Background: miR-21 expression is regulated by BMP4 and plays a critical role in vSMC phenotype regulation.
Results: Affinity purification of mRNAs associated with miR-21 yielded nearly all members of the DOCK superfamily.
Conclusion: miR-21 targets multiple members of the DOCK superfamily and modulates the activity of Rac1 small GTPase to regulate vSMC phenotype.
Significance: This study identified novel targets of miR-21 using a biochemical method.
Keywords: Bone Morphogenetic Protein (BMP), Cell Migration, Cell Signaling, MicroRNA, Vascular Smooth Muscle Cells
Abstract
The bone morphogenetic protein 4 (BMP4) signaling pathway plays a critical role in the promotion and maintenance of the contractile phenotype in vascular smooth muscle cell (vSMC). Misexpression or inactivating mutations of the BMP receptor gene can lead to dedifferentiation of vSMC characterized by increased migration and proliferation that is linked to vascular proliferative disorders. Previously we demonstrated that vSMCs increase microRNA-21 (miR-21) biogenesis upon BMP4 treatment, which induces contractile gene expression by targeting programmed cell death 4 (PDCD4). To identify novel targets of miR-21 that are critical for induction of the contractile phenotype by BMP4, biotinylated miR-21 was expressed in vSMCs followed by an affinity purification of mRNAs associated with miR-21. Nearly all members of the dedicator of cytokinesis (DOCK) 180-related protein superfamily were identified as targets of miR-21. Down-regulation of DOCK4, -5, and -7 by miR-21 inhibited cell migration and promoted cytoskeletal organization by modulating an activity of small GTPase. Thus, this study uncovers a regulatory mechanism of the vSMC phenotype by the BMP4-miR-21 axis through DOCK family proteins.
Introduction
MicroRNAs (miRNAs)2 regulate basic cellular activities, such as differentiation, proliferation, and development, by suppressing gene expression post-transcriptionally (1, 2). In mammals, the active strand miRNA sequence of ∼22 nucleotides recognizes partially complementary miRNA recognition element (MRE) or seed-matched sequences located mostly in the 3′-UTR of mRNAs (3, 4). Seed-matched sites are defined as Watson-Crick consecutive base pairing between mRNAs and the miRNA positioned at 2–7 nucleotides from the 5′-end. Gene suppression by miRNAs is thought to occur either by inhibiting translation, mediating mRNA destabilization, or a combination of both. Aberrant miRNA expression is associated with various developmental abnormalities and human diseases (5, 6). miR-21 is involved in tumorigenesis and cardiovascular function (7–12). Elevated expression of miR-21 is observed in various types of tumors, and suppression of miR-21 in tumors leads to reduced proliferation and migration, suggesting that miR-21 acts as an oncogene (13–22). Higher levels of miR-21 are observed after multiple cardiac stresses, including acute pressure overload by transaortic constriction, chronic calcineurin activation, infusion of angiotensin II, and myocardial infarction (10, 23, 24). Cardiac hypertrophy and fibrosis in response to pressure overload after transaortic constriction are prevented by knockdown of miR-21 using antisense RNA oligonucleotides against miR-21 (11), suggesting a critical role for miR-21 in cardiac hypertrophy and fibrosis.
One of the effects of BMP and transforming growth factor-β (TGF-β) signaling is to promote the contractile phenotype in vSMC. We identified miR-21 as a critical downstream mediator of this effect (8). Upon TGF-β or BMP treatment, the R-Smad proteins, which are signal transducers of TGF-β and BMPs, associate with primary transcripts of miR-21 (pri-miR-21) in complex with Drosha to promote pri-miR-21 to precursor miR-21 processing and elevated miR-21 expression (8, 25). By inducing miR-21, both BMP4 and TGF-β lead to knockdown of a negative regulator of contractile gene expression, programmed cell death 4 (PDCD4) (8). We have demonstrated previously that platelet-derived growth factor-BB (PDGF-BB), a potent inducer of the synthetic phenotype in vSMCs, induces miR-221, which in turn promotes (i) down-regulation of contractile genes and (ii) cell proliferation through down-regulation of two target genes, c-kit and p27Kip1, respectively (26). Although down-regulation of PDCD4 by miR-21 is critical for the regulation of vSMC contractile gene expression, it remained unclear whether other effects of TGF-β or BMP on vSMC are also mediated by miR-21 and which miR-21 targets are critical mediators of such activities.
There are several approaches to identify targets of miRNAs. The most common uses bioinformatics algorithms to predict potential targets based on the existence of partial complementary MREs in the 3′-UTR. Another approach is to identify mRNAs that are down-regulated when a miRNA is overexpressed (8, 27, 28). miRNA targets can also be enriched by co-immunoprecipitation with exogenously expressed tagged Argonaute (Ago) or GW182 proteins in cells overexpressing miRNA (29, 30). Some studies used stable isotope labeling with amino acids in culture to identify proteins that are down-regulated upon overexpression of specific miRNAs (31, 32).
In this study, we expressed biotinylated miR-21 in vSMCs and isolated mRNAs that are associated with miR-21 using streptavidin beads followed by microarray analysis for identification of targets. Multiple members of the DOCK family, which are guanine nucleotide exchange factors for Rac and/or Cdc42 (33), were found to be associated with miR-21 in cells. DOCK family genes are not predicted as targets of miR-21 by conventional target prediction algorithms, and little is known about their function in vSMC. Identification of 3′-UTR sequences, which are critical for recognition by miR-21, revealed that miR-21 regulates DOCK genes by base pairing at sites other than the seed sequence. We also demonstrate a function for DOCK family members in the regulation of vSMC phenotype by BMP4.
EXPERIMENTAL PROCEDURES
Cell Culture
Human primary pulmonary artery smooth muscle cells (PASMCs) were purchased from Lonza (CC-2581) and maintained in Sm-GM2 medium (Lonza) containing 5% fetal bovine serum (FBS). COS7 and rat pulmonary artery smooth muscle PAC1 cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% FBS. Cells were cultured at 37 °C in the presence of 5% CO2.
Isolation of miR-21 Targets
3′-Biotinylated miR-21 mimic (bio-miR-21) and 3′-biotinylated control Caenorhabditis elegans miR-67 mimic (bio-cel-miR-67) were synthesized by Dharmacon. PASMCs were transfected with bio-miR-21 or bio-cel-miR-67 mimic at a final concentration of 30 nm using RNAiMax (Invitrogen). Twenty-four hours later, the cells were trypsinized and lysed in TNE buffer (20 mm Tris (pH 7.5), 100 mm KCl, 5 mm MgCl2, 0.3% Nonidet P-40, RNase OUT (Invitrogen), protease inhibitor mixture (Roche Applied Science)). The streptavidin-coated magnetic beads (Invitrogen) were incubated with cell lysates at 4 °C for 4 h followed by washing with a washing buffer (same as TNE buffer except containing 200 mm KCl). Total mRNAs bound to the streptavidin beads (pulldown RNA) were isolated using TRIzol LS (Invitrogen) according to the manufacturer's instructions. For microarray analysis, isolated mRNAs were subjected to Affymetrix human U133plus 2.0 array at the Clinical Microarray Core, University of California, Los Angeles. Fold enrichment of mRNAs by miR-21 over control cel-miR-67 was calculated.
Quantitative Reverse Transcription-PCR (qRT-PCR)
Total RNAs were isolated using TRIzol (Invitrogen) and reverse transcribed using a first strand cDNA synthesis kit (Invitrogen) according to the manufacturer's instructions. qRT-PCR analysis was performed in triplicates using the SYBR Green Master Mix (Applied Biosystems). The mRNA level was normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH). For a quantitation of miRNAs, a TaqMan microRNA assay kit (Applied Biosystems) was used according to the manufacturer's instructions, and results were normalized to U6 snRNA. Primers for qRT-PCR analysis are listed in the supplemental information.
