A., Melchior F. (2002) The nucleoporin RanBP2 has SUMO1 E3 ligase activity. Cell 108, 109–120 [DOI] [PubMed] [Google Scholar]
- 13. Pichler A., Knipscheer P., Saitoh H., Sixma T. K., Melchior F. (2004) The RanBP2 SUMO E3 ligase is neither HECT- nor RING-type. Nat. Struct. Mol. Biol. 11, 984–991 [DOI] [PubMed] [Google Scholar]
- 14. Reverter D., Lima C. D. (2005) Insights into E3 ligase activity revealed by a SUMO-RanGAP1-Ubc9-Nup358 complex. Nature 435, 687–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Matunis M. J., Wu J., Blobel G. (1998) SUMO-1 modification and its role in targeting the Ran GTPase-activating protein, RanGAP1, to the nuclear pore complex. J. Cell Biol. 140, 499–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Matunis M. J., Coutavas E., Blobel G. (1996) A novel ubiquitin-like modification modulates the partitioning of the Ran-GTPase-activating protein RanGAP1 between the cytosol and the nuclear pore complex. J. Cell Biol. 135, 1457–1470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mahajan R., Delphin C., Guan T., Gerace L., Melchior F. (1997) A small ubiquitin-related polypeptide involved in targeting RanGAP1 to nuclear pore complex protein RanBP2. Cell 88, 97–107 [DOI] [PubMed] [Google Scholar]
- 18. Zhu S., Goeres J., Sixt K. M., Békés M., Zhang X. D., Salvesen G. S., Matunis M. J. (2009) Protection from isopeptidase-mediated deconjugation regulates paralog-selective sumoylation of RanGAP1. Mol. Cell 33, 570–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Olsen S. K., Capili A. D., Lu X., Tan D. S., Lima C. D. (2010) Active site remodeling accompanies thioester bond formation in the SUMO E1. Nature 463, 906–912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lois L. M., Lima C. D. (2005) Structures of the SUMO E1 provide mechanistic insights into SUMO activation and E2 recruitment to E1. EMBO J. 24, 439–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bernier-Villamor V., Sampson D. A., Matunis M. J., Lima C. D. (2002) Structural basis for E2-mediated SUMO conjugation revealed by a complex between ubiquitin-conjugating enzyme Ubc9 and RanGAP1. Cell 108, 345–356 [DOI] [PubMed] [Google Scholar]
- 22. Reverter D., Lima C. D. (2006) Structural basis for SENP2 protease interactions with SUMO precursors and conjugated substrates. Nat. Struct. Mol. Biol. 13, 1060–1068 [DOI] [PubMed] [Google Scholar]
- 23. Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 24. Collaborative Computational Project, Number 4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 25. Emsley P., Cowtan K. (2004) COOT: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 26. Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 27. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Shen L. N., Dong C., Liu H., Naismith J. H., Hay R. T. (2006) The structure of SENP1-SUMO-2 complex suggests a structural basis for discrimination between SUMO paralogues during processing. Biochem. J. 397, 279–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tatham M. H., Jaffray E., Vaughan O. A., Desterro J. M., Botting C. H., Naismith J. H., Hay R. T. (2001) Polymeric chains of SUMO-2 and SUMO-3 are conjugated to protein substrates by SAE1/SAE2 and Ubc9. J. Biol. Chem. 276, 35368–35374 [DOI] [PubMed] [Google Scholar]
- 30. Meulmeester E., Kunze M., Hsiao H. H., Urlaub H., Melchior F. (2008) Mechanism and consequences for paralog-specific sumoylation of ubiquitin-specific protease 25. Mol. Cell 30, 610–619 [DOI] [PubMed] [Google Scholar]
- 31. Sekiyama N., Ikegami T., Yamane T., Ikeguchi M., Uchimura Y., Baba D., Ariyoshi M., Tochio H., Saitoh H., Shirakawa M. (2008) Structure of the small ubiquitin-like modifier (SUMO)-interacting motif of MBD1-containing chromatin-associated factor 1 bound to SUMO-3. J. Biol. Chem. 283, 35966–35975 [DOI] [PubMed] [Google Scholar]
- 32. Ouyang J., Shi Y., Valin A., Xuan Y., Gill G. (2009) Direct binding of CoREST1 to SUMO-2/3 contributes to gene-specific repression by the LSD1/CoREST1/HDAC complex. Mol. Cell 34, 145–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chang P. C., Izumiya Y., Wu C. Y., Fitzgerald L. D., Campbell M., Ellison T. J., Lam K. S., Luciw P. A., Kung H. J. (2010) Kaposi sarcoma-associated herpesvirus (KSHV) encodes a SUMO E3 ligase that is SIM-dependent and SUMO-2/3-specific. J. Biol. Chem. 285, 5266–5273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wickliffe K. E., Lorenz S., Wemmer D. E., Kuriyan J., Rape M. (2011) The mechanism of linkage-specific ubiquitin chain elongation by a single-subunit E2. Cell 144, 769–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Saha A., Lewis S., Kleiger G., Kuhlman B., Deshaies R. J. (2011) Essential role for ubiquitin-ubiquitin-conjugating enzyme interaction in ubiquitin discharge from Cdc34 to substrate. Mol. Cell 42, 75–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Rodrigo-Brenni M. C., Foster S. A., Morgan D. O. (2010) Catalysis of lysine 48-specific ubiquitin chain assembly by residues in E2 and ubiquitin. Mol. Cell 39, 548–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yunus A. A., Lima C. D. (2006) Lysine activation and functional analysis of E2-mediated conjugation in the SUMO pathway. Nat. Struct. Mol. Biol. 13, 491–499 [DOI] [PubMed] [Google Scholar]
- 38. DeLano W. L. (2010) The PyMOL Molecular Graphics System, version 1.3r1, Schrödinger, LLC, New York [Google Scholar]
- 39. Chang C. C., Naik M. T., Huang Y. S., Jeng J. C., Liao P. H., Kuo H. Y., Ho C. C., Hsieh Y. L., Lin C. H., Huang N. J., Naik N. M., Kung C. C., Lin S. Y., Chen R. H., Chang K. S., Huang T. H., Shih H. M. (2011) Structural and functional roles of Daxx SIM phosphorylation in SUMO paralog-selective binding and apoptosis modulation. Mol. Cell 42, 62–74 [DOI] [PubMed] [Google Scholar]
- 40. Chen V. B., Arendall W. B., 3rd, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., Richardson D. C. (2010) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.