Luciferase Reporter Constructs
The full-length 3′-UTR sequences were cloned by RT-PCR from mRNAs isolated from PASMCs. Predicted MRE sequences were cloned into the pIS0 vector (Addgene) containing the luciferase gene. Primers for cloning are listed in the supplemental information.
Luciferase Assay
COS7 cells were transfected with luciferase reporter constructs using FuGENE 6 (Roche Applied Science) and a β-galactosidase (β-gal) expression plasmid as an internal transfection control. Twenty-four hours later, cells were transfected using RNAiMax (Invitrogen) with 5 nm miR-21 mimic or control miRNA mimic. Luciferase assays were carried out, and luciferase activities were normalized to β-gal activities.
RNA Interference
Small interfering RNA (siRNA) duplexes were synthesized by Thermo Scientific. The target nucleotide sequences for DOCK4 siRNA (5′-GUGUGAUGGCUGGUACAGAUU-3′), DOCK5 siRNA (5′-AUCCAUUGCUAUAGAAGAAUU-3′), and DOCK7 siRNA (5′-AAGACGUUCAAUGUCAAUAGAUU-3′) were used. Negative control siRNA (Qiagen) was used as a control.
In Vitro Migration Assay
PASMCs transfected with miR-21 mimic, antisense oligonucleotides against miR-21 (anti-miR-21), or siRNAs were plated in 6-well plates, and three scratch wounds were generated with a 200-μl disposable pipette tip. Scratch wounds were photographed over 9 h with a Nikon inverted microscope with an attached digital camera, and their widths were quantitated with ImageJ software.
DOCK7 Expression Plasmid
DOCK7 transcript with deleted 3′-UTR (DOCK7wo3′-UTR) was cloned into pcDNA3.1(+) by RT-PCR from mRNAs isolated from PASMCs. For PCR amplification, primers 5′-ATGGCTAGCATGGCCGAGCGCCGCGCC-3′ and 5′-CGTTCTAGATTAGAGATCCATTTTGCG-3′ were used.
Collagen Matrix Contraction Assay
A collagen matrix contraction assay was performed as described (34). PASMCs transfected with miR-21 mimic, control miRNA mimic, anti-miR-21, or siRNAs were embedded in attached collagen matrices followed by 3 nm BMP4 treatment for 24 h. Twenty-four hours after detachment of the gel from the plate, the gel surface area was quantitated with the ImageJ program.
Detection of Active Rac1
To detect active GTP-bound Rac1, cell lysates of PASMCs transfected with control miRNA mimic or miR-21 mimic were incubated with 20 μg of GST-Pak1-PBD (Thermo Scientific), which binds specifically to the GTP-bound form of Rac1, in the presence of glutathione-agarose beads. To measure the total amount of Rac1, Western blot was also performed with anti-Rac1 (Thermo Scientific). The band intensity in the Western blot was quantified, and the levels of Rac1-GTP were normalized to the amount of total Rac1.
Immunoblotting
Cells were lysed in TNE buffer, and total cell lysates were separated by SDS-PAGE, transferred to PVDF membranes (Millipore), immunoblotted with antibodies, and visualized using an enhanced chemiluminescence detection system (Amersham Biosciences). Antibodies used for immunoblotting were: anti-GAPDH antibody (2E3-2E10, Abnova) and anti-human DOCK7 antibody (35). Protein bands were quantitated with the imaging analysis software ImageJ.
Statistical Analysis
The results presented are an average of at least three experiments, each performed in triplicate with standard errors. Statistical analyses were performed by analysis of variance followed by Tukey's multiple comparison test or by Student's t test as appropriate using Prism 4 (GraphPad Software Inc.). p values of <0.05 were considered significant and are indicated with asterisks.
RESULTS
Identification of Target mRNAs for miR-21
To further understand the physiological role of miR-21 in vSMCs, targets of miR-21 were isolated using a modified method developed previously (36, 37). PASMCs were transfected with bio-miR-21 or bio-cel-miR-67 mimic as a negative control. mRNAs associated with bio-miRNA mimic were isolated following affinity purification using streptavidin beads. mRNA targets were identified by comparing isolated pulldown RNAs from cells transfected with bio-miR-21 mimic with those of cells transfected with control mimic by human gene microarray analysis. There are currently 33 genes that are validated as targets of miR-21 according to the miRecords web site. 30 of 33 validated miR-21 targets were enriched more than 2-fold by miR-21 pulldown (supplemental Table S1). None of the housekeeping gene transcripts were enriched (supplemental Table S2). Among previously validated miR-21 targets, E2F1, GLCCI1, and SLC16A10 were not enriched significantly in PASMCs (supplemental Table S1). However, GLCCI1 cannot be found on the microarray, and SLC16A10 is not expressed in PASMCs. Thus, the bio-miR-21 pulldown technique was able to detect all validated targets except one and has a potential to identify novel targets.
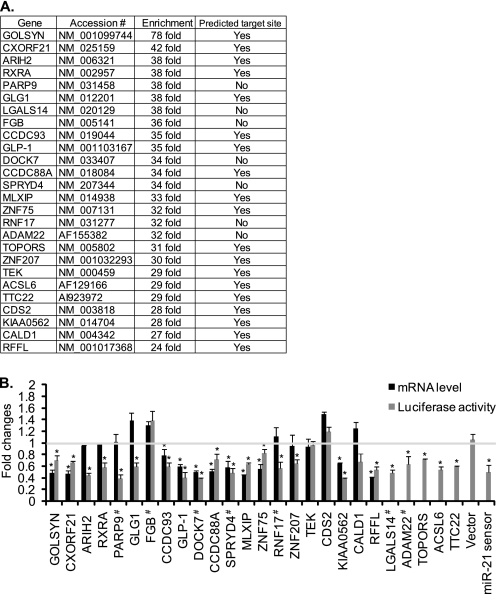
Of >9000 gene transcripts that were enriched by ≥2-fold over control in bio-miR-21 pulldown samples, we examined 26 genes that were not identified previously as miR-21 targets and had the highest enrichment in bio-miR-21 pulldown samples (≥24-fold over bio-cel-miR-67 pulldown samples) (Fig. 1A). To investigate whether expression of these genes is down-regulated by miR-21, the full-length 3′-UTRs or predicted MREs of all 26 genes was inserted at the 3′-end of the luciferase gene, and luciferase reporter activity was measured upon overexpression of miR-21 or control cel-miR-67 mimic. As miR-21 MRE or seed sequence is not predicted by target prediction algorithms in the 3′-UTR of transcripts encoding PARP9, LGALS14, FGB, DOCK7, SPRYD4, RNF17, and ADAM22, full-length 3′-UTRs were examined. For the rest of the genes, predicted miR-21 MREs or seed regions were examined. As a positive control, a miR-21 sensor luciferase construct, which contains two copies of a sequence complementary to miR-21, was used. The miR-21 sensor activity was reduced to 47 ± 6% when miR-21 mimic was expressed (Fig. 1B). Luciferase reporter activities of 22 genes were significantly reduced, suggesting that ∼85% (22 of 26) of miR-21 pulldown mRNAs, which are enriched ≥24-fold over control, are miR-21 targets (Fig. 1B, gray bars). Among these 22 genes, qRT-PCR analysis demonstrated that mRNAs for 11 genes (GOLSYN, CXORF21, CCDC93, GLP-1, DOCK7, CCDC88A, SPRYD4, MLXIP, ZNF75, KIAA0562, and RFFL) were significantly reduced by miR-21, indicating that these transcripts undergo destabilization of mRNA by miR-21 (Fig. 1B, black bars). Levels of expression of four genes (FGB, TEK, CDS2, and CALD1) were unchanged both by luciferase assay and at the mRNA level (Fig. 1B). Thus, using this method, we identified 22 novel targets of miR-21 in PASMCs (Fig. 1B).
FIGURE 1.
Genes enriched by miR-21 pulldown. A, the 26 most highly enriched genes by bio-miR-21 pulldown in PASMCs. The presence of predicted target sites for miR-21 was identified by using PITA and RNA22. Accession indicates NCBI Reference Sequence accession numbers. B, overexpression of miR-21 down-regulates genes that are highly enriched by bio-miR-21 pulldown. Total RNAs were harvested from PASMCs transfected with control or miR-21 mimic, and mRNA levels of the indicated genes relative to GAPDH were measured by qRT-PCR. Fold change of mRNA levels of each gene in cells overexpressing miR-21 mimic in comparison with control (cel-miR-67) mimic-transfected cells is presented (black bars). COS7 cells transfected with luciferase construct containing full-length 3′-UTR or MRE of the indicated genes were transfected with control (cel-miR-67) or miR-21 mimic. # indicates the luciferase reporter constructs containing a full-length 3′-UTR, whereas the rest of the constructs contain only predicted miR-21 MREs. A luciferase vector carrying two copies of a sequence complementary to miR-21 (miR-21 sensor) and a luciferase vector without the 3′-UTR sequence (Vector) were used as positive and negative controls, respectively. Luciferase activity was then measured in triplicates. Fold changes by miR-21 mimic transfection in comparison with cel-miR-67 mimic transfection are presented (gray bars). Data represent mean ± S.E. *, p < 0.01.
miR-21 Regulates Expression of DOCK Family Proteins
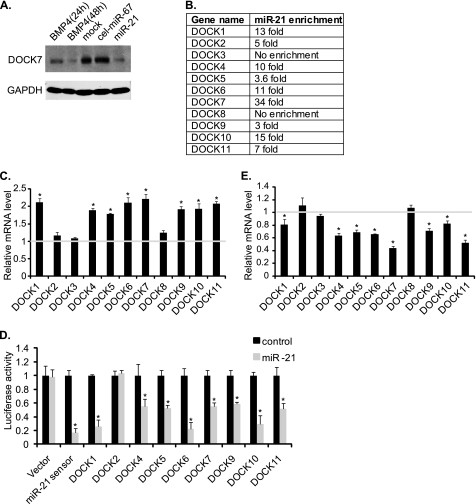
One of the newly identified miR-21 targets is DOCK7, a member of the DOCK family of guanine nucleotide exchange factors for the Rho family of GTPases (Fig. 1A). Immunoblot analysis indicated that the endogenous DOCK7 protein in PASMCs was down-regulated upon miR-21 overexpression in comparison with control cel-miR-67 (Fig. 2A). Furthermore, cells stimulated with BMP4 for 48 h exhibited reduced DOCK7 levels, similar to what was seen in miR-21-transfected cells, confirming that miR-21 leads to down-regulation of DOCK7 in PASMCs (Fig. 2A). Other members of the DOCK family genes, DOCK1, -2, -4, -5, -6, -9, -10, and -11, were also significantly enriched in miR-21 pulldowns (Fig. 2B). To determine whether miR-21 also targets other members of the DOCK family and mediates destabilization of their mRNAs similarly to DOCK7 (Fig. 1B), qRT-PCR analysis was performed upon overexpression of anti-miR-21, which inhibits miR-21 activity in PASMCs (Fig. 2C). Significant up-regulation of the mRNA levels of DOCK1, -4, -5, -6, -7, -9, -10, and -11 was observed after anti-miR-21 transfection, suggesting that these DOCK family members are down-regulated by endogenous miR-21 (Fig. 2C). To confirm that miR-21 targets members of the DOCK family, we measured the expression of constructs containing the 3′-UTR of DOCK1, -2, -4, -5, -6, -7, -9, -10, or -11 downstream of the luciferase reporter gene in the presence of miR-21 mimic (Fig. 2D). Consistent with the results of anti-miR-21 in Fig. 2C, the luciferase activity of the DOCK1, -4, -5, -6, -7, -9, -10, and -11 3′-UTR constructs were significantly reduced upon expression of miR-21 mimic, demonstrating that, in addition to DOCK7, DOCK1, -4, -5, -6, -9, -10, and -11 are targets of miR-21 (Fig. 2D). Although DOCK2 transcripts were enriched in miR-21 pulldowns 5-fold over cel-miR-67 (Fig. 2B), the luciferase assay indicated that DOCK2 is not a target of miR-21 (Fig. 2D). Finally, treatment of PASMCs with BMP4, which is known to induce miR-21 by ∼2-fold (8), resulted in decreases in DOCK1, -4, -5, -6, -7, -9, -10, and -11 transcripts, further confirming that these members of the DOCK family are regulated by the BMP4-miR-21 axis (Fig. 2E and supplemental Fig. S4).
FIGURE 2.
miR-21 regulates multiple members of DOCK family proteins. A, PASMCs were treated with 3 nm BMP4 (24 or 48 h) or transfected with control mimic (cel-miR-67) or miR-21 mimic. Total cell lysates were subjected to immunoblotting with anti-DOCK7 or anti-GAPDH (loading control) antibody. B, all members of the DOCK family except DOCK3 and DOCK8 were enriched 3–34-fold upon bio-miR-21 pulldown in comparison with control (bio-cel-miR-67) pulldown. C, total RNA was harvested from PASMCs transfected with control or anti-miR-21 oligonucleotides, and mRNA levels of the indicated DOCK genes relative to GAPDH were measured by qRT-PCR. Data represent fold changes of the mRNA levels in anti-miR-21-transfected cells in comparison with control oligonucleotide-transfected cells ±S.E. from triplicates. *, p < 0.01. D, COS7 cells were transfected with a luciferase reporter containing 3′-UTR of DOCK1, -2, -4, -5, -6, -7, -9, -10, or -11 gene transcript and cel-miR-67 (control) or miR-21 mimic. Relative luciferase activities were examined. The miR-21 sensor and an empty luciferase vector (Vector) were used as controls. Data represent fold changes in the luciferase activity in miR-21-mimic-transfected cells compared with control-mimic-transfected cells ±S.E. from triplicates. *, p < 0.01. E, PASMCs were treated with 3 nm BMP4 for 24 h. mRNA levels of the indicated DOCK genes relative to GAPDH were measured by qRT-PCR. Data represent fold changes of the mRNA level in BMP4-treated cells compared with mock-treated cells ±S.E. from triplicates. *, p < 0.01.
Identification of miR-21 Recognition Sequence in 3′-UTR of DOCK4, -5, and -7 Transcripts
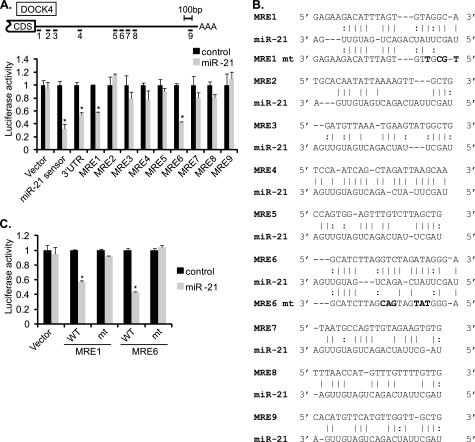
DOCK4, -5, and -7 are known as guanine nucleotide exchange factors for Rac1 GTPases and play a role in cell migration and cytoskeletal organization in neuronal and tumor cells (33, 38). DOCK4, -5, and -7 were not predicted as miR-21 targets by the algorithms whose prediction is based on a seed-matched sequence and cross-species conservation of the seed sequence. RNA22, which predicts targets based on a pattern-based method (39), found nine potential MREs in the 3′-UTR of DOCK4 that are partially complementary to the miR-21 sequence (Fig. 3, A and B). To determine whether any of these predicted MREs are responsible for knockdown by miR-21, luciferase reporters containing individual MREs were transfected into COS7 cells together with miR-21 or control cel-miR-67 mimic. miR-21 significantly reduced the luciferase activity of MRE1 (43 ± 1%) and MRE6 (58 ± 1%) (Fig. 3A). Mutations in MRE1 or MRE6 (Fig. 3B, mt), which disrupted critical base pairing with miR-21, abrogated the inhibition of luciferase activity by miR-21 (Fig. 3C), suggesting that MRE1 and MRE6 are critical target sites for recognition of DOCK4 mRNAs by miR-21.
FIGURE 3.
DOCK4 is direct target of miR-21. A, schematic diagram of predicted miR-21 MREs in the 3′-UTR of DOCK4 transcripts (top panel). CDS and AAA stand for protein coding sequence and poly(A) tail, respectively. Luciferase activity was examined in COS7 cells using the luciferase vector containing each of nine MREs or the full-length 3′-UTR of DOCK4 mRNAs. Luciferase activity of each construct upon overexpression of miR-21 mimic relative to control mimic is shown (bottom panel). Data represent mean ± S.E. *, p < 0.001. B, sequences of miR-21 MREs predicted by the RNA22 program in the 3′-UTR of DOCK4. Perfect base matches are indicated by a vertical line; G:U pairs are indicated by a colon (:). Mutations introduced in MRE1 or MRE6 to disrupt a base paring with the miR-21 sequence are indicated in bold letters. C, luciferase activities of constructs with wild type or mutated DOCK4 MRE1 (MRE1 mt) or MRE6 (MRE6 mt) were examined in COS7 cells by transfecting cel-miR-67 (control) or miR-21 mimic. Data represent mean ± S.E. *, p < 0.0001.
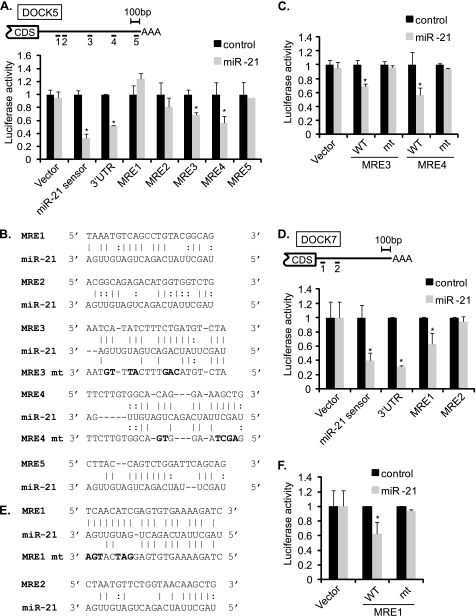
There are five potential miR-21 MREs in the DOCK5 3′-UTR based on prediction by RNA22 (Fig. 4, A and B). miR-21 significantly reduced the luciferase activity of MRE3 (32 ± 4%) and MRE4 (44 ± 10%) (Fig. 4A). Mutations in MRE3 or MRE4 abolished the regulation by miR-21 (Fig. 4B, mt), indicating an essential role of these two MREs in the regulation of DOCK5 mRNA by miR-21 (Fig. 4C). Although DOCK7 does not contain a seed match or predicted MRE in the 3′-UTR, it contains two sequence elements that are highly complementary to miR-21; MRE1 and MRE2 (Fig. 4, D and E). Luciferase activity of the construct containing MRE1, but not MRE2, was significantly reduced by miR-21 (38 ± 2%) (Fig. 4D). Mutations in MRE1 (Fig. 4E, MRE1 mt) blocked down-regulation by miR-21, suggesting a critical role of MRE1 for recognition of DOCK7 by miR-21 (Fig. 4F). Thus, we identified sequence element(s) critical for recognition by miR-21 in all three DOCK gene transcripts.
FIGURE 4.
DOCK5 and DOCK7 are miR-21 targets. A, schematic diagram of predicted miR-21 MREs in the 3′-UTR of DOCK5 transcripts (top panel). CDS and AAA stand for protein coding sequence and poly(A) tail, respectively. Luciferase activity was examined in COS7 cells using the luciferase vector containing each of five MREs or the full-length 3′-UTR of DOCK5 mRNAs. miR-21 sensor and an empty luciferase vector (Vector) were examined as positive and negative controls, respectively. Luciferase activity of each construct upon overexpression of miR-21 mimic relative to control mimic is shown (bottom panel). Data represent mean ± S.E. *, p < 0.001. B, sequences of miR-21 MREs predicted by the RNA22 program in the 3′-UTR of DOCK5. Perfect base matches are indicated by a vertical line; G:U pairs are indicated by a colon (:). Mutations introduced in the MRE3 or MRE4 to disrupt a base paring with miR-21 sequence are indicated in bold letters. C, luciferase activities of constructs with wild type or mutated DOCK5 MRE3 (MRE3 mt) or MRE4 (MRE4 mt) were examined in COS7 cells by transfecting cel-miR-67 or miR-21 mimic. Data represent mean ± S.E. *, p < 0.001. D, schematic diagram of potential miR-21 MREs in the 3′-UTR of DOCK7 transcripts (top panel). Luciferase activity was examined in COS7 cells using the luciferase vector containing each of two MREs or the full-length 3′-UTR of DOCK7 mRNAs. miR-21 sensor and an empty luciferase vector (Vector) were examined as controls, respectively. Luciferase activity of each construct upon overexpression of miR-21 mimic relative to control mimic is shown (bottom panel). Data represent mean ± S.E. *, p < 0.01. E, sequences of miR-21 MREs in the 3′-UTR of DOCK7. Perfect base matches are indicated by a vertical line; G:U pairs are indicated by a colon (:). Mutations introduced in the MRE1 to disrupt a base paring with the miR-21 sequence are indicated in bold letters (MRE1 mt). F, luciferase activities of constructs with wild type or mutated DOCK7 MRE1 were examined in COS7 cells by transfecting cel-miR-67 or miR-21 mimic. *, p < 0.01.
Down-regulation of DOCKs by miR-21 Inhibits Rac1 Activity and Functions
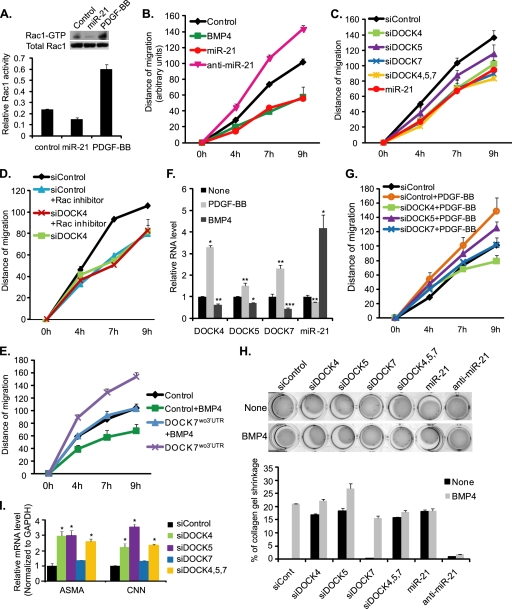
As DOCK4, -5 and -7 are linked to the activation of the Rac1 GTPase (33, 38), we hypothesized that miR-21 might modulate vSMC migration and cytoskeletal organization by inhibiting Rac1 activity. To examine whether miR-21 modulates Rac1 GDP/GTP exchange activity in vSMCs, the amount of active Rac1 was measured by pulldown assays using the GST-Pak1-PBD domain fusion protein, which specifically binds active Rac1. Treatment with PDGF-BB, which is known to activate Rac1, was included as a positive control and triggered a ∼3-fold increase in active Rac1 (Fig. 5A). miR-21 reduced the level of active Rac1 by ∼50% compared with control cel-miR-67-transfected cells without affecting the amount of total Rac1 (Fig. 5A), suggesting that elevation of miR-21 decreases active GTP-bound Rac1. Next, we examined a role for miR-21 in cell migration by performing a scratch wound assay (34). To this end, a wound was created in a monolayer of PASMCs, and images were captured at the beginning and at regular intervals during cell migration to close the wound. Similarly to the cells treated with BMP4 (Fig. 5B, green line), the migration distance was reduced to 47 ± 3% 9 h after a scratch wound was generated in miR-21-transfected cells (Fig. 5B, red line) compared with control miRNA-transfected cells (Fig. 5B, black line). In contrast with the miR-21 mimic, transfection of anti-miR-21 increased cell migration (Fig. 5B, magenta line), indicating that a differential level of expression of miR-21 determines vSMC migration. When DOCK4, -5, and -7 were down-regulated individually or simultaneously by siRNAs by 54 ± 3, 59 ± 15, and 64 ± 4%, respectively (supplemental Fig. S1A), cell migration was significantly reduced comparably with that of miR-21-overexpressing cells (Fig. 5C), suggesting that DOCK4, -5, and -7 regulate vSMC motility. This result also indicates a redundant function among DOCK4, -5, and -7 for the regulation of cell migration. When PASMCs were treated with NSC23766, an inhibitor of Rac1 GDP/GTP exchange activity (38), cell migration was reduced similarly to that in DOCK4, -5, and -7 knockdown cells (Fig. 5D). Furthermore, exogenous expression of DOCK7 mRNAs (DOCK7wo3′-UTR), which have the 3′-UTR deleted and are resistant to miR-21, in rat pulmonary artery smooth muscle PAC1 cells blocked BMP4-induced inhibition of migration (Fig. 5E, green square versus blue triangle). Thus, regulation of DOCK4, -5, and -7 levels is critical for the migratory function in vSMCs. Altogether, these results indicate that the GDP/GTP exchange activity of Rac1 is critical for vSMC migration, and the BMP4-miR-21 axis suppresses vSMC migration by down-regulation of DOCK proteins.
FIGURE 5.
miR-21-mediated inhibition of DOCKs modulates vSMC motility. A, PASMCs were transfected with control cel-miR-67 mimic or miR-21 mimic. Active GTP-bound Rac1 was pulled down by GST-Pak1-PBD fusion protein and visualized by immunoblotting with anti-Rac1 antibody (top panel, Rac1-GTP). Immunoblotting of total cell lysates with anti-Rac1 antibody is shown to demonstrate total amount of Rac1 (top panel, Total Rac1). The level of Rac1-GTP relative to total Rac1 is presented as a relative Rac1 activity (bottom panel). B, PASMCs transfected with control cel-miR-67 mimic (Control; black diamonds), miR-21 mimic (miR-21; red circles), or anti-miR-21 (magenta triangles) were subjected to the scratch wound assay. As a control, PASMCs were treated with 3 nm BMP4 (green squares). The distance of migration was measured using ImageJ over 9 h after a scratch wound was introduced. C, PASMCs transfected with 25 nm non-targeting control siRNA (siControl); siRNA against DOCK4 (siDOCK4), DOCK5 (siDOCK5), or DOCK7 (siDOCK7); or miR-21 mimic (miR-21) were subjected to the scratch wound assay. D, PASMCs transfected with siControl or siDOCK4 were subjected to the scratch wound migration assay in the presence or absence of 50 μm Rac1 inhibitor (NSC23766). E, rat PASMC PAC1 cells were transfected with control vector (Control) or DOCK7 expression construct carrying the DOCK7 cDNA deleted in the 3′-UTR (DOCK7wo3′-UTR) followed by the scratch wound assay in the presence or absence of 3 nm BMP4. F, levels of DOCK4, -5, or -7 mRNA relative to GAPDH or miR-21 relative to U6 snRNA were measured by qRT-PCR in PASMCs stimulated with 20 ng/ml PDGF-BB or 3 nm BMP4 for 24 h. Data represent mean ± S.E. *, p < 0.0001; **, p < 0.01; ***, p < 0.001. G, PASMCs transfected with siRNA (siControl, siDOCK4, siDOCK5, or siDOCK7) were subjected to the scratch wound assay in the presence or absence of 20 ng/ml PDGF-BB. Data represent mean ± S.E. H, PASMCs were transfected with siControl, siDOCK4, siDOCK5, siDOCK7, miR-21 mimic, or anti-miR-21 followed by stimulation with 3 nm BMP4 or no stimulation (None) for 24 h. The PASMC embedded collagen lattices were photographed by using a digital camera to measure gel contraction. The relative size of collagen gel was quantitated using ImageJ. Data represent the mean percentage of the collagen gel shrinkage compared with unstimulated control. I, levels of contractile genes. α-Smooth muscle actin (ASMA) and calponin1 (CNN) mRNA levels relative to GAPDH were measured by qRT-PCR in PASMCs transfected with siControl (siCont), siDOCK4, siDOCK5, or siDOCK7. Data represent mean ± S.E. *, p < 0.01.
As PDGF-BB antagonizes BMP4 activity and promotes vSMC migration (34), we hypothesized that activation of Rac1 by PDGF-BB in PASMCs (see Fig. 5A) might be in part due to down-regulation of miR-21, leading to increased expression of DOCK. qRT-PCR analysis demonstrated that miR-21 expression was reduced by 50% at 9 h (supplemental Fig. S3) and 26% at 24 h of PDGF-BB treatment and that DOCK4, -5, and -7 mRNAs were induced by ∼1.5–3-fold by PDGF-BB (Fig. 5F). Furthermore, knockdown of DOCK4, -5, or -7 greatly reduced PDGF-BB-mediated cell migration (Fig. 5G), suggesting that regulation of DOCK4, -5, and -7 levels is critical for the promigratory function of PDGF-BB in vSMCs.
Next, in vitro collagen gel contraction assays were performed in PASMCs transfected with miR-21 mimic, anti-miR-21, or siDOCKs to evaluate the effect of miR-21-mediated down-regulation of DOCKs in the cytoskeletal organization of PASMCs. Similarly to BMP4-treated cells, transfection of miR-21 mimic reduced gel size by ∼20% compared with control samples (Fig. 5H). Overexpression of miR-21 mimicked the effect of BMP4 and induced contraction, whereas transfection of anti-miR-21 abolished BMP4-induced contraction (Fig. 5H), indicating a critical role of miR-21 expression in contraction of the collagen gel. Down-regulation of DOCK4 or DOCK5 by siRNA also reduced the size of collagen lattice by 18 ± 3 and 20 ± 4%, respectively (Fig. 5H), which is similar to that of miR-21-transfected cells, suggesting that down-regulation of DOCK4 or DOCK5 by miR-21 is sufficient to mediate vSMC contraction. Knockdown of DOCK7 showed no effect on contraction (Fig. 5H). Consistently, knockdown of DOCK4 or DOCK5 elevated the levels of contractile genes (ASMA and CNN), whereas siDOCK7 had no effect (Fig. 5I), suggesting a specific role of DOCK4 and -5 in the regulation of contractile gene expression and contractility. Treatment of cells with the Rac1 inhibitor NSC23766 did not affect the contraction of the collagen lattice (supplemental Fig. S2). Thus, DOCK4 and DOCK5 promote cytoskeletal remodeling in a Rac1-independent manner. These results indicate that miR-21 modulates cell migration and contractility through down-regulation of DOCK family members in vSMCs. Therefore, the miR-21-DOCK axis regulates critical characteristics of the vSMC phenotype, such as motility and cytoskeletal organization, in response to BMP4 and PDGF signaling (Fig. 6).
FIGURE 6.
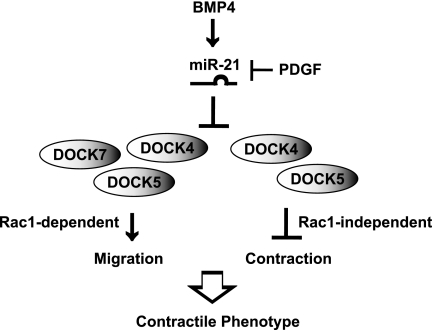
Schematic representation of cellular effects of miR-21-mediated down-regulation of DOCK4, -5, or -7 in vascular smooth muscle cells. BMP4 induces miR-21 and inhibits migration and increases contractility of vSMCs. miR-21 targets DOCK4, -5, and -7 to control vSMC migration by modulating the activity of Rac1, whereas DOCK4 and -5 modulate contractility of vSMC via a Rac1-independent mechanism.
DISCUSSION
In this study, we identified the DOCK family members as miR-21 targets downstream of BMP4 and provided evidence that their down-regulation by miR-21 is critical for promoting the contractile vSMC phenotype through inhibiting cell migration and promoting contractility (Fig. 6). The role of DOCK proteins in the regulation of cell motility, cytoskeletal organization, phagocytosis, and polarity has been elucidated in various cell types, especially in the nervous system (33). However, no study on DOCK proteins has been performed in vascular cells previously. We previously reported that DOCK7 protein is asymmetrically distributed in unpolarized hippocampal neurons and becomes enriched in axons during the development of this process (35). However, the mechanism underlying the regulation of DOCK7 in hippocampal neurons is unclear. It is interesting to speculate that such specific localization of DOCK7 might be due to selective expression of miR-21.
Using a miRNA pulldown method, we were able to identify DOCK family genes as targets of miR-21. Over 29,000 genes are predicted as targets of miR-21 by at least one of 11 commonly used target prediction algorithms according to the miRecords web site. Thus, experimental methods to purify/identify targets of specific miRNAs are needed. In vivo study of targets of several miRNAs, including miR-1, miR-16, miR-30a, miR-155, and let-7b, using pulsed stable isotope labeling with amino acids in culture analyses in HeLa cells that overexpress these miRNAs indicates that roughly 300 proteins are down-regulated by at least 30% (32). The same study estimated that about 60% of the 300 down-regulated proteins contain seed-matched sites (Watson-Crick consecutive base pairing between mRNAs and the miRNA at positions 2–7 counted from the 5′-end of the miRNA) in the 3′-UTR regions of transcripts (32). Among the 33 previously validated miR-21 targets, only eight targets are predicted by a seed sequence-based algorithm. Non-seed-based algorithms tend to predict too many sites (on average ∼3,000–11,000 MREs per miRNA), which defeats the purpose of using target prediction algorithms. Thus, the biochemical enrichment of miRNA targets presented in this study is useful. We identified ∼26 genes that are ≥24-fold enriched in miR-21 pulldowns over control cel-miR-67 in PASMCs. We tested 28 predicted MREs located in the 3′-UTR of mRNAs associated with bio-miR-21, including 16 predicted MREs or sequences partially complementary to miR-21 localized in the 3′-UTR of DOCK4, -5, or -7 transcripts. Independently, miR-34a target analysis was performed in HCT116 and K562 cells using the bio-miR-34a pulldown method and confirmed that genes enriched in bio-miR-34a pulldowns by as little as 2.5-fold over control miRNA pulldowns in both cells have 3′-UTRs that are regulated directly by miR-34a as measured by the luciferase reporter activity assay (40). Thus, additional miR-21-regulated genes are likely, and further verification and investigation of genes that are enriched in bio-miR-21 pulldowns in PASMC are warranted to understand the biological functions of miR-21 in vSMCs.
Six of the 22 miR-21-associated mRNAs that are enriched >24-fold over control miRNAs (PARP9, DOCK7, ADAM22, RNF17, LGALS14, and SPRYD4) do not contain miR-21 seed sequence or predicted MREs. RNAhybrid predicted some of the DOCK genes; however, this algorithm predicts on average ∼11,000 target sites per miRNA. MRE sites in DOCK4, DOCK5, and DOCK7 3′-UTRs identified in this study contain complementary sequences of 6–11-mers starting somewhere between position 1 and 15. These results suggest that base pairing between a miRNA and target mRNA can be important downstream of the seed site.
It is intriguing that miR-21 targets multiple transcripts belonging to the same family. There are a few other examples of a single miRNA targeting multiple genes of the same family. miR-200 targets the zinc finger homeodomain enhancer-binding protein (ZEB) family of transcription repressors; ZEB1 and ZEB2 regulate the epithelial-to-mesenchymal transition (41). miR-164 targets the NAC domain transcription factors CUP-SHAPED COTYLEDON1 (CUC1) and CUC2 that establish axillary meristems of leaves in plant (42). In both cases, products of the same gene family share a redundant function as in the case of DOCK4, -5, and -7 and Rac1 regulation in the regulation of cell migration (33). Although examples of a single miRNA targeting multiple genes in the same signaling pathway or with related biological functions have been reported previously, our results are striking because they suggest that a single miRNA targets multiple transcripts of the same gene family. In the case of miR-21 MREs identified in the 3′-UTR of human DOCK7 (MRE1), an 8-mer at the 5′-end is 100% conserved in mouse. For human DOCK4, a 6-mer complementary sequence starting at position 3 in MRE1 is also 100% conserved in mouse, whereas in MRE6, only six of the 11 residues complementary to miR-21 starting at position 3 are conserved in the mouse 3′-UTR sequence. However, two MREs found in DOCK5 are not conserved in mouse, suggesting that only some MREs are evolutionally conserved.
Increased proliferation and migration of vSMCs are associated with various stimuli, including activation of PDGF signaling, inhibition of BMP/TGFβ or Notch signaling, hypoxia treatment, mechanical stretch, and activation of Rac1 (43, 44). However, a role for DOCK4, -5, or -7 has not been characterized in vSMCs previously. Our study illustrates both common and distinct roles of DOCK family of proteins in the regulation of vSMC phenotype (Fig. 6). DOCK4 is known to mediate cell migration promoted by the PDGF signal in NIH3T3 cells (45). In vSMCs, BMP and PDGF signals antagonize each other and modulate vSMC phenotype during vascular injury repair or pathogenesis of vascular proliferative diseases (46). Thus, the BMP4-miR-21-DOCK4 axis is a novel mechanism of antagonism between BMP and PDGF signaling pathways in addition to the miR-24-Trb3 axis, which has been described previously (34). Interestingly, cytoskeletal remodeling is mediated by DOCK4 and DOCK5, but not by DOCK7, through a mechanism that is independent of Rac1. As DOCK4 and -5 contain multiple Src homology 3 domains that are not found in DOCK7 (33), we speculate that the Src homology 3 domain might play a critical role in the cytoskeletal remodeling through docking other signaling molecule(s).
miR-21 is elevated in most human cancers and has been shown to function as an oncogene by promoting cell growth and migration (17). Indeed, many validated targets of miR-21 are tumor suppressors, including PDCD4 (47), PTEN (48), and tropomyosin 1 (TPM1) (21). Homozygous deletion of DOCK4 gene occurs during tumor progression in the TP53-NF2 mouse model and is associated with increased invasion of tumor cells (49). Furthermore, a missense mutation (Pro-1718 to Leu) of DOCK4 that results in defective activation of the Rap1 GTPase (49) has been identified in human prostate and ovarian cancers. Thus, in this context, DOCK4 is a novel target of miR-21 with tumor suppressor function. It is intriguing, however, that miR-21, which promotes invasion and migration of cancer cells (50), inhibits migration in vSMCs. This observation is indicative of a cell type-specific function of miR-21. A potential mechanism of the cell type-specific function of miRNAs involves how different sets of genes are targeted by a miRNA due to (i) a cell type-specific expression of mRNA targets and/or (ii) a cell type-specific recognition/targeting of mRNA targets by a miRNA. A potential advantage of the miRNA target pulldown approach is that it allows identification of miRNA targets in a context-dependent manner. In addition to the relevance to vSMC biology, further investigation of the targets identified by the miR-21 pulldown may provide insights for understanding the sequence requirements for specific recognition of mRNA targets by miRNA and facilitate the development of more accurate target prediction algorithms.
Supplementary Material
Acknowledgments
We thank Dr. Xinmin Li for microarray analysis of miRNA pulldowns. We also thank all members of the Hata and Lagna laboratories for helpful suggestions and critical discussion.
This work was supported, in whole or in part, by National Institutes of Health Grants HD042149, HL093154, and HL082854 (to A. H.). This work was also supported by American Heart Association Grant 0940095N and a LeDucq Foundation transatlantic network grant (to A. H.).

This article contains supplemental Figs. S1–S4, Tables S1 and S2, and a list of primers.
- miRNA
- microRNA
- DOCK
- dedicator of cytokinesis
- miR-21
- microRNA-21
- vSMC
- vascular smooth muscle cell
- BMP
- bone morphogenetic protein
- PDCD4
- programmed cell death 4
- MRE
- miRNA recognition element
- pri-miR-21
- primary transcripts of miR-21
- PASMC
- pulmonary artery smooth muscle cell
- cel-miR-67
- control C. elegans miR-67 mimic
- bio-miR-21
- 3′-biotinylated miR-21 mimic
- bio-cel-miR-67
- 3′-biotinylated control C. elegans miR-67 mimic
- qRT
- quantitative reverse transcription
- anti-miR-21
- antisense oligonucleotides against miR-21
- ZEB
- zinc finger homeodomain enhancer-binding protein.
REFERENCES
- 1. Siomi H., Siomi M. C. (2009) On the road to reading the RNA-interference code. Nature 457, 396–404 [DOI] [PubMed] [Google Scholar]
- 2. Bartel D. P. (2009) MicroRNAs: target recognition and regulatory functions. Cell 136, 215–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lewis B. P., Burge C. B., Bartel D. P. (2005) Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120, 15–20 [DOI] [PubMed] [Google Scholar]
- 4. Doench J. G., Sharp P. A. (2004) Specificity of microRNA target selection in translational repression. Genes Dev. 18, 504–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Small E. M., Frost R. J., Olson E. N. (2010) MicroRNAs add a new dimension to cardiovascular disease. Circulation 121, 1022–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Slack F. J., Weidhaas J. B. (2006) MicroRNAs as a potential magic bullet in cancer. Future Oncol. 2, 73–82 [DOI] [PubMed] [Google Scholar]
- 7. Asangani I. A., Rasheed S. A., Nikolova D. A., Leupold J. H., Colburn N. H., Post S., Allgayer H. (2008) MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene 27, 2128–2136 [DOI] [PubMed] [Google Scholar]
- 8. Davis B. N., Hilyard A. C., Lagna G., Hata A. (2008) SMAD proteins control DROSHA-mediated microRNA maturation. Nature 454, 56–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ji R., Cheng Y., Yue J., Yang J., Liu X., Chen H., Dean D. B., Zhang C. (2007) MicroRNA expression signature and antisense-mediated depletion reveal an essential role of microRNA in vascular neointimal lesion formation. Circ. Res. 100, 1579–1588 [DOI] [PubMed] [Google Scholar]
- 10. Roy S., Khanna S., Hussain S. R., Biswas S., Azad A., Rink C., Gnyawali S., Shilo S., Nuovo G. J., Sen C. K. (2009) MicroRNA expression in response to murine myocardial infarction: miR-21 regulates fibroblast metalloprotease-2 via phosphatase and tensin homologue. Cardiovasc. Res. 82, 21–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Thum T., Gross C., Fiedler J., Fischer T., Kissler S., Bussen M., Galuppo P., Just S., Rottbauer W., Frantz S., Castoldi M., Soutschek J., Koteliansky V., Rosenwald A., Basson M. A., Licht J. D., Pena J. T., Rouhanifard S. H., Muckenthaler M. U., Tuschl T., Martin G. R., Bauersachs J., Engelhardt S. (2008) MicroRNA-21 contributes to myocardial disease by stimulating MAP kinase signalling in fibroblasts. Nature 456, 980–984 [DOI] [PubMed] [Google Scholar]
- 12. Volinia S., Calin G. A., Liu C. G., Ambs S., Cimmino A., Petrocca F., Visone R., Iorio M., Roldo C., Ferracin M., Prueitt R. L., Yanaihara N., Lanza G., Scarpa A., Vecchione A., Negrini M., Harris C. C., Croce C. M. (2006) A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl. Acad. Sci. U.S.A. 103, 2257–2261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Busacca S., Germano S., De Cecco L., Rinaldi M., Comoglio F., Favero F., Murer B., Mutti L., Pierotti M., Gaudino G. (2010) MicroRNA signature of malignant mesothelioma with potential diagnostic and prognostic implications. Am. J. Respir. Cell Mol. Biol. 42, 312–319 [DOI] [PubMed] [Google Scholar]
- 14. Chan J. A., Krichevsky A. M., Kosik K. S. (2005) MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 65, 6029–6033 [DOI] [PubMed] [Google Scholar]
- 15. Chang S. S., Jiang W. W., Smith I., Poeta L. M., Begum S., Glazer C., Shan S., Westra W., Sidransky D., Califano J. A. (2008) MicroRNA alterations in head and neck squamous cell carcinoma. Int. J. Cancer 123, 2791–2797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hatley M. E., Patrick D. M., Garcia M. R., Richardson J. A., Bassel-Duby R., van Rooij E., Olson E. N. (2010) Modulation of K-Ras-dependent lung tumorigenesis by microRNA-21. Cancer Cell 18, 282–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Krichevsky A. M., Gabriely G. (2009) miR-21: a small multi-faceted RNA. J. Cell. Mol. Med. 13, 39–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ladeiro Y., Couchy G., Balabaud C., Bioulac-Sage P., Pelletier L., Rebouissou S., Zucman-Rossi J. (2008) MicroRNA profiling in hepatocellular tumors is associated with clinical features and oncogene/tumor suppressor gene mutations. Hepatology 47, 1955–1963 [DOI] [PubMed] [Google Scholar]
- 19. Moriyama T., Ohuchida K., Mizumoto K., Yu J., Sato N., Nabae T., Takahata S., Toma H., Nagai E., Tanaka M. (2009) MicroRNA-21 modulates biological functions of pancreatic cancer cells including their proliferation, invasion, and chemoresistance. Mol. Cancer Ther. 8, 1067–1074 [DOI] [PubMed] [Google Scholar]
- 20. Si M. L., Zhu S., Wu H., Lu Z., Wu F., Mo Y. Y. (2007) miR-21-mediated tumor growth. Oncogene 26, 2799–2803 [DOI] [PubMed] [Google Scholar]
- 21. Zhu S., Si M. L., Wu H., Mo Y. Y. (2007) MicroRNA-21 targets the tumor suppressor gene tropomyosin 1 (TPM1). J. Biol. Chem. 282, 14328–14336 [DOI] [PubMed] [Google Scholar]
- 22. Zhu S., Wu H., Wu F., Nie D., Sheng S., Mo Y. Y. (2008) MicroRNA-21 targets tumor suppressor genes in invasion and metastasis. Cell Res. 18, 350–359 [DOI] [PubMed] [Google Scholar]
- 23. Cheng Y., Ji R., Yue J., Yang J., Liu X., Chen H., Dean D. B., Zhang C. (2007) MicroRNAs are aberrantly expressed in hypertrophic heart: do they play a role in cardiac hypertrophy? Am. J. Pathol. 170, 1831–1840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Silvestri P., Di Russo C., Rigattieri S., Fedele S., Todaro D., Ferraiuolo G., Altamura G., Loschiavo P. (2009) MicroRNAs and ischemic heart disease: towards a better comprehension of pathogenesis, new diagnostic tools and new therapeutic targets. Recent Pat. Cardiovasc. Drug Discov. 4, 109–118 [DOI] [PubMed] [Google Scholar]
- 25. Davis B. N., Hilyard A. C., Nguyen P. H., Lagna G., Hata A. (2010) Smad proteins bind a conserved RNA sequence to promote microRNA maturation by Drosha. Mol. Cell 39, 373–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Davis B. N., Hilyard A. C., Nguyen P. H., Lagna G., Hata A. (2009) Induction of microRNA-221 by platelet-derived growth factor signaling is critical for modulation of vascular smooth muscle phenotype. J. Biol. Chem. 284, 3728–3738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lal A., Navarro F., Maher C. A., Maliszewski L. E., Yan N., O'Day E., Chowdhury D., Dykxhoorn D. M., Tsai P., Hofmann O., Becker K. G., Gorospe M., Hide W., Lieberman J. (2009) miR-24 Inhibits cell proliferation by targeting E2F2, MYC, and other cell-cycle genes via binding to “seedless” 3′UTR microRNA recognition elements. Mol. Cell 35, 610–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chang T. C., Wentzel E. A., Kent O. A., Ramachandran K., Mullendore M., Lee K. H., Feldmann G., Yamakuchi M., Ferlito M., Lowenstein C. J., Arking D. E., Beer M. A., Maitra A., Mendell J. T. (2007) Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol. Cell 26, 745–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Easow G., Teleman A. A., Cohen S. M. (2007) Isolation of microRNA targets by miRNP immunopurification. RNA 13, 1198–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhang L., Ding L., Cheung T. H., Dong M. Q., Chen J., Sewell A. K., Liu X., Yates J. R., 3rd, Han M. (2007) Systematic identification of C. elegans miRISC proteins, miRNAs, and mRNA targets by their interactions with GW182 proteins AIN-1 and AIN-2. Mol. Cell 28, 598–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Baek D., Villén J., Shin C., Camargo F. D., Gygi S. P., Bartel D. P. (2008) The impact of microRNAs on protein output. Nature 455, 64–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Selbach M., Schwanhäusser B., Thierfelder N., Fang Z., Khanin R., Rajewsky N. (2008) Widespread changes in protein synthesis induced by microRNAs. Nature 455, 58–63 [DOI] [PubMed] [Google Scholar]
- 33. Miyamoto Y., Yamauchi J. (2010) Cellular signaling of Dock family proteins in neural function. Cell. Signal. 22, 175–182 [DOI] [PubMed] [Google Scholar]
- 34. Chan M. C., Hilyard A. C., Wu C., Davis B. N., Hill N. S., Lal A., Lieberman J., Lagna G., Hata A. (2010) Molecular basis for antagonism between PDGF and the TGFβ family of signalling pathways by control of miR-24 expression. EMBO J. 29, 559–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Watabe-Uchida M., John K. A., Janas J. A., Newey S. E., Van Aelst L. (2006) The Rac activator DOCK7 regulates neuronal polarity through local phosphorylation of stathmin/Op18. Neuron 51, 727–739 [DOI] [PubMed] [Google Scholar]
- 36. Ørom U. A., Nielsen F. C., Lund A. H. (2008) MicroRNA-10a binds the 5′UTR of ribosomal protein mRNAs and enhances their translation. Mol. Cell 30, 460–471 [DOI] [PubMed] [Google Scholar]
- 37. Orom U. A., Lund A. H. (2007) Isolation of microRNA targets using biotinylated synthetic microRNAs. Methods 43, 162–165 [DOI] [PubMed] [Google Scholar]
- 38. Gao Y., Dickerson J. B., Guo F., Zheng J., Zheng Y. (2004) Rational design and characterization of a Rac GTPase-specific small molecule inhibitor. Proc. Natl. Acad. Sci. U.S.A. 101, 7618–7623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Miranda K. C., Huynh T., Tay Y., Ang Y. S., Tam W. L., Thomson A. M., Lim B., Rigoutsos I. (2006) A pattern-based method for the identification of microRNA binding sites and their corresponding heteroduplexes. Cell 126, 1203–1217 [DOI] [PubMed] [Google Scholar]
- 40. Lal A., Thomas M. P., Altschuler G., Navarro F., O'Day E., Li X. L., Concepcion C., Han Y. C., Thiery J., Rajani D. K., Deutsch A., Hofmann O., Ventura A., Hide W., Lieberman J. (2011) Capture of microRNA-bound mRNAs identifies the tumor suppressor miR-34a as a regulator of growth factor signaling. PLoS Genet. 7, e1002363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Adam L., Zhong M., Choi W., Qi W., Nicoloso M., Arora A., Calin G., Wang H., Siefker-Radtke A., McConkey D., Bar-Eli M., Dinney C. (2009) miR-200 expression regulates epithelial-to-mesenchymal transition in bladder cancer cells and reverses resistance to epidermal growth factor receptor therapy. Clin. Cancer Res. 15, 5060–5072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Raman S., Greb T., Peaucelle A., Blein T., Laufs P., Theres K. (2008) Interplay of miR164, CUP-SHAPED COTYLEDON genes and LATERAL SUPPRESSOR controls axillary meristem formation in Arabidopsis thaliana. Plant J. 55, 65–76 [DOI] [PubMed] [Google Scholar]
- 43. Qi Y. X., Qu M. J., Yan Z. Q., Zhao D., Jiang X. H., Shen B. R., Jiang Z. L. (2010) Cyclic strain modulates migration and proliferation of vascular smooth muscle cells via Rho-GDIα, Rac1, and p38 pathway. J. Cell. Biochem. 109, 906–914 [DOI] [PubMed] [Google Scholar]
- 44. Owens G. K., Kumar M. S., Wamhoff B. R. (2004) Molecular regulation of vascular smooth muscle cell differentiation in development and disease. Physiol. Rev. 84, 767–801 [DOI] [PubMed] [Google Scholar]
- 45. Kawada K., Upadhyay G., Ferandon S., Janarthanan S., Hall M., Vilardaga J. P., Yajnik V. (2009) Cell migration is regulated by platelet-derived growth factor receptor endocytosis. Mol. Cell. Biol. 29, 4508–4518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lagna G., Ku M. M., Nguyen P. H., Neuman N. A., Davis B. N., Hata A. (2007) Control of phenotypic plasticity of smooth muscle cells by bone morphogenetic protein signaling through the myocardin-related transcription factors. J. Biol. Chem. 282, 37244–37255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Frankel L. B., Christoffersen N. R., Jacobsen A., Lindow M., Krogh A., Lund A. H. (2008) Programmed cell death 4 (PDCD4) is an important functional target of the microRNA miR-21 in breast cancer cells. J. Biol. Chem. 283, 1026–1033 [DOI] [PubMed] [Google Scholar]
- 48. Meng F., Henson R., Wehbe-Janek H., Ghoshal K., Jacob S. T., Patel T. (2007) MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology 133, 647–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Yajnik V., Paulding C., Sordella R., McClatchey A. I., Saito M., Wahrer D. C., Reynolds P., Bell D. W., Lake R., van den Heuvel S., Settleman J., Haber D. A. (2003) DOCK4, a GTPase activator, is disrupted during tumorigenesis. Cell 112, 673–684 [DOI] [PubMed] [Google Scholar]
- 50. Cho W. C. (2007) OncomiRs: the discovery and progress of microRNAs in cancers. Mol. Cancer 6, 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.